Summary
The human mtHSP60/HSPD1-mtHSP10/HSPE1 system prevents protein misfolding and maintains proteostasis in the mitochondrial matrix. Altered activities of this chaperonin system have been implicated in human diseases, such as cancer and neurodegeneration. However, how defects in HSPD1 and HSPE1 affect mitochondrial structure and dynamics remains elusive. In the current study, we address this fundamental question in a human cell line, HEK293T. We found that the depletion of HSPD1 or HSPE1 results in fragmentation of mitochondria, suggesting a decrease in mitochondrial fusion. Supporting this notion, HSPE1 depletion led to proteolytic inactivation of OPA1, a dynamin-related GTPase that fuses the mitochondrial membrane. This OPA1 inactivation was mediated by a stress-activated metalloprotease, OMA1. In contrast, HSPD1 depletion did not induce OMA1 activation or OPA1 cleavage. These data suggest that HSPE1 controls mitochondrial morphology through a mechanism separate from its chaperonin activity.
Subject areas: Biological sciences, Molecular biology, Cell biology
Graphical abstract

Highlights
-
•
Depletion of HSPE1 results in the cleavage of OPA1 independent of HSPD1
-
•
HSPD1 and HSPE1 depletion decreases respiration activity
-
•
OMA1 plays a role in OPA1 cleavage from HSPE1 depletion
-
•
The mobile loop region of HSPE1 is required to rescue OPA1 cleavage
Biological sciences; Molecular biology; Cell biology
Introduction
Human mitochondria contain approximately 1,000–1,500 proteins, more than 99% encoded in nuclear DNA, synthesized in the cytosol, and imported to mitochondria.1 Only 13 proteins are encoded in mtDNA.2 Since the protein translocases that import proteins into mitochondria have small pores, nuclear-encoded mitochondrial proteins must be translocated into mitochondria in an unfolded conformation.3 Once imported into mitochondria, protein folding is aided by chaperones via an oxidative protein folding pathway in the intermembrane space and an ATP-driven protein folding pathway in the matrix.4 After maturation, correct folding of mitochondrial proteins is ensured by chaperonins.5 Defects in mitochondrial chaperones and chaperonins have been linked to many human diseases.6,7
In the mitochondrial matrix, two major types of machinery mediate protein folding: the mtHSP70-mtHSP40 system (chaperone) and mtHSP60-mtHSP10 system (chaperonin).5,7,8 The mtHSP70-mtHSP40 system, along with other proteins, facilitates client proteins to be correctly folded during import into the matrix through the translocase of the inner membrane.7,9,10,11 The mtHSP60-mtHSP10 system counteracts protein misfolding and unfolding under physiological conditions and in response to different types of stress.7,9 The pioneering studies on the bacterial GroEL-GroES system, the founding members of the mtHSP60-mtHSP10 system, revealed that GroEL forms a barrel structure with two ring-shaped GroEL oligomers.5,8,10 The GroEL barrel accommodates substrate proteins and mediates protein folding reactions using ATP hydrolysis. GroES binds to the top and bottom of the barrel and closes a lid to optimize reactions. GroES also regulates the cycle of ATP hydrolysis by GroEL and faciliates GroEL to bind and fold substrate proteins.5,8,10 While GroEL and GroES are essential for bacterial cell viability, mtHSP60 and mtHSP10 are vital for mitochondrial function.10
Mitochondria are dynamic organelles and change their morphology through membrane fusion and division.12,13,14,15,16,17,18 These dynamic processes play important roles in maintaining mitochondrial health.19,20,21,22 For example, bioenergetic stress could lead to fragmentation of mitochondria due to decreased mitochondrial fusion relative to division, facilitating efficient mitophagy.12 Three dynamin-related GTPases mediate mitochondrial fusion.12,23 An inner membrane GTPase, OPA1, fuses the inner membrane through interactions with a mitochondria-specific phospholipid, cardiolipin.23 Inner membrane fusion is coupled to outer membrane fusion, which is controlled by two homologous GTPases, MFN1 and MFN2. A regulatory mechanism for mitochondrial fusion is proteolytic controls of these three GTPases.12,13,14 For example, OPA1 produces membrane-anchored long forms that mediate mitochondrial fusion, while its cleavage by a metalloprotease, OMA1, greatly inactivates OPA1’s ability to fuse the membrane (Figures 1A and 1B).23,24,25,26,27,28,29 MFN1 and MFN2 are subjected to degradation to decrease their fusion activity via different ubiquitin E3 ligases and proteosomes.27,29,30 Various stresses and bioenergetic deficits can activate these regulatory systems for mitochondrial fusion.12,13,14,30 However, it remains unclear whether defects in the mtHSP60-mtHSP10 system induce changes in OPA1, MFN1, or MFN2.
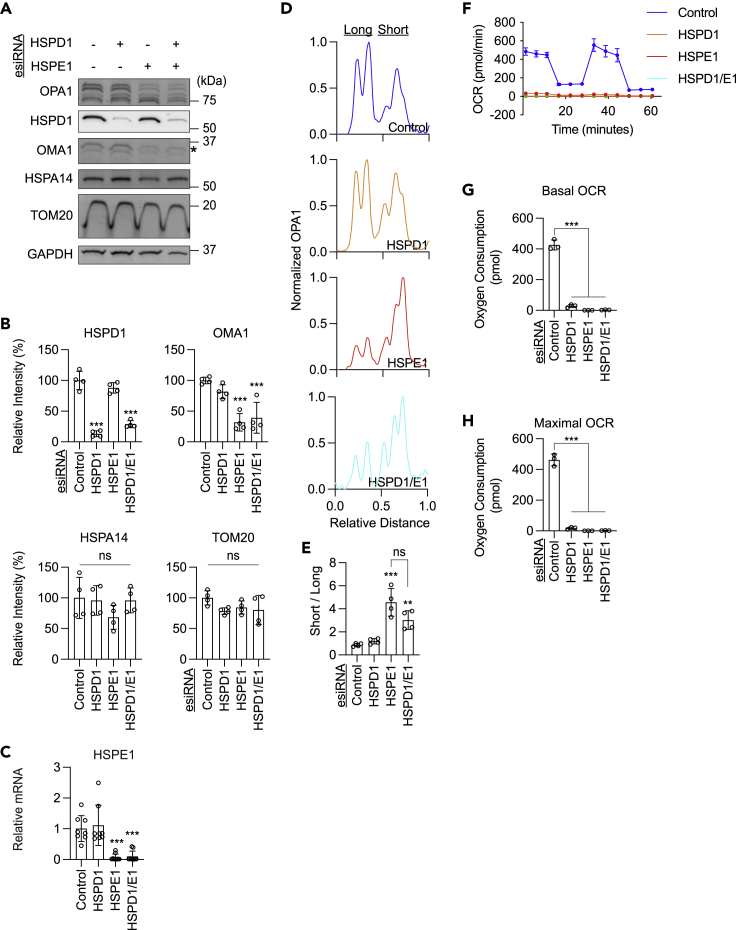
Figure 1.
