Abstract
Background
Accurate assessment of coronavirus disease 2019 (COVID-19) lung involvement through chest radiograph plays an important role in effective management of the infection. This study aims to develop a two-step feature merging method to integrate image features from deep learning and radiomics to differentiate COVID-19, non-COVID-19 pneumonia and normal chest radiographs (CXR).
Methods
In this study, a deformable convolutional neural network (deformable CNN) was developed and used as a feature extractor to obtain 1,024-dimensional deep learning latent representation (DLR) features. Then 1,069-dimensional radiomics features were extracted from the region of interest (ROI) guided by deformable CNN’s attention. The two feature sets were concatenated to generate a merged feature set for classification. For comparative experiments, the same process has been applied to the DLR-only feature set for verifying the effectiveness of feature concatenation.
Results
Using the merged feature set resulted in an overall average accuracy of 91.0% for three-class classification, representing a statistically significant improvement of 0.6% compared to the DLR-only classification. The recall and precision of classification into the COVID-19 class were 0.926 and 0.976, respectively. The feature merging method was shown to significantly improve the classification performance as compared to using only deep learning features, regardless of choice of classifier (P value <0.0001). Three classes’ F1-score were 0.892, 0.890, and 0.950 correspondingly (i.e., normal, non-COVID-19 pneumonia, COVID-19).
Conclusions
A two-step COVID-19 classification framework integrating information from both DLR and radiomics features (guided by deep learning attention mechanism) has been developed. The proposed feature merging method has been shown to improve the performance of chest radiograph classification as compared to the case of using only deep learning features.
Keywords: Coronavirus disease 2019 (COVID-19), radiomics, deep learning, chest radiograph, classification
Introduction
Coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has caused a devastating effect on the health and well-being of the global population. Chest radiograph imaging is a main radiological investigation performed on patients who are suspected or confirmed to have COVID-19. It is an essential tool for auxiliary diagnosis, evaluating complications, severity assessment and prognosis monitoring (1-4). Former studies indicated patients infected with COVID-19 present certain lung region abnormalities in the chest radiography images (4-7), such as the bilateral and peripheral ground-glass opacity (GGO) in the early stage and the pulmonary consolidation opacity in the later stage. The effectiveness of chest radiograph in COVID-19 clinical applications, however, has been hampered by their relatively low sensitivity for early detection and diagnosis.
In recent years, deep learning has been successfully applied to both medical imaging analysis and automatic diseases diagnosis (8-10). For the COVID-19 classification of chest radiograph images, most research is based on modifications to classification models to improve COVID-19 detection accuracy. The categories can be divided into binary and multiple classification tasks (11-13). For binary classification task, it is mainly based on the discrimination between COVID-19 and normal or the COVID-19 and other pneumonias. Abbas et al. validated a deep convolutional neural network (CNN) called Decompose, Transfer, and Compose (DeTraC), for the binary classification of COVID-19 chest radiograph images. The experimental results showed that the high accuracy of 93.1% was achieved by DeTraC in the detection of COVID-19 from normal, and severe acute respiratory syndrome cases (14). Karakanis et al. proposed a ResNet8-based CNN model to classifier the COVID-19 and normal chest radiograph images. It was designed in a form to support a lightweight architecture without transfer learning, whilst performing well. The improved model attained an accuracy of up to 98.7% (15). The multiple classification is to classify chest radiograph images of three or more categories. Khan et al. proposed a classification model called CB-STM-RENet to exploit channel boosting and learn textural variations to effectively screen the chest radiograph images of COVID-19 infection. The three public datasets with normal, viral or bacterial pneumonia, COVID-19 chest radiograph images were employed for training and testing. The extensive experimentations provided satisfactory detection performance with accuracy of 98.53% (CoV-Healthy-6k dataset), 97.48% (CoV-NonCoV-10k dataset) and 96.53% (CoV-NonCoV-15k dataset) (16). Hussain et al. adopted a 22 layered CoroDet model to classify four different classes, including 800 normal, 400 viral pneumonia, 400 bacteria pneumonia and 500 COVID-19. The improved model showed good classification results with the accuracy of 91.2% (17). Ghosh et al. presented an ENResNet model based on deep residual CNN architecture to do the classification task of normal, pneumonia and COVID-19 chest radiograph images. It achieved the overall accuracy of 98.42%, and the sensitivity and specificity were 99.46% and 98.27% respectively (18).
With the widespread application of artificial intelligence in COVID-19 detection, deep learning models have proven to be the powerful tools to augment radiologists. However, most deep learning models lack interpretability due to their black-box nature. Therefore, the understanding of the decision-making process of deep learning is still limited. Radiomics provides a convenient and reliable way to mine quantitative features from medical images and provide extensive information for diagnosis and prognosis (19-22). Some attempts have also been made to utilize radiomics features to predict COVID-19 from chest radiograph images and CT images (23-26). Previous studies suggest that the combination of radiomics features and deep learning features could achieve improved diagnostic performance compared to the case of using only one feature space (27,28). Mangalagiri et al. extracted 7 radiomic texture features from the lung regions and developed a 3D CNN model for obtaining the deep learning features. He performed the random forest classification of COVID-19 using the two classes of extracted features. The proposed model achieved a higher true positive rate with the average area under the receiver curve (AUC) of 0.9768 (29). Hu et al. proposed a deep learning model combined with radiomics analysis for enhancing the accuracy of COVID-19 classification. He extracted two radiomic feature maps based on cross-correlation analysis from pre-defined region of interest (ROI). The radiomics-boosted deep-learning model improved the AUC of VGG-16 from 0.918 to 0.969 (30). However, manually delineating lesions is a labor-intensive and time-consuming task.
In this study, we propose a novel classification method using the attention mechanism of a deep learning neural network to combine the information from deep learning with that from radiomics to differentiate COVID-19, non-COVID-19 pneumonia and normal chest radiographs (CXR). The effectiveness of the proposed combined method is validated by extensive comparative experiments. Our paper makes the following major contributions. First, a deformable CNN was developed by combining a deformable convolution layer with DenseNet121 (31), which can help the network adapt to the varying shapes of lung abnormalities and better aggregate local and global information. Second, a gradient based visualization method Grad-CAM++ (32) was utilized to generate the lung region class activation map (LCAM) for performing radiomics analysis on the attentional lung region. We present the following article in accordance with the STARD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-531/rc).
