A platform combining stimulated Raman scattering microscopy and a convolutional neural network provides rapid histopathology and automated Gleason scoring on fresh prostate core needle biopsies without complex tissue processing.
Abstract
Focal therapy (FT) has been proposed as an approach to eradicate clinically significant prostate cancer while preserving the normal surrounding tissues to minimize treatment-related toxicity. Rapid histology of core needle biopsies is essential to ensure the precise FT for localized lesions and to determine tumor grades. However, it is difficult to achieve both high accuracy and speed with currently available histopathology methods. Here, we demonstrated that stimulated Raman scattering (SRS) microscopy could reveal the largely heterogeneous histologic features of fresh prostatic biopsy tissues in a label-free and near real-time manner. A diagnostic convolutional neural network (CNN) built based on images from 61 patients could classify Gleason patterns of prostate cancer with an accuracy of 85.7%. An additional 22 independent cases introduced as external test dataset validated the CNN performance with 84.4% accuracy. Gleason scores of core needle biopsies from 21 cases were calculated using the deep learning SRS system and showed a 71% diagnostic consistency with grading from three pathologists. This study demonstrates the potential of a deep learning–assisted SRS platform in evaluating the tumor grade of prostate cancer, which could help simplify the diagnostic workflow and provide timely histopathology compatible with FT treatment.
Significance:
A platform combining stimulated Raman scattering microscopy and a convolutional neural network provides rapid histopathology and automated Gleason scoring on fresh prostate core needle biopsies without complex tissue processing.
Introduction
Prostate cancer is the second most diagnosed cancer and the fifth leading cause of cancer-related deaths among men worldwide (1). Over the past decades, focal therapy (FT) has been an emerging novel option for selected patients with clinically localized cancer like prostate cancer, aiming to eradicate the targeted lesion while preserving the surrounding key functional structures (2–5) However, many urologists are concerned that FT could fail in cancer control for the inability of imaging biopsy to detect multifocal lesions in prostate cancer and the inaccuracy of tumor location during operation (3). The main concern was derived from the incomplete tumor ablation due to the lack of real-time intraoperative pathologic guidance. The conventional intraoperative frozen section–based workflow requires highly effective cooperation during tissue transport and slide processing, which is not only time consuming, but also lacks the diagnostic accuracy for actual needs (6, 7). Therefore, developing a real-time solution that could collect histologic images from fresh unprocessed tissues for grading prostate cancer with high accuracy is critically important for FT in oncological control.
As a novel chemical imaging technology, stimulated Raman scattering (SRS) microscopy is capable of providing histologic images of tissue specimens in a label-free manner with submicrometer spatial resolution (8, 9). SRS images produce chemical contrast based on the intrinsic vibrational spectroscopy of biomolecules (such as lipid and protein). As a result, SRS could reveal essential histologic features in near-perfect agreement with traditional hematoxylin and eosin (H&E)-stained images (10, 11), while avoid the processes of tissue fixation/freezing, sectioning, and staining. It has already shown success in rapid histopathology for various types of human diseases, including brain tumors, laryngeal squamous cell carcinoma, gastrointestinal tumors, pancreatic cancer, and neurodegenerative diseases (12–23). In the case of timely prostate cancer detection, SRS may provide the foundation for intraoperative diagnosis and guided FT.
On the other hand, histopathologic interpretation of prostate cancer for FT faces several challenges. First, the shortage of pathologists remains a problem of medical system worldwide. Second, prostate cancer tissues are highly heterogenous with diverse histologic patterns. Recommended by the World Health Organization, Gleason score system was widely adapted to evaluate the histopathologic stages and determine the most suitable treatment modalities (24, 25). On the basis of the histoarchitectural patterns, five Gleason patterns were assigned from 1 to 5 with decreasing differentiation. The final Gleason score is reported as the sum of the two patterns that represents the two most predominant histologic patterns in the specimen. Such criteria for multiple classifications rendered high variation in interpathologist diagnosis due to the subjectivity (26, 27). In principle, artificial intelligence (AI) assistance could be a possible solution to release labor of pathologists and provide more objective diagnosis (28–33). In fact, leveraging the advances of deep learning–aided image identification, histologic images obtained by SRS have been further fed to machines to generate reliable diagnostic results on surgical specimens (14, 21). However, the huge histologic heterogeneity of prostate core needle biopsy yields significant challenges for both imaging and diagnosis. Specifically, most previous works only focused on the binary differentiation between cancer and benign tissues (14, 20), while Gleason grading requires more elaborated deep learning algorithms for the classification and segmentation of multiple tumor subtypes.
In this study, we demonstrated the capability of deep learning–based SRS microscopy for rapid histopathology and automated diagnosis of prostate cancer tissues that was able to predict Gleason scores on core needle biopsy. Label-free images containing the distribution of lipids, protein, and collagen fibers in fresh prostate tissues were taken to reveal the key diagnostic features of prostate cancer. In addition, convolutional neural network (CNN) based deep learning model was tailored to classify the images into benign and different Gleason patterns (G3–G5) with an accuracy of 85.7%. Furthermore, semantic segmentation was realized to visualize and quantify the intratumor heterogeneity of prostate needle biopsy and generated the overall score for individual specimen. Our approach provides a quantitative measure of tumor grading scores that holds potential for rapid and accurate diagnosis on fresh prostate core needle biopsy.
Materials and Methods
Study design and tissue collection
The study was approved by the Institutional Ethics Committee of the Ren Ji Hospital affiliated to the Shanghai Jiao Tong University, School of Medicine with written informed consent (approval no. KY2021-030). All tissue samples were collected from patients underwent radical prostatectomy or prostate core needle biopsy [18 gauge (G)]. The 18G biopsy needles were of approximately 0.8 mm core diameter and approximately 15 mm length. Total 104 patients were recruited and have been divided into three parts according to the recruitment time. The first part contained 61 patients and the generated images were used to train and test the CNN network. The second part included 22 patients and collected data were considered as an external test dataset to verify the CNN classification performance. The third part involved 21 patients and this part was used to simulate the clinical scene of Gleason scoring.
For frozen sections, fresh samples were snap frozen in liquid nitrogen and then immersed in optimal cutting temperature compound and stored at −80°C. Frozen tissues were sectioned to 20-μm- thick sections for SRS imaging and adjacent 5-μm-thick sections for H&E staining. To prepare unprocessed tissues, the fresh samples were collected and transferred to the imaging laboratory under 4°C within 2 hours, sealed and slightly squeezed between two coverslips with a spacer to generate uniform thickness (∼400 μm) slices for direct SRS imaging. The thickness between two coverslips could be easily changed with different spacers. We then trained and validated a CNN model to provide rapid and automated diagnosis and grading of fresh prostatic specimens imaged with SRS. In a further step, the performance of CNN was tested in an external, pilot study conducted in our medical center with fresh needle biopsy. All patients’ information was summarized in Supplementary Table S1.
H&E staining and histologic examination
The prepared frozen sections were stained with H&E and the standard staining procedure was performed as described previously (34). A team of three pathologists were participated in this study and adapted to SRS images of prostate tissue for histologic examination, using the adjacent H&E images that served as the “ground truth.” The pathologists diagnosed and graded prostatic specimens according to the International Society of Urological Pathology (ISUP) grading classification (24). The study team of pathologists reviewed all the digital images separately and then voted to reach a consensus on final pathologic results.