Knockdown of HSPE1 or HSPA9 induces OPA1 cleavage in HEK293T cells
(A) Cleavage of OPA1 long isoforms by OMA1 and YME1L to produce short isoforms.
(B) OPA1 isoforms were quantified by measuring band intensity along a uniform line. The images come directly from the Western blot for OPA1 (C).
(C) The indicated mitochondrial chaperones and chaperonins were knocked down in HEK293T cells for 5 days using esiRNAs. Whole-cell lysates were analyzed by Western blotting with the indicated antibodies. The asterisks indicate the target band.
(D) Quantifications of band intensity normalized relative to GAPDH. Bars indicate averages ±SD (n = 4 and 6).
(E) Relative mRNA levels of HSPE1 were determined by qPCR in cells treated with the indicated esiRNAs. Bars indicate averages ±SD (n = 6).
(F) A representative band intensity of OPA1 isoforms in cells with the indicated knockdown.
(G) The ratio of short isoforms of OPA1 over long isoforms is quantified ± SD (n = 4). Statistical analysis was performed by one-way ANOVA with post hoc Tukey: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
In the current study, we depleted mtHSP60 and mtHSP10 in a human cell line and analyzed the impact on the three GTPases. We found that the knockdown of mtHSP60 or mtHSP10 induced fragmentation of mitochondria. To our surprise, mtHSP10 knockdown increased levels of inactive forms of OPA1 through activation of OMA1 without affecting MFN1 and MFN2 levels. In contrast, mtHSP60 knockdown did not affect OPA1, MFN1, or MFN2. These findings suggest that mtHSP10 and mtHSP60 affect mitochondrial morphology through multiple pathways independent of their chaperonin activities.
Results
Knockdown of HSPE1 enhances OPA1 processing independent of HSPD1
To understand the role of mitochondrial chaperones and chaperonins for mitochondrial fusion, we knocked down four matrix-located heat shock proteins — HSPD1 (mtHSP60), HSPE1 (mtHSP10), HSPA9 (mtHSP70), and DNAJA3 (mtHSP40) — using esiRNAs for 5 days in HEK293T cells. We then examined OPA1, MFN1, and MFN2 using Western blotting. Knockdown of each protein was confirmed by Western blotting of whole-cell lysates except for HSPE1 (Figures 1C and 1D). Since we could not detect HSPE1 by Western blotting using five commercially available HSPE1 antibodies (Figure S1), we confirmed its knockdown by qPCR (Figure 1E). The lack of HSPE1 detection by Western blotting may be due to its small size (102 amino acids, 10,932 Da); when ectopically expressed in HEK293T cells, we were able to detect HSPE1-GFP (GFP consists of 238 amino acids, 27 kDa) using two out of the five anti-HSPE1 antibodies on Western blotting, but not HSPE1-FLAG (FLAG consists of 8 amino acids, 1 kDa) using anti-FLAG antibodies (Figure S1). In addition, the expression level may also contribute to the efficiency of detection.
OPA1 is encoded by a single gene and generates multiple isoforms through alternative mRNA splicing and proteolytic processing.23,24,27 In HEK293T cells, five isoforms, including two long forms (L1 and L2) and three short forms (S3, S4, and S5) (Figures 1A and 1B), were detected by Western blotting (Figure 1C). S3 and S5 are produced via cleavage of L1 and L2, respectively, by the inner membrane metalloprotease OMA1 (Figure 1A).23,24,25,26,27 However, in HEK293T cells (unlike in mouse embryonic fibroblasts),27 we noticed that the distances between L1 and L2 and between S4 and S5 in Western blots were sometimes too close to clearly separate them. Therefore, to analyze the conversion of L1 and L2 to S3 and S5, we measured the ratio of short forms relative to long forms (Figures 1F and 1G). Interestingly, we found that the ratio of short forms over long forms increased after the depletion of HSPE1 or HSPA9 (Figures 1F and 1G). In contrast, we found no significant changes in the amounts of MFN1 or MFN2 after depletion of HSPD1, HSPE1, HSPA9, and DNAJA3 (Figures 1C and 1D). Since the role of HSPA9 and DNAJA3 has been reported previously,31 we focused on HSPE1 in the current study.
Previous studies have shown that yeast mtHSP60 and mtHSP10 function together in protein folding in the mitochondrial matrix as chaperonins.5,8,10 mtHSP60 creates oligomeric rings and assembles into a barrel in which protein folding reactions occur using ATP binding and hydrolysis.5,8,10 mtHSP10 covers the apical sides of the mtHSP60 barrel structure as a cap and regulates ATP hydrolysis. Both mtHSP60 and mtHSP10 are required for this protein folding mechanism in the matrix. Therefore, we were surprised by our finding that knockdown of HSPE1, but not HSPD1, stimulates the conversion of long OPA1 isoforms to short isoforms (Figures 1F and 1G). These data suggest that HSPE1 has a role in OPA1 processing distinct from its protein folding function together with HSPD1. Alternatively, since the loss of HSPE1 could halt the function of HSPD1, the changes in OPA1 processing may be caused by a dominant-negative effect of HSPD1 in the absence of HPSE1. This second model predicts that an additional knockdown of HSPD1 could mitigate the effect of HSPE1 knockdown on OPA1 processing. In the third model, if a decrease in protein folding function resulted in increased OPA1 processing, double knockdown of HSPD1 and HSPE1 may exacerbate enhanced OPA1 processing. To distinguish these models, we simultaneously knocked down HSPD1 and HSPE1 using esiRNAs (Figures 2A and 2B). We found that additional knockdown of HSPD1 did not significantly change the OPA1 conversion in HSPE1-knockdown cells (Figures 2C and 2D). These data suggest that HSPE1 depletion stimulates OPA1 processing independently of its chaperonin function with HSPD1.
Figure 2.
OPA1 cleavage induced by HSPE1 knockdown is independent of HSPD1
(A) Western blot of HEK293T cells after individual or simultaneous knockdown of HSPD1 and HSPE1 using esiRNA. The asterisks indicate the target band.
(B) Quantitation of band intensity normalized relative to GAPDH. Bars indicate averages ±SD (n = 4).
(C) Relative mRNA levels of HSPE1 were determined by qPCR. Bars indicate averages ±SD (n = 8).
