Abstract
Proteins mainly perform their functions by interacting with other proteins. Protein–protein interactions underpin various biological activities such as metabolic cycles, signal transduction, and immune response. However, due to the sheer number of proteins, experimental methods for finding interacting and non-interacting protein pairs are time-consuming and costly. We therefore developed the ProtInteract framework to predict protein–protein interaction. ProtInteract comprises two components: first, a novel autoencoder architecture that encodes each protein’s primary structure to a lower-dimensional vector while preserving its underlying sequence attributes. This leads to faster training of the second network, a deep convolutional neural network (CNN) that receives encoded proteins and predicts their interaction under three different scenarios. In each scenario, the deep CNN predicts the class of a given encoded protein pair. Each class indicates different ranges of confidence scores corresponding to the probability of whether a predicted interaction occurs or not. The proposed framework features significantly low computational complexity and relatively fast response. The contributions of this work are twofold. First, ProtInteract assimilates the protein’s primary structure into a pseudo-time series. Therefore, we leverage the nature of the time series of proteins and their physicochemical properties to encode a protein’s amino acid sequence into a lower-dimensional vector space. This approach enables extracting highly informative sequence attributes while reducing computational complexity. Second, the ProtInteract framework utilises this information to identify protein interactions with other proteins based on its amino acid configuration. Our results suggest that the proposed framework performs with high accuracy and efficiency in predicting protein-protein interactions.
Keywords: Protein–Protein interaction; Autoencoder; Long short-term memory; Temporal convolutional, Network; Convolutional neural network; Sequential pattern; Feature selection
Graphical Abstract
1. Introduction
A protein is a macromolecule constituted by chains of amino acids, with side-chain groups extending from a connected backbone [1]. Protein–protein interactions (PPI) play a vital role in almost every intracellular biological and biochemical activity [2], [3], [4]. Most proteins execute their functions by interacting with other proteins. These functions include nutrient transport, enzymatic reactions, immune response, communication between cells, and tissue regeneration. Thus, accurately identifying PPIs is essential for understanding cell physiology [5]. We now understand the close relationship between abnormal PPIs and a range of pathologies such as neurodegenerative conditions, cancer, and infectious diseases [6], [7], [8]: identifying functional PPIs has proved to be a turning point in how technology can instaurate the foundations for target, and individualised therapies [9], [10], [11].
Many have addressed the PPI identification problem via experimental methods such as yeast two-hybrid screening [12], [13], [14], mass spectrometry [15], [16], [17], [18], [19], protein microarrays [20], [21], [22], [23]. These approaches are often costly and time-consuming [24], [25], [26]. Computational methods aim, therefore, to reduce false-positive and false-negative results by providing preliminary testing prior to the experiments [27], [28], [29].
Recent advances in deep learning frameworks, fuelled by the large amount of data available for training, have revolutionised scientific studies [30]. In particular, it has become possible, to some extent, to predict PPI [31], [32].
Nevertheless, the performance of deep learning algorithms is affected by multiple factors such as network structure, which may involve multiple sub-networks and the feature selection mechanism along with the activation and pooling functions, noise, and imbalanced datasets [33], [34]. To handle problems such as noise, wavelet transforms have been employed [35], while more informative representations and dimensionality reduction have been implemented using methods such as Boltzmann machines and autoencoders [36]. For problems with high feature dimensionality, learning can be optimised by extracting uncorrelated features while keeping highly informative features using convolutional and stacked autoencoders [33], [34], [35], [37]. In biological contexts, modelling protein structures includes prediction from the amino acid sequence, designing proteins with specific functionalities, and predicting properties or behaviour of a protein, which require a broad knowledge at the molecular level [38].
Deep learning architectures for PPI prediction may operate on different inputs, including primary structure [39], [40], [41], [42], domain composition [43], [44], gene expression [45], [46], [47], and the proteins’ 3-D structure [48], to mention a few. It is often believed that 3-D structure delivers a relatively more complete set of information for PPI prediction [49]. However, all the vital information required to identify PPIs is encoded in the amino acid sequence, i.e. the primary structure [50], so sequence-based methods have gained particular attention [51]. DeepPPI [52], for example, aims to improve PPI prediction using a variety of statistical descriptors to learn the protein characteristics. TransPPI [53] captures latent patterns in the protein sequence using a Siamese-like architecture of convolutional layers. This method predicts interaction probabilities by concatenating a pair of protein representations from two identical sub-networks. CAMP is another PPI prediction method based on convolutional layers; it receives an integration of multiple features such as primary and second structures, physicochemical properties, and protein evolutionary information as input [54]. This method includes a self-attention layer to learn the long-range dependencies between residues in protein sequences. The D-SCRIPT framework [55] comprises a pre-trained, bidirectional long short-term memory (BiLSTM) model [56] that generates structurally informative features for each protein and a projection module to reduce the dimensionality of embeddings. These low-dimensional embeddings include the protein’s residue contact map; these maps are used to predict the interaction probabilities. Another convolution-based framework, called DeepTrio [57], performs binary PPI prediction by employing multiple parallel convolutional neural networks. PIPR [58] is a sequence-based framework which integrates a convolution-pooling layer and bidirectional residual gated recurrent units (GRU) [59], extracting the local features and global sequential information of proteins, and thus capturing the mutual effects of protein pairs in PPI prediction tasks. S-VGAE [60] uses a variational graph autoencoder [61] to learn the latent features of proteins. A PPI identification method using GCNs [5] integrates the one-hot encoding representation of protein sequences and the positional information of each protein in the PPI network.
In this paper, we propose a sequence-based method called ProtInteract that predicts protein–protein interactions through two main tasks: encoding and prediction. A novel autoencoder architecture encodes each protein to remedy the computational complexity posed by the high dimensionality of protein representation while extracting sequential attributes of the protein’s primary structure. This autoencoder is composed of 2-D convolutional and temporal convolutional network (TCN) layers (see Section 5.1).
ProtInteract is trained on the STRING database, which contains PPI datasets for various organisms. In each dataset, protein pairs are associated with an interaction score which shows the confidence of STRING assessment on an interaction being valid, given the available evidence [62], [63]. Those interaction scores are the basis for ProtInteract’s prediction process. These interaction scores are then normalised between zero (non-interacting pairs) and one (interacting pairs with the highest confidence). We formulate the prediction task as three different multiclass classification scenarios comprising, respectively two, three and five classes [64]. For the two-class scenario, interactions are divided into non-interacting (0 ≤ interaction score ≤ 0.5) and interacting pairs (0.5 < interaction score ≤ 1). For the three-class scenario, a range for erroneous results (0.4 < interaction score ≤ 0.7) is considered as well as non-interacting (0 ≤ interaction score ≤ 0.4), and interacting pairs (0.7 < interaction score ≤ 1). In the STRING database, an interaction score of 0.5 may suggest that, on average, every second interaction is labelled incorrectly (a false positive) [63], [65], [66]; we consider a wider span for such interactions. The final scenario comprises five classes, where the non-interacting and interacting spans are divided into high- and low-confidence spans (see Table 4), to define five different classes. In each scenario, a deep convolutional neural network (deep CNN) predicts the probability of correctly classifying an interaction. Additionally, many other researchers employed an LSTM architecture as the baseline model when dealing with time series and sequential datasets [67], [68], [69], [70], which inspired to leverage LSTM as a baseline model to demonstrate the comparative advantages of the TCN architecture. The rest of the paper is organised as follows: In Section 2, some of the most recent methods for addressing the PPI problem are briefly discussed. Section 3 outlines our proposed framework. The novel autoencoder architecture will be presented in Section 5.1, and the baseline model in section 10.2. The multi-class classification scenarios are discussed in Section 6. The architecture of the deep CNN is presented in Section 7. Section 8 will discuss the experimental results, through the examples of Homo sapiens and Mus musculus. Finally, Section 9 concludes this work.
Table 4.
onfidence score thresholds for five classes of protein–protein interactions.
| Classes | Confidence Score | Description |
|---|---|---|
| class 1 | 0 ≥ – ≤ 0.15 | The confidence of interaction is very low. |
| class 2 | > 0.15 – ≤ 0.4 | This score range shows low confidence in the occurrence of an interaction between a protein pair. |
| class 3 | > 0.4 – ≤ 0.7 | A score in this range may indicate a falsely labelled pair. |
| class 4 | > 0.7 – ≤ 0.9 | Interaction scores within this range indicate high confidence of interaction between two proteins. |
| class 5 | > 0.9 – ≤ 1 | This range indicates highly interactive protein pairs, based on experimental and theoretical evidence. |
2. PPI prediction
Among the computational methods developed for PPI prediction, some aim to extract new features from protein information while others use the extracted features to learn new models [71]. For instance, the approach proposed by Guo et al. [72] to obtain non-interacting protein pairs has been used to develop a SVM-based method in [41], and a PIPR method in [58]. A multimodal approach based on LSTM is proposed in [71], which predicts PPI by integrating structural and sequential information of proteins as additional features to the input feature set. The physiochemical attributes of amino acids, such as hydropathy [73], isoelectric point, and charge, play crucial roles in identifying interactions between protein sequences [74]. The PPI prediction is conducted based on sequences in [75] by defining units of three adjacent amino acids and measuring the frequencies of those units in a protein sequence. Other methods such as amino acid index distribution [76], the conjoint triad method [75], and autocovariance [72] extract features such as locations of amino acids, frequencies and physicochemical properties aiming to represent a protein sequence.
In order to reduce the high dimensionality of protein structures, several techniques have been developed, such as random forest [41] and support vector machine (SVM) and its derivatives [77], [78]. Another SVM-based approach named ACT-SVM [79] extracts features from protein sequences for input vector to the classifier. A sequence-based human PPI prediction, proposed in [80], is based on a stacked autoencoder (SAE). Another sequence-based PPI prediction approach, Deep Sequence Contact Residue Interaction Prediction Transfer (D-SCRIPT) [81], models protein structure using the pre-trained language model in [56]. A heterogeneous network is used for PPI prediction in [82]. They represent proteins by concatenating local and global features. They used a k-mer method 1 [83] to extract local features. To extract global features, LINE (Large-scale Information Network Embedding) is employed [84]. Finally, a random forest classifies the potential protein pairs.
A deep learning method for PPI prediction, called ordinal regression and recurrent convolutional neural network (OR-RCNN) [85], comprises two recurrent convolutional neural networks (RCNNs) to extract local features and sequential information from the protein pairs, and ordinal regression to construct multiple sub-classifiers. The viability of these techniques is yet to be verified experimentally. Therefore, to understand the potential of generative models in protein engineering, more needs to be done, such as analysing the strengths and limitations of different models and the possibilities for incorporation into existing engineering operations. First and foremost, representation of the input to the network is a matter of great importance [86]. Given the high dimensionality of protein sequence datasets—which include amino acid physicochemical features and large chains of amino acids— deep learning methods may suffer from extended training times. On the other hand, reducing the number of amino acid features may lead to underfitting [87], [88].
We propose, here, a framework for learning a meaningful representation of protein sequence while reducing its dimensionality using a novel autoencoder. PPIs are characterised based on their associated score from the STRING database. This score, a value from zero and one, expresses the confidence of an interaction existence based on the available evidence. A score of zero indicates the highest confidence that a pair of proteins do not interact. In contrast, interaction with the highest confidence is characterised by a score of one. In this work, we composed the PPI prediction as three separate classification problems by defining various ranges of interaction scores. The first scenario is a classification with two classes, where the interaction score range is bisected, indicating only non-interacting and interacting pairs (Table 2). The second and third scenarios designate multiple levels of confidence: a score in (0.4,0.7] indicates medium confidence, which may include false-positive samples, with the extremes indicating one or two levels of confidence in the interacting and non-interacting pairs (Table 3, Table 4). The concept of defining three scenarios for ProtInteract is rooted in the idea of identifying high-affinity protein-protein interactions (PPIs) with scores within the range of (0.9,1] (the probability of interactions falling within the most extreme confidence ranges). This expands the potential applications of ProtInteract to include the identification of protein–ligand pairs [89]. Furthermore, by establishing these scenarios, we are able to eliminate uncertainty present in the datasets, specifically regarding interactions falsely labelled as positive within the range of [0.4,0.7). This allows for more accurate classification of interactions within higher confidence ranges. These scenarios will be discussed in more detail in Section 6.
