Abstract
Introduction
Polycystic Ovarian Syndrome (PCOS) has been identified as a gynecological, hormonal, and metabolic condition in women of reproductive age. Genetic studies can contribute to understand the pathogenesis of PCOS; which can be beneficial in early diagnosis and long-term management of the disease. Apurinic/apyrimidinic endonuclease 1 (APE1) has been related in the literature to polycystic ovarian syndrome.
Aim
The purpose of this study was to investigate the effects of −656 T > G and 1349 T > G single nucleotide polymorphisms (SNPs) in the APE1 gene in Saudi women with PCOS.
Methods
This study includes 100 PCOS women and 100 healthy controls were genotyped for −656 T > G and 1349 T > G SNPs using PCR-RFLP method. Serum sample was used for FBG and lipid profile tests. The obtained biochemical and genotypes data were entered into Excel and utilized for statistical analysis.
Results
Clinical data presented in Table 1 was used to calculate the t-tests between PCOS and control subjects and results indicate age, weight, BMI, TG, LDLC and PCOS family history was associated (p < 0.0001). Genotype and allele frequencies showed the negative association in −656 T > G SNP (GG vs TT: OR-1.15 (0.61–2.17); p = 0.65 and GG + TG vs TT: OR-1.17 (0.67–2.04); p = 0.57) and positive association in 1349 T > G SNP (GG vs TT: OR-3.52 (1.48–8.36); p = 0.003 and GG + TG vs TT: OR-2.84 (1.27–6.31); p = 0.008) in APE1 gene. Anova analysis was not associated with any one of the involved parameters (p > 0.05).
Conclusion
This study found that the 1349 T > G SNP was related with PCOS in Saudi women. However, the −656SNP had no favorable effect on the APE1 gene.
Keywords: APE1 gene, Single Nucleotide Polymorphisms (SNPs), −656T>G and 1349T>G
1. Introduction
PCOS is a condition that affects women of childbearing age and is caused by a combination of genetic, hormonal, and environmental factors. Symptoms include acne, amenorrhea, hyperandrogenism, hirsutism, obesity, oligomenorrhea, insulin resistance, and infertility. The prevalence of PCOS in female infertility has been confirmed to be 40 % (Nautiyal et al., 2022). Between 40 and 80 % of PCOS women are overweight or obese. Clustering of PCOS in families provides compelling evidence for a genetic predisposition to the condition. Insulin resistance was found to be common pathway between PCOS and obesity (Henry 2022). PCOS has become very common in global women and is identified as one of the potential risk factors for infertility and has been linked to acne and hirsutism (Abraham Gnanadass et al., 2021). Androgen levels are elevated, ovulation dysfunction is prevalent, and PCOS morphological abnormalities are also present. “Hyperandrogenism with ovulatory dysfunction,” as described by the National Institutes of Health. The prevalence of PCOS in women in their reproductive years is from 6 to 10 %, although it can be as high as double that (Xu and Qiao 2022). The global prevalence of PCOS was found to be between 4 and 21 %, whereas frequency in adolescents ranges from 9.1 to 36 % depending on the diagnostic criteria utilized (Rashid et al., 2022). Stein and Leventhal identified hirsutism, amenorrhea, infertility, enlarged ovaries, obesity, and prolonged anovulation in 1935. In 1990, the World Health Organization included PCOS to the 10th version of the International Classification of Diseases. Unapproved pharmaceuticals were used to treat PCOS patients based on their symptoms (Bogari 2020). Large prospective cohorts have indicated development to either pre-diabetes or type 2 diabetic mellitus (T2DM) over the course of years. Because insulin resistance and β-cell dysfunction are common in PCOS women, it is possible to predict the development of T2DM (Livadas et al., 2022). PCOS symptoms are not limited to the gynecological sphere, but also include endocrinological disorders such as metabolic syndrome (MetS), T2DM, hypertension, dyslipidemia, and obesity, which are more common in women with the disease than in those without it (Shetty et al., 2022).
