Summary
Human mesenchymal stem cells (hMSCs) are an appealing cell type for therapeutic applications but remain limited by poor efficacy in clinical trials. Here, we describe a conditioning technique that enhances the vascular regenerative properties of hMSCs and increases their expression of endothelial cell and pericyte markers. We also describe an alginate gel encapsulation protocol for delivering the conditioned cells.
For complete details on the use and execution of this protocol, please refer to Lee et al. (2021).1
Subject areas: Cell Biology, Cell Culture, Flow Cytometry/Mass Cytometry, Health Sciences, Model Organisms, Stem Cells, Tissue Engineering, Biotechnology and Bioengineering
Graphical abstract

Highlights
-
•
This is a conditioning protocol to enhance the regenerative properties of hMSCs
-
•
The protocol uses mechanical stretch to enhance the vascular phenotype of hMSCs
-
•
We also describe a cell encapsulation protocol to deliver the conditioned cells
-
•
We describe methods to assess cell phenotype and tests in animal models of ischemia
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Human mesenchymal stem cells (hMSCs) are an appealing cell type for therapeutic applications but remain limited by poor efficacy in clinical trials. Here, we describe a conditioning technique that enhances the vascular regenerative properties of hMSCs and increases their expression of endothelial cell and pericyte markers. We also describe an alginate gel encapsulation protocol for delivering the conditioned cells.
Before you begin
Here, we describe a validated treatment involving the use of biomechanical forces and pharmacological conditioning to drive MSC differentiation into a novel hybrid endothelial cell and pericyte phenotype. This phenotype demonstrates expression of both endothelial cell and pericyte markers and enhanced vascular regenerative properties.
We previously conducted a high-throughput screening of mechanical loading regimes and pharmacological treatments on MSCs, and evaluated treatment efficacy by measuring angiogenic signaling.1 We identified maximum angiogenic signal responsiveness in MSCs with a specific mechanical loading regime using a physiological waveform similar to the stretch of the brachial artery during the cardiac cycle (brachial loading) in combination with an EGFR/ErbB-2/4 (E/E) inhibitor. These findings were consistent for MSCs isolated from three independent patient donors.
The protocol outlined below begins with a description of MSC sample preparation and stretch machine preparation. Procedures for conditioning the cells with the angiogenic conditioning regime, validation of phenotype with flow cytometry, and evaluation of MSC vascular regenerative properties with an in vivo animal study are also described. Potential problems and proposed troubleshooting of these problems has been highlighted at the end of each key protocol step. Detailed troubleshooting information can be found here.
Institutional permissions
All experiments were approved by the Institutional Animal Care and Use Committee of the University of Texas at Austin and conform to the relevant regulatory standards. Researchers who would be applying this protocol for their own work would need to acquire permission from their respective institutions.
Preparation of hMSC media
Timing: as needed for cell culture expansion (for steps 1 to 4)
Here, culture media for human mesenchymal stem cells is prepared. Use sterile culture techniques in the preparation of media and culture of cells to prevent contamination (Figure 1A).
-
1.
Thaw Fetal Bovine Serum (FBS), L-Glutamine, and Penicillin-Streptomycin in a water bath at 37°C.
-
2.Add FBS, L-Glutamine, and Penicillin-Streptomycin in 410 mL of DMEM.
-
a.Use sterile technique throughout the culture of the cells.
-
a.
-
3.
Dispense the hMSC media in 50 mL aliquots.
-
4.
Store aliquots at 4°C.
Figure 1.
Preparation of mesenchymal stem cell culture and stretch plates
(A) hMSCs are cultured in T75 flasks until Passage 5.
(B) Stretch plates are assembled in the sterile cell culture hood.
(C) Stretch plate silicone membranes are coated with fibronectin.
(D) Coated silicone membranes are washed, dried, and sterilized under UV light for 24 h.
(E) MSCs are seeded on the 96 well stretch plates.
Preparation of stretch plates
Timing: 1 day (for steps 5 to 9)
Here, custom cell culture plates are prepared for the mechanical conditioning of the MSCs. If alternative equipment is used for the mechanical conditioning of cells, commercially available stretch plates can be sourced from Flexcell.
-
5.
Autoclave stainless steel plate top and bottom pieces, silicone gaskets, and #2–56 stainless steel screws in gravity cycle for 1 h.
-
6.
Move plate parts and silicone membranes to sterile cell culture hood.
-
7.
Assemble stretch plates (Figure 1B).
-
8.
Test stretch plates for leaks by adding 1 mL of sterile PBS to each well and leaving plate on a shaker for 30 min. If no liquid leaks out of the sides or bottom of the plates, the plates can be used.
-
9.
Remove PBS from leak-tested plates and leave in a sterile cell culture hood under UV light overnight (∼12 h).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PECAM-1 | BD Biosciences | Cat#562855 |
| VE-Cadherin | BD Biosciences | Cat#565671 |
| CD146 | BD Biosciences | Cat#564644 |
| PDGFR-β | BD Biosciences | Cat#558821 |
| CD105 | BD Biosciences | Cat#563466 |
| Nestin | BD Biosciences | Cat#561231 |
| CD90 | R&D Systems | Cat#967542 |
| CD73 | R&D Systems | Cat#967544 |
| CD105 | R&D Systems | Cat#967546 |
| Biological samples | ||
| Nu/J Mice, 10 weeks, male and female | Jackson Labs | Cat#002019 |
| Chemicals, peptides, and recombinant proteins | ||
| Trypsin-EDTA 0.05% | Thermo Fisher | Cat#25300120 |
| DMEM, low glucose, pyruvate, no glutamine, no phenol red | Thermo Fisher Scientific | Cat#11054001 |
| Fetal bovine serum (FBS) | Thermo Fisher Scientific | Cat#A3160602 |
| L-Glutamine | Thermo Fisher Scientific | Cat#25030081 |