(D) A representative band intensity of OPA1 isoforms in cells with the indicated knockdown.
(E) The ratio of short isoforms of OPA1 over long isoforms is quantified ± SD (n = 4).
(F) OCRs were analyzed in cells with the indicated knockdown.
(G) The basal OCRs are quantified. Bars indicate averages ±SD (n = 3).
(H) The maximal OCRs are quantified. Bars indicate averages ±SD (n = 3). Statistical analysis was performed by one-way ANOVA with post hoc Tukey: ∗∗p < 0.01, ∗∗∗p < 0.001.
Another member of the HSP60 family, HSPA14, is located in the cytosol (www.uniprot.org/uniprotkb/Q0VDF9/entry). We tested whether HSPA14 is upregulated to compensate for the loss of HSPD1. Western blotting showed that the level of HSPA14 was not significantly changed in HSPD1-, HSPE1-, and HSPD1/E1-knockdown cells (Figures 2A and 2B).
To determine whether the depletion of HSPD1 and HSPE1 differently affects mitochondrial function, we measured oxygen consumption rates in the single and double knockdown cells. Results show that these knockdown cells similarly decreased oxygen consumption rates at the basal and maximal levels (Figures 2F–2H). These data suggest that the distinct effects of HPSD1 and HSPE1 depletion on OPA1 processing do not result from their impacts on mitochondrial respiration.
OMA1 mediates increased OPA1 processing in HSPE1-knockdown cells
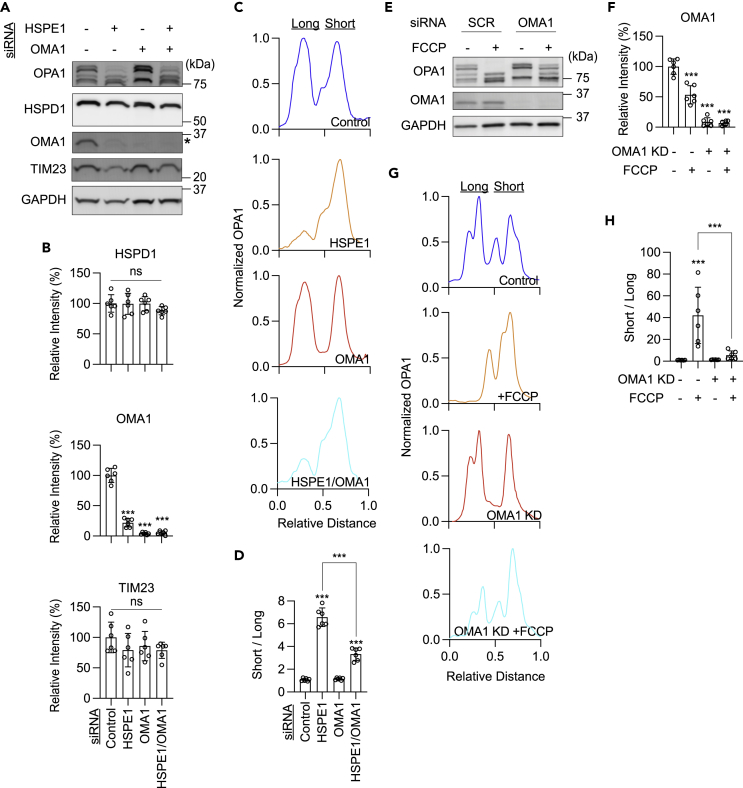
OPA1 is cleaved by a metalloprotease located in the inner membrane, OMA1.23,24,25,26,27,32,33,34,35,36 In response to mitochondrial stress, OMA1 is activated via its proteolytic cleavage followed by degradation. This stimulation mechanism assures a transient induction of its enzymatic activity.23,24,25,26,27 After activation, OMA1 cleaves OPA1 long forms and generates its short forms. Therefore, decreased levels of OMA1 have been used as a readout for its activation.27 In HSPE1-knockdown cells, we found that OMA1 levels are decreased by Western blotting (Figures 1C, 1D, 2A, 2B, 3A, and 3B). Based on these results, we hypothesized that OMA1 is activated upon HSPE1 depletion.
Figure 3.
Knockdown of OMA1 shows a reduction in OPA1 cleavage induced by HSPE1 knockdown
(A) Western blotting of HEK293T cells depleted for HSPE1 and OMA1 using siRNAs.
(B) Quantitation of band intensity. Bars indicate averages ±SD (n = 6).
(C) A representative band intensity of OPA1 isoforms in cells with the indicated knockdown.
(D) Quantifying OPA1 isoforms using the ratio of short isoforms over long isoforms in C ± SD (n = 6).
(E) Western blot with or without OMA1 siRNA knockdown in the presence or absence of 10 μM FCCP treatment with the indicated antibodies.
(F) Quantitation of band intensity. Bars indicate averages ±SD (n = 6).
(G) A representative band intensity of OPA1 isoforms in cells with the indicated knockdown.
(H) The ratio of short isoforms of OPA1 over long isoforms is quantified ± SD (n = 6). Statistical analysis was performed by one-way ANOVA with post hoc Tukey: ∗∗p < 0.01, ∗∗∗p < 0.001.
To test whether OMA1 is responsible for converting OPA1 long forms to short forms in HSPE1-depleted cells, we knocked down OMA1 along with HSPE1 using siRNAs in HEK293T cells (Figures 3A and 3B). Western blotting of whole-cell lysates showed that OMA1 knockdown partially rescued the ratio of the short forms over the long forms (Figures 3C and 3D). As a control experiment, we treated HEK293T cells with a proton ionophore, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP), which dissipates the membrane potential across the inner membrane. FCCP treatments activate OMA1 and induce OPA1 processing.27 We found that FCCP treatments decrease OMA1 levels and that OMA1 knockdown partially rescues FCCP-induced OPA1 processing (Figures 3E–3G). These data, taken together, suggest that the loss of HSPE1 activates OMA1 and induces proteolytic cleavage of OPA1 from long forms to short forms.
Although we observed a significant decrease in OPA1 processing in HSPE1/OMA1-knockdown cells compared to HSPE1-knockdown cells, some processing still remained in HSPE1/OMA1-knockdown cells (Figures 3A and 3D). Similarly, FCCP treatments still produced processed forms of OPA1 in OMA1-knockdown cells (Figures 3E and 3H). To ask if OPA1 cleavage events occur independently of OMA1, we knocked out OMA1 in HEK293T cells using CRISPR (Figure S2). We found a complete block of OPA1 processing in the presence or absence of FCCP in OMA1-KO cells (Figures S2A–S2D), as expected.23,24,25,26,27,32,33,34,35,36 These data suggest that the residual OPA1 cleavage activities result from remaining OMA1 after OMA1 knockdown. However, it is still possible that other mitochondrial proteases, such as YME1L, cleave OPA1 when OMA1 and/or HSPE1 are depleted.
