Abstract
Honey bees play an important role in the pollination of crops and wild plants and provide important products to humans. Pathogens and parasites are the main factors that threaten beekeeping in South Korea. Therefore, a nationwide detection of 14 honey bee pathogens, including parasites (phorid flies, Nosema ceranae, and Acarapis woodi mites), viruses, bacteria, and fungal pathogens, was conducted from 2017 to 2021 in the country. The infection rate and the trend of detection of each pathogenic agent were determined. A total of 830 honey bee samples from Apis cerana (n = 357) and A. mellifera (n = 473) were examined. N. ceranae (35.53%), deformed wing virus (52.63%), sacbrood virus (SBV) (52.63%), and black queen cell virus (55.26%) were the most prevalent honey bee pathogens, and their prevalence rapidly increased from 2017 to 2021. The prevalence of Paenibacillus larvae, Israeli acute paralysis virus, Ascosphaera apis, A. woodi, Melissococcus plutonius, and chronic bee paralysis virus remained stable during the surveillance period, with infection rates ranging from 5.26% to 16.45% in 2021. Other pathogens, including acute bee paralysis virus, phorid flies, Kashmir bee virus, and Aspergillus flavus, had low infection rates that gradually declined during the detection period. The occurrence of honeybee pathogens peaked in July. SBV was the most common pathogen in A. cerana, whereas N. ceranae was predominant in A. mellifera. This study provides information regarding the current status of honey bee pathogens and presents the trend of the occurrence of each pathogen in South Korea. These data are important for predicting outbreaks of honey bee diseases in the country.
Keywords: Apis cerana, Apis mellifera, Honey bee pathogens, SBV, Nosema, DWV, BQCV
Highlights
-
•
Fourteen honey bee pathogens and parasites were recorded in Korea from 2017 to 2021.
-
•
N. ceranae, DWV, SBV, and BQCV were the most prevalent honey bee pathogens.
-
•
SBV and N. ceranae were dominantly detected in A. cerana and A. mellifera, respectively.
-
•
BQCV and DWV were prevalently found in both honey bee species.
-
•
The pathogens developed rapidly in summer from May and peaked in July.
1. Introduction
Honey bees are important pollinators of crops and wild plants and provide important products to humans, including honey, royal jelly, pollen, beeswax, and propolis. Notable losses of honey bee colonies worldwide have received considerable attention from beekeepers and researchers. Of the various factors assumed to be associated with the decline of honey bees, parasites and pathogens are significant [[1], [2], [3], [4]].
Viral infections pose a serious threat to honey bee health, and viral diseases cause damage to honey bees at all stages, including eggs, larvae, pupae, and adults [5]. More than 30 viruses have been identified in honey bees [6,7], of which seven, including the acute bee paralysis virus (ABPV), black queen cell virus (BQCV), chronic bee paralysis virus (CBPV), deformed wing virus (DWV), Israeli acute paralysis virus (IAPV), Kashmir bee virus (KBV), and sacbrood virus (SBV), are the most prevalent and threatening in apiculture [4]. The identification of viral infections based on specific honey bee symptoms is inefficient and ineffective due to asymptomatic bees with low infection levels, similar symptoms of different viral infections, and co-infection with several pathogens [6]. Therefore, molecular methods using specific primers should be used to accurately detect and differentiate viral diseases in honey bees [8].
In addition, fungal and bacterial pathogens that cause brood diseases significantly affect the health of honey bees. Fungal brood disease, chalkbrood, and stonebrood are caused by the fungal species Ascosphaera apis and Aspergillus spp., including Aspergillus flavus. Gram-positive bacteria Melissococcus plutonius and Paenibacillus larvae are pathogens of European foulbrood and American foulbrood, respectively. Brood diseases have spread throughout the world and are a significant cause of death of honey bee larvae, which may result in weakening and even death of infected colonies [[9], [10], [11], [12], [13]]. The accurate identification of brood diseases is important to determine the appropriate treatment method. Polymerase chain reaction (PCR)-based detection is recommended to accurately identify causative pathogens [12,14,15].
In contrast, parasites contribute to honey bee colony loss [16], including the mites Varroa destructor, Tropilaelaps clareae, and Acarapis woodi, which are considered to be the most harmful parasites. Mites feed on honey bees and cause winter colony loss [17]. V. destructor and T. clareae mites are important vectors of honey bee pathogens [[18], [19], [20]]. In addition, microsporidia such as Nosema ceranae shorten the lifespan of adult honey bees, reduce brood rearing and pollen collection, and alter honey bee behavior, resulting in decreased productivity and survival of honey bee colonies [2,11,21]. The phorid fly (Apocephalus borealis) is a new threat to honey bees, as they parasitize honey bees by laying eggs on the bee's abdomen. The hatching larvae feed on the bees, ultimately killing them [22]. V. destructor and T. clareae mites can be directly observed in honey bee hives [[22], [23], [24]]. However, smaller parasites or those inside honey bees are difficult to detect by direct observation, and microscopic detection is time-consuming [25,26]. Therefore, molecular detection with specific primers is an alternative method for the rapid identification of parasites, including A. woodi, Nosema, and A. borealis [22,27,28].
Beekeeping has an important role in the economy of South Korea. The estimated annual honey bee production and pollination values are approximately 5.5 billion USD [20]. In 2020, there were 2,697,842 colonies and 27,532 apiaries in South Korea [29]. However, parasites and pathogens have become major challenges for beekeepers in South Korea [30]. Viral diseases, brood diseases, and parasites have been reported in Korean apiculture [31]. SBV has been identified as the major pathogen causing the loss of A. cerana colonies [32,33]. The number of apiaries in South Korea declined from 29,026 apiaries in 2019 to 27,532 apiaries in 2020 [29], and serious colony loss due to unclear reasons has been reported in many regions of the country. Therefore, it is necessary to identify the factors that adversely affect beekeeping. A significant V. destructor and T. clareae mite infestation is affecting Korean apiculture [20]. However, the identities of other viral pathogens, brood pathogens, and parasites are unclear. Therefore, 14 major honey bee viruses, brood disease pathogens, and parasites were investigated from 2017 to 2021 in this study. Distribution of the pathogens in the last five years were determined. It is important for predicting outbreaks of honey bee diseases and selecting the appropriate methods for mitigating the propagation of honey bee diseases in the country.