| Penicillin-Streptomycin | Thermo Fisher Scientific | Cat#15140163 |
| PBS | Corning | Cat#46-013-CM |
| HEPES | VWR | Cat#J848-100ML |
| Fibronectin bovine plasma | Sigma-Aldrich | Cat#F1141-5MG |
| EGFR/ErbB-2/4 inhibitor | Santa Cruz | Cat#HDS 029 |
| Accutase® Cell Dissociation Solution | Sigma-Aldrich | Cat#A6964 |
| Stain Buffer (BSA) | BD Biosciences | Cat#554657 |
| BD Pharmingen™ Transcription-Factor Buffer Set (fix/perm buffer, perm/wash buffer) | BD Biosciences | Cat#562574 |
| Anti-Mouse Ig, κ/Negative Control Compensation Particles Set (mouse IG beads) | BD Biosciences | Cat#552843 |
| Sodium alginate, pharmaceutical grade | Sigma-Aldrich | Cat#PHR1471-1G |
| Novatach RGD | DuPont | Cat#4270321 |
| Type 1 collagen | VWR | Cat#354249 |
| Calcium chloride | Sigma-Aldrich | Cat#C8106-500G |
| Heparin sodium porcine mucosa | Sigma-Aldrich | Cat#SRE0027-250KU |
| Compressed argon gas | Airgas | Cat#AR 200 |
| Experimental models: Cell lines | ||
| Human bone marrow mesenchymal stem cells: donor 1 (Asian, male, aged 22 years), donor 2 (white, male, aged 24 years), donor 3 (white, female, aged 32 years) | MilliporeSigma | Cat#SCC034 |
| Software and algorithms | ||
| MATLAB | MathWorks | N/A |
| Speckle Pylon | N/A | N/A |
| FlowJo | FlowJo | N/A |
| Other | ||
| Custom mechanical loading system (See Table Alternative Equipment. Steps 13-25: Biomechanical Conditioning for alternative mechanical loading system) | N/A | N/A |
| Coaxial Airflow Encapsulator (See Table Alternative Equipment. Steps 48-71: Alginate Gel Encapsulation & Laser Speckle Imaging for alternative system) | N/A | N/A |
| Custom Laser Speckle Imaging Setup (See Table Alternative Equipment. Steps 89-103: Alginate Gel Encapsulation & Laser Speckle Imaging for alternative system) | N/A | N/A |
| Custom Laser Speckle Imaging Analysis Code (See Table Alternative Equipment. Steps 104-108: Alginate Gel Encapsulation & Laser Speckle Imaging for alternative system) | MATLAB | Dunn et al.2 |
| Retractor - Blunt/5 mm wide | Fine Science Tools | Cat#18200-11 |
| Adson Forceps – Wide Grip | Fine Science Tools | Cat#11006-12 |
| Dumont #5/45 Forceps – Standard/Dumoxel | Fine Science Tools | Cat#11251-35 |
| Student Dumont #7 Forceps – Standard/Inox | Fine Science Tools | Cat#91197-00 |
| Round handled suture tying forceps – Curved/10.5 cm | Fine Science Tools | Cat#18026-10 |
| Harman Hemostat – Straight | Fine Science Tools | Cat#13002-10 |
| Bonn Scissors – Straight, sharp point, 15 mm blade length, 3.5″ overall length | Roboz | Cat#RS-5840 |
| Vannas Spring Scissors – Straight/2 mm cutting edge | Fine Science Tools | Cat#15000-03 |
| Q-Tips – Cotton head, wood shaft, non-sterile | Puritan Medical | Cat#806-WC |
Materials and equipment
hMSC media
DMEM containing low glucose and pyruvate without glutamine and phenol red is supplemented with FBS, L-glutamine, and Penicillin-Streptomycin. The medium is stored at 4°C.
| Reagent | Final concentration | Total quantity |
|---|---|---|
| DMEM, low glucose, pyruvate, no glutamine, no phenol red | 82% v/v | 410 mL |
| Fetal Bovine Serum (FBS) | 15% v/v | 75 mL |
| L-Glutamine(200 mM) | 4 mM | 10 mL |
| Penicillin-Streptomycin | 100 U/mL | 5 mL |
Alternative Equipment. Steps 13-25: Biomechanical Conditioning
| Used equipment | Equipment alternative | Category number |
|---|---|---|
| Custom Mechanical Loading System1,3,4 |
Flexcell Mechanical Loading Tension System | FX-6000T |
Surface flow antibody cocktail: Add 5 μL VE-CAD antibody, 5 μL CD105 antibody, 5 μL CD146 antibody, 5 μL NG2 antibody, 5 μL PECAM antibody, 5 μL PDGFRβ antibody in 55 μL of BD stain buffer, and mix gently by pipetting up and down.
| Reagent | Final concentration | Total quantity |
|---|---|---|
| BD stain buffer | 55% v/v | 55 μL |
| VE-CAD (BV421-A) | 5% v/v | 5 μL |
| CD105 (BV650-A) | 5% v/v | 5 μL |
| CD146 (BB515) | 5% v/v | 5 μL |
| NG2 (AF647-A) | 5% v/v | 5 μL |
| PECAM (BV605-A) | 5% v/v | 5 μL |
| PDGFRβ (PE-A) | 20% v/v | 20 μL |
Alternative Equipment. Steps 89-103: Alginate Gel Encapsulation & Laser Speckle Imaging
| Used equipment | Equipment alternative | Category number |
|---|---|---|
| Coaxial Airflow Encapsulator | Coaxial Airflow Encapsulator | Nisco Engineering AG |
| Custom Laser Speckle Imaging Setup | RWD Laser Speckle Imaging System | RFLSI III |
| Custom Laser Speckle Imaging Analysis Code | MATLAB | Mathworks |
CRITICAL: BSL2 level safety precautions should be used when culturing hMSCs. Standard safety precautions should be observed when handling compressed gasses and lasers.
Step-by-step method details
Preparation of hBM-MSCs
Timing: 30 min + 10 days (for steps 1 to 12)
In this step, hMSCs will be cultured until passage 5 to expand the cell population while maintaining healthy morphology and characteristics of a stem cell. Higher passages alter phenotypic properties that can affect experimental reproducibility. Using the recommended seeding density and confluency at passaging is also crucial to maintain cell genotype and phenotype. After proliferation under standard culture conditions, cells will be seeded on plates with flexible culture surfaces for mechanical conditioning. This step expands the cell population for use in experiments (Figure 1A).
-
1.
Thaw hMSCs at 37°C.
-
2.
Add 10 mL of warm hMSC media.
-
3.
Centrifuge hMSC at 300G for 3 min at room temperature (20°C–25°C).
-
4.
Aspirate culture media.
-
5.Add 10 mL of warm hMSC media.
-
a.Resuspend cells thoroughly.
-
a.
-
6.
Seed hMSC in T75 flask.
-
7.Passage hMSC at 70%–80% confluence.
-
a.Check cell confluency under a microscope.
-
b.Warm cell culture media, PBS, and 0.05% Trypsin-EDTA at 37°C.
-
c.Aspirate culture media.
-
d.Wash cells with 5 mL sterile PBS with 30μM of HEPES.
-
e.Add 5 mL of warm 0.05% Trypsin-EDTA and incubate for 3 min at 37°C.
-
f.Add 5 mL of warm cell culture media to deactivate trypsin-EDTA.
-
g.Centrifuge at 300 G for 3 min at room temperature (20°C–25°C).
-
h.Aspirate culture media.