Methods
Datasets
In this retrospective study, chest radiograph images were obtained from several online public sources. For COVID-19 cases, 1,570 COVID-19 chest radiograph images were collected from several limited datasets (33-37). The collected dataset contained the medical images, while no demographic information was included. To avoid the possibility of date repetitiveness, manual check was conducted within the collected COVID-19 cases. Additional manual examination was also performed to assure the independence between training set and testing set. For non-COVID-19 pneumonia and normal cases, 1,700 chest radiograph images were randomly selected for each category from the RSNA Pneumonia Detection Challenge dataset (38). All radiograph images were acquired from a non-specific age group from various medical institutions. This minimized the bias caused by age-related characteristics and imaging protocols.
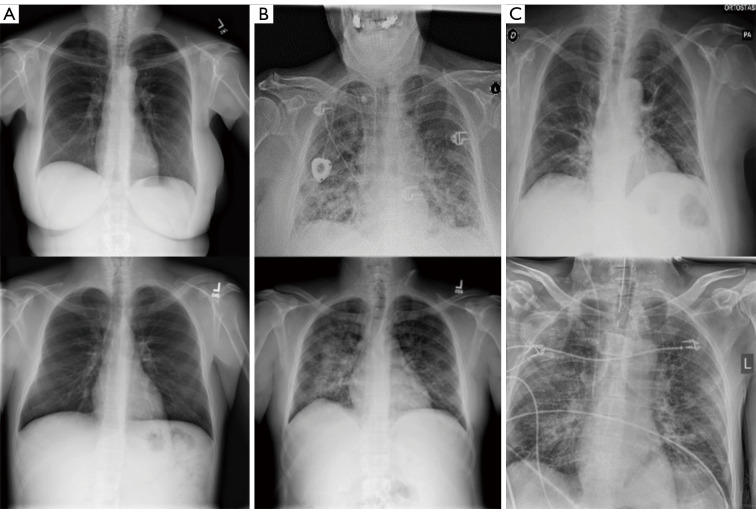
To preserve the model’s performance and prevent overfitting, 3,770 cases (1,300 normal, 1,300 non-COVID-19 pneumonia, 1,170 COVID-19 pneumonia) were randomly selected from the whole dataset as the training dataset. Another 300 random cases (100 normal, 100 non-COVID-19 pneumonia, 100 COVID-19 pneumonia) were selected as the validation set for hyperparameters’ fine-tuning and training monitoring. The remaining 900 cases (300 normal, 300 non-COVID-19 pneumonia, 300 COVID-19 pneumonia) were then used as the testing set to evaluate proposed model’s performance. To prevent the testing dataset’s information from being included in the training process, the same dataset splits were used for both the deep learning feature extractor training and the subsequent merged model training, while the testing dataset was not used for training any model throughout this study. Figure 1 shows some representative samples of Chest radiograph images from the dataset.
Figure 1.
Examples of chest radiograph images. (A) Normal lungs; (B) non-COVID-19 pneumonia; (C) COVID-19. COVID-19, coronavirus disease 2019.
Study design
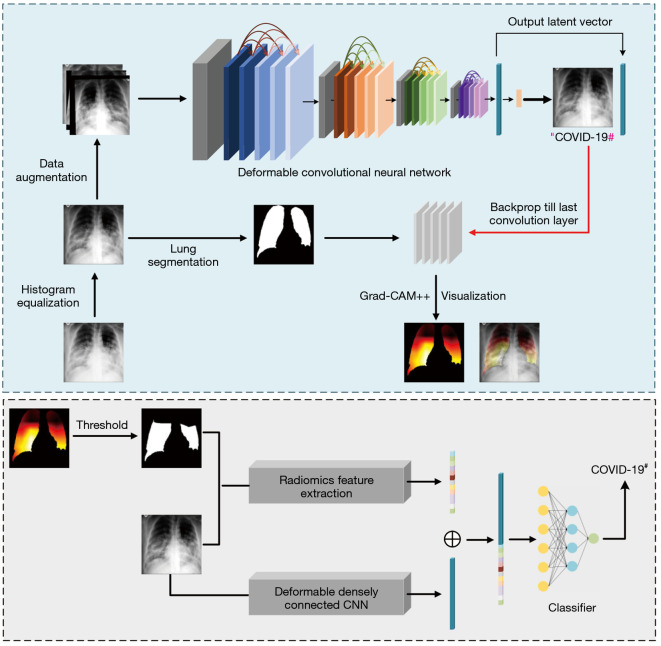
In this section, an automatic COVID-19 chest radiograph diagnostic method with merged information is proposed. The overall framework of our method is illustrated in Figure 2. The details of corresponding parts are presented in the following subsections.
Figure 2.
Illustration of our proposed COVID-19 classification method. Before training the classification network, images are preprocessed to improve data quality and augmented to increase the training dataset size. When a chest Radiograph image is fed into the deformable CNN, the network outputs a preliminary classification result and a 1,024-dimensional latent representation of the input image. Then, a LCAM is generated by lung region segmentation and the Grad-CAM++ method. A ROI mask is generated by thresholding the LCAM. Radiomics features extracted from the ROI are concatenated with the latent representation and then fed into different machine learning classifiers for classification. #, predicted class. CNN, convolutional neural network; COVID-19, coronavirus disease 2019; LCAM, lung region class activation map; ROI, region of interest.
Data preprocessing
Due to the various sources of the collected dataset, all images were unified by being resized to 256×256 with a bit depth of 8. Most of the radiograph images have large area opacification and blurred lung boundaries. The histogram equalization is a technique used to distribute the gray levels within a CXR, during which the image histogram is converted into a uniform histogram and the edges and borders between different regions are highlighted. This enhanced texture information will promote the feature extraction process by CNNs. It has been proved in previous research (39,40), histogram equalization could increase the contrast of low intensity regions and improve classification performance. Therefore, global-level histogram equalization was applied on the whole dataset for preprocessing. After histogram equalization, a lung segmentation module was implemented using a pre-trained model (41) to generate lung region masks, which were later used to generate lung region class activation maps (LCAMs). To exclude small erroneous regions, connected component analysis was applied after mask generation, and binary morphological closing was performed to fill any small holes of radius up to 10 pixels in the segmentation.