There is a strong correlation between environmental and genetic factors, and vice versa, when it comes to disease. Both environmental and genetic factors have a role in PCOS. Although the genetic etiology of PCOS varies within and within families, a common pathway can be found. Patients from the same family have varying levels of genetic vulnerability to disease. It is therefore possible to uncover possible correlations by conducting large population-based case-control studies and genome-wide association studies (GWAS). In order to uncover possible correlations in which PCOS is known to be polygenic and multifactorial syndromic disorder, population-based case-control studies and GWAS are beneficial (Khan et al., 2019a, Khan et al., 2019b). The human APE1 gene is engaged in the base excision repair process and is found on chromosome 14q11.2-q12. The APE1 gene is made up of five exons totaling 2.21 kb. A total of 18 SNPs were found in the APE1 gene, with Asp148Glu (rs3136820) being the most prevalent. APE1 generates normal 39-hydroxyl nucleotide termini from oxidized DNA by hydrolyzing 39-blocking fragments (Gu et al., 2011). Another significant ApE1 gene SNP, 656 T > G (rs1760944), is found in the promoter region (Yousefi et al., 2015). Limited studies have addressed the relation between specific SNPs in gynecological complications such as female infertility (Mashayekhi et al., 2016) and PCOS (Gulbay et al., 2017). Apart from this, the prevalence of PCOS was found to be 53.7 % in Saudi Arabia (Guraya 2013). As a result, the current study sought to investigate the relationship between −656 T > G and 1349 T > G polymorphisms in the APE1 gene and PCOS in Saudi women.
2. Materials and methods
2.1. Ethical concerns
Ethical committee has approved this study within hospital premises. Two hundred Saudi women who took part in this study signed an informed consent form before taking part. This study protocol complied with both original and updated version of the Helsinki Declaration.
2.2. Deigned of Saudi women
Based on predetermined inclusion and exclusion criteria, two hundred Saudi women participated in this study. The Saudi women were extracted into 100 PCOS patients and remaining 100 women as control participants. These 200 Saudi women came from the KKUH Gynec department. This study recruitment was carried out for 10 months starting from January 2021-November 2021 in the hospital premises. Inclusion criteria of PCOS women is based on Rotterdam criteria (Eshre and Group 2004), oligo-or-anovulation and polycystic ovaries and women who does not meet under Rotterdam criteria is confirmed as exclusion criteria of PCOS women (Kawwass et al., 2010). Saudi women with normal ovulation are confirmed as inclusion criteria of control subjects and exclusion criteria will be confirmed based on fertile subjects, ovarian lesions and family history associated with PCOS. Pregnant women and women with other metabolic diseases were excluded from both the PCOS and control groups in this study. Participant information was acquired.
2.3. Ergonomic assessments
Age and body mass index (BMI) (Alshammary and Khan 2021)data were collected for 200 women. A blood sample of 4 mL was obtained in a plain tube and a blood sample of 2 mL was collected in an EDTA tube.
2.4. Biochemical and genotyping analysis
The FBG and lipid profile were measured using a plain blood sample (Alharbi et al., 2021), and the genomic DNA was isolated using a Qiagen kit and methodology. The genomic DNA was converted into 20 ng using NanoDrop spectrophotometer. PCR-RFLP analysis was implemented in this study. A couple of SNPs (-656 T > G and 1349 T > G) was adopted from the previous studies among PCOS women (Gulbay et al., 2017). Using a PCR master kit, double-distilled purified water, primers (+/-), and DNA template, genotyping analysis and PCR conditions was performed (Khan et al., 2015). PCR products were electrophoretically separated on a 2 % agarose gel stained with ethidium bromide. RFLP analysis was carried out. Electrophoresis was carried out. Table 1 lists the primers, band sizes, and restriction enzymes.
Table 1.
