Abstract
Mosquitoes are vectors for emerging and re-emerging infectious viral diseases of humans, livestock and other animals. In addition to these arthropod-borne (arbo)viruses, mosquitoes are host to an array of insect-specific viruses, collectively referred to as the mosquito virome. Mapping the mosquito virome and understanding if and how its composition modulates arbovirus transmission is critical to understand arboviral disease emergence and outbreak dynamics. In recent years, next-generation sequencing as well as PCR and culture-based methods have been extensively used to identify mosquito-associated viruses, providing insights into virus ecology and evolution. Until now, the large amount of mosquito virome data, specifically those acquired by metagenomic sequencing, has not been comprehensively integrated. We have constructed a searchable database of insect-specific viruses associated with vector mosquitoes from 175 studies, published between October 2000 and February 2022. We identify the most frequently detected and widespread viruses of the Culex, Aedes and Anopheles mosquito genera and report their global distribution. In addition, we highlight the challenges of extracting and integrating published virome data and we propose that a standardized reporting format will facilitate data interpretation and re-use by other scientists. We expect our comprehensive database, summarizing mosquito virome data collected over 20 years, to be a useful resource for future studies.
Keywords: Virome, Insect-specific viruses, Mosquito, Aedes, Culex, Anopheles, Metagenomics
1. Introduction
Hematophagous mosquitoes are vectors for the transmission of arthropod-borne viruses (arboviruses) to humans, livestock and wild animals. Particularly, mosquitoes of the genera Aedes and Culex transmit epidemic arboviruses, including dengue virus, yellow fever virus, Zika virus and West Nile virus [[1], [2], [3]]. Mosquitoes of the genus Anopheles are the main vector for O'nyong-nyong virus as well as for malaria parasites [4]. Besides arboviruses, which have a dual host range alternating between vertebrates and arthropods, mosquitoes carry viruses with an insect-restricted host range (insect-specific viruses, ISVs), as well as viruses that infect microbes such as the bacteria and fungi that colonize the mosquito host [5,6]. These mosquito-associated viruses are collectively referred to as the mosquito virome.
Virome studies of mosquitoes, and invertebrates in general, have shed light on the vast diversity of viruses on earth [7,8]. In recent years, next-generation sequencing, PCR-based detection, and virus culture approaches have been extensively used to map the virome across mosquito genera, ecological environments, and geographical locations. These studies expanded the host range of virus families to include arthropods (e.g., in the Totiviridae and Partitiviridae families [[9], [10], [11]]), introduced new clades within existing viral families or orders (e.g., Artivirus in the Totiviridae [9] and Goukovirus, Herbevirus, Jonvirus and Feravirus in the Bunyavirales [[12], [13], [14]]), and necessitated the creation of novel viral families and genera (e.g., Mesoniviridae and Negevirus [[15], [16], [17], [18]]). Additionally, fundamental insights into virus evolution may be obtained from these studies, as exemplified by the discovery of Nam Dinh virus (or alphamesonivirus 1) in mosquitoes, which led to the establishment of a new family Mesoniviridae in the order Nidovirales, containing viruses with a genome size intermediate between the small-sized Arteriviridae and the large-sized Coronaviridae and Roniviridae [16].
The mosquito virome has raised significant interest because of its potential impact on the transmission of arboviruses or malarial parasites [[19], [20], [21], [22], [23], [24]]. Correlating spatiotemporal virome data with vector-borne disease incidence may provide insights into the impact of ISVs on pathogen transmission. Moreover, for many ISVs, the host range and the potential to cross the species barrier and infect other (vertebrate or invertebrate) animals remains to be established. Virus infection may impact mosquito physiology and development, which is almost completely uncharacterized thus far but may, directly or indirectly, affect vectorial capacity. Thus, for a One Health perspective on arbovirus transmission, a systematic overview of the prevalence of mosquito-specific viruses is essential. Such an overview may also inform biotechnological applications of ISVs, such as the development of novel vaccine platforms, or their use as biological agents to prevent arbovirus transmission by mosquitoes [22,[25], [26], [27]].
Arboviruses are mainly transmitted horizontally between mosquito and vertebrate hosts. In contrast, during adverse conditions such as cold winters or drought, it is hypothesized that arboviruses are vertically transmitted, even if it may be relatively inefficient [28,29]. ISVs are often assumed to be transmitted vertically from parent to offspring, but direct experimental support for this transmission mode is scarce and both vertical and horizontal transmission routes have been proposed [30,31]. Virome studies could be used to deduce transmission modes. For example, frequent recovery of an ISV from early life stages such as eggs or larvae could be indicative of vertical transmission, whereas recovery of the same virus from different mosquito species would suggest a horizontal transmission mode via the environment, such as shared food sources.
It is likely that the virome differs between mosquito species, between populations of the same species of mosquitoes, and between individual mosquitoes within populations, which may depend on the transmission mode as well as on viral and host genetics, mosquito ecology, and environmental and climatic conditions. Yet, some ISVs may be present in mosquito populations across the globe or have a broad mosquito host range. For those viruses, it will be particularly relevant to determine their impact on mosquito physiology, development and pathogen transmission.
The large amount of mosquito virome information has thus far been integrated at different levels of analysis. Some studies compared their acquired metagenomic data with sequencing data from other studies [[32], [33], [34], [35]] and several reviews have collated lists of (insect-specific) viruses detected in mosquitoes [5,25,36]. However, an exhaustive analysis of mosquito-associated viruses, including their location and associated mosquito hosts, is lacking. In this study, we performed a comprehensive review of 175 mosquito virome studies, published between October 2000 and February 2022, to construct a searchable database of mosquito-associated viruses. We present the most widespread and frequently detected insect-specific viruses within the Culex, Aedes and Anopheles mosquito genera and highlight viruses with a particularly broad mosquito host range. We expect our database to be a useful resource for further study of insect-specific viruses.
2. Methods
2.1. Search strategy
A PubMed search was performed on January 26, 2022, using a combination of title/abstract (Tiab) search terms and Medical Subject Headings (MeSH) terms. The search strategy combined the following terms for (insect-specific) virus discovery with terms for mosquito research:
(“Virome”[MeSH Terms] OR “Metagenomics”[MeSH Terms] OR “Insect Viruses”[MeSH Terms] OR “Metatranscriptomic*”[Title/Abstract] OR “Meta transcriptomic*”[Title/Abstract] OR “Metagenom*”[Title/Abstract] OR “Insect Specific Virus*”[Title/Abstract] OR “ISV”[Title/Abstract] OR “Virus Discovery”[Title/Abstract] OR “Virom*”[Title/Abstract] OR “Insect Specific Flavivirus*”[Title/Abstract] OR “Insect Specific Alphavirus*”[Title/Abstract] AND (“Culicidae”[MeSH Terms] OR “Culicid*”[Title/Abstract] OR “Aedes”[Title/Abstract] OR “Anophel*”[Title/Abstract] OR “Culex”[Title/Abstract] OR “Mosquit*”[Title/Abstract]).
The search strategy retrieved 743 articles, which were manually screened. All articles written in English and reporting primary data on virus detection or identification in wild-caught mosquitoes were eligible for the analysis. Articles that only detected arboviruses in mosquitoes were excluded, leading to a final selection of 175 articles (references in Supplementary file 1).
2.2. Database assembly
Information on mosquito-associated viruses was extracted from the articles at the level of individual samples, containing either a single mosquito or a pool of mosquitoes, to construct a sample-structured database (Supplementary Table S1). Known arboviruses were not included in the table. Each entry in the database constitutes a virus detected in a mosquito sample. Samples tested negative for viruses were not included in the database. For each virus-positive sample, virus taxonomy at the family level, the mosquito species in which the virus was detected, sampling location, blood-feeding status, method for virus detection, material used for sequencing (RNA, DNA or both), the number of mosquitoes in the sample, and the developmental stage (larva, pupa, adult) was extracted from the articles, if this information could be unambiguously deduced. For consistency, Culex pipiens was used for studies reporting Culex pipiens complex and Culex pipiens sensu lato [37,38]. Likewise, Ochlerotatus caspius and Ochlerotatus scapularis were denoted as Aedes caspius and Aedes scapularis, respectively, as both genus names were used in the literature [39]. Virus detection methods were classified into four categories: 1) sequencing, for samples that were directly analyzed by next-generation sequencing, 2) PCR, for samples in which viruses of a particular virus taxon or species were detected by PCR, 3) culture-sequencing, when mosquito homogenate was cultured on mosquito cell lines, after which viruses were detected by next-generation sequencing, and 4) culture-PCR, when mosquito homogenate was cultured on cell lines and virus was detected by PCR using virus taxon or species specific PCR primers.
Information for the database was extracted from the relevant (supplemental) figures or tables as reported. When viruses were not assigned to the species level, but only the closest viral match was reported, these were included in the database. No thresholds were taken into account for the minimal number of reads and genome coverage required for the accurate detection of viruses, with the exception of the study by Hameed et al. [40] (see below). In addition, the authors' assessments were accepted for considering an identified viral sequence novel and giving it a new name. To allow comparison between studies, a column named ‘Virus (clean)’ was defined, in which strain or isolate names from the ‘Virus (reported)’ column were removed, virus abbreviations were written out, and consistent spelling was used.
