Abstract
Osteoporosis is a chronic skeletal condition characterized by low bone mass and deteriorated microarchitecture of bone tissue and puts tens of millions of people at high risk of fractures. New therapeutic agents like i-bodies, a class of next-generation single-domain antibodies, are needed to overcome some limitations of conventional treatments. An i-body is a human immunoglobulin scaffold with two long binding loops that mimic the shape and position of those found in shark antibodies, the variable new antigen receptors of sharks. Its small size (∼12 kDa) and long binding loops provide access to drug targets, which are considered undruggable by traditional monoclonal antibodies. Here, we have successfully identified a human receptor activator of nuclear factor-κB ligand (RANKL) i-body, ADR3, which demonstrates a high binding affinity to human RANKL (hRANKL) with no adverse effect on the survival or proliferation of bone marrow–derived macrophages. Differential scanning fluorimetry suggested that ADR3 is stable and able to tolerate a wide range of physical environments (including both temperature and pH). In addition, in vitro studies showed a dose-dependent inhibitory effect of ADR3 on osteoclast differentiation, podosome belt formation, and bone resorption activity. Further investigation on the mechanism of action of ADR3 revealed that it can inhibit hRANKL-mediated signaling pathways, supporting the in vitro functional observations. These clues collectively indicate that hRANKL antagonist ADR3 attenuates osteoclast differentiation and bone resorption, with the potential to serve as a novel therapeutic to protect against bone loss.
Keywords: i-body, RANKL, osteoclast, bone resorption, osteoporosis
Abbreviations: α-MEM, minimum essential medium; AP1, activator protein 1; BMMs, bone marrow-derived macrophages; BSA, bovine serum albumin; CDR, complementarity determining-like binding region; DSF, differential scanning fluorimetry; FBS, fetal bovine serum; HO-1, heme oxygenase-1; hRANKL, human RANKL; HRP, horseradish peroxidase; IgG, immunoglobulin G; NCAM, neural cell adhesion molecule; NFATc1, nuclear factor of activated T-cells, cytoplasmic 1; PDB, Protein Data Bank; P/S, Penicillin-Streptomycin; RANK, receptor activator of nuclear factor-κB; RANKL, RANK ligand; ROS, reactive oxygen species; rRANKL, rat RANKL; sdAbs, single-domain antibodies; TNF, tumor necrosis factor; TRAcP, tartrate-resistant acid phosphatase; VNARs, variable new antigen receptors; V-ATPase, Vacuolar-type H+ ATPase
It is reported that the human skeleton is the second largest component of the total body, comprising about 14.84% of total weight (1). It allows locomotion, protects vital organs, stores minerals, and produces blood cells as well as endocrine factors. Bone constantly undergoes life-time remodeling which is a metabolic process of bone breakdown and bone formation (2). Interrupting the balance of this dynamic interaction between bone-resorptive cells, osteoclasts, and bone-forming cells, osteoblasts, leads to skeletal disorders like osteoporosis, osteopetrosis, Paget’s disease, etc (3, 4). Osteoporosis is defined as a chronic skeletal condition characterized by reduced bone mass and deteriorated microarchitecture of bony tissues, resulting in undermined bone quality and increased risk of fractures, especially in the hip, spine, and wrist (5). In Australia, the total cost relating to osteoporosis was $7.4 billion per annum, and it was estimated that in 2022, 6.2 million Australians over 50 years old suffered from osteoporosis or osteopenia, aggravating the health and socioeconomic burden (6). The prevalence of osteoporosis in China shows a similar trend with a total of more than 60 million people aged over 50 (6.46% men and 29.13% women, respectively) estimated to suffer from osteoporosis (7).
Common antiresorptive agents against osteoporosis include bisphosphonates and Denosumab (Prolia). Denosumab is the first U.S. Food and Drug Administration (FDA)–approved humanized monoclonal antibody (Immunoglobulin G (IgG) 2) that antagonizes the receptor activator of nuclear factor-κB (RANK) ligand (RANKL) and inhibits osteoclast differentiation (8). RANKL, a type II transmembrane glycoprotein produced by the mesenchymal lineage, binds to its receptor, RANK, and induces osteoclast differentiation and bone resorption (9). Excessive RANKL causes hyperactive osteoclasts and enhanced osteolysis, commonly observed in osteoporosis, Rheumatoid arthritis, and cancer treatment–induced bone loss, suggesting that it is a druggable target (9, 10, 11). In addition, the RANKL-RANK signal is found to regulate mammary gland maturation, tumorigenesis, and bone metastasis, suggesting that RANKL inhibitors could be beneficial to stop disease progression (12, 13). Denosumab was proven to reduce fracture incidence; however, limitations such as its large molecular weight, difficulty and high cost of manufacture, higher risk of severe infections, accelerated bone loss, and spontaneous fractures following treatment discontinuation were reported, providing a strong need for next-generation biological therapeutic goods (14, 15, 16, 17, 18).
The variable new antigen receptors (VNARs), single-domain antibody-like molecules, are the variable regions of shark antibodies which possess an exquisite stability and a high affinity against specific antigens (19). The structure of VNARs shares a high similarity with the i-set family of immunoglobulin domains (Igs), for example, the immunoglobulin domains of human neural cell adhesion molecule 1 (NCAM1), suggesting that NCAM Ig domain can be potentially refitted as a human-compatible scaffold. Based on this concept, a new class of next-generation antibodies, i-body, was designed by engineering two long binding loops upon a modified Ig domain of NCAM, which mimics the shape of VNARs, allowing for high specificity and strong affinity against a particular disease-causing target (17, 19, 20). There are several advantages of an i-body compared to a conventional antibody: (1) high accessibility to targets, (2) extended paratopes (up to twice the length of typical human antibody complementarity determining regions), (3) high stability, (4) low cost of manufacture, (5) tailored half-life in humans, and (6) low potential immunogenicity (17, 19). Preclinical studies have shown positive results regarding the safety and efficacy of i-body AD-114 against pulmonary fibrosis in the murine bleomycin model (17, 20). ADR3-Im7-FH (ADR3) is a novel i-body with a high affinity to human and murine RANKL. In order to evaluate its curative potential against hRANKL-induced bone loss, we tested the biological functions of ADR3 in our well-established in vitro system.
Results
The construction of i-body library and the identification of i-body ADR3
In general, i-bodies mimic shark antibodies by inserting two extended loops onto a human NCAM immunoglobulin domain 1 scaffold, obtaining a stable recombinant protein incorporating two complementarity determining–like binding regions (CDRs) in the equivalent positions as found in the VNARs. An i-body library displayed on phage was previously constructed and used to select binders for the G protein–coupled receptor C-X-C chemokine receptor type 4 (19). This library was used to screen against immobilized recombinant human RANKL (hRANKL), yielding a number of i-body binders that had an affinity for hRANKL (Fig. 1A). Potential candidates were then expressed and purified into recombinant ADRs for further investigation. Based on phage ELISA and surface plasmon resonance results, we identified the best candidate, ADR3, which exhibits the highest binding affinity to hRANKL (KD = 13 nM) (Fig. 1B). ADR3 expressed in Escherichia coli was shown to have an approximate molecular weight of 21 kDa (including the Im7 Flag and a His tag) and to be predominantly monomeric (Fig. S1A). Flow cytometry analysis indicated a high specificity of ADR3 with hRANKL, as ADR3 only generates a fluorescence signal on hRANKL-expressing osteoblast-like cells versus control cells (Fig. S1, B and C). Consistently, our binding affinity test showed no interaction between ADR3 and irrelevant proteins, such as apical membrane antigen 1 and bovine serum albumin (BSA) (data not shown). The crystal structure of 21H5, Ig domain of NCAM (Protein Data Bank (PDB): 5AEA), is shown in Figure 1C-a, along with two color-indicated regions (CDR1 and CDR3) where the long binding loops were inserted, enabling a tight binding to disease-causing targets. We further provided a best-fit computational model (Fig. S1D) for the interaction between the predicted ADR3 (Fig. S1E) and hRANKL (PDB: 5BNQ) on the basis of thermodynamics and homology modeling, in which the model clearly shows that CDR1 and CDR3 in ADR3 are intensively involved in the interaction (Fig. 1C-b). Further characterization of the predicted model showed that the close contact interface between hRANKL and the CDR1 and CDR3 loops of ADR3 involves the GH loop of hRANKL (P301 to Y307), equivalent to mouse RANKL residues P300 to Y306, where this region was precisely indicated to play an important role in RANKL–RANK interaction based on the crystal structure (PDB ID code: 3NZY) (Fig. S1F) (21).