Mutations in the mobile loop inhibit the role of HSPE1 in OPA1 processing
The structure of the bacterial homolog of GroEL (HSPD1) together with GroES (HSPE1) has been solved.5,8,37,38 Oligomeric GroEL rings form barrel structures, and GroES caps the top and bottom of barrels. A mobile loop in GroES with 20 amino acids mediates its interaction with the apical region of the GroEL barrels. Mutations in three amino acids (Ile 25, Val 26, and Leu 27) in bacterial GroES have been shown to block its binding to HSPD1 and protein folding.38,39 The structure and amino acid sequence of the mobile loop are conserved in bacterial GroES and human HSPE1 (Figure 4A).40 To test the functional importance of the mobile loop, we mutated the corresponding three residues (Ile 31, Met 32, and Leu 33) to alanine in a siRNA-resistant version of HSPE1. WT and AAA mutant HSPE1 proteins fused to GFP were expressed in HSPE1-knockdown cells. We found that WT HSPE1-GFP rescued increased OPA1 cleavage in HSPE1-knockdown cells (Figures 4B–4D). In contrast, the AAA mutant HSPE1-GFP failed to do so. These data demonstrate that the mobile loop is important for the function of HSPE1 in the control of OPA1 processing. Since HSPD1 is not involved in the enhanced OPA1 processing observed in HSPE1-knockdown cells (Figures 4B–4D), the mobile loop has a critical role that is separable from its interaction with HSPD1 and protein folding.
Figure 4.
HSPE1 carrying mutations in the mobile loop fails to rescue increased OPA1 processing in HSPE1-knockdown cells
(A) Amino acid sequence of bacterial GroES and human HSPE1. The mobile loop is highlighted. Mutated residues are indicated in red.
(B) HSPE1-knockdown HEK293T cells were transfected with siRNA-resistant forms of HSPE1 fused GFP. WT and mutant HSPE1-GFP carrying the AAA mutations are used.
(C) A representative band intensity of OPA1 isoforms in cells with the indicated knockdown.
(D) Quantifying OPA1 isoforms using the ratio of short isoforms over long isoforms ± SD (n = 5).Significance was calculated using one-way ANOVA with post hoc Tukey: ∗∗p < 0.01, ∗∗∗p < 0.001.
Fragmentation of mitochondria is induced by the depletion of mitochondrial chaperones and chaperonins
To test the role of HSPD1 and HSPE1 for mitochondrial morphology, we examined mitochondria using laser scanning confocal immunofluorescence microscopy using antibodies to TOM20 (a mitochondrial outer membrane protein) and pyruvate dehydrogenase (PDH, a matrix protein) in HEK293T cells knocked down for HSPD1, HSPE1, HSPA9, and DNAJA3. We found that individual knockdown of all four proteins produced shorter mitochondria (Figures 5A and 5B). These data show that these proteins are important for maintaining mitochondrial morphology. Since the ratio of OPA1 short forms and long forms was only altered in HSPE1- or HSPA9-knockdown cells, mitochondrial fragmentation in HSPD1- or DNAJA3-knockdown cells likely resulted from other mechanisms. To test whether a decrease in mitochondria size in HSPE1-knockdown cells depends on OMA1, we simultaneously knocked down HSPE1 and OMA1 in HEK293T cells. We found that the OMA1 depletion slightly increased the size of mitochondria in HSPE1-knockdown cells, although the difference was not statistically significant (Figures 6A and 6B). These data may result from partial depletion of OMA1 using siRNA. Alternatively, mitochondrial fragmentation by HSPE1 depletion may involve a mechanism independent of OMA1 and OPA1. The latter is supported by the fact that depletion of HSPD1 and DNAJA3 led to mitochondrial fragmentation without increased OPA1 cleavage (Figures 1C, 1D, 5A, and 5B).
Figure 5.
Mitochondria length decreases with the knockdown of mitochondrial chaperones and chaperonins
(A) Confocal immunofluorescence microscopy of the indicated knockdown HEK293T cells using antibodies to pyruvate dehydrogenase (PDH, a matrix protein) and TOM20 (an outer membrane protein). Boxed regions are enlarged.
(B) Quantitation of mitochondria length using TOM20 staining. Bars indicate averages ±SD (n = 150, 150 mitochondria in 30 cells were quantified for each knockdown).Significance was calculated using one-way ANOVA with post hoc Tukey: ∗∗∗p < 0.0001.
Figure 6.
Mitochondria length was not rescued by the depletion of OMA1 in HSPE1-knockdown cells
(A) Confocal immunofluorescence microscopy of the indicated knockdown HEK293T cells using antibodies to PDH and TOM20. Boxed regions are enlarged.
(B) Quantitation of mitochondria length using TOM20 staining. Bars indicate averages ±SD (n = 150, 150 mitochondria in 30 cells were quantified for each knockdown). Significance was calculated using one-way ANOVA with post hoc Tukey: ∗∗∗p < 0.001.
Discussion
Here, we report that the loss of HSPE1 (mtHSP10) leads to the activation of the stress-responsive metalloprotease OMA1 and promotes the cleavage of OPA1. In contrast, the depletion of HSPD1 (mtHSP60), a functional partner of HSPE1 in protein folding, did not induce OMA1 activation or OPA1 cleavage. These data suggest that HSPE1 plays a role in protection from mitochondrial stress independently of HSPD1. At this moment, we do not know how the loss of HSPE1 activates OMA1. If HSPE1 depletion directly activates OMA1, one possibility is that a pool of HSPE1 is associated with OMA1 as a negative regulator, suppressing OMA1 activation at the steady state. In this model, dissociation of HSPE1 from OMA1 can activate this metalloprotease. However, our immunoprecipitation experiments found no stable association between HSPE1 and OMA1.
OMA1 is activated by various stressors, such as reactive oxygen species.23,24,25,26,27 Given that HSPE1 deficiency induces OMA1 activation, some mitochondrial stress may promote OMA1 activation through changes in HSPE1 levels or function. Indeed, a previous study has suggested that sequestration of HSPE1 by pathogenic alpha-synuclein in the cytosol results in mitochondrial dysfunction and contributes to Parkinson disease.41 Parkinson disease has been strongly associated with aberrant mitochondrial structure and function.42 In future studies, it would be interesting to test if alpha-synuclein accelerates OPA1 conversion.
