Abstract
Radioresistance is the major reason for the failure of radiotherapy in esophageal squamous cell carcinoma (ESCC). Previous evidence indicated that stanniocalcin 2 (STC2) participates in various biological processes of malignant tumors. However, researches on its effect on radioresistance in cancers are limited. In this study, STC2 was screened out by RNA-sequencing and bioinformatics analyses as a potential prognosis predictor of ESCC radiosensitivity and then was determined to facilitate radioresistance. We found that STC2 expression is increased in ESCC tissues compared to adjacent normal tissues, and a higher level of STC2 is associated with poor prognosis. Also, STC2 mRNA and protein expression levels were higher in radioresistant cells than in their parental cells. Further investigation revealed that STC2 could interact with protein methyltransferase 5 (PRMT5) and activate PRMT5, thus leading to the increased expression of symmetric dimethylation of histone H4 on Arg 3 (H4R3me2s). Mechanistically, STC2 can promote DDR through the homologous recombination and non-homologous end joining pathways by activating PRMT5. Meanwhile, STC2 can participate in SLC7A11-mediated ferroptosis in a PRMT5-dependent manner. Finally, these results were validated through in vivo experiments. These findings uncovered that STC2 might be an attractive therapeutic target to overcome ESCC radioresistance.
Keywords: STC2, PRMT5, Radioresistance, Esophageal squamous cell carcinoma, DNA damage Repair, Ferroptosis
Abbreviations: ESCC, Esophageal squamous cell carcinoma; STC2, Stanniocalcin 2; DDR, DNA damage repair; ROS, Reactive oxygen species; MDA, Malonic dialdehyde; PRMT5, Protein methyltransferase 5; H4R3me2s, Symmetric dimethylation of histone H4 on Arg; IR, Ionizing radiation; DSBs, DNA double-strand breaks; HR, Homologous recombination; NHEJ, Non-homologous end joining; RCD, Regulated cell death; FINs, Ferroptosis inducers; SLC7A11, Solute carrier family 7 member 11; GPX4, Glutathione peroxidase 4; RT-qPCR, Real-time quantitative PCR; GSH, Glutathione; IHC, Immunohistochemistry; MS, Mass spectrometry; co-IP, co-immunoprecipitation; DEGs, Differentially expressed genes; TCGA, The Cancer Genome Atlas; SDMA, Symmetrical dimethylarginine arginine; SLC3A2, Solute carrier family 3 member 2; ATF4, Activating transcription factor 4
1. Introduction
Esophageal cancer (EC) is one of the most frequently occurring malignancies and ranks as the seventh leading cause of cancer-related deaths globally [1]. It is estimated that half of all EC cases are diagnosed in China, with esophageal squamous cell carcinoma (ESCC) constituting the majority of cases [2,3]. Despite tremendous efforts to develop new therapeutic approaches, the outcome of ESCC patients remains dismal. Unfortunately, early diagnosis of ESCC is hard to achieve, resulting in that most ESCC patients being diagnosed at advanced stages. For patients with advanced ESCC, the standard treatment is neoadjuvant chemotherapy or chemoradiotherapy followed by radical surgery [4]. However, the intrinsic radioresistance of ESCC cells limits the clinical efficacy of radiotherapy, leading to tumor recurrence and distant metastasis [5]. Hence, elucidating the molecular mechanisms and discovering potential targets to overcome the radioresistance in ESCC are urgently needed.
Resistance to Ionizing radiation (IR) is polymodal and associated with various biological mechanisms. DNA double-strand breaks (DSBs) are primary factors and the most lethal lesions in response to IR, inducing the subsequent DNA damage repair (DDR) [6]. Homologous recombination (HR) and non-homologous end joining (NHEJ) are the most prevalent pathways involved in IR-induced DDR [7]. In recent years, targeting involved proteins in HR and NHEJ pathways has been considered a promising strategy to enhance sensitivity to IR, and some remarkable progress has been made. For instance, inhibition of DNA-PKcs has been proposed as an effective method to overcome radioresistance in cancers [8,9]. Some specific and potent PARP-1 inhibitors have been developed to sensitize cancers, including ESCC [[10], [11], [12], [13]]. Overall, the enhancement of sensitization to radiotherapy through DDR pathways has been a focus for the past few decades. Besides its direct effects on DSBs, IR can also exert its indirect effects via eliciting radiolysis of cellular water, which then subsequently targets nucleic, lipids, etc. These direct and indirect effects would trigger distinct forms of regulated cell death (RCD). Ferroptosis, a novel form of RCD, was first described in 2012 [14]. Recent studies revealed that IR induces potent ferroptosis, and ferroptosis can also modulate radiosensitivity in cancers [[15], [16], [17]]. Many preclinical trials presented that ferroptosis inducers (FINs) can enhance radiosensitivity when combined with IR. Class I FINs targeting solute carrier family 7 member 11 (SLC7A11), class II FINs targeting glutathione peroxidase 4 (GPX4), and class III FINs depleting CoQ and GPX4, were all confirmed to sensitize cancer cells to IR in vitro and vivo [16]. Thus, targeting traditional DDR or novel ferroptosis might be an effective therapeutic strategy to overcome radioresistance.
The stanniocalcin (STC) family consists of stanniocalcin 1 (STC1) and stanniocalcin 2 (STC2), which are glycoproteins functioning as hormones to regulate calcium and phosphate secretion [18]. STC2 is an ortholog of fish STCs and is widely expressed in various organs and tissues [19]. Previous studies suggested that STC2 is broadly up-regulated in multiple malignant tumors, including colon cancer [20,21], cervical cancer [22], nasopharyngeal cancer [23,24], liver cancer [25,26] etc. Accumulating evidence has indicated that STC2 participates in a wide variety of biological processes, such as cell proliferation, migration, invasion, immune response, and drug resistance. Regarding ESCC, STC2 was up-regulated in ESCC tissues compared with corresponding normal tissues and can facilitate cell proliferation, invasion, and metastasis [27]. However, no investigation has been conducted to elucidate the relationship between STC2 and radiosensitivity in ESCC.
In this study, we confirmed the expression of STC2 and its regulation on proliferation and metastasis in ESCC. Our research focuses on determining the critical role of STC2 on radioresistance and investigating the underlying mechanism involved in ESCC. We demonstrated that STC2 could interact with protein methyltransferase 5 (PRMT5) and activate PRMT5, thus inducing radioresistance through DDR and ferroptosis pathways. The evidence in our study indicated that targeting STC2 might be a promising therapeutic strategy for ESCC treatment.
2. Results
2.1. STC2 negatively correlates with the radiosensitivity of ESCC
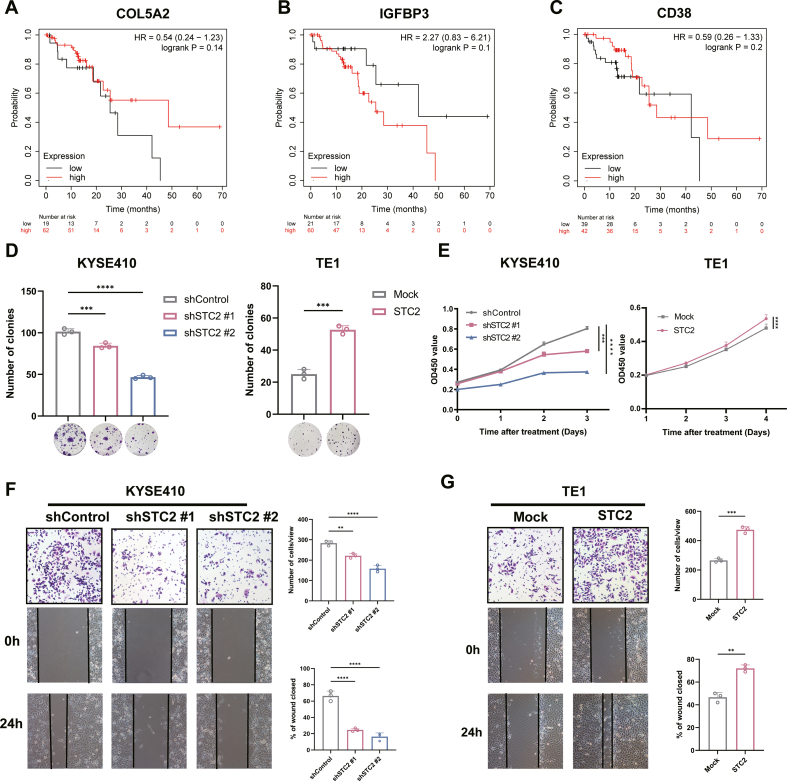
In our previous study, two radioresistant ESCC cell lines KYSE150R and TE1R were constructed from their parental cell lines KYSE150 and TE1, respectively [28]. To investigate the potential genes involved in the radioresistance of ESCC, we used RNA-seq to explore the differentially expressed genes (DEGs) between these two radioresistant ESCC cells and their parental cells (Fig. 1A). Moreover, we extracted and analyzed 3 datasets (GSE100942, GSE149609, GSE164158) containing gene expression profiles of ESCC as well as normal esophageal tissues from GEO database to determine DEGs. We then combined up-regulated genes of each database into a Venn diagram analysis, which revealed that only 4 genes (STC2, COL5A2, IGFBP3, CD38) were up-regulated simultaneously (Fig. 1B). Particularly, Kaplan-Meier survival analysis indicated that high expression of STC2 referred to poor prognosis of ESCC patients (Fig. 1E). In comparison, there is no statistically significant relationship between expression of the other 3 genes and prognosis (Figs. S1A–C). Herein, we focused on STC2 in the present study based on the above results and analyses.