Figure 1.
The affinity-based selection of hRANKL i-body.A, the brief illustration of i-body selection processes. B, determination of the affinity of the ADR3–hRANKL interaction by SPR assay. C, a, the crystal structure of 21H5 (human NCAM1, Ig1 domain, and PDB: 5AEA). Red and purple colors highlight the inserted binding loops (CDR1 and CDR3, respectively). b, the predicted ADR3 structure in silico docked to hRANKL (PDB: 5BNQ). In the best-fit computational model, red and purple colors indicate the CDR1 and CDR3 binding loops, which are shown to be involved in ADR3–hRANKL interaction. CDR, complementarity determining-like binding region; hRANKL, human RANKL; NCAM1, neural cell adhesion molecule 1; PDB, Protein Data Bank; RANKL, RANK ligand; SPR, surface plasmon resonance.
The thermal stability of i-body ADR3
We used a thermal shift assay (differential scanning fluorimetry, DSF) to evaluate the thermal stability of ADR3. As shown in Figure 2, A and B, DSF results indicated that ADR3 exhibits good thermal stability across different temperatures and pHs. ADR3 is more stable from pH 7.0 to 9.0 (Fig. 2C). Lower stability from pH 4.0 to 5.0 may be due to the fact that this range is near the theoretical pI of the molecule (4.96, calculated based on sequence using ProtParam (https://web.expasy.org/protparam/)) (22). At this pI, the net charge of the protein is zero, resulting in aggregation and lower stability of the protein. Given the physiological temperature and pH range in human subjects, it is predicted that ADR3 will be sufficiently stable as a potential therapeutic agent in the human body.
Figure 2.
Thermal stability of ADR3.A, differential scanning fluorimetry (DSF) curves showing the thermal stability of ADR3 in a wide range of pHs. Quadruplicates were recorded for each group and are presented as a line chart. B, derivative curves displaying a thermal shift at different pHs, suggesting that ADR3 favors a neutral or more alkaline environment over a more acidic environment. C, heatmap displaying the correlation between pH and the thermal stability of ADR3, where white color represents a lower Tm and red color indicates a higher Tm.
i-body ADR3 inhibits osteoclast formation in both RAW264.7 cells and BMMs
Cell proliferation assay (MTS) showed no cytotoxicity of ADR3 in bone marrow–derived macrophages (BMMs) within a range of concentrations (0.5–2 μM) (Fig. S2A), and ADR3 effectively inhibits hRANKL-induced osteoclastogenesis in RAW264.7 cells (IC50 = 4 nM) (Fig. S2B). Consistently, ADR3 dose-dependently suppresses osteoclast differentiation and podosome belt formation in freshly isolated BMMs (Figs. 3, A–C and S2C), in which the inhibition effectiveness (IC50) reaches 0.17 μM. ADR3 also evidently inhibits rat RANKL (rRANKL)-stimulated osteoclastogenesis in BMMs (Fig. S2D). The predicted region of hRANKL that binds to CDR1 and CDR3 loops of ADR3 is conserved among humans, mouses, and rats; thus, it is not surprising to observe the interaction between ADR3 and rRANKL. Nuclear factor of activated T-cells, cytoplasmic 1 (NFATc1) is a master transcriptional factor induced by RANKL for the terminal differentiation of osteoclasts, so we next examined whether the activity of NFATc1 could also be inhibited by ADR3. A robust reduction in the luciferase activity of RAW264.7 cells stably transfected with NFATc1 luciferase reporter gene was observed in ADR3 treatment groups in comparison to the positive control, suggesting that ADR3 interrupts the activation of NFATc1 by hRANKL in a dose-dependent manner (Fig. 3D).
Figure 3.
i-body ADR3 inhibitshRANKL-induced osteoclastogenesis. TRAcP staining (A) and podosome belt staining (B) of noninduced or hRANKL-induced BMMs treated with or without 1 μM i-body ctrl, 0.5 μM ADR3, and 1 μM ADR3, respectively. The scale bar represents 200 μm. C, quantification of TRAcP-positive cells in each group (n = 4). All data are presented as the mean ± SD. ∗∗∗∗p < 0.0001 compared to the positive control. ####p < 0.0001 compared between two groups of ADR3. D, RAW264.7 cells stably transfected with an NFATc1 luciferase reporter construct were treated with hRANKL alone or with 1 μM i-body control, 0.5 μM and 1 μM ADR3, respectively. The luciferase activity induced by hRANKL treatment is dose-dependently inhibited by ADR3 (n = 4). The bar graph is constructed as the mean ± SD. ∗∗∗∗p < 0.0001 compared to the positive control. ####p < 0.0001 compared between two doses of ADR3. hRANKL, human RANKL; NFATc1, nuclear factor of activated T-cells, cytoplasmic 1; RANKL, RANK ligand; TRAcP, tartrate-resistant acid phosphatase.
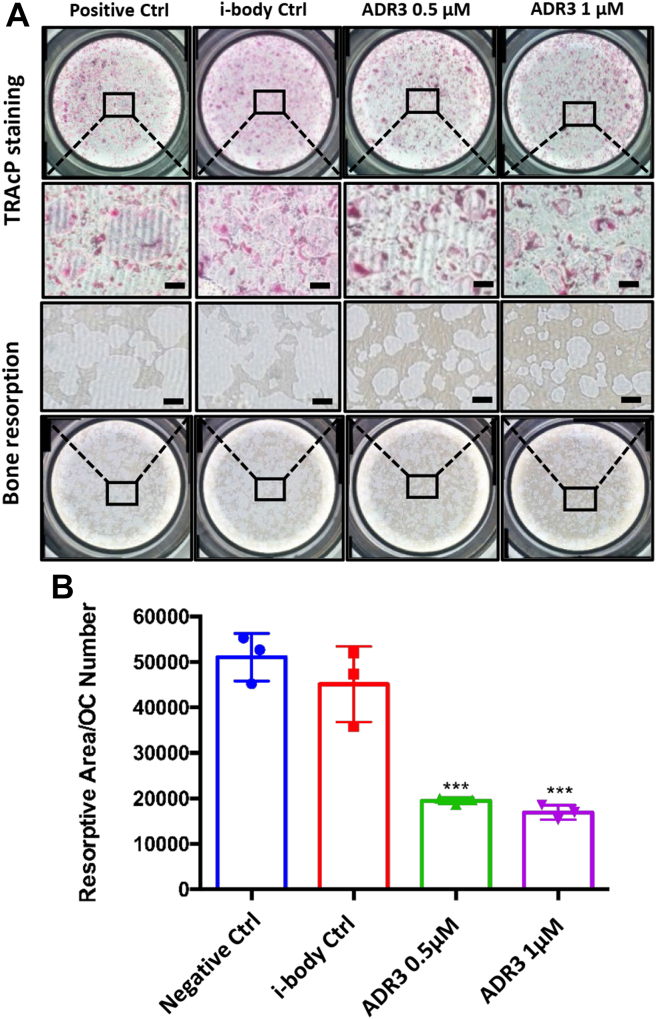
i-body ADR3 suppresses osteoclast bone resorption
RANKL is involved not only in the formation of prefusion osteoclasts but also in the maintenance of mature osteoclast survival and bone-resorbing activity (23). We therefore examined the in vitro effect of ADR3 on bone resorption via osteo assay plates. Compared to the no-treatment group, bone resorptive area is significantly reduced after 48-h treatment with ADR3 in two doses (0.5 μM and 1 μM), along with a subtle change in the number of tartrate-resistant acid phosphatase (TRAcP)-positive cells (Fig. 4A). The ratio of total resorptive area to osteoclast number showed an evident regress of resorbing capacity for each osteoclast (Fig. 4B), indicating that ADR3 hampers osteoclasts’ resorptive functionality.
Figure 4.