In addition to the chaperonin function of HSPD1 in mitochondria, previous studies have also reported the roles of HSPD1 outside mitochondria in protein degradation and tumor necrosis factor alpha-mediated activation of the IKK/NF-κB survival pathway in the cytosol.43,44,45 Interestingly, HSPD1 is further exported from cells and regulates the innate and adaptive immune system.46 Like HSPD1, HSPE1 might also play non-chaperonin roles inside or outside mitochondria or even in the extracellular space beyond protein folding.
Mitochondria are equipped with various mechanisms for stress response and quality control to protect their bioenergetic, metabolic, and signaling functions.12,47,48 For example, superoxide dismutases and glutathione peroxidases convert toxic reactive oxygen species to non-toxic molecules.49 Intramitochondrial proteases degrade misfolded or unfolded protein. In addition, mitochondrial-derived vesicles and mitophagy sense damage to mitochondria, isolate damaged regions, remove such damaged portions from the rest of the mitochondria or even eliminate the whole mitochondria.12,47,48 Our finding of a role for HSPE1 independent of protein folding for OMA1 activation and OPA1 processing may add another layer of defense mechanisms that maintain mitochondrial health against physiological changes and pathological insults. It would be of great interest to define the underlying molecular mechanism by which HSPE1 senses and eliminates damage in mitochondria.
Limitations of the study
We discovered that the loss of HSPE1, but not HSPD1, activates OMA1 and promotes the proteolytic processing of OPA1. These findings indicate that HSPE1 ensures mitochondrial health independently of HSPD1 and beyond its function in protein folding. Although the underlying mechanism of this was not determined in the current study, it would be of great importance to how HSPE1 controls OMA1, OPA1 processing, and mitochondrial structure and dynamics. We believe such information would help better understand mitochondrial quality control mechanisms and mitochondrial diseases.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-HSPD1 (D6F1) | Cell Signaling Technology | Cat# 12165; RRID:AB_2636980 |
| Rabbit polyclonal anti-TID1 | Proteintech | Cat# 11088-1-AP; RRID:AB_2293102 |
| Rabbit polyclonal anti-GRP75 | Proteintech | 14887-1-AP; RRID:AB_2120458 |
| Rabbit monoclonal anti-HSPA14 | Thermo Fisher Scientific | Cat# MA5-32413; RRID:AB_2809691 |
| Mouse monoclonal anti-OPA1 | BD Biosciences | Cat# 612607; RRID:AB_399889 |
| Mouse monoclonal anti-GAPDH (GA1R) | Thermo Fisher Scientific | MA5-15738; RRID:AB_10977387 |
| Rabbit polyclonal anti-TOM20 (FL-145) | Santa Cruz Biotechnology | Cat# sc-11415; RRID:AB_2207533 |
| Mouse monoclonal anti-OMA1 (H-11) | Santa Cruz Biotechnology | Cat# sc-515788; RRID:AB_2905488 |
| Mouse monoclonal anti-MFN1 | Abcam | ab57602; RRID:AB_2142624 |
| Mouse monoclonal anti-TIM23 | BD Biosciences | Cat# 611223; RRID:AB_398755 |
| Rabbit anti-GFP | Gift from Dr. Peter Devreotes | N/A |
| Alexa Fluor 568 anti-mouse IgG | Thermo Fischer Scientific | Cat# A10037; RRID:AB_2534013 |
| Alexa Fluor 488 anti-rabbit IgG | Thermo Fischer Scientifc | Cat# A-21206; RRID:AB_2535792 |
| Chemicals, peptides, and recombinant proteins | ||
| Rotenone | Sigma | Cat# R8875 |
| Antimycin A | Sigma | Cat# A8674 |
| FCCP | Sigma | Cat# C2920 |
| Oligomycin | Sigma | Cat# O4876 |
| BioRad Assay Dye | BioRad | Cat# 5000006 |
| Fetal Bovine Serum | Corning | Cat# 35-011-CV |
| Dulbecco’s Modified Eagle’s Medium | Corning | Cat# 10-017-CV |
| Dulbecco’s Phosphate-Buffered Saline | Corning | Cat# 21-031-CV |
| 0.05% Trypsin-EDTA | Gibco | Cat# 25300-054 |
| RIPA Buffer (10x) | Cell Signaling Technology | Cat# 9806 |
| cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail | Roche | Cat# 11836170001 |
| Paraformaldehyde | Sigma | Cat# P6148 |
| Lipofectamine RNAiMax Reagent | Invitrogen | Cat# 13778-150 |
| Lipofectamine 2000 Reagent | Invitrogen | Cat# 11668-019 |
| Critical commercial assays | ||
| RNAeasy Mini Kit | Qiagen | Cat# 74106 |
| Pure Yield Plasmid Mini Prep System | Promega | Cat# A1222 |
| PowerUp SYBR Green Master Mix | Applied Biosystems | Cat# A25742 |
| ReadyScript cDNA Synthesis Mix | Sigma-Aldrich | Cat# RDRT |
| GeneArt CRISPR Nuclease (OFP Reporter) Vector Kit | Thermo Fisher Scientific | Cat# A21174 |
| Seahorse XF96 Cell Culture Microplates | Agilent | Cat# 101085-004 |
| Experimental models: Cell lines | ||
| Human: HEK293T cells | ATCC | Cat# CRL-3216 |
| Oligonucleotides | ||
| qRT-PCR for HSPE1 | This paper | N/A |
| qRT-PCR for GAPDH | This paper | N/A |
| gRNA OMA1 targeting sequence: TCCACGGTTTCAAGCTGCTCCGG |
This paper | N/A |
| esiRNA HSPD1 | Millipore Sigma | Cat# EHU113521 |
| esiRNA HSPE1 | Millipore Sigma | Cat# EHU112631 |
| esiRNA HSPA9 | Millipore Sigma | Cat# EHU011841 |
| esiRNA DNAJA3 | Millipore Sigma | Cat# EHU091601 |
| siRNA HSPD1 targeting sequence: CGAUUUCUGCAAACGGAGAtt |
Thermo Fisher Scientific | Cat# S7003 |
| siRNA HSPE1 targeting sequence: AGAAAAAUCUCAAGGAAAAtt |
Thermo Fisher Scientific | Cat# S7005 |
| siRNA OMA1: GACCCUCGAUUA CUAUUCAtt |
Thermo Fisher Scientific | Cat# S41775 |
| Scrambled siRNA Negative Control | Thermo Fisher Scientific | Cat# 4390843 |
| Recombinant DNA | ||
| Human HSPE1 in pcDNA3.1+/C-(K)DYK | Genscript | Cat# OHu17870D |
| Software and algorithms | ||
| Fiji | Fiji | https://fiji.sc/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hiromi Sesaki (hsesaki@jhmi.edu).