The proposed deformable CNN
In this section, we proposed a deformable CNN for COVID-19 latent representation extraction. The backbone model of the network was DenseNet121 (31), comprised of four Dense Blocks and three transition layers. The Dense Blocks were composed of 6, 12, 24, 16 densely connected layers, respectively. In each Dense Block, each layer was connected to every other layer in a feed-forward fashion to ensure information reuse. The transition layers (consisting of a convolutional layer and a pooling layer) were responsible for down-sampling and increasing receptive fields. To improve the network’s ability to adapt to different shapes and irregular boundaries of lung tissues and lesions, a deformable convolutional layer (42) was used to replace the last convolutional layer in the last Dense Block, and a fully connected layer outputting a 3-vector was added at the very end of the model to act as a 3-class classifier.
The principle of deformable convolution, as illustrated in Figure 3, is adding 2D offsets to the regular grid sampling locations from standard convolution, which enables the sampling grid to deform away from rectangular shapes. Additional convolutional layers are used to generate both horizontal and vertical offsets from the preceding feature maps. Thus, the deformation is conditioned on the input features in a local, dense, and adaptive manner.
Figure 3.
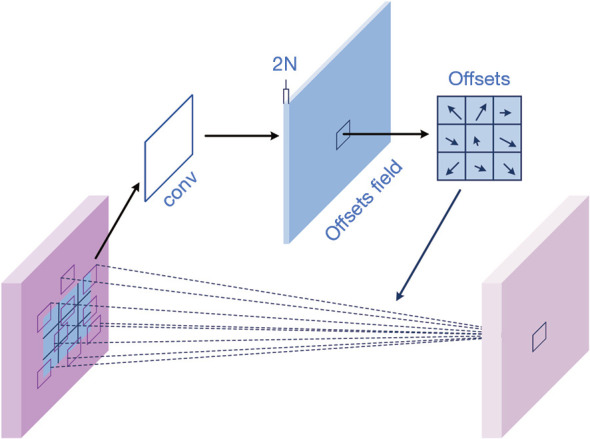
Illustration of 3×3 deformable convolution. Horizontal and vertical offsets generated from additional convolutional layers are added to the regular grid sampling locations from standard convolution, which enables the sampling grid to deform away from rectangular shapes. conv, convolution.
A standard 2D convolution consists of two steps: (I) sampling using a regular grid R over the input feature map x, and (II) summation of sampled values weighted by w. In a basic 2D convolutional layer, the sampling location grid R of a 3x3 convolution kernel with dilation 1 can be expressed as:
| [1] |
Correspondingly, the output feature map y can be expressed as
| [2] |
Where p0 enumerates the location in y, and pn enumerates the location in R.
Instead of using a fixed sampling grid, deformable convolution added 2D offsets ∆pn to R to augment the sampling process. The feature map after deformable convolution can be expressed as
| [3] |
Since the location of (p0 + pn + ∆pn) is typically fractional, the value of x (p0 + pn + ∆pn) was obtained through bilinear interpolation.
A standard convolutional layer is applied over the preceding feature map and outputs the offset fields with the same spatial resolution as the input feature map. The dimension 2N corresponds to N 2D offsets.
The deformable convolutional layer was implemented after the last convolutional layer in Dense Block 4. The output of the deformable convolution layer was average-pooled and flattened to a 1,024-dimensional vector—the latent representation of the input image. After that, a fully-connected layer outputting three classes was implemented to output the predicted class. The cross-entropy loss between the predicted and true classes was used to optimize the network parameters.
Training, validation, and testing procedures
In this study, we implemented our deformable CNN using Python 3.7 and Pytorch 1.7 with CUDA 11.1 on a workstation equipped with an NVIDIA GeForce RTX 2080 Ti GPU.
To prevent overfitting during training, after data preprocessing, data augmentation was applied to the training set, while no augmentation was applied to the testing set. The augmentation techniques used are listed as follows: (I) random horizontal flipping (probability =0.5); (II) random affine transformation [the range of degrees was (−10, +10), the range of translation was 0.1 for both horizontal and vertical directions, the range of scale factors was (0.9, 1.1)]; (III) random brightness change [the range was (0.9, 1.1)]. All augmentation techniques were implemented with PyTorch. The backbone model was initialized with ImageNet (43) pretrained weights. The deformable convolutional layer was then implemented, with its offset field initialized with zeros, while the other learnable parameters of the deformable convolutional layer and the parameters of the fully connected layer were initialized with default PyTorch initialization methods.
During DCNN training, the layers before 15th convolutional layer in Dense Block four were frozen to avoid damaging the pre-learned feature extraction information during the re-training. The 15th and 16th convolutional layers in Dense Block were unfrozen and fine-tuned to adapt the pre-trained features to new medical image data, while the deformable convolutional layer and fully connected layer were trained from sketch. The number of unfrozen layers was set to three to balance overfitting issue and training performance. The Adam optimizer (44) was used, with hyperparameter values of β1=0.9 and β2=0.999. The learning rate for the three convolutional layers was set to 5e-5, and the learning rate for the fully connected layer was set to 5e-2. Training was performed using a batch size of 64. We trained 60 epochs to get convergence.
After convergence, the trained deformable CNN was used to generate a 1,024 deep learning latent representation (DLR) features of the training set and validation set. The LCAMs of training set and validation set were also generated for radiomics feature extraction.
LCAM generation
Grad-CAM++ (32) is a state-of-the-art CNN interpretation method. It assesses the importance of each pixel in a feature map towards the overall classification decision. After the deformable CNN was trained, Grad-CAM++ was used to generate the saliency map for the original image mw. The lung region binary mask ml was combined with mw by element-wise multiplication to obtain the LCAM. The areas of the map with relatively high importance score were the critical factors for why the neural network classified the input into a particular class.