Details of SNPs, Primers and RFLPs used in this study.
| Gene | Mutation | rsnumber | Forward Primers | Reverse Primers | PCR | Enzyme | Band Size |
|---|---|---|---|---|---|---|---|
| APE1 | −656 T > G | rs1760944 | CTCCTAACCCGAGCACAAAG | CTAACTGCCAGCGAGGCCA | 371 bp | HpyCH4III | T-280/91; G-258/91/22 |
| APE1 | 1349 T > G | rs1130409 | CTGTTTCATTTCTATAGGCTA | AGGAACTTGCGAAAGGCTTC | 164 bp | BfaI | T-164 bp; G-144/20 bp |
2.5. Statistical analysis
SPSS software was used for statistical analysis (v.25). When applicable, baseline features of PCOS women and controls were examined using independent sample t-tests (Table 2). χ2 goodness of fit tests were used to look for evidence of a departure from Hardy Weinberg equilibrium (HWE) at the −656 T > G and 1349 T > G SNPs in the APE1 gene (Table 3). Adjusted odds ratios (ORs) with 95 % confidence intervals (CIs) were calculated using multiple logistic regression analysis to determine the correlation between allele, genotype, and various genetic model frequencies for APE1 SNPs and the risk of PCOS (Table 4). ANOVA (one-way; Table 5) analysis was done between baseline parameters and the APE1 gene SNPs −656 T > G and 1349 T > G (Khan et al., 2019a, Khan et al., 2019b). A p value of < 0.05 was deemed statistically significant (p < 0.05).
Table 2.
Demographic factors between PCOS women and control subjects.
| PCOS (n = 100) | Controls (n = 100) | P value | |
|---|---|---|---|
| Age (Years) | 29.63 ± 5.42 | 29.14 ± 5.18 | 0.51 |
| Weight (kgs) | 77.63 ± 9.40 | 62.14 ± 8.18 | <0.0001 |
| Height (cms) | 156.79 ± 4.76 | 156.58 ± 4.58 | 0.75 |
| BMI (Kg/m2) | 31.37 ± 3.90 | 25.30 ± 3.01 | <0.0001 |
| FBG (mmol/L) | 5.12 ± 0.76 | 5.08 ± 0.67 | 0.69 |
| TC (mmol/L) | 1.79 ± 1.07 | 1.61 ± 0.98 | 0.21 |
| TG (mmol/L) | 5.06 ± 1.06 | 2.24 ± 0.26 | <0.0001 |
| HDLC (mmol/L) | 0.63 ± 0.23 | 0.61 ± 0.21 | 0.52 |
| LDLC (mmol/L) | 3.61 ± 0.90 | 2.58 ± 0.64 | <0.0001 |
| Family History | 33 (33 %) | 00 (0 %) | <0.0001 |
Table 3.
Output of HWE analysis between T656G and T1349G variants in APE1 gene.
| rs1760944 | HWE | χ2 | P value |
|---|---|---|---|
| Controls | 0.13 | 0.07 | 0.78 |
| Cases | 0.42 | 37.05 | 0.00001 |
| rs1130409 | HWE | χ2 | P value |
| Controls | 0.52 | 1.61 | 0.20 |
| Cases | 0.66 | 0.48 | 0.48 |
Table 4.
Genotype and allele distribution between T656G and T1349G variants in APE1 gene.
| rs1760944 (-T656G) | PCOS (n = 100) | Controls (n = 100) | ORs (95 %CI) | P value |
|---|---|---|---|---|
| TT Genotype | 49 (49 %) | 53 (53 %) | Reference | Reference |
| TG Genotype | 19 (19 %) | 17 (17 %) | 1.20 (0.56–2.58) | 0.62 |
| GG Genotype | 32 (32 %) | 30 (30 %) | 1.15 (0.61–2.17) | 0.65 |
| Dominant Model | 51 (51 %) | 47 (47 %) | 1.17 (0.67–2.04) | 0.57 |
| Co-dominant Model | 19 (19 %) | 17 (17 %) | 1.14 (0.55–2.35) | 0.71 |
| Recessive Model | 32 (32 %) | 30 (30 %) | 1.09 (0.60–2.00) | 0.75 |
| T allele | 117 (58.5 %) | 123 (61.5 %) | Reference | Reference |
| G allele | 83 (41.5 %) | 77 (38.5 %) | 1.13 (0.75–1.69) | 0.54 |
| rs1130409 (T1349G) | PCOS (n = 100) | Controls (n = 100) | ORs (95 %CI) | P value |
| TT Genotype | 10 (10 %) | 26 (26 %) | Reference | Reference |
| TG Genotype | 48 (48 %) | 44 (44 %) | 2.83 (1.22–6.54) | 0.01 |
| GG Genotype | 42 (42 %) | 31 (31 %) | 3.52 (1.48–8.36) | 0.003 |
| Dominant Model | 90 (90 %) | 76 (76 %) | 2.84 (1.27–6.31) | 0.008 |
| Co-dominant Model | 48 (48 %) | 44 (44 %) | 1.17 (0.67–2.05) | 0.57 |
| Recessive Model | 42 (42 %) | 31 (31 %) | 1.61 (0.90–2.88) | 0.10 |
| T allele | 68 (34 %) | 96 (48 %) | Reference | Reference |
| G allele | 132 (66 %) | 104 (52 %) | 1.79 (1.19–2.68) | 0.004 |
Table 5.