Table 2.
Confidence score thresholds for two classes of protein–protein interactions.
| Classes | Confidence Score | Description |
|---|---|---|
| class 1 | 0 ≥ – ≤ 0.5 | The score threshold is assigned to non-interacting protein pairs. |
| A score of zero suggests a non-interactive pair with the highest confidence | ||
| class 2 | > 0.5 – ≤ 1 | This range indicates interacting protein pairs based on experimental and theoretical evidence. |
| A score of one indicates full confidence in the protein pair’s interactivity. |
Table 3.
Confidence score thresholds for three classes of protein–protein interactions.
| Classes | Confidence Score | Description |
|---|---|---|
| class 1 | 0 ≥ – ≤ 0.4 | The interaction confidence threshold is low. |
| class 2 | > 0.4 – ≤ 0.7 | A score in this range may indicate a falsely labelled pair.(for instance, a score of 0.5 suggests that nearly every second interaction may be incorrectly labelled as an interacting pair). |
| class 3 | > 0.7 – ≤ 1 | This range shows highly interactive protein pairs based on experimental and theoretical evidence. A score of one indicates full confidence in the protein pair’s interactivity. |
3. ProtInteract outline
Most protein design problems require profound knowledge to analyse difficulties and obtain optimal design [90]. However, the emergence of deep neural networks has provided the computational capacity to exploit the rich reserves of existing historical data. The proposed framework comprises two deep neural networks, each responsible for one task. First, a deep learning architecture reduces the input data size by extracting meaningful sequential patterns from the protein sequence. Second, a deep learning architecture performs the PPI prediction task using the extracted features.
For the first task, we propose two different architectures, namely a convolutional-TCN autoencoder and a convolutional-LSTM autoencoder, and compare their accuracies by using the latter as a baseline model. The main reason for this autoencoder framework is to reduce the dimensionality and to remove redundancies from the input tensor, which accelerates the training of the following network for PPI prediction. The intuition behind adopting TCN and LSTM in the framework is their ability to extract sequential patterns. As first introduced in [91], TCN allows a hierarchical, bi-level analysis of sequential data such as time series [92]. TCNs tend to outperform and have a longer long-term memory than recurrent neural networks (RNNs) such as LSTM and Gated Recurrent Units (GRU) when sequential data are involved [93], [94]. Here, we show the advantage of using TCN over LSTM in our architecture.
In the following step, a deep convolutional neural network (CNN) architecture receives encoded proteins and predicts their interaction according to the three defined different scenarios: two, three and five classes, according to Table 2, Table 3, Table 4, respectively. For each scenario, the deep CNN predicts the class of each interaction given the encoded protein pairs. A schematic representation of these steps is shown in Fig. 1. The network structure is described in greater detail below.
Fig. 1.
The ProtInteract framework comprises a convolutional TCN autoencoder that extracts highly informative sequential patterns encoded by each protein and lowers its dimension. A deep CNN predicts the probability of an interaction falling within the range of a defined class. The two proteins form a complex, which is represented in this architecture by the stacking of the two proteins.
The following section discusses our approach to encoding proteins using the physicochemical properties of their amino acids.
4. Protein representation
The first step toward developing a deep learning algorithm for PPI prediction is preprocessing the datasets to encode the categorical amino acids into a set of numerical values [95]. A common technique is one-hot encoding [95], [96], which may not be well-suited to problems with high cardinality, such as encoding proteins of significant length. For example, for a dataset comprising the twenty standard amino acids and an unknown amino acid (X), Alanine (A) might be represented as [10000000000000000000] using one-hot encoding, which is a simple vector with twenty zeros. Here, one-hot encoding results in sparse vectors with many non-informative zeroes representing a protein of some length [97], [98]. Such a high proportion of zeroes in the encodings results in a neural network that struggles to learn because it observes mostly zero-valued inputs. To eliminate this difficulty, each amino acid is first characterised by ten physicochemical features, as reported in Table 1. These features are highly informative and represent various aspects of the protein. For inter-protein bindings to form, amino acids should chemically complement each other, allowing hydrophobic, polar, or charged interactions [99]. The hydropathy properties of amino acids, i.e., hydrophobicity and hydrophilicity, are the dominant properties of protein-protein binding [100], [101]. The helix and sheet probabilities are two statistically driven parameters [102]. The protein’s isoelectric point pI (i.e., the pH at which its net charge is zero), its side-chain net charge number NCN [103], [104], and its solvent-accessible surface area (SASA) are key factors in protein stability and folding studies [105]. The features of unknown amino acids (X) are calculated by the median of each known feature from twenty standard amino acids. With the varied ranges of the physicochemical features, min–max normalisation is performed for each feature reported in Table 1. These ten attributes have proven efficient in determining the potential for protein interactions.
Table 1.
Physicochemical properties of 20 amino acids: a) volume; b) isoelectric point; c) helix probability; d) sheet probability; e) hydrophobicity; f) hydrophilicity; g) polarity; h) polarisability) [102], [104], [106], [107].
| Amino acid | Symbol | a | b | c | d | e | f | g | h | SASA | NCN |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alanine | A | 1.00 | 6.11 | 0.42 | 0.23 | 0.62 | -0.50 | 8.10 | 0.046 | 1.181 | 0.007187 |
| Cysteine | C | 2.43 | 6.35 | 0.17 | 0.41 | 0.29 | -1.00 | 5.50 | 0.128 | 1.461 | -0.03661 |
| Aspartate | D | 2.78 | 2.95 | 0.25 | 0.20 | -0.90 | 3.00 | 13.00 | 0.105 | 1.587 | -0.02382 |
| Glutamate | E | 3.78 | 3.09 | 0.42 | 0.21 | -0.74 | 3.00 | 12.30 | 0.151 | 1.862 | 0.006802 |
| Phenylalanine | F | 5.89 | 5.67 | 0.30 | 0.38 | 1.19 | -2.50 | 5.20 | 0.290 | 2.228 | 0.037552 |
| Glycine | G | 0.00 | 6.07 | 0.13 | 0.15 | 0.48 | 0.00 | 9.00 | 0.000 | 0.881 | 0.179052 |
| Histidine | H | 4.66 | 7.69 | 0.27 | 0.30 | -0.40 | -0.50 | 10.40 | 0.230 | 2.025 | -0.01069 |
| Isoleucine | I | 4.00 | 6.04 | 0.30 | 0.45 | 1.38 | -1.80 | 5.20 | 0.186 | 1.810 | 0.021631 |
| Lysine | K | 4.77 | 9.99 | 0.32 | 0.27 | -1.50 | 3.00 | 11.30 | 0.219 | 2.258 | 0.017708 |
| Leucine | L | 4.00 | 6.04 | 0.39 | 0.31 | 1.06 | -1.80 | 4.90 | 0.186 | 1.931 | 0.051672 |
| Methionine | M | 4.43 | 5.71 | 0.38 | 0.32 | 0.64 | -1.30 | 5.70 | 0.221 | 2.034 | 0.002683 |
| Asparagine | N | 2.95 | 6.52 | 0.21 | 0.22 | -0.78 | 2.00 | 11.60 | 0.134 | 1.655 | 0.005392 |
| Proline | P | 2.72 | 6.80 | 0.13 | 0.34 | 0.12 | 0.00 | 8.00 | 0.131 | 1.468 | 0.23953 |
| Glutamine | Q | 3.95 | 5.65 | 0.36 | 0.25 | -0.85 | 0.20 | 10.50 | 0.180 | 1.932 | 0.049211 |
| Arginine | R | 6.13 | 10.74 | 0.36 | 0.25 | -2.53 | 3.00 | 10.50 | 0.291 | 2.560 | 0.043587 |
| Serine | S | 1.60 | 5.70 | 0.20 | 0.28 | -0.18 | 0.30 | 9.20 | 0.062 | 1.298 | 0.004627 |
| Threonine | T | 2.60 | 5.60 | 0.21 | 0.36 | -0.05 | -0.40 | 8.60 | 0.108 | 1.525 | 0.003352 |
| Valine | V | 3.00 | 6.02 | 0.27 | 0.49 | 1.08 | -1.50 | 5.90 | 0.140 | 1.645 | 0.057004 |
| Tryptophan | W | 8.08 | 5.94 | 0.32 | 0.42 | 0.81 | -3.40 | 5.40 | 0.409 | 2.663 | 0.037977 |
| Tyrosine | Y | 6.47 | 5.66 | 0.25 | 0.41 | 0.26 | -2.30 | 6.20 | 0.298 | 2.368 | 0.023599 |
Given a protein dataset, such as that of Homo sapiens, the number of amino acids in each protein may be substantially different. For instance, TRP-Cage protein [PDB code: 2M7D] has the shortest length in the H. sapiens dataset, comprising twenty amino acids, while Titin2 is composed of more than 34,000 amino acids, thus making it difficult to represent to the neural network. Since nearly 98% of H. sapiens and M. musculus sequence datasets from the STRING database are proteins with a length of 2048 amino acids or fewer; we ignore the longer proteins in this paper with only a small loss of content. Additionally, to equalise the variety of lengths, all protein encodings are padded, with zeroes, to 2048 units. Consequently, each protein may be encoded as a tensor with the unwieldy dimensionality of 2048 × 10, requiring slow and computationally expensive training [33], [36].
The underlying sequential structure and orientation of proteins are dictated by the well-defined order of amino acids in the peptide chain: the amino end of each amino acid (its N-terminus) links to the carboxyl end (C-terminus) of the growing peptide chain [108], [109], and the position of each amino acid along a specific chain determines the chain’s final three-dimensional configuration and its binding ability [110]. The arrangement of amino acids along a protein determines a causal directionality which is contrary to an assumption on which traditional neural networks are based, viz. the independence of all the input vector units [35], [111]. Thus, traditional neural networks lack the ability to capture sequential information. For this reason, the recurrent neural network (RNN) and its variants, such as the gated recurrent unit (GRU) [59], and LSTM [112], [113] incorporate hidden states which are generated by sequential information, thus capturing the underlying distribution of such datasets [91], [114], [115]. However, these methods are often computationally expensive, and training is slow [93], [116], [117]. This led to the development of the temporal convolutional network (TCN) for sequence modelling based on dilated causal 1-D convolutional layers [93]. TCN outperforms LSTM in a diverse range of tasks and datasets [93]. Owing to those advantages, we employ TCN layers as the autoencoder architecture’s main framework, and we propose a baseline model using LSTM layers for comparison (see Section 10). The TCN architecture is explained in the following section.
4.1. Temporal convolutional network (TCN)
The many advantages of LSTM come at a high computational cost, consequently, and it is slow to train [93], [116]. We overcome these difficulties using the temporal convolutional network.
The TCN architecture leverages two basic principles. Firstly, there is no information leakage from future to past, owing to causal directionality in the convolution, i.e. for an input sequence with length t as {0,...,t − 1} the ith element of the output sequence depends only on the elements that precede it in the input sequence [118], i.e. the input sequence with indices {0,...,i}. The unique bond directionality (i.e. sequentiality) in the peptide backbone imposed by the N-terminus to-C-terminus translation of amino acids is analogous to that of time-series data [108], [109], [119]. Therefore, one may consider proteins as pseudo-time series as illustrated in Fig. 2, where the time is a proxy for the amino acid positions in the polypeptide chain, which determines the functionality and properties of that protein [38].