2. Materials and methods
2.1. Honey bee sample collection
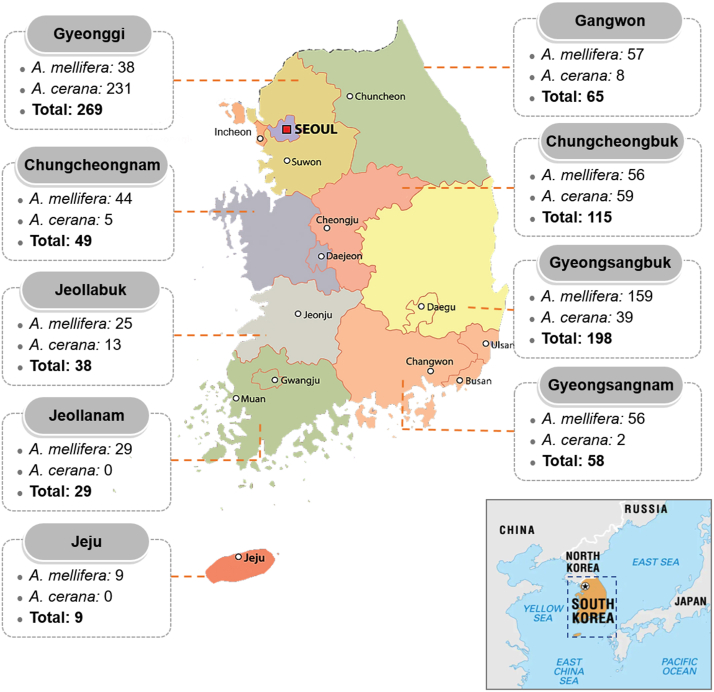
A total of 830 honey bee samples, including adults (n = 1414) and larvae (n = 615), were collected throughout South Korea (Fig. 1) from 2017 to 2021 for the detection of honey bee pathogens. Overall, 357 A. cerana and 473 A. mellifera samples were collected from apiaries where paralysis and dead adult bees or symptoms of honey bee brood diseases in larvae were observed (Figs. S1–S3). The samples were collected and stored in a dry ice box, then carried to the Honey Bee Disease Laboratory, Animal and Plant Quarantine Agency, Republic of Korea for pathogen detection.
Fig. 1.
Collection of honey bee samples in South Korea. A total of 830 samples of Apis mellifera (n = 473) and A. cerana (n = 357) honey bees were collected from 2017 to 2021. The numbers of samples collected in each of nine provinces are shown.
2.2. Nucleic acid extraction
The nucleic acids were extracted using a Maxwell® RSC viral total nucleic acid purification kit (Promega, Madison, WI, USA). Two adult or five larvae samples were combined with 600 μL PBS solution in a tissue-homogenizing tube with steel beads (diameter = 2.381 mm; SNC, Hanam, South Korea) and homogenized using a Precellys 24 tissue homogenizer (Bertin Instruments, Montigny-le-Bretonneux, France) for four 15-s cycles at 5000 rpm. Then, 300 μL homogenate, 300 μL lysis buffer, and 30 μL proteinase K solution were combined in a 1.5-mL microcentrifuge tube and incubated at 56 °C for 10 min. The nucleic acids were then purified automatically using the automated Maxwell® RSC instrument according to the manufacturer's instructions. Finally, 100 μL total nucleic acids was used to detect honey bee pathogens.
2.3. Detection of honey bee pathogens
The LiliF™ SBV/KSBV/DWV/BQCV reverse transcription real-time polymerase chain reaction (RT-qPCR) kit and LiliF™ ABPV/KBV/IAPV/CBPV RT-qPCR kit (iNtRON Biotechnology, Inc., Seongnam, Korea) were used to detect DWV, ABPV, BQCV, CBPV, IAPV, KBV, and SBV (Table 1). RT-qPCR consisted of reverse transcription at 45 °C for 30 min, incubation at 95 °C for 10 min, and 40 PCR cycles of 15 s at 95 °C and 1 min at 62 °C, positive detection was identified with the cycle threshold (Ct) value ≤ 35. The POBGEN™ Bee Pathogen Detection Kits (DB-A2 and DB-B2) (POSTBIO Inc., Guri, Korea) were used to detect fungal pathogens (A. flavus, the causative agent of stonebrood, and A. apis, the causative agent of chalkbrood), microsporidians (N. ceranae), bacterial pathogens (P. larvae, the causative agent of American foulbrood and M. plutonius, the causative agent of European foulbrood), and parasites (A. woodi mites and A. borealis) (Table 2). PCR detection was performed at 95 °C for 5 min for initial denaturation followed by 40 PCR cycles of 10 s at 95 °C and 30 s at 60 °C, positive detection was identified with Ct value ≤ 35.
Table 1.
Primer and probe sequences used to detect viral pathogens.
| No. | Target | Primer | Sequence (5′ → 3′) | Target gene | Reference |
|---|---|---|---|---|---|
| 1 | SBV | SBV-F | AGAAGACATTTGATACAGTGGACTC | Polyprotein gene, 131 bp | This study |
| SBV-R | GGAATTCCAGATTCTTCGTCCAC | ||||
| Probe | FAM–GATTTGTTTAATGGTTGGGTTTCTGGTA–BHQ-1 | ||||
| 2 | DWV | DWV-F | TTCAACTCGGCTTTCTACGG | Polyprotein gene, 170 bp | This study |
| DWV-R | GTGTCTTTTTCTCTTTCTGACACC | ||||
| Probe | ROX-ATGTCAACATTGGTATGCTCCGTTGAC-BHQ-2 | ||||
| 3 | BQCV | BQCV-F | CCTTTGGCAATAGAACAAATACC | Capsid, 143 bp | This study |
| BQCV-R | GTGGCTATATCGAGATTATTCCG | ||||
| Probe | Cy5-AGTCGCAGAGTTCCAAATACCGTACTATG-BHQ-3 | ||||
| 4 | ABPV | ABPV-F | TGCCCTATTTAGGGTGAGGAG | Capsid, 239 bp | This study |
| ABPV-R | GGAGTTTCCACATCATGAAAGG | ||||
| Probe | FAM-CTCTGAAGAAAACTCAGTTGAAACGGAAC-BHQ-1 | ||||
| 5 | KBV | KBV-F | ACCAGGAAGTATTCCCATGGTAAG | Capsid, 79 bp | This study |
| KBV-R | TGGAGCTATGGTTCCGTTCAG | ||||
| Probe | HEX-CCGCAGATAACTTAGGACCAGATCAATCACA-BHQ-1 | ||||
| 6 | IAPV | IAPV-F | TGCCCTATTTAGGGTGAGGAG | Capsid, 245 bp | This study |
| IAPV-R | GGAGTTTCCACATCATGAAAGG | ||||
| Probe | ROX-ACTAGTGAGAACTCGGTTGAGACCCAAG-BHQ-2 | ||||
| 7 | CBPV | CBPV-F | CGCAAGTACGCCTTGATAAAGAAC | RdRp, 101 bp | This study |
| CBPV-R | ACTACTAGAAACTCGTCGCTTCG | ||||
| Probe | Cy5-TCAAGAACGAGACCACCGCCAGTTC-BHQ-3 |
Abbreviations: ABPV: acute bee paralysis virus; BQCV: black queen cell virus; CBPV: chronic bee paralysis virus; DWV: deformed wing virus; IAPV: Israeli acute paralysis virus; KBV: Kashmir bee virus; SBV: sacbrood virus.