Anova analysis between clinical factors and T656G and T1349G variants.
|
rs1760944 (-T656G) |
rs1130409 (T1349G) |
|||||||
|---|---|---|---|---|---|---|---|---|
| TT (n = 49) | TG (n = 19) | GG (n = 32) | P value | TT (n = 10) | TG (n = 48) | GG (n = 42) | P Value | |
| Age | 29.34 ± 5.55 | 29.36 ± 5.41 | 30.21 ± 5.34 | 0.76 | 30.20 ± 6.09 | 29.60 ± 5.73 | 29.52 ± 5.00 | 0.93 |
| Weight | 77.21 ± 9.24 | 76.57 ± 9.00 | 78.96 ± 10.00 | 0.61 | 78.17 ± 6.45 | 76.82 ± 11.11 | 78.47 ± 7.82 | 0.70 |
| Height | 156.42 ± 5.14 | 157.31 ± 3.92 | 157.05 ± 4.70 | 0.73 | 157.40 ± 4.67 | 156.10 ± 4.41 | 157.44 ± 5.15 | 0.37 |
| BMI | 31.37 ± 2.88 | 30.76 ± 2.72 | 31.71 ± 3.59 | 0.57 | 31.39 ± 1.63 | 31.25 ± 3.54 | 31.49 ± 2.83 | 0.93 |
| FBG | 5.16 ± 0.74 | 5.11 ± 0.52 | 5.05 ± 0.91 | 0.81 | 4.87 ± 0.61 | 5.10 ± 0.63 | 5.19 ± 0.91 | 0.48 |
| TC | 1.78 ± 0.97 | 1.70 ± 1.12 | 1.85 ± 1.23 | 0.89 | 2.20 ± 1.28 | 1.66 ± 0.88 | 1.84 ± 1.22 | 0.32 |
| TG | 4.95 ± 1.11 | 5.03 ± 0.96 | 5.24 ± 1.07 | 0.49 | 5.12 ± 0.67 | 5.03 ± 1.00 | 5.08 ± 1.22 | 0.95 |
| HDLC | 0.64 ± 0.25 | 0.68 ± 0.21 | 0.59 ± 0.21 | 0.38 | 0.65 ± 0.25 | 0.64 ± 0.22 | 0.62 ± 0.25 | 0.89 |
| LDLC | 3.50 ± 0.97 | 3.58 ± 0.82 | 3.80 ± 0.84 | 0.34 | 3.47 ± 0.65 | 3.64 ± 0.89 | 3.62 ± 0.98 | 0.86 |
3. Results
3.1. Basic information of Saudi women
One hundred PCOS women and one hundred control subjects were collected from KKUH for this study. The 200 women participating in this study were all of Saudi origin. Table 2 provides the clinical and biochemical information. The mean age was 29.63 ± 5.42 in PCOS women and 29.14 ± 5.18 in controls. No significant difference was found among PCOS women and controls in terms of age (p = 0.51), height (p = 0.75) FBG (p = 0.69), TC (n = 0.21) and HDLC (p = 0.52). A strong significant association was observed in weight, BMI, TG and LDLC (p < 0.0001) when compared between PCOS and control women.
3.2. HWE and genotyping analysis of APE1 gene
The findings indicate that both rs1760944 (-656 T > G) and rs1130409 (1349 T > G) SNPs were found on chromosome 14q11.2. The 1349 T > G SNP is involved in protein coding. Except for PCOS women with-656 T > G, significant difference in rs1760944 (-656 T > G) and rs1130409 (1349 T > G) SNPs in the APE1 gene was detected between cases and controls (Table 3).