Fig. 1.
STC2 was identified as a potential prognosis predictor of ESCC radiosensitivity.
a Volcano plot of differentially expressed genes (DEGs) between radioresistant cells and their normal parental cells. b Venn diagram of the co-upregulated DEGs among 2 RNA-sequencing datasets (TE1R/TE1, KYSE150R/KYSE150) and 3 datasets from the GEO database (GSE100942, GSE149609, GSE164158). c STC2 mRNA expression level in adjacent normal tissues and ESCC from the database. d Immunohistochemistry of STC2 expression in 24 ESCC tissues and their adjacent non-tumor tissues. Scale bar: 20 μm e Kaplan-Meier curves of ESCC survival based on the expression level of STC2 according to the TCGA database. f-g qRT-PCR and western blotting were performed to detect STC2 expression in radioresistant cancer cells compared with their normal cancer cells. ***p < 0.001; ****p < 0.0001.
Consistent with the analysis of paired samples from the GEO database, the STC2 mRNA expression was up-regulated in ESCC tissue compared to that in normal tissue from The Cancer Genome Atlas (TCGA) and GTEx datasets (Fig. 1C). To further validate the expression of STC2 in ESCC patients, immunohistochemistry (IHC) was performed in ESCC tissues and their paired adjacent non-tumor tissues. It was revealed that the protein level of STC2 was overexpressed in ESCC tissues (Fig. 1D). We also detected the differential expression of STC2 in radioresistant cells and parental cells by real-time quantitative PCR (RT-qPCR) and western blot (WB) (Fig. 1F–G), which was consistent with RNA-seq results. Taken together, these results strongly suggested that STC2 play a crucial role in tumor progression and radioresistance in ESCC.
2.2. STC2 facilitates ESCC progression
We first examined the expression levels of STC2 in various ESCC cells. The expression levels of STC2 are variable in different cancer cell lines (Fig. 2A). Based on the expression level, STC2 was knocked down in KYSE410 and KYSE510 cells as well as overexpressed in KYSE150 and TE1 cells for the following assays (Fig. 2B). Colony formation and cell counting kit-8 (CCK8) assays indicated that inhibition of STC2 remarkably suppressed cell proliferation. In contrast, elevated expression of STC2 enhanced proliferation (Figs. S1D–E). We also found that the expression of STC2 was related to cell migration of ESCC (Figs. S1F–G). The above results implied that STC2 acted as an oncogene and facilitated ESCC progression.
Fig. 2.
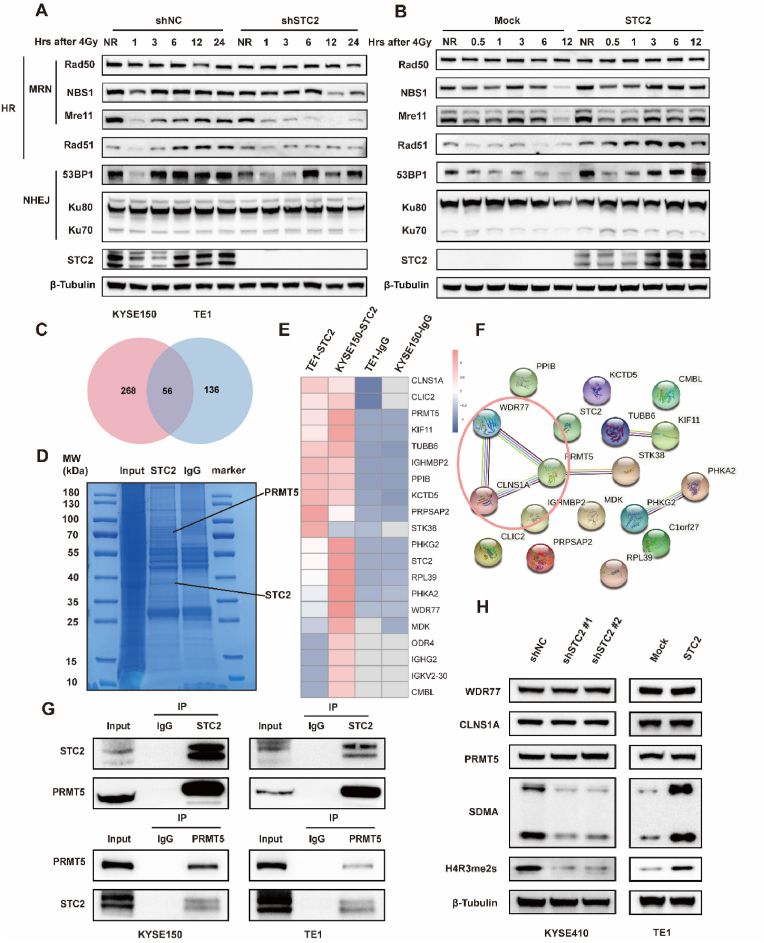
STC2 facilitates radioresistance and promotes DNA damage repair (DDR) of ESCC after IR exposure. a Examination of STC2 protein levels in different ESCC cell lines. b The protein level of STC2 was detected in STC2-KD and -overexpressing cells transfected with the related lentivirus. c Colony formation assays and survival fraction of cells after irradiation with 0, 2, 4, and 8 Gy d The values of D0, Dq, N in each group. e Representative images of γH2AX-positive nuclei in STC2-KD and -overexpressing groups at different times post IR. The 0h means no IR. γH2AX foci in red, nuclear counterstaining with 4′,6-diamidino-2-phenylindole in blue. Scale bar: 20 μm f Comet assay was carried out in STC2-KD and -overexpressing cells at the indicated time points after IR treatment. Scale bar: 20 μm *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.3. STC2 induces radioresistance and promotes DDR through the NHEJ and HR pathways
Referring to the effect of STC2 expression on radiosensitivity in ESCC, we first performed colony formation assays in ESCC cells, which were exposed to a series of single radiation doses. Overexpression of STC2 improved the colony formation after IR while the surviving fractions of ESCC cells with STC2 knockdown were diminished conversely in response to IR, suggesting STC2 conferred radioresistance in ESCC cells (Fig. 2C–D, Figs. S2A–B).
γ-H2AX is identified as a marker of DSBs and correlates with radiosensitivity, and we evaluated the level of γ-H2AX in ESCC cells after IR. Overall, the number of γ-H2AX-positive nuclei significantly increased following IR and gradually reduced. As shown in Fig. 2E, KYSE410-shSTC2 cells exhibited elevated levels of γ-H2AX foci compared to control cells. On the contrary, there were fewer γ-H2AX foci in TE1-STC2 cells after IR. We also acquired similar results in KYSE510-shSTC2 cells and KYSE150-STC2 cells (Fig. S2C). Moreover, a comet assay was also conducted to measure the DSBs after 4Gy IR. Consistent with the immunofluorescence (IF) of γ-H2AX foci, we revealed that the olive tail moment is higher in STC2 deficient cells than in control cells, while STC2-overexpressed cells exhibited shorter comet tails (Fig. 2F, Fig. S2D).
It has been reported that HR and NHEJ are two critical pathways involved in DDR. Therefore, we analyzed expression levels of essential proteins involved in HR and NHEJ in ESCC cells. Notably, we observed lower expression of HR-related proteins, including NBS1, Mre11, and RAD51 in KYSE410-shSTC2 cells, whereas STC2 overexpressing resulted in higher expression of those proteins in TE1-STC2 cells. Moreover, STC2 altered the expression levels of 53BP1, Ku70, and Ku80, indicating that STC2 can also regulate NHEJ (Fig. 3A–B). Collectively, these data demonstrated that STC2 could simultaneously promote DDR through NHEJ and HR pathways, thus inducing radioresistance in ESCC cells.
Fig. 3.