Table 2.
Primers and probe sequences used to detect brood diseases and parasites.
| No. | Target | Primer | Sequence (5’→3′) | Target gene | Reference |
|---|---|---|---|---|---|
| 1 | Paenibacillus larvae (AFB) | AFB-F | AAATCATCATGCCCCTTATG | 16S rRNA, 158 bp | This study |
| AFB-R | CGATTACTAGCAATTCCGACT | ||||
| Probe | FAM-CGTACTACAATGGCCGGTACAACG–BHQ-1 | ||||
| 2 | Melissococcus plutonius (EFB) | EFB-F | TGTTGTTAGAGAAGAATAGGGGAA | 16S rRNA, 69 bp | This study |
| EFB-R | CGTGGCTTTCTGGTTAGA | ||||
| Probe | Cy5-AGAGTAACTGTTTTCCTCGTGACGGT-BHQ-3 | ||||
| 3 | Apocephalus borealis (phorid fly) | Phorid-F | CCTCTGTTCTACTTTCATTGGTTTAT | 18S rRNA, 255 bp | This study |
| Phorid-R | GAGRGCCATAAAAGTAGCTACACC | ||||
| Probe | JOE-GGCATTAGTATTACGACGCGAGAGGTG–BHQ-1 | ||||
| 4 | Acarapis woodi mite | ACAR-F | CAGTAGGGCTAGATATCGATACCCGAGCTT | COI, 247 bp | This study |
| ACAR-R | TGAGCTACAACATAATATCTGTCATGAAGA | ||||
| Probe | TexasRed-CAATCCACCTACAGAAAATAAAAATAAAAATCC–BHQ-2 | ||||
| 5 | Nosema cerana | Nosema-F | CGGATAAAAGAGTCCGTTACC | LSU rRNA, 249 bp | This study |
| Nosema-R | GAGCAGGGTTCTAGGGAT | ||||
| Probe | FAM-CGTTACCCTTCGGGGAATCTTC–BHQ-1 | ||||
| 6 | Aspergillus flavus (stonebrood) | ASP-F | GCTGCCCATCAAGCACGG | ITS2, 127 bp | This study |
| ASP-R | CCTACAGAGCGGGTGACAAAG | ||||
| Probe | JOE-TGTGTGTTGGGTCGTCGTCCCCTCTC–BHQ-1 | ||||
| 7 | Ascosphaera apis (chalkbrood) | ASCO-F | ATTGCGCCCTCTGGTATTC | ITS2, 215 bp | This study |
| ASCO-R | CCACTAGAAGTAAATGATGGTTAGA | ||||
| Probe | TexasRed-GCTTGAGGGTTGCAATGACGCTCG BHQ-2 |
Abbreviations: AFB: American foulbrood; EFB: European foulbrood.
2.4. Data analysis
Each sample was analyzed for 14 honey bee parasites and pathogens. The infection rates were calculated by dividing the number of samples with positive results by the total number of collected samples and multiplying by 100%. The Mann-Whitney U test was used to compare the results of two independent groups, and the one-way ANOVA test was used to compare more than two groups. Statistical significance was set at p < 0.05. All analyses were conducted using PAST version 4.03 software [34].
3. Results
3.1. Detection of honey bee pathogens
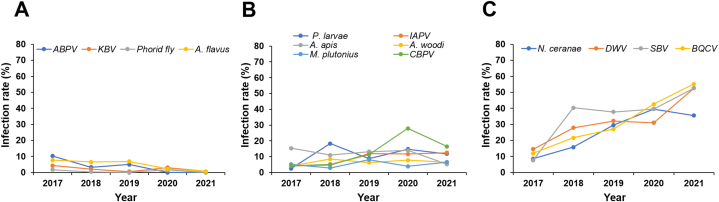
ABPV, phorid flies, KBV, and A. flavus had a low infection rate that gradually declined throughout the study period. In 2017, the infection rates of ABPV, phorid flies, KBV, and stonebrood were 10.00%, 1.71%, 4.27%, and 7.69%, respectively. In 2021, only stonebrood (0.66%) and KBV (0.66%) were detected (Fig. 2A; Table S1).
Fig. 2.
Infection rates of 14 honey bee pathogens. The detection trends of the 14 honey bee pathogens differed. The infection rates of acute bee paralysis virus (ABPV), Kashmir bee virus (KBV), A. borealis, and A. flavus gradually declined during the study period (A). The infection rates of P. larvae, A. apis, M. plutonius, Israeli acute paralysis virus (IAPV), A. woodi, and CBPV were moderate throughout the study period (B). The infection rates of N. ceranae, deformed wing virus (DWV), sacbrood virus (SBV), and black queen cell virus (BQCV) increased during the study period (C).
P. larvae, IAPV, A. apis, A. woodi, M. plutonius, and CBPV had moderate infection rates during the surveillance period. The infection rate of A. apis decreased from 15.38% in 2017 to 5.26% in 2021. The infection rate of CBPV rapidly increased from 4.27% in 2017 to 27.91% in 2020, and decreased to 16.45% in 2021. The infection rates of P. larvae, IAPV, A. woodi, and M. plutonius decreased during the study period (Fig. 2B; Table S1).
The most prevalent honey bee pathogens in South Korea were N. ceranae, DWV, SBV, and BQCV. The infection rates of these pathogens increased significantly during the study period (p = 0.0293). The infection rates of N. ceranae, DWV, SBV, and BQCV increased from 8.55%, 14.53%, 7.69%, and 11.97% in 2017 to 35.53%, 52.63%, 52.63%, and 55.26%, respectively, in 2021 (Fig. 2C; Table S1).
