Abstract
The wood-inhabiting fungi play an integral role in wood degradation and the cycle of matter in the ecological system. They are considered as the “key player” in wood decomposition, because of their ability to produce all kinds of enzymes that break down woody lignin, cellulose and hemicellulose. In the present study, three new wood-inhabiting fungal species, Steccherinum fissurutum, S. punctatum and S. subtropicum spp. nov., collected from southern China, are proposed based on a combination of morphological features and molecular evidence. Steccherinum fissurutum is characterized by the resupinate, subceraceous basidiomata with cracked hymenophore, a monomitic hyphal system with clamped generative hyphae and cylindrical basidiospores; S. punctatum is characterized by the annual, punctate basidiomata with leathery hymenophore, cylindrical, strongly encrusted cystidia and ellipsoid basidiospores (3.6–4.5 ×2.6–3.4 µm); S. subtropicum is characterized by its effuse-reflexed basidiomata, a odontioid hymenophore with pink to lilac hymenial surface and ellipsoid basidiospores measuring as (2.8–3.4 × 2.0–2.7 µm). Sequences of ITS and nLSU rRNA markers of the studied samples were generated, and phylogenetic analyses were performed with maximum likelihood, maximum parsimony, and Bayesian inference methods. The ITS+nLSU analysis of the family Steccherinaceae indicated that the three new species clustered into the genus Steccherinum. Based on further analysis of ITS+nLSU dataset, the phylogenetic analysis confirmed that S. subtropicum was sister to S. enuispinum; S. fissurutum formed a monophyletic lineage; S. punctatum grouped with a clade comprised S. straminellum and S. ciliolatum.
Keywords: biodiversity, molecular systematics, Steccherinaceae, wood-inhabiting fungi, Yunnan Province
Introduction
The phylum Basidiomycota constitute a major group of the kingdom Fungi and is second in species numbers to the phylum Ascomycota (Wijayawardene et al., 2017; Wijayawardene et al., 2018a; Wijayawardene et al., 2018b). Wood-inhabiting fungal is a large group of Basidiomycota with simpler basidiomata with the diverse morphological features, but the phylogenetic diversity of this group is less intensively studied (Larsson et al., 2004; Bernicchia and Gorjón, 2010).
The genus Steccherinum Gray (Steccherinaceae, Polyporales), typified by S. ochraceum (Pers. ex J.F. Gmel.) Gray, was established by Gray (1821). It is a cosmopolitan genus characterized by a combination of resupinate to effused-reflexed or pileate basidiome with a membranaceous consistencey, hymenophore odontioid to hydnoid, a dimitic hyphal structure with clamp connections or simple-septate generative hyphae, cystidia numerous, strongly encrusted in the obtuse apex, basidia subclavate and basidiospores hyaline, thin-walled, smooth, ellipsoid to subcylindrical, acyanophilous and negative in Melzer’s reagent (Fries, 1821; Gray, 1821; Bernicchia and Gorjón, 2010). So far, about 80 species have been accepted in this genus worldwide (Fries, 1821; Banker, 1906; Banker, 1912; Cunningham, 1958; Snell and Dick, 1958; Lindsey and Gilbertson, 1977; Ryvarden, 1978; Lindsey and Gilbertson, 1979; Burdsall and Nakasone, 1981; Melo, 1995; Legon and Roberts, 2002; Yuan and Dai, 2005a; Spirin et al., 2007; Hjortstam and Ryvarden, 2008; Bernicchia and Gorjón, 2010; Miettinen et al., 2012; Yuan and Wu, 2012; Miettinen and Ryvarden, 2016; Westphalen et al., 2018; Liu and Dai, 2021; Westphalen et al., 2021; Wu et al., 2021a; Wu et al., 2021b; Dong et al., 2022). In recent years, several new Steccherinum species were described in China, S. fragile Z.B. Liu & Y.C. Dai, S. hirsutum Y.X. Wu & C.L. Zhao, S. puerense Y.X. Wu, J.H. Dong & C.L. Zhao, S. rubigimaculatum Y.X. Wu, J.H. Dong & C.L. Zhao, S. subcollabens (F. Wu, P. Du & X.M. Tian) Z.B. Liu & Y.C. Dai, S. tenuissimum C.L. Zhao & Y.X. Wu and S. xanthum C.L. Zhao & Y.X. Wu, and S. yunnanense Y.X. Wu & C.L. Zhao (Liu and Dai, 2021; Wu et al., 2021a; Wu et al., 2021b; Dong et al., 2022).
Molecular phylogenies have provided increased knowledge concerning the evolution of Steccherinum (Miettinen et al., 2012; Binder et al., 2013; Justo et al., 2017; Westphalen et al., 2018; Westphalen et al., 2021). Utilizing sequences of the gene regions ITS, nLSU, mtSSU, atp6, rpb2, and tef1, Miettinen et al. (2012) revealed that the phylogeny of the poroid and hydnoid genera Antrodiella Ryvarden and I. Johans., Junghuhnia Corda and Steccherinum (Polyporales, Basidiomycota) grouped together and Steccherinum was shown to contain both hydnoid and poroid species. Using of whole genome sequence data in comparison to extensively sampled multigene datasets indicated that Steccherinum species belonged to the residual polyporoid clade and the generic type (S. ochraceum) was grouped with Junghuhnia nitida (Pers.) Ryvarden (Binder et al., 2013). Justo et al. (2017) clarified family-level classification of eighteen families within the order Polyporales (Basidiomycota), which showed that Steccherinum belonged to family Steccherinaceae Parmasto. Westphalen et al. (2018) worked on morphological and multigene analyses of Junghuhnia s.lat., in which a new species Steccherinum neonitidum Westphalen & Tomšovský and three new combinations, S. meridionale (Rajchenb.) Westphalen, Tomšovský & Rajchenberg, S. polycystidiferum (Rick) Westphalen, Tomšovský & Rajchenb. and S. undigerum (Berk. & M.A. Curtis) Westphalen & Tomšovský were reported. Westphalen et al. (2021) provided the morphological and phylogenetic analyses on hydnoid specimens of Steccherinaceae, in which four genera as Cabalodontia Piatek, Etheirodon Banker, Metuloidea G. Cunn., and Steccherinum were introduced and three new neotropical species was found.
Scientific names are important link to communicate biological information across many spheres of use, in which how to publish a new fungal species is recommended to provide DNA barcode sequences in a public repository for the holotype specimen with the barcode locus (ITS) as well as any additional taxa specific secondary barcode loci (Aime et al., 2021). In order to allow BLAST searches to work optimally, sequences of DNA barcodes should include the generally used region for that marker (Aime et al., 2021). Sometimes, this genus Steccherinum for the barcoding gene ITS is less than 97% of nucleotide difference between different species.
The aim of this study is to explore the diversity and phylogeny of Steccherinum in China. During our investigations on the diversity of wood-inhabiting fungi in southern China, three undescribed species were collected from Yunnan Province, and their morphology corresponds to the concept of Steccherinum. To confirm their placement in Steccherinum, morphological examination and phylogenetic analyses based on the internal transcribed spacer (ITS) and large subunit nuclear ribosomal RNA (nLSU) genens, were carried out.