STC2 promotes IR-induced HR and NHEJ and PRMT5 are identified as a target of STC2.
a-b WB analysis of homologous recombination (HR) and non-homologous end joining (NHEJ) related proteins influenced by STC2 expression following 4Gy IR at different time points. NR means no IR. c Venn diagram of differential expressed gene between the anti-IgG group and anti-STC2 group in 2 overexpressing cells. d Coomassie blue-stained gel with protein. e The heatmap of top 20 proteins with significant fold change. f Protein-protein network of these top 20 significant genes. g Co-IP experiments were performed using either an STC2 antibody to pull down PRMT5 or a PRMT5 antibody to pull down STC2 in TE1-STC2 and KYSE150-STC2 cells. h The protein levels of WDR77, CLNS1A, PRMT5, SDMA and H4R3me2s were determined by western blotting in KYSE410-shSTC2 cells and TE1-STC2 cells. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.4. STC2 interacts with and activates PRMT5
To further explore the mechanism of how STC2 regulated DDR, we performed immunoprecipitation (IP)-mass spectrometry (MS) in TE1-STC2 cells and KYSE150-STC2 cells. We first conducted IP with anti-STC2 antibody and stained the co-immunoprecipitation (co-IP) samples with Coomassie Brilliant Blue after electrophoresis (Fig. 3D). Then, MS analysis identified the DEGs between the anti-STC2 group and anti-IgG group. The potential targets of STC2 were proteins just exported in the anti-STC2 group or which met the criteria with a log fold change≥3 (Fig. 3C). According to their fold change, we ranked and subjected to the top 20 proteins (Fig. 3E), which were imported into Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://cn.string-db.org/). Referring to the protein-protein network of these DEGs, we found that PRMT5 was the core of the network (Fig. 3F). Since previous studies have identified that PRMT5 cooperated with CLNS1A or WDR77 as a master epigenetic activator of DDR through HR and NHEJ [29], we hypothesized that STC2 might bind to PRMT5 in ESCC cells. To validate this hypothesis, we performed co-IP assays with an anti-STC2 antibody to pull down PRMT5 and then with an anti-PRMT5 antibody to pull down STC2 reciprocally. The interaction between STC2 and PRMT5 was validated in TE1-STC2 cells and KYSE150-STC2 cells (Fig. 3G). Indeed, we also found that STC2 can interact with CLNS1A and WDR77 (Fig. S3A). Unexpectedly, the manipulation of STC2 did not change the protein level of PRMT5, CLNS1A, and WDR77. However, we discovered that symmetrical dimethylarginine arginine (SDMA) and symmetric dimethylation of histone H4 on Arg 3 (H4R3me2s), which were mediated by PRMT5 and indicated the activity of PRMT5, altered with the change of STC2 (Fig. 3H). To sum up, these findings implied that STC2 not only interacted with PRMT5 but also activated PRMT5.
2.5. STC2 enhances DDR of ESCC by activating PRMT5
GSK3326595(GSK) has been widely reported to specifically inhibit PRMT5 enzymatic activity [30,31]. Inhibition of PRMT5 with GSK decreased expression levels of HR and NEHJ-related proteins in a concentration- and time-dependent manner (Fig. S3B). Based on these results, we treated ESCC cells with GSK (20 μM) for 48 h to carry out the following experiments. As we can see from Fig. 4A–D, the addition of GSK inhibited DDR and sensitized cells to IR, resulting in increased γ-H2AX foci and tail moment compared with control cells. More importantly, GSK specifically inhibited the methyltransferase activity of PRMT5 and partially abolished the alleviation of the STC2 overexpression-mediated γ-H2AX and olive tail effects. In addition, the survival curves under IR also indicated that PRMT5 inhibitor could recover radiosensitivity in STC2-overexpressed cells (Fig. 4E). The exact values of D0, Dq, and N in each group were presented in Fig. S3C. Similar results were observed by WB that pharmacological inhibition of PRMT5 decreased the expression of STC2-induced proteins involved in HR and NHEJ (Fig. 4F).
Fig. 4.
STC2 enhances DNA damage repair of ESCC by activating PRMT5
a-b Mock and overexpressing-STC2 cells were treated with the indicated drugs. Representative pictures and summary of γH2AX foci formation after 12h when exposed to IR (4 Gy) Scale bar: 20 μm. c-d Neutral comet assay performed after exposure to irradiation (12h). Scale bar: 20 μm e Dose responses of survival factions of mock and overexpressing-STC2 cells (KYSE150, TE1) treated with indicated drugs. f Western blot assay of involved proteins in overexpressing-STC2 KYSE150 and TE1 cells treated with indicated drugs. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
2.6. Depletion of STC2 promotes ferroptosis in ESCC with or without IR
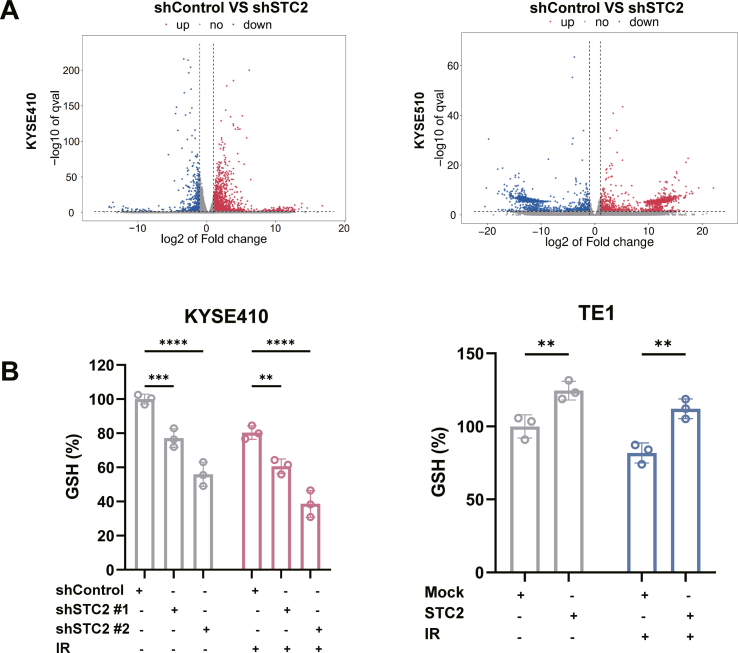
We also employed RNA-seq screening in STC2-depleted cells to further identify the molecular pathways. The DEGs were evaluated in ESCC cells without IR using standard criteria, including |logFC| > 1 and p-value <0.05 (Fig. S4A). Taking the intersection of these DEGs, we screened out a total of 211 DEGs (Fig. 5A). These DEGs were mapped to KEGG analysis and the top 15 significantly enriched pathways were shown (Fig. 5B). We also presented a heatmap of ferroptosis-related gene enriched in KEGG (Fig. 5C). It is obvious that ferroptosis signaling pathway was altered, indicating STC2 may affect the process of ferroptosis.
Fig. 5.
STC2 is investigated to participate in ferroptosis with or without IR
a Differential gene expression between shSTC2 cells and control cancer cells from RNA-Seq. b Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of differentially expressed proteins between STC2-depleted and control KYSE410 cells. c The heatmap of ferroptosis-related gene enriched in KEGG. d Transmission electron microscopy of STC2-depleted and control cells. e Representative images of intracellular levels of Fe2+ in STC2-depleted and -overexpressed cells. f The relative MDA level of STC2 depleted and overexpressing cells with or without IR (4Gy). g The relative total ROS levels were assayed via DCFH-DA fluorescence with or without IR (4Gy). h The relative lipid ROS levels were assayed via C11-BODIPY fluorescence with or without IR (4Gy). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Ferroptosis has recently been revealed to play an essential role in IR-induced cell death. Also, many studies suggested that combining FINs and IR is a promising strategy for radiosensitization in cancers [16,17]. Therefore, we hypothesized that the effect of STC2 in radioresistance might partly result from ferroptosis. We conducted the following assays to clarify the relationship between STC2 and ferroptosis. First, transmission electron microscopy was carried out to identify the typical morphological characteristics of ferroptosis in STC2-depleted ESCC cells without IR. As expected, the depletion of STC2 led to decreased mitochondrial volume and increased membrane density (Fig. 5D). Then, the intracellular level of Fe2+ was evaluated with the specific fluorescent probe FerroOrange under confocal microscopy. The fluorescence intensity was increased in STC2-depleted cells and decreased in STC2-overexpressed cells (Fig. 5E). We then measured the effect of STC2 on malonic dialdehyde (MDA) and glutathione (GSH), which reflects the occurrence of ferroptosis. Indeed, inhibition of STC2 resulted in up-regulated MDA and depleted GSH with or without IR, while STC2 overexpression occurred with contrary results (Fig. 5F, Fig. S4B). Flow cytometry was also performed to detect intracellular reactive oxygen species (ROS) and lipid peroxidation, whether treated with IR or not. Knock-down of STC2 caused increased levels of ROS and lipid peroxidation, while STC2-overexpression resulted in a decreased level of peroxidation (Fig. 5G–H). Besides, as previous studies reported, we also observed that IR could induce ferroptosis with elevated ROS, lipid peroxidation, MDA, and GSH depletion. Our results indicated that STC2 might also modulate ferroptosis and induce radioresistance in ESCC cells.
2.7. STC2 induces ferroptosis by targeting SLC7A11
Coinciding with ferroptosis-related genes enriched in the heatmap of our research, SLC7A11 was recognized as a strongly associated gene with STC2 in esophageal cancer when resorting to the online bioinformatics websites UALCAN (http://ualcan.path.uab.edu/) and GEPIA (http://gepia.cancer-pku.cn/) (Figure S5A and Fig. 6A). We also examined the potential relationship between STC2 and SLC7A11 mRNA expression in ESCC using data from TCGA database (Fig. 6B). Evidence can be acquired that SLC7A11 is strongly related to STC2 with high R ratio and significant p value. As we all know, solute carrier family 3 member 2 (SLC3A2) is a chaperone protein of SLC7A11 and, together with it to encode the cystine/glutamate antiporter system XC-. Notably, SLC3A2 was also involved in related genes and confirmed by further analysis in ESCC (Fig. 6C–D).
Fig. 6.