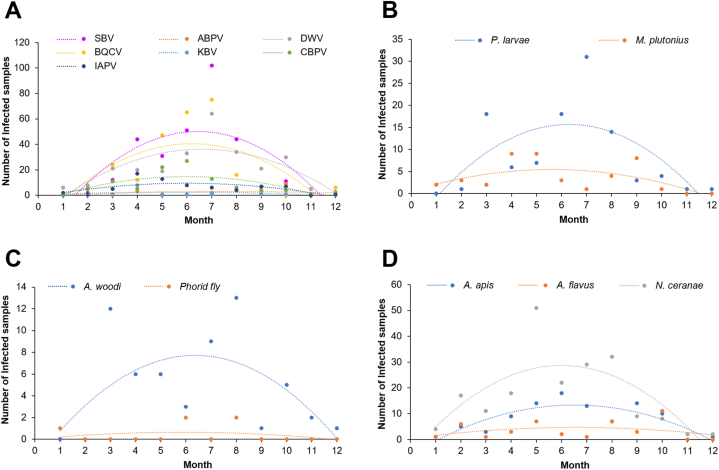
3.2. Seasonal occurrence of honey bee disease
The number of honey bee samples infected with at least one pathogen was significantly different between the seasons (p < 0.001). The number of infected samples increased to a peak infection rate in July, then declined from August to December (Fig. 3A–D; Table S2). The period of pathogen development was February to October, and pathogen propagation occurred from May to July. The variation of prevalent pathogens in viral (SBV, BQCV, DWV, and CBPV), bacterial (P. larvae), parasitic (A. woodi), and fungal group (N. ceranae and A. apis) were seen with significant difference among the seasons (p ≤ 0.01). These pathogens showed the highest number of infected samples from June to July (Fig. 3A–D). Meanwhile, other less common pathogens (IAPV, ABPV, KBV, M. plutonius, Phorid fly, A. flavus) were seen with low number of infected samples in the whole year (Fig. 3A–D; Table S2).
Fig. 3.
Seasonal fluctuation of honey bee pathogen infection in South Korea. The number of honey bee samples carrying viral (A), bacterial (B), parasitic (C), and fungal group (D) of 14 pathogens in each month of the study period are shown.
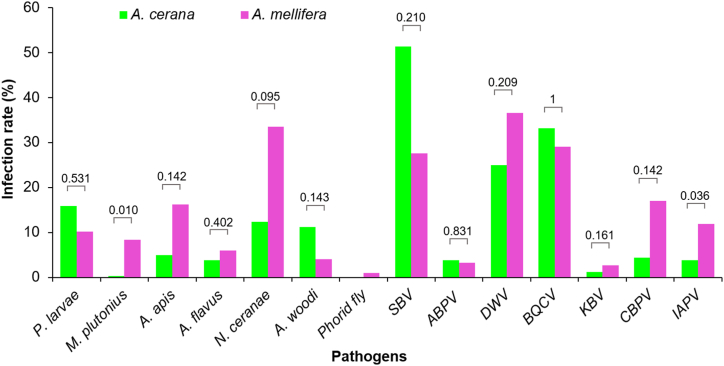
3.3. Prevalence of pathogens in different honey bee species
The infection rates of the 14 pathogens were not significantly different between A. cerana and A. mellifera honey bees during the study period (p > 0.05), except for M. plutonius (p = 0.010) and IAPV (p = 0.036; Fig. 4), the two pathogens were mainly detected in A. mellifera. However, a significant difference (p < 0.001) was seen in infection rate among the pathogens detected in each honey bee species. SBV was the most common pathogen in A. cerana with an infection rate of 51.32%, followed by BQCV (33.14%), DWV (24.93%), P. larvae (15.84%), N. ceranae (12.32%), and A. woodi (11.14%). Other pathogens were seen with infection rate lower than 5%, and phorid fly was not detected in A. cerana (Fig. 4). Meanwhile, A. mellifera was majorly infected with four pathogens DWV (36.61%), N. ceranae (33.54%), BQCV (29.04%), and SBV (27.61%). The pathogens with moderate infection level include CBPV (16.97%), A. apis (16.16%), IAPV (11.86%), and P. larvae (10.22%). Other pathogens were seen with infection rate lower than 10% (Fig. 4; Table S3).
Fig. 4.
Infection rates of honey bee pathogens in different honey bee species. The infection rates of 14 honey bee pathogens in Apis cerana and A. mellifera are shown. Mann-Whitney U test was performed to compare infection rate of each pathogen between two honey bee species, P values shown. Phorid fly was detected only in A. mellifera.
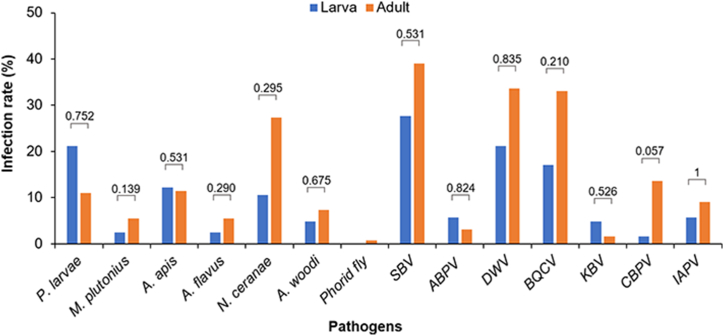
3.4. Infection rate of pathogens in larva and adult honey bees
Infection rate of the 14 pathogens and parasites was not significantly different (p > 0.05) between larvae and adult bees (Fig. 5). However, a great difference (p < 0.001) was seen in the infection rate among the pathogens and parasites detected in each living stage of honey bee. SBV was the most prevalent pathogen detected in larvae whose infection rate was 27.64%, followed by P. larvae and DWV with 21.14% of each pathogen, BQCV (17.07%), A. apis (12.20%), and N. ceranae (10.57%). Other targets were seen with low infection rate (<10%), and phorid fly was not detected in larval samples. Meanwhile, adult bees were prevalently infected with SBV (39.04%), followed by DWV (33.66%), BQCV (33.10%), N. ceranae (27.30%), CBPV (13.58%), P. larvae (11.03%), and A. apis (11.46%). Other pathogens and parasites were seen with infection rate lower than 10% (Fig. 5; Table S4).
Fig. 5.
Infection rates of honey bee pathogens in larvae and adult honey bee. The infection rates of 14 honey bee pathogens and parasites in larvae and adult bees from 2017 to 2021 are shown.
4. Discussion
In this study, the infection rates of 14 honey bee pathogens in South Korea were detected from 2017 to 2021. N. ceranae, DWV, SBV, and BQCV were the most prevalent pathogens adversely affect beekeeping in the country. These pathogens rapidly propagated in summer and mainly distributed in Gyeonggi, Gyengsangbuk, and Chungcheongbuk provinces.