Fig. 2.
Proteins may be represented as pseudo-time series such as financial instruments in stock market.
Second, the TCN architecture can map an input sequence of any length to an output sequence with the same length, just as RNNs can. To accomplish this, the TCN employs a 1-D fully convolutional architecture, in which the input and hidden layers have the same length.
In a simple causal convolution network, the size of the receptive field determines how far into the past the network can access. This size grows linearly with the network depth. This precludes causal convolution from being applied to sequential tasks requiring a long history. To avoid this obstacle, the TCN architecture uses dilated convolutions that provide an exponentially large receptive field [120]. The dilated convolution operator F(s) on element s of a 1-D input sequence x ∈ Rn and filter f: {0, ,k − 1} → R is defined as
where d is the dilation factor, k is the filter size, and s − i.d indicates the direction of the past [93]. In addition to network depth, the receptive field depends on filter size k and the dilation factor d [118], so, to expand the receptive field, one may increase the dilation factor, which includes a wider range of inputs, i.e. it captures long range sequential patterns [118], [121]. Therefore, the dilation factor adjusts the effective history of the TCN. Fig. 3 depicts a TCN with three dilated causal convolutional layers.
Fig. 3.
a) A temporal convolution with dilation factors d = 1,2,4 and a filter size of k = 3; b) Architecture of the residual block associated with the TCN.
The network depth is essential for learning robust representations, but it leads to vanishing gradients. In order to circumvent this issue and build deep networks, the residual block has been developed [93], [122]. Residual blocks are employed between TCN layers, allowing for training much deeper models and accelerating convergence [123]. Essentially, as the name suggests, the residual blocks learn residual mapping with reference to the input. The intuition behind such a design is that optimising the residual mapping is easier than optimising the desired underlying mapping (mapping function from input to output) [122]. Let H(x) and F(x) respectively denote the desired underlying mapping and residual mapping. We may represent the original mapping as H(x) = F(x) + x for input x. If identity mapping is desired, as shown in Fig. 3(b), it would be easier to push the residual mapping F(x) to zero rather than to fit an identity mapping by a stack of nonlinear layers [122], [124]. The residual module for TCN consists of a dilated causal convolution, followed by weight normalisation [125] and a nonlinear activation function (a rectified linear unit, ReLU) [126].
In the RNN timeline, the later frames are computed once their predecessors are complete. In contrast, convolutional networks can perform the computations in parallel owing to the independent use of the same kernel (or similar kernel) in each layer repeatedly [117]. TCN offers a flexible receptive field that may be expanded to cover the entire sequence length and captures longer effective history by integrating dilated convolutions and residual layers [121]. TCNs tend to have a more extended long-term memory than RNNs, making them more suitable for capturing long-range dependencies as a common binding mechanism [93], [94], [127], [128].
Given the high-dimensional tensor representing each protein sequence and the size of protein datasets, training a neural network may be slow and computationally expensive [33], [36], [117]. To address this issue, a novel autoencoder framework is proposed, which extracts informative abstract features that characterise each protein in a lower dimension.
In the next section, the fundamentals of autoencoder structure are discussed.
5. Autoencoder
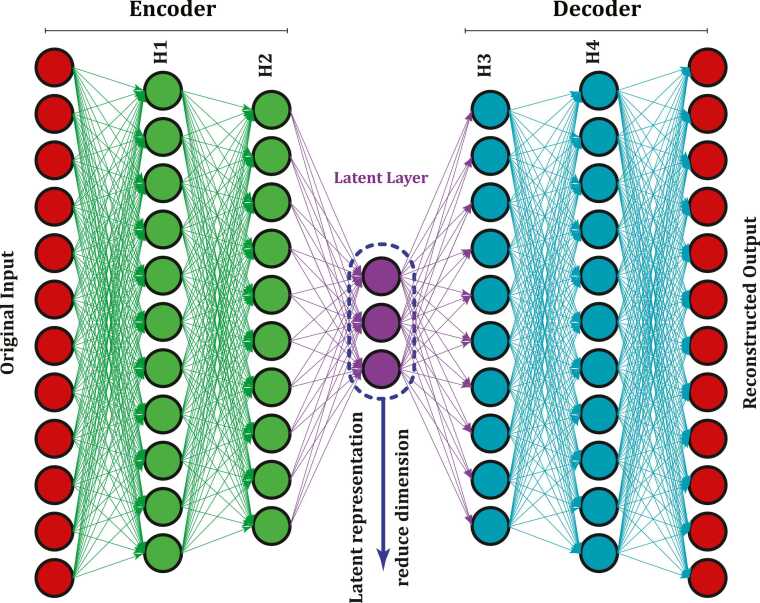
Autoencoders, as depicted in Fig. 4, are a type of unsupervised, feedforward neural network that reconstructs the output from the input [129]. They consist of two parts, namely, the encoder and the decoder. The encoder maps the input data into a lower-dimensional space in the so-called latent layer (latent representation), while the decoder reconstructs the input data from the latent layer; such a network is said to be under-complete because of the constriction that the latent layer imposes [130]. Autoencoders are capable of learning nonlinear dimension reduction functions, removing redundancies and correlations while extracting highly informative features [130]. The input layer represents a high-dimensional and possibly correlated feature space, and this network is used to extract low-dimensional and uncorrelated features via the latent layer (the encoder output).
Fig. 4.
Autoencoder architecture.
The intention behind using autoencoders is to transform the high-dimensional input tensor into a low-dimensional, informative uncorrelated feature [33], [34].
The proposed autoencoder architecture is composed of 2-D convolutional layers that reduce the input tensor dimension, while TCN layers extract sequential patterns. These patterns are extracted from the latent layer and fed into the deep CNN to predict the probability of PPI (interacting vs non-interacting). Let us now examine the proposed autoencoder architecture more closely.
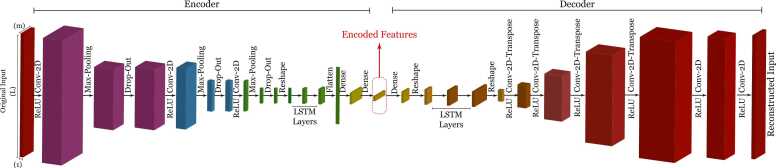
5.1. Convolutional TCN Autoencoder
The architecture of the convolutional TCN autoencoder (Conv-TCN-AE) is shown in Fig. 5, comprising an encoder and a decoder. This architecture is inspired by convolutional autoencoders, which learn the low-dimensional representation of the input tensor [131], [132]. However, the standard convolutional layer fails to comply with causal directionality so to extract the underlying sequential attributes encoded by each protein and long-range dependencies, TCN layers are employed after the 2-D convolutional layers [94].
Fig. 5.
Conv-TCN-AE architecture that reduces the dimension of input data and extracts informative and uncorrelated features.
Initially, the encoder receives a tensor of 2048 × 10 dimensions, representing a protein sequence plus the physicochemical features of its amino acids. This tensor is passed through 2-D convolutional and max-pooling layers to reduce its dimensionality. The dropout layers are employed to reduce over-fitting and to improve generalisation error [133]. Since the TCN layers are based on causal 1-D convolutional layers, a Reshape layer is placed between the 2-D-convolutional layers and TCN layers. Next, the densely connected layer extracts these features with the desired encoding size. The encoder generates a fixed-sized vector in the latent layer, which captures the informative sequential attributes delivered by the input tensor. These latent features may provide information on the short- and long-range interactions that may occur after protein folding. These features will represent a protein in a lower-dimensional space, which reduces the computational complexity of predicting interactions.
Following encoding, the decoder reconstructs the input data from the latent features. The features are transformed into a 2-D tensor using a dense layer and a Reshape layer. Next, two TCN layers and then a Reshape layer transform the tensor to 3-D. Then, four 2-D transposed convolutional layers restore the original input tensor. Two 2-D convolution layers fully reconstruct the input with its original dimensions. The pseudocode of this architecture is presented in Alg. 1.
Algorithm 1
Convolutional-TCN Autoencoder [134].
 |
6. Classifying interactions
The datasets used in this work are retrieved from the publicly available databases STRING [63] and Negatome 2.0 [135]. The Negatome 2.0 includes experimentally curated negative (non-interacting protein pairs, i.e., with the lowest confidence score of zero for the interaction) and positive interactions (pairs with the highest confidence score of one for the interaction) for various organisms, such as Homo sapiens, Mus musculus, Saccharomyces cerevisiae, Escherichia coli, and Drosophila melanogaster. The STRING database includes a broader range of organisms compared to Negatome 2.0, such as Caenorhabditis elegans, Danio rerio, and Arabidopsis thaliana. To facilitate access to these datasets, we provided a GitHub repository with all data incorporated into this study. This repository includes the Negatome 2.0 datasets for Homo sapiens and Mus musculus (231 KB) and the protein sequence datasets with the amino acid sequence of each protein for Homo sapiens and Mus musculus (12.6 MB) used to train the autoencoders and PPI datasets (700 MB), which predict the PPIs.
The protein network dataset retrieved from the STRING database comprises pairs of proteins, and their respective interaction scores computed based on seven evidence channels [62], [63], as follows:
-
1.
Experiments channel: evidence originating from laboratory experiments (including biophysical, biochemical and genetic experiments). The primary interaction databases organised in the IMEx consortium [136], [137], plus BioGRID [138], are the main source for this channel.
-
2.
Database channel: evidence that has been confirmed by a human expert curator using information imported from pathway databases [139].
-
3.
Text mining channel: evidence collected by searching through all PubMed abstracts, through an in-house collection of more than three million full-text articles, and through other text collections related to proteins [140], [141]. An association score is defined based on how frequently a pair of proteins are mentioned in the same paper. When parsing one or more sentences via natural language processing, if a concept connecting the two proteins is discovered (such as ‘binding’ or ‘phosphorylation by’), the association score is increased.
-
4.
Co-expression channel: for this channel, gene expression data resulting from various expression experiments are normalised, pruned and then correlated [62], [63]. A high association score is assigned to protein pairs with consistent similarity in their expression patterns under different conditions.
-
5.
Neighbourhood channel: this is a genome-based prediction channel where an association score is given to protein pairs consistently observed in each other’s genome neighbourhood. This and the next two channels are generally most relevant for Bacteria and Archaea.
-
6.
Fusion channel: an association score is given to a protein pair when there are one or more organisms whose respective orthologs have fused into a single, protein-coding gene.
-
7.
Co-occurrence channel: in a given organism, the phylogenetic distribution of orthologs of all proteins is evaluated. An association score is assigned if two proteins show high resemblance in this distribution, i.e. their orthologs tend to be observed as ‘present’ or ‘absent’ in the same subsets of organisms [142].
Each technique has its own disadvantages leading to biases, false positives and false negatives. The STRING dataset quantifies these uncertainties by allocating scores to proposed protein interactions indicating the probability of the interaction existence according to the nature and quality of the supporting evidence [62], [63]. All these scores are integrated into a final “combined score” that reflects the confidence on whether an interaction between two non-identical proteins is biologically meaningful, given all the contributing evidence in the STRING database [63]. Consequently, the interactions are associated with a score between zero and one (zero for non-interacting pairs and one for pairs whose interaction is indicated by strong evidence).
In this work, the prediction of protein–protein interactions is formulated as multi-class classification problems. Three separate scenarios are defined to evaluate the interactions from different perspectives. Firstly, we begin with a common scenario where interactions are divided into interacting and non-interacting pairs. As described in Table 2, interactions with the score within the [0,0.5] range are labelled non-interacting, and those belonging to (0.5,1] range are labelled interacting pairs. In the STRING database, interactions with a score of 0.5 are indicated as false positives, suggesting erroneous labelling for roughly every second interaction [65], [66].