Materials and methods
Morphological studies
The studied specimens are deposited at the herbarium of Southwest Forestry University (SWFC), Yunnan Province, P.R. China (Herbarium numbers: Steccherinum fissurutum: SWFCF00021634, SWFCF00021673, SWFCF00021675, SWFCF00021680, SWFCF00021703, SWFCF00021744, SWFCF00021754, SWFCF00020803, SWFCF00021808, SWFCF00021811, SWFCF00021826, SWFCF00021841; S. punctatum: SWFCF00009181, SWFCF00009184; S. subtropicum: SWFCF00011059, SWFCF00016901). Macromorphological descriptions are based on field notes. Petersen (1996) was followed for the colour terms. Micromorphological data were obtained from the dried specimens and observed under a light microscope Eclipse E 80i (Nikon, Tokyo) following Dai (2012). The following abbreviations were used for the micro characteristics description: KOH = 5% potassium hydroxide, CB = Cotton Blue, CB– = acyanophilous, IKI = Melzer’s reagent, IKI– = both non-amyloid and non-dextrinoid, L = mean spore length (arithmetic average of all spores), W = mean spore width (arithmetic average of all spores), Q = variation in the L/W ratios between the specimens studied, n (a/b) = number of spores (a) measured from given number (b) of specimens.
Molecular procedures and phylogenetic analyses
CTAB rapid plant genome extraction kit-DN14 (Aidlab Biotechnologies Co., Ltd, Beijing) was used to obtain genomic DNA from dried specimens, according to the manufacturer’s instructions. ITS region was amplified with primer pairs ITS5 and ITS4 (White et al., 1990). Nuclear LSU region was amplified with primer pairs LR0R and LR7 (https://sites.duke.edu/vilgalyslab/rdna_primers_for_fungi/ ) Table 1 .
Table 1.
A list of genes, primers and primer sequences used in this study.
| Fragment of amplification | Name of primer | Primer base sequence (5′-3′) b | References |
|---|---|---|---|
| ITS | ITS5 | GGA AGT AAA AGT CGT AAC AAG G | White et al., 1990 |
| ITS4 | TCC TCC GCT TAT TGA TAT GC | ||
| nLSU | LR0R | ACC CGC TGA ACT TAA GC | http://www.biology.duke.edu/fungi/mycolab/primers.htm |
| LR7 | TAC TAC CAC CAA GAT CT |
degenerate base: R = A or G, Y = C or T, N = A or T or C or G, V = G or A.
The PCR procedure for ITS was as follows: initial denaturation at 95°C for 3 min, followed by 35 cycles at 94°C for 40 s, 58°C for 45 s and 72°C for 1 min, and a final extension of 72°C for 10 min. The PCR procedure for nLSU was as follows: initial denaturation at 94°C for 1 min, followed by 35 cycles at 94°C for 30 s, 48°C 1 min and 72°C for 1.5 min, and a final extension of 72°C for 10 min. The PCR products were purified and sequenced at Kunming Tsingke Biological Technology Limited Company, Kunming, Yunnan Province, P.R. China. All newly generated sequences were deposited at GenBank ( Table 2 ).
Table 2.
List of species, specimens and GenBank accession numbers of sequences used in this study. * is shown type material, holotype.
| Species Name | Sample No. | GenBank Accession No. | References | |
|---|---|---|---|---|
| ITS | nLSU | |||
| Antella americana | KHL 11949 | JN710509 | JN710509 | Cao et al., 2021 |
| A. americana | HHB-4100 | KP135316 | KP135196 | Cao et al., 2021 |
| A. chinensis | Dai 8874 | JX110843 | KC485541 | Yuan, 2013 |
| A. chinensis | Dai 9019 | JX110844 | KC485542 | Yuan, 2013 |
| A. niemelaei | Renvall 3218 | AF126876 | — | Cao et al., 2021 |
| A. niemelaei | Haikonen 14727 | AF126877 | — | Cao et al., 2021 |
| Antrodiella onychoides | Miettinen 2312 | JN710517 | JN710517 | Miettinen et al., 2012 |
| A. pallescens | Nordén 8.8.2008 | JN710518 | JN710518 | Miettinen et al., 2012 |
| A. romellii | Miettinen 7429 | JN710520 | JN710520 | Miettinen et al., 2012 |
| A. semisupina | Labrecque & Labbé 372 | JN710521 | JN710521 | Miettinen et al., 2012 |
| A. stipitata | FD-136 | KP135314 | KP135197 | Westphalen et al., 2021 |
| A. stipitata | Yuan 5640 | KC485525 | KC485544 | Yuan, 2014 |
| Atraporiella neotropica | Miettinen X1021 | HQ659221 | HQ659221 | Cao et al., 2021 |
| A. yunnanensis | CLZhao 604 | MF962482 | MF962485 | Wu et al., 2017 |
| A. yunnanensis | CLZhao 605 | MF962483 | MF962486 | Wu et al., 2017 |
| Butyrea japonica | MN 1065 | JN710556 | JN710556 | Cao et al., 2021 |
| B. luteoalba | FP-105786 | KP135320 | KP135226 | Dong et al., 2022 |
| B. luteoalba | KHL 13238b | JN710558 | JN710558 | Dong et al., 2022 |
| Climacocystis borealis | KHL 13318 | JN710527 | JN710527 | Cao et al., 2021 |
| Elaphroporia ailaoshanensis | CLZhao 596 | MG231572 | MG748855 | Wu et al., 2018 |
| E. ailaoshanensis | CLZhao 597 | MG231847 | MG748856 | Wu et al., 2018 |
| Etheirodon fimbriatum | KHL 11905 | JN710530 | JN710530 | Cao et al., 2021 |
| E. fimbriatum | HR 98811 | MT849300 | — | Westphalen et al., 2021 |
| E. purpureum | MCW 642/18 | MT849301 | MT849301 | Westphalen et al., 2021 |
| Flaviporus brownii | MCW 362/12 | KY175008 | KY175008 | Westphalen et al., 2018 |
| F. brownie | X 462 | JN710538 | JN710538 | Cao et al., 2021 |
| F. liebmannii | X 249 | JN710539 | JN710539 | Cao et al., 2021 |
| F. liebmannii | Yuan 1766 | KC502914 | — | Yuan, 2014 |
| F. subundatus | MCW 367/12 | KY175004 | KY175004 | Westphalen et al., 2018 |
| F. subundatus | MCW 457/13 | KY175005 | KY175005 | Westphalen et al., 2018 |
| F. tenuis | MCW 442/13 | KY175001 | KY175001 | Westphalen et al., 2018 |
| F. tenuis | MCW 356/12 | KY175002 | KY175002 | Westphalen et al., 2018 |
| Frantisekia fissiliformis | CBS 435.72 | MH860521 | MH872232 | Vu et al., 2019 |
| F. mentschulensis | BRNM 710170 | FJ496670 | FJ496728 | Dong et al., 2022 |
| F. mentschulensis | AH 1377 | JN710544 | JN710544 | Dong et al., 2022 |
| F. ussurii | Wei 3081 | KC485527 | KC485545 | Yuan, 2014 |
| F. ussurii | Dai 8249 | KC485526 | — | Yuan, 2014 |
| Irpex lacteus | DO 421/951208 | JX109852 | JX109852 | Dong et al., 2022 |
| Junghuhnia crustacea | X 262 | JN710553 | JN710553 | Miettinen et al., 2012 |
| J. delicate | MCW 564/17 | MT849295 | MT849295 | Du et al., 2020 |
| J. delicate | MCW 693/19 | MT849297 | MT849297 | Du et al., 2020 |