-
i.Resuspend cells in 1 mL of warm hMSC media.
-
a.
-
8.
Count cells using hemacytometer.
-
9.
Prepare cell density of 40,000 cells/mL.
-
10.
Seed cell in T75 flask.
-
11.
Monitor cells daily for growth.
-
12.
Change media completely every other day.
Biomechanical conditioning
Timing: 9 days (for steps 13 to 25)
In this step, a mechanical stretching device is used to deliver physiologically mimetic patterns of mechanical strain to cells in culture. A pharmaceutical compound, an EGFR/ERBB-2/4 (E/E) inhibitor (CAS#: 881001-19-0), is also delivered to the cells during the weeklong biomechanical conditioning, and together these treatments result in the generation of enhanced vascular regenerative phenotypes. We used a custom-built device, but commercial devices are available that could be programmed to produce similar stretch patterns.
Note: A commercially available Flexcell FX-6000T Tension System can be used in place of the custom-built High Throughput Biaxial Oscillatory Stretch System described here. The custom-built HT-BOSS has an advantage over the Flexcell in experimental throughput, as the HT-BOSS can apply equibiaxial strain to up to 36 6-well (9.6 cm2) culture wells, or 576 96-well (0.32 cm2) culture wells simultaneously. The Flexcell FX-6000T can apply equibiaxial strain to up to 24 6-well (9.6 cm2) culture wells and enables programming of custom mechanical waveforms across a range of strain amplitudes and frequencies of loading that enable the mechanical loading regime detailed in this protocol. See the Flexcell FX-6000T user manual for more details regarding the technical specifications of this commercially available system.
-
13.
UV sterilize completed stretch plates overnight (∼12 h).
-
14.
Coat plates with 50 μg/mL fibronectin overnight (∼12 h) at 37°C.
-
15.Remove fibronectin solution gently (Figure 1B).
-
a.Wash plates once with 1 mL sterile PBS.
-
a.
-
16.Trypsinize hMSCs.
-
a.Aspirate culture media and wash once with 4 mL of sterile PBS.
-
b.Add 3 mL of warm 0.05% Trypsin-EDTA and incubate for 3 min at 37°C.
-
c.Add 10 mL of warm MSC media to deactivate trypsin-EDTA.
-
d.Waterfall cells thoroughly and collect cell suspension in 15 mL conical tube.
-
e.Centrifuge 300G for 3 min.
-
a.
-
17.Prepare cell suspension.
-
a.Aspirate media from 15 mL tube.
-
b.Resuspend cells in 1 mL MSC media.
-
c.Count cells using hemacytometer.
-
d.Adjust the concentration of the cell suspension by adding additional media to generate required seeding density.
-
i.For a 6 well plate: 95,000 cells/mL.
-
ii.For a 96 well plate: 43,000 cells/mL.
-
i.
-
a.
-
18.Seed cells into stretch plates at a density of 20,000 cells/cm2.
-
a.For a 6 well plate: Seed 2 mL/well at density of 95,000 cells/mL.
-
b.For a 96 well plate: Seed 150 μL/well at density of 43,000 cells/mL.
-
a.
-
19.
Let cells attach and grow overnight (∼12 h) in 37°C 5% CO2 cell culture incubator.
-
20.
Aspirate media and replace with fresh MSC media + 1 μM EGFR/ERBB-2/4 inhibitor.
-
21.Attach stretch plate to machine (Figure 2A).
-
a.First tighten outer screws.
-
b.Then tighten inner screws.
-
a.
-
22.Start water coolant system (Figure 2B).
-
a.Run at 15C during stretching of cells.
-
a.
-
23.Stretch cells.
-
a.Apply brachial waveform mechanical loading at 7.5% maximal strain, 0.1 Hz for 4 h per day for 7 days (Figure 2C).
-
a.
-
24.
Every other day, replace culture media with fresh MSC media and E/E inhibitor.
-
25.
Troubleshooting 1: Stretch machine does not remain balanced in zero position.
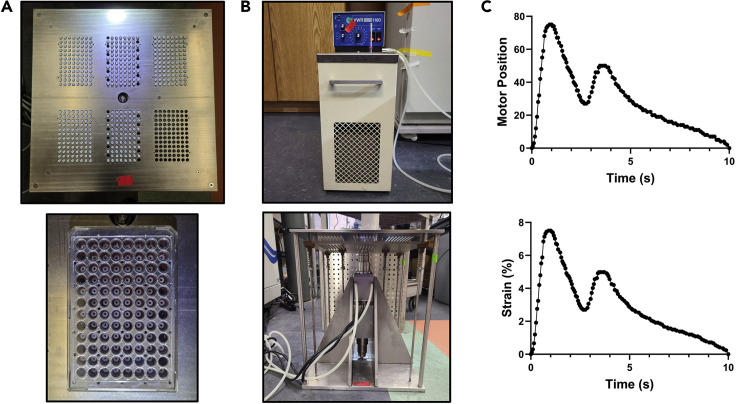
Figure 2.
The custom-built high throughput biaxial oscillatory stretch system (HT-BOSS)
(A) The completed 96 well stretch plates are attached to the HT-BOSS.
(B) The coolant system and HT-BOSS are turned on.
(C) The brachial waveform at 7.5% maximal strain is applied to the hMSCs at a frequency of 0.1 Hz.
Flow cytometry analysis
Timing: 5 h (for steps 26 to 47)
In this step, conditioned cells are fixed and permeabilized followed by immunostaining for endothelial cell and pericyte markers. MSC marker expression is evaluated by flow cytometry analysis.
-
26.Collect hMSCs from stretch plates.
-
a.Thaw accutase at 4°C overnight (∼12 h).
-
b.Rinse cells with PBS.
-
c.Aspirate PBS.
-
d.Add accutase to cells.
-
e.Incubate cells with accutase at RT for 5–10 min.
-
f.Use cell scraper to detach cells.
-
g.Collect cells into a FACS tube.
-
i.Use cell strainer on a tube.
-
ii.Keep tubes on ice.
-
i.
-
a.
-
27.
Centrifuge cells at 4°C for 4 min at 400 G.
-
28.
Aspirate supernatant.
-
29.
Count cells using hemacytometer.
-
30.
Add 150,000 cells to each flow cytometry tube.
-
31.Add surface staining antibodies to cells.
-
a.Add surface staining antibodies in BD stain buffer.
-
i.Include 1 tube of unstained cells as a negative control.
-
i.
-
b.Add BD stain buffer with antibodies to cells.
-
c.Pipette up and down to resuspend cell pellet.
-
a.
-
32.
Incubate cells at 4°C for 30 min.
-
33.
Add 1 mL of stain buffer to the tube.