STC2 inhibits ferroptosis by targeting SLC7A11.
a The relationship between STC2 and SLC7A11 in esophageal cancer (EC) was extracted from the GEPIA website. b The correlation between STC2 and SLC7A11 in ESCC was confirmed in the TCGA dataset. c The relationship between STC2 and SLC3A2 in EC was extracted from the GEPIA website. d The correlation between STC2 and SLC3A2 in ESCC was confirmed in the TCGA dataset. e Western blot of SLC7A11, SLC3A2, and GPX4 in KYSE410-shSTC2 cells and TE1-STC2 cells with or without IR (4Gy). f-h The relative MDA level of TE1-STC2 cells treated with RSL3, Erastin, and sulfasalazine. i-k The relative lipid ROS level of TE1-STC2 cells treated with RSL3, Erastin, and sulfasalazine. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
To investigate whether STC2 regulates ferroptosis through SLC7A11, we examined the related protein expression in ESCC cells. As a result of STC2 knockdown, SLC7A11, SLC3A2, and GPX4 expression were markedly reduced. In contrast, overexpression of STC2 resulted in increased protein levels (Fig. 6E). Subsequently, MDA assays and flow cytometry were performed with several FINs in TE1-STC2 cells. I FINs (Erastin and sulfasalazine), which can inhibit SLC7A11 activity, and II FIN (RSL3), which decreases GPX4 activity, can elevate lipid peroxidation in ESCC cells in a dose-dependent manner. However, when these three FINs were used in TE1-STC2 cells, only I FINs could rescue the STC2-mediated inhibition of ferroptosis with significance. At the same time, the level of lipid peroxidation in STC2-TE1 cells treated with RSL3 presented no statistical significance (Fig. 6F–K). In summary, the above characteristic phenotypes of ferroptosis confirmed that STC2 regulates SLC7A11-dependent ferroptosis.
2.8. STC2 participates in ferroptosis in a PRMT5-dependent manner
As mentioned above, STC2 can interact with PRMT5 and activate the activity of PRMT5. Resorting to previous research, we found that inhibiting the activity of PRMT5 can reduce expression levels of ATF4, SLC7A11 and SLC3A2 [30]. Therefore, we explored whether the role of STC2 in ferroptosis was related to PRMT5 activity.
We determined whether inhibition of PRMT5 by GSK can lead to altered expressions of activating transcription factor 4 (ATF4), SLC7A11 and SLC3A2. The results revealed that GSK inhibited the protein level of ATF4, SL7A11 and SLC3A2 in a time-dependent and concentration-dependent manner (Fig. 7A–B). Then, we also investigated that PRMT5 activity can induce alteration of SLC3A2 and SLC7A11 through ATF4 in ESCC cells (Fig. S5B). Furthermore, the ferroptosis-related analyses were carried out to confirm the vital role of PRMT5. Cells treated with GSK and combined with IR would acquire higher lipid peroxidation and MDA than those control cells only exposed to IR. More importantly, the inhibition of ferroptosis in ESCC cells induced by overexpressing STC2 could be reversed when introducing the PRMT5 inhibitor GSK after IR (Fig. 7C–E). Lipid peroxidation was also measured and presented similar results without IR (Fig. S5B). Similar effects in protein level can be observed from WB, as expected. The protein levels of ATF4, SLC7A11 and SLC3A2 were increased when STC2 was overexpressed. In contrast, it was decreased when GSK was applied (Fig. 7F). These results showed that STC2 can modulate the activity of PRMT5 to suppress ferroptosis, resulting in radioresistance.
Fig. 7.
STC2 inhibits ferroptosis in a PRMT5-dependent manner.
a-b PRMT5 inhibitor GSK332659 decreased the protein level of ATF4, SL7A11 and SLC3A2 in a concentration-dependent and time-dependent manner. c-d Lipid ROS of STC2-overexpressing cells treated with GSK332659(20 μM) when exposed to IR (4Gy). e The relative MDA level of cells treated with GSK332659(20 μM) and subjected to IR (4Gy). f The protein expression of ATF4, SLC7A11 and SLC3A2 were measured in cells treated with GSK332659(20 μM) and exposed to IR. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
2.9. STC2 modulates radioresistance by activating PRMT5 activity in vivo
To further investigate whether STC2 facilitates radioresistance of ESCC in vivo, we constructed a subcutaneous xenograft tumor model, therapeutic modalities shown in Fig. 8A. Compared with the control group, overexpression of STC2 can significantly increase the volume and weight of xenograft tumors with or without IR. In addition, the IR group tumors exhibited lower tumor volumes and weights than those in the non-IR groups. More importantly, the combination therapy showed better therapeutic efficacy, overcoming the radioresistance induced by STC2 (Fig. 8B–D). The protein levels of STC2 and H4R3me2s in each group after IR were presented in Fig. 8E, which is in line with the tendency illustrated above. Altogether, STC2 can activate PRMT5 to facilitate radioresistance in vivo.
Fig. 8.
STC2 facilitates radioresistance in ESCC in vivo.
a Schematic representation of the xenograft study design and experimental workflow. b General view of tumor mass in each indicated group at 15 days after treatment. c Growth curves of xenograft tumors in each group. d Tumor weights were measured in each group. e Representative immunohistochemistry images of the expressions of STC2 and H4R3me2s protein in the xenograft tumors. Scale bar: 20 μm.
3. Discussion
In the current study, we first combined the RNA-seq data of our radioresistant ESCC cells and bioinformatic analysis to identify the critical genes. Then, STC2, our study's target, was discovered to induce radioresistance in ESCC. Mechanistically, STC2 can interact with PRMT5 and activate PRMT5, thus promoting DDR and inhibiting SLC7A11-dependent ferroptosis (Fig. 9).
Fig. 9.
The mechanism of the study.
Accumulating evidence indicated that STC2 played an indispensable role in the occurrence and development of tumors. For instance, STC2 was found to promote cell EMT and glycolysis via activating ITGB2/FAK/SOX6 signaling pathway in nasopharyngeal carcinoma [23]. In non-small cell lung cancer, STC2 overexpression was correlated with acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors [31]. STC2 was also identified as an interacting partner of the plasma-protein histidine-rich glycoprotein, regulating glioma growth through the modulation of antitumor immunity [32]. Regarding ESCC, some bioinformatic studies strongly suggested that STC2 was a potential prognostic biomarker without further validation [[33], [34], [35]]. One study explored and validated that STC2 showed higher expression in ESCC tissues than in corresponding normal tissues. They also found that STC2 transfected cells had a significantly higher proliferation rate and were more invasive than control cells, but they did not elaborate on the specific molecular mechanism. Consistent with previous studies, our results revealed that STC2 was overexpressed in tumor tissues compared with normal tissues, combining public data and tumor samples in our center. We further validated the effects of STC2 in promoting ESCC proliferation and migration. There has been no study concerning the specific mechanisms for STC2-mediated radioresistance and exploring the role of STC2 in the radiosensitivity of ESCC. Therefore, in the present study, the radioresistance effects of STC2 in ESCC were examined, and the underlying mechanisms were investigated.
IR formed various biochemical changes and induced a variety of RCDs. DNA damage is crucial in response to IR, and DDR determines the fate of cells. Accurate and efficient repair of DNA is conducive to maintaining genome stability and keeping cells alive. DDR relies on HR and NHEJ, previous studies have proved hyperactivated HR and NHEJ markers indicate poor IR efficacy in ESCC [[36], [37], [38]]. IR increases lipid peroxidation levels and induced ferroptosis. Targeting ferroptosis can also facilitate radiosensitivity in turn [16,39]. It is indicated that SLC7A11-induced ferroptosis can be identified as a future therapeutic target against radioresistance in ESCC [40]. We demonstrated that STC2 indeed held an essential place in the radioresistance of ESCC. In particular, STC2 can mediate HR and NHEJ concurrently to promote DDR, which had never been reported before. Cells with overexpressed STC2 presented enhanced DNA repair capacity, and altered expression levels of proteins, including NBS1, MRE11, and RAD51 involved in HR and NHEJ-related proteins Ku70, Ku80, and 53BP1 proved the notions. We also determined the role of STC2 in ferroptosis through a series of functional experiments. STC2 can influence ferroptosis with or without IR. Our study found the relationship between STC2 and SLC7A11 based on RNA-seq and bioinformatic analysis. And then, we focused on that STC2 inhibited the SLC7A11-induced ferroptosis. Thus, STC2 transferred ESCC cells acquired radioresistance via the dual effects of enhanced DDR and decreased ferroptosis.
To determine the specific mechanism of STC2-induced radioresistance, we conducted IP combined with MS and then identified PRMT5 as a pivotal target of STC2. PRMT5 is a type II protein arginine methyltransferase that methylates histone or non-histone proteins. Usually, PRMT5 forms a complex with its partner protein, such as WDR77 and CLIS1A, for catalytic activity and specifically mediate SDMA of arginine residues in histones H4R3 (H4R3me2s), H3R2 (H3R2me2s), H3R8 (H3R8me2s), and H2AR3 (H2AR3me2s) [[41], [42], [43], [44]]. In this study, we observed that changes in the expression level of STC2 could affect the PRMT5 activity by regulating H4R3me2s. Many studies emphasized that PRMT5 was up-regulated in multiple cancer types, including ESCC, and was associated with poor prognosis [[45], [46], [47], [48]]. Also, emerging evidence suggests that PRMT5 is a critical regulator of cellular proliferation, apoptosis, migration, cell cycle progression, cell metabolism, DNA damage response, and cell death [[49], [50], [51], [52], [53], [54]].