| J. pseudocrustacea | Yuan 6160 | MF139551 | — | Yuan et al., 2019 |
| J. pseudocrustacea | Zhou 283 | MF139552 | — | Yuan et al., 2019 |
| Loweomyces fractipes | X 1149 | JN710570 | JN710570 | Cao et al., 2021 |
| L. fractipes | MT 13/2012 | KX378866 | KX378866 | Cao et al., 2021 |
| L. spissus | MCW 488/14 | KX378869 | KX378869 | Cao et al., 2021 |
| L. tomentosus | MCW 366/12 | KX378870 | KX378870 | Cao et al., 2021 |
| L. wynneae | X 1215 | JN710604 | JN710604 | Cao et al., 2021 |
| Metuloidea cinnamomea | X 1228 | KU926963 | — | Cao et al., 2021 |
| M. fragrans | LE 295277 | KC858281 | — | Cao et al., 2021 |
| M. murashkinskyi | X 449 | JN710588 | JN710588 | Cao et al., 2021 |
| M. reniformis | MCW 542/17 | MT849303 | MT849303 | Westphalen et al., 2021 |
| M. reniformis | MCW 523/17 | MT849302 | MT849302 | Westphalen et al., 2021 |
| M. rhinocephala | X 460 | JN710562 | JN710562 | Cao et al., 2021 |
| Mycorrhaphium hispidum | MCW 363/12 | MH475306 | MH475306 | Cao et al., 2021 |
| M. hispidum | MCW 429/13 | MH475307 | MH475307 | Cao et al., 2021 |
| M. subadustum | Yuan 12976 | MW491378 | MW488040 | Cao et al., 2021 |
| M. subadustum | Dai 10173 | KC485537 | KC485554 | Cao et al., 2021 |
| Nigroporus stipitatus | KaiR 116 | MT110231 | MT110231 | Piepenbring et al., 2020 |
| N. vinosus | MQN 015 | AB811861 | AB811861 | Hai Bang et al., 2014 |
| N. vinosus | X 839 | JN710575 | JN710575 | Cao et al., 2021 |
| Steccherinum autumnale | Spirin 2957 | JN710549 | JN710549 | Liu and Dai, 2021 |
| S. bourdotii | HR99893 | MT849311 | Westphalen et al., 2021 | |
| S. bourdotii | Saarenoksa 10195 | JN710584 | JN710584 | Miettinen et al., 2012 |
| S. ciliolatum | Ryvarden 47033 | JN710585 | JN710585 | Miettinen et al., 2012 |
| S. collabens | KHL 11848 | JN710552 | JN710552 | Liu and Dai, 2021 |
| S. fissurutum | CLZhao 21803 * | OP799385 | OP799397 | Present study |
| S. fissurutum | CLZhao 21841 | OP799388 | OP799400 | Present study |
| S. fissurutum | CLZhao 21808 | OP799386 | OP799398 | Present study |
| S. fissurutum | CLZhao 21675 | OP799380 | OP799392 | Present study |
| S. fissurutum | CLZhao 21811 | OP799389 | OP799399 | Present study |
| S. fissurutum | CLZhao 21680 | OP799381 | OP799393 | Present study |
| S. fissurutum | CLZhao 21703 | OP799382 | OP799394 | Present study |
| S. fissurutum | CLZhao 21744 | OP799383 | OP799395 | Present study |
| S. fissurutum | CLZhao 21826 | OP799387 | — | Present study |
| S. fissurutum | CLZhao 21634 | OP799378 | — | Present study |
| S. fissurutum | CLZhao 21673 | OP799379 | — | Present study |
| S. fissurutum | CLZhao 21754 | OP799384 | OP799396 | Present study |
| S. fragile | Dai 19972 | MW364629 | MW364627 | Liu and Dai, 2021 |
| S. fragile | Dai 20479 | MW364628 | MW364626 | Liu and Dai, 2021 |
| S. hirsutum | CLZhao 4222 | MW290040 | MW290054 | Dong et al., 2022 |
| S. hirsutum | CLZhao 4523 | MW290041 | MW290055 | Dong et al., 2022 |
| S. larssonii | MCW 593/17 | MT849306 | MT849306 | Westphalen et al., 2021 |
| S. larssonii | MCW 594/17 | MT849307 | MT849307 | Westphalen et al., 2021 |
| S. meridionalis | MR 10466 | KY174994 | KY174994 | Westphalen et al., 2018 |
| S. meridionalis | MR 284 | KY174992 | KY174992 | Westphalen et al., 2018 |
| S. neonitidum | MCW 371/12 | KY174990 | KY174990 | Westphalen et al., 2018 |
| S. neonitidum | RP 79 | KY174991 | KY174991 | Westphalen et al., 2018 |
| S. nitidum | KHL 11903 | JN710560 | JN710560 | Westphalen et al., 2018 |
| S. nitidum | MT 33/12 | KY174989 | KY174989 | Westphalen et al., 2018 |
| S. ochraceum | KHL11902 | JN710590 | JN710590 | Westphalen et al., 2021 |
| S. ochraceum | 2060 | JN710589 | JN710589 | Liu and Dai, 2021 |
| S. polycystidiferum | RP 140 | KY174996 | KY174996 | Westphalen et al., 2018 |
| S. polycystidiferum | MCW 419/12 | KY174995 | KY174995 | Westphalen et al., 2018 |
| S. pseudozilingianum | Kulju 1004 | JN710561 | JN710561 | Liu and Dai, 2021 |
| S. puerense | CLZhao 3122 | MW682341 | — | Wu et al., 2021a |
| S. puerense | CLZhao 3644 | MW682342 | MW682338 | Wu et al., 2021a |
| S. punctatum | CLZhao 9181 | OP799375 | OP799401 | Present study |
| S. punctatum | CLZhao 9184 * | OP799376 | OP799402 | Present study |
| S. robustius | G1195 | JN710591 | JN710591 | Cao et al., 2021 |
| S. rubigimaculatum | CLZhao 4069 | MW682343 | MW682339 | Wu et al., 2021a |
| S. rubigimaculatum | CLZhao 10638 | MW682344 | MW682340 | Wu et al., 2021a |
| S. straminellum | KHL 13849 | JN710597 | JN710597 | Cao et al., 2021 |
| S. subcollabens | Dai 19344 | MN871758 | MN877771 | Liu and Dai, 2021 |
| S. subcollabens | Dai 19345 | MN871759 | MN877772 | Liu and Dai, 2021 |
| S. subtropicum | CLZhao 16901 | OP799391 | — | Present study |
| S. subtropicum | CLZhao 11059 * | OP799390 | OP799377 | Present study |
| S. tenue | FP-102082 | KY948817 | — | Liu and Dai, 2021 |
| S. tenue | KHL 12316 | JN710598 | JN710598 | Liu and Dai, 2021 |
| S. tenuispinum | Spirin 2116 | JN710600 | JN710600 | Miettinen et al., 2012 |
| S. tenuispinum | Miettinen 8065 | JN710599 | JN710599 | Miettinen et al., 2012 |
| S. undigerum | MCW 472/13 | KY174987 | KY174987 | Westphalen et al., 2018 |
| S. undigerum | MCW 426/13 | KY174986 | KY174986 | Westphalen et al., 2018 |
| S. xanthum | CLZhao 5030 | MW204588 | MW204577 | Wu et al., 2021b |
| S. xanthum | CLZhao 5032 | MW204589 | MW204578 | Wu et al., 2021b |
| S. yunnanense | CLZhao 1445 | MW290042 | MW290056 | Dong et al., 2022 |
| S. yunnanense | CLZhao 2822 | MW290043 | MW290057 | Dong et al., 2022 |
| Trullella conifericola | Cui 2851 | MT269764 | — | Cao et al., 2021 |
| T. conifericola | Yuan 12655 | MT269760 | MT259326 | Cao et al., 2021 |
| T. dentipora | X 200 | JN710512 | JN710512 | Cao et al., 2021 |
| T. duracina | MCW 410/13 | MH475309 | MH475309 | Cao et al., 2021 |
| T. duracina | RP 96 | MH475310 | MH475310 | Cao et al., 2021 |
| Xanthoporus syringae | Jeppson 2264 | JN710607 | JN710607 | Cao et al., 2021 |
| X. syringae | AFTOL-ID 774 | AY789078 | AY684166 | Cao et al., 2021 |
* is shown type material, holotype.