-
34.
Centrifuge cells at 4°C for 4 min at 400 G.
-
35.
Aspirate supernatant.
-
36.Fix cells with 1 mL of fix/perm solution.
-
a.Prepare fix/perm solution by diluting the solution according to the manufacturer’s protocol.
-
b.Add 1 mL of fix/perm solution.
-
c.Pipette up and down to break up pellet.
-
d.Incubate the solution at 4°C for 50 min.
-
a.
-
37.Wash cells with 1 mL of perm/wash buffer.
-
a.Prepare perm/wash buffer by diluting the solution according to the manufacturer’s protocol.
-
b.Add 1 mL of perm/wash buffer in the tube.
-
c.Centrifuge cells at 4°C for 4 min at 400 G.
-
d.Aspirate supernatant.
-
a.
-
38.Add Nestin antibody to cells.
-
a.Add 5 μL of nestin antibody in BD stain buffer.
-
b.Add BD stain buffer with the antibody to cells.
-
c.Pipette up and down to break pellet.
-
a.
-
39.
Incubate cells at 4°C for 50 min.
-
40.Wash cells with 1 mL of perm/wash buffer.
-
a.Add 1 mL of perm/wash buffer in the tube.
-
b.Centrifuge cells at 4°C for 4 min at 400 G.
-
c.Aspirate supernatant.
-
a.
-
41.Wash cells again with 1 mL of perm/wash buffer.
-
a.Add 1 mL of perm/wash buffer in the tube.
-
b.Centrifuge cells at 4°C for 4 min at 400 G.
-
c.Aspirate supernatant.
-
a.
-
42.Resuspend cells in 500 μL of stain buffer.
-
a.Samples can be stored in a cooler with ice at 4°C overnight (∼12 h) to be analyzed by flow cytometry the next day. Cover samples with aluminum foil to prevent exposure to light.
-
a.
-
43.Analyze MSC phenotype by flow cytometry.
-
a.Prepare compensation controls.
-
i.Mix one droplet of positive mouse IG beads and one droplet of negative mouse IG beads in each flow tube.
-
ii.Add 80 μL of stain buffer to each tube.
-
iii.Add 20 μL of each mouse IG fluorescence to each tube.
-
iv.Pipette up and down to mix thoroughly.
-
v.Incubate beads at 4°C for 30 min.
-
vi.Add 1 mL of stain buffer to each tube.
-
vii.Centrifuge beads at 4°C for 4 min at 400 G.
-
viii.Aspirate supernatant.
-
ix.Add 1 mL of fix/perm solution to each tube.
-
x.Pipette up and down to break pellet.
-
xi.Incubate beads at 4°C for 50 min.
-
xii.Add 1 mL of stain buffer to wash beads.
-
xiii.Centrifuge beads at 4°C for 4 min at 400 G.
-
xiv.Aspirate supernatant.
-
xv.Add 1 mL of stain buffer to wash beads.
-
xvi.Centrifuge beads at 4°C for 4 min at 400 G.
-
xvii.Aspirate supernatant.
-
xviii.Add 1 mL of stain buffer to wash beads.
-
xix.Centrifuge beads at 4°C for 4 min at 400 G.
-
xx.Aspirate supernatant.
-
xxi.Resuspend beads in 500 μL of stain buffer.
-
i.
-
b.Set fluorescence in compensation window.
-
c.Change filter configuration to be matched with dyes (Figure 3).
-
d.Adjust voltages for FSC and SSC channels using an unstained MSC control sample to set the position of the cell population.
-
e.Adjust voltages for fluorescence channels using an unstained MSC control sample so that the intensity of the unstained group is lower than 103.
-
f.Adjust voltages for each fluorescence channels using each IG control beads so that the peak of each channel is around 104.
-
g.Set gate around the peak.
-
h.Mix in unstained beads to assure that the background peak is well spaced from the positive peak.
-
i.Measure samples to analyze phenotype. Count at least 10,000 events per tube.
-
a.
-
44.
Troubleshooting: There is spectral overlap between different fluorophores during compensation step.
-
45.Perform gating analysis in Flowjo software for control groups including endothelial cells, vSMCs, and MSCs to find an optimal gating strategy (Figure 4).
-
a.Make scatterplot and set FSC-A on X-axis and SSC-A on Y-axis with endothelial cell data (Figure 4A).
-
b.Gate for the population of cells to exclude debris and large particles.
-
c.Select gated population, then plot SSC-A on X-axis versus SSC-H on Y-axis.
-
d.Gate for the population of single cells to exclude doublets and large particles.
-
e.Select gated population, then plot FSC-A on X-axis versus FSC-H on Y-axis.
-
f.Gate for the population of single cells to exclude doublets.
-
g.Select gated population, then plot PDGFRβ on X-axis versus PECAM-1 on Y-axis.
-
h.Gate for the population of cells to separate endothelial phenotype from vSMCs, pericyte, undifferentiated MSCs, and the hybrid phenotype.
-
i.Select gated population, then plot CD105 on X-axis versus CD144 on Y-axis.
-
j.Repeat steps A-G with vSMC data to confirm vSMC phenotype (Figure 4B).
-
k.Repeat steps A-G with pericyte data to separate pericyte phenotype from the other phenotypes (Figure 4C).
-
l.Gate for the population of cells that only express PDGFRβ.
-
m.Plot NG2 on X-axis versus CD146 on Y-axis.
-
n.Gate for the population of cells that express both markers.
-
o.Plot FSC-A on X-axis versus Nestin on Y-axis.
-
p.Gate for the population of cells to confirm pericyte phenotype.
-
a.
-
46.Perform gating analysis in Flowjo software for mechanically conditioned cells (Figure 4D).
-
a.Make scatterplot and set FSC-A on X-axis and SSC-A on Y-axis.
-
b.Gate for the population of cells to exclude debris and large particles.
-
c.Select gated population, then plot SSC-A on X-axis versus SSC-H on Y-axis.
-
d.Gate for the population of single cells to exclude doublets and large particles.
-
e.Select gated population, then plot FSC-A on X-axis versus FSC-H on Y-axis.
-
f.Gate for the population of single cells to exclude doublets.
-
g.Select gated population, then plot PDGFRβ on X-axis versus PECAM-1 on Y-axis.
-
h.Gate for the population of cells that express both markers to separate the hybrid phenotype from endothelial cells, vSMCs, pericyte, and undifferentiated MSCs.
-
i.Select gated population, then plot CD105 on X-axis versus CD144 on Y-axis.
-
j.Gate for the population of cells that express both markers.
-
k.Plot Nestin on X-axis versus CD146 on Y-axis.
-
l.Gate for the population of cells that express both markers.