PRMT5 was found to be an essential protein involved in DDR and identified as a master epigenetic activator of DSB repair through HR and NHEJ [29,51,55,56]. Additionally, it was previously found that PRMT5 regulated DNA repair by controlling the alternative splicing of histone-modifying enzymes [57], and SDMA of histone H4 (producing H4R3me2s) serves as a bridge between DNA damage and subsequent repair upon oxidative stress [58]. H4R3me2s can be recognized as a reader protein that senses DNA damage and a writer protein that promotes DNA repair. We revealed that ESCC cells with a decreased level of H4R3me2s induced by inhibiting STC2 were more susceptible to IR. Meanwhile, inhibition of PRMT5 can increase the intracellular ROS and decrease mRNA expression levels of SLC7A11 and SLC3A2 [30]. PRMT5 has been reported as a regulator of ATF4, and ATF4 can promote ferroptosis by mediating SLC7A11 [30,59,60]. We confirmed that PRMT5 activity can regulate SLC7A11 and SLC3A2 through ATF4, determining the role of PRMT5-ATF4-SLC3A2/SLC7A11 axis in ferroptosis of ESCC. Combining the above results that STC2 can interact with PRMT5, we speculated if PRMT5 participates in STC2-mediated ferroptosis. We also found that regulated PRMT5 inhibited the expression of SLC7A11 in STC2-overexpressed cells, resulting in the accumulation of lipid peroxidation and ferroptosis. The documented evidence and our results [47,63] indicate that inhibition of PRMT5-ATF4-SLC3A2/SLC7A11 axis regulated by STC2 leads to radiosensitization in ESCC.
Several PRMT5 inhibitors have been used as potential novel therapeutic approaches in clinical trials [[61], [62], [63]]. GSK3326595, a typical PRMT5 inhibitor, was first applied in ESCC cells. We observed that introducing the GSK3326595 can promote DDR, inhibit ferroptosis, sensitize ESCC cells to radiation, and even reverse the radioresistance of STC2-overexpressed cells. The efficacy and safety of GSK3326595 were also verified through in vivo assays. Our results revealed that due to the lack of a specially targeted molecule to STC2, inhibition of PRMT5 might be a potential therapeutic method in STC2-overexpressed ESCC patients.
In conclusion, STC2 activates PRMT5 to induce radioresistance in ESCC via promoting DDR and inhibiting ferroptosis. Our research suggests that STC2 might be an attractive therapeutic target to overcome ESCC radioresistance.
4. Materials and methods
4.1. Cell lines and clinical specimens
The ESCC cell lines (KYSE150, KYSE410, KYSE70, KYSE510, KYSE450, EC1, TE7) were obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, Braunschweig, Germany). TE1 cells were purchased from the Cell Bank of the Chinese Academy (Shanghai, China). All ESCC cells were cultured in RPMI-1640 supplemented with 10% FBS and 100IU/ml Penicillin-Streptomycin solution at 37 °C in 5% CO2. TE1R and KYSE150R were previously established in our laboratory [28].
Twenty-four pairs of ESCC tumor samples and adjacent nontumor tissues were collected from patients who had undergone surgery and were histologically diagnosed at the First Affiliated Hospital of the Zhejiang University of China.
4.2. Cell and tumor irradiation
Cells were irradiated with a Precision X-RAD 225 machine operating at 225 kV and 13.3 mA with a 2-mm Al filter (source-to-skin distance: 36 cm; dose rate: 1.3 Gy/min). For in vivo, mice were irradiated with a total dose of 20 Gy (10 Gy*2) using an X- RAD225 small animal irradiator with a 0.95 Gy/min dose rate.
4.3. Xenograft tumor model
Four-to six-week-old BALB/c female nude mice were allocated into our study and injected with 5*106 stable cells (Mock, OE-STC2). All these mice were randomly distributed into three groups: ctrl, IR, and IR combined with inhibitor. Following treatments were performed when the tumors reached an average volume of 80 mm3. GSK3326595(50 mg/kg) was administered to nude mice every two days by oral gavage, and 10Gy IR was delivered on the second and the 8th day after medication. The tumor volume was measured every three days using a standard formula: length × width2/2. All animal experiments were approved by the Tab of Animal Experimental Ethical Inspection of the First Affiliated Hospital, Zhejiang University School of Medicine.
4.4. RNA sequencing analysis and RT-qPCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen). RNA sequencing was performed by Lc-bio Technologies (Hangzhou, China), including RNA quantification, library preparation, clustering, and sequencing. Differentially expressed genes between groups were analyzed by R package edgeR according to the criteria with corrected P-values of 0.05 and absolute fold-changes of 2. According to the manufacturer's protocol, reverse transcription of total RNA to cDNA was carried out using qPCR RT Master Mix (Takara, Japan). The relative expression levels of mRNA were determined by the △Ct method using the SYBR Green qPCR Kit (Takara, Japan). The primers for STC2 were: forward, 5′-GGGAATGCTACCTCAAGCAC-3′ and reverse, 5′-GGTCCACGTAGGGTTCGT-3′.
4.5. WB analysis
Cells were lysed in denaturing lysis buffer (RIPA buffer) and then measured protein concentrations using Bicinchoninic Acid protein assays kit (Thermo, USA). Proteins were boiled in 1X SDS loading buffer. Total protein was loaded and separated by SDS-PAGE gel and then transferred on polyvinylidene fluoride membranes (Millipore, USA). After blocking with 5% skim milk diluted by 0.05% Tris-buffered saline/Tween, membranes were incubated overnight at 4 °C with primary antibodies. Following incubation with HRP-conjugated secondary antibodies, the membranes were detected by the ChemiScopeTouch series fluorescence and chemiluminescence imaging system (CLINX, Shanghai, China). The detailed information on relevant antibodies is listed in Supplementary Table S1.
4.6. Establishment of stable cell lines
Lentivirus-containing STC2 and ATF4 or shRNA targeting STC2 were purchased from GeneChem Company (Shanghai, China). Lentiviruses were transfected into cells according to the manufacturer's protocol. Stable cells were selected based on antibiotic resistance with 2 μg/ml puromycin (Solarbio, Beijing, China), and verified by WB. The following shRNAs against STC2 were used:
shSTC2 #1: 5′-GUGGAGAUGAUCCAUUUCATT-3′
shSTC2 #2: 5′-GACGAACAGTCTGAGTATTCT-3′
4.7. Colony formation assay
Cells (500-10000/well) were seeded into 6-well plates and exposed to different doses of IR (0, 2, 4, and 8 Gy) the next day. After being cultured for about 10–14 days, plates were fixed with methanol for 10 min and stained with crystal violet for 30 min. Then the number of colonies was calculated and analyzed by image J software. The determined value and surviving fraction were obtained by Prism 9 software according to the single-hit multi-target model (Y = 1-(1-exp(−n*X))^m).
4.8. Cell viability assay
The cells (2500/well) were seeded in 96-well plates and cultured for 24 h, 48 h, and 72 h, respectively. 10 μL CCK8 reagent (NCM Biotech, Suzhou, China) was added to each well and incubated for 1 h to measure cell viability. We measured the absorbance values at 450 nm with a microplate reader (SpectraMax i3x, USA).
4.9. MS and co-IP assay
Cells were washed with phosphate buffered saline (PBS) and lysed with IP lysis buffer (Epizyme Biotech, Shanghai, China). For the IP assay, Protein G Magnetic Beads (Bio-rad, USA) were washed with IP buffer and crosslinked to the antibodies with rotation for 1 h at room temperature. Then, a portion of the lysate was used as input, while the other part was incubated with bead-antibody complex overnight at 4 °C. The next day, immunocomplexes were separated from the beads using a magnetic rack and then boiled for 10 min. Finally, purified protein complexes were separated by SDS–PAGE and stained by colloidal Coomassie Blue (Epizyme Biotech, Shanghai, China). The stained gel slices were excised for the subsequent MS analysis in Nanjing jiangbei new area biopharmaceutical public service platform (Nanjing, China). IgG (Cell Signaling Technology, Danvers, MA) was used as a negative control. The proteins of interest in the Co-IP were detected by WB assay.
4.10. Comet assay
Comet assay was performed using reagents from the Trevigen Comet Assay kit (Bio-Techne, USA) according to the manufacturer's instructions. In brief, cells mixed with comet low melting agarose gel at the proper proportion and spread onto comet slides. Then the slides were subjected to electrophoresis at 21 V for 25 min after immersing into a series of the buffer. We finally stained the slides with SYBR Green, and images were captured with Olympus BX53 inverted microscope (Tokyo, Japan). The quantitation of tail moments was analyzed by CASP Software, and at least 20 cells were measured in each group.
4.11. IF
Cells were plated on glass slides that were put in 24-well plates. After treatment, cells were washed three times with PBS and fixed with 4% paraformaldehyde for 20 min. Then cells were permeabilized with 0.5% Triton-X-100 in PBS for 20 min and blocked with 5% BSA in PBS for 1h. Further, we use an antibody against γH2AX to incubate cells overnight at 4 °C, followed by a fluorescence secondary antibody and an anti-fluorescence quenching agent with DAPI (Solarbio, Beijing, China). At last, the images were captured by Olympus FV3000 confocal microscope.
4.12. Flow cytometry
The level of Lipid ROS and intracellular total ROS were detected by C11-BODIPY 581/591 (Thermo Fisher Scientific) and 2′,7′-Dichlorofluorescin diacetate (DCFH-DA) (Beyotime, China) fluorescent probe respectively. Add BODIPY-C11 to the culture medium at a final concentration of 5 μM, and the final concentration of DCFH-DA is 10 μM. These cells were incubated for 30 min inside the incubator at 37 °C. Cells were washed twice before detection by flow cytometry.