The NCBI Taxonomy database was used as a reference to define unique viruses, as many ISVs are not yet formally classified by the International Committee on Taxonomy of Viruses (ICTV) and therefore absent from the ICTV Master Species List 2021 [41]. For entries without unambiguous reference to a unique virus, ‘unknown’ was used, except in occasional cases in which the virus name could be deduced from the NCBI taxonomy database using the reported GenBank accession numbers. Virus taxonomy was obtained from the NCBI Taxonomy Database (resourced March through June 2022; [42]) for entries that lack a definition of the virus family in the original article and for viruses with inconsistent taxonomy between articles. In the absence of virus taxonomy at the family level, ‘unknown’ was used.
2.3. Database curation
Initial analysis of the contribution of individual publications to the database indicated that one study dominated the dataset, supplying 4169 of the 8378 (50%) total unique virus entries [40] (Supplemental Fig. 1A). This overrepresentation could not be accounted for by the sampling size or sequence depth in this study as only ten pools of mosquitoes were sequenced, which contributed between 121 and 836 virus entries per pool to the database [40]. Instead, for the majority of the reported viruses only a single or few reads were detected and the percentage identity to the viral reference genomes was unreported, providing limited evidence for the presence of these viruses [40]. To prevent a disproportionally large influence of this study on our dataset, a threshold on the minimal number of sequencing reads was applied and only virus entries supported by ≥100 reads were included in the database for this particular study. After this curation, this study supplied 382 of the 4591 (8%) total unique virus entries in our database.
2.4. Analyses
Unique virus entries were defined by unique combinations of the columns ‘Study’, ‘Virus family’, ‘Virus (reported)’, ‘Location (Specific)’ and ‘Species’ for the analyses of virus families, or the columns ‘Study’, ‘Virus (clean)’, ‘Location (Specific)’ and ‘Species’ for the analyses at the virus species level. In-house R-scripts were used for data analyses.
3. Results
We performed a review of the literature on virus identification in wild-caught mosquitoes. Based on a total of 175 publications, we generated a database consisting of 11,261 rows, each entry representing a virus detected in a specific sample (Supplementary Table S1). The number of virus entries in this database is biased towards studies that acquire a large number of samples from the same location, in particular PCR studies that often test multiple mosquito pools sampled at the same site. To account for these biases, we used a transformed database for our analyses, which only included rows with unique combinations of Study - Virus - Mosquito species - Location. We refer to the rows of this database as unique virus entries (n = 4591) and use it as a metric for the abundance of viruses and virus families.
3.1. Overview of the literature
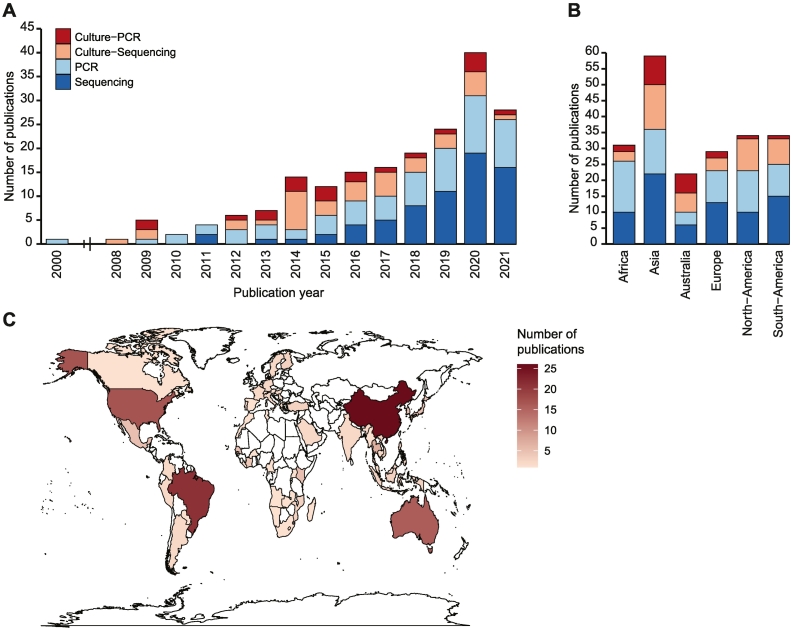
The number of mosquito virome studies has gradually risen over the years, with two studies published between 2000 and 2008 and a total of 27 studies published in 2021 (Fig. 1A). A slight majority of these studies (n = 70) used next-generation sequencing-based approaches to characterize the virome, whereas PCR-based approaches (n = 67) were frequently used to specifically detect viruses from genera known to contain arboviruses and/or ISVs, such as flaviviruses, alphaviruses, phleboviruses, orthobunyaviruses, densoviruses and rhabdoviruses (Fig. 1A). In fewer studies, mosquito homogenate was first cultured on mosquito cell lines, often the RNA interference (RNAi)-deficient Aedes albopictus C6/36 cell line, followed by PCR (n = 21) or next-generation sequencing (n = 38) to detect in vitro replicating viruses.
Fig. 1.
Characteristics of mosquito virome studies.
(A-B) Number of mosquito virome publications over time (A), and across continents (B), with fill color indicating the study approach. The sum of the categories within each bar may exceed the actual number of publications as some studies used multiple virus detection methods. (C) Geographic distribution of countries in which mosquitoes were sampled for virome studies.
Unique virus entries were not equally distributed across virome studies, with the majority of entries (88.1%) derived from metagenomic sequencing studies and only a small percentage from PCR studies (7.2%) and culturing studies (4.7%). Furthermore, over 50% of the total number unique virus entries were derived from only 10 out of 175 studies (Supplementary Fig. S1A).
Mosquitoes were sampled across the globe (Fig. 2B,C) with a relatively even distribution of PCR and sequencing-based methods (Fig. 1B). Sampling was, however, not uniform across continents, as China, Brazil, and the USA were the main sources of virome information from Asia, South-America and North-America, respectively (Fig. 1C).
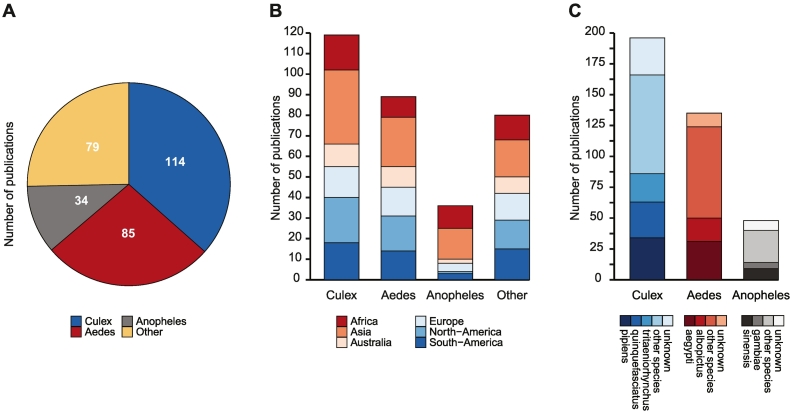
Fig. 2.
Characteristics of mosquitoes sampled for virome studies.
(A) Pie chart indicating the number of publications detecting viruses for each mosquito genus. (B) Number of publications for each mosquito genus with fill color indicating the continent in which mosquitoes were sampled. (C) Number of publications for each mosquito genus, with fill color indicating the most frequently sampled species. Some studies sampled mosquitoes from (A) multiple genera, (B) multiple continents, or (C) multiple species within one mosquito genus, and the sum of publications in each panel therefore exceeds the total number of publications.
3.2. Mosquito species sampled for virome studies
The 175 studies collectively detected viruses in 128 different mosquito species from 14 mosquito genera (Supplementary Table S2). Mosquitoes from the Culex, Aedes, and Anopheles genera were most frequently found to harbor viruses, likely because these vector mosquitoes are sampled more often for their importance in pathogen transmission (Fig. 2A). Culex mosquitoes contributed most unique virus entries in a total of 114 studies, whereas 85 studies detected viruses in Aedes mosquitoes, and 34 studies reported viruses in Anopheles mosquitoes (Fig. 2A, Table 1). As few studies detected viruses in other mosquito genera, we did not further analyze the data from those mosquitoes (Supplementary Table S1).
Table 1.
Mosquito genera in which viruses were detected with the corresponding number of species, continents, countries and studies. Sorted on the number of studies.
| Genus | Species | Continents | Countries | Studies |
|---|---|---|---|---|
| Culex | 33 | 6 | 42 | 114 |
| Aedes | 42 | 6 | 35 | 85 |
| Anopheles | 23 | 6 | 21 | 34 |
| Mansonia | 4 | 5 | 11 | 14 |
| Armigeres | 2 | 1 | 5 | 8 |
| Coquillettidia | 5 | 4 | 5 | 7 |
| Culiseta | 5 | 3 | 5 | 7 |
| Psorophora | 5 | 2 | 4 | 6 |
| Ochlerotatus | 3 | 4 | 5 | 5 |
| Uranotaenia | 1 | 2 | 2 | 3 |
| Sabethes | 2 | 1 | 1 | 2 |
| Aedeomyia | 1 | 1 | 1 | 1 |
| Heamagogus | 1 | 1 | 1 | 1 |
| Wyeomyia | 1 | 1 | 1 | 1 |
Mosquitoes were collected on every continent except Antarctica, with most extensive sampling in Asia and a clear overrepresentation of Anopheles sampling in Asia and Africa (Fig. 2B). Almost half of the studies reporting viruses in Culex, detected these viruses in Culex pipiens, Culex quinquefasciatus and Culex tritaeniorhynchus. For Aedes and Anopheles mosquitoes, most studies detected virus in Aedes aegypti and Aedes albopictus, and in Anopheles sinensis and Anopheles gambiae, respectively (Fig. 2C, Supplementary Table S2). Noteworthy, the mosquito species or even the genus was unknown for some virus positive pools, due to studies not defining their species and/or genus [[43], [44], [45]], or due to the use of pooled samples containing multiple species [[46], [47], [48]]. Only 13 studies evaluated the virome in immature developmental stages, such as eggs, larvae, or pupa [32,[49], [50], [51], [52], [53], [54], [55], [56], [57], [58], [59], [60]].