SRS microscopy system
In our SRS imaging apparatus, a femtosecond optical parametric oscillator (Insight DS+, Newport) laser with a fixed Stokes beam (1,040 nm, ∼200 fs) and a tunable pump beam (680 to 1,300 nm, ∼150 fs) served as the light source. Both the pump and Stokes beams were linearly chirped to picoseconds through SF57 glass rods to provide sufficient spectral and/or chemical resolution. The Stokes beam was modulated by an electro-optical modulator at 20 MHz repetition rate. After spatially and temporally overlapping, the two laser beams were delivered into a laser scanning microscope (FV1200, Olympus) and tightly focused onto the tissues through a water immersion objective lens (UPLSAPO 60XWIR, NA 1.2 water, Olympus). The transmitted stimulated Raman loss signal was optically filtered (CARS ET890/220, Chroma) and detected by a homemade back-biased photodiode. The electronic signal was further demodulated with a lock-in amplifier (HF2LI, Zurich Instruments) to feed the analog input of the microscope to form images. The target Raman frequency was selected by adjusting the time delay between the two pulses. For histologic imaging, we imaged at the two delay positions corresponding to 2,845 cm−1 and 2,930 cm−1 channels. Then the raw images were decomposed into the distributions of lipid and protein using the numerical algorithm to yield two-color SRS images. In addition, second harmonic generation (SHG) signal was simultaneously harvested using a narrow band pass filter (FF01-405/10, Semrock) and a photomultiplier tube in the epi mode. All the images used the same setting of 512 × 512 pixels with a pixel dwell time of 2 μs. The spatial resolution of our system is approximately 350 nm. To image a large area of tissue, mosaicking and stitching were performed to merge the small fields of view into a large flattened image. Laser powers at the sample were kept as: pump 50 mW and Stokes 50 mW.
CNN for histopathologic diagnosis
To realize the multilevel diagnosis of pathology, we design a deep learning algorithm based on CNN to: (i) implement multilevel classification; (ii) generate probabilities of different levels in a mosaic. Both the diagnosis algorithms and CNN named “Inception-Resnet-V2” uses the Pytorch framework compiled by Python language. The network “Inception-Resnet-V2” was the core of our deep learning algorithm consists of convolution layer and full connection layer with a depth of 572, including an input layer of 3 × 299 × 299 units and an output layer of five units because of image size and diagnostic categories. There are many more substructures inside the convolution layers, including: (i) Inception-ResNet-A; (ii) Reduction-A; (iii) Inception-ResNet-B; (iv) Reduction-B; (v) Inception-ResNet-C, and vi) average pooling layer.
Each large SRS image was sliced to 299 × 299 sized tiles to generate dataset that matched the input layer of the CNN. Then, the dataset was divided into five parts randomly, four of which were used for training, and the remaining was used for validation. The CNN was initialized in several aspects, including the following parts: (i) Set the output units to 5; (ii) Select Adam as the optimizer with the parameters: learning rate = 0.001, betas = (0.9, 0.999), eps = 1e-8, weight_decay = 1e-4; (iii) Select CrossEntropyLoss as the loss function.
Analysis of the CNN feature space
The high-resolution probability image based on semantic segmentation is generated by trained neural network and designed algorithm, which is performed as follows:
(i) The SRS image was sliced from the upper left corner to select out a tile with the standard size of 299 × 299 pixels.
(ii) Feed the tile to the CNN and generate a tensor with five numbers that correspond to prediction probabilities of the five categories.
(iii) The highest value of the five numbers was selected as the determined classification and marked in the probability matrix.
(iv) Move the selection range to the right by 25 pixels. If it has been moved to the end of the line, move down by 25 pixels and start from the beginning of the line. Repeat steps 1–3 for the entire image.
(v) Normalize the probability matrix to generate probability heatmap by color coding.
Gleason scoring criteria
Generally, the Gleason score is reported as the sum of the primary and secondary patterns of tumor. Specifically, if there is no secondary component or the secondary component has a lower pattern with the proportion less than 5%, then the Gleason score = primary component pattern + primary component pattern, such as 3 + 3 = 6; otherwise, if the secondary component's pattern is higher than 5%, then Gleason score = primary component pattern + secondary component pattern, as usual; if the tumor component has more than two hierarchical forms and the component of the highest level pattern is >5%, then Gleason score = primary component pattern + highest level pattern, no matter what the secondary component pattern is.
On basis of above criteria, algorithm for Gleason scoring was developed. The area of each category was first calculated and input to logic code as parameter. The diagnosis process goes through the following steps:
(i) Determine whether the benign tissue area is much larger than the cancer area. Considering the potential misjudgment of grading, we set the boundary as 95%, that is, if the area of benign accounts for more than 95% of total tissue, then the biopsy would be diagnosed as benign. Otherwise go to the next step.
(ii) Determine the primary and secondary patterns based on the area proportions. If the secondary pattern is less than 5% of total tumor area, then the scoring of the biopsy would be twice the primary pattern. Otherwise go to the next step.
(iii) Determine whether the least pattern is G5 and its area accounts for more than 5% of total tumor tissue. If it is, the scoring of the biopsy would be the primary pattern + G5. If it is not, the scoring of the biopsy would be the primary pattern + secondary pattern.
Statistical analysis
On the basis of the prediction and labeling of the trained network for the test set, we draw the ROC for four effective classification groups. When drawing the ROC of each classification, the corresponding value of the output five values is used to calculate specificity and sensitivity, respectively. The analyses were run on Matlab for MacOS (https://www.mathworks.com/products/matlab.html).
Data availability
The data that support the findings of this study are available online at Zenodo (https://github.com/Zhijie-Liu/Gleason-scoring-of-prostate). Raw image data are available from the corresponding author upon request.
Code availability
The codes for CNN-based results presented in this article can be found at (https://github.com/Zhijie-Liu/Gleason-scoring-of-prostate).
Results
Validation of SRS histology on thin frozen sections
To correctly define the ablative region during FT for localized prostate cancer, it is crucial to develop an image-guided method with high speed and accuracy. SRS microscopy provides promise to achieve this goal with the unique capability of chemical-specific imaging on specimens without complex tissue preparations. As illustrated in Fig. 1A, after tissue collection via either surgical dissection or core needle biopsy, multicolor SRS images with lipid and protein channels were decomposed from the raw images of CH2 (2,845 cm−1) and CH3 (2,930 cm−1) vibrations, via the linear decomposition method developed previously (10, 35, 36). The main contrast of cellular morphology was resolved on the basis of that cell nucleus contained less lipids than the surrounding tissues. An additional channel of SHG was detected to image collagen fibers in extracellular matrix, which were often seen surrounding the prostatic glands to form the prostatic stroma (Supplementary Fig. S1). Although collagen fibers are not considered as the key histologic feature for direct cancer diagnosis in conditional H&E interpretation, they are closely related to tumor genesis and metastasis (37–40), and contribute to a more complete view of tissue histology.