The sequences were aligned in MAFFT version 7 (Katoh et al., 2019) using the G-INS-i strategy. The alignment was adjusted manually using AliView version 1.27 (Larsson, 2014). The dataset was aligned first and then ITS and nLSU sequences were combined with Mesquite version 3.51. Alignment datasets were deposited in TreeBASE (submission ID 29889). Sequence of Climacocystis borealis (Fr.) Kotl. & Pouzar obtained from GenBank was used as an outgroup to root trees in the ITS+nLSU analysis in the family Steccherinaceae ( Figure 1 ), and Irpex lacteus (Fr.) Fr. was used as an outgroup in the ITS+nLSU analysis in the genus Steccherinum ( Figure 2 ) (Dong et al., 2022).
Figure 1.
Maximum parsimony strict consensus tree illustrating the phylogeny of three new species and related species in the family Steccherinaceae based on ITS+nLSU sequences. Branches are labeled with maximum likelihood bootstrap values higher than 70%, parsimony bootstrap values higher than 50% and Bayesian posterior probabilities more than 0.95 respectively.
Figure 2.

Maximum parsimony strict consensus tree illustrating the phylogeny of three new species and related species in Steccherinum based on ITS+nLSU sequences. Branches are labeled with maximum likelihood bootstrap values higher than 70%, parsimony bootstrap values higher than 50% and Bayesian posterior probabilities more than 0.95 respectively.
Maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI) analyses were applied to the combined three datasets following previous study (Zhao and Wu, 2017), and the tree construction procedure was performed in PAUP* version 4.0b10 (Swofford, 2002). All characters were equally weighted and gaps were treated as missing data. Trees were inferred using the heuristic search option with TBR branch swapping and 1000 random sequence additions. Max-trees were set to 5000, branches of zero length were collapsed, and all parsimonious trees were saved. Clade robustness was assessed using bootstrap (BT) analysis with 1000 replicates (Felsenstein, 1985). Descriptive tree statistics-tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated for each maximum parsimonious tree generated. The multiple sequence alignment was also analyzed using maximum likelihood (ML) in RAxML-HPC2 through the Cipres Science Gateway (Miller et al., 2012). Branch support (BS) for ML analysis was determined by 1000 bootstrap replicates.
MrModeltest 2.3 (Nylander, 2004) was used to determine the best-fit evolution model for each data set for Bayesian inference (BI), which was performed using MrBayes 3.2.7a with a GTR+I+G model of DNA substitution and a gamma distribution rate variation across sites (Ronquist et al., 2012). A total of 4 Markov chains were run for 2 runs from random starting trees for 2.8 million generations for ITS+nLSU in Steccherinaceae ( Figure 1 ), and 1.7 million generations for ITS+nLSU in Steccherinum ( Figure 2 ) with trees and parameters sampled every 1000 generations. The first one-fourth of all generations was discarded as burn-in. The majority rule consensus tree of all remaining trees was calculated. Branches were considered as significantly supported if they received maximum likelihood bootstrap value (BS) >70%, maximum parsimony bootstrap value (BT) >70%, or Bayesian posterior probabilities (BPP) >0.95.
Results
Molecular phylogeny
The ITS+nLSU dataset ( Figure 1 ) included sequences from 82 fungal specimens representing 50 species. The dataset had an aligned length of 2257 characters, of which 1304 characters are constant, 237 are variable and parsimony uninformative, and 716 are parsimony informative. Maximum parsimony analysis yielded 36 equally parsimonious trees (TL = 3992, CI = 0.3885, HI = 0.6115, RI = 0.6621, and RC = 0.2572). The best model for the ITS+nLSU dataset estimated and applied in the Bayesian analysis was GTR+I+G (lset nst = 6, rates = invgamma; prset statefreqpr = dirichlet (1,1,1,1)). Bayesian analysis and ML analysis resulted in a similar topology to MP analysis with an average standard deviation of split frequencies = 0.007830 (BI), and the effective sample size (ESS) across the two runs is the double of the average ESS (avg ESS) = 182.
The phylogram inferred from the ITS+nLSU rDNA gene regions ( Figure 1 ) showed that sixteen genera nested into the family Steccherinaceae as Antella Miettinen, Antrodiella Ryvarden & I.johans, Atraporiella Ryvarden, Butyrea Miettinen, Elaphroporia Z.Q. Wu & C.L. Zhao, Etheirodon Banker, Flaviporus Murrill, Frantisekia Spirin & Zmitr, Junghuhnia Corda, Loweomyces (Kotl. & Pouzar) Jülich, Metuloidea G. Cunn, Mycorrhaphium Maas Geest, Nigroporus Murrill, Steccherinum, Trullella Zmitr and Xanthoporus Audet, in which three new species Steccherinum fissurutum, S. punctatum and S. subtropicum grouped into genus Steccherinum.
The ITS+nLSU dataset ( Figure 2 ) included sequences from 57 fungal specimens representing 27 species. The dataset had an aligned length of 2068 characters, of which 1465 characters are constant, 168 are variable and parsimony-uninformative, and 435 are parsimony-informative. Maximum parsimony analysis yielded 5000 equally parsimonious trees (TL = 1640, CI = 0.5213, HI = 0.4787, RI = 0.7996, RC = 0.4169). Best model for the ITS+nLSU dataset estimated and applied in the Bayesian analysis was GTR+I+G (lset nst = 6, rates = invgamma; prset statefreqpr = dirichlet (1,1,1,1). Bayesian analysis and ML analysis resulted in a similar topology to MP analysis with an average standard deviation of split frequencies = 0.009192 (BI), and the effective sample size (ESS) across the two runs is the double of the average ESS (avg ESS) = 198.
The phylogenetic tree ( Figure 2 ) inferred from ITS+nLSU sequences covered 26 species of Steccherinum, which demonstrated that S. subtropicum was sister to S. enuispinum; S. fissurutum formed a monophyletic lineage; S. punctatum grouped with a clade comprised S. straminellum (Bres.) Melo and S. ciliolatum (Berk. & M.A. Curtis) Gilb. & Budington.
Taxonomy
Steccherinum fissurutum J.H. Dong & C.L. Zhao, sp. nov. Figures 3 , 4 .
Figure 3.
Basidiomata of Steccherinum fissurutum (holotype). Bars: (A) 1 cm; (B) 0.5 mm.
Figure 4.