-
m.Plot FSC-A on X-axis versus NG2 on Y-axis.
-
n.Gate for the population of cells that express the pericyte marker NG2.
-
o.Export the frequency of the population and statistics in the table editor to draw graphs.
-
a.
-
47.
Troubleshooting: The cell populations are appearing off the edge of the chart, likely due to incorrect FSC and SSC voltage parameters.
CRITICAL: It is essential to run cell lines with known phenotype when preparing the gating strategy for the flow cytometry to account for the specific flow cytometer. Mature endothelial cells and pericytes should be run at the same time as the hMSCs to guide accurate gating of the cell populations.
Figure 3.
Setup of flow cytometry hMSC phenotype analysis
The filter configuration of flow cytometry system is adjusted to match fluorescent spectra and minimize spectral overlap (upper panel).
The spectral profiles of fluorescent dyes (lower panel).
Figure 4.
Overview of flow cytometry phenotype gating strategy
The optimal gating strategy for identifying endothelial cell and pericyte markers of mechanically conditioned MSCs in Flowjo software.
Alginate gel encapsulation and testing in in vivo model
Timing: 1 day foralginate gel encapsulation
Timing: 1 day forhindlimb ischemia surgery
Timing: 28 days forlaser speckle imaging
In this step, conditioned cells are encapsulated in alginate-RGD peptide beads, and angiogenic potential of the MSCs is evaluated in a hindlimb ischemia mouse model. Blood flow recovery through the limb is assessed over time with laser speckle imaging. A coaxial airflow encapsulator (Nisco Engineering AG Var J1) is used to prepare the alginate beads.
Alginate gel encapsulation
1 day before surgery:
-
48.
Prepare alginate solution 1 day in advance: 2% alginate solution (2g alginate per 100 mL sterile saline) mixed 1:5 with 2% alginate:RGD.
-
49.Supplement alginate solution with 0.045% type 1 collagen. Prepare 250 μL per mouse even though only 200 μL per mouse will be used.
-
a.Mix extremely well. Keep on ice or in the fridge at 4°C.
-
a.
-
50.
Prepare calcium chloride solution: 1.1% CaCl2 in ultrapure water.
-
51.
Autoclave sterile water and sterile surgical saline solutions.
-
52.Prepare gel extruder parts following manufacturer protocol:
-
a.Rinse and autoclave feeder tubes.
-
b.Soak extruder parts in 70% ethanol.
-
a.
-
53.
Leave parts in fridge overnight (∼12 h).
Day of surgery:
-
54.Prepare gel extruder:
-
a.Assemble gel extruder pieces (Figure 5A).
-
b.Load device with 70% ethanol.
-
c.Let air dry for 30 min.
-
a.
-
55.
Cover syringe feeder tube with ice and place syringes in 4°C refrigerator before beginning.
-
56.
Using the syringe, pump air through entire system to ensure it is unclogged.
-
57.Prepare cells for encapsulation.
-
a.Remove culture media from 6 well stretch plates.
-
b.Wash cells once with PBS.
-
c.Treat cells with warmed 0.05% Trypsin-EDTA for less than 5 min to detach cells.
-
a.
-
58.
Centrifuge cells 300G for 5 min.
-
59.
Resuspend cells in 1 mL room temperature (20°C–25°C) PBS and count them with a hemacytometer.
-
60.
Centrifuge cells 300G for 5 min and resuspend 1 × 106 cells in 200 μL (5 × 106 cells/mL) of alginate solution. Prepare slightly more than 200 μL in syringe, at least 250 μL or more per mouse.
-
61.
Prime encapsulator tube with alginate and cell solution, be sure to keep tube on ice. Prime until alginate solution drips into waste dish.
-
62.
Fill syringe with air to push alginate solution through tube, load syringe onto syringe pump and lock in place.
-
63.
Replace waste dish with clean dish containing calcium chloride solution.
-
64.
Begin flowing argon gas by opening main gas tank valve, then opening secondary valve slowly until flow reaches 3 L/min.
-
65.
Begin flowing syringe pump at 0.1 mL/min, be sure that syringe is locked in place (Figure 5B).
-
66.
Collect alginate beads in calcium chloride solution, beads should have a diameter of 1,200 μm. Pause syringe pump once 200 μL of solution is extruded.
-
67.
Remove CaCl2 and wash beads with sterile saline 2–3 times. Collect beads using 50 mL serological pipette and transport to animal.
-
68.
Tip: Prep separate saline syringe to flush extruder if it becomes clogged.
-
69.
Troubleshooting: Gel beads do not fall straight out of encapsulator.
-
70.
Troubleshooting: Gel solution solidifies before encapsulation.
-
71.
Troubleshooting: Bead flow is turbulent.
Figure 5.
Preparation of gel extruder and fabrication of alginate beads
(A) Gel extruder parts are assembled in the sterile cell culture hood.
(B) Mechanically conditioned MSCs are encapsulated in alginate gel beads.
Hindlimb ischemia model
Timing: 1 day
In this step, surgical techniques involving the ligation of the femoral artery are used to generate a hindlimb ischemia mouse model. The conditioned MSCs encapsulated in alginate beads are implanted directly to the region surrounding the wounded femoral artery. This step also includes IACUC-approved practices for inducing anesthesia, applying analgesics, and post-operative monitoring of the animal subjects.
Necessary Reagents/Tools for Surgical Procedure
| Item | Vendor | Catalog # |
|---|---|---|
| Retractor - Blunt/5 mm wide | Fine Science Tools | 18200-11 |
| Adson Forceps – Wide Grip | Fine Science Tools | 11006-12 |
| Dumont #5/45 Forceps – Standard/Dumoxel | Fine Science Tools | 11251-35 |
| Student Dumont #7 Forceps – Standard/Inox | Fine Science Tools | 91197-00 |
| Round Handled Suture Tying Forceps – Curved/10.5 cm | Fine Science Tools | 18026-10 |
| Harman Hemostat – Straight | Fine Science Tools | 13002-10 |
| Bonn Scissors – Straight, sharp point, 15 mm blade length, 3.5″ overall length | Roboz | RS-5840 |
| Vannas Spring Scissors – Straight/2 mm cutting edge | Fine Science Tools | 15000-03 |
| Q-Tips – Cotton head, wood shaft, non-sterile | Puritan Medical | 806-WC |
| Heparin Sodium porcine mucosa | Sigma Aldrich | SRE0027-250KU |
| Sodium Alginate, Pharmaceutical Grade | Sigma Aldrich | PHR1471-1G |
| Collagen Type 1, Sterile | VWR | 354249 |
| Novatach RGD | DuPont | 4270321 |
1 Day Before Surgery:
-
72.