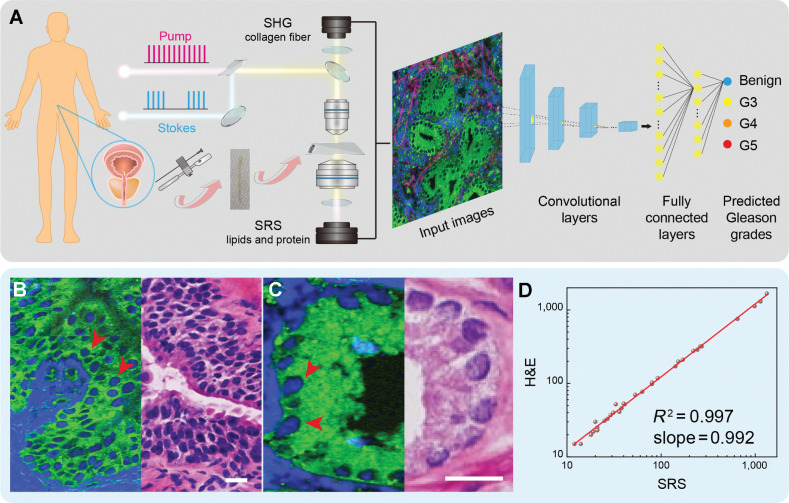
Figure 1.
Experimental design and workflow. A, Illustration of prostate FT and taking fresh biopsy for SRS imaging and CNN-based Gleason grading and scoring. B and C, SRS (left) and H&E (right) images of adjacent frozen sections from prostate tissues. D, Linear correlation plot of cell counting from SRS and H&E images (correlation factor R2 = 0.997). SRS images are color coded. Green, lipid; blue, protein; red, collagen (SHG @ 405 nm). Scale bars, 25 μm.
For the standard-of-care histologic evaluation of SRS by direct comparison with traditional H&E, we prepared 27 pairs of adjacent frozen sections. In each pair of the tissue sections, one was imaged under SRS microscope without further processing, whereas the other was sent for H&E staining, detailed procedures could be found in Materials and Methods. As shown in Supplementary Figs. S1 and S2, SRS images demonstrated near-perfect agreement with adjacent H&E in revealing key histologic features among benign and various patterns of prostate cancer. For benign prostatic tissues (benign), each prostatic gland unit presented large, plum-like acinar structure, occasionally accompanied with secretions inside the cavity (Supplementary Fig. S1A, stars). Zoom-in SRS images visualized papillary glandular cavity and glandular tube, and the surrounding basal cells were also clearly identifiable (Supplementary Fig. S1B; Fig. 1B, arrows). For neoplastic tissues of Gleason pattern 3 (G3), the gland units shrank and degenerated as small and discrete glandular structure with only single layer of epithelial cells (Supplementary Fig. S2A; Fig. 1C, arrows). As the degree of malignancy increased, fused, irregular cribriform and ill-defined glands with poorly formed glandular lumen were shown in Gleason pattern 4 (G4; Supplementary Fig. S2B). Eventually, tumor nests or solid sheets composed by numerous cancer cells, formed patterns of Gleason pattern 5 (G5; Supplementary Fig. S2C). It is also noteworthy that Gleason pattern 1 and pattern 2 were not considered in our work because those two types were not used in contemporary Gleason grading system on the basis of that G1 and G2 tumors have very low malignancy and have negligible impact to prognosis (24).
We also noticed that the high-resolution SRS images displayed near perfect cell-to-cell correlation with H&E (Fig. 1B and C). To further characterize this relationship, quantitative cell counting was performed and showed high linearity (R2 = 0.99) between the cell densities extracted from the two modalities (Fig. 1D). The linearity of cell density shows the consistency between SRS and H&E, and validates the ability of SRS to provide similar histologic information of prostate cancer tissues to conventional H&E. Furthermore, the above 27 pairs of large adjacent frozen sections were segmented into 450 pairs of images with specific Gleason patterns for three professional pathologists to perform diagnostic rating based on their own clinical professions. Responses were collected regarding the classification of Gleason patterns based on cytology and histoarchitecture, and rated results by comparing with standard histology are shown in Table 1. Statistical analysis of the pathologists’ diagnostic results on SRS and H&E images yielded high concordance (Cohen kappa) between them (κ = 0.944–0.967). Pathologists were highly accurate in distinguishing the prostatic Gleason patterns based on SRS images (G4 is the lowest with 88.6%; Table 1).
Table 1.
Diagnostic comparison of three pathologists based on SRS and H&E images of adjacent frozen sections. A total of 450 pairs of images with specific Gleason patterns (benign, G3, G4, and G5) were segmented from 27 pairs of sections.
| P1 | P2 | P3 | Accuracy % | |||||
|---|---|---|---|---|---|---|---|---|
| Diagnosis | Correct | Incorrect | Correct | Incorrect | Correct | Incorrect | ||
| Benign | H&E | 202 | 6 | 208 | 0 | 208 | 0 | 99.0 |
| SRS | 198 | 10 | 203 | 5 | 205 | 3 | 97.1 | |
| G3 | H&E | 174 | 7 | 177 | 4 | 181 | 0 | 98.0 |
| SRS | 165 | 16 | 173 | 8 | 172 | 9 | 93.9 | |
| G4 | H&E | 34 | 4 | 30 | 8 | 38 | 0 | 89.5 |
| SRS | 34 | 4 | 32 | 6 | 35 | 3 | 88.6 | |
| G5 | H&E | 20 | 3 | 21 | 2 | 23 | 0 | 92.8 |
| SRS | 22 | 1 | 23 | 0 | 23 | 0 | 98.6 | |
| Total | H&E | 430 | 20 | 436 | 14 | 450 | 0 | 97.5 |
| SRS | 419 | 31 | 431 | 19 | 435 | 15 | 95.2 | |
| Accuracy % | 94.3 | 96.3 | 98.3 | |||||
| Concordance | 0.956 | 0.944 | 0.967 | |||||
SRS reveals key diagnostic features in fresh prostate tissues
We then demonstrated the unique advantage of our label-free imaging system to produce histopathologic images on fresh prostate tissue specimens without thin sectioning. The fresh specimens of approximately 400 μm thickness were imaged with SRS directly without any further processing. The results of typical SRS images of fresh prostate tissues were classified into the same categories as in frozen sections: benign, G3, G4, and G5, according to the ISUP 2014 Gleason grading definition (24).
Representative images of the four categories are shown in Fig. 2A–I to demonstrated the well-preserved histologic features without freezing or sectioning artefacts. In the benign group, normal prostate tissues with regular glands and fibromuscular stroma were well resolved. As shown in Fig. 2A, the papillary infoldings and the formation of corpora amylacea (stars) in normal gland lumen could be visualized. Magnified images also clearly revealed the double-layered cellular structure of the normal prostatic ducts: a basal cell layer and a secretory cell layer (Fig. 2B, arrows). In prostate cancer tissues, SRS revealed the typical histoarchitectures of the cancerous tissues graded as G3–G5. As shown in Fig. 2C–E, the G3 featured the individual, discrete glands with complete circumference of cells forming lumia. The G3 glands mainly appeared two typical patterns with different shapes and sizes, including the small glands (microacinar glands in Fig. 2C), the large gland with a microcystic appearance (Fig. 2D) and the branched ducts (Fig. 2E). For G4, SRS clearly depicted the key features of fused glands and invasion of intervening stroma between adjacent glands. Specifically, the detailed structures of the G4 glands were shown in Fig. 2F–H, including the fused pattern of cords or chains (Fig. 2F), the glomeruloid pattern (Fig. 2G), and the cribriform pattern (Fig. 2H). In Fig. 2I, the major patterns of G5 could be readily seen: the central necrosis within papillary and cribriform spaces.