Materials availability
All unique reagents generated in this study are available from the lead contact with a completed materials transfer agreement.
Experimental model and subject details
Cells
HEK293T cells were cultured in Dulbecco’s Modified Eagle Medium containing 10% fetal bovine serum in a 37°C incubator with 5% CO2. esiRNAs and Silencer Select siRNAs were obtained from Millipore Sigma and Thermo Fisher, respectively (esiRNA: HSPD1 [EHU113521], HSPE1 [EHU112631], HSPA9 [EHU011841], DNAJA3 [EHU091601]; siRNA: Negative control [4390843], HSPD1 [S7003], HSPE1 [S7005], OMA1 [S41775]). These RNAs were transfected into HEK293T using Lipofectamine RNAiMAX (13778150, Themo Fisher).
Method details
Plasmids
Human HSPE1-GFP was cloned into pcDNA3.1 from the HSPE1 (NM_002157.2) ORF Clone (OHu17870D, Genscript). The HSPE1-GFP constructs were transfected into cells using Lipofectamine 2000 (11668019, Themo Fisher). OMA1-KO HEK293T cells were generated using a GeneArt CRISPR Nuclease (OFP Reporter) Vector Kit (A21174; Thermo Fisher Scientific) in accordance with manufacturer’s instructions.27 The target gRNA sequence was 5′- TCCACGGTTTCAAGCTGCTCCGG -3′.
Western blotting
Cells were harvested and lysed in RIPA buffer (9806S, Cell Signaling Technology) supplemented with cOmplete Mini EDTA-free Protease Inhibitor Cocktail (11836170001, Roche) on ice.27 The lysates were centrifuged at 16,000 g for 10 min at 4°C, and the supernatants were collected. Proteins were separated by SDS-PAGE and transferred onto Immobilon-FL Transfer Membrane (Millipore). The membranes were blocked in PBS-T (PBS containing 0.05% Tween 20) containing 3% BSA at room temperature for 1 h and then incubated with primary antibodies in PBS-T containing 3% BSA at 4°C overnight. The antibodies used were HSPD1 (1:1,000 dilution, 12165, Cell Signaling), DNAJA3 (1:1,000 dilution, 11088-1-AP; Proteintech), HSPA9 (1:5,000 dilution, 14887-1-AP; Proteintech), HSPA14 (1:1,000 dilution, MA5-32413, Invitrogen), OPA1 (1:1,000 dilution, 612607; BD Biosciences), GAPDH (1:1,000 dilution, MA5-15738; Invitrogen), TOM20 (1:2,000 dilution, sc-11415; Santa Cruz Biotechnology), OMA1 (1:500 dilution, sc-515788; Santa Cruz Biotechnology), MFN1/2 (1:1,000 dilution, ab57602; Abcam), PDH (1:1,000 dilution, ab110338; Abcam), TIM23 (1:5,000 dilution, 611223; BD), and GFP (1:5,000 dilution, a gift from Dr. Peter Devreotes).50 The membranes were washed three times in PBS-T, followed by incubation with fluorescently labeled appropriate secondary antibodies at room temperature for 1 h. After washing the membranes three times in PBS-T, fluorescence signals were detected using a Typhoon laser-scanner platform (Amersham). Band intensity was quantified using the NIH FIJI software.27
Real-time qPCR
Total RNAs were purified from HEK293T cells using an RNeasy Mini Kit (74106; Qiagen) and reverse-transcribed using a ReadyScript cDNA Synthesis Mix (RDRT; Sigma-Aldrich).28,51 PCR was performed using Quant Studio 3 (Thermo Scientific) and PowerUp SYBR Green Master Mix (A25741; Thermo Scientific). The DNA oligos are CAGTAGTCGCTGTTGGATCG and TCCAACTTTCACGCTAACTGG.
Mitochondrial respiration
Mitochondrial OCRs were measured using an XF96 Extracellular Flux Analyzer (Seahorse Bioscience).27,52 Cells were seeded at 20,000 cells/well in an XF 96-well culture microplate and cultured for 48 h. Cells were washed twice in an XF base medium supplemented with 25 mM glucose, and 4 mM L-glutamine. The culture medium was replaced with the XF base medium and then cells were incubated at 37°C in a CO2-free incubator for 1 h. OCR measurement was performed according to the manufacturer's instructions. Baseline OCR was recorded three times, and then, 1.6 μg/ml oligomycin, 1 μM FCCP, and 0.5 μM rotenone/antimycin A were sequentially injected into each well. OCRs were normalized relative to the amount of protein in each well.
Immunofluorescence microscopy
HEK293T cells were fixed in pre-warmed (37°C) PBS containing 4% paraformaldehyde for 20 min, washed three times in PBS, permeabilized with PBS containing 0.1% Triton X-100 for 8 min, washed again three times in PBS, and blocked in PBS containing 0.5% BSA at room temperature for 30 min.27,53 Cells were then incubated with antibodies to PDH (1:400 dilution) and TOM20 (1:300 dilution) at 4°C overnight. Cells were washed three times in PBS and incubated with Alexa 568-conjugated anti-mouse IgG (A10037, Thermo Fisher Scientific) and Alexa 488-conjugated anti-rabbit IgG (A21206, Thermo Fisher Scientific) at room temperature for 1 h. Cells were washed again, three times in PBS. The samples were observed using an LSM800 GaAsP laser scanning confocal microscope with a 100× objective.27
Quantification of mitochondrial size
To measure the mitochondria length, a circle with a 10 μm diameter was randomly drawn on each cell, and the length of at least five randomly selected mitochondria was measured in the long axis based on an anti-TOM20 signal using the NIH FIJI software.50 Each condition had a total of 30 cells that were analyzed, totaling 150 mitochondria per condition.
Quantification and statistical analysis
Statistical analysis is performed using GraphPad Prism 7 and described in corresponding figure legends.
Acknowledgments
We are grateful to past and present members of the Iijima and Sesaki labs for helpful discussions and technical assistance. This work was supported by NIH grants to M.I. (GM131768) and H.S. (GM144103), and the Isaac Morris Hay and Lucille Elizabeth Hay Graduate Fellowship to N.Y.
Author contributions
N.Y., M.I., and H.S. conceived the project and designed the study. N.Y., D.M., and M.I. performed experiments and analyzed data. N.Y., M.I., and H.S. wrote the manuscript.
Declaration of interests
The authors declare no competing financial interests.
Published: January 26, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106067.
Contributor Information
Miho Iijima, Email: miijima@jhmi.edu.