3.3. Genotype distribution between PCOS women and control subjects
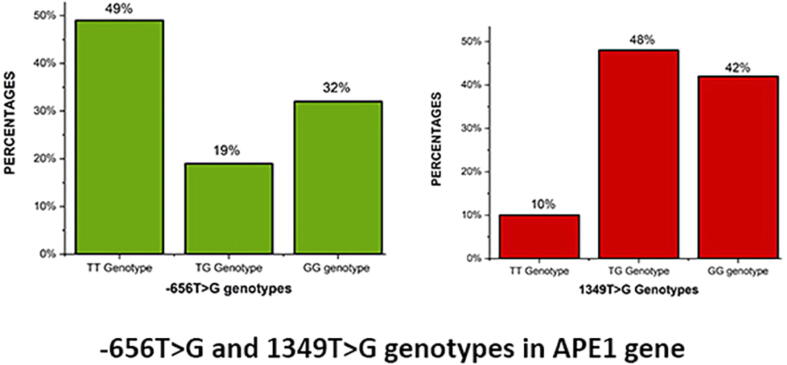
Table 4 shows the genotype distribution and allele frequencies of the −656 T > G and 1349 T > G SNPs in PCOS women and controls. In the −656 T > G SNP, PCOS women had genotype frequencies of 49 %, 19 %, and 32 % as TT, TG, and GG, respectively Fig. 1, while controls had genotype frequencies of 53 %, 17 %, and 30 % as TT, TG, and GG, respectively. The dominant allele (T) was found to be present in 58.5 % of PCOS women and 61.5 % of controls, whereas the minor allele (G) was found to be present in 41.5 % of PCOS women and 38.5 % of controls. There was no correlation between PCOS women and control participants in terms of allele frequency (OR-1.13 [95 %CI:0.75–1.69]; p = 0.54), genotype (TG vs TT: OR-1.20 [95 %CI: 0.56–2.58]; p = 0.62 and GG vs TT: OR-1.15 [95 %CI: 0.61–2.17]; p = 0.65), or genetic models such as dominant (OR-1.17 [95 %CI: 0.67–2.17]; p = 0.57), co-dominant (OR-1.14 [95 %CI: 0.55–2.35]; p = 0.71), and recessive models (OR-1.09 [95 %CI: 0.60–200]; p = 0.75).
Fig. 1.
Genotype frequencies between −656 T > G and 1349 SNPs in PCOS women.
The 1349 T > G SNP was found to be associated to allele (OR-1.79 [95 %CI: 1.19–2.68]; p = 0.004), genotype (TG vs TT: OR-2.83 [95 %CI: 1.22–6.54]; p = 0.01 and GG vs TT: OR-3.52 [95 %CI: 1.48–8.36]; p = 0.003), and dominant models (OR-2.84 [95 %CI: 1.27–6.31); p = 0.008). However, neither co-dominant (OR-1.17 [95 %CI: 0.67–2.05]; p = 0.57) nor recessive models were (OR-1.61 [95 %CI: 0.90–2.88]; p = 0.10) associated. PCOS women had 66 % of the G allele and 34 % of the T allele, while controls had 52 % of the G allele and 48 % of the T allele. In PCOS women, the genotype frequencies were 10 %, 48 %, and 42 % as TT, TG, and GG genotypes (Figure-1), respectively, whereas in controls, the genotype frequencies were 26 % as TT, 44 % as TG, and 31 % as GG genotypes.
3.4. Anova analysis between APE1 variants and Table 1
Anova analysis was conducted in Table 5 in this study between genotypes involving TT, TG, and GG in-656 T > G and 1349 T > G SNPs and clinical and biochemical factors include age, height, weight, BMI, FBG, TC, TG, HDLC, and LDLC levels in PCOS women. No significant association was found between −656 and 1349 SNPs (p < 0.05). FBG (5.16 ± 0.74) was found to be higher in the TT genotype at the −656 SNP, whereas height (157.31 ± 3.92) and HDLC (0.68 ± 0.21) were found to be elevated in the TG genotype and age (30.21 ± 5.34), weight (78.96 ± 10.00), BMI (31.71 ± 3.59), TC (1.85 ± 1.23), TG (5.24 ± 1.07), and LDLC (3.80 ± 0.84) were found to be elevated in the GG genotype. Among 1349 T > G SNP, age (30.20 ± 6.09), TC (30.20 ± 6.09), TG (5.12 ± 0.67), and HDLC (0.65 ± 0.25) were found to be higher values in the TT genotype, LDLC (3.64 ± 0.89) was found to be higher in TG genotype, and weight (78.47 ± 7.82), height (157.44 ± 5.15), BMI (31.49 ± 2.83), and FBG (5.19 ± 0.91) were found to be significantly higher in the GG genotype.