The next scenarios take a more nuanced approach: interactions scoring 0.4 in STRING are considered mediumconfidence, and interactions scoring above 0.7 are high-confidence interactions (i.e. more likely to interact, based on the available evidence) [62], [63], [65]. We hence propose a score range of (0.4,0.7], indicating medium confidence, for the second and third scenarios.
This mid-range may include falsely labelled (false-positive) pairs. The second scenario comprises three classes – non-interacting, interacting, and the aforementioned medium-confidence interactions, as shown in Table 3.
Finally, the third scenario comprises five classes. The non-interacting and interaction ranges from the second scenario are further subdivided into two, as reported in Table 4. The interactions scoring in [0, 0.15] are noninteracting with the highest confidence, while those in (0.15, 0.4] are non-interacting with relatively less confidence. At the upper end, (0.7, 0.9] and (0.9,1] represent interacting pairs with high and highest confidence, respectively [65], [66], [143]. One advantage of defining these thresholds is that one may find high-affinity PPIs among those protein pairs with interaction scores in (0.9,1] range, which extends the applications of ProtInteract to identifying protein–ligand pairs [89].
The next section illustrates the deep CNN architecture.
7. Predicting PPI using a Deep CNN
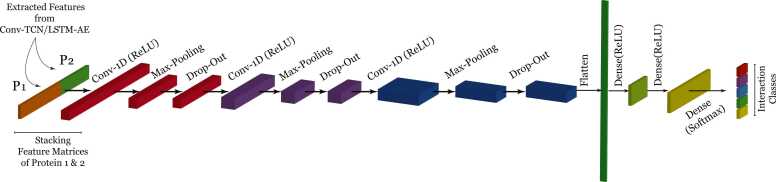
A deep CNN is designed, as shown in Fig. 6, to predict the probability of an interaction belonging to a class according to three defined scenarios. Initially, each protein is encoded using a Conv-TCN-AE (in ProtInteract) or Conv-LSTM-AE (for the baseline model). Then, protein pairs are constructed by stacked encoded proteins in tandem, as depicted in Fig. 1, to create the input tensor for the deep CNN.
Fig. 6.
Deep CNN architecture outputs the interaction probabilities in the defined classes. The output dimension changes for each scenario, where we have two neurons (two classes) for the first scenario, three neurons (three classes) for the second scenario and five neurons (five classes) for the final scenario.
The CNN architecture is described in Alg. 2. The CNN comprises three 1-D-convolutional layers, max-pooling layers to downsample the tensor, dropout layers to avoid overfitting and data leakage, and dense layers to map the tensor from the preceding layer to a desirable size. Finally, the output is obtained from a densely connected layer using a softmax activation function, which yields the probability of an interaction falling within the defined range of classes for each scenario according to Table 2, Table 3, Table 4. As described above, the first scenario entails a simple yes–no prediction of whether a protein pair is interacting or non-interacting. The second scenario includes a medium-confidence range in addition to interacting and non-interacting pairs. Finally, in the third scenario, the deep CNN outputs five values indicating the probability of given interaction belonging to each of the classes described in Table 4.
Algorithm 2
CNN – PPI prediction [134].
 |
8. Experimental results
Training the proposed framework requires two types of datasets: protein sequences and protein–protein interactions. In the first stage, dimensionality is reduced on a protein sequence dataset, extracting an abstract representation while preserving the sequential structure information in each protein. This is followed by encoding the PPI dataset and performing PPI prediction based on the scenarios in Table 2, Table 3, Table 4. The data processing and the training of the neural networks were performed on an HP Apollo System with 40 Dual Intel Xeon Gold 6149 processors, one Nvidia Tesla V100 (16 GB) graphics processor, and 378 GB of memory. The following sections explain these stages further in detail.
8.1. Dimensionality Reduction
Training and evaluation datasets are retrieved from the STRING database, which contains multiple datasets, including protein sequences, protein network data, and association scores between orthologous groups, amongst others. Here, we use the protein sequence to train the networks at the encoding stage and protein network datasets to train the deep CNN. This work is conducted on two species, namely, Homo sapiens and Mus musculus. The protein sequence dataset comprises protein IDs and their amino acid sequence from multiple species, including those reported in Table 5. Here, we consider proteins with a length of 2048 amino acids or less in order to constrain the overall data dimensionality.
Table 5.
The number of protein sequence instances retrieved from STRING database for H. sapiens and M. musculus.
| Species | Samples |
|---|---|
| H. sapiens | 19542 |
| M. musculus | 22024 |
The training hyper-parameters for both Conv-TCN-AE and Conv-LSTM-AE are acquired by grid search for the TCN and LSTM architectures. Both architectures are optimised in terms of hyperparameters and can be fairly compared. Grid search is an exhaustive method for finding an optimal combination of hyperparameters. This method divides the domain of hyperparameters into a discrete grid and calculates the performance metrics of every combination of hyperparameters using k-fold cross-validation. The part of the grid that maximises the average value in cross-validation yields the optimal combination of hyperparameters [144], as shown in Table 6. The training is conducted based on five-fold cross-validation. Five-fold cross-validation is an evaluation method that helps discover a machine learning model configuration that can best predict the outcome of unseen data. With this approach, the data is randomly shuffled and divided into five subsets of equal size, called ”folds”. The process is repeated five times, each using a different fold as the testing set (20%) and the combined other four folds (80%) as the training set. This allows for a robust evaluation of the model’s performance, as each data point is used for testing exactly once. The model’s performance is determined by averaging the results from the five iterations. First, this method supports reducing overfitting by training and evaluating the model on different subsets of the data. Second, data leakage is avoided by ensuring the model is not trained on or exposed to the test data, which provides a more realistic estimate of its ability to generalise to new data [145]. Finally, the validation set used during training is created by randomly selecting 20% of the data from the training set. The accuracy of each network corresponds to its ability to reconstruct encoded data after 100 epochs.
Table 6.
Training and validation hyperparameters for Conv-TCN-AE and Conv-LSTM-AE.
| Parameter | Value | Description |
|---|---|---|
| Padding Length | 2048 | Padding length of all amino acid sequences |
| Epochs | 100 | Training epochs per protein |
| Batch Size | 16 | Size of training batch |
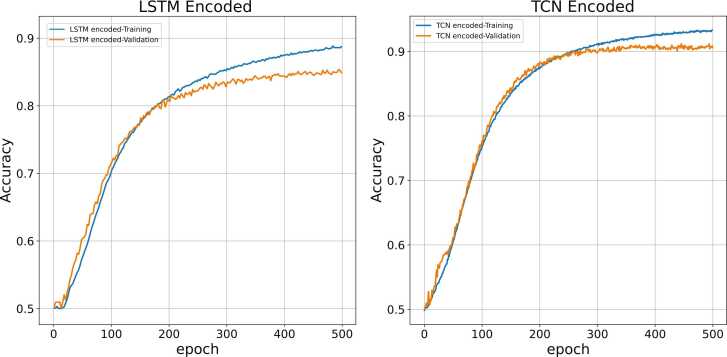
The Conv-TCN-AE and Conv-LSTM-AE reconstruction accuracy (similarity of the input tensor to reconstructed output tensor by reducing mean squared error) are compared for H. sapiens and M. musculus in Fig. 7, Fig. 8 respectively.
Fig. 7.
The accuracy of Conv-TCN-AE and Conv-LSTM-AE for H. sapiens.
Fig. 8.
The accuracy of Conv-TCN-AE and Conv-LSTM-AE for M. musculus.
It may be concluded that the TCN architecture is slightly more accurate. It is worth mentioning that ConvTCN-AE is noticeably faster and less computationally demanding to train compared to Conv-LSTM-AE [111]. In our tests, it took 2 h 37 min to train Conv-TCN-AE through five-fold cross-validation, whereas ConvLSTM-AE took 72 h 41 min to train, with the same number of LSTM and TCN layers. Moreover, both Conv-TCN-AE and Conv-LSTM-AE obtain better accuracy for H. sapiens than for M. musculus.(Table 7).
Table 7.
The five-fold cross-validation training times for Conv-TCN-AE and Conv-LSTM-AE.
| Architecture | Training Time |
|---|---|
| Conv-TCN-AE | 2 h 37 min |
| Conv-LSTM-AE | 72 h 41 min |
The PPI prediction stage is discussed thoroughly in the next section.
8.2. PPI prediction
As mentioned earlier, ProtInteract’s performance is assessed using two organisms, H. sapiens and M. musculus. Subsequently, a deep CNN predicts the probability of an interaction belonging to the defined classes according to three defined scenarios (see Table 2, Table 3, Table 4). The dataset for this task is created by merging the STRING and the Negatome 2.0 datasets and removing the duplicated samples. As mentioned earlier, Negatome 2.0 only comprises interactions with a score of zero non-interacting with the highest confidence) and one (interacting with the highest confidence) [135], whereas STRING includes protein pairs with interaction scores distributed on a continuum from zero and one [62], [63], [65]. Both STRING and Negatome 2.0 datasets are imbalanced in the sense of having uneven numbers of non-interacting, and interacting pairs [146].
For instance, let us consider the H. sapiens PPI dataset from STRING. Nearly 83% of the dataset belongs to interactions in class 1 from Table 2 or class 1 from Table 3 or classes 1 and 2 from Table 4; showing considerable imbalance in the dataset, which has an adverse effect on training the deep CNN [147]. To tackle this issue, we randomly downsample the classes with a higher number to the class with the least number of instances. For instance, the H. sapiens dataset that is distributed into three classes contains 858726 samples in class 1, 112706 in class 2 and 74814 in class 3. Therefore, we randomly sample 74814 pairs from classes 1 and 2 to obtain three balanced classes. Thus, a balanced dataset is created for each scenario, and the number of samples is reported in Table 8. Furthermore, the training, testing and validation are performed using five-fold cross-validation.
Table 8.
The balanced protein–protein interaction datasets for H. sapiens and M. musculus include a different number of protein pairs for each class, corresponding to each classification scenario.
| Species/Scenario (samples) | Two classes | Three classes | Five classes |
|---|---|---|---|
| H. sapiens | 160113 | 74813 | 27128 |
| M. musculus | 75501 | 42905 | 11650 |
The hyperparameters for training the CNN are reported in Table 9. These parameters were obtained by grid search [144].
Table 9.
Hyperparameters of training and validation of CNN for PPI prediction.
| Parameters | Value | Description |
|---|---|---|
| Epochs | 500 | Training epochs |
| Batch Size | 64 | Size of training batch |
| α | 5 × 10−5 | Learning rate |
8.2.1. H. sapiens PPI prediction results
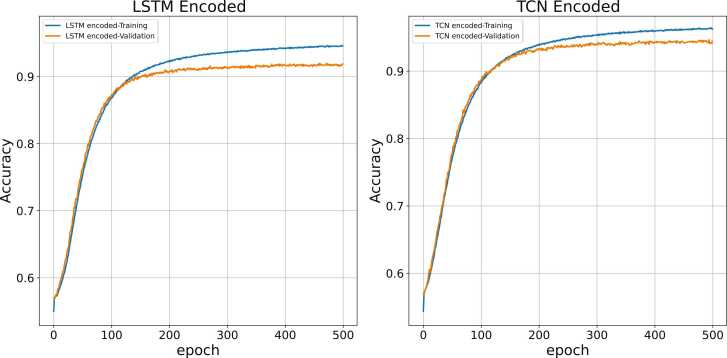
The classification accuracies (correctly labelling an interaction) using the deep CNN are illustrated in Fig. 9, Fig. 10, Fig. 11 for two, three and five classes respectively. Accordingly, the deep CNN achieves relatively higher accuracy in all three scenarios for those proteins encoded by Conv-TCN-AE than Conv-LSTM-AE. This may be due to the extended long-term memory of TCN layers, which allows better retention of information on long-range interactions compared to LSTM layers [93], [94], [127]. Therefore, it can be concluded that encoded features of Conv-TCN-AE encapsulate more informative attributes.