4.13. Measurement of intracellular Fe2+ content
The level of intracellular Fe2+ ions was measured with a Ferro-orange kit (Dojingo, Japan) according to the manufacturer's instructions. Cells were plated on laser confocal dishes and exposed to the indicated treatments. After treatment, FerroOrange (1:1000) was added to the cells, and the cells were incubated at 37 °C for 30 min. All images were acquired using a fluorescence confocal microscope.
4.14. Transwell migration assay
After transfection, cells were selected for transwell assay. The upper chamber was supplemented with 200 μL serum-free medium containing 1 × 105 cells, and the lower chamber was supplemented with 600 μL medium containing 10% FBS. After 24 h of incubation at 37 °C, chambers were washed with PBS and fixed with 4% paraformaldehyde for 20 min, then stained with 0.4% crystal violet. Images were captured in a microscope (Olympus, Japan) with 200 × magnification. Each experiment was conducted three times.
4.15. Wound healing assay
Cells were cultured in a 6-well plate after growing to 100% density. A 10-μL pipette tip was used to scratch the plate, then washed the plate with PBS. Cells cultured in serum-free medium, after 0 h, 24 h, and 48 h, plates were captured images with a microscope (Olympus, Japan) with 200 × magnification. Each experiment was conducted three times.
4.16. GSH and MDA assays
Cells were treated with radiation for 24 h, then collected cell for quantitative detection of GSH and MDA. The glutathione assay kit (Nanjing Jiancheng, China) and MDA assay kit (Dojindo, Japan) were used according to the manufacturer's instructions.
4.17. Transmission electron microscope
Cells treated with radiation were collected and fixed by 2.5% glutaraldehyde for 24 h at 4 °C. Then, the samples were washed three times with PBS, and 1% osmium tetroxide was used to fix them for 2 h. Samples were dehydrated in a graded series of ethanol and acetone, followed by embedding in Spurr resin. Ultrathin sections were obtained and stained with uranyl acetate and lead citrate. Images were captured by Tecnai G2 (120 kV) transmission electron microscope (FEI, USA).
4.18. IHC
Tissues were formalin-fixed and paraffin-embedded, and tissues were sliced into 4 μm thick sections. After dewaxing and dehydration, slides were treated with antigen retrieval solution. Then the slides were incubated with H2O2 and goat serum. Slides were incubated with primary antibody at 4 °C overnight. After incubation with a secondary biotinylated antibody and DAB stain, the slides were scanned by a Motic EasyScan (Motic, USA).
4.19. Statistical analysis
All statistical analyses were conducted using SPSS 23.0 (SPSS Inc., Chicago, USA) and GraphPad Prism 9.0 (GraphPad Software Inc, USA). Data are presented as the means ± standard deviation. P values < 0.05 indicate statistical significance. Each experiment was replicated thrice.
Funding
This study was supported by grants from the Fundamental Research Funds for the Central Universities (Grant No.2021FZZX005-32), the Natural Science Foundation of Zhejiang Province of China (Grant No.LSY19H160004, Grant No.LQ23H160024), the Key Research and Development Projects of Zhejiang Provincial Science and Technology Department (2021C03122).
Authors' contributions
SXY and KJ designed the experiments. KJ, XY, and QYZ performed most of the experiments and wrote the manuscript. JY, QYT, MYX, LYW and YFS participated in part of the experiments and helped to draft the manuscript. ZYZ and HY carried out the in vivo studies. All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
We thank the excellent technical assistant of microscopy core facility, Central Laboratory, the First Affiliated Hospital, Zhejiang University School of Medicine with the confocal laser microscopy.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102626.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Fig. S1.
Fig. S2.
Fig. S3.
Fig. S4.
Fig. S5.
Data availability
Data will be made available on request.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA A Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Zhou M., Wang H., Zeng X., Yin P., Zhu J., Chen W., Li X., Wang L., Wang L., Liu Y., Liu J., Zhang M., Qi J., Yu S., Afshin A., Gakidou E., Glenn S., Krish V.S., Miller-Petrie M.K., Mountjoy-Venning W.C., Mullany E.C., Redford S.B., Liu H., Naghavi M., Hay S.I., Wang L., Murray C.J.L., Liang X. Mortality, morbidity, and risk factors in China and its provinces, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2019;394:1145–1158. doi: 10.1016/S0140-6736(19)30427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eyck B.M., van Lanschot J.J.B., Hulshof M., van der Wilk B.J., Shapiro J., van Hagen P., van Berge Henegouwen M.I., Wijnhoven B.P.L., van Laarhoven H.W.M., Nieuwenhuijzen G.A.P., Hospers G.A.P., Bonenkamp J.J., Cuesta M.A., Blaisse R.J.B., Busch O.R., Creemers G.M., Punt C.J.A., Plukker J.T.M., Verheul H.M.W., Spillenaar Bilgen E.J., van der Sangen M.J.C., Rozema T., Ten Kate F.J.W., Beukema J.C., Piet A.H.M., van Rij C.M., Reinders J.G., Tilanus H.W., Steyerberg E.W., van der Gaast A., Group C.S. Ten-year outcome of neoadjuvant chemoradiotherapy plus surgery for esophageal cancer: the randomized controlled CROSS trial. J. Clin. Oncol. 2021;39:1995–2004. doi: 10.1200/JCO.20.03614. [DOI] [PubMed] [Google Scholar]
- 5.van Hagen P., Hulshof M.C., van Lanschot J.J., Steyerberg E.W., van Berge Henegouwen M.I., Wijnhoven B.P., Richel D.J., Nieuwenhuijzen G.A., Hospers G.A., Bonenkamp J.J., Cuesta M.A., Blaisse R.J., Busch O.R., ten Kate F.J., Creemers G.J., Punt C.J., Plukker J.T., Verheul H.M., Spillenaar Bilgen E.J., van Dekken H., van der Sangen M.J., Rozema T., Biermann K., Beukema J.C., Piet A.H., van Rij C.M., Reinders J.G., Tilanus H.W., van der Gaast A., Group C. Preoperative chemoradiotherapy for esophageal or junctional cancer. N. Engl. J. Med. 2012;366:2074–2084. doi: 10.1056/NEJMoa1112088. [DOI] [PubMed] [Google Scholar]
- 6.Santivasi W.L., Xia F. Ionizing radiation-induced DNA damage, response, and repair. Antioxidants Redox Signal. 2014;21:251–259. doi: 10.1089/ars.2013.5668. [DOI] [PubMed] [Google Scholar]
- 7.Huang R.X., Zhou P.K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Targeted Ther. 2020;5:60. doi: 10.1038/s41392-020-0150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mamo T., Mladek A.C., Shogren K.L., Gustafson C., Gupta S.K., Riester S.M., Maran A., Galindo M., van Wijnen A.J., Sarkaria J.N., Yaszemski M.J. Inhibiting DNA-PKCS radiosensitizes human osteosarcoma cells. Biochem. Biophys. Res. Commun. 2017;486:307–313. doi: 10.1016/j.bbrc.2017.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willoughby C.E., Jiang Y., Thomas H.D., Willmore E., Kyle S., Wittner A., Phillips N., Zhao Y., Tudhope S.J., Prendergast L., Junge G., Lourenco L.M., Finlay M.R.V., Turner P., Munck J.M., Griffin R.J., Rennison T., Pickles J., Cano C., Newell D.R., Reeves H.L., Ryan A.J., Wedge S.R. Selective DNA-PKcs inhibition extends the therapeutic index of localized radiotherapy and chemotherapy. J. Clin. Invest. 2020;130:258–271. doi: 10.1172/JCI127483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryu H., Kim H.J., Song J.Y., Hwang S.G., Kim J.S., Kim J., Bui T.H.N., Choi H.K., Ahn J. A small compound KJ-28d enhances the sensitivity of non-small cell lung cancer to radio- and chemotherapy. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20236026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirai T., Saito S., Fujimori H., Matsushita K., Nishio T., Okayasu R., Masutani M. Radiosensitization by PARP inhibition to proton beam irradiation in cancer cells. Biochem. Biophys. Res. Commun. 2016;478:234–240. doi: 10.1016/j.bbrc.2016.07.062. [DOI] [PubMed] [Google Scholar]
- 12.Qin C., Ji Z., Zhai E., Xu K., Zhang Y., Li Q., Jing H., Wang X., Song X. PARP inhibitor olaparib enhances the efficacy of radiotherapy on XRCC2-deficient colorectal cancer cells. Cell Death Dis. 2022;13:505. doi: 10.1038/s41419-022-04967-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lourenco L.M., Jiang Y., Drobnitzky N., Green M., Cahill F., Patel A., Shanneik Y., Moore J., Ryan A.J. PARP inhibition combined with thoracic irradiation exacerbates esophageal and skin toxicity in C57BL6 mice. Int. J. Radiat. Oncol. Biol. Phys. 2018;100:767–775. doi: 10.1016/j.ijrobp.2017.10.051. [DOI] [PubMed] [Google Scholar]