3.3. General overview of the mosquito virome
In total, viruses from 102 virus families were reported in all mosquito species combined, although the number of unique virus entries was very low for the majority of these families (Supplementary Fig. S1B). As expected, virus families known to contain ISVs and/or arboviruses were among the top 10 most frequently observed RNA virus families, including Flaviviridae, Rhabdoviridae, Iflaviridae, Nodaviridae, Mesoniviridae, Orthomyxoviridae and Totiviridae (Supplementary Fig. S1C, discussed below). In addition, two DNA virus families were found in the top 10 of multiple mosquito genera, Parvoviridae and Genomoviridae.
Metagenomic surveys have the power to identify viruses of every organism present in the sample. Indeed, bacteriophages were frequently detected, predominantly from the tailed dsDNA bacteriophage families Siphoviridae [40,43,45,[61], [62], [63], [64]], Myoviridae [34,40,45,62,[64], [65], [66], [67], [68]], and Autographiviridae [40,65,67,69] (Fig. S1B). These data underline that metagenomic sequencing can detect viral sequences of bacteria that colonize mosquitoes. Although it remains possible that mosquito physiology is affected by phage infection of bacterial symbionts [70], we have excluded phage families from further analyses.
3.4. Positive-stranded RNA viruses
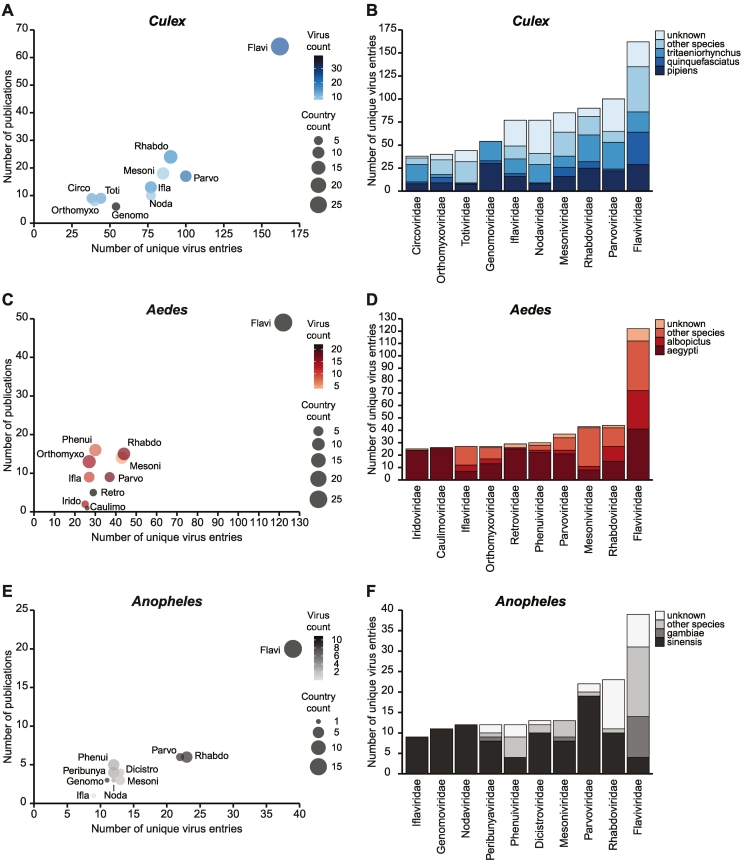
We analyzed the contribution of virus families to the virome of Culex, Aedes and Anopheles mosquitoes, specifically. The Flaviviridae family, and specifically the Flavivirus genus, contains many mosquito-associated viruses including both arboviruses and ISVs [71,72]. In our dataset, Flaviviridae was the most abundant virus family in all mosquito genera (Fig. 3). A significant percentage of these entries derived from a few highly abundant viruses, including Culex flavivirus (51%) for the Culex genus, Aedes flavivirus (25%) and Cell fusing agent virus (20%) for the Aedes genus, and Anopheles flavivirus (23%) for the Anopheles genus. These viruses were among the most abundant and widespread in our dataset and have been detected in multiple species within and across mosquito genera (Fig. 4). Strikingly, Culex flavivirus seems to have a particularly broad host tropism, as it was detected in 12 species of Culex mosquitoes as well as three Aedes mosquito species and one species of Anopheles mosquitoes (Table 2). While flaviviruses are clearly highly prevalent, the family is overrepresented due to the frequent use of PCR studies to detect mosquito-associated flaviviruses, accounting for approximately 60% of Flaviviridae unique virus entries (Fig. S1C).
Fig. 3.
Most frequently reported virus families in mosquitoes.
(A, C, E) Top 10 most frequently reported virus families for (A) Culex, (C) Aedes, and (E) Anopheles mosquitoes. The X-axis represents the number of unique virus entries for each family as a measure of virus abundance. The Y-axis indicates the number of studies that reported at least one virus from that family. Fill color indicates the total number of unique viruses detected for each family. Symbol size indicates the total number of countries for each virus family. (B, D, F) Top 10 most frequently reported virus families for (B) Culex, (D) Aedes, and (F) Anopheles mosquitoes with fill color indicating the mosquito species.
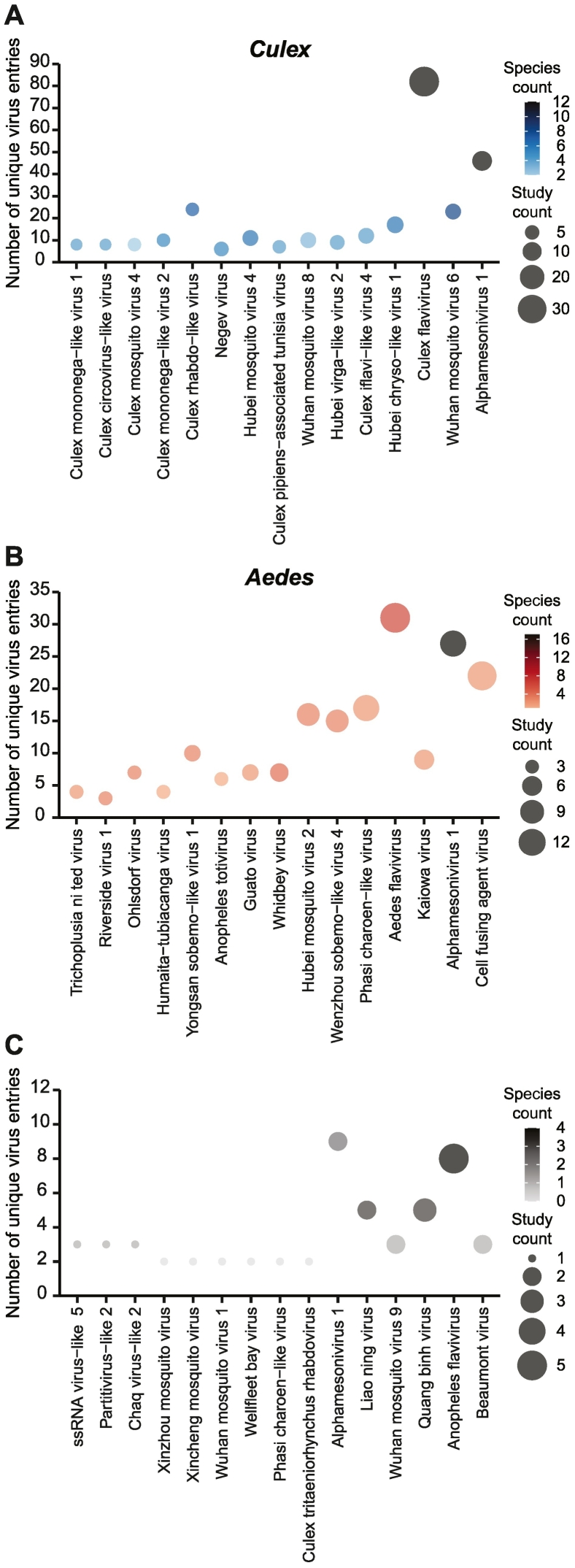
Fig. 4.
Most widespread mosquito viruses.
Top 15 most widespread viruses for (A) Culex, (B) Aedes, and (C) Anopheles mosquitoes. Virus names were ordered according to the number of continents in which the virus was detected. Fill color indicates the number of mosquito species in which the virus has been found within the genus. Symbol size indicates the number of studies in which the virus was found.
Table 2.