Several pathogens that have been reported in Asia have affected beekeeping in South Korea [35] including viruses, bacteria, fungi, and parasites [20,36]. N. ceranae, DWV, SBV, and BQCV were the most prevalent pathogens detected in this study. The prevalence of honey bee pathogens in South Korea has changed in the last decade with the occurrence of DWV, SBV, and BQCV [31]. V. destructor and T. mercedesae mites were reported to have high infestation rates in 2012 (75%) [31] that increased to 93.6% in 2019 [20]. These mites feed directly on honey bees and are an important natural reservoir contributing to the transmission of honey bee viral pathogens [19,20,37]. The role of mites in the high propagation of DWV, SBV, and BQCV requires additional research.
Previous studies have reported that the observed increase in SBV is correlated with an increase in temperature and that the risk of SBV disease increases temporally [38,39]. In this study, the infection rates of SBV and other viral and bacterial pathogens increased from June to August and peaked in July. Therefore, there is a high risk of honey bee colony loss during the summer months in South Korea. During the summer season, worker bees increase their foraging activities as the temperature increases. As foragers invade other colonies to steal honey, the propagation of pathogens among colonies increases [38,40]. Increased temperature also result in increased honey bee mites [38] that serve as a vector for 12 honey bee pathogens [20]. Infestations of Varroa mites result in drifting behaviors in adult honey bees, increasing the movements of foraging honey bees into other colonies [19], allowing for the transfer of mites and the spread of honey bee pathogens.
In South Korea, A. cerana suffers from three major pathogens: SBV, BQCV, and DWV. These pathogens were also prevalently detected in A. cerana in other Asian countries, including China [41,42] and Vietnam [43], and SBV was seen with the highest infection rate. SBV was first detected in A. mellifera [44], and different geographical genotypes of SBV were identified in Asian honey bee, A. cerana [45]. These genotypes have more devastating effects on A. cerana than on A. mellifera [46], and become the major pathogens threatening A. cerana in different countries such as China [47], Vietnam [48], Korea [32], and Thailand [49]. The Korean genotype of SBV poses a threat to A. cerana since it was first recorded in 2008, and up to 90% of the A. cerana colonies in South Korea have been affected by SBV disease [32,33]. The international trade of living honey bees may lead to the spread of geographical SBV variants to other countries, resulting in outbreaks of the disease in native honey bee strains. Therefore, an efficient method for the prevention and treatment of SBV disease is necessary. RNA interference [50] or artificial breeding with selection for disease-resistant honey bee strains may be effective methods to prevent SBV disease [51].
Four major pathogens were prevalent in A. mellifera honey bees in this study, including N. ceranae, DWV, BQCV, and SBV. These were also the major pathogens of A. mellifera in other countries, such as China [52], the United States [53,54], European countries, Italy [55], Belgium [56], and Austria [57]. SBV and BQCV adversely affect honey bee by causing the death of larvae [58,59]. Meanwhile, N. ceranae and DWV have been associated with honey bee colony losses over winter [[60], [61], [62]]. Co-infections of N. ceranae and BQCV have also been reported to significantly affect honey bee colonies [63]. Therefore, these pathogens may be linked to winter colony loss in A. mellifera apiaries in South Korea.
The BQCV, SBV, and DWV were majorly detected pathogens among the 14 targets in both adult and larval stages. Although infection rate of the three viral pathogens was higher in adult compared to larval stage, the pathogens adversely influence brood stage of the honey bee [64,65]. SBV was the most damaging pathogen of A. cerana by causing the death of larvae in Asian countries [32,41,43,49]. Meanwhile, DWV infection in developing pupae causes the pupal death or deformed wings in adult bee [66]. Varroa destructor mite was the most important vector of this pathogen, and the widespread of the mite in A. mellifera apiaries could result in the prevalence of DWV [20,67]. These pathogens infected all stages of honey bee. However, the symptoms of infection in adult bees are not clear [59,66,68], and the identification of each pathogen infection by directly observing the bees is difficult. Therefore, the molecular identification using specific primer is important. In addition, CBPV and N. ceranae are the common parasite and pathogen affecting adult bees [21,69]. Therefore, the two targets were predominantly detected in adult bees in this study. Another brood pathogen, P. larvae, the causative agent of American foulbrood disease was highly detected in larvae. This pathogen adversely affects young larvae when the spores of P. larvae were ingested [70].
N. ceranae and viral pathogens including DWV, SBV, and BQCV were dominantly detected among the 14 selected targets in this study. Beekeeping in South Korea also suffers from heavy infestation of V. destructor and T. clareae mite [20]. However, other pathogens such as Varroa destructor virus-1, Lake Sinai virus, and Nosema apis were not targeted for detection in this study. These pathogens were identified to increase winter mortality or colony loss of honey bee [21,71,72]. In addition, small hive beetle (Aethina tumida), a serious parasite of honey bees, was also recorded in South Korea [73,74]. However, the prevalence of this parasite in Korean bee keeping still remain unknown. Therefore, further study needs to be conducted to understand the prevalence of the remaining pathogens and parasites in South Korea. It could be important to identify the causative agents of honey bee colony losses recently occurred in the country (https://www.newstree.kr/newsView/ntr202205120006; https://www.nst.com.my/world/region/2022/01/761016/millions-honey-bees-mysteriously-killed-or-missing).
5. Conclusion
Fourteen honey bee pathogens were detected in South Korea between 2017 and 2021, of which N. ceranae, DWV, SBV, and BQCV were the most prevalent. The most common pathogens of A. cerana include SBV, DWV, and BQCV. The most common pathogens of A. mellifera are SBV, BQCV, N. ceranae, and DWV. The pathogens develop during the summer months, peaking in July. Therefore, appropriate management practices to control the propagation of pathogens during the summer months should be established. The results of this study are important for identifying the occurrence patterns of honey bee pathogens and the major factors affecting apiculture in South Korea. These results can also be used to predict the outbreak of related diseases and estimate the economic loss caused by pathogens in the beekeeping industry. Furthermore, the result provided important information on the geographical distribution of each honey bee pathogens by which the appropriate methods to control spread of pathogens via trading of living honey bee could be selected.
Author contribution statement
A-Tai Truong: Analyzed and interpreted the data; Wrote the paper.
Mi-Sun Yoo: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Soo Kyoung Seo: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Tae Jun Hwang: Performed the experiments.
Soon-Seek Yoon: Analyzed and interpreted the data.
Yun Sang Cho: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
Dr. Yun Sang Cho was supported by Animal and Plant Quarantine Agency [N-1543081-2021-25-03], Republic of Korea.
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interest's statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e13494.