ADR3 suppresseshRANKL-induced bone resorption.A, fresh BMMs were induced by 50 ng/ml hRANKL to form TRAcP-positive osteoclasts on osteo assay surface plates. Upon the formation of mature osteoclasts, cells were treated with i-body control or ADR3. Representative resorption pits (n = 3) were visualized in parallel with TRAcP staining (n = 3). The scale bar represents 200 μm. B, quantification of resorbed hydroxyapatite area per cell in each group. The bar graph is presented by the mean ± SD. ∗∗∗p < 0.001 compared to the negative control. BMMs, bone marrow–derived macrophages; hRANKL, human RANKL; RANKL, RANK ligand; TRAcP, tartrate-resistant acid phosphatase.
i-body ADR3 decreases the expression of osteoclast markers induced by hRANKL both at the gene and protein levels
During osteoclastogenesis, the formation of RANKL–RANK complexes induces c-Fos (proto-oncogene c-Fos) expression, initiating the induction of NFATc1, which promotes the synthesis of fusion proteins, such as vacuolar (H+) ATPase V0 domain d2 isoform (encoded by Atp6v0d2 gene), and lysosomal enzymes, including TRAcP type 5 (encoded by Acp5 gene), cathepsin K (encoded by Ctsk gene), and matrix metalloproteinase-9 (encoded by Mmp9 gene) that help to dissolve bone matrix (24). Quantitative PCR showed a considerably lower expression of calcitonin gene-related peptide type 1 receptor (Calcrl), c-Fos, Nfatc1, Atp6v0d2, Acp5, Ctsk, and Mmp9 in ADR3-treated groups than the control group (Fig. 5). This is supported by Western blotting results which showed that protein levels of NFATc1, c-Fos, Atp6v0d2, and Ctsk also decrease in groups treated by ADR3 (Fig. 6A), consolidating the anticatabolic effect observed in vitro. Interestingly, RANKL was reported to activate reactive oxygen species (ROS) during osteoclast differentiation (25). Heme oxygenase-1 (HO-1), an antioxidant enzyme, was suggested to negatively regulate osteoclast formation by alleviating the elevated ROS activity (26, 27). Similarly, another antioxidant enzyme, Catalase, was also reported to inhibit ROS production (25). Therefore, we measured the levels of HO-1 and Catalase in hRANKL-stimulated BMMs after ADR3 treatment and found that ADR3 robustly increases HO-1 expression (Fig. 6B). The same trend but to a lesser effect was observed in the levels of Catalase (Fig. 6B). Collectively, i-body ADR3 was shown to effectively suppress the expression of osteoclast makers, bone-resorptive enzymes, transcriptional factors, and ROS activity both at the gene and protein levels.
Figure 5.
Osteoclastic markers (Calcrl, c-Fos, Nfatc1, Atp6v0d2, Acp5, Ctsk, and Mmp9) are downregulated by ADR3 treatment in a dose-dependent manner. The bar graphs are presented as the mean ± SD (n = 3). ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 compared to the positive control. #p < 0.05, ##p < 0.01, and ####p < 0.0001 compared between the two doses of ADR3. NFATc1, nuclear factor of activated T-cells, cytoplasmic 1.
Figure 6.
hRANKL-mediated downstream signaling is disrupted dose-dependently by ADR3.A, left panel, representative Western blotting images showing the effect of ADR3 on hRANKL-induced NFATc1, c-Fos, D2, and Ctsk at the protein level. Right panel, the ratios of the densities of NFATc1, c-Fos, D2, and Ctsk bands relative to β-actin bands were generated through ImageJ. B, left panel, representative images displaying the protective effect of ADR3 on hRANKL-regulated antioxidant enzymes HO-1 and Catalase. Right panel, protein expression levels were normalized to β-actin and quantified through measuring the gray values in ImageJ. All data are presented by the mean ± SD (n = 3). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 compared to the positive control. #p < 0.05 compared between two doses of ADR3. hRANKL, human RANKL; HO-1, heme oxygenase-1; NFATc1, nuclear factor of activated T-cells, cytoplasmic 1; RANKL, RANK ligand.
i-body ADR3 treatment dose-dependently impairs hRANKL-induced Ca2+ oscillations
It is known that RANKL-triggered Ca2+ oscillation activates CAM-dependent enzymes which facilitate the auto-amplification and translocation of NFATc1 into the nucleus (28, 29). Thus, we labeled the calcium with the fluorescence indicator Fluo-4 AM and investigated the effect of ADR3 on Ca2+ transport during hRANKL-mediated osteoclastogenesis. As shown in Figure 7A, 24-h treatment of hRANKL significantly increases Ca2+ oscillation in BMMs, while 0.5 μM and 1 μM ADR3 treatment clearly inhibits the oscillation signal, flattening the fluctuation of the fluorescence signal. The quantification of maximum fluorescence change also showed a dose-dependent reduction in peak to peak values in the ADR3 treatment group, along with a decrease in the percentage of oscillations of BMMs, suggesting that both the Ca2+ flux intensity of single cell and the percentage of oscillated cells were reduced (Fig. 7B).
Figure 7.
ADR3 interruptsintracellular calcium oscillation.A, Ca2+ oscillation induced by hRANKL is inhibited by ADR3 in a dose-dependent manner. B, the intensity of fluorescence and the percentage of oscillated cells in each group were quantified into bar charts presented as the mean ± SD (n = 3). ∗∗p < 0.01 and ∗∗∗p < 0.001 compared to the positive control. #p < 0.05 compared between the two doses of ADR3. hRANKL, human RANKL.
Discussion
The health and socioeconomic impact of osteoporosis is not limited to Australia but rather occurs globally. Annually, 1.5 million fractures in the US are attributed to osteoporosis with most occurrences amongst postmenopausal women. It was also reported that more than 40 million women have low bone mineral density (BMD) and a 50% lifetime risk of any kind of osteoporotic fractures (30). As a key component of bone metabolism, osteoclasts coordinate with other bone cells (e.g., osteoblasts and osteocytes) to maintain the balance of bone turnover, representing a therapeutic target which is not limited to osteoporosis, but also for other skeletal diseases like periprosthetic osteolysis, Rheumatoid arthritis, bone tumors, and so on (31). Anti-RANKL monoclonal antibody, Denosumab, has been shown to effectively suppress osteoclast formation and reduce the chance of fractures but with inevitable limitations and adverse side effects. The objectives of this study are to (1) evaluate the concept and technology platform of i-bodies, modified human proteins, by library screening against disease-causing targets and (2) investigate the potential of an hRANKL i-body, ADR3, in preventing excessive bone resorption.
Single-domain antibodies (sdAbs) with alternative scaffolds belong to next-generation antibodies which can offer some advantages over conventional monoclonal antibodies, such as larger epitopes, higher accessibility, lower cost of production, and increased robustness (19, 32). Immunoglobulin new antigen receptors, a sdAbs-like molecule derived from sharks, were reported having high stability and are structurally similar to the i-set family members including NCAM (19, 33). Through the engineering of two extended loops with random sequences to a modified NCAM protein, we constructed an i-body library which contains over 20 billion candidates ready for rapid screening, yielding i-bodies that specifically interact with designated targets. In recent years, disease targets that were refractory to conventional monoclonal antibodies are being overcome with the support of well-established screening platforms that can generate sdAbs designed based on alternative scaffolds. In 2009, a scaffold-based drug, Kalbitor (Dyax), succeeded in reaching the market, and at least 15 other candidates are currently being investigated (34). The design of i-bodies is intended to achieve a high level of stability, effectiveness, and accessibility with low immunogenicity, aiming to provide a new solution for a wide range of diseases (19).