4. Discussion
The purpose of this study is to examine the effects of the APE1 −656 T > G and −1349 T > G SNPs in the predisposition to PCOS women in Saudi Arabia. The APE1 gene is a key regulator of the frontline base excision repair pathway, which may be employed to repair DNA damage processes. The APE1 gene has been linked to a variety of human diseases, including cancer (Liu et al., 2021), male (Yousefi et al., 2015) and female infertilities (Mashayekhi et al., 2016) and PCOS (Gulbay et al., 2017). The human APE1 gene is 318 amino acid protein and 3 kb in size, found on chromosome 14, with 4 introns and 5 exons. APE1 is a numerous oxidation reduction factors as well as other repair pathways such as DNA repair, transcription factor activity modulation, apoptosis, redox homeostasis, and maturation related multifunctional protein. DNA repair systems are damaged in monocytes, and APE1 plays an important function in those cells in other ways (Betlej et al., 2020). Kaur was identified Asp148Glu SNP (Kaur and Kaur 2018).
The currents study results confirmed age, weight, BMI, TG, LDLC and family history are associated in PCOS women (p < 0.05) and HWE analysis showed that −656 T > G and 1349 T > G SNPs in the APE1 gene were the same as in the control group (p > 0.05). None of the allele and genotype frequencies in the −656 T > G SNP were linked to each other. (p > 0.05), whereas, 1349 T > G SNP was found to be associated with allele (OR-1.79 [95 %CI: 1.19–2.68]; p = 0.004), genotypes (TG vs TT: OR-2.83 [95 %CI: 1.22–6.54]; p = 0.01 and GG vs TT: OR-3.52 [95 %CI: 1.48–8.36]; p = 0.003) and dominant model (OR-2.84 [95 %CI: 1.27–6.31); p = 0.008). However, Anova analysis failed to show the association between Table 1 and a couple of SNPs involved in this study (p > 0.05). The genotyping analysis of 1349 T > G SNP of current study results was found to be associated with previous studies carried out in PCOS (Gulbay et al., 2017) and female infertility (Mashayekhi et al., 2016). APE1 gene studies was carried out in Saudi Population and confirmed the positive association with 1349 T > G SNP (AlMutairi, et al., 2015, Nemer et al., 2018). Unfortunately, limited studies in female populations have been conducted in the APE1 gene, and this is the first study documented in PCOS disease in Saudi Arabia, and it was examined with −656 T > G and 1349 T > G SNPs, and Asp148Glu (1349 T > G) variant was strongly associated with PCOS women. Having a 1349 T > G SNP in APE1 can lead to decreased activity of APE1 in the DNA base excision repair pathway, which increases the risk of polycystic ovary syndrome in carriers. APE1 individuals with the 1349 T > G SNP have reduced APE1 activity in the DNA base excision repair pathway, which contributes to an increased risk of polycystic ovary syndrome. A greater prevalence of human diseases among Asian women is suggested by a previous meta-analysis study (Mohammed Nawi et al., 2021). This study analysis supports the observation by demonstrating that the Glu residue at 148 locations in APE1 enhances the risk of PCOS in Saudi women. The current study discovered and demonstrated that the Glu residue at APE1 position 148 significantly increases the incidence of PCOS in Saudi women.