Autoclave surgical tools (see table above) and absorbent surgical pads.
-
73.
Anesthetize mice with 2%–5% isoflurane for 1–2 min.
-
74.Prepare mice for surgery:
-
a.Perform all steps in sterile cell culture hood (Figure 6A).
-
b.Anesthetize mice with 2%–5% isoflurane for 1–2 min.
-
c.Mask and maintain mouse on 2%–5% isoflurane for duration of procedure.
-
d.Shave surgical site: left hindlimb.
-
e.Scrub surgical site with three cycles of betadine scrub followed by alcohol rinse.
-
a.
Figure 6.
The hindlimb ischemia surgical model and laser speckle imaging analysis
(A) Preparation of surgical workplace and surgical tools.
(B) The left hindlimb femoral artery is ligated without damaging adjacent veins and nerves.
(C) Setup of laser speckle imaging device.
(D) Representative laser speckle images of control and mechanically conditioned mice.
(E) The laser speckle images are converted to greyscale and blood flow intensity is analyzed with Photoshop.
(F) Representative quantification of relative ischemic to contralateral hindlimb blood perfusion for control and mechanically conditioned mice.
Day of Surgery
-
75.
Lay down fresh autoclaved absorbent surgical pads, these should be used as a sterile field for the placement of autoclaved surgical instruments and other sterile materials throughout the procedure.
-
76.Prepare mice for surgery:
-
a.Perform all steps in sterile cell culture hood (Figure 6A).
-
b.Anesthetize mice with 2%–5% isoflurane for 1–2 min.
-
c.Mask and maintain mouse on 2%–5% isoflurane for duration of procedure.
-
d.Test adequate anesthesia with withdrawal to simulation in toe pinch test.
-
e.Pre-emptively inject 10 mg/kg carprofen subcutaneously at least 30 min before the end of the procedure.
-
a.
-
77.During the procedure, check adequate anesthesia every 15 min.
-
a.Normal breathing.
-
b.Normal skin color.
-
c.Lack of response to toe pinch stimulus.
-
a.
-
78.
Make a skin incision on the media side of the thigh of the left hindlimb.
-
79.
Dissect femoral artery free without damaging the vein and nerve (Figure 6B).
-
80.
Ligate the left femoral artery with two sutures using 3-0 woven silk, just distal to the inguinal ligament (Figure 6B).
-
81.
Deliver 200 μL (1 × 106 cells) of alginate beads directly to the region surrounding the femoral artery.
-
82.
Close wound with 4-0 Vicryl resorbable sutures.
-
83.
Monitor the mouse for 10 min as it recovers from anesthesia.
Post-operative monitoring and recovery
-
84.Monitor the mouse for the following criteria:
-
a.Normal breathing.
-
b.Normal skin color.
-
c.Normal posture.
-
d.Weight loss or gain.
-
e.Appropriately healing wound site. No bleeding, wound dehiscence, signs of inflammation or infection.
-
f.Signs of pain including tearing and changes in movement or activity level.
-
a.
-
85.Follow recommended post-operative monitoring schedule:
-
a.Twice daily for two days following surgery.
-
b.Once per day for the next three days.
-
c.Every other day following the first five days.
-
a.
-
86.Inject 10 mg/kg carprofen subcutaneously every 24 h for 48 h following the surgery.
-
a.After the first 48 post-operative hours, extend 10 mg/kg carprofen treatment if evidence of pain is observed.
-
a.
-
87.
Troubleshooting: Mouse chews sutures and reopens wound.
-
88.Troubleshooting: Mouse shows inadequate wound healing or signs of infection. Consult your facility’s veterinarian to determine if euthanasia is necessary if any of the following occur:
-
a.Weight loss: loss of 10%–15% or if not measured, characterized by cachexia and muscle wasting.
-
b.Weakness/inability to obtain feed or water: Inability or extreme reluctance to stand which persists for 24 h, assuming that the animal has recovered from anesthesia.
-
c.Moribund state: measured by a lack of sustained purposeful response to gentle stimuli.
-
d.Infection: infection involving any organ system which fails to respond to antibiotic therapy within an appropriate time and is accompanied by systemic signs of illness.
-
e.Non-healing wounds, repeated self-trauma, or self-mutilation of toes or limbs.
-
f.CNS depression, seizures, paralysis of one or more extremities; pain unresponsive to analgesic therapy.
-
a.
Laser speckle imaging
Timing: 28 days
In this step, a custom-built laser speckle imaging system2 was used to monitor blood flow recovery in murine hindlimbs as described in our previous studies.5,6,7,8 Alternative laser speckle imaging equipment can be sourced from RWD RFLSI III. Speckle Pylon software was used for laser speckle image acquisition. Alternative image acquisition software can also be sourced from the RWD RFLSI III suite. A second alternative is to use laser doppler imaging as described in previous study using a similar surgical model.9
-
89.
Plug in the heating pad and maintain it at a constant temperature of 37°C.
-
90.
Connect laser box to power (Figures 6A and 6C) and to camera (Figures 6B and 6C). Turn on laser power and adjust to 25%.
-
91.
Connect the camera to power and to the computer. Open the camera lid.
-
92.Open the speckle pylon software program.
-
a.If the full-size window doesn’t pop out, the power is not connected.
-
a.
-
93.Edit the following parameters in the speckle control panel:
-
a.Acquisition settings: change # sequence from 2 to 10.
-
b.Display settings: check on “enable overlay”, and then change raw min = 2.00, max = 250.00; sc min = 0.025, max = 0.11.
-
a.
-
94.Edit file name to reflect mouse sample number and time point (e.g., “m400_d14”).
-
a.Change file name for each sample mouse.
-
b.Default file name is “test.”
-
a.
-
95.Click on “home” icon to choose a path to save file.
-
a.Example: USB → day14 group A→ m400.
-
b.Check the files after each mouse imaging session to ensure proper acquisition and saving.
-
a.
-
96.
Once the equipment is set up, turn on isoflurane to 2%–5% and oxygen to 1 L/min flow rate. Make sure to check on the level of isoflurane. Put the mouse in the induction box for anesthesia and then transfer it to the nose cone for speckle imaging (Figure 6C).
-
97.
When imaging, laser detection card to test laser coverage. Adjust mouse position to expose both paws and make them face up (Figure 6D).
-
98.Click on camera icon to look at live image, wait 20 s–40 s to stabilize, then click on stop icon, and click on the green triangle icon to start acquisition. You should see the progress of acquisition on the bottom of the control panel window.
-
a.Troubleshooting: image is frozen.
-
b.Before proceeding to the next mouse, ensure that all 10 images are properly saved after 100% progress bar is complete.
-
a.
-
99.