In this article, we briefly described the construction of an i-body ADR library and the selection of i-body ADR3 which resulted in high binding affinity and specificity to hRANKL. To examine the specificity of ADR3, besides testing with some irrelevant proteins, we also compared the binding affinity of ADR3 between MG63 control cells, a human osteoblast-like cell line that presents an undetectable level of hRANKL, and MG63 hRANKL cells which overexpress hRANKL using flow cytometry. Our results indicated that ADR3 binds only to MG63 cells overexpressing hRANKL. A protein structure modeling software, ModWeb (https://modbase.compbio.ucsf.edu/modweb/), was used to predict the structure of ADR3 based on the crystal structure of human protein scaffold 21H5 (human NCAM1, Ig1 domain, PDB: 5AEA). The predicted ADR3 structure was then docked to hRANKL (PDB: 5BNQ) via the protein-protein docking program ClusPro 2.0 (https://cluspro.bu.edu/login.php), in which the best-fit model clearly showed that the two extended loops (CDR1 and CDR3) are extensively involved in the interaction between ADR3 and hRANKL. Further study of the predicted model showed that the GH loop of hRANKL (P301 to Y307) interacts with the CDR1 and CDR3 loops of ADR3, consistent with the findings of the crystal structure of mouse RANKL–RANK complex (21). It is known that a healthy body temperature ranges from 36.1 to 37.2 °C with a pH value between 7.35 and 7.45. To work effectively in humans, therapeutic goods must be stable while traveling through the body. Hence, we exposed ADR3 to a diverse environment to test its stability via a DSF assay. Sypro Orange dye was used in this assay to monitor the thermal denaturation of ADR3. As proteins unfold, their hydrophobic core residues are exposed to the solution, where the dye binds to those residues and emits a fluorescent signal (35). We found that ADR3 is well-tolerated across a wide range of pHs and temperatures, satisfying the need for storage, shipment, and delivery in the human body. In vitro cell proliferation assays (MTS) showed no cytotoxicity from ADR3 that affects BMMs’ viability. To study the potential of ADR3 on the inhibition of excessive bone loss, we first examined whether ADR3 could suppress osteoclast differentiation. An in vitro osteoclastogenesis assay showed that TRAcP-positive osteoclast-like cells are significantly inhibited by ADR3 treatment in a dose-dependent manner. WP9QY (W9) is a small peptide structurally similar to the cysteine-rich domain of tumor necrosis factor (TNF) receptor type I and can inhibit the activity of TNFα (36). This peptide was also found to suppress hRANKL-induced osteoclastogenesis and is being investigated as an alternative for Denosumab due to its innate merits over conventional monoclonal antibodies (37). According to the previous study, 0.5 μM ADR3 has achieved an inhibitory effect of osteoclastogenesis equivalent to 100 μM W9, suggesting that ADR3 is approximately 200 times more effective than W9 (38). This in vitro result was repeated and consistent in RAW264.7 cells and primary BMMs. Next, we measured the activation of transcriptional factor NFATc1 in RAW264.7 cells stably transfected with a luciferase reporter gene and found that the NFATc1 activity is inhibited in a dose-dependent manner by ADR3 treatment. Consistently, we observed that the gene and protein levels of c-Fos and NFATc1 are also downregulated in the ADR3-treated groups. Osteoclast formation and bone resorption require multistep biological processes initiated by the formation of a RANKL–RANK complex. The complex induces a series of signaling cascades, starting with the recruitment of TNF receptor-associated factor 6. Recruited TNF receptor-associated factor 6 activates a complex comprised of IκB kinase (IKKα, IKKβ, and IKKγ (NEMO)) which phosphorylates and promotes the degradation of IκB, releasing NF-κB, and facilitating its translocation into the nucleus (39, 40). Intranuclear NF-κB activates the transcription of target genes such as c-Fos and NFATc1 through binding to their promoter motifs (40). Intracellular calcium oscillation induced by RANKL is another critical process to regulate NFATc1 expression, in which the cytosolic Ca2+ binds to and activates calmodulin, leading to autoamplification and translocation of NFATc1 into the nucleus. Using live-cell imaging techniques with the calcium indicator Fluo-4 AM to detect the Ca2+ flux in BMMs, we observed a significant suppression of the fluorescence signal and oscillated cell number in ADR3-treated groups, further consolidating our previous results. NFATc1 was reported to cooperate with c-Fos, one critical component of the activator protein 1 (AP1), forming a complex essential for the activation of osteoclast marker genes (29). Studies showed that genes like Calcrl, Acp5, Ctsk, Mmmp9, and Atp6v0d2 which are being transcribed during osteoclast terminal differentiation contain multiple binding sites of NFAT and AP1 in their promoter regions, suggesting that NFATc1–c-Fos complex is the key for the transcription of these genes (41, 42, 43, 44, 45). Osteoclasts are bone resorbing cells which require an acidified microenvironment with a series of enzymes involved to perform bone resorption. Vacuolar-type H+ ATPase (V-ATPase) subunit d2 is an essential component of a lysosomal macromolecular complex proton pump V-ATPase which facilitates the establishment of an acidic environment for mineral dissolution and the support of protein-degradation enzymes such as TRAcP, cathepsin K, and matrix metallopeptidase 9 (46). The loss of Atp6v0d2 leads to high bone mass, indicating the role of this gene in osteoclast function (47). TRAcP (encoded by Acp5 gene), cathepsin K (encoded by Ctsk gene), and matrix metallopeptidase 9 (encoded by Mmp9 gene) are lysosomal enzymes required to degrade proteins, for example, collagen, in ECM, and the loss of each of them results in mild osteopetrosis, high bone mass, or reduced bone remodeling (47). As mentioned, the quantitative PCR result demonstrated that hRANKL-induced osteoclastic markers Calcrl, Atp6v0d2, Acp5, Ctsk, and Mmmp9 are inhibited in a dose-dependent manner by ADR3 treatment, as a rational consequence of interrupted expression and activation of NFATc1 and c-Fos. This was supported by Western blotting results which showed a diminished expression of NFATc1, c-Fos, and bone-digesting enzymes at the protein level. During bone resorption, to deliver the cocktail of matrix-dissolving enzymes and create a favorable condition for osteoclast activity, the membrane of osteoclasts polarizes to form a sealing zone called “ruffled border”. Using rhodamine–phalloidin, a F-actin fluorescence probe, we were able to visualize the formation of a podosome belt (ruffled border). As shown, the sealing zone is well-established in the hRANKL-treated group but not in the groups treated with ADR3, suggesting that ADR3 compromises the formation of the F-actin ring and leads to reduced capacity in bone resorption which was later confirmed in vitro. ROS are free radicals (also called oxygen radicals) and involved in many biochemical processes. At high concentrations, ROS can lead to oxidative stress which is considered deleterious to humans, while small amount of ROS may play a role as a secondary messenger in a variety of different signaling cascades (25). Notably, RANKL can activate ROS in BMMs to regulate osteoclast differentiation and bone resorption partially through reducing endogenous antioxidants, HO-1 and Catalase (25, 48). These two antioxidant enzymes were reported to attenuate a redox imbalance and maintain bone mass (25, 26). We found that hRANKL-dependent suppression of HO-1 and Catalase is fully or partially released by ADR3 treatment, further supporting the anticatabolic effect of ADR3 in osteoclasts. Taken together, ADR3 exhibits a strong inhibitory effect on osteoclast formation and bone resorption.
Up to date, researchers are still improving the pharmaceutical properties of monoclonal antibodies in fields like antigen-binding affinity, antigen-binding specificity, stability, pharmacokinetics, and pharmacodynamics. Some diseases are much more challenging to address, due to their undruggable targets, such as G protein–coupled receptors and iron channels. Modern medicine commonly develops drug targets via small molecules which are often not specific or effective enough, resulting in undesirable side effects. The development of an i-body library and a screening system offers a unique platform to modulate disease progress. i-body is less than 1/10 of the size of traditional monoclonal antibodies and has long binding loops, twice the length of human antibodies, allowing for more accessibility and specificity to drug targets.
To summarize, we designed and established a system to produce i-bodies and evaluate the inhibitory effect of ADR3 on osteoclast differentiation and bone resorption. Although the IC50 of ADR3 to hRANKL is not superior to Denosumab, more potent candidates will be achieved through structural modifications. The effect of ADR3 could be further explored on cancer-related conditions which require a high efficiency in tissue-penetrating delivery. Given the improved accessibility, stability, specificity, affordability, and low or equivalent immunogenicity of i-bodies, they showed long-term advantages and utilities over conventional monoclonal antibodies.