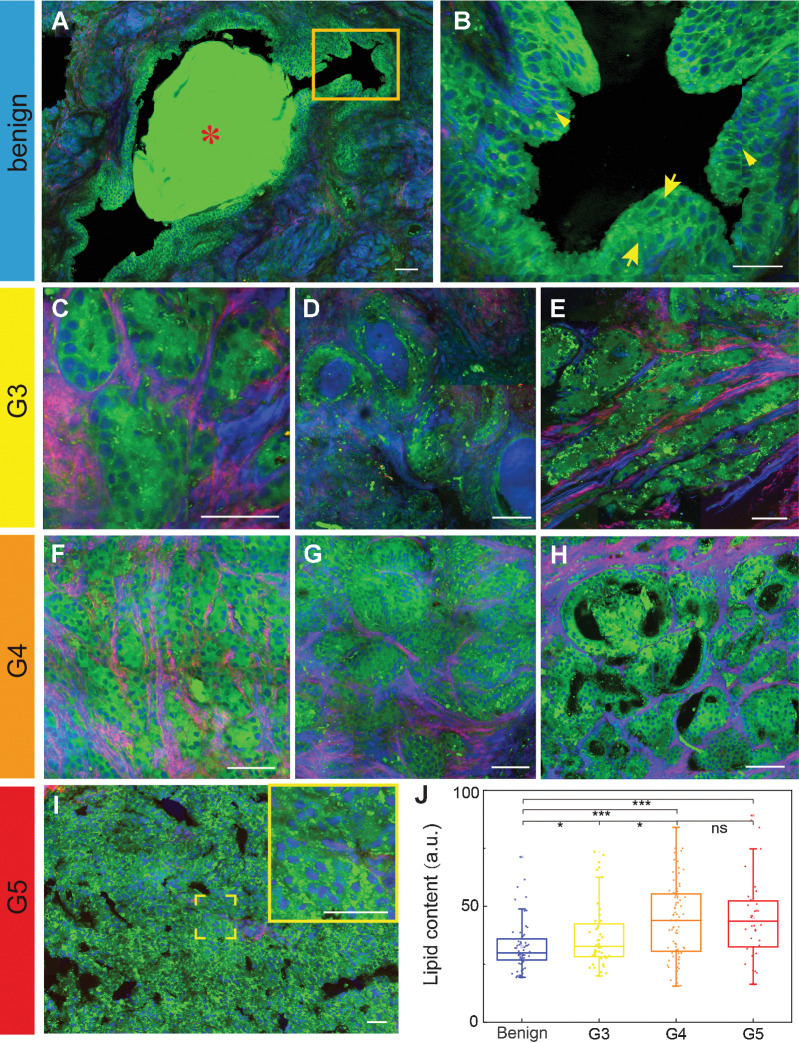
Figure 2.
SRS reveals diagnostic features of fresh prostate tissues. A–I, Key histologic hallmarks of different Gleason patterns, including: amylacea (A) and the double-layered cellular structure of normal prostatic ducts of the benign group (B), zoomed image from the yellow rectangle in A; small glands (C), large glands (D) and branched ducts of G3 (E); fused pattern of cords or chains (F), the glomeruloid pattern (G), and the cribriform pattern of G4 (H); and the central necrosis within papillary and cribriform spaces for G5 (I). J, Quantification of lipid contents in different patterns. *, P < 0.05, ***, P < 0.001; ns, not significant (P > 0.05) from two-tail unpaired t tests. Green, lipid; blue, protein; red, collagen. Scale bars, 50 μm.
In addition to tissue histopathology, SRS is also advantageous in providing quantitative information of biochemical compositions such as lipids. Lipid content in tissue is known to strongly correlate with tumor metabolism and malignancy, but is inaccessible with H&E because of the loss of lipids during tissue deparaffination process. It can be seen that tumorous glands tend to contain more lipid droplets (LD) than the benign ones (Supplementary Fig. S3). To be quantitative, we have calculated the total lipid contents of benign and G3–G5 grade prostate tissues based on the integrated SRS image intensity at 2,845 cm−1, because proteins contribute little to this spectral channel (35). Our results indicated that lipid content increases with tumor malignancy from benign to G3 and to G4, but kept similar between G4 and G5 (Fig. 2J). This finding is consistent with previous works on lipid metabolism in the development and progression of prostate cancer, which is featured by increased abundance of lipids in prostate cancer tissues (41–43). And from the extracted 20 spectra of LDs (Supplementary Fig. S4A), we also noticed that the intensity ratio of 2,870 cm−1 to 2,850 cm−1 was higher in the malignant specimens (Supplementary Fig. S4B), revealing an aberrant cholesteryl ester (CE) accumulation in tumorous tissues (44).
Deep learning–based classification and grading
To reduce the workload of pathologists and provide potentially more objective diagnosis for intraoperative histopathology, we proceeded to train the Inception-ResNet-V2, a deep CNN composed of an inception module and a ResNet module with a depth of 572, to achieve automated diagnosis and grading of prostate cancer. Compared with previous works on brain and larynx tissues (14), prostate tissues are histologically much more heterogeneous with key diagnostic features localized in the gland and/or lumen structures. Moreover, a large tissue slide usually contains multiple areas of different Gleason patterns, hence it could be hardly considered as a single uniform class for training the network as previously adapted in brain and larynx studies. In this work, digital SRS images from 61 patient cases were presegmented by two pathologists. The resulting 386 segmented and annotated smaller images (termed as “seg-images”; Supplementary Table S2) were used as the training and test datasets, categorized into five groups: benign (n = 65), G3 (n = 44), G4 (n = 80), G5 (n = 39), and nondiagnostic (n = 158; Fig. 3A; Supplementary Table S2). Here, the nondiagnostic group represents areas of interstitial substance such as collagen fibers and connective tissues without clear cellular structures.
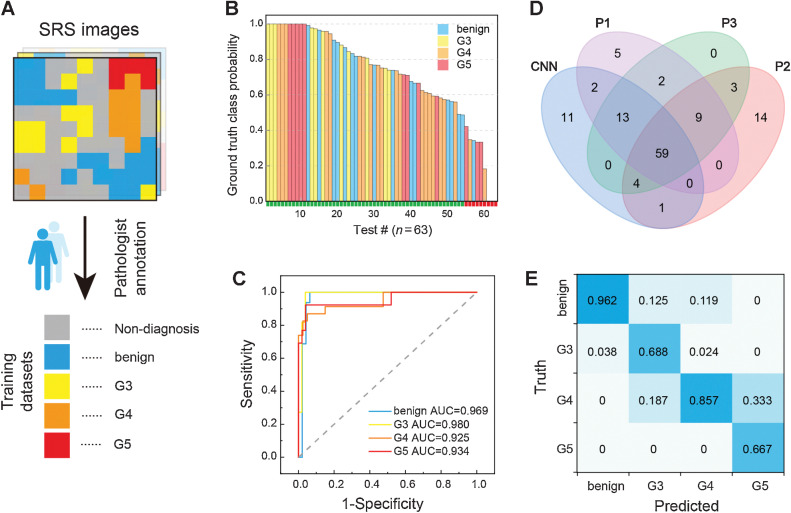
Figure 3.