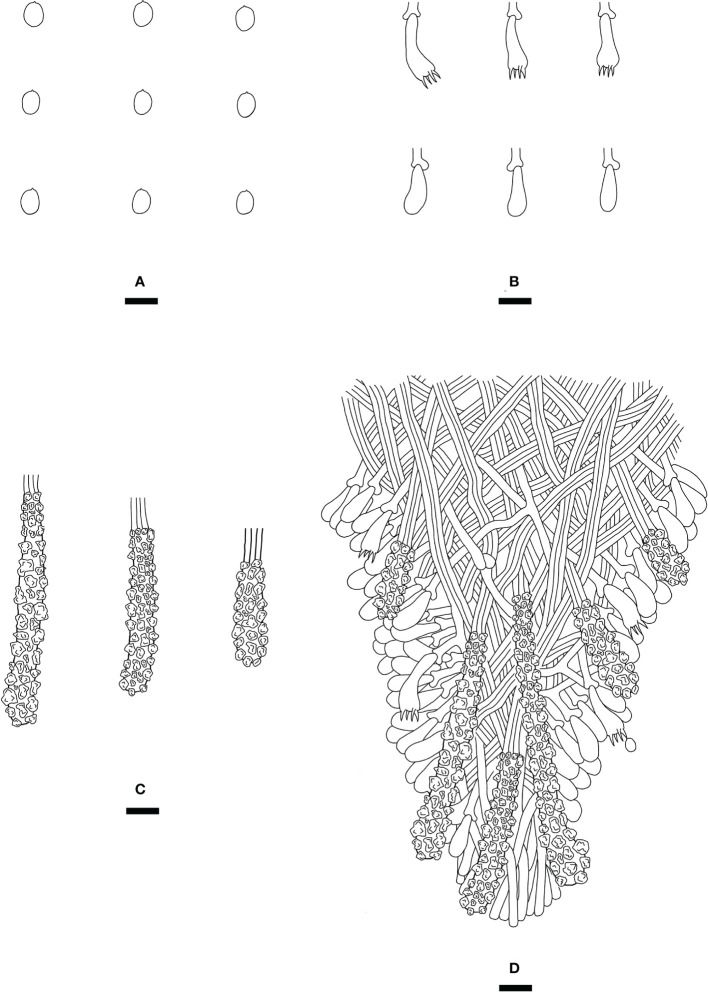
Microscopic structures of Steccherinum fissurutum (drawn from the holotype). (A) Basidiospores. (B) Basidia and basidioles. (C) Skeletocystidia. (D) A section of hymenium. Bars: (A) 5 µm; (B–D) 10 µm.
Hierarchical information: Fungi, Dikarya, Basidiomycota, Agaricomycotina, Agaricomycetes, Polyporales, Steccherinaceae, Steccherinum.
MycoBank no.: MB 846499.
Diagnosis: differs from other Steccherinum species by its white to buff, cracked, subceraceous, grandinoid hymenial surface, a monomitic hyphal system with clamped generative hyphae and cylindrical basidiospores measuring 4.5–6.0 × 2.5–3.0 µm.
Holotype—China. Yunnan Province, Lijiang, Heilongtan Park, Xiangshan, GPS coordinates 26°53′ N, 100°13′ E, altitude 2, 400 m asl., on the fallen branch of angiosperm, leg. C.L. Zhao, 21 July 2021, CLZhao 21803 (SWFC).
Etymology—fissurutum (Lat.): referring to the cracked hymenophore surface of the type specimens.
Basidiomata: Annual, resupinate, adnate, cracked, subceraceous, without odor or taste when fresh, becoming brittle upon drying, up to 10 cm long, up to 2 cm wide, 50–150 µm thick. Hymenial surface grandinoid, aculei 3–5 per mm, the length of aculei up to 0.2 mm, white (60) when fresh, turning to white (60) to buff (13) upon drying. Sterile margin white, 0.5 mm wide.
Hyphal system: Monomitic, generative hyphae with clamp connections, colorless, thin-walled, frequently branched, interwoven, 2.5–3.5 µm in diam. IKI–, CB–, tissues unchanged in KOH.
Hymenium: Skeletocystidia numerous in the aculei, strongly encrusted in the obtuse apex, 26.5–36 × 6.5–9.5 µm; cystidioles absent. Basidia clavate, with 4 sterigmata and a basal clamp connection, 12.5–16.5 × 4.5–7 µm; basidioles dominant, in shape similar to basidia, but slightly smaller.
Basidiospores: Cylindrical, colorless, thin-walled, with one oil drop inside, IKI–, CB–, 4.5–6.0 × 2.5–3.0 µm, L = 5.23 µm, W = 2.79 µm, Q = 1.75–1.98 (n = 180/6).
Type of rot: White rot.
Additional specimens examined (paratypes): CHINA, Yunnan Province, Lijiang, Heilongtan Park, Xiangshan, GPS coordinates 26°53′ N, 100°13′ E, altitude 2, 400 m asl., on the fallen branch of angiosperm, leg. C.L. Zhao, 21 July 2021, CLZhao 21634, 21673, 21675, 21680, 21703, 21744, 21754, 21808, 21811, 21826, 21841 (SWFC).
Steccherinum punctatum J.H. Dong & C.L. Zhao, sp. nov. Figures 5 , 6 .
Figure 5.
Basidiomata of Steccherinum punctatum (holotype). Bars: (A) 1 cm; (B) 0.5 mm.
Figure 6.
Microscopic structures of Steccherinum punctatum (drawn from the holotype). (A) Basidiospores. (B) Basidia and basidioles. (C) Skeletocystidia. (D) A section of hymenium. Bars: (A) 5 µm; (B–D) 10 µm.
Hierarchical information: Fungi, Dikarya, Basidiomycota, Agaricomycotina, Agaricomycetes, Polyporales, Steccherinaceae, Steccherinum.
MycoBank no.: MB 846500.
Diagnosis: differs from other Steccherinum species by its cream to buff, punctate, grandinoid hymenial surface, a monomitic hyphal system with clamped generative hyphae and ellipsoid basidiospores measuring 3.6–4.5 × 2.6–3.4 µm.
Holotype—China. Yunnan Province, Yuxi, Xinping County, Jinshan Primeval Forest Park, GPS coordinates 24°07′ N, 101°99′ E, altitude 2, 300 m asl., on the stump of angiosperm, leg. C.L. Zhao, 2 January 2019, CLZhao 9184 (SWFC).
Etymology—punctatum (Lat.): referring to the punctate hymenophore surface.
Basidiomata: Annual, resupinate, adnate, punctate, soft leathery, without odor or taste when fresh, becoming leathery upon drying, up to 15 cm long, up to 5 cm wide, 50–100 µm thick. Hymenial surface grandinoid, aculei 5–9 per mm, the length of aculei up to 0.1 mm, white (60) when fresh, turning to cream (21) to buff (13) upon drying. Sterile margin cream, 0.5 mm wide.
Hyphal system: Monomitic, generative hyphae with clamp connections, colorless, thin-walled, frequently branched, interwoven, 3–4.5 µm in diam. IKI–, CB–, tissues unchanged in KOH.
Hymenium: Skeletocystidia numerous, thin-walled, cylindrical, strongly encrusted in the surface and almost entirely, 36–47 × 7.5–12 µm; cystidioles absent. Basidia subclavate to barrel, with 4 sterigmata and a basal clamp connection, 23–27 × 5.5–7.5 µm; basidioles dominant, in shape similar to basidia, but slightly smaller.
Basidiospores: Ellipsoid, colorless, thin-walled, smooth, with one oil drop inside, IKI–, CB–, 3.6–4.5(–4.7) × 2.6–3.4 µm, L = 4.00 µm, W = 2.88 µm, Q = 1.37–1.42 (n = 60/2).
Type of rot: White rot.
Additional specimen examined (paratype): CHINA, Yunnan Province, Yuxi, Xinping County, Jinshan Primeval Forest Park, GPS coordinates 24°07′ N, 101°99′ E, altitude 2, 300 m asl., on the stump of angiosperm, leg. C.L. Zhao, 2 January 2019, CLZhao 9181 (SWFC).
Steccherinum subtropicum J.H. Dong & C.L. Zhao, sp. nov. Figures 7 , 8 .
Figure 7.