Experimental procedures
Materials and reagents
RAW264.7 cells were from the American Type Culture Collection. Minimum essential medium (α-MEM), L-glutamine, and Penicillin-Streptomycin (P/S) were from the media laboratory at Harry Perkins Institute of Medical Research. Fetal bovine serum (FBS, 16000044) and hRANKL recombinant protein (PHP0034) are from Gibco. Goat anti-mouse (ab6789) IgG H&L (horseradish peroxidase [HRP]) and Goat anti-rabbit (ab6721) IgG H&L (HRP) were purchased from Abcam. Antibodies to NFATc1 (sc-7294), V-ATPase subunit d 2 (Atp6v0d2) (sc-517031), cathepsin k (sc-48353), and β-actin (sc-47778) were obtained from Santa Cruz. Antibody c-Fos (CST2250s), HO-1 (D60G11), and Catalase (D5N7V) were purchased from Cell Signalling Technology. DYKDDDDK Epitope Tag Alexa Fluor 488–conjugated antibody was from R&D systems (IC8529G). Glutaraldehyde solution (25%) was purchased from Thermo Fisher Scientific. CellTiter 96 Aqueous One Solution Cell Proliferation Assay (MTS) kit (G3580), luciferase assay system (E1501), and the reverse transcription system were brought from Promega. Rhodamine-phalloidin, Fluo-4, AM (F14201), Purelink RNA mini kit (12183018A), Trizol (15596026), PowerUp SYBR Green Master Mix (A25918), centrifuge tubes, and flasks were brought from Thermo Fisher Scientific. Western Lightning Ultra (NEL112001EA) was purchased from PerkinElmer. SYPRO Orange Protein Gel Stain (S5692), cell culture plates, collagen-coated plates, osteo assay surface multiple well plates (CLS3987-4EA), primers, transwell polycarbonate membrane cell culture 8.0 μm inserts (CLS3422), and all other chemicals were purchased from Merck & Co, Inc.
i-body engineering
The i-body library, generated by AdAlta Limited, was used to generate binders against hRANKL recombinant protein. This library, derived from NCAM1, incorporates two patented binding regions (International Patent AU2005/000789; WO/2005/118629) and was cloned in-frame with the gene III of the bacteriophage M13KO7 into the pHENH6 vector. This phagemid was transformed into TG-1; E. coli was used to pan against hRANKL, according to the methods of Griffiths et al. (2016) (19). Briefly, at the beginning of each panning round, the library was amplified by adding 1 ml of phage library to 10 ml of 2YT medium and incubated for 1 h at 37 °C with shaking. The culture was then inoculated into 2YT (200 ml) containing ampicillin (100 μg/ml) and 1% w/v glucose. Culture was subsequently incubated in a shaking machine at 37 °C until the A600 reading reached to 0.4 to 0.6. Thereafter, M13KO7 helper phage particles (New England Biolabs) were added at a multiplicity of infection of 20:1 (phage to bacteria) based on the assumption that an A600 nm of 1 is equivalent to 8 × 108 E. coli cells/ml. The culture was incubated with no shaking for 1 h at 37 °C, followed by a centrifugation at 8000g to pellet the cells. The pellet was then resuspended into 200 ml of 2YT containing ampicillin (100 μg/ml) and kanamycin (70 μg/ml) (Sigma Aldrich) before incubated overnight at 30 °C, with shaking. Next day, supernatant was centrifuged for 10 min twice at 10,000g and phage particles were precipitated 100-fold by PEG/NaCl precipitation according to the method of Sambrook et al., (1989) (49) and resuspended in 2 ml of PBS.
Phage affinity panning
The phage-displayed i-body library prepared by AdAlta as described previously (19) was used to pan on immobilized carrier-free hRANKL (R&D Systems). Phage displaying the i-bodies was incubated with hRANKL absorbed to NI-NTA HisSorb plates via the Histidine tag on the hRANKL. The panning was carried out essentially as described (19) using phage from the final rounds of panning and demonstrated that there were clones specific for hRANKL (totally 5 rounds of panning were conducted). Seventeen i-bodies were expressed and characterized for binding to hRANKL and not to several irrelevant proteins. ADR3 was chosen as the clone with the highest specificity and affinity for hRANKL.
Phage ELISAs
To prepare loading samples, single-clone phage particles were eluted from panning rounds and precipitated into 1 ml resuspension buffer. For the ELISA assay, 96-well plates were coated with hRANKL (0.1 μg/well), irrelevant protein control (AMA-1; 1 μg/ml) or PBS. Diluted phage particles (≥1:10 in 5% milk powder in PBS) were added to precoated ELISA plates and incubated for 60 min followed by five-time washes to remove unbound phage particles. Bound phage particles were then visualized with an anti-M13-HRP antibody and the substrate 3,3′,5,5′-tetramethylbenzidine.
Surface plasmon resonance assay
A kinetic-binding assay of ADR3 with immobilized hRANKL was carried out by a BIAcore T200 instrument. Biotinylated hRANKL protein was diluted in a running buffer (1× HBS/BSA) and immobilized onto streptavidin-containing channels. A blank reference channel containing no hRANKL was used to control for nonspecific binding. To analyze binding kinetics, serial dilutions of candidates from ADR library were injected over the bound hRANKL, in which the association and dissociation phases were recorded for 60 and 600 s with the blank channel used for referencing. All bound i-bodies were dissociated within 600 s; therefore, no regeneration of the hRANKL surface was needed during this experiment.
Cell flow cytometry analysis
MG63 plvx-CTRL (low hRANKL expression cell line) and MG63 plvx-hRANKL cells (high hRANKL expression cell line) were gently washed by PBS containing 0.2% w/v BSA, followed by a 1-h incubation with i-body 21H5 (control) or ADR3 before being washed with 0.2% w/v BSA-PBS to remove excessive primary antibodies. Cells were then incubated with DYKDDDDK Epitope Tag Alexa Fluor 488–conjugated Antibody for another hour before being washed with 0.2% w/v BSA-PBS to reduce background. Stained cells were quantified by BD FACSCanto II flow cytometry, and the result was analyzed using the flowjo software (https://www.flowjo.com/solutions/flowjo/downloads).
Homology modeling, protein-protein docking, and the calculation of theoretical pI
ADR3 structure modeling was done using the Modweb server (Version: r265) in the default setting using the ADR3 sequence and the crystal structure of 21H5 (PDB: 5AEA). The predicted ADR3 structure was then docked to hRANKL (PDB: 5BNQ) using the protein-protein docking program ClusPro 2.0, mimicking the interaction between ADR3 and hRANKL (50, 51, 52). Balanced coefficient weights were generated for the top ten docking models, showing the cluster scores for evaluation. Coefficient weights: E = 0.40Erep + −0.40Eatt + 600Eelec + 1.00EDARS. Acquired in silico structures were visualized via the PyMOL Molecular Graphics System (Version: 2.2.0, Schrödinger, LLC). The ProtParam tool (ExPASy) was used to calculate the theoretical pI of the designated molecules (22). PyMOL (Version: 2.5.3, https://pymol.org/2/) was used to visualize protein structures and study the close contact interfaces.
Thermal shift assay (DSF)
Thermal shift assays were carried out in a LightCycler 480 real-time PCR system (Roche Life Science) using a 384-well PCR plate. The excitation and emission filters were set at 483 nm and 640 nm, respectively. The protocol for the assay was adapted from Niesen et al. (53). The protein was equilibrated in PBS buffer which had been adjusted to pH 4.0 to 9.0 in intervals of 1.0 using HCl or NaOH. The assay used 20 μM protein and 5x Sypro Orange per well with a total volume of 10 μl. The PCR plate was sealed with an optical seal and centrifuged at 1000g for 2 min after the protein and dye were added. The assays were carried out in quadruplicate. A temperature gradient of 20 to 95 °C with a ramp rate of 0.03 °C per second and 20 acquisitions per °C were used for the assay. The data was analyzed using the “Tm calling” feature of the LightCycler 480 software (https://diagnostics.roche.com/global/en/products/instruments/lightcycler-480-ins-445.html) which plots the temperature against the first derivative of fluorescence intensity over temperature. The unfolding temperature (Tm) of the molecule is indicated by the maximum value of the first derivative of fluorescence intensity over temperature.
Animal cell isolation
BMMs were flushed out from the long bones (femur and tibia) of 12-week C57BL/6J mice using the standard method under the approval of the University of Western Australia Animal Ethics Committee (RA/3/100/1601) and then maintained in α-MEM containing 10% v/v FBS, 1% P/S v/v, and M-CSF (complete α-MEM). To remove undesired fibroblasts, nonadherent cells were transferred to new flasks 6 h after the incubation.
MTS assay
Fresh BMMs were seeded in 96-well plates at the density of 6 × 103 cells/well for overnight incubation. Varying doses of ADR3 i-body were added to BMMs for 48-h incubation. Fifty microliters of MTS/phenazine methosulfate mixture were then added into each well, followed by a 2-h incubation. The absorbance was measured by an ELISA plate reader (BMG LABTECH GmbH).