Abbreviations
- ABPV
acute bee paralysis virus
- BQCV
black queen cell virus
- CBPV
chronic bee paralysis virus
- DWV
deformed wing virus
- IAPV
Israeli acute paralysis virus
- KBV
Kashmir bee virus
- SBV
sacbrood virus
- AFB
American foulbrood
- EFB
European foulbrood
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Genersch E. Honey bee pathology: current threats to honey bees and beekeeping. Appl. Microbiol. Biotechnol. 2010;87(1):87–97. doi: 10.1007/s00253-010-2573-8. [DOI] [PubMed] [Google Scholar]
- 2.Hristov P., Shumkova R., Palova N., Neov B. Factors associated with honey bee colony losses: a mini-review. Vet. Sci. 2020;7(4):166. doi: 10.3390/vetsci7040166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jordan A., Patch H.M., Grozinger C.M., Khanna V. Economic dependence and vulnerability of United States agricultural sector on insect-mediated pollination service. Environ. Sci. Technol. 2021;55(4):2243–2253. doi: 10.1021/acs.est.0c04786. [DOI] [PubMed] [Google Scholar]
- 4.Tantillo G., Bottaro M., Pinto A.D., Martella V., Pinto P.D., Terio V. Virus infections of honeybees Apis mellifera. Ital. J. Food Saf. 2015;4:5364. doi: 10.4081/ijfs.2015.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ullah A., Tlak Gajger I., Majoros A., Dar S.A., Khan S., Kalimullah, et al. Viral impacts on honey bee populations: a review. Saudi J. Biol. Sci. 2021;28(1):523–530. doi: 10.1016/j.sjbs.2020.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Miranda J.R., Bailey L., Ball B.V., Blanchard P., Budge G.E., Chejanovsky N., et al. Standard methods for virus research in Apis mellifera. J. Apicult. Res. 2013;52(4):1–56. doi: 10.3896/IBRA.1.52.4.22. [DOI] [Google Scholar]
- 7.Remnant E.J., Shi M., Buchmann G., Blacquière T., Holmes E.C., Beekman M., et al. A diverse range of novel RNA viruses in geographically distinct honey bee populations. J. Virol. 2017;91(16) doi: 10.1128/JVI.00158-17. e00158-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuc N.T.K., Phu N.V., Yoon B. Molecular detection of honey bee pathogenic microbes: recent advances and future perspective. Korean J. Apic. 2021;36(2):35–45. doi: 10.17519/apiculture.2021.06.36.2.35. [DOI] [Google Scholar]
- 9.Bailey L. The epizootiology of European foulbrood of the larval honey bee, Apis mellifera Linnaeus. J. Insect Pathol. 1960;2:67–83. [Google Scholar]
- 10.Brødsgaard C.J., Hansen H., Ritter W. Progress of Paenibacillus larvae larvae infection in individually inoculated honey bee larvae reared singly in vitro, in micro colonies, or in full-size colonies. J. Apicult. Res. 2000;39(1–2):19–27. doi: 10.1080/00218839.2000.11101017. [DOI] [Google Scholar]
- 11.Ellis J.D., Munn P.A. The worldwide health status of honey bees. Bee World. 2005;86:88–101. [Google Scholar]
- 12.Jensen A.B., Aronstein K., Flores J.M., Vojvodic S., Palacio M.A., Spivak M. Standard methods for fungal brood disease research. J. Apicult. Res. 2013;52(1):1–20. doi: 10.3896/IBRA.1.52.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rauch S., Ashiralieva A., Hedtke K., Genersch E. Negative correlation between individual-insect-level virulence and colony-level virulence of Paenibacillus larvae, the etiological agent of American foulbrood of honeybees. Appl. Environ. Microbiol. 2009;75(10):3344–3347. doi: 10.1128/AEM.02839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Graaf D.C., Alippi A.M., Antúnez K., Aronstein K.A., Budge G., Koker D.D., et al. Standard methods for American foulbrood research. J. Apicult. Res. 2013;52(1):1–28. doi: 10.3896/IBRA.1.52.1.11. [DOI] [Google Scholar]
- 15.Forsgren E., Budge G.E., Charrière J.D., Hornitzky M.A.Z. Standard methods for European foulbrood research. J. Apicult. Res. 2013;52(1):1–14. doi: 10.3896/IBRA.1.52.1.12. [DOI] [Google Scholar]
- 16.Coffey M.F. Crops Research Centre; Oak Park, Carlow: 2007. Parasites of the Honey Bee, Teagasc. [Google Scholar]
- 17.van Dooremalen C., Gerritsen L., Cornelissen B., van der Steen J.J., van Langevelde F., Blacquière T. Winter survival of individual honey bees and honey bee colonies depends on level of Varroa destructor infestation. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0036285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson D.L., Trueman J.W.H. Varroa jacobsoni (Acari: varroidae) is more than one species. Exp. Appl. Acarol. 2000;24:165–189. doi: 10.1023/a:1006456720416. [DOI] [PubMed] [Google Scholar]
- 19.Forfert N., Natsopoulou M.E., Frey E., Rosenkranz P., Paxton R.J., Moritz R.F.A. Parasites and pathogens of the honeybee (Apis mellifera) and their influence on inter-colonial transmission. PLoS One. 2015;10(10) doi: 10.1371/journal.pone.0140337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Truong A.-T., Yoo M.S., Yun B.R., Kang J.E., Noh J., Hwang T.J., et al. Prevalence and pathogen detection of Varroa and Tropilaelaps mites in Apis mellifera (Hymenoptera, Apidae) apiaries in South Korea. J. Apicult. Res. 2022 doi: 10.1080/00218839.2021.2013425. [DOI] [Google Scholar]
- 21.Botías C., Martín-Hernández R., Barrios L., Meana A., Higes M. Nosema spp. infection and its negative effects on honey bees (Apis mellifera iberiensis) at the colony level. Vet. Res. 2013;44(1):25. doi: 10.1186/1297-9716-44-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Core A., Runckel C., Ivers J., Quock C., Siapno T., Denault S., et al. A new threat to honey bees, the parasitic phorid fly Apocephalus borealis. PLoS One. 2012;7(1) doi: 10.1371/journal.pone.0029639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson D.L., Roberts J.M.K. Standard methods for Tropilaelaps mites research. J. Apicult. Res. 2013;52(4):1–16. doi: 10.3896/IBRA.1.52.4.21. [DOI] [Google Scholar]
- 24.Dietemann V., Nazzi F., Martin S.J., Anderson D.L., Locke B., Delaplane K.S., et al. Standard methods for varroa research. J. Apicult. Res. 2013;52(1):1–54. doi: 10.3896/IBRA.1.52.1.09. [DOI] [Google Scholar]
- 25.Ptaszyńska A.A., Borsuk G., Mułenko W., Demetraki-Paleolog J. Differentiation of Nosema apis and Nosema ceranae spores under scanning electron microscopy (SEM) J. Apicult. Res. 2014;53(5):537–544. doi: 10.3896/IBRA.1.53.5.02. [DOI] [Google Scholar]