The importance score within the lung region was normalized to (0, 1). Then, the thresholding technique proposed by Zhou et al. was used to segment the regions of the class activation map. In Zhou’s paper, the region with the value above 20% of the max value is regarded with the distinctive region. In this study, the region with a value greater than 20% of the maximum value (0.2) was segmented and termed as the “attentional lung region” (45).
Radiomics analysis and merged model construction
The attentional lung region was used as the ROI for radiomics feature extraction. To further analyze the differences in imaging phenotype between different classes, radiomics features widely used in tumor diagnosis were extracted using Python (version 3.7) and PyRadiomics (46) (version 3.0). Nine 2D image transforms in PyRadiomics (including the original image, Laplacian of Gaussian filter, wavelet filtering, Square transform, Square Root transform, Logarithm transform, Exponential transform, Gradient transform, and LocalBinaryPattern2D transform) were applied. Six series of features were extracted, including first order features, Gray Level Co-occurrence Matrix, Gray Level Size Zone Matrix, Gray Level Run Length Matrix, Neighboring Gray Tone Difference Matrix, and Gray Level Dependence Matrix. In total, 1,069 radiomics features were extracted and concatenated with the 1,024 DLR features as the merged feature set.
As common issue in radiomics studies with hundreds of features, many of the biomarkers (features) used as predictors were highly correlated with one another. This challenge necessitated feature selection in order to avoid collinearity, reduce dimensionality, and minimize noise (47,48).
Unsupervised and supervised feature selection methods were used successively. The training set and validation set were combined as the new training set for feature selection. First, an unsupervised feature selection method was applied by calculating Pearson correlation coefficients between each pair of features. Features with a correlation coefficient higher than 0.95 were regarded as redundant, and one feature was selected to represent those highly-correlated features. After unsupervised feature selection, supervised feature selection was performed using scikit-learn package (49). A transformer was built from a Random Forest composed of 1,000 decision trees and used to select features based on importance weights. Features whose importance was greater or equal to mean value were kept while the others are discarded. After applying the abovementioned feature selection methods to the merged feature set, 458 DLR features and 264 radiomics features were selected.
Four different classifiers (Decision Tree, Random Forest, LinearSVC, Multilayer Perception) were constructed based on the selected merged feature set. In order to quantify the effect of using radiomic data in classification, a DLR-only feature set was generated using the same procedures as described previously. This DLR-only feature set contained 489 out of 1,024 features. Classification results of each classifier using the DLR-only feature set and the merged feature set, respectively, were calculated and compared. For each classifier, the training and testing procedures were repeated 50 times. The mean and standard deviation of the accuracy were computed for each classifier. The relative standard deviation (RSD), defined in Eq. [4], was calculated to quantify the stability of the different classifiers:
| [4] |
Where σacc and µacc are the standard deviation and the mean of the accuracy, respectively. Lower RSD means higher stability of the corresponding model. The two-tailed t-test was used to measure the significance between different feature sets’ classification accuracy. P values <0.05 indicated statistical significance.
From intermediate results shown in Table 1, Random Forest was chosen as the classifier for our proposed model shown in Figure 2. A range of different evaluation metrics, including average accuracy for the testing dataset as well as recall, precision and F1-score for individual classes were used to quantitatively evaluate the classification performance using the two different feature sets (DLR + radiomics and DLR-only).
Table 1. Classification performance of different models.
| Models | Hyper-parameters | Features | Accuracy (mean ± SD) | RSD (%) | P value |
|---|---|---|---|---|---|
| Decision Tree | criterion=‘gini’, min_samples_split=2, min_samples_leaf=1 | DLR | 0.842±0.006 | 0.69 | <0.0001 |
| DLR + ARF | 0.854±0.007 | 0.84 | |||
| Random Forest | n_estimators=100, criterion=‘gini’ | DLR | 0.904±0.003 | 0.40 | <0.0001 |
| DLR + ARF | 0.910±0.003 | 0.40 | |||
| LinearSVC | penalty=‘l1’, loss=‘squared_hinge’, max_iter=10000 | DLR | 0.867±0.001 | 0.10 | <0.0001 |
| DLR + ARF | 0.881±0.001 | 0.11 | |||
| Multilayer Perceptron | hidden_layer_sizes=(100) activation=‘relu’, solver=‘adam’ |
DLR | 0.908±0.004 | 0.47 | <0.0001 |
| DLR + ARF | 0.911±0.004 | 0.42 |
SVC, support vector classification; DLR, deep learning latent representation; ARF, attentional lung region radiomics features; RSD, relative standard deviation.
This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Lung region activation visualization
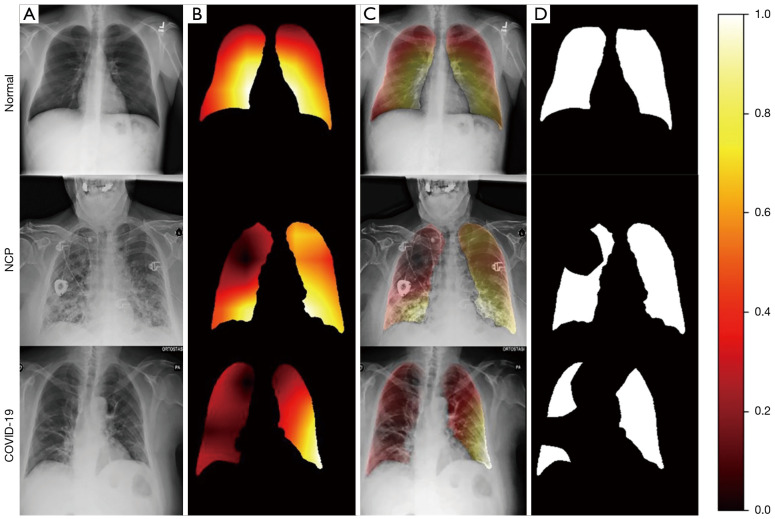
It takes about 120 min to train the deep learning feature extractor. After the proposed deformable CNN was trained, the Grad-CAM++ method was used to visualize the attention of the network and to extract attentional areas. Representative cases’ visualization results are presented in Figure 4. Figure 4A shows the original chest radiograph images, while Figure 4B shows the lung region heatmaps highlighting the activation region associated with the predicted class. In Figure 4C, the heatmaps are overlapped on the original images to show the attentional lung region. The ROI masks generated by thresholding are shown in Figure 4D.