PCOS is considered as hormonal and metabolic disorder affecting in reproductive aged women causing them ovarian problems. The definition of PCOS can be defined as a hormonal condition that affects women of reproductive age. PCOS, which often begins after adolescence, causes the emergence of numerous symptoms connected with the hormonal disorder, which causes ovarian issues and leads to egg underdevelopment or hinders its release during ovulation. The pathophysiology of PCOS is elusive, however it is thought to be connected to both inherited and environmental factors. Acne and irregular menstruation were considered to be frequent PCOS symptoms. Obesity and T2DM have been linked to PCOS. All human diseases are connected to a family health history that may be inherited, and genes can be transmitted and detected in individuals who have a greater than average risk of developing human disorders. Family members will inherit common traits via sharing their DNA, environment, lifestyles, and behaviors. The family history of PCOS was found to be 33 % in this study, indicating that one-third of Saudi women were diagnosed, and these women are at risk of developing future complications such as diabetes, cancer, MetS, and endometrial cancer (Helvaci and Yildiz 2020). There is currently no cure for PCOS, however it can be treated with restricted medicines and fertility treatment. The strength of this study was to enroll all Saudi women and studied the couple of important markers in APE1 gene. The limitations of this study were the selection of 100 PCOS women and 100 controls, as well as the omission of serum concentration.
5. Conclusion
Finally, this study confirms as 1349 T > G SNP was associated in PCOS women. However, −656 T > G was not associated. Future studies should be carried out in global population on male and female couples, and the impact of the −656 T > G and 1349 T > G polymorphisms in the APE1 gene should be investigated using meta-analysis.
Declaration of Competing Interest
The author declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
References
- Abraham Gnanadass S., Divakar Prabhu Y., Valsala Gopalakrishnan A. Association of metabolic and inflammatory markers with polycystic ovarian syndrome (PCOS): an update. Arch. Gynecol. Obstet. 2021;303(3):631–643. doi: 10.1007/s00404-020-05951-2. [DOI] [PubMed] [Google Scholar]
- Alharbi K.K., Abudawood M., Khan I.A. Amino-acid amendment of arginine-325-tryptophan in rs13266634 genetic polymorphism studies of the SLC30A8 gene with type 2 diabetes-mellitus patients featuring a positive family history in the Saudi population. Journal of King Saud University-Science. 2021;33(1) [Google Scholar]
- AlMutairi, F., Ali Khan Pathan, A., Alanazi, M. et al., 2015. Association of DNA repair gene APE1 Asp148Glu polymorphism with breast cancer risk. Disease Markers. 2015. [DOI] [PMC free article] [PubMed]
- Alshammary A.F., Khan I.A. Screening of obese offspring of first-cousin consanguineous subjects for the angiotensin-converting enzyme gene with a 287-bp Alu sequence. Journal of obesity & metabolic syndrome. 2021;30(1):63. doi: 10.7570/jomes20086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betlej G., Bator E., Pyrkosz A., et al. A dual face of APE1 in the maintenance of genetic stability in monocytes: an overview of the current status and future perspectives. Genes. 2020;11(6):643. doi: 10.3390/genes11060643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogari N.M. Genetic construction between polycystic ovarian syndrome and type 2 diabetes. Saudi Journal of Biological Sciences. 2020;27(10):2539–2543. doi: 10.1016/j.sjbs.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshre, R. and Group, A.-S. P. C. W. 2004. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS). Human Reproduction (Oxford, England). 19 (1), 41-47. [DOI] [PubMed]
- Gu D., Wang M., Wang S., et al. The DNA repair gene APE1 T1349G polymorphism and risk of gastric cancer in a Chinese population. PLoS One. 2011;6(12):e28971. doi: 10.1371/journal.pone.0028971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbay G., Yesilada E., Celik O., et al. The investigation of polymorphisms in DNA repair genes (XRCC1, APE1 and XPD) in women with polycystic ovary syndrome. Asian Pac. J. Cancer Prev. 2017;18(5):1219. doi: 10.22034/APJCP.2017.18.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guraya S.S. Prevalence and ultrasound features of polycystic ovaries in young unmarried Saudi females. Journal of Microscopy and ultrastructure. 2013;1(1–2):30–34. [Google Scholar]
- Helvaci N., Yildiz B.O. Polycystic ovary syndrome and aging: Health implications after menopause. Maturitas. 2020;139:12–19. doi: 10.1016/j.maturitas.2020.05.013. [DOI] [PubMed] [Google Scholar]
- Henry, L., 2022. Genetic Origins of Polycystic Ovarian Syndrome (PCOS): An Analysis of the Genetic Correlation Between PCOS and Insulin Receptor Mutations.