Hiromi Sesaki, Email: hsesaki@jhmi.edu.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request
-
•
This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Pfanner N., Warscheid B., Wiedemann N. Mitochondrial proteins: from biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 2019;20:267–284. doi: 10.1038/s41580-018-0092-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kopinski P.K., Singh L.N., Zhang S., Lott M.T., Wallace D.C. Mitochondrial DNA variation and cancer. Nat. Rev. Cancer. 2021;21:431–445. doi: 10.1038/s41568-021-00358-w. [DOI] [PubMed] [Google Scholar]
- 3.Araiso Y., Imai K., Endo T. Role of the TOM complex in protein import into mitochondria: structural views. Annu. Rev. Biochem. 2022;91:679–703. doi: 10.1146/annurev-biochem-032620-104527. [DOI] [PubMed] [Google Scholar]
- 4.Finka A., Mattoo R.U.H., Goloubinoff P. Experimental milestones in the discovery of molecular chaperones as polypeptide unfolding enzymes. Annu. Rev. Biochem. 2016;85:715–742. doi: 10.1146/annurev-biochem-060815-014124. [DOI] [PubMed] [Google Scholar]
- 5.Horwich A.L., Fenton W.A. Chaperonin-assisted protein folding: a chronologue. Q. Rev. Biophys. 2020;53:e4. doi: 10.1017/S0033583519000143. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez A., Von Salzen D., Holguin B.A., Bernal R.A. Complex destabilization in the mitochondrial chaperonin Hsp60 leads to disease. Front. Mol. Biosci. 2020;7:159. doi: 10.3389/fmolb.2020.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bahr T., Katuri J., Liang T., Bai Y. Mitochondrial chaperones in human health and disease. Free Radic. Biol. Med. 2022;179:363–374. doi: 10.1016/j.freeradbiomed.2021.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayer-Hartl M., Bracher A., Hartl F.U. The GroEL-GroES chaperonin machine: a nano-cage for protein folding. Trends Biochem. Sci. 2016;41:62–76. doi: 10.1016/j.tibs.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 9.Song J., Becker T. Fidelity of organellar protein targeting. Curr. Opin. Cell Biol. 2022;75:102071. doi: 10.1016/j.ceb.2022.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Horwich A.L., Neupert W., Hartl F.U. Protein-catalysed protein folding. Trends Biotechnol. 1990;8:126–131. doi: 10.1016/0167-7799(90)90153-o. [DOI] [PubMed] [Google Scholar]
- 11.Shin C.S., Meng S., Garbis S.D., Moradian A., Taylor R.W., Sweredoski M.J., Lomenick B., Chan D.C. LONP1 and mtHSP70 cooperate to promote mitochondrial protein folding. Nat. Commun. 2021;12:265. doi: 10.1038/s41467-020-20597-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murata D., Arai K., Iijima M., Sesaki H. Mitochondrial division, fusion and degradation. J. Biochem. 2020;167:233–241. doi: 10.1093/jb/mvz106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giacomello M., Pyakurel A., Glytsou C., Scorrano L. The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol. 2020;21:204–224. doi: 10.1038/s41580-020-0210-7. [DOI] [PubMed] [Google Scholar]
- 14.Kraus F., Roy K., Pucadyil T.J., Ryan M.T. Function and regulation of the divisome for mitochondrial fission. Nature. 2021;590:57–66. doi: 10.1038/s41586-021-03214-x. [DOI] [PubMed] [Google Scholar]
- 15.Kashatus D.F. The regulation of tumor cell physiology by mitochondrial dynamics. Biochem. Biophys. Res. Commun. 2018;500:9–16. doi: 10.1016/j.bbrc.2017.06.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman J.R., Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Youle R.J., van der Bliek A.M. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Widlansky M.E., Hill R.B. Mitochondrial regulation of diabetic vascular disease: an emerging opportunity. Transl. Res. 2018;202:83–98. doi: 10.1016/j.trsl.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy M., Reddy P.H., Iijima M., Sesaki H. Mitochondrial division and fusion in metabolism. Curr. Opin. Cell Biol. 2015;33:111–118. doi: 10.1016/j.ceb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itoh K., Nakamura K., Iijima M., Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 2013;23:64–71. doi: 10.1016/j.tcb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liesa M., Shirihai O.S. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serasinghe M.N., Chipuk J.E. Mitochondrial fission in human diseases. Handb. Exp. Pharmacol. 2017;240:159–188. doi: 10.1007/164_2016_38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kameoka S., Adachi Y., Okamoto K., Iijima M., Sesaki H. Phosphatidic acid and cardiolipin coordinate mitochondrial dynamics. Trends Cell Biol. 2018;28:67–76. doi: 10.1016/j.tcb.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacVicar T., Langer T. OPA1 processing in cell death and disease - the long and short of it. J. Cell Sci. 2016;129:2297–2306. doi: 10.1242/jcs.159186. [DOI] [PubMed] [Google Scholar]
- 25.Head B., Griparic L., Amiri M., Gandre-Babbe S., van der Bliek A.M. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 2009;187:959–966. doi: 10.1083/jcb.200906083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehses S., Raschke I., Mancuso G., Bernacchia A., Geimer S., Tondera D., Martinou J.C., Westermann B., Rugarli E.I., Langer T. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 2009;187:1023–1036. doi: 10.1083/jcb.200906084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murata D., Yamada T., Tokuyama T., Arai K., Quirós P.M., López-Otín C., Iijima M., Sesaki H. Mitochondrial Safeguard: a stress response that offsets extreme fusion and protects respiratory function via flickering-induced Oma1 activation. EMBO J. 2020;39:e105074. doi: 10.15252/embj.2020105074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada T., Murata D., Adachi Y., Itoh K., Kameoka S., Igarashi A., Kato T., Araki Y., Huganir R.L., Dawson T.M., et al. Mitochondrial stasis reveals p62-mediated ubiquitination in parkin-independent mitophagy and mitigates nonalcoholic fatty liver disease. Cell Metab. 2018;28:588–604.e5. doi: 10.1016/j.cmet.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamada T., Dawson T.M., Yanagawa T., Iijima M., Sesaki H. SQSTM1/p62 promotes mitochondrial ubiquitination independently of PINK1 and PRKN/parkin in mitophagy. Autophagy. 2019;15:2012–2018. doi: 10.1080/15548627.2019.1643185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorn G.W., 2nd Mitofusins as mitochondrial anchors and tethers. J. Mol. Cell. Cardiol. 2020;142:146–153. doi: 10.1016/j.yjmcc.