Figure 4.
Representative cases of (A) original chest Radiograph images; (B) lung region activation maps generated by the proposed network; (C) lung region activation maps overlaid on the chest Radiograph images; (D) binary mask of the region of interest. Colors represent the importance scores, which represent the importance of each pixel in a feature map towards the overall classification decision. The lung region which has a value greater than 0.2 is segmented as the ROI. From top to bottom, the three cases were from normal class, non-COVID-19 pneumonia class and COVID-19 class respectively. NCP, non-COVID-19 pneumonia; COVID-19, coronavirus disease 2019; ROI, region of interest.
Evaluation of multi-class classification
The accuracy statistics for different classifier models over 50 train-test iterations is shown in Figure 5. The iterations took approximate 4 hours in total. The results demonstrate that, for all tested classifiers, the performance of the merged feature set is better than that of the deep learning feature set only.
Figure 5.
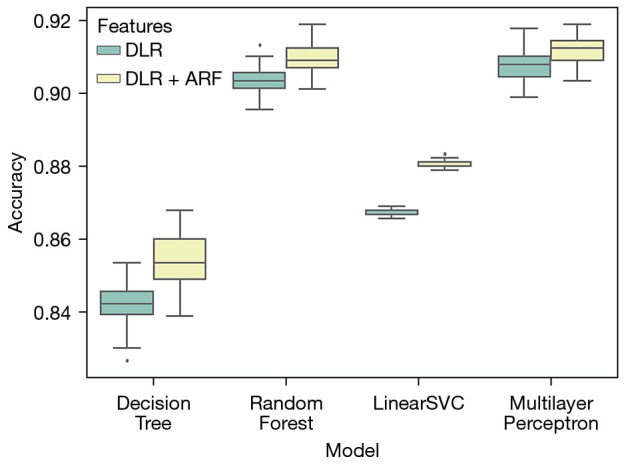
Box plot comparing the classification accuracy of four different classifiers with two different feature sets. The black dot indicates outliers. DLR, deep learning latent representation; ARF, attentional lung region radiomics features; SVC, support vector classification.
To further investigate the difference between different classification models, quantitative measurements of the classification framework with different classifiers and different feature sets are listed in Table 1.
The detailed hyper-parameters of the classifiers are listed in the table. Mean accuracy and standard deviation are also listed. For all classifiers, the classification models based on the merged feature set achieve significant better performance than those based on the DLR-only feature set (P value <0.0001).
The Random Forest and Multilayer Perception (MLP) methods have higher accuracy scores than the Decision Tree and LinearSVC methods. Moreover, Random Forest has lower RSD than MLP, suggesting that the Random Forest method is more stable. Hence, Random Forest was chosen as the classifier for further quantitative analysis.
The recall, precision, F1-score and overall accuracy of the Random Forest classifier for each class and each feature set are given in Table 2. The merged feature set achieves improved overall accuracy and a higher F1-score for all three classes compared to the DLR-only feature set.
Table 2. Random Forest’s Classification Performance with different feature sets.
| Methods | Class labels | Recall (95% CI) | Precision (95% CI) | F1-score (95% CI) | Overall accuracy (95% CI) |
|---|---|---|---|---|---|
| DLR | Normal | 0.922 (0.892–0.952) | 0.855 (0.815–0.895) | 0.887 (0.851–0.923) | 0.904 (0.870–0.937) |
| NCP | 0.877 (0.839–0.914) | 0.886 (0.851–0.922) | 0.881 (0.845–0.918) | ||
| COVID-19 | 0.913 (0.881–0.945) | 0.979 (0.963–0.995) | 0.945 (0.919–0.971) | ||
| DLR + ARF | Normal | 0.922 (0.892–0.953) | 0.864 (0.825–0.903) | 0.892 (0.857–0.927) | 0.910 (0.878–0.943) |
| NCP | 0.883 (0.847–0.919) | 0.898 (0.864–0.932) | 0.890 (0.855–0.926) | ||
| COVID-19 | 0.926 (0.896–0.956) | 0.976 (0.958–0.993) | 0.950 (0.925–0.975) |
DLR, deep learning latent representation; ARF, attentional lung region radiomics features; NCP, non-COVID-19 pneumonia; COVID-19, coronavirus disease 2019; CI, confidence interval.
Discussion
Several studies have illustrated the importance of integrating information from both radiomics and deep learning features (27,30) for COVID-19 detection from CT images.. This study proposed a novel method to classify COVID-19, non-COVID-19 pneumonia and normal lung by integrating information from deep learning features and attention-guided radiomics features. The results shown in Figure 5 demonstrated that the combination of DLR features and radiomics features led to improved classification performance as compared to DLR features alone. The two-tailed t-test was used to further verify the statistical significance of this performance difference on four classifiers. As shown in Table 1, P values of all classifiers are less than 0.0001, indicating that merged feature set could achieve better classification performance regardless of the classification algorithm. This may be because the radiomics features and DLR features provide complimentary information, i.e., radiomics features mainly contain low-level information while the DLR features mainly contain high-level information. As a result, the combination of these features describes the medical images in a more complete manner than a single feature set. Furthermore, the introduction of the radiomics features also improves the interpretability of the classification model, which is crucial for clinical adoption of AI technologies (50).