Basidiomata of Steccherinum subtropicum (holotype). Bars: (A) 1 cm; (B) 0.5 mm.
Figure 8.
Microscopic structures of Steccherinum subtropicum (drawn from the holotype). (A) Basidiospores. (B) Skeletocystidia. (C) Basidia and basidioles. (D) A section of hymenium. Bars: (A) 5 µm; (–D) 10 µm.
Hierarchical information: Fungi, Dikarya, Basidiomycota, Agaricomycotina, Agaricomycetes, Polyporales, Steccherinaceae, Steccherinum.
MycoBank no.: MB 846501.
Diagnosis: differs from other Steccherinum species by its pink to lilac, effuse-reflexed, odontioid hymenial surface, a dimitic hyphal system with clamped generative hyphae and ellipsoid basidiospores measuring 2.8–3.4 × 2.0–2.7 µm.
Holotype—China. Yunnan Province, Wenshan, Xichou County, Xiaoqiaogou National Nature Reserve, GPS coordinates 23°22′ N, 104°47′ E, altitude 1700 m asl., on the fallen branch of angiosperm, leg. C.L. Zhao, 15 January 2019, CLZhao 11059 (SWFC).
Etymology—subtropicum (Lat.): referring to distribution (subtropical zone) of the type specimens.
Basidiomata: Annual, effuse-reflexed, without odor or taste when fresh, becoming leathery upon drying, up to 6 cm long, up to 1.5 cm wide, 100–150 µm thick. Hymenial surface odontioid, aculei 5–7 per mm, the length of aculei 0.5–1 mm long, fresh pink (27) when fresh, turning to rose (28) to lilac (48) upon drying. Sterile margin cream, 0.5–1 mm wide.
Hyphal system: Dimitic, generative hyphae with clamp connections, colorless, thin-walled, branched, more or less interwoven, 2.3–3.5 µm in diam. Skeletal hyphae colorless, thick-walled, 3.5–4.5 µm diam; all hyphae IKI–, CB–, tissues unchanged in KOH.
Hymenium: Skeletocystidia numerous strongly encrusted in the obtuse apex, 20–82 × 5.5–10 µm; cystidioles absent. Basidia clavate, with 4 sterigmata and a basal clamp connection, 14.5–20 × 4–6 µm; basidioles dominant, in shape similar to basidia, but slightly smaller.
Basidiospores: Ellipsoid, colorless, thin-walled, IKI–, CB–, 2.8–3.4 × 2.0–2.7 µm, L = 3.00 µm, W = 2.31 µm, Q = 1.24–1.37 (n = 60/2).
Type of rot: White rot.
Additional specimen examined (paratype): CHINA, Yunnan Province, Wenshan, Xiaojie Town, Laojunshan National Nature Reserve, GPS coordinates 22°56′ N, 104°37′ E, altitude 2500 m asl., on the fallen branch of angiosperm, leg. C.L. Zhao, 15 January 2019, CLZhao 16901 (SWFC).
Discussion
In the present study, three new species, Steccherinum fissurutum, S. punctatum and S. subtropicum are described based on phylogenetic analyses and morphological characters.
Phylogenetically, seven clades were found in Polyporales: the residual polyporoid clade, the phlebioid clade, the antrodia clade, the tyromyces clade, the fragiliporia clade, the core polyporoid clade and the gelatoporia clade (Binder et al., 2005; Binder et al., 2013). Miettinen et al. (2012) employed the molecular systematics of Steccherinum and related genera Antrodiella, and Junghuhnia utilizing sequences of the gene regions ITS, nLSU, mtSSU, ATPase subunit 6 (atp6), RNA polymerase II second largest subunit (rpb2), and translation elongation factor 1-alpha (tef1), to reveal that at least 16 transitions have taken place between poroid and hydnoid hymenophore types within the family Steccherinaceae. In the present study, based on the sequences of the gene regions ITS and nLSU ( Figure 1 ), three new species, S. fissurutum, S. punctatum and S. subtropicum nested within the genus Steccherinum. Amplifying ITS and nLSU genes across genus Steccherinum ( Figure 2 ), S. fissurutum formed a monophyletic lineage; S. punctatum grouped with a clade comprised S. straminellum and S. ciliolatum; S. subtropicum was sister to S. tenuispinum Spirin, Zmitr. & Malysheva. However, morphologically, S. straminellum differs from S. punctatum by having the dimitic hyphal system and narrower basidiospores (3.5–4.5 × 2.0–2.2 µm; Melo, 1995); S. ciliolatum is distinguished from S. punctatum by having narrowly ellipsoid to cylindrical basidiospores (4–4.5 × 2.2–2.5 µm; Maas Geesteranus, 1974). S. tenuispinum differs from S. subtropicum by its fimbriate rhizomorphs and longer aculei (1–4 mm; Spirin et al., 2007).
Morphologically, Steccherinum fissurutum resembles S. litschaueri and S. ciliolatum in having cylindrical basidiospores. However, S. litschaueri is distinguished from S. fissurutum by its rhizomorphic margin and narrower basidiospores (4.5–5.5 × 2.0–2.2 µm; Bernicchia and Gorjón, 2010). Steccherinum ciliolatum differs in having longer aculei (up to 1.5 mm) and longer basidia (18–22 × 4.5–6 µm; Maas Geesteranus, 1974).
Steccherinum punctatum is similar to S. hydneum Rick ex Maas Geest., S. tenuispinum and S. yunnanense in having leathery hymenophore. However, S. hydneum differs from S. punctatum by its longer aculei (2–3 mm) and wider basidiospores (4.2–5.0 × 3.6–4.1 µm; Yuan and Dai, 2005b); S. tenuispinum differs from S. punctatum in having whitish to dirty-ochraceous hymenial surface and narrower basidia (12–24 × 3.5–4.8 µm; Spirin et al., 2007); S. yunnanense differs in its fimbriate margin and shorter basidia (10.5–15 × 5–6 µm; Dong et al., 2022). Steccherinum punctatum resembles S. aggregatum Hjortstam & Spooner, S. fragile and S. xanthum in having a monomitic hyphal system. However, S. aggregatum is distinguished from S. punctatum by having longer cystidia (100–150 × 10–12 µm) and smaller basidia (15–20 × 4–5 µm; Hjortstam et al., 1990); S. fragile differs in having the fragile basidiomata and smaller basidiospores (2.8–3.1 × 2.1–2.2 μm; Liu and Dai, 2021). Steccherinum xanthum is distinguished from S. punctatum in having smaller basidia (10–19.3 × 3–5.2 μm; Wu et al., 2021b).
Steccherinum subtropicum is similar to S. hydneum, S. oreophilum Lindsey & Gilb. and S. rubigimaculatum in the effuse-reflexed basidiomata. However, S. hydneum differs from S. subtropicum by its cinnamon buff hymenial surface and larger basidiospores (4.2–5.0 × 3.6–4.1 µm; Yuan and Dai, 2005b). Steccherinum oreophilum differs in its cottony hymenophore and larger basidiospores (5–6.5 × 3–3.2 µm; Bernicchia and Gorjón, 2010); S. rubigimaculatum differs in having rust hymenial surface and longer basidiospores (3.5–5 × 2.5–3.5 µm; Wu et al., 2021a); S. subtropicum resembles S. fragile, S. ochraceum and S. robustius (J. Erikss. & S. Lundell) J. Erikss. in having ellipsoid basidiospores. However, S. fragile is distinguished from S. subtropicum in having a monomitic hyphal system and shorter basidia (13–14 × 4.0–4.5 µm; Liu and Dai, 2021). S. ochraceum differs in its ocherous hymenial surface and longer cystidia (100 × 7–10 µm; Bernicchia and Gorjón, 2010). The species S. robustius is distinguished from S. subtropicum by its fimbriate margin and longer basidiospores (3.5–5 × 2.5–3 µm; Bernicchia and Gorjón, 2010).