In vitro osteoclastogenesis assay
RAW264.7 cells and fresh BMMs were maintained in α-MEM (10% v/v FBS and P/S) and complete α-MEM, respectively, under the culture condition of 37 °C with 5% CO2. For functional assay, RAW264.7 cells and BMMs were seeded to a 96-well plate at the density of 6 × 103 cells/well and induced by 50 ng/ml hRANKL or rRANKL for 5 days with or without i-bodies. The media was replaced every 2 days until day 5, when mature osteoclasts were formed in the positive control group. Cells were then fixed for 10 min by 2.5% v/v glutaraldehyde at room temperature (RT) and stained for TRAcP. TRAcP-positive cells were imaged by a Nikon microscope (Nikon Corporation) and quantified via imageJ (NIH). IC50 is calculated via an online calculation tool: https://www.aatbio.com/tools/ic50-calculator.
Podosome belt staining
BMMs were seeded in a 24-well plate with coverslips (13-mm) and induced by 50 ng/ml hRANKL with or without i-bodies, until the emergence of multinucleated osteoclasts. Cells were fixed in 4% w/v paraformaldehyde for 10 min at RT and permeabilized by 0.1% v/v Triton X-100 for 5 min before 1-h incubation with 3% w/v BSA PBS for blocking. After washing with 0.2% w/v BSA-PBS three times, cells were incubated with rhodamine–phalloidin (1:300 diluted in 0.2% w/v BSA-PBS) for 2 h at RT. Cells were then gently washed four times in 0.2% w/v BSA-PBS and PBS in sequence before staining with 4′,6-diamidino-2-phenylindole (1:10,000 diluted in PBS) for 5 min. After washing with PBS, stained cells were visualized via a Nikon A1Si Confocal Microscope (Nikon Corporation).
Luciferase reporter assay
To measure the activity of NFATc1, 5 × 104 RAW264.7 cells stably transfected with NFATc1 luciferase reporter gene described in (54) were seeded into 48-well plates overnight. Cells were then induced by 50 ng/ml hRANKL treated with or without i-bodies for 24 h. Luciferase activity was detected using the Promega Luciferase Assay System according to the manufacturer’s instructions.
In vitro hydroxyapatite resorption assay
BMMs were cultured on 6-well collagen-coated plates in the presence of 50 ng/ml hRANKL for 5 days. Upon the formation of a small number of osteoclasts, cells were trypsinized and transferred to 96-well osteo assay plates for a 2-day culture in the presence of 50 ng/ml hRANKL with or without i-bodies. At day 8, half the wells of each group were fixed in 2.5% v/v glutaraldehyde for TRAcP staining, while the other half were bleached for the visualization of resorptive pits. Images were captured by a Nikon microscope (Nikon Corporation) and quantified through ImageJ (NIH).
RNA extraction and cDNA synthesis
BMMs were cultured in 6-well plates with complete α-MEM at a density of 1 × 105 cells/well. Cells were treated with 50 ng/ml hRANKL with or without i-bodies for 5 days until osteoclasts were formed in positive control. Cells were then lysed for total RNA extraction using Trizol and Purelink RNA mini kit in accordance with the manufacturer’s protocol. For cDNA synthesis, real-time PCR was performed using Promega reverse transcription system.
Real-time PCR
SYBR Green PCR Master Mix was used for real-time PCR (rtPCR). The thermal cycler was programmed to 94 °C for 5 min, 30 cycles of 94 °C (40 s), 60 °C (40 s), and 72 °C (40 s), along with an elongation step for 5 min at 72 °C. rtPCR was performed using primers as described: calcitonin gene-related peptide type 1 receptor (Calcrl; Forward: 5′-TGGTTGAGGTTGTGCCCA-3′; Reverse: 5′-CTCGTGGGTTTGCCTCATC-3′), c-Fos (Forward: 5′-GCGAGCAACTGAGAAGAC-3′; Reverse: 5′-TTGAAACCCGAGAACATC-3′), Nfatc1 (Forward: 5′-CAACGCCCTGACCACCGATAG-3′; Reverse: 5′-GGCTGCCTTCCGTCTCATAGT-3′), ATPase, H+ transporting, lysosomal V0 subunit D2 (Atp6v0d2; Forward: 5′-GTGAGACCTTGGAAGACCTGAA-3′; Reverse: 5′-GAGAAATGTGCTCAGGGGCT-3′), TRAcP (Acp5; Forward: 5′-TGTGGCCATCTTTATGCT-3′; Reverse: 5′-GTCATTTCTTTGGGGCTT-3′), cathepsin K (Ctsk; Forward: 5′-GGGAGAAAAACCTGAAGC-3′; Reverse: 5′-ATTCTGGGGACTCAGAGC-3′), matrix metallopeptidase 9 (Mmp9; Forward: 5′-CGTGTCTGGAGATTCGACTTGA-3′; Reverse: 5′-TTGGAAACTCACACGCCAGA-3′), and b-actin (Actb; Forward: 5′-AAGATCAAGATCATTGCTCCTCCT-3′, Reverse: 5′-AGCTCAGTAACAGTCCGCCT-3′). The fluorescence signal of PCR amplification was read and monitored by a ViiA 7 rtPCR machine (Applied Biosystems). The Ct values of target genes were normalized to the Ct value of Actb to give a ΔCt value, in which the data of the experimental groups was further normalized to the control groups to obtain ΔΔCt.
Western blotting
Fresh BMMs were induced by 50 ng/ml hRANKL with or without i-bodies in 6-well plates (1 × 105 cells/well) for 5 days until the formation of mature osteoclasts in the positive control. Cells were lysed in RIPA buffer and boiled for 5 min with 4× loading buffer. Samples were resolved in 10% w/v SDS-denatured acrylamide gels and electroblotted onto 0.2 μm nitrocellulose membranes. Membranes were then blocked with 5% w/v nonfat milk powder in Tris buffered saline with Tween 20 (TBST) containing 10 mM Tris, 150 mM NaCl, as well as 0.1% v/v Tween-20, accompanied with the incubation of primary antibodies diluted (1:500∼1000) in TBST containing 1% w/v BSA. HRP-conjugated secondary antibodies were diluted (1:3300) with 1% BSA w/v in TBST. Proteins were visualized by Western Lightning Ultra from PerkinElmer and an ImageQuant LAS4000 (GE HealthCare).
Calcium flux assay
In general, starved fresh BMMs were seeded in 48-well plates (1 × 104 cells/well) overnight before being treated with 50 ng/ml hRANKL with or without i-bodies for 24 h. Cells were gently washed with assay buffer (Hanks’ buffer supplemented with 1 mM probenecid and 1% v/v FBS) and labeled by calcium indicator Fluo-4 AM (in assay buffer containing 20% w/v dimethyl sulfoxide–diluted pluronic-F127), according to the manufacturer’s guidance (Molecular Probes; Thermo Fisher Scientific). Intracellular Ca2+ was subsequently visualized through inverted fluorescent microscopy (Nikon) at 488 nm wavelength. Images were taken every 2 s, lasting for 3 min for each well. Further analysis was conducted using the Nikon Basic Research Software.
Statistical analysis and graph preparation
The statistics was carried out by Students' t test (p < 0.05). GraphPad Prism 6 (https://www.graphpad.com/support/prism-6-updates/) was used to generate plots and present data. At least three independent experiments were performed for statistical analysis.
Data availability
The data that support this study are available from the corresponding author upon a reasonable request.
Supporting information
This article contains supporting information.
Conflicts of interest
The authors declare that they have no conflicts of interest in this article. Michael Foley is a chief founding scientist and shareholder of AdAlta Limited. Christopher Hosking's and Kevin Lim's salaries are funded by AdAlta Ltd.
Acknowledgments
The study was supported by the Australian National Health and Medical Research Council (Grant No. APP1107828, APP1127156, APP1163933). The authors would also like to thank Prof Robin Anderson and Judy Doherty from the Peter MacCallum Cancer Centre who were involved in some initial in vitro experiments on ADR3.
Author contributions
J. X. conceptualization; C. H., K. L., and M. F. methodology; A. S., K. C., V. K., H. J., S. Z., and A. V. software; H. Q., C. H., K. L., and M. F. formal analysis; H. Q., C. H., E. R., A. S., A. V., K. L., and M. F. investigation; H. Q. writing – original draft; C. H., E. R., A. S., K. C., V. K., H. J., S. Z., A. V., K. L., M. F., and J. X. writing – review and editing; J. X. supervision.