In Table 2, the classification performance using the merged feature set and that using the DLR-only feature set were compared using more comprehensive evaluation metrics. It is noticed that the three categories achieved various performance in the recall and precision metrics. For recall, it is the measure of the correctly predicted positive cases over all positive cases in the data, while precision represents how many of the positive predictions made are correct. With or without the addition of radiomics features, COVID-19 achieved higher precision score than other two categories indicating that normal and non-COVID-19 pneumonia cases are less likely to be predicted as COVID-19. The COVID-19 recall score reflects the probability of a missed classification of COVID-19 (the higher the recall score, the less likely the wrong classification of COVID-19). The merged model achieved 0.926 for COVID-19 recall, which is 0.013 higher than the DLR-only model. Compared with non-COVID-19 pneumonia, normal category possessed a higher recall rate but a lower precision rate. It can be inferred that some non-COVID-19 pneumonia cases had relatively less obvious imaging features, which were likely to be misclassified as normal cases. In the further development, feature enhancement methods could be utilized to address this problem.
For radiomics features generation, previous studies usually extracted features from the lung volume or the manually delineated lesions (27,30). In this study, we used a CNNs’ attention mechanism to determine the ROI for radiomics feature extraction. The Grad-CAM++ visualization method was utilized to generate the lung region attentional map. It can be seen from Figure 4 that our proposed deep learning network was able to detect suspicious lung regions and make classification decisions based on these regions and their neighborhoods.
The novel two-step framework proposed in this study has various potential implications for medical image analysis. First, we demonstrated that combining radiomics features and DLR features was able to improve classification performance. Second, the attention-guided radiomics features extraction not only avoids the averaging effect caused by using the whole lung volume, but also removes the requirement for labor-intensive manual segmentation. The selection of threshold value also plays an important role in the process of generating the ROI mask. The change of threshold values could result in the variation of classification performance. Lower threshold value includes larger proportion of the lung region for feature extraction, while higher threshold value only focuses on the smaller region of highly activated region guided by deep learning model. The threshold level regulates the influence of the deep learning attention on the feature selection. In the future research, we plan to optimize the threshold value based on classification performance and physicians’ manually delineated ROI. The methods introduced in this study could be utilized for the computer-aided diagnosis of diseases aside from COVID-19, such as lung tumors.
Despite the promising results, the present study has several limitations. First, the datasets used here were collected from several online datasets. Image compression may have created artifacts in the source images. Second, the chest radiograph images from the online datasets lack relevant clinical information, which may include predictive information such as age and body mass index. The lack of dataset’s demographic information also may introduce risks of bias caused by ambiguous source and inauthentic data. Though the current research proposed a novel method to improve the classification performance, the clinical transferability of the model is restricted by the authenticity of the public dataset. Hence in future research, to ensure the dataset’s authenticity and quality, we planned to develop our own datasets based on local hospital’s data. Besides CXR, demographic information (e.g., age, sex) and other available modalities’ data will be also collected to conduct more comprehensive assessments. Further image synthesis method (51,52) and enhancement techniques (53) could be explored to improve the COVID-19 classification or severity assessment.
Conclusions
In this study, we have demonstrated a two-step COVID-19 classification framework integrating information from both DLR and radiomics features guided by deep learning attention mechanism. Merging deep learning and radiomics features in modeling has been shown to improve the chest radiograph COVID-19 classification performance as compared to the case of using deep learning features alone.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported in part by Health and Medical Research Fund (HMRF COVID190211), the Food and Health Bureau, The Government of the Hong Kong Special Administrative Region, and Shenzhen-Hong Kong-Macau S&T Program (Category C) (No. SGDX20201103095002019), Shenzhen Basic Research Program (No. JCYJ20210324130209023), Shenzhen Science and Technology Innovation Committee.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-531/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-531/coif). YXJW serves as the Editor-in-Chief of Quantitative Imaging in Medicine and Surgery. JC reports that he receives grants from Health and Medical Research Fund (HMRF COVID190211), the Food and Health Bureau, The Government of the Hong Kong Special Administrative Region, and Shenzhen-Hong Kong-Macau S&T Program (Category C) (No. SGDX20201103095002019), Shenzhen Basic Research Program (No. JCYJ20210324130209023), Shenzhen Science and Technology Innovation Committee. The other authors have no conflicts of interest to declare.
References
- 1.Inui S, Gonoi W, Kurokawa R, Nakai Y, Watanabe Y, Sakurai K, Ishida M, Fujikawa A, Abe O. The role of chest imaging in the diagnosis, management, and monitoring of coronavirus disease 2019 (COVID-19). Insights Imaging 2021;12:155. 10.1186/s13244-021-01096-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanne JP, Bai H, Bernheim A, Chung M, Haramati LB, Kallmes DF, Little BP, Rubin GD, Sverzellati N. COVID-19 Imaging: What We Know Now and What Remains Unknown. Radiology 2021;299:E262-79. 10.1148/radiol.2021204522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akl EA, Blažić I, Yaacoub S, Frija G, Chou R, Appiah JA, et al. Use of Chest Imaging in the Diagnosis and Management of COVID-19: A WHO Rapid Advice Guide. Radiology 2021;298:E63-9. 10.1148/radiol.2020203173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwan KEL, Tan CH. The humble chest radiograph: an overlooked diagnostic modality in the COVID-19 pandemic. Quant Imaging Med Surg 2020;10:1887-90. 10.21037/qims-20-771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Degerli A, Ahishali M, Yamac M, Kiranyaz S, Chowdhury MEH, Hameed K, Hamid T, Mazhar R, Gabbouj M. COVID-19 infection map generation and detection from chest X-ray images. Health Inf Sci Syst 2021;9:15. 10.1007/s13755-021-00146-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aggarwal S, Gupta S, Alhudhaif A, Koundal D, Gupta R, Polat K. Automated COVID-19 detection in chest X-ray images using fine-tuned deep learning architectures. Expert Syst 2022;39:e12749. 10.1111/exsy.12749 [DOI] [Google Scholar]