Most widespread viruses in Culex mosquitoes.
| Virus name | Virus family | Virus genus | Countries within each continent |
Studies | Mosquito species | Unique entries | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Africa | Asia | Australia | Europe | North-America | South-America | ||||||
| Alphamesonivirus 1 | Mesoniviridae | Alphamesonivirus | – | 2 | 1 | 3 | 2 | 1 | 11 | 12 | 46 |
| Wuhan mosquito virus 6 | Orthomyxoviridae | Quaranjavirus | – | 1 | 1 | 2 | 1 | 1 | 6 | 8 | 23 |
| Culex flavivirus | Flaviviridae | Flavivirus | 2 | 6 | – | – | 3 | 4 | 33 | 12 | 82 |
| Hubei chryso-like virus 1 | Unclassified | Unclassified | – | 2 | 1 | 2 | 1 | – | 7 | 6 | 17 |
| Culex iflavi-like virus 4 | Iflaviridae | Unclassified | – | 1 | – | 2 | 1 | 1 | 6 | 4 | 12 |
| Hubei virga-like virus 2 | Unclassified | Unclassified | – | 1 | – | 1 | 2 | 1 | 5 | 4 | 9 |
| Wuhan mosquito virus 8 | Chuviridae | Culicidavirus | – | 1 | – | 1 | 1 | 1 | 6 | 3 | 10 |
| Culex pipiens-associated tunisia virus | Unclassified | Unclassified | 1 | 1 | – | – | 1 | 1 | 4 | 4 | 7 |
| Hubei mosquito virus 4 | Unclassified | Unclassified | – | 2 | – | – | 1 | 1 | 6 | 6 | 11 |
| Negev virus | Unclassified | Unclassified | – | 1 | – | 1 | – | 1 | 5 | 5 | 6 |
| Culex rhabdo-like virus | Rhabdoviridae | Ohlsrhavirus | – | 1 | 1 | – | 1 | – | 4 | 7 | 24 |
| Culex mononega-like virus 2 | Unclassified | Unclassified | – | 1 | 1 | 1 | – | – | 4 | 5 | 10 |
| Culex mosquito virus 4 | Chuviridae | Culicidavirus | – | 1 | – | 1 | 1 | – | 4 | 2 | 8 |
| Culex circovirus-like virus | Circoviridae | Circovirus | – | 1 | – | – | 1 | 1 | 3 | 4 | 8 |
| Culex mononega-like virus 1 | Unclassified | Unclassified | – | 1 | 1 | 1 | – | – | 3 | 4 | 8 |
| Merida virus | Rhabdoviridae | Merhavirus | – | 1 | – | 1 | 1 | – | 3 | 4 | 4 |
| Culex bunyavirus 1 | Unclassified | Unclassified | – | 1 | – | – | 1 | 1 | 3 | 3 | 4 |
| Wenzhou sobemo-like virus 3 | Unclassified | Unclassified | – | 3 | – | 2 | – | – | 7 | 4 | 19 |
| Quang binh virus | Flaviviridae | Flavivirus | 1 | 4 | – | – | – | – | 7 | 4 | 15 |
| Hubei mosquito virus 2 | Unclassified | Unclassified | – | 3 | – | 1 | – | – | 6 | 4 | 17 |
The family of Mesoniviridae is a recently established taxon of mosquito-infecting positive-sense RNA viruses [17]. The large majority of Mesoniviridae virus entries in both Culex (74%) and Aedes (70%) mosquitoes were derived from alphamesonivirus 1, the founding species of the family that includes several closely related variants, such as Nam Dinh virus, Houston virus and Cavally virus [73,74]. Alphamesonivirus 1, with most entries from Nam Dinh virus and Houston virus, had a broad global distribution as it was detected in 11 countries across five continents (Table 2, Table 3). Strikingly, alphamesonivirus 1 also had the broadest host range of all viruses in our dataset, as it was detected in 34 mosquito species across five genera.
Table 3.
Most widespread viruses in Aedes mosquitoes.
| Virus name | Virus family | Virus genus | Countries within each continent |
Studies | Mosquito species | Unique entries | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Africa | Asia | Australia | Europe | North-America | South-America | ||||||
| Cell fusing agent virus | Flaviviridae | Flavivirus | 3 | 4 | 1 | – | 3 | 1 | 14 | 2 | 22 |
| Alphamesonivirus 1 | Mesoniviridae | Alphamesonivirus | – | 1 | 1 | 1 | 3 | 1 | 11 | 17 | 27 |
| Kaiowa virus | Unclassified | Unclassified | – | 2 | 1 | 1 | 1 | 1 | 6 | 2 | 9 |
| Aedes flavivirus | Flaviviridae | Flavivirus | 2 | 3 | – | 2 | – | 1 | 15 | 6 | 31 |
| Phasi Charoen-like virus | Phenuiviridae | Phasivirus | – | 3 | 1 | – | 3 | 1 | 11 | 2 | 17 |
| Wenzhou sobemo-like virus 4 | Unclassified | Unclassified | – | 2 | – | 2 | 2 | 1 | 8 | 3 | 15 |
| Hubei mosquito virus 2 | Unclassified | Unclassified | – | 2 | – | 2 | 1 | 1 | 8 | 3 | 16 |
| Whidbey virus | Orthomyxoviridae | Unclassified | – | 1 | 1 | 3 | – | 1 | 5 | 4 | 7 |
| Guato virus | Unclassified | Unclassified | – | 1 | – | 1 | 1 | 1 | 4 | 2 | 7 |
| Anopheles totivirus | Totiviridae | Unclassified | – | 1 | 1 | – | 1 | 1 | 3 | 1 | 6 |
| Yongsan sobemo-like virus 1 | Solemoviridae | Sobemovirus | – | 2 | – | 1 | – | 1 | 4 | 3 | 10 |
| Humaita-Tubiacanga virus | Unclassified | Unclassified | – | 1 | 1 | – | 2 | – | 3 | 1 | 4 |
| Ohlsdorf virus | Rhabdoviridae | Ohlsrhavirus | – | 1 | – | 1 | – | 1 | 3 | 3 | 7 |
| Riverside virus 1 | Rhabdoviridae | Unclassified | – | 1 | – | 1 | – | 1 | 3 | 3 | 3 |
| Trichoplusia ni ted virus | Metaviridae | Errantivirus | – | 1 | – | – | 1 | 1 | 3 | 2 | 4 |
| Hubei toti-like virus 10 | Unclassified | Unclassified | – | 1 | 1 | 1 | – | – | 3 | 2 | 3 |
| Menghai flavivirus | Flaviviridae | Flavivirus | – | 1 | – | – | 1 | 1 | 3 | 2 | 3 |
| dsRNA virus environmental sample | Unclassified | Unclassified | – | – | 1 | 1 | 1 | – | 3 | 2 | 3 |
| Croada virus | Unclassified | Unclassified | – | 1 | – | – | 1 | 1 | 3 | 1 | 3 |
| Blackford virus | Unclassified | Unclassified | – | 1 | 1 | – | 1 | – | 2 | 1 | 3 |
Nodaviridae and Iflaviridae are both well-established families containing insect-associated viruses [75,76], which were frequently detected in Culex and Aedes mosquitoes. The majority of these virus entries was derived from only two studies, that sampled in California and China [77,78]. In China, Nodaviridae and Iflaviridae sequences, although not classified at the virus species level, were detected in 12 mosquito species [77]. In California, four iflaviruses (Culex iflavi-like virus 1–4) and five viruses currently classified as Nodaviridae (Culex Noda-like virus 1 and Culex mosquito virus 1, 2, 3 and 6) were detected in several species of Culex mosquitoes, representing 29% of the unique virus entries for Iflaviridae and 51% for Nodaviridae [78]. Culex iflavi-like virus 4 has also been detected in Culex mosquitoes in Brazil, China, Belgium and Serbia [65,[79], [80], [81]] (Table 2).
3.5. Negative-stranded RNA viruses
Members of the Rhabdoviridae family were among the most frequently observed across mosquito genera in our dataset (Fig. 3) and included several of the most widespread viruses, including Culex rhabdo-like virus and Merida virus for Culex mosquitoes, Ohlsdorf virus and Riverside virus 1 for Aedes mosquitoes, and Beaumont virus and Wuhan mosquito virus 9 for Anopheles mosquitoes, which were each detected in at least three countries (Table 2, Table 3, Table 4). Notably, Merida virus has been found in four continents and in mosquito species of the genera Culex, Aedes and Heamagogus [32,33,40,66,79,82].
Table 4.
Most widespread viruses in Anopheles mosquitoes.
| Virus name | Virus family | Virus genus | Countries within each continent |
Studies | Mosquito species | Unique entries | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Africa | Asia | Australia | Europe | North-America | South-America | ||||||
| Beaumont virus | Rhabdoviridae | Unclassified | 1 | 1 | 1 | – | – | – | 2 | 1 | 3 |
| Anopheles flavivirus | Flaviviridae | Unclassified | 5 | – | – | 1 | – | – | 5 | 4 | 8 |
| Quang binh virus | Flaviviridae | Unclassified | 2 | 1 | – | – | – | – | 3 | 3 | 5 |
| Wuhan mosquito virus 9 | Rhabdoviridae | Unclassified | 1 | 2 | – | – | – | – | 2 | 1 | 3 |
| Liao ning virus | Reoviridae | Seadornavirus | – | 1 | 1 | – | – | – | 2 | 3 | 5 |
| Alphamesonivirus 1 | Mesoniviridae | Alphamesonivirus | – | 1 | 1 | – | – | – | 2 | 2 | 9 |
| Culex tritaeniorhynchus rhabdovirus | Rhabdoviridae | Merhavirus | 1 | 1 | – | – | – | – | 1 | n.d.⁎ | 2 |
| Phasi Charoen-like virus | Phenuiviridae | Phasivirus | 1 | 1 | – | – | – | – | 1 | n.d.⁎ | 2 |
| Wellfleet Bay virus | Orthomyxoviridae | Quaranjavirus | 1 | 1 | – | – | – | – | 1 | n.d.⁎ | 2 |
| Wuhan mosquito virus 1 | Phasmaviridae | Orthophasmavirus | 1 | 1 | – | – | – | – | 1 | n.d.⁎ | 2 |
| Xincheng mosquito virus | Xinmoviridae | Anphevirus | 1 | 1 | – | – | – | – | 1 | n.d.⁎ | 2 |
| Xinzhou mosquito virus | Peribunyaviridae | Unclassified | 1 | 1 | – | – | – | – | 1 | n.d.⁎ | 2 |
| Chaq virus-like 2 | Unclassified | Unclassified | 3 | – | – | – | – | – | 1 | 1 | 3 |
| Partitivirus-like 2 | Partitiviridae | Unclassified | 3 | – | – | – | – | – | 1 | 1 | 3 |
| ssRNA virus-like 5 | Unclassified | Unclassified | 2 | – | – | – | – | – | 1 | 1 | 3 |
| Bolahun virus | Xinmoviridae | Anphevirus | 2 | – | – | – | – | – | 1 | 1 | 2 |
| ssRNA virus-like 6 | Unclassified | Unclassified | 2 | – | – | – | – | – | 1 | 1 | 2 |
| Chaq virus-like 3 | Unclassified | Unclassified | 2 | – | – | – | – | – | 1 | 1 | 2 |
| Partitivirus-like 3 | Partitiviridae | Unclassified | 2 | – | – | – | – | – | 1 | 1 | 2 |
| Hubei mosquito virus 2 | Unclassified | Unclassified | – | 1 | – | – | – | – | 3 | 1 | 4 |
n.d., Anopheles mosquitoes not defined at the species level.