Fig. 9.
Prediction accuracy of CNN with input features encoded by Conv-LSTM-AE and Conv-TCN-AE for two-class classification (H. sapiens).
Fig. 10.
Prediction accuracy of CNN with input features encoded by Conv-LSTM-AE and Conv-TCN-AE for three-class classification (H. sapiens).
Fig. 11.
Prediction accuracy of CNN with input features encoded by Conv-LSTM-AE and Conv-TCN-AE for five-class classification (H. sapiens).
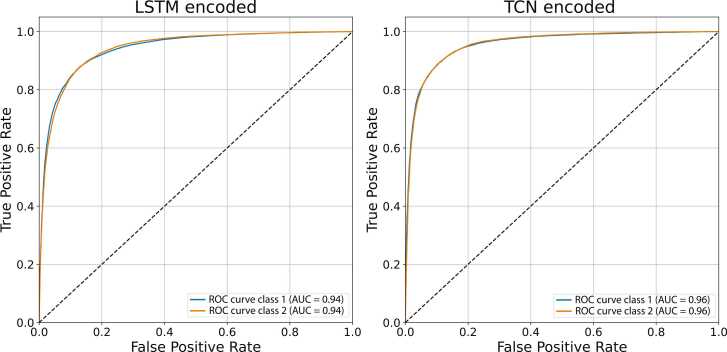
The Receiver Operator Characteristic (ROC) curve is a graphical plot visualising the ability of deep CNN to correctly predict the class of an interaction. A plot relates the true positive rate (TPR) on the y-axis versus the false positive rate (FPR) on the x-axis. Accordingly, the top left corner of the plot represents a false positive rate of zero and a true positive rate of one. Thus, a larger area under the curve (AUC) is usually better. Here, the deep CNN predicts the probability of an interaction falling within the range of the defined classes, reflecting the confidence level of an interaction between a pair of proteins.
Fig. 12 shows the ROC curve for the two-class classification scenario. The deep CNN obtains higher AUC using TCN-encoded features than LSTM-encoded features.
Fig. 12.
ROC curve for two-class classification using LSTM- and TCN-encoded features (H. sapiens).
Fig. 13 illustrates the ROC curve for the three-class classification scenario. For both TCN-encoded and LSTM-encoded features, the AUC diminishes compared to the two-class classification.
Fig. 13.
ROC curve for three-class classification using LSTM- and TCN-encoded features (H. sapiens).
The ROC curve for the five-class scenario is depicted in Fig. 14, where AUC is the lowest among all three scenarios. Note that the AUC for all three scenarios is higher for TCN-encoded features than LSTM-encoded features. This may once again show the advantage of the TCN architecture compared to LSTM architecture for encoding proteins. From AUC for all scenarios, it may be inferred that ProtInteract can accurately classify pairs with the lowest and highest interaction confidence.
Fig. 14.
ROC curve for five-class classification using LSTM- and TCN-encoded features (H. sapiens).
8.2.2. M. musculus PPI prediction results
The classification accuracies for M. musculus are shown in Fig. 15, Figs. 16 and 17, for all three scenarios, respectively.
Fig. 15.
Prediction accuracy of CNN with input features encoded by Conv-LSTM-AE and Conv-TCN-AE for two classes classification (M. musculus).
Fig. 16.
Prediction accuracy of CNN with input features encoded by Conv-LSTM-AE and Conv-TCN-AE for three classes classification (M. musculus).
Fig. 17.
Prediction accuracy of CNN with input features encoded by Conv-LSTM-AE and Conv-TCN-AE for five classes classification (M. musculus).
By comparing Fig. 9 to Fig. 15, classification accuracy for M. musculus is slightly lower than H. sapiens. This may be due to the smaller training set for M. musculus compared to H. sapiens (see Table 8).
On increasing the number of classes, the classification accuracy decreases.
The following figures, Fig. 12, Fig. 13, and Fig. 14, display the ROC curves for M. musculus for all three scenarios, respectively.
From Fig. 18, Fig. 19, Fig. 20, the average AUC for both TCN-encoded and LSTM-encoded cases show slight decreases with the increase of classes. However, classes three from the second scenario and five from the third scenario have shown equal or higher AUC compared to both classes from the two-class classification scenario. This means that ProtInteract can distinguish the classes with the highest confidence of interaction regardless of the scenario.
Fig. 18.
ROC curve for two-class classification using LSTM- and TCN-encoded features (M. musculus).
Fig. 19.
ROC curve for three-class classification using LSTM- and TCN-encoded features (M.musculus).
Fig. 20.
ROC curve for five-class classification using LSTM- and TCN-encoded features (M. musculus).
8.3. Discussion
By analysing the results obtained by ProtInteract, as reported in Table 10, one may infer that ProtInteract classifies the interaction more accurately for H. sapiens than for M. musculus. This may be due to the fact that H. sapiens has 112%, 74%, and 132.50% more training samples in the balanced datasets, as reported in Table 8, compared to the M. musculus balanced dataset for two, three and five classes, respectively. Additionally, the lower number of training samples among three scenarios results in a slight decline in classification accuracy (see Table 8).
Table 10.
ProtInteract performance of five-fold cross-validation for three different multi-class classification scenarios.
| Species | Scenario | Accuracy |
AUC |
AveragePrecision |
|||
|---|---|---|---|---|---|---|---|
| TCN Encoded | LSTM Encoded | TCN Encoded | LSTM Encoded | TCN Encoded | LSTM Encoded | ||
| H. sapiens | 2- classes | 0.9568 | 0.9315 | 0.9600 | 0.9400 | 0.9550 | 0.9400 |
| 3- classes | 0.9244 | 0.9182 | 0.9340 | 0.9200 | 0.8950 | 0.8600 | |
| 5 - classes | 0.9132 | 0.8756 | 0.9060 | 0.8940 | 0.7920 | 0.7620 | |
| M. musculus | 2- classes | 0.9145 | 0.8456 | 0.8600 | 0.8100 | 0.8450 | 0.7850 |
| 3- classes | 0.8983 | 0.8337 | 0.8600 | 0.8100 | 0.7870 | 0.7850 | |
| 5- classes | 0.8749 | 0.8216 | 0.8480 | 0.7980 | 0.7000 | 0.6800 | |
Let us consider H. sapiens: for TCN-encoded features, the deep CNN accuracy of correctly labelling interactions of two-class is 3.50% and 4.77% higher than three and five classes, respectively. For LSTM-encoded features, the classification accuracy of two-class is 1.40% than three and 6.30% higher than five classes.
TCN-encoded (LSTM-encoded) features are those encoded by Conv-TCN-AE (Conv-LSTM-AE), which serve as the input tensor of the deep CNN. The area under the ROC curve (AUC) portrays the discriminative ability of the deep CNN for each class [148]. Considering all three scenarios, AUC is evidently higher when the deep CNN is fed by TCN-encoded features. The deep CNN achieves higher accuracy, precision and AUC, where the input tensor is encoded by Conv-TCN-AE (TCN-encoded) compared to the baseline model, Conv-LSTM-AE (LSTM-encoded). Considering H. sapiens, the deep CNN achieves 2.7%, 0.6% and 4.3% higher accuracy when fed by TCN-encoded features than LSTM-encoded features.
A similar pattern appears for M. musculus, where, for TCN-encoded features, classification accuracy for two class is 1.80% and 4.50% higher than three and five classes, respectively. For LSTM-encoded features, the deep CNN obtains 1.42% and 2.92% higher accuracy for two-class compared to three and five classes. Finally, considering M. musculus, the deep CNN with TCN-encoded features obtains 6.70%, 7.70% and 4% higher accuracy than LSTM-encoded features for all three scenarios, respectively.
Consequently, one may conclude that Conv-TCN-AE architecture is not only considerably faster than Conv-TCN-AE (baseline model) to train but also that ProtInteract’s accuracy of correctly labelling interactions is noticeably higher for protein pairs encoded by Conv-TCN-AE than by Conv-LSTM-AE.
9. Conclusion
Various diseases such as neurodegenerative diseases, infectious diseases, and cancer are closely related to abnormal protein–protein interactions [6], [7], [8]. Therefore, identifying protein–protein interactions helps pave the way towards developing new drugs and targeted therapeutic approaches [9], [10], [11].
In this work, a deep learning framework is developed to predict protein–protein interactions corresponding to sequential patterns encoded for each protein. The prediction problem is expressed as a multi-class classification problem under three scenarios [64]. Each scenario is defined according to different confidence score thresholds of protein interactions [65]. This framework is composed of two components. The first component encodes the amino-acid sequences and reduces the data dimensionality representing each protein while extracting underlying sequential informative features. These tasks are implemented using a novel deep autoencoder architecture called Conv-TCN-AE. We proposed a similar architecture using LSTM layers called Conv-LSTM-AE as a baseline model to compare the performance of TCN layers with a well-known model such as LSTM [93], [94], [117], [127]. The second component is a deep CNN that predicts the class of a given interaction based on the confidence score thresholds from the STRING database.
One of the significant achievements of the proposed framework is its low computational complexity and relatively fast response. Training LSTM and TCN networks as an individual architecture is extremely time-consuming and requires comparably high computational capacity, preferably GPU [149]. However, the proposed autoencoder architecture, namely, Conv-TCN-AE, benefits from the sequential pattern recognition ability of TCN while considerably reducing the training time and computational complexity [117]. The Conv-TCN-AE encodes the dataset with relatively high dimensions into a sequential tensor with a limited number of informative features. Therefore, we can analyse large sets of data for far less time.
To further extend this work, we plan to improve the proposed framework by incorporating a bidirectional architecture and adding a generative architecture to accurately predict the interaction scores. In addition, we will test ProtInteract on other species, including Saccharomyces cerevisiae, Escherichia coli, Drosophila melanogaster and Caenorhabditis elegans.
Authorship contributions
-
•
Conception and design of study: Farzan Soleymani, Eric Paquet, Herna Lydia Viktor
-
•
Acquisition of data: Farzan Soleymani, Eric Paquet, Herna Lydia Viktor
-
•
Analysis and/or interpretation of data: Farzan Soleymani, Eric Paquet
-
•
Drafting the manuscript: Farzan Soleymani, Eric Paquet, Davide Spinello
-
•
Revising the manuscript critically for important intellectual content: Eric Paquet, Herna Lydia Viktor, Wojtek Michalowski, Davide Spinello
-
•
Approval of the version of the manuscript to be published (the names of all authors must be listed): Farzan Soleymani, Eric Paquet, Herna Lydia Viktor, Wojtek Michalowski, Davide Spinello
Acknowledgements
All persons who have made substantial contributions to the work reported in the manuscript (e.g., technical help, writing and editing assistance, general support), but who do not meet the criteria for authorship, are named in the Acknowledgements and have given us their written permission to be named. If we have not included an Acknowledgements, then that indicates that we have not received substantial contributions from non-authors.
Footnotes
In an amino acid sequence of length N, any k < N consecutive amino acids are called a k-mer. These are collected using a sliding window of length 1 ≤ k ≤ N.
https://pdb101.rcsb.org/motm/185
Contributor Information
Farzan Soleymani, Email: fsoleyma@uottawa.ca.
Eric Paquet, Email: Eric.Paquet@nrc-cnrc.gc.ca.
Herna Lydia Viktor, Email: hviktor@uottawa.ca.
Wojtek Michalowski, Email: wojtek@telfer.uottawa.ca.
Davide Spinello, Email: dspinell@uottawa.ca.
Appendix
The LSTM architecture is explained as follows.
A.1. Long Short-Term Memory
As mentioned earlier, RNNs were introduced in order to incorporate sequential information when mapping input to output sequences [35], [111]. This means RNNs have the capability of comprehending and interpreting input and output sequences by identifying the correlation and progression of elements within the sequence. This is achieved through the utilisation of a memory component within RNNs, thus enabling them to retain and recall previous information in the sequence. This allows for the formation of decisions based on a comprehensive understanding of the entire sequence, as opposed to just the current input. However, RNNs often suffer from vanishing gradients, limiting the range of context to which they can be applied [150], [151]. To cope with this issue, LSTM architecture was introduced [112], as shown in Fig. 21.