- 7.Roig-Marín N, Roig-Rico P. Ground-glass opacity on emergency department chest X-ray: a risk factor for in-hospital mortality and organ failure in elderly admitted for COVID-19. Postgrad Med 2022. [Epub ahead of print]. doi: . 10.1080/00325481.2021.2021741 [DOI] [PubMed] [Google Scholar]
- 8.Verma DK, Saxena G, Paraye A, Rajan A, Rawat A, Verma RK. Classifying COVID-19 and Viral Pneumonia Lung Infections through Deep Convolutional Neural Network Model using Chest X-Ray Images. J Med Phys 2022;47:57-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nasiri H, Hasani S. Automated detection of COVID-19 cases from chest X-ray images using deep neural network and XGBoost. Radiography (Lond) 2022;28:732-8. 10.1016/j.radi.2022.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Dahshan EA, Bassiouni MM, Hagag A, Chakrabortty RK, Loh H, Acharya UR. RESCOVIDTCNnet: A residual neural network-based framework for COVID-19 detection using TCN and EWT with chest X-ray images. Expert Syst Appl 2022;204:117410. 10.1016/j.eswa.2022.117410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam NFD, Sun H, Song L, Yang D, Zhi S, Ren G, Chou PH, Wan SBN, Wong MFE, Chan KK, Tsang HCH, Kong FS, Wáng YXJ, Qin J, Chan LWC, Ying M, Cai J. Development and validation of bone-suppressed deep learning classification of COVID-19 presentation in chest radiographs. Quant Imaging Med Surg 2022;12:3917-31. 10.21037/qims-21-791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Moura J, Novo J, Ortega M. Fully automatic deep convolutional approaches for the analysis of COVID-19 using chest X-ray images. Appl Soft Comput 2022;115:108190. 10.1016/j.asoc.2021.108190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattacharyya A, Bhaik D, Kumar S, Thakur P, Sharma R, Pachori RB. A deep learning based approach for automatic detection of COVID-19 cases using chest X-ray images. Biomed Signal Process Control 2022;71:103182. 10.1016/j.bspc.2021.103182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbas A, Abdelsamea MM, Gaber MM. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl Intell (Dordr) 2021;51:854-64. 10.1007/s10489-020-01829-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karakanis S, Leontidis G. Lightweight deep learning models for detecting COVID-19 from chest X-ray images. Comput Biol Med 2021;130:104181. 10.1016/j.compbiomed.2020.104181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan SH, Sohail A, Khan A, Lee YS. COVID-19 Detection in Chest X-ray Images Using a New Channel Boosted CNN. Diagnostics (Basel) 2022;12:267. 10.3390/diagnostics12020267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hussain E, Hasan M, Rahman MA, Lee I, Tamanna T, Parvez MZ. CoroDet: A deep learning based classification for COVID-19 detection using chest X-ray images. Chaos Solitons Fractals 2021;142:110495. 10.1016/j.chaos.2020.110495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh SK, Ghosh A. ENResNet: A novel residual neural network for chest X-ray enhancement based COVID-19 detection. Biomed Signal Process Control 2022;72:103286. 10.1016/j.bspc.2021.103286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu TT, Lam SK, To LH, Tse KY, Cheng NY, Fan YN, Lo CL, Or KW, Chan ML, Hui KC, Chan FC, Hui WM, Ngai LK, Lee FK, Au KH, Yip CW, Zhang Y, Cai J. Pretreatment Prediction of Adaptive Radiation Therapy Eligibility Using MRI-Based Radiomics for Advanced Nasopharyngeal Carcinoma Patients. Front Oncol 2019;9:1050. 10.3389/fonc.2019.01050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lafata K, Cai J, Wang C, Hong J, Kelsey CR, Yin FF. Spatial-temporal variability of radiomic features and its effect on the classification of lung cancer histology. Phys Med Biol 2018;63:225003. 10.1088/1361-6560/aae56a [DOI] [PubMed] [Google Scholar]
- 21.Attallah O. A computer-aided diagnostic framework for coronavirus diagnosis using texture-based radiomics images. Digit Health 2022;8:20552076221092543. 10.1177/20552076221092543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Won SY, Park YW, Ahn SS, Moon JH, Kim EH, Kang SG, Chang JH, Kim SH, Lee SK. Quality assessment of meningioma radiomics studies: Bridging the gap between exploratory research and clinical applications. Eur J Radiol 2021;138:109673. 10.1016/j.ejrad.2021.109673 [DOI] [PubMed] [Google Scholar]
- 23.Chaddad A, Hassan L, Desrosiers C. Deep Radiomic Analysis for Predicting Coronavirus Disease 2019 in Computerized Tomography and X-Ray Images. IEEE Trans Neural Netw Learn Syst 2022;33:3-11. 10.1109/TNNLS.2021.3119071 [DOI] [PubMed] [Google Scholar]
- 24.Shiri I, Sorouri M, Geramifar P, Nazari M, Abdollahi M, Salimi Y, Khosravi B, Askari D, Aghaghazvini L, Hajianfar G, Kasaeian A, Abdollahi H, Arabi H, Rahmim A, Radmard AR, Zaidi H. Machine learning-based prognostic modeling using clinical data and quantitative radiomic features from chest CT images in COVID-19 patients. Comput Biol Med 2021;132:104304. 10.1016/j.compbiomed.2021.104304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferreira JR, Junior, Cardona Cardenas DA, Moreno RA, de Sá Rebelo MF, Krieger JE, Gutierrez MA. Novel Chest Radiographic Biomarkers for COVID-19 Using Radiomic Features Associated with Diagnostics and Outcomes. J Digit Imaging 2021;34:297-307. 10.1007/s10278-021-00421-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rezaeijo SM, Abedi-Firouzjah R, Ghorvei M, Sarnameh S. Screening of COVID-19 based on the extracted radiomics features from chest CT images. J Xray Sci Technol 2021;29:229-43. 10.3233/XST-200831 [DOI] [PubMed] [Google Scholar]
- 27.Bae J, Kapse S, Singh G, Gattu R, Ali S, Shah N, Marshall C, Pierce J, Phatak T, Gupta A, Green J, Madan N, Prasanna P. Predicting Mechanical Ventilation and Mortality in COVID-19 Using Radiomics and Deep Learning on Chest Radiographs: A Multi-Institutional Study. Diagnostics (Basel) 2021;11:1812. 10.3390/diagnostics11101812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouchareb Y, Moradi Khaniabadi P, Al Kindi F, Al Dhuhli H, Shiri I, Zaidi H, Rahmim A. Artificial intelligence-driven assessment of radiological images for COVID-19. Comput Biol Med 2021;136:104665. 10.1016/j.compbiomed.2021.104665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangalagiri J, Sugumar JS, Menon S, Chapman D, Yesha Y, Gangopadhyay A, Yesha Y, Nguyen P. Classification of COVID-19 using Deep Learning and Radiomic Texture Features extracted from CT scans of Patients Lungs. Proc - 2021 IEEE Int Conf Big Data, Big Data 2021. 2021:4387-95. [Google Scholar]
- 30.Hu Z, Yang Z, Lafata KJ, Yin FF, Wang C. A radiomics-boosted deep-learning model for COVID-19 and non-COVID-19 pneumonia classification using chest x-ray images. Med Phys 2022;49:3213-22. 10.1002/mp.15582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. Proc - 30th IEEE Conf Comput Vis Pattern Recognition, CVPR 21-26 July 2017. doi: 10.1109/CVPR.2017.243. 10.1109/CVPR.2017.243 [DOI] [Google Scholar]
- 32.Chattopadhay A, Sarkar A, Howlader P, Balasubramanian VN. Grad-CAM++: Generalized gradient-based visual explanations for deep convolutional networks. Proc - 2018 IEEE Winter Conf Appl Comput Vision, WACV 12-15 March 2018. doi: 10.1109/WACV.2018.00097. 10.1109/WACV.2018.00097 [DOI] [Google Scholar]
- 33.Cohen JP, Morrison P, Dao L. COVID-19 Image Data Collection 2020. Available online: https://doi.org/10.48550/arxiv.2003.11597
- 34.Chung A. Figure1-COVID-chestxray-dataset: Figure 1 COVID-19 Chest X-ray Dataset Initiative 2020. Available online: https://github.com/agchung/Figure1-COVID-chestxray-dataset (accessed April 30,2021).