The Xinmoviridae family was established in 2017 to encompass the free-floating genus Anphevirus in the Mononegavirales order [83]. Anpheviruses were detected in multiple mosquito species, including Xincheng mosquito virus and Bolahun virus in Anopheles mosquitoes and Aedes aegypti anphevirus, Aedes albopictus anphevirus and Aedes anphevirus in Aedes mosquitoes [66,79,81,84,85,86]. Notably, the contribution of Xinmoviridae to the mosquito virome may be underestimated due to the recent establishment of this family.
Several segmented viruses of the Orthomyxoviridae and Phenuiviridae were among the most widespread in Culex and Aedes mosquitoes (Fig. 4A,B). For the Orthomyxoviridae, these included Wuhan mosquito virus 6 for Culex mosquitoes and Whidbey virus for Aedes mosquitoes (Table 2, Table 3). In particular, Wuhan mosquito virus 6 showed a near-global distribution and broad mosquito host range, as it was detected in eight countries across all continents except Antarctica, and in 12 mosquito species across four genera (Supplementary file 1). Phenuiviridae was among the most frequently detected virus families in Aedes mosquitoes due to the high prevalence of Phasi Charoen-like virus, which contributed 60% of the entries of this family. Phasi Charoen-like virus is one of the most widespread viruses in Aedes aegypti specifically (Table 3), although it was also detected in Aedes albopictus [32], Culex quinquefasciatus [86], Haemagogus janthinomys [82] and Anopheles mosquitoes [87].
3.6. Double-stranded RNA viruses
The Artivirus genus in the family Totiviridae is comprised of double-stranded RNA viruses of arthropods, including mosquitoes [9,88]. Totiviridae entries in our database corresponded to multiple totiviruses, with limited cross-detection between studies. Notably, the most frequently detected totivirus was Anopheles totivirus which, after its initial detection in Anopheles gambiae in Liberia [84], was found in Aedes aegypti in several countries across multiple continents [66,67,69] (Table 3).
3.7. DNA virus families
The Parvoviridae family of single-stranded DNA viruses is the most abundant DNA virus family in all three mosquito genera (Fig. 3). The family contains densoviruses, which have been studied as a potential biological control agent of insects and mosquitoes specifically [89]. A large proportion of the Parvoviridae entries for all three mosquito genera was derived from a single study [77], which detected parvovirus sequences in several Culex, Aedes and Anopheles species across China. An additional 38% of the Parvoviridae entries for Culex mosquitoes corresponded to Culex densovirus, which was detected in Culex pipiens and mixed pools of Culex mosquitoes across California [78].
Two families of circular Rep-encoding single-stranded DNA viruses (also referred to as CRESS-DNA viruses; [90]), Genomoviridae and Circoviridae, were frequently detected (Fig. 3; Fig. 4A). The high abundance of Genomoviridae in our dataset was mostly due to three metagenomic sequencing studies, each detecting sequences mapping to multiple genomoviruses [40,61,67]. For the Circoviridae, Culex circovirus-like virus was detected in three studies over three continents in species of all three genera [40,78,81].
For both Genomoviridae and Circoviridae, novel species have mostly been detected through metagenomic sequencing [91,92]. As active replication of these viruses has not been described in the animals sampled for sequencing, it is possible that these viruses are associated with food sources or pathogens of the host, precluding conclusions on the host range of these virus families [91,92]. However, for viruses from the Circoviridae, and specifically the Cyclovirus genus, arthropods (and mosquitoes specifically) have been suggested to be the primary host [67,93]. Moreover, Sclerotinia sclerotiorumhypovirulence-associated DNA virus 1, the founding species of the Genomoviridae viral family, was found to infect the mycophagous mosquito species Lycoriella ingenua under experimental conditions [94]. These studies suggest that vector mosquitoes could be a part of the host range of genomoviruses and circoviruses.
3.8. Unclassified viruses
The majority of virus entries in our database are unclassified at the family level (Fig. S1B). This large group included 114 unique virus entries from the genus Negevirus [18], a taxon of insect-specific, non-segmented, enveloped, positive-sense RNA viruses that has not been classified yet at the family level. Negeviruses are among the most abundant virus taxa in Culex (53 unique virus entries) and Aedes (32 unique virus entries) mosquitoes. Negeviruses have been reported in multiple Culex and Aedes species and across at least four continents. Among these, the most abundant virus was the eponymous Negev virus, which was detected in Aedes aegypti [81] and several Culex species across three continents [33,65,95,96] (Fig. 4A).
3.9. Most frequently detected viruses per mosquito genus
We collated lists of the top 20 most frequently detected viruses for each mosquito genus according to the number of continents and countries in which they were detected (Table 2, Table 3, Table 4, Fig. 4). For Aedes and Culex, the broad global distribution of these viruses was well supported, being detected in multiple countries in two to five continents in at least three independent studies, lending support to the validity of these observations. Surprisingly, all or nearly all top 20 viruses for Culex and Aedes respectively, were detected in multiple mosquito species, suggesting that vertical transmission is not the sole transmission route for these viruses. In line with the more limited sampling of Anopheles mosquitoes (Fig. 2), the top 20 Anopheles viruses were detected less frequently (between 2 and 9 unique virus entries) and at fewer places across the globe (Fig. 4C; Table 4).
A large number of unclassified viruses were among the top 20 (ten for Culex, nine for Aedes, and five for Anopheles; Table 2, Table 3, Table 4). Among these, some have a particularly broad global distribution, having been detected in at least four continents. These include Hubei chryso-like virus 1, Hubei virga-like virus 2 and Culex pipiens-associated Tunisia virus for Culex mosquitoes, and Kaiowa virus, Whenzhou Sobemo-like virus and Hubei mosquito virus 2 for Aedes mosquitoes (Table 2, Table 3). Notably, the detection of Kaiowa virus in Aedes aegypti metagenomic studies was proposed to be due to the presence of endogenous viral elements in mosquito genomes instead of replicating virus [71]. Metagenomic studies have also reported sequences with homology to Kaiowa virus in samples from Aedes albopictus, Culex quinquefasciatus and Heamagogus janthinomys [32,82,86,97] and the origin of these sequences merits further investigation.
4. Discussion
Growing scientific interest and increasing accessibility to deep-sequencing technology has led to a large body of literature on the mosquito virome. We have collated information from 175 research articles from the last 22 years to construct a comprehensive and searchable database of mosquito-associated viruses, along with the locations and hosts in which they have been detected. We found that RNA viruses from the families Flaviviridae and Rhabdoviridae are widespread in Culex, Aedes and Anopheles mosquitoes globally. We collated lists of the top 20 viruses with the widest global distribution for each of these mosquito genera and found that most of these viruses were detected in multiple mosquito species within, and sometimes across mosquito genera. The prevalence and overall stability of these viruses within mosquito populations and the transmission routes enabling them to persist and spread merits further investigation.
We collated virome data as reported, accepting the authors' assessment for assigning viral sequences to established virus species or taxons or for considering a virus novel. A limitation of this approach is that different thresholds for genome coverage, number of virus mapping reads or contigs, and nucleotide or amino acid sequence identity scores were used for virus identification between studies. More concerningly, some studies did not unambiguously report the criteria used for virus identification, which makes side-by-side comparisons of studies difficult. Consequently, the database contains some low-confidence virus entries due to insufficiently stringent thresholds for virus identification or misclassification of virus sequences. Due to these caveats and differences in sampling intensities, our database cannot be used to accurately infer the absence of a virus in specific mosquito species. Another limitation of our study is that we may have inevitably missed relevant articles that did not match our search terms, despite our best efforts to use a comprehensive literature search strategy. Despite these limitations, the most widespread and abundant viruses in our dataset have been found in multiple independent studies. As such, the collated top 20 most widespread viruses can be considered high-confidence constituents of the mosquito virome, especially for Aedes and Culex that have been sampled most extensively.