- 14.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S., Morrison B., 3rd, Stockwell B.R. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye L.F., Chaudhary K.R., Zandkarimi F., Harken A.D., Kinslow C.J., Upadhyayula P.S., Dovas A., Higgins D.M., Tan H., Zhang Y., Buonanno M., Wang T.J.C., Hei T.K., Bruce J.N., Canoll P.D., Cheng S.K., Stockwell B.R. Radiation-induced lipid peroxidation triggers ferroptosis and synergizes with ferroptosis inducers. ACS Chem. Biol. 2020;15:469–484. doi: 10.1021/acschembio.9b00939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei G., Zhang Y., Koppula P., Liu X., Zhang J., Lin S.H., Ajani J.A., Xiao Q., Liao Z., Wang H., Gan B. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res. 2020;30:146–162. doi: 10.1038/s41422-019-0263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lang X., Green M.D., Wang W., Yu J., Choi J.E., Jiang L., Liao P., Zhou J., Zhang Q., Dow A., Saripalli A.L., Kryczek I., Wei S., Szeliga W., Vatan L., Stone E.M., Georgiou G., Cieslik M., Wahl D.R., Morgan M.A., Chinnaiyan A.M., Lawrence T.S., Zou W. Radiotherapy and immunotherapy promote tumoral lipid oxidation and ferroptosis via synergistic repression of SLC7A11. Cancer Discov. 2019;9:1673–1685. doi: 10.1158/2159-8290.CD-19-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCudden C.R., James K.A., Hasilo C., Wagner G.F. Characterization of mammalian stanniocalcin receptors. Mitochondrial targeting of ligand and receptor for regulation of cellular metabolism. J. Biol. Chem. 2002;277:45249–45258. doi: 10.1074/jbc.M205954200. [DOI] [PubMed] [Google Scholar]
- 19.Li S., Huang Q., Li D., Lv L., Li Y., Wu Z. The significance of Stanniocalcin 2 in malignancies and mechanisms. Bioengineered. 2021;12:7276–7285. doi: 10.1080/21655979.2021.1977551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ieta K., Tanaka F., Yokobori T., Kita Y., Haraguchi N., Mimori K., Kato H., Asao T., Inoue H., Kuwano H., Mori M. Clinicopathological significance of stanniocalcin 2 gene expression in colorectal cancer. Int. J. Cancer. 2009;125:926–931. doi: 10.1002/ijc.24453. [DOI] [PubMed] [Google Scholar]
- 21.Hashemzadeh S., Arabzadeh A.A., Estiar M.A., Sakhinia M., Mesbahi N., Emrahi L., Ghojazadeh M., Sakhinia E. Clinical utility of measuring expression levels of Stanniocalcin 2 in patients with colorectal cancer. Med. Oncol. 2014;31:237. doi: 10.1007/s12032-014-0237-8. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., Gao Y., Cheng H., Yang G., Tan W. Stanniocalcin 2 promotes cell proliferation and cisplatin resistance in cervical cancer. Biochem. Biophys. Res. Commun. 2015;466:362–368. doi: 10.1016/j.bbrc.2015.09.029. [DOI] [PubMed] [Google Scholar]
- 23.Li J., Zhang Z., Feng X., Shen Z., Sun J., Zhang X., Bu F., Xu M., Tan C., Wang Z. Stanniocalcin-2 promotes cell EMT and glycolysis via activating ITGB2/FAK/SOX6 signaling pathway in nasopharyngeal carcinoma. Cell Biol. Toxicol. 2022;38:259–272. doi: 10.1007/s10565-021-09600-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He H., Qie S., Guo Q., Chen S., Zou C., Lu T., Su Y., Zong J., Xu H., He D., Xu Y., Chen B., Pan J., Sang N., Lin S. Stanniocalcin 2 (STC2) expression promotes post-radiation survival, migration and invasion of nasopharyngeal carcinoma cells. Cancer Manag. Res. 2019;11:6411–6424. doi: 10.2147/CMAR.S197607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu F., Li T.Y., Su S.C., Yu J.S., Zhang H.L., Tan G.Q., Liu J.W., Wang B.L. STC2 as a novel mediator for Mus81-dependent proliferation and survival in hepatocellular carcinoma. Cancer Lett. 2017;388:177–186. doi: 10.1016/j.canlet.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 26.Wang H., Wu K., Sun Y., Li Y., Wu M., Qiao Q., Wei Y., Han Z.G., Cai B. STC2 is upregulated in hepatocellular carcinoma and promotes cell proliferation and migration in vitro. BMB Rep. 2012;45:629–634. doi: 10.5483/BMBRep.2012.45.11.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kita Y., Mimori K., Iwatsuki M., Yokobori T., Ieta K., Tanaka F., Ishii H., Okumura H., Natsugoe S., Mori M. STC2: a predictive marker for lymph node metastasis in esophageal squamous-cell carcinoma. Ann. Surg Oncol. 2011;18:261–272. doi: 10.1245/s10434-010-1271-1. [DOI] [PubMed] [Google Scholar]
- 28.Tang Q., Wu L., Xu M., Yan D., Shao J., Yan S. Osalmid, a novel identified RRM2 inhibitor, enhances radiosensitivity of esophageal cancer. Int. J. Radiat. Oncol. Biol. Phys. 2020;108:1368–1379. doi: 10.1016/j.ijrobp.2020.07.2322. [DOI] [PubMed] [Google Scholar]
- 29.Owens J.L., Beketova E., Liu S., Tinsley S.L., Asberry A.M., Deng X., Huang J., Li C., Wan J., Hu C.D. PRMT5 cooperates with pICln to function as a master epigenetic activator of DNA double-strand break repair genes. iScience. 2020;23 doi: 10.1016/j.isci.2019.100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szewczyk M.M., Luciani G.M., Vu V., Murison A., Dilworth D., Barghout S.H., Lupien M., Arrowsmith C.H., Minden M.D., Barsyte-Lovejoy D. PRMT5 regulates ATF4 transcript splicing and oxidative stress response. Redox Biol. 2022;51 doi: 10.1016/j.redox.2022.102282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y.N., Tsai M.F., Wu S.G., Chang T.H., Tsai T.H., Gow C.H., Chang Y.L., Shih J.Y. Acquired resistance to EGFR tyrosine kinase inhibitors is mediated by the reactivation of STC2/JUN/AXL signaling in lung cancer. Int. J. Cancer. 2019;145:1609–1624. doi: 10.1002/ijc.32487. [DOI] [PubMed] [Google Scholar]
- 32.Roche F.P., Pietila I., Kaito H., Sjostrom E.O., Sobotzki N., Noguer O., Skare T.P., Essand M., Wollscheid B., Welsh M., Claesson-Welsh L. Leukocyte differentiation by histidine-rich glycoprotein/stanniocalcin-2 complex regulates murine glioma growth through modulation of antitumor immunity. Mol. Cancer Therapeut. 2018;17:1961–1972. doi: 10.1158/1535-7163.MCT-18-0097. [DOI] [PubMed] [Google Scholar]
- 33.Zhu C., Xia Q., Gu B., Cui M., Zhang X., Yan W., Meng D., Shen S., Xie S., Li X., Jin H., Wang S. Esophageal cancer associated immune genes as biomarkers for predicting outcome in upper gastrointestinal tumors. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.707299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z., Chen C., Fang Y., Li S., Wang X., Sun L., Zhou G., Ye J. Development of a prognostic signature for esophageal cancer based on nine immune related genes. BMC Cancer. 2021;21:113. doi: 10.1186/s12885-021-07813-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kashyap M.K., Pawar H.A., Keerthikumar S., Sharma J., Goel R., Mahmood R., Kumar M.V., Kumar K.V., Pandey A., Kumar R.V., Prasad T.S., Harsha H.C. Evaluation of protein expression pattern of stanniocalcin 2, insulin-like growth factor-binding protein 7, inhibin beta A and four and a half LIM domains 1 in esophageal squamous cell carcinoma. Cancer Biomarkers. 2012;12:1–9. doi: 10.3233/CBM-120289. [DOI] [PubMed] [Google Scholar]
- 36.Liu W., Miao C., Zhang S., Liu Y., Niu X., Xi Y., Guo W., Chu J., Lin A., Liu H., Yang X., Chen X., Zhong C., Ma Y., Wang Y., Zhu S., Liu S., Tan W., Lin D., Wu C. VAV2 is required for DNA repair and implicated in cancer radiotherapy resistance. Signal Transduct. Targeted Ther. 2021;6:2322. doi: 10.1038/s41392-021-00735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li M.Y., Fan L.N., Han D.H., Yu Z., Ma J., Liu Y.X., Li P.F., Zhao D.H., Chai J., Jiang L., Li S.L., Xiao J.J., Duan Q.H., Ye J., Shi M., Nie Y.Z., Wu K.C., Liao D.J., Shi Y., Wang Y., Yan Q.G., Guo S.P., Bian X.W., Zhu F., Zhang J., Wang Z. Ribosomal S6 protein kinase 4 promotes radioresistance in esophageal squamous cell carcinoma. J. Clin. Invest. 2020;130:4301–4319. doi: 10.1172/JCI134930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y., Wang J., Ma Y., Li J., Sun X., Zhao X., Shi X., Hu Y., Qu F., Zhang X. Radiation induces NORAD expression to promote ESCC radiotherapy resistance via EEPD1/ATR/Chk1 signalling and by inhibiting pri-miR-199a1 processing and the exosomal transfer of miR-199a-5p. J. Exp. Clin. Cancer Res. 2021;40:306. doi: 10.1186/s13046-021-02084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lei G., Mao C., Yan Y., Zhuang L., Gan B., Ferroptosis radiotherapy, and combination therapeutic strategies. Protein Cell. 2021;12:836–857. doi: 10.1007/s13238-021-00841-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng L., Zhao K., Sun L., Yin X., Zhang J., Liu C., Li B. SLC7A11 regulated by NRF2 modulates esophageal squamous cell carcinoma radiosensitivity by inhibiting ferroptosis. J. Transl. Med. 2021;19:367. doi: 10.1186/s12967-021-03042-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sengupta S., Kennemer A., Patrick K., Tsichlis P., Guerau-de-Arellano M. Protein arginine methyltransferase 5 in T lymphocyte biology. Trends Immunol. 2020;41:918–931. doi: 10.1016/j.it.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y., Shao X., Zhao X., Ji Y., Liu X., Li P., Zhang M., Wang Q. Targeting protein arginine methyltransferase 5 in cancers: roles, inhibitors and mechanisms. Biomed. Pharmacother. 2021;144 doi: 10.1016/j.biopha.2021.112252. [DOI] [PubMed] [Google Scholar]