Fig. 21.
LSTM architecture.
The LSTM architecture consists of a set of recurrently connected memory blocks and corresponding control gates, namely, the forget gate ft, the input gate it and the output gate ot which update and control the cell states [152]. The input and forget gates control the current network memory and a flow of new information. More specifically, as the new information flows into the network, the forget gate manages what information needs to be removed from cell states, while the input gate controls what information will be stored in the cell state. Finally, the output gate determines the encoded information to be forwarded as the input of the next time step.
| (A.1a) |
| (A.1b) |
where σ is the sigmoid activation function, W and b are the weight matrix and bias vector, and ⊗ denotes point-wise product. The initial operation is conducted by the forget gate ft, Eq. (A.1a), which controls which information to keep or to remove. The LSTM architecture contains the hidden state ht, Eq. (A.1b) that is formed by the sequential information. The next step is to store the new input information in the cell state via the input gate it, Eq. (A.2a). Accordingly, the cell state can be modified via candidate values by Eq. (A.2b) and Eq. (A.2c). Finally, the LSTM determines the output of each unit by Eq. (A.2d).
| it = σ(Wi·[ht−1,xt] + bi) | (A.2a) |
| (A.2b) |
| (A.2c) |
| ot = σ(Wo·[ht−1,xt] + bo) | (A.2d) |
The following section illustrates the architecture of the convolutional LSTM autoencoder as the baseline model in this work.
A.2. Convolutional LSTM Autoencoder
The architecture of the convolutional LSTM autoencoder (Conv-LSTM-AE) is shown in Fig. 22. This architecture comprises the well-known LSTM layer, which serves as a baseline model to compare with Conv-TCN-AE in the proposed framework. Similar to Conv-TCN-AE, Conv-LSTM-AE consists of two main components. The encoder receives high-dimensionality data to encode into a lower dimensionality and extracts the sequential patterns using two layers of LSTM. Next, the latent features are extracted from the output of a densely connected layer. The decoder reconstructs the input data via dense, LSTM, and 2-D transposed convolutional layers. Finally, a 2-D convolutional layer maps the data into the proper dimensionality. The pseudocode of this architecture is shown in Alg. 3.
Algorithm 3
Convolutional-LSTM Autoencoder [134].
 |
Fig. 22.
Conv-LSTM-AE architecture that reduces the dimensionality of input data and extracts informative and uncorrelated features.
References
- 1.Lodish H., Zipursky S.L. Molecular cell biology. Biochem Mol Biol Educ. 2001;29:126–133. [Google Scholar]
- 2.A.W. White, A.D. Westwell, G. Brahemi, Protein–protein interactions as targets for small-molecule therapeutics in cancer, Expert reviews in molecular medicine 10. [DOI] [PubMed]
- 3.Blazer L.L., Neubig R.R. Small molecule protein–protein interaction inhibitors as cns therapeutic agents: current progress and future hurdles. Neuropsychopharmacology. 2009;34(1):126–141. doi: 10.1038/npp.2008.151. [DOI] [PubMed] [Google Scholar]
- 4.Rosell M., Fern´andez-Recio J. Hot-spot analysis for drug discovery targeting protein-protein interactions. Expert Opin Drug Discov. 2018;13(4):327–338. doi: 10.1080/17460441.2018.1430763. [DOI] [PubMed] [Google Scholar]
- 5.Liu L., Zhu X., Ma Y., Piao H., Yang Y., Hao X., et al. Combining sequence and network information to enhance protein–protein interaction prediction. BMC Bioinforma. 2020;21(16):1–13. doi: 10.1186/s12859-020-03896-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.S. Ferrari, F. Pellati, M. Costi, Disruption of protein-protein interfaces (2013).
- 7.Rual J.-F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437(7062):1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 8.Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122(6):957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 9.F. Browne, H. Zheng, H. Wang, F. Azuaje, From experimental approaches to computational techniques: a review on the prediction of protein-protein interactions., Advances in Artificial Intelligence (16877470).
- 10.Skrabanek L., Saini H.K., Bader G.D., Enright A.J. Computational prediction of protein–protein interactions. Mol Biotechnol. 2008;38(1):1–17. doi: 10.1007/s12033-007-0069-2. [DOI] [PubMed] [Google Scholar]
- 11.Lu H., Zhou Q., He J., Jiang Z., Peng C., Tong R., et al. Recent advances in the development of protein– protein interactions modulators: mechanisms and clinical trials. Signal Transduct Target Ther. 2020;5(1):1–23. doi: 10.1038/s41392-020-00315-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwikowski B., Uetz P., Fields S. A network of protein–protein interactions in yeast. Nat Biotechnol. 2000;18(12):1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- 13.Ito T., Tashiro K., Muta S., Ozawa R., Chiba T., Nishizawa M., et al. Toward a protein–protein interaction map of the budding yeast: a comprehensive system to examine twohybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci. 2000;97(3):1143–1147. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavin A.-C., B¨osche M., Krause R., Grandi P., Marzioch M., Bauer A., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415(6868):141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 15.Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.-L., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 16.Pandey A., Mann M. Proteomics to study genes and genomes. Nature. 2000;405(6788):837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 17.Figeys D. Novel approaches to map protein interactions. Curr Opin Biotechnol. 2003;14(1):119–125. doi: 10.1016/s0958-1669(02)00005-8. [DOI] [PubMed] [Google Scholar]
- 18.Noor Z., Ahn S.B., Baker M.S., Ranganathan S., Mohamedali A. Mass spectrometry–based protein identification in proteomics—a review. Brief Bioinforma. 2021;22(2):1620–1638. doi: 10.1093/bib/bbz163. [DOI] [PubMed] [Google Scholar]
- 19.Garza K.Y., Feider C.L., Klein D.R., Rosenberg J.A., Brodbelt J.S., Eberlin L.S. Desorption electrospray ionization mass spectrometry imaging of proteins directly from biological tissue sections. Anal Chem. 2018;90(13):7785–7789. doi: 10.1021/acs.analchem.8b00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacBeath G., Schreiber S.L. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289(5485):1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 21.Bu¨ssow K., Nordhoff E., Lu¨bbert C., Lehrach H., Walter G. A human cdna library for high-throughput protein expression screening. Genomics. 2000;65(1):1–8. doi: 10.1006/geno.2000.6141. https://www.sciencedirect.com/science/article/pii/S088875430096141X (URL) [DOI] [PubMed] [Google Scholar]
- 22.Brizuela L., Braun P., LaBaer J. Flexgene repository: from sequenced genomes to gene repositories for high-throughput functional biology and proteomics. Mol Biochem Parasitol. 2001;118(2):155–165. doi: 10.1016/s0166-6851(01)00366-8. [DOI] [PubMed] [Google Scholar]
- 23.Brizuela L., Richardson A., Marsischky G., Labaer J. The flexgene repository: exploiting the fruits of the genome projects by creating a needed resource to face the challenges of the post-genomic era. Arch Med Res. 2002;33(4):318–324. doi: 10.1016/s0188-4409(02)00372-7. [DOI] [PubMed] [Google Scholar]
- 24.Piehler J. New methodologies for measuring protein interactions in vivo and in vitro. Curr Opin Struct Biol. 2005;15(1):4–14. doi: 10.1016/j.sbi.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 25.V.S. Rao, K. Srinivas, G. Sujini, G. Kumar, Protein-protein interaction detection: methods and analysis, International journal of proteomics 2014. [DOI] [PMC free article] [PubMed]
- 26.Wu J., Paquet E., Viktor H.L., Michalowski W. Paying attention: using a siamese pyramid network for the prediction of protein-protein interactions with folding and self-binding primary sequences. 2021 Int Jt Conf Neural Netw (IJCNN) 2021:1–8. [Google Scholar]
- 27.Wang T., Li L., Huang Y.-A., Zhang H., Ma Y., Zhou X. Prediction of protein-protein interactions from amino acid sequences based on continuous and discrete wavelet transform features. Molecules. 2018;23(4):823. doi: 10.3390/molecules23040823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding Z., Kihara D. Computational identification of protein-protein interactions in model plant proteomes. Sci Rep. 2019;9(1):1–13. doi: 10.1038/s41598-019-45072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.S. Tsukiyama, M.M. Hasan, S. Fujii, H. Kurata, Lstm-phv: Prediction of human-virus protein-protein interactions by lstm with word2vec, bioRxiv. [DOI] [PMC free article] [PubMed]
- 30.I. Goodfellow, Y. Bengio, A. Courville, Deep learning, MIT press, 2016.
- 31.Jangmin O., Lee J., Lee J.W., Zhang B.-T. Adaptive stock trading with dynamic asset allocation using reinforcement learning. Inf Sci. 2006;176(15):2121–2147. [Google Scholar]
- 32.Wang Y., You Z., Li L., Chen Z. A survey of current trends in computational predictions of protein-protein interactions. Front Comput Sci. 2020;14(4):1–12. [Google Scholar]
- 33.Soleymani F., Paquet E. Financial portfolio optimization with online deep reinforcement learning and restricted stacked autoencoder—deepbreath. Expert Syst Appl. 2020;156 [Google Scholar]
- 34.Soleymani F., Paquet E. Deep graph convolutional reinforcement learning for financial portfolio management– deeppocket. Expert Syst Appl. 2021;182 [Google Scholar]
- 35.Bao W., Yue J., Rao Y. A deep learning framework for financial time series using stacked autoencoders and long-short term memory. PloS One. 2017;12(7) doi: 10.1371/journal.pone.0180944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinton G.E., Salakhutdinov R.R. Reducing the dimensionality of data with neural networks. science. 2006;313(5786):504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 37.C.O.S. Sorzano, J. Vargas, A.P. Montano, A survey of dimensionality reduction techniques, arXiv preprint arXiv:1403.2877.