Edited by Qi-Qun Tang
Supporting information
A, size-exclusion chromatography (SEC) analysis of ADR3 showed that purified ADR3 is predominantly monomeric. B and C, flow cytometry analysis showed the binding specificity of ADR3 to human RANKL (hRANKL). D, information for the predicted ADR3 structure generated by a template-based prediction software ModWeb. E, coefficient weights for the top ten docking models of the ADR3-hRANKL complex are shown in the table. The best-fit model was selected to present. F, close contact residues in the predicted ADR3-hRANKL model match with previous studies.
A, MTS assay showed no cytotoxicity of ADR3 on murine-isolated bone marrow cells (BMMs) (n = 4). B, osteoclastogenesis is reduced by ADR3 in RAW264.7 cells. C, independent osteoclastogenesis assay demonstrated the dose-dependent inhibition of ADR3 on hRANKL-stimulated BMMs (n = 3). Scale bar = 200 μm IC50 was calculated basing on the number of TRAcP-positive osteoclasts. IC50 calculation tool: https://www.aatbio.com/tools/ic50-calculator. D, ADR3 effectively reduces rat RANKL (rRANKL)-stimulated osteoclast differentiation (n = 4). All graphs are presented as the mean ± SD. ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001 compared to the positive control. #p < 0.05 and ##p < 0.01 compared between different doses of ADR3.
References
- 1.Forbes R.M., Cooper A.R., Mitchell H.H. The composition of the adult human body as determined by chemical analysis. J. Biol. Chem. 1953;203:359–366. [PubMed] [Google Scholar]
- 2.Zhu S., Yao F., Qiu H., Zhang G., Xu H., Xu J. Coupling factors and exosomal packaging microRNAs involved in the regulation of bone remodelling. Biol. Rev. Camb. Philos. Soc. 2018;93:469–480. doi: 10.1111/brv.12353. [DOI] [PubMed] [Google Scholar]
- 3.Feng X., McDonald J.M. Disorders of bone remodeling. Annu. Rev. Pathol. 2011;6:121–145. doi: 10.1146/annurev-pathol-011110-130203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kular J., Tickner J., Chim S.M., Xu J. An overview of the regulation of bone remodelling at the cellular level. Clin. Biochem. 2012;45:863–873. doi: 10.1016/j.clinbiochem.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 5.Compston J.E., McClung M.R., Leslie W.D. Osteoporosis. Lancet. 2019;393:364–376. doi: 10.1016/S0140-6736(18)32112-3. [DOI] [PubMed] [Google Scholar]
- 6.Sambrook P.N., Seeman E., Phillips S.R., Ebeling P.R., Osteoporosis Australia. National Prescribing Service Preventing osteoporosis: outcomes of the Australian fracture prevention summit. Med. J. Aust. 2002;176:S1–16. doi: 10.5694/j.1326-5377.2002.tb04475.x. [DOI] [PubMed] [Google Scholar]
- 7.Zeng Q., Li N., Wang Q., Feng J., Sun D., Zhang Q., et al. The prevalence of osteoporosis in China, a nationwide, multicenter DXA survey. J. Bone Miner. Res. 2019;34:1789–1797. doi: 10.1002/jbmr.3757. [DOI] [PubMed] [Google Scholar]
- 8.Tu K.N., Lie J.D., Wan C.K.V., Cameron M., Austel A.G., Nguyen J.K., et al. Osteoporosis: a review of treatment options. P T. 2018;43:92–104. [PMC free article] [PubMed] [Google Scholar]
- 9.Ono T., Hayashi M., Sasaki F., Nakashima T. RANKL biology: bone metabolism, the immune system, and beyond. Inflamm. Regen. 2020;40:2. doi: 10.1186/s41232-019-0111-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steffen U., Schett G., Bozec A. How autoantibodies regulate osteoclast induced bone loss in rheumatoid arthritis. Front. Immunol. 2019;10:1483. doi: 10.3389/fimmu.2019.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taxel P., Faircloth E., Idrees S., Van Poznak C. Cancer treatment-induced bone loss in women with breast cancer and men with prostate cancer. J. Endocr. Soc. 2018;2:574–588. doi: 10.1210/js.2018-00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ming J., Cronin S.J.F., Penninger J.M. Targeting the RANKL/RANK/OPG axis for cancer therapy. Front. Oncol. 2020;10:1283. doi: 10.3389/fonc.2020.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asano T., Okamoto K., Nakai Y., Tsutsumi M., Muro R., Suematsu A., et al. Soluble RANKL is physiologically dispensable but accelerates tumour metastasis to bone. Nat. Metab. 2019;1:868–875. doi: 10.1038/s42255-019-0104-1. [DOI] [PubMed] [Google Scholar]
- 14.McDonald M.M., Khoo W.H., Ng P.Y., Xiao Y., Zamerli J., Thatcher P., et al. Osteoclasts recycle via osteomorphs during RANKL-stimulated bone resorption. Cell. 2021;184:1330–1347.e13. doi: 10.1016/j.cell.2021.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cummings S.R., San Martin J., McClung M.R., Siris E.S., Eastell R., Reid I.R., et al. Denosumab for prevention of fractures in postmenopausal women with osteoporosis. N. Engl. J. Med. 2009;361:756–765. doi: 10.1056/NEJMoa0809493. [DOI] [PubMed] [Google Scholar]
- 16.Bone H.G., Wagman R.B., Brandi M.L., Brown J.P., Chapurlat R., Cummings S.R., et al. 10 Years of denosumab treatment in postmenopausal women with osteoporosis: results from the phase 3 randomised FREEDOM trial and open-label extension. Lancet Diabetes Endocrinol. 2017;5:513–523. doi: 10.1016/S2213-8587(17)30138-9. [DOI] [PubMed] [Google Scholar]
- 17.Griffiths K., Binder U., McDowell W., Tommasi R., Frigerio M., Darby W.G., et al. Half-life extension and non-human primate pharmacokinetic safety studies of i-body AD-114 targeting human CXCR4. MAbs. 2019;11:1331–1340. doi: 10.1080/19420862.2019.1626652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diker-Cohen T., Rosenberg D., Avni T., Shepshelovich D., Tsvetov G., Gafter-Gvili A. Risk for infections during treatment with denosumab for osteoporosis: a systematic review and meta-analysis. J. Clin. Endocrinol. Metab. 2020;105 doi: 10.1210/clinem/dgz322. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths K., Dolezal O., Cao B., Nilsson S.K., See H.B., Pfleger K.D.G., et al. i-bodies, human single domain antibodies that antagonize chemokine receptor CXCR4. J. Biol. Chem. 2016;291:12641–12657. doi: 10.1074/jbc.M116.721050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths K., Habiel D.M., Jaffar J., Binder U., Darby W.G., Hosking C.G., et al. Anti-fibrotic effects of CXCR4-targeting i-body AD-114 in preclinical models of pulmonary fibrosis. Sci. Rep. 2018;8:3212. doi: 10.1038/s41598-018-20811-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ta H.M., Nguyen G.T., Jin H.M., Choi J., Park H., Kim N., et al. Structure-based development of a receptor activator of nuclear factor-kappaB ligand (RANKL) inhibitor peptide and molecular basis for osteopetrosis. Proc. Natl. Acad. Sci. U. S. A. 2010;107:20281–20286. doi: 10.1073/pnas.1011686107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkins M.R., Gasteiger E., Bairoch A., Sanchez J.C., Williams K.L., Appel R.D., et al. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 23.Suda T., Takahashi N., Udagawa N., Jimi E., Gillespie M.T., Martin T.J. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr. Rev. 1999;20:345–357. doi: 10.1210/edrv.20.3.0367. [DOI] [PubMed] [Google Scholar]
- 24.Matsuo K., Galson D.L., Zhao C., Peng L., Laplace C., Wang K.Z., et al. Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J. Biol. Chem. 2004;279:26475–26480. doi: 10.1074/jbc.M313973200. [DOI] [PubMed] [Google Scholar]
- 25.Lee N.K., Choi Y.G., Baik J.Y., Han S.Y., Jeong D.W., Bae Y.S., et al. A crucial role for reactive oxygen species in RANKL-induced osteoclast differentiation. Blood. 2005;106:852–859. doi: 10.1182/blood-2004-09-3662. [DOI] [PubMed] [Google Scholar]
- 26.Ke K., Safder M.A., Sul O.J., Kim W.K., Suh J.H., Joe Y., et al. Hemeoxygenase-1 maintains bone mass via attenuating a redox imbalance in osteoclast. Mol. Cell. Endocrinol. 2015;409:11–20. doi: 10.1016/j.mce.2015.03.022. [DOI] [PubMed] [Google Scholar]