- 43.Stopa N., Krebs J.E., Shechter D. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell. Mol. Life Sci. 2015;72:2041–2059. doi: 10.1007/s00018-015-1847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krzyzanowski A., Gasper R., Adihou H., Hart P., Waldmann H. Biochemical investigation of the interaction of pICln, RioK1 and COPR5 with the PRMT5-MEP50 complex. Chembiochem. 2021;22:1908–1914. doi: 10.1002/cbic.202100079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang X., Zeng Z., Jie X., Wang Y., Han J., Zheng Z., Li J., Liu H., Dong X., Wu G., Xu S. Arginine methyltransferase PRMT5 methylates and destabilizes Mxi1 to confer radioresistance in non-small cell lung cancer. Cancer Lett. 2022;532 doi: 10.1016/j.canlet.2022.215594. [DOI] [PubMed] [Google Scholar]
- 46.Liu S., Liu Z., Piao C., Zhang Z., Kong C., Yin L., Liu X. Flavokawain A is a natural inhibitor of PRMT5 in bladder cancer. J. Exp. Clin. Cancer Res. 2022;41:2293. doi: 10.1186/s13046-022-02500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y.R., Li H.N., Zhang L.J., Zhang C., He J.G. Protein arginine methyltransferase 5 promotes esophageal squamous cell carcinoma proliferation and metastasis via LKB1/AMPK/mTOR signaling pathway. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.645375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang L., Zhang X.O., Rozen E.J., Sun X., Sallis B., Verdejo-Torres O., Wigglesworth K., Moon D., Huang T., Cavaretta J.P., Wang G., Zhang L., Shohet J.M., Lee M.M., Wu Q. PRMT5 activates AKT via methylation to promote tumor metastasis. Nat. Commun. 2022;13:3955. doi: 10.1038/s41467-022-31645-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beketova E., Fang S., Owens J.L., Liu S., Chen X., Zhang Q., Asberry A.M., Deng X., Malola J., Huang J., Li C., Pili R., Elzey B.D., Ratliff T.L., Wan J., Hu C.D. Protein arginine methyltransferase 5 promotes pICln-dependent androgen receptor transcription in castration-resistant prostate cancer. Cancer Res. 2020;80:4904–4917. doi: 10.1158/0008-5472.CAN-20-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rengasamy M., Zhang F., Vashisht A., Song W.M., Aguilo F., Sun Y., Li S., Zhang W., Zhang B., Wohlschlegel J.A., Walsh M.J. The PRMT5/WDR77 complex regulates alternative splicing through ZNF326 in breast cancer. Nucleic Acids Res. 2017;45:11106–11120. doi: 10.1093/nar/gkx727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clarke T.L., Sanchez-Bailon M.P., Chiang K., Reynolds J.J., Herrero-Ruiz J., Bandeiras T.M., Matias P.M., Maslen S.L., Skehel J.M., Stewart G.S., Davies C.C. PRMT5-Dependent methylation of the TIP60 coactivator RUVBL1 is a Key regulator of homologous recombination. Mol. Cell. 2017;65 doi: 10.1016/j.molcel.2017.01.019. 900-916 e907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai W.W., Niessen S., Goebel N., Yates J.R., 3rd, Guccione E., Montminy M. PRMT5 modulates the metabolic response to fasting signals. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8870–8875. doi: 10.1073/pnas.1304602110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kryukov G.V., Wilson F.H., Ruth J.R., Paulk J., Tsherniak A., Marlow S.E., Vazquez F., Weir B.A., Fitzgerald M.E., Tanaka M., Bielski C.M., Scott J.M., Dennis C., Cowley G.S., Boehm J.S., Root D.E., Golub T.R., Clish C.B., Bradner J.E., Hahn W.C., Garraway L.A. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science. 2016;351:1214–1218. doi: 10.1126/science.aad5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mavrakis K.J., McDonald E.R., 3rd, Schlabach M.R., Billy E., Hoffman G.R., deWeck A., Ruddy D.A., Venkatesan K., Yu J., McAllister G., Stump M., deBeaumont R., Ho S., Yue Y., Liu Y., Yan-Neale Y., Yang G., Lin F., Yin H., Gao H., Kipp D.R., Zhao S., McNamara J.T., Sprague E.R., Zheng B., Lin Y., Cho Y.S., Gu J., Crawford K., Ciccone D., Vitari A.C., Lai A., Capka V., Hurov K., Porter J.A., Tallarico J., Mickanin C., Lees E., Pagliarini R., Keen N., Schmelzle T., Hofmann F., Stegmeier F., Sellers W.R. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science. 2016;351:1208–1213. doi: 10.1126/science.aad5944. [DOI] [PubMed] [Google Scholar]
- 55.Wei X., Yang J., Adair S.J., Ozturk H., Kuscu C., Lee K.Y., Kane W.J., O'Hara P.E., Liu D., Demirlenk Y.M., Habieb A.H., Yilmaz E., Dutta A., Bauer T.W., Adli M. Targeted CRISPR screening identifies PRMT5 as synthetic lethality combinatorial target with gemcitabine in pancreatic cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2020;117:28068–28079. doi: 10.1073/pnas.2009899117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hwang J.W., Kim S.N., Myung N., Song D., Han G., Bae G.U., Bedford M.T., Kim Y.K. PRMT5 promotes DNA repair through methylation of 53BP1 and is regulated by Src-mediated phosphorylation. Commun Biol. 2020;3:428. doi: 10.1038/s42003-020-01157-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hamard P.J., Santiago G.E., Liu F., Karl D.L., Martinez C., Man N., Mookhtiar A.K., Duffort S., Greenblatt S., Verdun R.E., Nimer S.D. PRMT5 regulates DNA repair by controlling the alternative splicing of histone-modifying enzymes. Cell Rep. 2018;24:2643–2657. doi: 10.1016/j.celrep.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma Z., Wang W., Wang S., Zhao X., Ma Y., Wu C., Hu Z., He L., Pan F., Guo Z. Symmetrical dimethylation of H4R3: a bridge linking DNA damage and repair upon oxidative stress. Redox Biol. 2020;37 doi: 10.1016/j.redox.2020.101653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao R., Kalathur R.K.R., Coto-Llerena M., Ercan C., Buechel D., Shuang S., Piscuoglio S., Dill M.T., Camargo F.D., Christofori G., Tang F. YAP/TAZ and ATF4 drive resistance to Sorafenib in hepatocellular carcinoma by preventing ferroptosis. EMBO Mol. Med. 2021;13 doi: 10.15252/emmm.202114351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen D., Fan Z., Rauh M., Buchfelder M., Eyupoglu I.Y., Savaskan N. ATF4 promotes angiogenesis and neuronal cell death and confers ferroptosis in a xCT-dependent manner. Oncogene. 2017;36:5593–5608. doi: 10.1038/onc.2017.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan-Penebre E., Kuplast K.G., Majer C.R., Boriack-Sjodin P.A., Wigle T.J., Johnston L.D., Rioux N., Munchhof M.J., Jin L., Jacques S.L., West K.A., Lingaraj T., Stickland K., Ribich S.A., Raimondi A., Scott M.P., Waters N.J., Pollock R.M., Smith J.J., Barbash O., Pappalardi M., Ho T.F., Nurse K., Oza K.P., Gallagher K.T., Kruger R., Moyer M.P., Copeland R.A., Chesworth R., Duncan K.W. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol. 2015;11:432–437. doi: 10.1038/nchembio.1810. [DOI] [PubMed] [Google Scholar]
- 62.Bonday Z.Q., Cortez G.S., Grogan M.J., Antonysamy S., Weichert K., Bocchinfuso W.P., Li F., Kennedy S., Li B., Mader M.M., Arrowsmith C.H., Brown P.J., Eram M.S., Szewczyk M.M., Barsyte-Lovejoy D., Vedadi M., Guccione E., Campbell R.M. LLY-283, a potent and selective inhibitor of arginine methyltransferase 5, PRMT5, with antitumor activity. ACS Med. Chem. Lett. 2018;9:612–617. doi: 10.1021/acsmedchemlett.8b00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan Y., Nie H. Protein arginine methyltransferase 5: a potential cancer therapeutic target. Cell. Oncol. 2021;44:33–44. doi: 10.1007/s13402-020-00577-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.