- 38.Gao W., Mahajan S.P., Sulam J., Gray J.J. Deep learning in protein structural modeling and design. Patterns. 2020 doi: 10.1016/j.patter.2020.100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X.-w, Jeong J.C. Sequence-based prediction of protein interaction sites with an integrative method. Bioinformatics. 2009;25(5):585–591. doi: 10.1093/bioinformatics/btp039. [DOI] [PubMed] [Google Scholar]
- 40.Zahiri J., Yaghoubi O., Mohammad-Noori M., Ebrahimpour R., Masoudi-Nejad A. Ppievo: Protein–protein interaction prediction from pssm based evolutionary information. Genomics. 2013;102(4):237–242. doi: 10.1016/j.ygeno.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 41.You Z.-H., Chan K.C., Hu P. Predicting protein-protein interactions from primary protein sequences using a novel multi-scale local feature representation scheme and the random forest. PloS One. 2015;10(5) doi: 10.1371/journal.pone.0125811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.F. Soleymani, E. Paquet, H. Viktor, W. Michalowski, D. Spinello, Protein–protein interaction prediction with deep learning: A comprehensive review, Computational and Structural Biotechnology Journal. [DOI] [PMC free article] [PubMed]
- 43.Chen H., Zhou H.-X. Prediction of interface residues in protein–protein complexes by a consensus neural network method: test against nmr data, Proteins: Structure. Funct, Bioinforma. 2005;61(1):21–35. doi: 10.1002/prot.20514. [DOI] [PubMed] [Google Scholar]
- 44.Rodgers-Melnick E., Culp M., DiFazio S.P. Predicting whole genome protein interaction networks from primary sequence data in model and non-model organisms using ents. BMC Genom. 2013;14(1):1–17. doi: 10.1186/1471-2164-14-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emamjomeh A., Goliaei B., Torkamani A., Ebrahimpour R., Mohammadi N., Parsian A. Protein-protein interaction prediction by combined analysis of genomic and conservation information. Genes Genet Syst. 2014;89(6):259–272. doi: 10.1266/ggs.89.259. [DOI] [PubMed] [Google Scholar]
- 46.Kotlyar M., Pastrello C., Pivetta F., Lo Sardo A., Cumbaa C., Li H., et al. In silico prediction of physical protein interactions and characterization of interactome orphans. Nat Methods. 2015;12(1):79–84. doi: 10.1038/nmeth.3178. [DOI] [PubMed] [Google Scholar]
- 47.Chang J.-W., Zhou Y.-Q., Ul Qamar M.T., Chen L.-L., Ding Y.-D. Prediction of protein–protein interactions by evidence combining methods. Int J Mol Sci. 2016;17(11):1946. doi: 10.3390/ijms17111946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.B.-Q. Li, K.-Y. Feng, L. Chen, T. Huang, Y.-D. Cai, Prediction of protein-protein interaction sites by random forest algorithm with mrmr and ifs, PloS one. [DOI] [PMC free article] [PubMed]
- 49.X. Hu, C. Feng, T. Ling, M. Chen, Deep learning frameworks for protein-protein interaction prediction, Computational and Structural Biotechnology Journal. [DOI] [PMC free article] [PubMed]
- 50.Jia J., Li X., Qiu W., Xiao X., Chou K.-C. ippi-pseaac (cgr): Identify protein-protein interactions by incorporating chaos game representation into pseaac. J Theor Biol. 2019;460:195–203. doi: 10.1016/j.jtbi.2018.10.021. [DOI] [PubMed] [Google Scholar]
- 51.Pan J., Li L.-P., Yu C.-Q., You Z.-H., Guan Y.-J., Ren Z.-H. Sequence-based prediction of plant proteinprotein interactions by combining discrete sine transformation with rotation forest. Evolut Bioinforma. 2021;17 doi: 10.1177/11769343211050067. 11769343211050067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Du X., Sun S., Hu C., Yao Y., Yan Y., Zhang Y. Deepppi: boosting prediction of protein–protein interactions with deep neural networks. J Chem Inf Model. 2017;57(6):1499–1510. doi: 10.1021/acs.jcim.7b00028. [DOI] [PubMed] [Google Scholar]
- 53.Yang X., Yang S., Lian X., Wuchty S., Zhang Z. Transfer learning via multi-scale convolutional neural layers for human–virus protein–protein interaction prediction. Bioinformatics. 2021;37(24):4771–4778. doi: 10.1093/bioinformatics/btab533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lei Y., Li S., Liu Z., Wan F., Tian T., Li S., et al. A deep-learning framework for multi-level peptide–protein interaction prediction. Nat Commun. 2021;12(1):1–10. doi: 10.1038/s41467-021-25772-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sledzieski S., Singh R., Cowen L., Berger B. D-script translates genome to phenome with sequence-based, structure-aware, genome-scale predictions of protein-protein interactions. Cell Syst. 2021;12(10):969–982. doi: 10.1016/j.cels.2021.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bepler T., Berger B. Learning protein sequence embeddings using information from structure. arXiv Prepr arXiv. 1902:08661. [Google Scholar]
- 57.Hu X., Feng C., Zhou Y., Harrison A., Chen M. Deeptrio: a ternary prediction system for protein–protein interaction using mask multiple parallel convolutional neural networks. Bioinformatics. 2022;38(3):694–702. doi: 10.1093/bioinformatics/btab737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen M., Ju C.J.-T., Zhou G., Chen X., Zhang T., Chang K.-W., et al. Multifaceted protein–protein interaction prediction based on siamese residual rcnn. Bioinformatics. 2019;35(14):i305–i314. doi: 10.1093/bioinformatics/btz328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.K. Cho, B. VanMerri¨enboer, C. Gulcehre, D. Bahdanau, F. Bougares, H. Schwenk, Y. Bengio,Learning phrase representations using rnn encoder-decoder for statisticalmachine translation, arXiv preprint arXiv:1406.1078.
- 60.Yang F., Fan K., Song D., Lin H. Graph-based prediction of protein-protein interactions with attributed signed graph embedding. BMC Bioinforma. 2020;21(1):1–16. doi: 10.1186/s12859-020-03646-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kipf T.N., Welling M. Variational graph auto-encoders. arXiv Prepr arXiv. 1611:07308. [Google Scholar]
- 62.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., et al. The string database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D. Szklarczyk, R. Kirsch,M. Koutrouli, K. Nastou, F. Mehryary, R. Hachilif, A. L. Gable, T. Fang, N. T.Doncheva, S. Pyysalo, et al., The string database in 2023: protein–proteinassociation networks and functional enrichment analyses for any sequencedgenome of interest, Nucleic Acids Research. [DOI] [PMC free article] [PubMed]
- 64.M. Grandini, E. Bagli, G. Visani, Metrics for multi-class classification: an overview, arXiv preprint arXiv:2008.05756.
- 65.Crosara K.T.B., Moffa E.B., Xiao Y., Siqueira W.L. Merging in-silico and in vitro salivary protein complex partners using the string database: a tutorial. J Proteom. 2018;171:87–94. doi: 10.1016/j.jprot.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 66.Tran L., Hamp T., Rost B. Profppidb: pairs of physical protein-protein interactions predicted for entire proteomes. Plos One. 2018;13(7) doi: 10.1371/journal.pone.0199988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ju J., Liu F.-A. Multivariate time series data prediction based on att-lstm network. Appl Sci. 2021;11(20):9373. [Google Scholar]
- 68.Bandara K., Bergmeir C., Hewamalage H. Lstm-msnet: leveraging forecasts on sets of related time series with multiple seasonal patterns. IEEE Trans Neural Netw Learn Syst. 2020;32(4):1586–1599. doi: 10.1109/TNNLS.2020.2985720. [DOI] [PubMed] [Google Scholar]
- 69.Bandara K., Bergmeir C., Smyl S. Forecasting across time series databases using recurrent neural networks on groups of similar series: a clustering approach. Expert Syst Appl. 2020;140 [Google Scholar]
- 70.Bandara K., Hewamalage H., Liu Y.-H., Kang Y., Bergmeir C. Improving the accuracy of global forecasting models using time series data augmentation. Pattern Recognit. 2021;120 [Google Scholar]
- 71.Jha K., Saha S. Amalgamation of 3d structure and sequence information for protein–protein interaction prediction. Sci Rep. 2020;10(1):1–14. doi: 10.1038/s41598-020-75467-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo Y., Yu L., Wen Z., Li M. Using support vector machine combined with auto covariance to predict protein–protein interactions from protein sequences. Nucleic Acids Res. 2008;36(9):3025–3030. doi: 10.1093/nar/gkn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 74.Biro J. Amino acid size, charge, hydropathy indices and matrices for protein structure analysis. Theor Biol Med Model. 2006;3(1):1–12. doi: 10.1186/1742-4682-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shen J., Zhang J., Luo X., Zhu W., Yu K., Chen K., et al. Predicting protein–protein interactions based only on sequences information. Proc Natl Acad Sci. 2007;104(11):4337–4341. doi: 10.1073/pnas.0607879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang S.-W., Hao L.-Y., Zhang T.-H. Prediction of protein–protein interaction with pairwise kernel support vector machine. Int J Mol Sci. 2014;15:3220–3233. doi: 10.3390/ijms15023220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Y.-A. Huang, Z.-H. You, X. Gao, L. Wong, L. Wang, Using weighted sparse representation model combined with discrete cosine transformation to predict protein-protein interactions from protein sequence, BioMed research international 2015. [DOI] [PMC free article] [PubMed]
- 78.You Z.-H., Yu J.-Z., Zhu L., Li S., Wen Z.-K. A mapreduce based parallel svm for large-scale predicting protein–protein interactions. Neurocomputing. 2014;145:37–43. [Google Scholar]
- 79.W. Ma, Y. Cao, W. Bao, B. Yang, Y. Chen, Act-svm: Prediction of protein-protein interactions based on support vector basis model, Scientific Programming 2020.
- 80.Sun T., Zhou B., Lai L., Pei J. Sequence-based prediction of protein interaction using a deep-learning algorithm. BMC Bioinforma. 2017;18(1):1–8. doi: 10.1186/s12859-017-1700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.S. Sledzieski, R. Singh, L. Cowen, B. Berger, Sequence-based prediction of protein-protein interactions: a structure-aware interpretable deep learning model, bioRxiv. [DOI] [PMC free article] [PubMed]
- 82.Su X.-R., You Z.-H., Chen Z.-H., Yi H.-C., Guo Z.-H. International Conference on Intelligent Computing. Springer; 2021. Protein-protein interaction prediction by integrating sequence information and heterogeneous network representation; pp. 617–626. [Google Scholar]
- 83.Das S., Deb T., Dey N., Ashour A.S., Bhattacharya D., Tibarewala D. Optimal choice of k-mer in composition vector method for genome sequence comparison. Genomics. 2018;110(5):263–273. doi: 10.1016/j.ygeno.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 84.Tang J., Qu M., Wang M., Zhang M., Yan J., Mei Q. Line: large-scale information network embedding. Proc 24th Int Conf World wide web. 2015:1067–1077. [Google Scholar]
- 85.Xu W., Gao Y., Wang Y., Guan J. Protein–protein interaction prediction based on ordinal regression and recurrent convolutional neural networks. BMC Bioinforma. 2021;22(6):1–21. doi: 10.1186/s12859-021-04369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hawkins-Hooker A., Depardieu F., Baur S., Couairon G., Chen A., Bikard D. Generating functional protein variants with variational autoencoders. PLoS Comput Biol. 2021;17(2) doi: 10.1371/journal.pcbi.1008736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.J. Cao, L. Xiong, Protein sequence classification with improved extreme learning machine algorithms, BioMed research international 2014. [DOI] [PMC free article] [PubMed]
- 88.Y. G¨ormez, Dimensionality reduction for protein secondary structure prediction, Master’s thesis, Abdullah Gu¨l Universitesi, Fen Bilimleri Enstitu¨su¨ (2017).¨
- 89.Du X., Li Y., Xia Y.-L., Ai S.-M., Liang J., Sang P., et al. Insights into protein–ligand interactions: mechanisms, models, and methods. Int J Mol Sci. 2016;17(2):144. doi: 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gupta A., Mu¨ller A.T., Huisman B.J., Fuchs J.A., Schneider P., Schneider G. Generative recurrent networks for de novo drug design. Mol Inform. 2018;37(1–2) doi: 10.1002/minf.201700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lea C., Vidal R., Reiter A., Hager G.D. European Conference on Computer Vision. Springer; 2016. Temporal convolutional networks: a unified approach to action segmentation; pp. 47–54. [Google Scholar]
- 92.Soleymani F., Paquet E. Long-term financial predictions based on feynman–dirac path integrals, deep bayesian networks and temporal generative adversarial networks. Mach Learn Appl. 2022;7 [Google Scholar]
- 93.Bai S., Kolter J.Z., Koltun V. An empirical evaluation of generic convolutional and recurrent networks for sequence modeling. arXiv Prepr arXiv. 1803:01271. [Google Scholar]
- 94.Wan R., Mei S., Wang J., Liu M., Yang F. Multivariate temporal convolutional network: a deep neural networks approach for multivariate time series forecasting. Electronics. 2019;8(8):876. [Google Scholar]
- 95.ElAbd H., Bromberg Y., Hoarfrost A., Lenz T., Franke A., Wendorff M. Amino acid encoding for deep learning applications. BMC Bioinforma. 2020;21(1):1–14. doi: 10.1186/s12859-020-03546-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu Z., Cui Y., Xiong Z., Nasiri A., Zhang A., Hu J. Deepseqpan, a novel deep convolutional neural network model for pan-specific class i hla-peptide binding affinity prediction. Sci Rep. 2019;9(1):1–10. doi: 10.1038/s41598-018-37214-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Prokhorenkova L., Gusev G., Vorobev A., Dorogush A.V., Gulin A. Catboost: unbiased boosting with categorical features. arXiv Prepr arXiv. 1706:09516. [Google Scholar]
- 98.Johnson J.M., Khoshgoftaar T.M. 2021 IEEE 22nd International Conference on Information Reuse and Integration for Data Science (IRI) IEEE; 2021. Encoding techniques for high-cardinality features and ensemble learners; pp. 355–361. [Google Scholar]
- 99.Cohen M., Reichmann D., Neuvirth H., Schreiber G. Similar chemistry, but different bond preferences in inter versus intra-protein interactions. Proteins. 2008;72(2):741–753. doi: 10.1002/prot.21960. [DOI] [PubMed] [Google Scholar]
- 100.Tsai C.-J., Lin S.L., Wolfson H.J., Nussinov R. Studies of protein-protein interfaces: a statistical analysis of the hydrophobic effect. Protein Sci. 1997;6(1):53–64. doi: 10.1002/pro.5560060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Desantis F., Miotto M., Di Rienzo L., Milanetti E., Ruocco G. Spatial organization of hydrophobic and charged residues affects protein thermal stability and binding affinity. Sci Rep. 2022;12(1):1–13. doi: 10.1038/s41598-022-16338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Meiler J., Mu¨ller M., Zeidler A., Schm¨aschke F. Generation and evaluation of dimension-reduced amino acid parameter representations by artificial neural networks. Mol Model Annu. 2001;7(9):360–369. [Google Scholar]
- 103.Xu P.-S., Luo J., Dou T.-Y. Predict protein-protein interactions from protein primary sequences: using wavelet transform combined with stacking algorithm. PeerJ Prepr. 2017;5:e2964v1. [Google Scholar]
- 104.L. Yang, Y. Han, H. Zhang, W. Li, Y. Dai, Prediction of protein-protein interactions with local weight-sharing mechanism in deep learning, BioMed Research International 2020. [DOI] [PMC free article] [PubMed]
- 105.Ausaf Ali S., Hassan I., Islam A., Ahmad F., et al. A review of methods available to estimate solventaccessible surface areas of soluble proteins in the folded and unfolded states. Curr Protein Pept Sci. 2014;15(5):456–476. doi: 10.2174/1389203715666140327114232. [DOI] [PubMed] [Google Scholar]
- 106.Jha K., Saha S., Tanveer M. Prediction of protein-protein interactions using stacked auto-encoder. Trans Emerg Telecommun Technol. 2021 [Google Scholar]
- 107.S. Debnath, A.F. Mollah, A supervised machine learning approach for sequence based protein-protein interaction (ppi) prediction, arXiv preprint arXiv:2203.12659.