- Kaur K., Kaur R. Absence of APE1 (Asp148Glu) gene polymorphism in North-West Indian population: A comparison with world population. Meta gene. 2018;16:208–212. [Google Scholar]
- Kawwass J.F., Loucks T.L., Berga S.L. An algorithm for treatment of infertile women with polycystic ovary syndrome. Middle East Fertility Society Journal. 2010;15(4):231–239. [Google Scholar]
- Khan I., Jahan P., Hasan Q., et al. Relationship between PTEN and gestational diabetes in Asian Indians womens. Journal of Health Specialties. 2015;3(3):184. [Google Scholar]
- Khan I.A., Jahan P., Hasan Q., et al. Genetic confirmation of T2DM meta-analysis variants studied in gestational diabetes mellitus in an Indian population. Diabetes Metab. Syndr. 2019;13(1):688–694. doi: 10.1016/j.dsx.2018.11.035. [DOI] [PubMed] [Google Scholar]
- Khan M.J., Ullah A., Basit S. Genetic basis of polycystic ovary syndrome (PCOS): current perspectives. Appl. Clin. Genet. 2019;12:249. doi: 10.2147/TACG.S200341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zheng J., Guo Y., et al. Association between APE1 rs1760944 and rs1130409 polymorphism with prostate cancer risk: A systematic review and meta-analysis. Medicine. 2021;100(46) doi: 10.1097/MD.0000000000027630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livadas S., Anagnostis P., Bosdou J.K., et al. Polycystic ovary syndrome and type 2 diabetes mellitus: A state-of-the-art review. World J. Diabetes. 2022;13(1):5. doi: 10.4239/wjd.v13.i1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashayekhi F., Yousefi M., Salehi Z., et al. The association of− 656T> G and 1349T> G polymorphisms of ApE1 gene and the risk of female infertility. J. Obstet. Gynaecol. 2016;36(4):544–547. doi: 10.3109/01443615.2015.1127903. [DOI] [PubMed] [Google Scholar]
- Mohammed Nawi, A., Mohammad, Z., Jetly, K., et al., 2021. The prevalence and risk factors of hypertension among the urban population in southeast asian countries: a systematic review and meta-analysis. International Journal of Hypertension. 2021. [DOI] [PMC free article] [PubMed]
- Nautiyal H., Imam S.S., Alshehri S., et al. Polycystic Ovarian Syndrome: A Complex Disease with a Genetics Approach. Biomedicines. 2022;10(3):540. doi: 10.3390/biomedicines10030540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemer A.O., Al Anazi M.S., Bhat R.S., et al. Association between preterm birth risk and polymorphism and expression of the DNA repair genes OGG1 and APE1 in Saudi women. Biocell. 2018;42(1):1. [Google Scholar]
- Rashid R., Mir S.A., Kareem O., et al. Polycystic ovarian syndrome-current pharmacotherapy and clinical implications. Taiwan. J. Obstet. Gynecol. 2022;61(1):40–50. doi: 10.1016/j.tjog.2021.11.009. [DOI] [PubMed] [Google Scholar]
- Shetty S.S., Kumari N.S., Hegde P. Leptin Gene Polymorphism Rs7799039; G2548A, Metabolic and Oxidative Stress Markers in Polycystic Ovarian Syndrome. Journal of King Saud University-Science. 2022;102222 [Google Scholar]
- Xu, Y. and Qiao, J. 2022. Association of Insulin Resistance and Elevated Androgen Levels with Polycystic Ovarian Syndrome (PCOS): A Review of Literature. Journal of Healthcare Engineering. 2022. [DOI] [PMC free article] [PubMed]
- Yousefi M., Salehi Z., Mashayekhi F., et al. The association of ApE1− 656T> G and 1349T> G polymorphisms and idiopathic male infertility risk. Int. Urol. Nephrol. 2015;47(6):921–926. doi: 10.1007/s11255-015-0979-z. [DOI] [PubMed] [Google Scholar]