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee B., Ahn Y., Kang S.M., Park Y., Jeon Y.J., Rho J.M., Kim S.W. Stoichiometric expression of mtHsp40 and mtHsp70 modulates mitochondrial morphology and cristae structure via Opa1L cleavage. Mol. Biol. Cell. 2015;26:2156–2167. doi: 10.1091/mbc.E14-02-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker M.J., Lampe P.A., Stojanovski D., Korwitz A., Anand R., Tatsuta T., Langer T. Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. EMBO J. 2014;33:578–593. doi: 10.1002/embj.201386474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quirós P.M., Ramsay A.J., Sala D., Fernández-Vizarra E., Rodríguez F., Peinado J.R., Fernández-García M.S., Vega J.A., Enríquez J.A., Zorzano A., López-Otín C. Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. EMBO J. 2012;31:2117–2133. doi: 10.1038/emboj.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Acin-Perez R., Lechuga-Vieco A.V., Del Mar Muñoz M., Nieto-Arellano R., Torroja C., Sánchez-Cabo F., Jiménez C., González-Guerra A., Carrascoso I., Benincá C., et al. Ablation of the stress protease OMA1 protects against heart failure in mice. Sci. Transl. Med. 2018;10:eaan4935. doi: 10.1126/scitranslmed.aan4935. [DOI] [PubMed] [Google Scholar]
- 35.Zhang K., Li H., Song Z. Membrane depolarization activates the mitochondrial protease OMA1 by stimulating self-cleavage. EMBO Rep. 2014;15:576–585. doi: 10.1002/embr.201338240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rainbolt T.K., Lebeau J., Puchades C., Wiseman R.L. Reciprocal degradation of YME1L and OMA1 adapts mitochondrial proteolytic activity during stress. Cell Rep. 2016;14:2041–2049. doi: 10.1016/j.celrep.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunt J.F., Weaver A.J., Landry S.J., Gierasch L., Deisenhofer J. The crystal structure of the GroES co-chaperonin at 2.8 A resolution. Nature. 1996;379:37–45. doi: 10.1038/379037a0. [DOI] [PubMed] [Google Scholar]
- 38.Xu Z., Horwich A.L., Sigler P.B. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 39.Nojima T., Ikegami T., Taguchi H., Yoshida M. Flexibility of GroES mobile loop is required for efficient chaperonin function. J. Mol. Biol. 2012;422:291–299. doi: 10.1016/j.jmb.2012.05.026. [DOI] [PubMed] [Google Scholar]
- 40.Bie A.S., Fernandez-Guerra P., Birkler R.I.D., Nisemblat S., Pelnena D., Lu X., Deignan J.L., Lee H., Dorrani N., Corydon T.J., et al. Effects of a mutation in the HSPE1 gene encoding the mitochondrial Co-chaperonin HSP10 and its potential association with a neurological and developmental disorder. Front. Mol. Biosci. 2016;3:65. doi: 10.3389/fmolb.2016.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szegő É.M., Dominguez-Meijide A., Gerhardt E., König A., Koss D.J., Li W., Pinho R., Fahlbusch C., Johnson M., Santos P., et al. Cytosolic trapping of a mitochondrial heat shock protein is an early pathological event in synucleinopathies. Cell Rep. 2019;28:65–77.e6. doi: 10.1016/j.celrep.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 42.Panicker N., Ge P., Dawson V.L., Dawson T.M. The cell biology of Parkinson's disease. J. Cell Biol. 2021;220:e202012095. doi: 10.1083/jcb.202012095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalderon B., Kogan G., Bubis E., Pines O. Cytosolic Hsp60 can modulate proteasome activity in yeast. J. Biol. Chem. 2015;290:3542–3551. doi: 10.1074/jbc.M114.626622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chun J.N., Choi B., Lee K.W., Lee D.J., Kang D.H., Lee J.Y., Song I.S., Kim H.I., Lee S.H., Kim H.S., et al. Cytosolic Hsp60 is involved in the NF-kappaB-dependent survival of cancer cells via IKK regulation. PLoS One. 2010;5:e9422. doi: 10.1371/journal.pone.0009422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi B., Choi M., Park C., Lee E.K., Kang D.H., Lee D.J., Yeom J.Y., Jung Y., Kim J., Lee S., Kang S.W. Cytosolic Hsp60 orchestrates the survival and inflammatory responses of vascular smooth muscle cells in injured aortic vessels. Cardiovasc. Res. 2015;106:498–508. doi: 10.1093/cvr/cvv130. [DOI] [PubMed] [Google Scholar]
- 46.Grundtman C., Kreutmayer S.B., Almanzar G., Wick M.C., Wick G. Heat shock protein 60 and immune inflammatory responses in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2011;31:960–968. doi: 10.1161/ATVBAHA.110.217877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eldeeb M.A., Thomas R.A., Ragheb M.A., Fallahi A., Fon E.A. Mitochondrial quality control in health and in Parkinson's disease. Physiol. Rev. 2022;102:1721–1755. doi: 10.1152/physrev.00041.2021. [DOI] [PubMed] [Google Scholar]
- 48.Sugiura A., McLelland G.L., Fon E.A., McBride H.M. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 2014;33:2142–2156. doi: 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphy M.P., Bayir H., Belousov V., Chang C.J., Davies K.J.A., Davies M.J., Dick T.P., Finkel T., Forman H.J., Janssen-Heininger Y., et al. Guidelines for measuring reactive oxygen species and oxidative damage in cells and in vivo. Nat. Metab. 2022;4:651–662. doi: 10.1038/s42255-022-00591-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adachi Y., Kato T., Yamada T., Murata D., Arai K., Stahelin R.V., Chan D.C., Iijima M., Sesaki H. Drp1 tubulates the ER in a GTPase-independent manner. Mol. Cell. 2020;80:621–632.e6. doi: 10.1016/j.molcel.2020.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamada T., Murata D., Kleiner D.E., Anders R., Rosenberg A.Z., Kaplan J., Hamilton J.P., Aghajan M., Levi M., Wang N.Y., et al. Prevention and regression of megamitochondria and steatosis by blocking mitochondrial fusion in the liver. iScience. 2022;25:103996. doi: 10.1016/j.isci.2022.103996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kageyama Y., Hoshijima M., Seo K., Bedja D., Sysa-Shah P., Andrabi S.A., Chen W., Höke A., Dawson V.L., Dawson T.M., et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 2014;33:2798–2813. doi: 10.15252/embj.201488658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adachi Y., Itoh K., Yamada T., Cerveny K.L., Suzuki T.L., Macdonald P., Frohman M.A., Ramachandran R., Iijima M., Sesaki H. Coincident phosphatidic acid interaction restrains Drp1 in mitochondrial division. Mol. Cell. 2016;63:1034–1043. doi: 10.1016/j.molcel.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request
-
•
This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.