- 26.Truong A.T., Sevin S., Kim S., Yoo M.S., Cho Y.S., Yoon B. Rapidly quantitative detection of Nosema ceranae in honeybees using ultra-rapid real-time quantitative PCR. J. Vet. Sci. 2021;22(3):e40. doi: 10.4142/jvs.2021.22.e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fries I., Chauzat M.P., Chen Y.P., Doublet V., Genersch E., Gisder S., et al. Standard methods for Nosema research. J. Apicult. Res. 2013;52(1):1–28. doi: 10.3896/IBRA.1.52.1.14. [DOI] [Google Scholar]
- 28.Sammataro D., de Guzman L., George S., Ochoa R., Otis G. Standard methods for tracheal mite research. J. Apicult. Res. 2013;52(4):1–20. doi: 10.3896/IBRA.1.52.4.20. [DOI] [Google Scholar]
- 29.Korean Beekeeping Association Statistical survey of Korean beekeeping industry. 2020. https://korapis.or.kr/jsp/sub3-1_01.jsp [cited 2022 May 27]; Available from:
- 30.Jung C., Lee M. In: Asian Beekeeping in the 21st Century. Chantawannakul P., Williams G., Neumann P., editors. Springer; Singapore: 2018. Beekeeping in Korea: past, present, and future challenges; pp. 175–197. [DOI] [Google Scholar]
- 31.Kang S.W., Yoo M.S., Noh J.H., Park H.S., Min I., Jeon D.M., et al. Occurrence and prevalence of honeybee disease in Apis mellifera and Apis cerana in Korea. Korean J. Apic. 2012;27(3):187–195. [Google Scholar]
- 32.Choe S.E., Nguyen L.T., Noh J.H., Kweon C.H., Reddy K.E., Koh H.B., et al. Analysis of the complete genome sequence of two Korean sacbrood viruses in the honey bee, Apis mellifera. Virology. 2012;432:155–161. doi: 10.1016/j.virol.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 33.Choi Y.S., Lee M.Y., Hong I.P., Kim N.S., Kim H.K., Lee K.G., et al. Occurrence of sacbrood virus in Korean apiaries from Apis cerana (Hymenoptera: Apidae) Korean J. Apic. 2010;25(3):187–191. [Google Scholar]
- 34.Hammer Ø., Harper D.A.T., Ryan P.D. Past: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001;4(1):9. [Google Scholar]
- 35.Chantawannakul P., de Guzman L.I., Li J., Williams G.R. Parasites, pathogens, and pests of honeybees in Asia. Apidologie. 2016;47(3):301–324. doi: 10.1007/s13592-015-0407-5. [DOI] [Google Scholar]
- 36.Namin S.M., Koh Y., Osabutey A.F., Jung C. Invasion pathway of the honeybee pest, small hive beetle, Aethina tumida (Coleoptera: nitidulidae) in the Republic of Korea inferred by mitochondrial DNA sequence analysis. J. Asia Pac. Entomol. 2019;22(3):963–968. doi: 10.1016/j.aspen.2019.07.008. [DOI] [Google Scholar]
- 37.Martin S.J., Highfield A.C., Brettell L., Villalobos E.M., Budge G.E., Powell M., et al. Global honey bee viral landscape altered by a parasitic mite. Science. 2012;336(6086):1304–1306. doi: 10.1126/science.1220941. [DOI] [PubMed] [Google Scholar]
- 38.Rowland B.W., Rushton S.P., Shirley M.D.F., Brown M.A., Budge G.E. Identifying the climatic drivers of honey bee disease in England and Wales. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-01495-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tentcheva D., Gauthier L., Zappulla N., Dainat B., Cousserans F., Colin M.E., et al. Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France. Appl. Environ. Microbiol. 2004;70(12):7185–7191. doi: 10.1128/AEM.70.12.7185-7191.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szabo T.I. Effect of weather factors on honeybee flight activity and colony weight gain. J. Apicult. Res. 1980;19(3):164–171. doi: 10.1080/00218839.1980.11100017. [DOI] [Google Scholar]
- 41.Li J., Qin H., Wu J., Sadd B.M., Wang X., Evans J.D., et al. The prevalence of parasites and pathogens in Asian honeybees Apis cerana in China. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0047955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hassanyar A.K., Huang S., Li Z., Rizwan M., Mehmood S., Raza M.F., et al. Prevalence of bee viruses in Apis cerana cerana populations from different locations in the Fujian Province of China. Microbiologyopen. 2019;8(9) doi: 10.1002/mbo3.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thu H.T., Thi N., Lien K., Linh M.T., Le T.H., Thi N., et al. Prevalence of bee viruses among Apis cerana populations in Vietnam. J. Apicult. Res. 2016;55(5):379–385. doi: 10.1080/00218839.2016.1251193. [DOI] [Google Scholar]
- 44.White G.F. US Department of Agriculture, Bureau of Entomology; Washington, DC, USA: 1913. Sacbrood, a Disease of Bees. U.S. Department of Agriculture, Bulletin of Entomology. [Google Scholar]
- 45.Truong A.-T., Kim J.M., Lim S.J., Yoo M.S., Cho Y.S., Yoon B.S. Development of ultra-rapid PCR system for genotyping of Sacbrood virus. J. Apic. 2018;33(2):83–98. doi: 10.17519/apiculture.2018.06.33.2.83. [DOI] [Google Scholar]
- 46.Shan L., Liuhao W., Jun G., Yujie T., Yanping C., Jie W., et al. Chinese sacbrood virus infection in Asian honey bees (Apis cerana cerana) and host immune responses to the virus infection. J. Invertebr. Pathol. 2017;150:63–69. doi: 10.1016/j.jip.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 47.Mingxiao M., Ming L., Jian C., Song Y., Shude W., Pengfei L. Molecular and biological characterization of Chinese sacbrood virus LN isolate. Comp. Funct. Genom. 2011;2011 doi: 10.1155/2011/409386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen N.T., Le T.H. Complete genome sequence of sacbrood virus strain SBM2, isolated from the honeybee Apis cerana in Vietnam. Genome Announc. 2013;1(1) doi: 10.1128/genomeA.00076-12. e00076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rao K.M., Katna S., Rana B.S., Rana R. Thai sacbrood and sacbrood viruses versus European foulbrood of hive bees in India – a review. J. Apicult. Res. 2016;54(3):192–199. doi: 10.1080/00218839.2016.1145417. [DOI] [Google Scholar]
- 50.Liu X., Zhang Y., Yan X., Han R. Prevention of Chinese sacbrood virus infection in Apis cerana using RNA interference. Curr. Microbiol. 2010;61(5):422–428. doi: 10.1007/s00284-010-9633-2. [DOI] [PubMed] [Google Scholar]