Automated diagnosis with deep learning on SRS images. A, Presegmentation and annotation of the whole-slide SRS images for training. B and C, Test results of the CNN model (B) and its ROC curves and AUC for the classification of benign and G3–G5 (C). D, Venn diagram of the diagnostic results of CNN model and three pathologists. E, Confusion matrix of the four diagnostic subtypes between CNN and the consensus of three pathologists.
For training the CNN model, each presegmented image (seg-images) was further sliced into tiles of standard image size of 299 × 299 pixels, and all the tiles in each group were divided into training and test datasets with a ratio of 5:2. Within the training set, 5-fold cross-validation was applied with the ratio between the training and validation subsets regulated as 4:1. The number of seg-images and tiles for all the data subsets are shown in Supplementary Table S3. The CNN input a patch of training tiles, and output the prediction value of five groups for each tile (Supplementary Fig. S5). The subsequent prediction probabilities for all the 63 test seg-images (without nondiagnostic class) are displayed in descending order with classification results of correct (green) or incorrect (red; Fig. 3B; ref. 45). The ROC curves are plotted in Fig. 3C, with the AUC of 0.969 for benign, 0.980 for G3, 0.925 for G4, and 0.934 for G5, indicating the high performance of the trained model in grading each diagnostic category. We further analyzed the misclassified images and found that among the 63 test cases, nine cases were misclassified, including one benign case, four G4 cases, and four G5 cases. Specifically, one benign case was misclassified as G5, four G4 cases were misclassified as G3, two G5 cases were misclassified as benign, one G5 case was misclassified as G4, and the rest one case was unable to be classified because each class possess similar probability. Overall, the trained CNN network achieved an accuracy of 85.7% (54/63).
Aside from the above test dataset, which was preverified by pathologists (Fig. 3A), we also introduced an additional external test set to evaluate the performance of our CNN model. The external test dataset was collected from another 22 independent cases. SRS image of each tissue was sliced into tiles in the same way as before, and packed as independent patches without preannotation to feed the CNN network. On the basis of the CNN grading results, the SRS images were then segmented into 90 smaller images, each of which contained one type of Gleason pattern. These 90 images were further rated by three professional pathologists and compared with AI grading results, summarized in the Venn diagram (Fig. 3D) and Table 2. We noticed that for Gleason grading, interpathologist variations appeared quite significant. Both pathologist 1 and pathologist 2 produced a few results (5 and 14 images for each) that were unable to match with others. The final consensus of pathologists was determined as at least two of the three pathologists sharing the same results and was taken as the ground truth for the evaluation of diagnostic results. The generated multiclass confusion matrix was shown in Fig. 3E, indicating the concordance (Cohen kappa) level between CNN and pathologists was approximately 0.763.
Table 2.
Diagnostic comparison between CNN and pathologists on external dataset containing 90 SRS images of fresh biopsy tissues.
| CNN | Pathologist 1 | Pathologist 2 | Pathologist 3 | Combined accuracy (%) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Correct | Incorrect | Correct | Incorrect | Correct | Incorrect | Correct | Incorrect | ||
| Benign | 25 | 7 | 32 | 0 | 29 | 3 | 32 | 0 | 92.2 |
| G3 | 11 | 2 | 6 | 7 | 13 | 0 | 13 | 0 | 82.7 |
| G4 | 36 | 5 | 34 | 7 | 37 | 4 | 41 | 0 | 90.2 |
| G5 | 4 | 0 | 3 | 1 | 4 | 0 | 4 | 0 | 93.8 |
| Total | 76 | 14 | 75 | 15 | 83 | 7 | 90 | 0 | 90.0 |
| Accuracy (%) | 84.4 | 83.3 | 92.2 | 100.0 | |||||
| Concordance | 0.763 | 0.738 | 0.883 | 1.000 | |||||
Visualizing the CNN feature representations
To gain insights into the image features based on which our CNN model had made decisions, we performed relevance backpropagation using default parameters for a number of examples. For each Gleason pattern, two representative SRS images were selected to generate the probability distribution heatmap of the corresponding diagnostic classes (Supplementary Fig. S6A–S6D). Interestingly but not surprisingly, the regions with glandular structure showed higher activation intensities. Particularly noted, in the benign tissues the thin luminal parts of the ducts were clearly activated (Supplementary Fig. S6A); and in tumor tissues the cellular regions were highlighted (Supplementary Fig. S6B–S6D). It is known that these regions appear rich in cellular morphology and patterns, which offer essential diagnostic features for pathologists. Hence such observations imply that the prediction of CNN model is indeed closely related to the key histoarchitectures for grading prostate cancer. By recoding the prediction probability of each class in cold-warm tones color (Supplementary Fig. S7), we preformed grading semantic segmentation to reveal the spatial characteristics of the diagnostic histologic heterogeneity on a prostate core needle biopsy imaged by SRS (Fig. 4A and B). The blue color represents the high probability regions of predicted benign with plum-like gland units and myofiber-like interstitial cells. The yellow color represents G3 region with moderately differentiated carcinoma. The orange and red color predicted the G4 with fused glands and G5 with cell clusters formed tumor nests, respectively. The gray area consisted with empty or collagen rich regions. For this particular specimen, the segmentation results showed the tumorous biopsy was mainly composed of G4 pattern with a few sites of G3 and G5 pattern, which agreed well with the ground truth segmentation labeled by a pathologist (Fig. 4C). Magnified images of the four predicted Gleason patterns showed the detailed histologic features to confirm the prediction accuracy (Fig. 4D). In addition, a visualization of the feature space learned by the model was pictured using t-distributed stochastic neighbor embedding (t-SNE) method (Supplementary Fig. S8; ref. 21). Each datapoint in t-SNE plot represents a single tile from test datasets stratified by Gleason pattern (including benign, G3, G4, and G5). As we can see in the feature space visualization, most tiles within each category tend to cluster together, further indicating a logical representation has learned by the model.
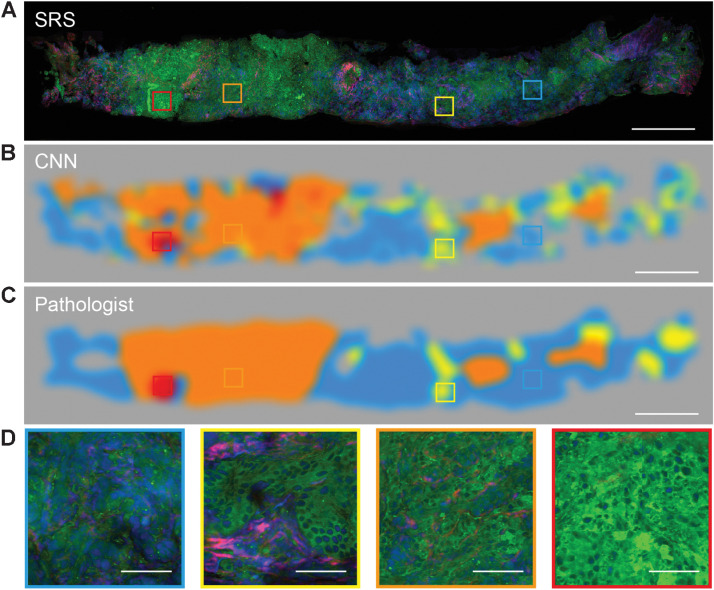
Figure 4.