Fungi are one of the most diverse groups of organisms on Earth and play a crucial role in ecosystem processes and functions (Hyde, 2022). New DNA sequencing techniques have revolutionized the researches of fungal taxonomy and diversity, in which about 150 thousand species of fungi have been described (Hyde, 2022). Wood decaying fungi have been studied intensively in recent years (Bernicchia and Gorjón, 2010; Dai, 2011; Cui et al., 2019; Guan et al., 2020; Wang and Zhao, 2021; Westphalen et al., 2021; Wu et al., 2021a; Wu et al., 2021b; Wu et al., 2021b; Luo and Zhao, 2022; Luo et al., 2022; Qu et al., 2022; Wu et al., 2022a; Wu et al., 2022b), but the hydnoid species in the order Polyporales are still not well investigated in China, especially in the subtropics and tropics. In the present study, three new species, Steccherinum fissurutum, S. punctatum and S. subtropicum spp. nov. were found in subtropics, which enriches the fungal diversity of East Asia.
Key to species of Steccherinum sensu lato from China
1. Hyphal system monomitic in subiculum······························2
1. Hyphal system dimitic in subiculum····································8
2. Basidiospores <2 μm wide··········Mycorrhaphium adustum
2. Basidiospores >2 μm wide······················································3
3. Skeletocystidia absent····························Steccherinum fragile
3. Skeletocystidia present·····························································4
4. Aculei >1mm long·············································S. aggregatum
4. Aculei <1 mm long···································································5
5. Aculei <0.3 mm long, basidiospores with oil drops··········6
5. Aculei >0.3 mm long, basidiospores without oil drops···································································Cabalodontia queletii
6. Basidia >20 μm long··········································S. punctatum
6. Basidia <20 μm long································································7
7. Cystidia>35 μm long, basidiospores ellipsoid····S. xanthum
7. Cystidia<35 μm long, basidiospores Cylindrical········································································ S. fissurutum
8. Skeletocystidia absent···········································S. hirsutum
8. Skeletocystidia present····························································9
9. Skeletocystidia subulate, apex acute···································10
9. Skeletocystidia clavate, apex blunt······································12
10. Basidiospores >5 μm wide, aculei >1.5 mm long···················································································S. oreophilum
10. Basidiospores <5 μm wide, aculei <1.5 mm long·········11
11. Basidiomata surface reddish to brick, basidiospores <2 μm wide················································································S. laeticolor
11. Basidiomata surface white to buff, basidiospores >2 μm wide····················································································S. subulatum
12. Basidiomata resupinate························································13
12. Basidiomata effused-reflexed··············································16
13. Basidiomata with broom-like rhizomorphs···················································Etheirodon fimbriatum
13. Basidiomata without broom-like rhizomorphs···············14
14. Basidiospores <2 μm wide·································S. mukhinii
14. Basidiospores >2 μm wide··················································15
15. Aculei <0.5 mm long, aculei <4 per mm··················································································S. tenuissimum
15. Aculei >0.5 mm long, aculei >4 per mm·····S. ochraceum
16. Sterile margin fimbriate······················································17
16. Sterile margin not fimbriate··············································18
17. Basidiospores <3.5 μm wide···························S. yunnanense
17. Basidiospores >3.5 μm wide····························S. elongatum
18. Basidiospores <4 μm long·················································19
18. Basidiospores >4 μm long·················································25
19. Aculei <2 mm long·····························································20
19. Aculei >2 mm long·····························································23
20. Aculei >0.5 mm long··························································21
20. Aculei <0.5 mm long··························································22
21 Basidiospores <2 μm wide····························S. subcollabens
21 Basidiospores >2 μm wide····························S. subtropicum
22. Basidiospores subcylindrical to allantoid················································································S. puerense
22. Basidiospores ellipsoid······································S. cremicolor
23. Aculei 3–4 mm long, pileus margin sharp ····································································Metuloidea murashkinskyi
23. Aculei up to 2 mm long, pileus margin blunt···············24
24. Basidiospores >1.5 μm wide····························S. rawakense
24. Basidiospores <1.5 μm wide························S. confragosum
25. Basidiospores subglobose····················································26
25. Basidiospores ellipsoid························································27
26. Aculei <2 mm long, basidiospores with a normal guttule or not··············································································S. subglobosum
26. Aculei >2 mm long, basidiospores with a distinct guttule···················································································S. hydneum
27. Basidia <11 μm long····························S. rubigimaculatum
27. Basidia >11 μm long····························································28
28. Basidiospores >3 μm wide································S. bourdotii
28. Basidiospores <3 μm wide···················································29
29. Aculei >0.5 mm long, pinkish buff to clay buff························································································S. robustius
29. Aculei <0.5 mm long, cream to pale buff·······················································································S. ciliolatum
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
Conceptualization, C-LZ; methodology, C-LZ and J-HD; software, C-LZ and J-HD; validation, C-LZ and J-HD; formal analysis, C-LZ and J-HD; investigation, C-LZ, Z-LZ, and J-HD; resources C-LZ; writing—original draft preparation, C-LZ, J-HD, X-CZ, and J-JC; writing—review and editing, C-LZ and J-HD; visualization, C-LZ and J-HD; supervision, C-LZ; project administration, C-LZ; funding acquisition, C-LZ and Z-LZ. All authors have read and agreed to the published version of the manuscript.
Funding Statement
The research was supported by the National Natural Science Foundation of China (Project No. 32170004), Natural Science Foundation of the Jiangsu Higher Education Institutions of China (Grant 20KJB220003), Yunnan Fundamental Research Project (Grant No. 202001AS070043), the High-level Talents Program of Yunnan Province (YNQR-QNRC-2018-111).