During our study, we noticed that virome data are often impractically reported for interpretation and re-use by other scientists, due to unreported critical variables or an impractical format to present results (e.g., in heat-maps) without accompanying presentation in a reusable data format. We propose that reporting can be improved by the standard inclusion of a supplementary table containing per virus positive sample, i) the viruses identified along with accession numbers, nucleotide and amino acid identity scores, genome coverage, numbers of reads/contigs mapping to the viral genome, ii) information on the sample, such as pool identifier, number of mosquitoes per sample, mosquito species, sex, and life stage, iii) the date and location of sampling, along with geographic coordinates and type of habitat, and iv) sequencing information, including the sequencing platform, method for library preparation, sequencing depth per library, and accession number of the repository in which the raw sequence data have been deposited.
Next-generation sequencing is a relatively unbiased approach that successfully detects both RNA and DNA viruses in mosquitoes. However, the detection of sequences of well-known mammalian viruses of the Retroviridae [40,65,66,82] (e.g., murine leukemia virus), Herpesviridae [40,45,65] (e.g., herpes simplex virus) and Hepadnaviridae (e.g., hepatitis B virus) [61] indicates that some datasets contain considerable amount of noise. The origin of these sequences is unclear but may be due to insufficiently stringent thresholds for virus identification, to biological contaminants such as sequences derived from blood meals, or to experimental contamination during library preparation and sequencing. Alternatively, although no insect viruses have currently been described in the family Retroviridae, it remains possible that these sequences derive from unidentified mosquito retroviruses or from retroelements in the mosquito genome.
Detection of viral sequences does not provide direct support of active replication in the mosquito host and, indeed, bacteriophage sequences were frequently detected in mosquito virome studies [34,40,43,45,65,67,69]. Isolation of a virus in mosquito cell culture would provide strong support for active virus replication in the mosquito host [98,99]. Alternatively, small RNA-sequencing approaches may be used to distinguish sequences of actively replicating viruses from contaminating sequences. Viral double-stranded RNA produced during replication of both DNA and RNA viruses are processed into 21-nt small interfering RNAs [100] that can be readily distinguished in small RNA size profiles. Indeed, some researchers have used small RNA sequencing as an alternative or complement to conventional mRNA sequencing for virome studies in insects [[101], [102], [103], [104]].
Small RNAs may also help to distinguish replicating viruses from another source of viral sequences, EVEs. The Aedes aegypti and Aedes albopictus genomes contain a large number of non-retroviral EVEs [[105], [106], [107]], transcripts of which may be detected in RNA-seq experiments. Next-generation sequencing studies therefore require careful analysis to differentiate between EVE-derived sequences and virus-derived sequences. The removal of contigs mapping to mosquito genomes may be impossible for species lacking reference genomes and, even for species with high-quality reference genomes, this may be insufficient as the EVE repertoire differs between mosquito populations [108]. Small RNA sequencing may help to distinguish EVE-derived sequences from sequences of replicating viruses as EVEs may primarily give rise to PIWI-interacting RNAs (piRNAs) with a typical size of 25–30 nt and strong strand biases. These can be readily distinguished from replication-dependent siRNAs of 21 nt, which are usually derived from both positive and negative-sense viral RNAs [102,108].
The composition of the mosquito virome is likely shaped by the environment, virus transmission modes, and restrictive factors in specific mosquito species or genera. Moreover, changing biotic and abiotic factors associated with global warming and increasing globalization may further affect the mosquito virome. Our database is an up-to-date, comprehensive overview of primary literature on mosquito-associated viruses from the last 22 years. As such, our study forms a solid foundation to study inter- and intra-species pathogen transmission from a One Health perspective. A future challenge will be to understand how virome dynamics affect mosquito-borne disease outbreaks.
Note added in proof
In agreement with our analyses, Olmo et al. recently reported that Phasi Charoen-like virus and Humaita Tubiacanga virus were highly abundant and widespread in Ae. aegypti worldwide (Nat Microbiol. 2023, 8:135-149).
The following are the supplementary data related to this article.
Final selection of 175 articles used for the analyses and their references.
Sample-structured database of viruses identified in mosquito virome studies.
Virus positive mosquito species, with the corresponding number of continents, countries and studies in which they are sampled.
Unique virus entries in mosquito virome studies. (A) Number of unique virus entries reported for the top 50 studies that contributed most entries in our database. Unique virus entries for Hameed et al. (2020) before (dark red) and after (salmon) data curation are indicated. (B) Number of unique virus entries corresponding to the individual virus families for all mosquito species in our database. Bacteriophage families are indicated with salmon fill color; viruses not classified at the family level in light blue. (C) Number of unique virus entries for the top 10 most frequently detected families, according to detection method.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank members of the laboratory for discussions. This publication is financially supported by the project ‘Preparing for vector-borne virus outbreaks in a changing world: A One Health Approach’ (NWA.1160.18.210), financed by the Dutch Research Council (NWO) and a grant from Radboud Institute for Molecular Life Science. We thank Alice Tillema (library Radboud University Medical Center) for advice on the PubMed search strategy.
Data availability
This is a review of published literature. The data are available in the supplementary files.
References
- 1.Weaver S.C., Reisen W.K. Present and future arboviral threats. Antivir. Res. 2010;85(2):328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciota A.T. West Nile virus and its vectors. Curr. Opin. Insect Sci. 2017;22:28–36. doi: 10.1016/j.cois.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Kauffman E.B., Kramer L.D. Zika virus mosquito vectors: competence, biology, and vector control. J. Infect. Dis. 2017;216(suppl_10):S976–s990. doi: 10.1093/infdis/jix405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pezzi L., et al. GloPID-R report on chikungunya, o’nyong-nyong and Mayaro virus, part 5: entomological aspects. Antivir. Res. 2020;174 doi: 10.1016/j.antiviral.2019.104670. [DOI] [PubMed] [Google Scholar]
- 5.Agboli E., et al. Mosquito-specific viruses-transmission and interaction. Viruses. 2019;11:873. doi: 10.3390/v11090873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Y.S., Higgs S., Vanlandingham D.L. Arbovirus-mosquito vector-host interactions and the impact on transmission and disease pathogenesis of arboviruses. Front. Microbiol. 2019;10:22. doi: 10.3389/fmicb.2019.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li C.X., et al. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife. 2015;4 doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi M., et al. Redefining the invertebrate RNA virosphere. Nature. 2016;540(7634):539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 9.Zhai Y., et al. Isolation and full-length sequence analysis of Armigeres subalbatus totivirus, the first totivirus isolate from mosquitoes representing a proposed novel genus (Artivirus) of the family Totiviridae. J. Gen. Virol. 2010;91(Pt 11):2836–2845. doi: 10.1099/vir.0.024794-0. [DOI] [PubMed] [Google Scholar]
- 10.Isawa H., et al. Identification and molecular characterization of a new nonsegmented double-stranded RNA virus isolated from Culex mosquitoes in Japan. Virus Res. 2011;155(1):147–155. doi: 10.1016/j.virusres.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Cross S.T., et al. Partitiviruses infecting Drosophila melanogaster and Aedes aegypti exhibit efficient Biparental vertical transmission. J. Virol. 2020;94(20):e01070–20. doi: 10.1128/JVI.01070-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marklewitz M., et al. Gouleako virus isolated from west African mosquitoes constitutes a proposed novel genus in the family Bunyaviridae. J. Virol. 2011;85(17):9227–9234. doi: 10.1128/JVI.00230-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marklewitz M., et al. Evolutionary and phenotypic analysis of live virus isolates suggests arthropod origin of a pathogenic RNA virus family. Proc. Natl. Acad. Sci. U. S. A. 2015;112(24):7536–7541. doi: 10.1073/pnas.1502036112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marklewitz M., et al. Discovery of a unique novel clade of mosquito-associated bunyaviruses. J. Virol. 2013;87(23):12850–12865. doi: 10.1128/JVI.01862-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zirkel F., et al. An insect nidovirus emerging from a primary tropical rainforest. mBio. 2011;2(3) doi: 10.1128/mBio.00077-11. e00077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nga P.T., et al. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 2011;7(9) doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauber C., et al. Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Arch. Virol. 2012;157(8):1623–1628. doi: 10.1007/s00705-012-1295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasilakis N., et al. Negevirus: a proposed new taxon of insect-specific viruses with wide geographic distribution. J. Virol. 2013;87(5):2475–2488. doi: 10.1128/JVI.00776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuwata R., et al. Analysis of mosquito-borne Flavivirus superinfection in Culex tritaeniorhynchus (Diptera: Culicidae) cells persistently infected with Culex Flavivirus (Flaviviridae) J. Med. Entomol. 2015;52(2):222–229. doi: 10.1093/jme/tju059. [DOI] [PubMed] [Google Scholar]