Continue to next mouse: repeat steps 7–10.
-
100.When the imaging session is complete, turn off isoflurane and oxygen, and return mice to animal housing. Close speckle windows. Turn the laser power to 0 and power device off. Disconnect equipment in the following order:
-
a.Laser power.
-
b.Laser camera connection.
-
c.Camera power.
-
d.Camera computer connection.
-
a.
-
101.
Troubleshooting: Observed blood flow is not constant over the course of imaging.
-
102.
Troubleshooting: Observed blood flow is drastically different between separate days of imaging.
-
103.
Troubleshooting: High amounts of external noise are observed in speckle images.
Laser speckle imaging analysis
Timing: 4 h
Laser speckle imaging analysis was performed using instruments and analysis tools that are extensively described in previously published work.2 These methods can be repeated with MathWorks MATLAB software.
-
104.
After acquiring laser speckle images, import .sc files into MATLAB.
-
105.
Run MATLAB script to process speckle data into grayscale, following previously described methods.2
-
106.
Export grayscale images into Photoshop, or a similar image processing tool, such as ImageJ.
-
107.
Circle mouse feet to measure the blood flow pixel intensity (Figure 6E).
-
108.
Export data as .csv files and organize into a spreadsheet for each mouse and time point measured. See example data (Figure 6F).
Expected outcomes
This protocol describes a way to apply a novel biomechanical conditioning regime to drive hMSC differentiation into a hybrid endothelial cell and pericyte phenotype with enhanced vascular regenerative properties. This protocol can be used to further study MSC differentiation, mechanobiology signaling pathways, and potentially be combined with additional strategies to further enhance the vascular regenerative characteristics of these cells.
Figure 4 demonstrates the upregulated expression of endothelial and pericyte markers that is demonstrated by MSCs after the week-long biomechanical treatment regime outlined in this protocol. Figure 6 illustrates the enhanced vascular regenerative properties of the conditioned MSCs after implantation in a hindlimb ischemia murine model. Our previous work demonstrated that these effects persist across multiple hMSC donor lines,1 and we expect that other researchers who follow this protocol will yield reproducible results.
Quantification and statistical analysis
All of the results are shown as mean ± S.E.M. All experiments used biological replicates that consisted of cells in non-repeated, independent cell culture wells or tissue samples from different animals, unless specified otherwise. Multiple comparisons between groups were analyzed using two-way ANOVA followed by a Tukey post hoc or a Dunnett post hoc test when testing multiple comparisons versus a control group. For nonparametric data, multiple comparisons were performed using Kruskal–Wallis tests followed by post hoc testing with the Conover–Iman procedure. P < 0.05 was considered to be statistically significant for all of the tests.
Limitations
Although mechanical stimulus is a powerful regulator of MSC differentiation, the expression of markers on MSCs can vary depending on waveform, frequency, direction of the mechanical strain, and surface coating of flexible stretch membranes. Since the described methods for applying mechanical strain are novel and only recently explored, mechanical strains by different methods or devices could lead to the variation of marker expression.
Furthermore, MSCs exhibit significant donor-to-donor and intra-population heterogeneity, which may lead to inconsistent results.10
MSC differentiation is sensitive to cell confluence. Accurate cell counting, regular maintenance, and consistent cell culture techniques are necessary to generate reproducible results. The in vitro age or passage number of the MSCs is also highly important to their multipotency and overall regenerative potential.11 The MSCs used in our published study were between passage 3–6, and we recommend following these criteria to obtain reproducible results.1
The threshold of each marker expression may be changed depending on the type of flow cytometer used as well as antibody batch efficacy. We recommend the use of “gold standard” endothelial and pericyte cell lines to set optimal flow cytometry gating settings for each specific flow cytometry system.
The hindlimb ischemia surgical mouse model relies upon well-trained and consistent surgical practices, regularly timed and dosed analgesia administration, and well-defined surgical and post-operative end-points.12 Variations in these practices can lead to altered vascular remodeling and neovascularization and confound the results. We recommend ensuring that personnel are well-trained in animal surgery, anesthesia administration, and post-operative monitoring before attempting this procedure. Consult with your facility’s animal research center for specific guidance on surgical practices and euthanasia criteria.
Troubleshooting
Problem 1
When attaching stretch plates to the stretch machine, the piston platen may become imbalanced, causing stretch to be disproportionately applied (step 23 in biomechanical conditioning).
Potential solution
Rebalance piston platen by removing the top plate, then adding washers to the corner posts until the top plate lays flat. A level can be used to manually verify that the plate is balanced. Once balanced, screw the top plate back on and try running the waveform once more.
Problem 2
When multiple fluorescent dyes are used to stain different markers, compensation is necessary to correct spectral overlap (steps 44-47 in flow cytometry analysis).
Potential solution
Make sure that all voltages are the same for all controls or compensation. Ensure that appropriate compensation controls are used for each fluorophore. After performing compensation, calculate and apply the offset to mitigate spectral overlap.
Problem 3
When the voltage of SSC or FSC is too high, cell populations can be pushed out of the chart, and the data is lost. Similarly, if SSC or FSC voltages are too low, cell populations will appear clustered on the lower left region of the chart (steps 44-47 in flow cytometry analysis).
Potential solution
Adjust the voltage of SSC or FSC so that most cells are on the chart.
Problem 4
If the beads do not fall straight out of the encapsulator, gas vents are likely clogged (step 69 in alginate gel encapsulation).
Potential solution
Pause gas flow and clean excess gel from the outflow vent. Try unclogging gas vents with a small surgical needle.
Problem 5
If the gel solution solidifies before encapsulation, it is likely that the collagen in the alginate gel solution solidified due to the solution being too warm (step 70 in alginate gel encapsulation).
Potential solution
Keep all gel tubes on ice for as long as possible to prevent collagen from gelling. Cool Eppendorf tubes and pipettes to 4°C before beginning.
Problem 6
If bead flow is turbulent, gas flow rate may be too high (step 71 in alginate gel encapsulation).
Potential solution
Try reducing the gas flow rate from 3L/min to 2L/min.
Problem 7
Mice may chew the sutures on their hindlimb wound and cause wound dehiscence in the days following the surgery (step 87 in hindlimb ischemia model).
Potential solution
If wound dehiscence is observed, regardless of severity, contact your facilities’ veterinarian. If wound dehiscence is deemed minor, with no signs of major bleeding or inflammation at the wound site, surgeon will conduct repair procedure to re-suture the wound. After observing minor wound dehiscence during post-op monitoring, surgeon will anesthetize mice using 2%–5% isoflurane gas. Animal will also be administered 10 mg/kg carprofen at the beginning of the repair procedure, so that analgesic has time to take effect before the animal wakes up. Next, wound site will be flushed with sterile saline to clean the wound. Next, tissue edges will be trimmed to ensure fresh blood flow following wound closure. Then, wound will be re-sutured with resorbable sutures (4-0 vicryl). Following repair procedure, surgeon will contact facility veterinarian, and the monitoring and analgesic plan will be followed the same as for the initial surgery. If repair procedure fails to close wound and/or wound dehiscence is recurring, surgeon will consult with facility veterinarian.