- 27.Florczyk-Soluch U., Jozefczuk E., Stepniewski J., Bukowska-Strakova K., Mendel M., Viscardi M., et al. Various roles of heme oxygenase-1 in response of bone marrow macrophages to RANKL and in the early stage of osteoclastogenesis. Sci. Rep. 2018;8:10797. doi: 10.1038/s41598-018-29122-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang J.Y., Kang N., Yang Y.M., Hong J.H., Shin D.M. The role of Ca(2+)-NFATc1 signaling and its modulation on osteoclastogenesis. Int. J. Mol. Sci. 2020;21:3646. doi: 10.3390/ijms21103646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 30.Black D.M., Rosen C.J. Postmenopausal osteoporosis. N. Engl. J. Med. 2016;374:2096–2097. doi: 10.1056/NEJMc1602599. [DOI] [PubMed] [Google Scholar]
- 31.Bi H., Chen X., Gao S., Yu X., Xiao J., Zhang B., et al. Key triggers of osteoclast-related diseases and available strategies for targeted therapies: a review. Front. Med. (Lausanne) 2017;4:234. doi: 10.3389/fmed.2017.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Y., Jiang S., Ying T. Single-domain antibodies as therapeutics against human viral diseases. Front. Immunol. 2017;8:1802. doi: 10.3389/fimmu.2017.01802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nuttall S.D. Overview and discovery of IgNARs and generation of VNARs. Methods Mol. Biol. 2012;911:27–36. doi: 10.1007/978-1-61779-968-6_3. [DOI] [PubMed] [Google Scholar]
- 34.Wurch T., Pierre A., Depil S. Novel protein scaffolds as emerging therapeutic proteins: from discovery to clinical proof-of-concept. Trends Biotechnol. 2012;30:575–582. doi: 10.1016/j.tibtech.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 35.Lo M.C., Aulabaugh A., Jin G., Cowling R., Bard J., Malamas M., et al. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal. Biochem. 2004;332:153–159. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 36.Takasaki W., Kajino Y., Kajino K., Murali R., Greene M.I. Structure-based design and characterization of exocyclic peptidomimetics that inhibit TNF alpha binding to its receptor. Nat. Biotechnol. 1997;15:1266–1270. doi: 10.1038/nbt1197-1266. [DOI] [PubMed] [Google Scholar]
- 37.Aoki K., Saito H., Itzstein C., Ishiguro M., Shibata T., Blanque R., et al. A TNF receptor loop peptide mimic blocks RANK ligand-induced signaling, bone resorption, and bone loss. J. Clin. Invest. 2006;116:1525–1534. doi: 10.1172/JCI22513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furuya Y., Inagaki A., Khan M., Mori K., Penninger J.M., Nakamura M., et al. Stimulation of bone formation in cortical bone of mice treated with a receptor activator of nuclear factor-kappaB ligand (RANKL)-binding peptide that possesses osteoclastogenesis inhibitory activity. J. Biol. Chem. 2013;288:5562–5571. doi: 10.1074/jbc.M112.426080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyce B.F., Xiu Y., Li J., Xing L., Yao Z. NF-kappaB-mediated regulation of osteoclastogenesis. Endocrinol. Metab. (Seoul) 2015;30:35–44. doi: 10.3803/EnM.2015.30.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wada T., Nakashima T., Hiroshi N., Penninger J.M. RANKL-RANK signaling in osteoclastogenesis and bone disease. Trends Mol. Med. 2006;12:17–25. doi: 10.1016/j.molmed.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 41.Kim K., Lee S.H., Ha Kim J., Choi Y., Kim N. NFATc1 induces osteoclast fusion via up-regulation of Atp6v0d2 and the dendritic cell-specific transmembrane protein (DC-STAMP) Mol. Endocrinol. 2008;22:176–185. doi: 10.1210/me.2007-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy S.V., Hundley J.E., Windle J.J., Alcantara O., Linn R., Leach R.J., et al. Characterization of the mouse tartrate-resistant acid phosphatase (TRAP) gene promoter. J. Bone Miner. Res. 1995;10:601–606. doi: 10.1002/jbmr.5650100413. [DOI] [PubMed] [Google Scholar]
- 43.Anusaksathien O., Laplace C., Li X., Ren Y., Peng L., Goldring S.R., et al. Tissue-specific and ubiquitous promoters direct the expression of alternatively spliced transcripts from the calcitonin receptor gene. J. Biol. Chem. 2001;276:22663–22674. doi: 10.1074/jbc.M007104200. [DOI] [PubMed] [Google Scholar]
- 44.Motyckova G., Weilbaecher K.N., Horstmann M., Rieman D.J., Fisher D.Z., Fisher D.E. Linking osteopetrosis and pycnodysostosis: regulation of cathepsin K expression by the microphthalmia transcription factor family. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5798–5803. doi: 10.1073/pnas.091479298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.David J.P., Rincon M., Neff L., Horne W.C., Baron R. Carbonic anhydrase II is an AP-1 target gene in osteoclasts. J. Cell Physiol. 2001;188:89–97. doi: 10.1002/jcp.1099. [DOI] [PubMed] [Google Scholar]
- 46.Qin A., Cheng T.S., Pavlos N.J., Lin Z., Dai K.R., Zheng M.H. V-ATPases in osteoclasts: structure, function and potential inhibitors of bone resorption. Int. J. Biochem. Cell Biol. 2012;44:1422–1435. doi: 10.1016/j.biocel.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 47.Lacombe J., Karsenty G., Ferron M. Regulation of lysosome biogenesis and functions in osteoclasts. Cell Cycle. 2013;12:2744–2752. doi: 10.4161/cc.25825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakai E., Shimada-Sugawara M., Nishishita K., Fukuma Y., Naito M., Okamoto K., et al. Suppression of RANKL-dependent heme oxygenase-1 is required for high mobility group box 1 release and osteoclastogenesis. J. Cell. Biochem. 2012;113:486–498. doi: 10.1002/jcb.23372. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J., Fritsch E.F., Maniatis T. 2nd ed. CSH Laboratory Press; Long Island, NY: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 50.Desta I.T., Porter K.A., Xia B., Kozakov D., Vajda S. Performance and its limits in rigid body protein-protein docking. Structure. 2020;28:1071–1081.e3. doi: 10.1016/j.str.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vajda S., Yueh C., Beglov D., Bohnuud T., Mottarella S.E., Xia B., et al. New additions to the ClusPro server motivated by CAPRI. Proteins. 2017;85:435–444. doi: 10.1002/prot.25219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., et al. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niesen F.H., Berglund H., Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 54.Qiu H., Qin A., Cheng T., Chim S.M., Smithers L., Chen K., et al. A missense mutation sheds light on a novel structure-function relationship of RANKL. J. Cell Physiol. 2021;236:2800–2816. doi: 10.1002/jcp.30045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A, size-exclusion chromatography (SEC) analysis of ADR3 showed that purified ADR3 is predominantly monomeric. B and C, flow cytometry analysis showed the binding specificity of ADR3 to human RANKL (hRANKL). D, information for the predicted ADR3 structure generated by a template-based prediction software ModWeb. E, coefficient weights for the top ten docking models of the ADR3-hRANKL complex are shown in the table. The best-fit model was selected to present. F, close contact residues in the predicted ADR3-hRANKL model match with previous studies.
A, MTS assay showed no cytotoxicity of ADR3 on murine-isolated bone marrow cells (BMMs) (n = 4). B, osteoclastogenesis is reduced by ADR3 in RAW264.7 cells. C, independent osteoclastogenesis assay demonstrated the dose-dependent inhibition of ADR3 on hRANKL-stimulated BMMs (n = 3). Scale bar = 200 μm IC50 was calculated basing on the number of TRAcP-positive osteoclasts. IC50 calculation tool: https://www.aatbio.com/tools/ic50-calculator. D, ADR3 effectively reduces rat RANKL (rRANKL)-stimulated osteoclast differentiation (n = 4). All graphs are presented as the mean ± SD. ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001 compared to the positive control. #p < 0.05 and ##p < 0.01 compared between different doses of ADR3.
Data Availability Statement
The data that support this study are available from the corresponding author upon a reasonable request.