- 20.Hobson-Peters J., et al. A new insect-specific flavivirus from northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0056534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall-Mendelin S., et al. The insect-specific Palm Creek virus modulates West Nile virus infection in and transmission by Australian mosquitoes. Parasit. Vectors. 2016;9(1):414. doi: 10.1186/s13071-016-1683-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nasar F., et al. Eilat virus induces both homologous and heterologous interference. Virology. 2015;484:51–58. doi: 10.1016/j.virol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schultz M.J., Frydman H.M., Connor J.H. Dual insect specific virus infection limits arbovirus replication in Aedes mosquito cells. Virology. 2018;518:406–413. doi: 10.1016/j.virol.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Öhlund P., Lundén H., Blomström A.L. Insect-specific virus evolution and potential effects on vector competence. Virus Genes. 2019;55(2):127–137. doi: 10.1007/s11262-018-01629-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atoni E., et al. The discovery and global distribution of novel mosquito-associated viruses in the last decade (2007-2017) Rev. Med. Virol. 2019;29(6) doi: 10.1002/rmv.2079. [DOI] [PubMed] [Google Scholar]
- 26.Reynaud J.M., et al. IFIT1 differentially interferes with translation and replication of alphavirus genomes and promotes induction of type I interferon. PLoS Pathog. 2015;11(4) doi: 10.1371/journal.ppat.1004863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patterson E.I., et al. Exploiting insect-specific viruses as a novel strategy to control vector-borne disease. Curr. Opin. Insect Sci. 2020;39:50–56. doi: 10.1016/j.cois.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lequime S., Lambrechts L. Vertical transmission of arboviruses in mosquitoes: a historical perspective. Infect. Genet. Evol. 2014;28:681–690. doi: 10.1016/j.meegid.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 29.Lequime S., Paul R.E., Lambrechts L. Determinants of arbovirus vertical transmission in mosquitoes. PLoS Pathog. 2016;12(5) doi: 10.1371/journal.ppat.1005548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altinli M., Schnettler E., Sicard M. Symbiotic interactions between mosquitoes and mosquito viruses. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.694020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nanfack-Minkeu F., et al. Interaction of RNA viruses of the natural virome with the African malaria vector, Anopheles coluzzii. Sci. Rep. 2019;9(1):6319. doi: 10.1038/s41598-019-42825-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi C., et al. Stability of the Virome in lab- and field-collected Aedes albopictus mosquitoes across different developmental stages and possible Core viruses in the publicly available Virome data of Aedes mosquitoes. mSystems. 2020;5(5):e00640–20. doi: 10.1128/mSystems.00640-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersson J.H., et al. Meta-transcriptomic comparison of the RNA Viromes of the mosquito vectors Culex pipiens and Culex torrentium in northern Europe. Viruses. 2019;11(11):1033. doi: 10.3390/v11111033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia H., et al. Comparative metagenomic profiling of Viromes associated with four common mosquito Species in China. Virol. Sin. 2018;33(1):59–66. doi: 10.1007/s12250-018-0015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi M., et al. High-resolution Metatranscriptomics reveals the ecological dynamics of mosquito-associated RNA viruses in Western Australia. J. Virol. 2017;91(17):e00680–17. doi: 10.1128/JVI.00680-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nanfack Minkeu F., Vernick K.D. A systematic review of the natural Virome of Anopheles mosquitoes. Viruses. 2018;10(5):222. doi: 10.3390/v10050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aardema M.L., Olatunji S.K., Fonseca D.M. The enigmatic Culex pipiens (Diptera: Culicidae) Species complex: phylogenetic challenges and opportunities from a notoriously tricky mosquito group. Ann. Entomol. Soc. Am. 2021;115(1):95–104. [Google Scholar]
- 38.Haba Y., McBride L. Origin and status of Culex pipiens mosquito ecotypes. Curr. Biol. 2022;32(5):R237–r246. doi: 10.1016/j.cub.2022.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferreira de Freitas L., Bartholomay L.C. The taxonomic history of Ochlerotatus Lynch Arribálzaga, 1891 (Diptera: Culicidae) Insects. 2021;12(5):452. doi: 10.3390/insects12050452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hameed M., et al. A metagenomic analysis of mosquito Virome collected from different animal farms at Yunnan-Myanmar border of China. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.591478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.ICTV Master Species List 2021 Version 1. 2022. https://talk.ictvonline.org/files/master-species-lists/m/msl/13425 [cited 2022 April 8]; Available from:
- 42.NCBI Taxonomy Database. 2022. https://www.ncbi.nlm.nih.gov/taxonomy [cited 2022 June 7]; Available from:
- 43.Birnberg L., et al. Viromics on honey-baited FTA cards as a new tool for the detection of circulating viruses in mosquitoes. Viruses. 2020;12(3):274. doi: 10.3390/v12030274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L., et al. Comparative viromes of Culicoides and mosquitoes reveal their consistency and diversity in viral profiles. Brief. Bioinform. 2021;22(4):bbaa323. doi: 10.1093/bib/bbaa323. [DOI] [PubMed] [Google Scholar]
- 45.Ng T.F., et al. Broad surveys of DNA viral diversity obtained through viral metagenomics of mosquitoes. PLoS One. 2011;6(6) doi: 10.1371/journal.pone.0020579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coffey L.L., et al. Enhanced arbovirus surveillance with deep sequencing: identification of novel rhabdoviruses and bunyaviruses in Australian mosquitoes. Virology. 2014;448:146–158. doi: 10.1016/j.virol.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frey K.G., et al. Bioinformatic characterization of mosquito Viromes within the eastern United States and Puerto Rico: discovery of novel viruses. Evol. Bioinformatics Online. 2016;12(Suppl. 2):1–12. doi: 10.4137/EBO.S38518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newton N.D., et al. Genetic, morphological and antigenic relationships between Mesonivirus isolates from Australian mosquitoes and evidence for their horizontal transmission. Viruses. 2020;12(10):1159. doi: 10.3390/v12101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ajamma Y.U., et al. Vertical transmission of naturally occurring Bunyamwera and insect-specific flavivirus infections in mosquitoes from islands and mainland shores of lakes Victoria and Baringo in Kenya. PLoS Negl. Trop. Dis. 2018;12(11) doi: 10.1371/journal.pntd.0006949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bennouna A., et al. Identification of Eilat virus and prevalence of infection among Culex pipiens L. populations, Morocco, 2016. Virology. 2019;530:85–88. doi: 10.1016/j.virol.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 51.de Araujo Coutinho C.J., et al. Occurrence and phylogenetic characterization of a baculovirus isolated from Culex quinquefasciatus in São Paulo state, Brazil. Arch. Virol. 2012;157(9):1741–1745. doi: 10.1007/s00705-012-1372-1. [DOI] [PubMed] [Google Scholar]
- 52.Evangelina M., Victoria M.M., José G.J. Culex pipiens affected by joint infection of a mosquito iridescent virus and Strelkovimermis spiculatus. J. Invertebr. Pathol. 2013;114(3):295–297. doi: 10.1016/j.jip.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 53.Kawakami K., et al. Characterization of a novel negevirus isolated from Aedes larvae collected in a subarctic region of Japan. Arch. Virol. 2016;161(4):801–809. doi: 10.1007/s00705-015-2711-9. [DOI] [PubMed] [Google Scholar]
- 54.Misencik M.J., et al. Isolation of a novel insect-specific Flavivirus from Culiseta melanura in the northeastern United States. Vect. Borne Zoonot. Dis. 2016;16(3):181–190. doi: 10.1089/vbz.2015.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morais P., et al. Insect-specific flaviviruses and densoviruses, suggested to have been transmitted vertically, found in mosquitoes collected in Angola: genome detection and phylogenetic characterization of viral sequences. Infect. Genet. Evol. 2020;80 doi: 10.1016/j.meegid.2020.104191. [DOI] [PubMed] [Google Scholar]
- 56.Muttis E., et al. First record of a mosquito iridescent virus in Culex pipiens L. (Diptera: Culicidae) Arch. Virol. 2012;157(8):1569–1571. doi: 10.1007/s00705-012-1302-2. [DOI] [PubMed] [Google Scholar]
- 57.Ramos-Nino M.E., et al. High prevalence of Phasi Charoen-like virus from wild-caught Aedes aegypti in Grenada, W.I. as revealed by metagenomic analysis. PLoS One. 2020;15(1) doi: 10.1371/journal.pone.0227998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rwegoshora R.T., Baisley K.J., Kittayapong P. Seasonal and spatial variation in natural densovirus infection in Anopheles minimus S.L. in Thailand. Southeast. Asian J. Trop. Med. Publ. Health. 2000;31(1):3–9. [PubMed] [Google Scholar]
- 59.Yamao T., et al. Novel virus discovery in field-collected mosquito larvae using an improved system for rapid determination of viral RNA sequences (RDV ver4.0) Arch. Virol. 2009;154(1):153–158. doi: 10.1007/s00705-008-0285-5. [DOI] [PubMed] [Google Scholar]
- 60.Zhang X., et al. Discovery and high prevalence of Phasi Charoen-like virus in field-captured Aedes aegypti in South China. Virology. 2018;523:35–40. doi: 10.1016/j.virol.2018.07.021. [DOI] [PubMed] [Google Scholar]
- 61.He W., et al. Virome in adult Aedes albopictus captured during different seasons in Guangzhou City, China. Parasit. Vectors. 2021;14(1):415. doi: 10.1186/s13071-021-04922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nebbak A., et al. Virome diversity among mosquito populations in a sub-urban region of Marseille, France. Viruses. 2021;13(5):768. doi: 10.3390/v13050768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thannesberger J., et al. Viral metagenomics reveals the presence of novel Zika virus variants in Aedes mosquitoes from Barbados. Parasit. Vectors. 2021;14(1):343. doi: 10.1186/s13071-021-04840-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiao P., et al. Metagenomic sequencing from mosquitoes in China reveals a variety of insect and human viruses. Front. Cell. Infect. Microbiol. 2018;8:364. doi: 10.3389/fcimb.2018.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He X., et al. Metagenomic sequencing reveals viral abundance and diversity in mosquitoes from the Shaanxi-Gansu-Ningxia region, China. PLoS Negl. Trop. Dis. 2021;15(4) doi: 10.1371/journal.pntd.0009381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ribeiro G.O., et al. Aedes aegypti from Amazon Basin harbor high diversity of novel viral Species. Viruses. 2020;12(8):866. doi: 10.3390/v12080866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thannesberger J., et al. Highly sensitive Virome characterization of Aedes aegypti and Culex pipiens Complex from Central Europe and the Caribbean reveals potential for interspecies viral transmission. Pathogens. 2020;9(9):686. doi: 10.3390/pathogens9090686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gil P., et al. A library preparation optimized for metagenomics of RNA viruses. Mol. Ecol. Resour. 2021;21(6):1788–1807. doi: 10.1111/1755-0998.13378. [DOI] [PubMed] [Google Scholar]