Visualizing the CNN feature representations. A and B, The typical SRS images of core needle biopsy (A) and corresponding activation heatmaps of benign and G3, G4, G5 patterns (B). C, The Gleason grading map merged from recoded color activation heatmaps (B). D, Zoom-in SRS images represent four classes of benign and G3, G4, and G5 patterns. Scale bars, 500 μm in A–C; 50 μm in D.
Gleason scoring of fresh core needle biopsy
We next simulated deep learning–based Gleason scoring on 21 cases of fresh core needle biopsy to evaluate the feasibility of automated grading and scoring of prostate cancer for FT. To achieve this goal, the trained CNN model first performed semantic segmentation on the whole tissue to map the spatial inhomogeneity of different diagnostic patterns as shown in Fig. 4, and then calculated their occupation percentage to yield the final score under the Gleason scoring criteria (see Materials and Methods).
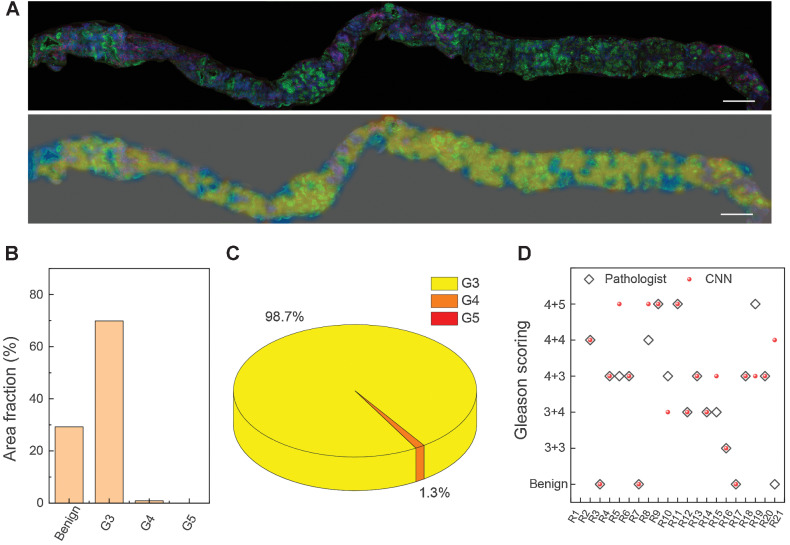
Fresh core needle biopsy tissues harvested from FT surgeries were imaged with SRS directly. A typical biopsy of 1 × 15 mm2 size was imaged within 8 minutes and then sent to the CNN model for prediction, which could be executed within 60 seconds to generate the color-coded segmentation heatmap (Fig. 5A). The heatmap interpreted the distributions of all the classified patterns of benign, G3, G4, and G5, with the same color codes as in Fig. 4. Moreover, in contrast to the pathologists’ subjective diagnosis, our deep learning system could quantitatively calculate the area partition ratio of each pattern (Fig. 5B and C), and offer the final scoring results based on Gleason scoring criteria. In the example case, the overall tumor occupies 70.7% of the entire sample (Fig. 5B), among which, the primary pattern was G3 (98.7%), the secondary pattern was G4 (1.3%), less than 5% of the total tumor area (Fig. 5C). Therefore, the final Gleason score of this biopsy was predicted to be “3+3.” We also demonstrated another case with 90.8% of the tissue composed of tumor area (Supplementary Fig. S9A), among which, the primary pattern was G4 (87.8%), the secondary pattern was G5 (9.2%), and G3 was notably less than 5% of the total tumor area (Supplementary Fig. S9B and S9C). Therefore, the final Gleason score of this biopsy was graded to be “4+5.” The CNN predicted scores of the total investigated 21 cases of core needle biopsy specimens are shown in Fig. 5D, along with the diagnostic consensus of three pathologists on the same SRS image (Supplementary Fig. S10), resulting in a diagnostic consistency of approximately 71.4% (15/21). It is worth noting that most discrepancies were originated from the secondary patterns, which is also commonly inconsistent between pathologists.
Figure 5.
Color-coded and scoring fresh FT core needle biopsy. A, The presented SRS image of fresh FT core needle biopsy and the corresponding color-coded grading image. B, The quantification analysis of the area fraction of benign and G3–G5. C, The proportion of each Gleason pattern (G3–G5) in the whole tumor region. D, Diagnostic scoring results of CNN-based SRS, compared with the consensus of three pathologists on 21 cases. Scale bars, 500 μm.
Discussion
On the basis of the premise that the significant cancer foci tend to drive the progression of prostate cancer, FT adopts a minimally invasive approach to destroy the significant cancer foci that would benefit cancer control (46). Currently, multiparametric MRI (mpMRI) is regard as a standard tool for FT in identifying cancer locations, because mpMRI has a high negative predictive value to detect clinically significant diseases (47). However, solely relying on MRI may face the challenges of accurate disease localization and guidance in the intraoperative settings. Recent attentions have been drawn to the precise evaluation and localization of diseases (48). For example, it would be of great help to develop a real-time solution to collect histologic images from fresh biopsy tissues for grading prostate cancer during the ablation needle insertions. However, the current intraoperative pathologic examination workflow is rather difficult to meet with the requirements of FT: First, the acquisition of histologic images is time consuming due to the complex procedure of tissue preparation and H&E staining; Second, the pathologic reporting of histologic images is not always timely. Given the lack of an intraoperative diagnostic tool for prostate biopsy, we have demonstrated the capability of deep learning–assisted SRS microscopy to provide rapid detection and grading of prostate cancer with relatively high accuracy in this work.
SRS presented advantages in rapid and high-resolution imaging of tissue morphology without any sample processing. Direct imaging of fresh specimens in a nondestructive way could avoid the artefacts introduced by sample process including freezing and staining and allows the specimens to be reusable for subsequent histopathologic tests to reduce duplicated sampling. Similar to the noticed recent works in exploring the value of three-dimensional (3D) pathology of clinical specimens, which were based on advanced light-sheet microscopy and optical clearing technique (49), nondestructive 3D pathology of prostate biopsy could also be developed to improve prostate cancer risk stratification, leveraging the intrinsic 3D optical sectioning ability of SRS (50). Meanwhile, the chemical specificity inherited from spontaneous Raman scattering endowed SRS with the ability to identify biochemical components in tissues, such as lipids, protein, and DNA (35). From the lipid-channel SRS images, we noted the lipid content was increases with tumor malignancy and the further hyperspectral analysis indicated the abundance in LDs might come from CE accumulation (Supplementary Figs. S3 and S4). This spectral differentiation could be a potential prostate cancer marker and we are working on combining spectral information with traditional histomorphology to construct high-dimensional data for tumor diagnosis. From the perspective of AI diagnosis, although the current automatic diagnosis accuracy is incomparable with our previous binary classification works (>90%), it is clinically acceptable at multiple classifications in cancers grading with actual diagnostic criteria. When the size of dataset became huge, weakly supervised learning, which could achieve classification based on single label in slide level (51), might be suited to achieve diagnosis without labored manual annotations as we have performed in Fig. 3A. Nonetheless, in the case of limited datasets, supervised learning with patch level labeling is still the optimal solution.