- 51.Vung N.N., Choi Y.S., Kim I. High resistance to Sacbrood virus disease in Apis cerana (Hymenoptera: Apidae) colonies selected for superior brood viability and hygienic behavior. Apidologie. 2020;51:61–74. doi: 10.1007/s13592-019-00708-6. [DOI] [Google Scholar]
- 52.Diao Q., Yang D., Zhao H., Deng S., Wang X., Hou C., et al. Prevalence and population genetics of the emerging honey bee pathogen DWV in Chinese apiculture. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-48618-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cornman R.S., Tarpy D.R., Chen Y., Jeffreys L., Lopez D., Pettis J.S., et al. Pathogen webs in collapsing honey bee colonies. PLoS One. 2012;7(8) doi: 10.1371/journal.pone.0043562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cavigli I., Daughenbaugh K.F., Martin M., Lerch M., Banner K., Garcia E., et al. Pathogen prevalence and abundance in honey bee colonies involved in almond pollination. Apidologie. 2016;47(2):251–266. doi: 10.1007/s13592-015-0395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bordin F., Zulian L., Granato A., Caldon M., Colamonico R., Toson M., et al. Presence of known and emerging honey bee pathogens in apiaries of Veneto region (northeast of Italy) during spring 2020 and 2021. Appl. Sci. 2022;12:2134. doi: 10.3390/app12042134. [DOI] [Google Scholar]
- 56.Matthijs S., DeWaele V., Vandenberge V., Verhoeven B., Evers J., Brunain M., et al. Nationwide screening for bee viruses and parasites in Belgian honey bees. Viruses. 2020;12(8):890. doi: 10.3390/v12080890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morawetz L., Köglberger H., Griesbacher A., Derakhshifar I., Crailsheim K., Brodschneider R., et al. Health status of honey bee colonies (Apis mellifera) and disease-related risk factors for colony losses in Austria. PLoS One. 2019;14(7) doi: 10.1371/journal.pone.0219293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bailey L. Recent research on honey bee viruses. Bee World. 1975;56:55–64. [Google Scholar]
- 59.Spurny R., Přidal A., Pálková L., Kiem H.K.T., de Miranda J.R., Plevka P. Virion structure of black queen cell virus, a common honey bee pathogen. J. Virol. 2017;91(6):e02100–e02116. doi: 10.1128/JVI.02100-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Higes M., Martín-Hernández R., Botías C., Bailón E.G., González-Porto A.V., Barrios L., et al. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 2008;10(10):2659–2669. doi: 10.1111/j.1462-2920.2008.01687.x. [DOI] [PubMed] [Google Scholar]
- 61.Highfield A.C., El Nagar A., Mackinder L.C., Noël L.M., Hall M.J., Martin S.J., et al. Deformed wing virus implicated in overwintering honeybee colony losses. Appl. Environ. Microbiol. 2009;75(22):7212–7220. doi: 10.1128/AEM.02227-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Natsopoulou M.E., McMahon D.P., Doublet V., Frey E., Rosenkranz P., Paxton R.J. The virulent, emerging genotype B of deformed wing virus is closely linked to overwinter honeybee worker loss. Sci. Rep. 2017;7(1):5242. doi: 10.1038/s41598-017-05596-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gajda A.M., Mazur E.D., Bober A.M., Czopowicz M. Nosema ceranae interactions with Nosema apis and black queen cell virus. Agriculture. 2021;11(10):963. doi: 10.3390/agriculture11100963. [DOI] [Google Scholar]
- 64.Grabensteiner E., Ritter W., Carter M.J., Davison S., Pechhacker H., Kolodziejek J., et al. Sacbrood virus of the honeybee (Apis mellifera): rapid identification and phylogenetic analysis using reverse transcription-PCR. Clin. Diagn. Lab. Immunol. 2001;8(1):93–104. doi: 10.1128/CDLI.8.1.93-104.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bailey L., Woods R.D. Two more small RNA viruses from honey bees and further observations on sacbrood and acute bee-paralysis viruses. J. Gen. Virol. 1977;25:175–186. doi: 10.1099/0022-1317-37-1-175. [DOI] [Google Scholar]
- 66.de Miranda J.R., Genersch E. Deformed wing virus. J. Invertebr. Pathol. 2010;103:S48–S61. doi: 10.1016/j.jip.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 67.Traynor K.S., Mondet F., de Miranda J.R., Techer M., Kowallik V., Oddie M.A.Y., et al. Varroa destructor: a complex parasite, crippling honey bees worldwide. Trends Parasitol. 2020;36(7):592–606. doi: 10.1016/j.pt.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 68.Bailey L., Fernando E.F.W. Effects of sacbrood virus on adult honey-bees. Ann. Appl. Biol. 1972;72(1):27–35. doi: 10.1111/j.1744-7348.1972.tb01268.x. [DOI] [Google Scholar]
- 69.Seitz K., Buczolich K., Dikunová A., Plevka P., Power K., Rümenapf T., et al. A molecular clone of chronic bee paralysis virus (CBPV) causes mortality in honey bee pupae (Apis mellifera) Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-52822-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Genersch E. American foulbrood in honeybees and its causative agent, Paenibacillus larvae. J. Invertebr. Pathol. 2010;103:S10–S19. doi: 10.1016/j.jip.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 71.Runckel C., Flenniken M.L., Engel J.C., Ruby J.G., Ganem D., Andino R., et al. Temporal analysis of the honey bee microbiome reveals four novel viruses and seasonal prevalence of known viruses, Nosema, and Crithidia. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ryabov E.V., Childers A.K., Chen Y., Madella S., Nessa A., vanEngelsdorp D., et al. Recent spread of Varroa destructor virus-1, a honey bee pathogen, in the United States. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-17802-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee S., Hong K., Cho Y.S., Choi Y.S., Yoo M., Lee S. Review of the subgenus Aethina Erichson s. str. (Coleoptera: nitidulidae: Nitidulinae) in Korea, reporting recent invasion of small hive beetle, Aethina tumida. J. Asia Pac. Entomol. 2017;20(2):553–558. doi: 10.1016/j.aspen.2017.03.006. [DOI] [Google Scholar]
- 74.Yoo M.S., Truong A.T., Choi Y.S., Hong K.J., Hwang T.J., Seo S.K., et al. Pathogen detection and phylogenetic analysis of Aethina tumida Murray in South Korea. J. Apicult. Sci. 2022;66(1):45–55. doi: 10.2478/jas-2022-0004. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.