- 69.Zakrzewski M., et al. Mapping the virome in wild-caught Aedes aegypti from Cairns and Bangkok. Sci. Rep. 2018;8(1):4690. doi: 10.1038/s41598-018-22945-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Metcalf J.A., Bordenstein S.R. The complexity of virus systems: the case of endosymbionts. Curr. Opin. Microbiol. 2012;15(4):546–552. doi: 10.1016/j.mib.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Almeida J.P., et al. The virome of vector mosquitoes. Curr. Opin. Virol. 2021;49:7–12. doi: 10.1016/j.coviro.2021.04.002. [DOI] [PubMed] [Google Scholar]
- 72.Simmonds P., et al. ICTV virus taxonomy profile: Flaviviridae. J. Gen. Virol. 2017;98(1):2–3. doi: 10.1099/jgv.0.000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vasilakis N., et al. Mesoniviruses are mosquito-specific viruses with extensive geographic distribution and host range. Virol. J. 2014;11:97. doi: 10.1186/1743-422X-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cigarroa-Toledo N., et al. Complete genome sequence of Houston virus, a newly discovered mosquito-specific virus isolated from Culex quinquefasciatus in Mexico. Microbiol. Resour. Announc. 2018;7(10):e00808–18. doi: 10.1128/MRA.00808-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sahul Hameed A.S., et al. ICTV Virus Taxonomy Profile: Nodaviridae. J. Gen. Virol. 2019;100(1):3–4. doi: 10.1099/jgv.0.001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Valles S.M., et al. ICTV virus taxonomy profile: Iflaviridae. J. Gen. Virol. 2017;98(4):527–528. doi: 10.1099/jgv.0.000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Du J., et al. Characterization of viromes within mosquito species in China. Sci. China Life Sci. 2020;63(7):1089–1092. doi: 10.1007/s11427-019-1583-9. [DOI] [PubMed] [Google Scholar]
- 78.Sadeghi M., et al. Virome of > 12 thousand Culex mosquitoes from throughout California. Virology. 2018;523:74–88. doi: 10.1016/j.virol.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 79.Batson J., et al. Single mosquito metatranscriptomics identifies vectors, emerging pathogens and reservoirs in one assay. Elife. 2021;10 doi: 10.7554/eLife.68353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stanojević M., et al. Depicting the RNA Virome of hematophagous arthropods from Belgrade, Serbia. Viruses. 2020;12(9):975. doi: 10.3390/v12090975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.da Silva Ferreira R., et al. Insect-specific viruses and arboviruses in adult male culicids from Midwestern Brazil. Infect. Genet. Evol. 2020;85 doi: 10.1016/j.meegid.2020.104561. [DOI] [PubMed] [Google Scholar]
- 82.Ali R., et al. Characterization of the virome associated with Haemagogus mosquitoes in Trinidad, West Indies. Sci. Rep. 2021;11(1):16584. doi: 10.1038/s41598-021-95842-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maes P., et al. Taxonomy of the order Mononegavirales: a second update 2018. Arch. Virol. 2019;164(4):1233–1244. doi: 10.1007/s00705-018-04126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fauver J.R., et al. West African Anopheles gambiae mosquitoes harbor a taxonomically diverse virome including new insect-specific flaviviruses, mononegaviruses, and totiviruses. Virology. 2016;498:288–299. doi: 10.1016/j.virol.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 85.Manni M., Zdobnov E.M. A novel Anphevirus in Aedes albopictus mosquitoes is distributed worldwide and interacts with the host RNA interference pathway. Viruses. 2020;12(11):1264. doi: 10.3390/v12111264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shi C., et al. Stable distinct core eukaryotic viromes in different mosquito species from Guadeloupe, using single mosquito viral metagenomics. Microbiome. 2019;7(1):121. doi: 10.1186/s40168-019-0734-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Belda E., et al. De novo profiling of RNA viruses in Anopheles malaria vector mosquitoes from forest ecological zones in Senegal and Cambodia. BMC Genomics. 2019;20(1):664. doi: 10.1186/s12864-019-6034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dantas M.D., et al. New insights about ORF1 coding regions support the proposition of a new genus comprising arthropod viruses in the family Totiviridae. Virus Res. 2016;211:159–164. doi: 10.1016/j.virusres.2015.10.020. [DOI] [PubMed] [Google Scholar]
- 89.Johnson R.M., Rasgon J.L. Densonucleosis viruses ('densoviruses') for mosquito and pathogen control. Curr. Opin. Insect Sci. 2018;28:90–97. doi: 10.1016/j.cois.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krupovic M., et al. Cressdnaviricota: a virus phylum unifying seven families of rep-encoding viruses with single-stranded, circular DNA genomes. J. Virol. 2020;94(12):e00582–20. doi: 10.1128/JVI.00582-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Varsani A., Krupovic M. Family Genomoviridae: 2021 taxonomy update. Arch. Virol. 2021;166(10):2911–2926. doi: 10.1007/s00705-021-05183-y. [DOI] [PubMed] [Google Scholar]
- 92.Breitbart M., et al. ICTV virus taxonomy profile: Circoviridae. J. Gen. Virol. 2017;98(8):1997–1998. doi: 10.1099/jgv.0.000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dennis T.P.W., et al. Insights into circovirus host range from the genomic fossil record. J. Virol. 2018;92(16):e00145–18. doi: 10.1128/JVI.00145-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu S., et al. Fungal DNA virus infects a mycophagous insect and utilizes it as a transmission vector. Proc. Natl. Acad. Sci. U. S. A. 2016;113(45):12803–12808. doi: 10.1073/pnas.1608013113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fujita R., et al. Bustos virus, a new member of the negevirus group isolated from a Mansonia mosquito in the Philippines. Arch. Virol. 2017;162(1):79–88. doi: 10.1007/s00705-016-3068-4. [DOI] [PubMed] [Google Scholar]
- 96.da Silva Ribeiro A.C., et al. Negeviruses isolated from mosquitoes in the Brazilian Amazon. Virol. J. 2022;19(1):17. doi: 10.1186/s12985-022-01743-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kubacki J., et al. Viral metagenomic analysis of Aedes albopictus mosquitos from southern Switzerland. Viruses. 2020;12(9):929. doi: 10.3390/v12090929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kallies R., et al. Genetic characterization of goutanap virus, a novel virus related to negeviruses, cileviruses and higreviruses. Viruses. 2014;6(11):4346–4357. doi: 10.3390/v6114346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hermanns K., et al. Discovery of a novel alphavirus related to Eilat virus. J. Gen. Virol. 2017;98(1):43–49. doi: 10.1099/jgv.0.000694. [DOI] [PubMed] [Google Scholar]
- 100.Bronkhorst A.W., et al. A DNA virus-encoded immune antagonist fully masks the potent antiviral activity of RNAi in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2019;116(48):24296–24302. doi: 10.1073/pnas.1909183116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cook S., et al. Novel virus discovery and genome reconstruction from field RNA samples reveals highly divergent viruses in dipteran hosts. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Aguiar E.R., et al. Sequence-independent characterization of viruses based on the pattern of viral small RNAs produced by the host. Nucleic Acids Res. 2015;43(13):6191–6206. doi: 10.1093/nar/gkv587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Obbard D.J., et al. A new lineage of segmented RNA viruses infecting animals. Virus Evol. 2020;6(1) doi: 10.1093/ve/vez061. vez061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Webster C.L., et al. The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS Biol. 2015;13(7) doi: 10.1371/journal.pbio.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Whitfield Z.J., et al. The diversity, structure, and function of heritable adaptive immunity sequences in the Aedes aegypti genome. Curr. Biol. 2017;27(22):3511–3519.e7. doi: 10.1016/j.cub.2017.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Palatini U., et al. Comparative genomics shows that viral integrations are abundant and express piRNAs in the arboviral vectors Aedes aegypti and Aedes albopictus. BMC Genomics. 2017;18(1):512. doi: 10.1186/s12864-017-3903-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Suzuki Y., et al. Uncovering the repertoire of endogenous Flaviviral elements in Aedes Mosquito genomes. J. Virol. 2017;91(15):e00571–17. doi: 10.1128/JVI.00571-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Crava C.M., et al. Population genomics in the arboviral vector Aedes aegypti reveals the genomic architecture and evolution of endogenous viral elements. Mol. Ecol. 2021;30(7):1594–1611. doi: 10.1111/mec.15798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Final selection of 175 articles used for the analyses and their references.
Sample-structured database of viruses identified in mosquito virome studies.
Virus positive mosquito species, with the corresponding number of continents, countries and studies in which they are sampled.
Unique virus entries in mosquito virome studies. (A) Number of unique virus entries reported for the top 50 studies that contributed most entries in our database. Unique virus entries for Hameed et al. (2020) before (dark red) and after (salmon) data curation are indicated. (B) Number of unique virus entries corresponding to the individual virus families for all mosquito species in our database. Bacteriophage families are indicated with salmon fill color; viruses not classified at the family level in light blue. (C) Number of unique virus entries for the top 10 most frequently detected families, according to detection method.
Data Availability Statement
This is a review of published literature. The data are available in the supplementary files.