We believe this AI assisted SRS microscopy could be an innovative option in rapid pathological diagnosis of prostate cancer, which could largely change the landscape of prostate cancer diagnosis and therapy. In the procedure of prostate biopsy, this technique could improve the accuracy of tumor detection, incidentally limiting the number of nondiagnostic biopsy samples. For FT, this technique could help the urologists to know the histology information nearby the ablation needle during insertion, helping the urologists to eradicate the tumor precisely. Our method could also be used to rapidly and accurately detect the surgical margin during radical prostatectomy, ensuring the safety of dissection. However, to fulfill the clinical translation, the technique should be further improved. First, the current system should be miniaturized and integrated (52). To simplify optical configuration, a U-net–based femto-SRS imaging method could be adapted (23), and compact fiber lasers could be used to replace bulky solid-state lasers (53). Second, the size of training dataset could be largely increased in a multicenter, prospective clinical trial to improve the accuracy of AI-based diagnosis. Because the pathology report was used as the ground truths for AI evaluation, the sensitivity of AI system may not exceed the employed pathologists. And considering the subjectivity reflected in interpathologist variability, increasing the dataset annotated by different pathologists may help reduce the effect of such variation (28).
In summary, we have presented the use of deep learning–assisted SRS system on prostate core needle biopsy for the imaging and diagnosis of prostate cancer. Our technique could potentially provide timely histologic information with high accuracy during the ablation needle insertion, which meets the demands of FT improvement in several aspects. It could help characterize the cancer foci in the targeted area of prostate; accurately guide and ensure the ablation needle into the cancer foci; and finally evaluate the oncologic efficacy in near real time. Therefore, the incorporation of our method may allow more accurate FT and facilitate the advancement of FT.
Supplementary Material
Supplementary information
Acknowledgments
M. Ji acknowledges financial support from the National Key R&D Program of China (2021YFF0502900), the National Natural Science Foundation of China (61975033), Shanghai Municipal Science and Technology Major Project No. 2018SHZDZX01, and ZJLab. J. Pan acknowledges financial support from the National Natural Science Foundation of China (82003148) and Shanghai Municipal Science and Technology Project (21Y11904100). L. Qi acknowledges funding from Shanghai Municipal Science and Technology Project (21Y11910500).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
M. Ji reports grants from the National Key R&D Program of China and the National Natural Science Foundation of China during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
J. Ao: Formal analysis, investigation, writing–original draft. X. Shao: Resources, investigation, writing–original draft. Z. Liu: Software, formal analysis. Q. Liu: Investigation. J. Xia: Resources, data curation. Y. Shi: Resources, data curation. L. Qi: Validation, visualization. J. Pan: Conceptualization, supervision, writing–review and editing. M. Ji: Conceptualization, supervision, writing–review and editing.
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2. Ashrafi AN, Tafuri A, Cacciamani GE, Park D, de Castro Abreu AL, Gill IS, et al. Focal therapy for prostate cancer: concepts and future directions. Curr Opin Urol 2018;28:536–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Valerio M, Cerantola Y, Eggener SE, Lepor H, Polascik TJ, Villers A, et al. New and established technology in focal ablation of the prostate: a systematic review. Eur Urol 2017;71:17–34. [DOI] [PubMed] [Google Scholar]
- 4. Ahmed HU, Hindley RG, Dickinson L, Freeman A, Kirkham AP, Sahu M, et al. Focal therapy for localised unifocal and multifocal prostate cancer: a prospective development study. Lancet Oncol 2012;13:622–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Valerio M, Ahmed HU, Emberton M, Lawrentschuk N, Lazzeri M, Montironi R, et al. The role of focal therapy in the management of localised prostate cancer: a systematic review. Eur Urol 2014;66:732–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gal AA, Cagle PT. The 100-year anniversary of the description of the frozen section procedure. JAMA 2005;294:3135–7. [DOI] [PubMed] [Google Scholar]
- 7. Eissa A, Zoeir A, Sighinolfi MC, Puliatti S, Bevilacqua L, Del Prete C, et al. Real-time" assessment of surgical margins during radical prostatectomy: state-of-the-art. Clin Genitourin Cancer 2020;18:95–104. [DOI] [PubMed] [Google Scholar]
- 8. Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, He C, et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 2008;322:1857–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng JX, Xie XS. Vibrational spectroscopic imaging of living systems: an emerging platform for biology and medicine. Science 2015;350:aaa8870. [DOI] [PubMed] [Google Scholar]
- 10. Ji M, Orringer DA, Freudiger CW, Ramkissoon S, Liu X, Lau D, et al. Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci Transl Med 2013;5:201ra119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hollon TC, Lewis S, Pandian B, Niknafs YS, Garrard MR, Garton H, et al. Rapid intraoperative diagnosis of pediatric brain tumors using stimulated Raman histology. Cancer Res 2018;78:278–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ji M, Lewis S, Camelo-Piragua S, Ramkissoon SH, Snuderl M, Venneti S, et al. Detection of human brain tumor infiltration with quantitative stimulated Raman scattering microscopy. Sci Transl Med 2015;7:309ra163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ji M, Arbel M, Zhang L, Freudiger CW, Hou SS, Lin D, et al. Label-free imaging of amyloid plaques in Alzheimer's disease with stimulated Raman scattering microscopy. Sci Adv 2018;4:eaat7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang L, Wu Y, Zheng B, Su L, Chen Y, Ma S, et al. Rapid histology of laryngeal squamous cell carcinoma with deep-learning based stimulated Raman scattering microscopy. Theranostics 2019;9:2541–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu F-K, Calligaris D, Olubiyi OI, Norton I, Yang W, Santagata S, et al. Label-free neurosurgical pathology with stimulated Raman imaging. Cancer Res 2016;76:3451–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang Y, Yang Y, Liu Z, Guo L, Li S, Sun X, et al. Microcalcification-based tumor malignancy evaluation in fresh breast biopsies with hyperspectral stimulated Raman scattering. Anal Chem 2021;93:6223–31. [DOI] [PubMed] [Google Scholar]
- 17. Sarri B, Canonge R, Audier X, Simon E, Wojak J, Caillol F, et al. Fast stimulated Raman and second harmonic generation imaging for intraoperative gastro-intestinal cancer detection. Sci Rep 2019;9:10052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shin KS, Francis AT, Hill AH, Laohajaratsang M, Cimino PJ, Latimer CS, et al. Intraoperative assessment of skull base tumors using stimulated Raman scattering microscopy. Sci Rep 2019;9:20392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bentley JN, Ji M, Xie XS, Orringer DA. Real-time image guidance for brain tumor surgery through stimulated Raman scattering microscopy. Expert Rev Anticancer Ther 2014;14:359–61. [DOI] [PubMed] [Google Scholar]