Problem 8
If recurring wound dehiscence, infection, or clinical or behavioral changes outlined above are observed (step 88 in hindlimb ischemia model).
Potential solution
Contact your facility’s veterinarian to advise on the necessity of euthanasia. If criteria are deemed serious, follow your facility’s guidance on appropriate euthanasia methods.
Problem 9
The live image becomes frozen during laser speckle imaging (step 98 in laser speckle imaging).
Potential solution
Click “stop” and then “camera live” twice, after clicking on the stop icon, wait to see the information: “de-registering buffer”. This will reset the live image and enable the resumption of imaging.
Problem 10
During laser speckle imaging, the observed blood flow may appear to vary significantly over the course of an imaging session (step 101 in laser speckle imaging).
Potential solution
Mouse may not be kept at a constant body temperature. Ensure mouse is well placed on heating pad set to 37°C. Allow pad to heat up to 37°C before beginning procedure.
Problem 11
It is expected to observe some differences in hindlimb blood flow between days of speckle imaging, due to natural healing of the vasculature. However, if blood flow appears to be drastically different, this can confound the results (step 102 in laser speckle imaging).
Potential solution
Ensure that each mouse is placed in the same orientation on the heating pad and speckle imaging set up. It is helpful to roughly outline the mouse position with sterile lab tape so that the experiment can be repeated between each mouse and day of imaging.
Problem 12
During laser speckle imaging, high amounts of external noise may be observed (step 103 in laser speckle imaging).
Potential solution
It is best to run the speckle imaging when it is dark outside, for example at the end of the day. All lights should be turned off and sources of light (e.g., windows) should be blocked out with aluminum foil. Lastly, run the imaging procedure at the same time each day to ensure consistent and reliable results.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Aaron Baker (abbaker@austin.utexas.edu).
Materials availability
All materials are commercially available, or an alternative is commercially available.
Acknowledgments
The authors gratefully acknowledge funding through the DOD CDMRP (W81XWH-16-1-0580; W81XWH-16-1-0582) and the National Institutes of Health (1R01HL141761-01) to A.B.B. Figures 1 and 2 are created with BioRender.com.
Author contributions
M.W.M., B.I., J.L., and A.B.B. performed experiments and processed and analyzed data. M.W.M., B.I., J.L., and A.B.B. wrote and edited the manuscript. A.B.B. initiated the project and oversaw all aspects of the project. All authors reviewed and approved the manuscript before publication.
Declaration of interests
A.B.B. and J.L. have a patent on the technology described in this work (US Patent US20200268801A1).
Data and code availability
The protocol includes all datasets generated or analyzed during this study. Software code is commercially available or available on request.
References
- 1.Lee J., Henderson K., Massidda M.W., Armenta-Ochoa M., Im B.G., Veith A., Lee B.-K., Kim M., Maceda P., Yoon E., et al. Mechanobiological conditioning of mesenchymal stem cells for enhanced vascular regeneration. Nat. Biomed. Eng. 2021;5:89–102. doi: 10.1038/s41551-020-00674-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunn A.K., Bolay H., Moskowitz M.A., Boas D.A. Dynamic imaging of cerebral blood flow using laser speckle. J. Cereb. Blood Flow Metab. 2001;21:195–201. doi: 10.1097/00004647-200103000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Lee J., Armenta Ochoa M., Maceda P., Yoon E., Samarneh L., Wong M., Baker A.B. A high throughput screening system for studying the effects of applied mechanical forces on reprogramming factor expression. Sci. Rep. 2020;10:15469. doi: 10.1038/s41598-020-72158-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee J., Wong M., Smith Q., Baker A.B. A novel system for studying mechanical strain waveform-dependent responses in vascular smooth muscle cells. Lab Chip. 2013;13:4573–4582. doi: 10.1039/c3lc50894c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monteforte A.J., Lam B., Das S., Mukhopadhyay S., Wright C.S., Martin P.E., Dunn A.K., Baker A.B. Glypican-1 nanoliposomes for potentiating growth factor activity in therapeutic angiogenesis. Biomaterials. 2016;94:45–56. doi: 10.1016/j.biomaterials.2016.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das S., Singh G., Majid M., Sherman M.B., Mukhopadhyay S., Wright C.S., Martin P.E., Dunn A.K., Baker A.B. Syndesome therapeutics for enhancing diabetic wound healing. Adv. Healthc. Mater. 2016;5:2248–2260. doi: 10.1002/adhm.201600285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das S., Majid M., Baker A.B. Syndecan-4 enhances PDGF-BB activity in diabetic wound healing. Acta Biomater. 2016;42:56–65. doi: 10.1016/j.actbio.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Das S., Monteforte A.J., Singh G., Majid M., Sherman M.B., Dunn A.K., Baker A.B. Syndecan-4 enhances therapeutic angiogenesis after hind limb ischemia in mice with type 2 diabetes. Adv. Healthc. Mater. 2016;5:1008–1013. doi: 10.1002/adhm.201500993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jang E., Albadawi H., Watkins M.T., Edelman E.R., Baker A.B. Syndecan-4 proteoliposomes enhance fibroblast growth factor-2 (FGF-2)-induced proliferation, migration, and neovascularization of ischemic muscle. Proc. Natl. Acad. Sci. USA. 2012;109:1679–1684. doi: 10.1073/pnas.1117885109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phinney D.G. Functional heterogeneity of mesenchymal stem cells: implications for cell therapy. J. Cell. Biochem. 2012;113:2806–2812. doi: 10.1002/jcb.24166. [DOI] [PubMed] [Google Scholar]
- 11.Zaim M., Karaman S., Cetin G., Isik S. Donor age and long-term culture affect differentiation and proliferation of human bone marrow mesenchymal stem cells. Ann. Hematol. 2012;91:1175–1186. doi: 10.1007/s00277-012-1438-x. [DOI] [PubMed] [Google Scholar]
- 12.Aref Z., de Vries M.R., Quax P.H.A. Variations in surgical procedures for inducing hind limb ischemia in mice and the impact of these variations on neovascularization assessment. Int. J. Mol. Sci. 2019;20:3704. doi: 10.3390/ijms20153704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The protocol includes all datasets generated or analyzed during this study. Software code is commercially available or available on request.