- 35.Chung A. Actualmed-COVID-chestxray-dataset 2020. Available online: https://github.com/agchung/Actualmed-COVID-chestxray-dataset (accessed April 30,2021).
- 36.Chowdhury MEH, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub Z Bin, Islam KR, Khan MS, Iqbal A, Emadi N Al, Reaz MBI, Islam MT. Can AI Help in Screening Viral and COVID-19 Pneumonia? IEEE Access 2020;8:132665-76.
- 37.Rahman T, Khandakar A, Qiblawey Y, Tahir A, Kiranyaz S, Abul Kashem SB, Islam MT, Al Maadeed S, Zughaier SM, Khan MS, Chowdhury MEH. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Comput Biol Med 2021;132:104319. 10.1016/j.compbiomed.2021.104319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radiological Society of North America, R. S. RSNA pneumonia detection challenge. Available online: https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data(2019) (accessed April 30, 2021).
- 39.Rahman T, Khandakar A, Qiblawey Y, Tahir A, Kiranyaz S, Abul Kashem SB, Islam MT, Al Maadeed S, Zughaier SM, Khan MS, Chowdhury MEH. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Comput Biol Med 2021;132:104319. 10.1016/j.compbiomed.2021.104319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lafraxo S, El Ansari M. CoviNet: Automated COVID-19 detection from X-rays using deep learning techniques. 2020 6th IEEE Congress on Information Science and Technology (CiSt), 2020:489-94. doi: 10.1109/CiSt49399.2021.9357250. 10.1109/CiSt49399.2021.9357250 [DOI] [Google Scholar]
- 41.Selvan R, Dam EB, Rischel S, Sheng K, Nielsen M, Pai A. Lung Segmentation from Chest X-rays using Variational Data Imputation. ArXiv 2020. [Google Scholar]
- 42.Dai J, Qi H, Xiong Y, Li Y, Zhang G, Hu H, Wei Y. Deformable Convolutional Networks. 2017 IEEE International Conference on Computer Vision (ICCV), 2017:764-73. doi: 10.1109/ICCV.2017.89. 10.1109/ICCV.2017.89 [DOI] [Google Scholar]
- 43.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L. ImageNet Large Scale Visual Recognition Challenge. Int J Comput Vis 2015;115:211-52. 10.1007/s11263-015-0816-y [DOI] [Google Scholar]
- 44.Kingma DP, Ba JL. Adam: A Method for Stochastic Optimization. 3rd Int Conf Learn Represent ICLR 2015 - Conf Track Proc 2014. Available online: 10.48550/arxiv.1412.6980.10.48550/arxiv.1412.6980 [Google Scholar]
- 45.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning Deep Features for Discriminative Localization. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2016;2016-December:2921-9.
- 46.van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts HJWL. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-7. 10.1158/0008-5472.CAN-17-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parmar C, Leijenaar RT, Grossmann P, Rios Velazquez E, Bussink J, Rietveld D, Rietbergen MM, Haibe-Kains B, Lambin P, Aerts HJ. Radiomic feature clusters and prognostic signatures specific for Lung and Head & Neck cancer. Sci Rep 2015;5:11044. 10.1038/srep11044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Oikonomou A, Wong A, Haider MA, Khalvati F. Radiomics-based Prognosis Analysis for Non-Small Cell Lung Cancer. Sci Rep 2017;7:46349. 10.1038/srep46349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit-learn: Machine Learning in Python. J Mach Learn Res 2011;12:2825-30. [Google Scholar]
- 50.Jia X, Ren L, Cai J. Clinical implementation of AI technologies will require interpretable AI models. Med Phys 2020;47:1-4. 10.1002/mp.13891 [DOI] [PubMed] [Google Scholar]
- 51.Zunair H, Hamza AB. Synthesis of COVID-19 chest X-rays using unpaired image-to-image translation. Soc Netw Anal Min 2021;11:23. 10.1007/s13278-021-00731-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zunair H, Hamza A, Ben. Synthetic COVID-19 Chest X-ray Dataset for Computer-Aided Diagnosis 2021. Available online: https://arxiv.org/abs/2106.09759
- 53.Ren G, Xiao H, Lam SK, Yang D, Li T, Teng X, Qin J, Cai J. Deep learning-based bone suppression in chest radiographs using CT-derived features: a feasibility study. Quant Imaging Med Surg 2021;11:4807-19. 10.21037/qims-20-1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as