- 20. Orringer DA, Pandian B, Niknafs YS, Hollon TC, Boyle J, Lewis S, et al. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat Biomed Eng 2017;1:0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hollon TC, Pandian B, Adapa AR, Urias E, Save AV, Khalsa SSS, et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat Med 2020;26:52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang L, Zou X, Huang J, Fan J, Sun X, Zhang B, et al. Label-free histology and evaluation of human pancreatic cancer with coherent nonlinear optical microscopy. Anal Chem 2021;93:15550–8. [DOI] [PubMed] [Google Scholar]
- 23. Liu Z, Su W, Ao J, Wang M, Jiang Q, He J, et al. Instant diagnosis of gastroscopic biopsy via deep-learned single-shot femtosecond stimulated Raman histology. Nat Commun 2022;13:4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA, et al. The 2014 international society of urological pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma. Am J Surg Pathol 2016;40:244–52. [DOI] [PubMed] [Google Scholar]
- 25. Sauter G, Steurer S, Clauditz TS, Krech T, Wittmer C, Lutz F, et al. Clinical utility of quantitative Gleason grading in prostate biopsies and prostatectomy specimens. Eur Urol 2016;69:592–8. [DOI] [PubMed] [Google Scholar]
- 26. Nagpal K, Foote D, Tan F, Liu Y, Chen P-HC, Steiner DF, et al. Development and validation of a deep learning algorithm for Gleason grading of prostate cancer from biopsy specimens. JAMA Oncol 2020;6:1372–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bulten W, Pinckaers H, van Boven H, Vink R, de Bel T, van Ginneken B, et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol 2020;21:233–41. [DOI] [PubMed] [Google Scholar]
- 28. Ström P, Kartasalo K, Olsson H, Solorzano L, Delahunt B, Berney DM, et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. Lancet Oncol 2020;21:222–32. [DOI] [PubMed] [Google Scholar]
- 29. Yang Q, Xu Z, Liao C, Cai J, Huang Y, Chen H, et al. Epithelium segmentation and automated Gleason grading of prostate cancer via deep learning in label-free multiphoton microscopic images. J Biophotonics 2020;13:e201900203. [DOI] [PubMed] [Google Scholar]
- 30. Marginean F, Arvidsson I, Simoulis A, Christian Overgaard N, Åström K, Heyden A, et al. An artificial intelligence-based support tool for automation and standardisation of Gleason grading in prostate biopsies. Eur Urol Focus 2021;7:995–1001. [DOI] [PubMed] [Google Scholar]
- 31. Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, et al. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc 2019;89:25–32. [DOI] [PubMed] [Google Scholar]
- 32. Nagpal K, Foote D, Liu Y, Chen P-HC, Wulczyn E, Tan F, et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit Med 2019;2:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fu Y, Jung AW, Torne RV, Gonzalez S, Vöhringer H, Shmatko A, et al. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat Cancer 2020;1:800–10. [DOI] [PubMed] [Google Scholar]
- 34. Fischer AH, Jacobson KA, Rose J, Zeller R. Hematoxylin and eosin staining of tissue and cell sections. CSH Protoc 2008;2008:pdb prot4986. [DOI] [PubMed] [Google Scholar]
- 35. Lu F-K, Basu S, Igras V, Hoang MP, Ji M, Fu D, et al. Label-free DNA imaging in vivo with stimulated Raman scattering microscopy. Proc Natl Acad Sci U S A 2015;112:11624–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He R, Xu Y, Zhang L, Ma S, Wang X, Ye D, et al. Dual-phase stimulated Raman scattering microscopy for real-time two-color imaging. Optica 2017;4:44–7. [Google Scholar]
- 37. Brown E, McKee T, diTomaso E, Pluen A, Seed B, Boucher Y, et al. Dynamic imaging of collagen and its modulation in tumors in vivo using second-harmonic generation. Nat Med 2003;9:796–800. [DOI] [PubMed] [Google Scholar]
- 38. Burke K, Smid M, Dawes RP, Timmermans MA, Salzman P, van Deurzen CHM, et al. Using second harmonic generation to predict patient outcome in solid tumors. BMC Cancer 2015;15:929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Y, Yi Y, Li Z, Zhan Z, Li L, Zheng L, et al. Visualization of collagen morphological changes in transition from tumor to normal tissue in breast cancer by multiphoton microscopy [abstract]. In:Luo Q, Li X, Gu Y, Zhu D, editors.Optics in health care and biomedical optics XI. Proceedings of SPIE; 2021.
- 40. Burns-Cox N, Avery NC, Gingell JC, Bailey AJ. Changes in collagen metabolism in prostate cancer: a host response that may alter progression. J Urol 2001;166:1698–701. [DOI] [PubMed] [Google Scholar]
- 41. Watt MJ, Clark AK, Selth LA, Haynes VR, Lister N, Rebello R, et al. Suppressing fatty acid uptake has therapeutic effects in preclinical models of prostate cancer. Sci Transl Med 2019;11:eaau5758. [DOI] [PubMed] [Google Scholar]
- 42. Yue S, Li J, Lee S-Y, Lee HJ, Shao T, Song B, et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab 2014;19:393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Randall EC, Zadra G, Chetta P, Lopez BGC, Syamala S, Basu SS, et al. Molecular characterization of prostate cancer with associated Gleason score using mass spectrometry imaging. Mol Cancer Res 2019;17:1155–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen X, Cui S, Yan S, Zhang S, Fan Y, Gong Y, et al. Hyperspectral stimulated Raman scattering microscopy facilitates differentiation of low-grade and high-grade human prostate cancer. J Phys D Appl Phys 2021;54:484001. [Google Scholar]
- 45. Arvaniti E, Fricker KS, Moret M, Rupp N, Hermanns T, Fankhauser C, et al. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci Rep 2018;8:12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med 2009;15:559–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sathianathen NJ, Omer A, Harriss E, Davies L, Kasivisvanathan V, Punwani S, et al. Negative predictive value of multiparametric magnetic resonance imaging in the detection of clinically significant prostate cancer in the prostate imaging reporting and data system era: a systematic review and meta-analysis. Eur Urol 2020;78:402–14. [DOI] [PubMed] [Google Scholar]
- 48. Nassiri N, Chang E, Lieu P, Priester AM, Margolis DJA, Huang J, et al. Focal therapy eligibility determined by magnetic resonance imaging/ultrasound fusion biopsy. J Urol 2018;199:453–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xie W, Reder NP, Koyuncu C, Leo P, Hawley S, Huang H, et al. Prostate cancer risk stratification via nondestructive 3D pathology with deep learning-assisted gland analysis. Cancer Res 2022;82:334–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Qi J, Li J, Liu R, Li Q, Zhang H, Lam JWY, et al. Boosting fluorescence-photoacoustic-Raman properties in one fluorophore for precise cancer surgery. Chem 2019;5:2657–77. [Google Scholar]
- 51. Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck Krauss Silva V, Busam KJ, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med 2019;25:1301–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liao C-S, Wang P, Huang CY, Lin P, Eakins G, Bentley RT, et al. In vivo and in situ spectroscopic imaging by a handheld stimulated Raman scattering microscope. ACS Photonics 2017;5:947–54. [Google Scholar]
- 53. Freudiger CW, Yang W, Holtom GR, Peyghambarian N, Xie XS, Kieu KQ. Stimulated Raman scattering microscopy with a robust fibre laser source. Nat Photonics 2014;8:153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Data Availability Statement
The data that support the findings of this study are available online at Zenodo (https://github.com/Zhijie-Liu/Gleason-scoring-of-prostate). Raw image data are available from the corresponding author upon request.