- 108.Ellis J.J., Huard F.P., Deane C.M., Srivastava S., Wood G.R. Directionality in protein fold prediction. BMC Bioinforma. 2010;11(1):1–16. doi: 10.1186/1471-2105-11-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.J. Wang, B. Wicher, V. Maurizot, I. Huc, Directing the self-assembly of aromatic foldamer helices using acridine appendages and metal coordination, Chemistry–A European Journal. [DOI] [PMC free article] [PubMed]
- 110.Idowu A.O., Alashi A.M., Nwachukwu I.D., Fagbemi T.N., Aluko R.E. Functional properties of sesame (Sesamum indicum linn) seed protein fractions, Food Production. Process Nutr. 2021;3(1):1–16. [Google Scholar]
- 111.S. Gopali, F. Abri, S. Siami-Namini, A.S. Namin, A comparative study of detecting anomalies in time series data using lstm and tcn models, arXiv preprint arXiv:2112.09293.
- 112.Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 113.Sagheer A., Kotb M. Unsupervised pre-training of a deep lstm-based stacked autoencoder for multivariate time series forecasting problems. Sci Rep. 2019;9(1):1–16. doi: 10.1038/s41598-019-55320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Palangi H., Deng L., Shen Y., Gao J., He X., Chen J., et al. Deep sentence embedding using long short-term memory networks: Analysis and application to information retrieval. IEEE/ACm Trans. Audio, Speech, Lang Process. 2016;24(4):694–707. [Google Scholar]
- 115.Palangi H., Ward R., Deng L. Distributed compressive sensing: a deep learning approach. IEEE Trans Signal Process. 2016;64(17):4504–4518. [Google Scholar]
- 116.Gopali S., Abri F., Siami-Namini S., Namin A.S. 2021 IEEE International Conference on Big Data (Big Data) IEEE; 2021. A comparison of tcn and lstm models in detecting anomalies in time series data; pp. 2415–2420. [Google Scholar]
- 117.Nan M., Tr˘asc˘au M., Florea A.M., Iacob C.C. Comparison between recurrent networks and temporal convolutional networks approaches for skeleton-based action recognition. Sensors. 2021;21(6):2051. doi: 10.3390/s21062051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Y. He, J. Zhao, Temporal convolutional networks for anomaly detection in time series, in: Journal of Physics: Conference Series, Vol. 1213, IOP Publishing, 2019, p. 042050.
- 119.Zerze G.H., Stillinger F.H., Debenedetti P.G. Computational investigation of retro-isomer equilibrium structures: Intrinsically disordered, foldable, and cyclic peptides. FEBS Lett. 2020;594(1):104–113. doi: 10.1002/1873-3468.13558. [DOI] [PubMed] [Google Scholar]
- 120.F. Yu, V. Koltun, Multi-scale context aggregation by dilated convolutions, arXiv preprint arXiv:1511.07122.
- 121.Zhao W., Gao Y., Ji T., Wan X., Ye F., Bai G. Deep temporal convolutional networks for short-term traffic flow forecasting. IEEE Access. 2019;7:114496–114507. [Google Scholar]
- 122.He K., Zhang X., Ren S., Sun J. Deep residual learning for image recognition. Proc IEEE Conf Comput Vis Pattern Recognit. 2016:770–778. [Google Scholar]
- 123.Duc T.N., Minh C.T., Xuan T.P., Kamioka E. Convolutional neural networks for continuous qoe prediction in video streaming services. IEEE Access. 2020;8:116268–116278. [Google Scholar]
- 124.Zhu R., Liao W., Wang Y. Short-term prediction for wind power based on temporal convolutional network. Energy Rep. 2020;6:424–429. [Google Scholar]
- 125.Salimans T., Kingma D.P. Weight normalization: a simple reparameterization to accelerate training of deep neural networks. Adv Neural Inf Process Syst. 2016;29:901–909. [Google Scholar]
- 126.V. Nair, G.E. Hinton, Rectified linear units improve restricted boltzmann machines, in: Icml, 2010.
- 127.A. v. d. Oord, S. Dieleman,H. Zen, K. Simonyan, O. Vinyals, A. Graves, N. Kalchbrenner, A. Senior, K.Kavukcuoglu, Wavenet: A generative model for raw audio, arXiv preprintarXiv:1609.03499.
- 128.Bouatta N., Sorger P., AlQuraishi M. Protein structure prediction by alphafold2: are attention and symmetries all you need? Acta Crystallogr Sect D: Struct Biol. 2021;77(8):982–991. doi: 10.1107/S2059798321007531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Y. Bengio, L. Yao, G. Alain, P. Vincent, Generalized denoising auto-encoders as generative models, arXiv preprint arXiv:1305.6663.
- 130.Bengio Y., Goodfellow I.J., Courville A. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 131.Y. Zhang, A better autoencoder for image: Convolutional autoencoder, in: ICONIP17-DCEC. Available online: http://users. cecs. anu. edu. au/Tom. Gedeon/conf/ABCs2018/paper/ABCs2018 paper 58. pdf (accessed on 23 March 2017), 2018.
- 132.Polic M., Krajacic I., Lepora N., Orsag M. Convolutional autoencoder for feature extraction in tactile sensing. IEEE Robot Autom Lett. 2019;4(4):3671–3678. [Google Scholar]
- 133.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15(1):1929–1958. [Google Scholar]
- 134.D.P. Kingma, J. Ba, Adam: A method for stochastic optimization, arXiv preprint arXiv:1412.6980.
- 135.Blohm P., Frishman G., Smialowski P., Goebels F., Wachinger B., Ruepp A., et al. Negatome 2.0: a database of non-interacting proteins derived by literature mining, manual annotation and protein structure analysis. Nucleic Acids Res. 2014;42(D1):D396–D400. doi: 10.1093/nar/gkt1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Orchard S., Kerrien S., Abbani S., Aranda B., Bhate J., Bidwell S., et al. Protein interaction data curation: the international molecular exchange (imex) consortium. Nat Methods. 2012;9(4):345–350. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., et al. The mintact project—intact as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42(D1):D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Oughtred R., Stark C., Breitkreutz B.-J., Rust J., Boucher L., Chang C., et al. The biogrid interaction database: 2019 update. Nucleic Acids Res. 2019;47(D1):D529–D541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bauer-Mehren A., Furlong L.I., Sanz F. Pathway databases and tools for their exploitation: benefits, current limitations and challenges. Mol Syst Biol. 2009;5(1):290. doi: 10.1038/msb.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. Omim. org: Online mendelian inheritance in man (omim®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(D1):D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., et al. Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40(D1):D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Franceschini A., Lin J., von Mering C., Jensen L.J. Svd-phy: improved prediction of protein functional associations through singular value decomposition of phylogenetic profiles. Bioinformatics. 2016;32(7):1085–1087. doi: 10.1093/bioinformatics/btv696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jarazo J., Barmpa K., Modamio J., Saraiva C., Sabat´e-Soler S., Rosety I., et al. Parkinson’s disease phenotypes in patient neuronal cultures and brain organoids improved by 2-hydroxypropyl-β-cyclodextrin treatment. Mov Disord. 2022;37(1):80–94. doi: 10.1002/mds.28810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Alibrahim H., Ludwig S.A. 2021 IEEE Congress on Evolutionary Computation (CEC) IEEE; 2021. Hyperparameter optimization: comparing genetic algorithm against grid search and bayesian optimization; pp. 1551–1559. [Google Scholar]
- 145.Wong T.-T., Yeh P.-Y. Reliable accuracy estimates from k-fold cross validation. IEEE Trans Knowl Data Eng. 2019;32(8):1586–1594. [Google Scholar]
- 146.J. Ingraham, V. Garg, R. Barzilay, T. Jaakkola, Generative models for graph-based protein design, Advances in Neural Information Processing Systems 32.
- 147.Kim J., Kim J. The impact of imbalanced training data on machine learning for author name disambiguation. Scientometrics. 2018;117(1):511–526. [Google Scholar]
- 148.van den Hout W.B. The area under an roc curve with limited information. Med Decis Mak. 2003;23(2):160–166. doi: 10.1177/0272989X03251246. [DOI] [PubMed] [Google Scholar]
- 149.Zhang Y., Wang C., Gong L., Lu Y., Sun F., Xu C., et al. 2017 IEEE International Conference on Cluster Computing (CLUSTER) IEEE; 2017. A power-efficient accelerator based on fpgas for lstm network; pp. 629–630. [Google Scholar]
- 150.Bengio Y., Simard P., Frasconi P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans Neural Netw. 1994;5(2):157–166. doi: 10.1109/72.279181. [DOI] [PubMed] [Google Scholar]
- 151.S. Hochreiter, Y. Bengio, P. Frasconi, J. Schmidhuber, et al., Gradient flow in recurrent nets: the difficulty of learning long-term dependencies (2001).
- 152.James J., Lam A.Y., Hill D.J., Li V.O. Delay aware intelligent transient stability assessment system. IEEE Access. 2017;5:17230–17239. [Google Scholar]