Abbreviations
ITS, internal transcribed spacer; nLSU, large subunit; SWFC, herbarium of Southwest Forestry University, Kunming, China; KOH, 5% potassium hydroxide; CB, Cotton Blue; CB–, acyanophilous; IKI, Melzer’s reagent; IKI–, both inamyloid and indextrinoid; L, mean spore length (arithmetic average for all spores); W, mean spore width (arithmetic average for all spores); Q, variation in the L/W ratios between The studied specimens, n (a/b), number of spores (a) measured from given number (b) of specimens, spore measurements do not include ornamentation; CTAB, cetyltrimethylammonium bromide; DNA, deoxyribonucleic acid; PCR, polymerase chain reaction; MP, maximum parsimony; ML, maximum likelihood; BI, Bayesian inference; TBR, tree-bisection reconnection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Aime M. C., Miller A. N., Aoki T., Bensch K., Cai L., Crous P. W., et al. (2021). How to publish a new fungal species, or name, version 3.0. IMA Fungus 12, 11. doi: 10.1186/s43008-021-00063-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker H. J. (1906). A contribution to a revision of the north American hydnaceae. Memoirs Torrey Botanical Club 12, 99–194. doi: 10.5962/bhl [DOI] [Google Scholar]
- Banker H. J. (1912). Type studies in the hydnaceae II. the genus Steccherinum . Mycologia 4, 309–318. doi: 10.1080/00275514.1912.12017921 [DOI] [Google Scholar]
- Bernicchia A., Gorjón S. P. (2010). Fungi europaei 12: Corticiaceae s.l. Alassio: Edizioni Candusso., 1–1007. [Google Scholar]
- Binder M., Hibbett D. S., Larsson K. H., Larsson E., Langer E., Langer G. (2005). The phylogenetic distribution of resupinate forms across the major clades of mushroom-forming fungi (Homobasidiomycetes). Syst. Biodivers. 3, 113–157. doi: 10.1017/S1477200005001623 [DOI] [Google Scholar]
- Binder M., Justo A., Riley R., Salamov A., Lopez-Giraldez F., Sjokvist E., et al. (2013). Phylogenetic and phylogenomic overview of the polyporales. Mycologia 105, 1350–1373. doi: 10.3852/13-003 [DOI] [PubMed] [Google Scholar]
- Burdsall H. H., Jr., Nakasone K. K. (1981). New or little known lignicolous aphyllophorales (Basidiomycotina) from southeastern united states. Mycologia 73, 454–476. doi: 10.1080/00275514.1981.12021368 [DOI] [Google Scholar]
- Cao T., Yu J. R., Yuan H. S. (2021). Multiple-marker phylogeny and morphological evidence reveal two new species in steccherinaceae (Polyporales, basidiomycota) from Asia. Mycokeys 78, 169–186. doi: 10.3897/mycokeys.78.57823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B. K., Li H. J., Ji X., Zhou J. L., Song J., Si J., et al. (2019). Species diversity, taxonomy and phylogeny of polyporaceae (Basidiomycota) in China. Fungal Divers. 97, 137–392. doi: 10.1007/s13225-019-00427-4 [DOI] [Google Scholar]
- Cunningham G. H. (1958). Hydnaceae of new zealand. i. the pileate genera Beenakia, Dentinum, Hericium, Hydnum, Phellodon and Steccherinum . Trans. Proc. R. Soc. New Z. 85, 585–601. [Google Scholar]
- Dai Y. C. (2011). A revised checklist of corticioid and hydnoid fungi in China for 2010. Mycoscience 52, 69–79. doi: 10.1007/S10267-010-0068-1 [DOI] [Google Scholar]
- Dai Y. C. (2012). Polypore diversity in China with an annotated checklist of Chinese polypores. Mycoscience 53, 49–80. doi: 10.1007/s10267-011-0134-3 [DOI] [Google Scholar]
- Dong J. H., Wu Y. X., Zhao C. L. (2022). Two new species of Steccherinum (Polyporales, basidiomycota) from southern China based on morphology and DNA sequence data. Mycoscience 63, 65–72. doi: 10.47371/mycosci.2022.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P., Wu F., Tian X. M. (2020). Three new species of Junghuhnia (Polyporales, basidiomycota) from China. MycoKeys 72, 1–16. doi: 10.3897/mycokeys.72.51872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence intervals on phylogenetics: An approach using bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
- Fries E. M. (1821). Systema mycologicum vol. 1 (Gryphiswaldia: Sumtibus Ernesti Mauritii; ). [Google Scholar]
- Gray S. F. (1821). A natural arrangement of British plants Vol. 1 (London: Baldwin, Cradock, and Joy; ). [Google Scholar]
- Guan Q. X., Liu C. M., Zhao T. J., Zhao C. L. (2020). Heteroradulum yunnanense sp. nov. (Auriculariales, basidiomycota) evidenced by morphological characters and phylogenetic analyses in China. Phytotaxa 437, 51–59. doi: 10.11646/phytotaxa.437.2.1 [DOI] [Google Scholar]
- Hai Bang T., Suhara H., Doi K., Ishikawa H., Fukami K., Parajuli G. P., et al. (2014). Wild mushrooms in Nepal: Some potential candidates as antioxidant and ACE-inhibition sources. Evidence-Based Complementary Altern. Med. 2014, 1–11. doi: 10.1155/2014/195305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjortstam K., Ryvarden L. (2008). Some corticioid fungi (Basidiomycotina) from Ecuador. Synopsis Fungorum 25, 14–27. [Google Scholar]
- Hjortstam K., Spooner B. M., Oldridge S. G. (1990). Some aphyllophorales and heterobasidiomycetes from sabah, Malaysia. Kew Bull. 45, 303–322. doi: 10.2307/4115688 [DOI] [Google Scholar]
- Hyde K. D. (2022). The numbers of fungi. Fungal Divers. 114, 1. doi: 10.1007/s13225-022-00507-y [DOI] [Google Scholar]
- Justo A., Miettinen O., Floudas D., Ortiz-Santana B., Sjökvist E., Lindner D., et al. (2017). A revised family-level classification of the polyporales (Basidiomycota). Fungal Biol. 121, 798–824. doi: 10.1016/j.funbio.2017.05.010 [DOI] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K. D. (2019). MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A. (2014). AliView: A fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 30, 3276–3278. doi: 10.1093/bioinformatics/btu531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson K. H., Larsson E., Kõljalg U. (2004). High phylogenetic diversity among corticioid homobasidiomycetes. Mycol. Res. 108, 983–1002. doi: 10.1017/S0953756204000851 [DOI] [PubMed] [Google Scholar]
- Legon N. W., Roberts P. (2002). Steccherinum albidum: A new species from southern England. Czech Mycology 54, 7–9. doi: 10.33585/cmy.54102 [DOI] [Google Scholar]
- Lindsey J. P., Gilbertson R. L. (1977). A new Steccherinum (Aphyllophorales, steccherinaceae) on quaking aspen. Mycologia 69, 193–197. doi: 10.1080/00275514.1977.12020045 [DOI] [Google Scholar]
- Lindsey J. P., Gilbertson R. L. (1979). A new Steccherinum (Aphyllophorales, steccherinaceae) from Alaska. Mycologia 71, 1264–1266. doi: 10.1080/00275514.1979.12021141 [DOI] [Google Scholar]
- Liu Z. B., Dai Y. C. (2021). Steccherinum fragile sp. nov. and S. subcollabens comb. nov. (Steccherinaceae, polyporales), evidenced by morphological characters and phylogenetic analysis. Phytotaxa 483, 106–116. doi: 10.11646/phytotaxa.483.2.3 [DOI] [Google Scholar]
- Luo K. Y., Chen Z. Y., Zhao C. L. (2022). Phylogenetic and taxonomic analyses of three new wood-inhabiting fungi of Xylodon (Basidiomycota) in a forest ecological system. J. Fungi 8, 405. doi: 10.3390/jof8040405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K. Y., Zhao C. L. (2022). Morphology and multigene phylogeny reveal a new order and a new species of wood-inhabiting basidiomycete fungi (Agaricomycetes). Front. Microbiol. 13. doi: 10.3389/fmicb.2022.970731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas Geesteranus R. A. (1974). Studies in the genera Irpex and. Steccherinum. Persoonia 7, 443–581. [Google Scholar]
- Melo I. (1995). Steccherinum straminellum comb. nov. Mycotaxon 54, 125–127. [Google Scholar]
- Miettinen O., Larsson E., Sjökvist E., Larsson K. H. (2012). Comprehensive taxon sampling reveals unaccounted diversity and morphological plasticity in a group of dimitic polypores (Polyporales, basidiomycota). Cladistics 28, 251–270. doi: 10.1111/j.1096-0031.2011.00380.x [DOI] [PubMed] [Google Scholar]
- Miettinen O., Ryvarden L. (2016). Polypore genera Antella, Austeria, Butyrea, Citripora, Metuloidea and Trulla (Steccherinaceae, polyporales). Ann. Bot. Fenn. 53, 157–172. doi: 10.5735/085.053.0403 [DOI] [Google Scholar]
- Miller M. A., Pfeiffer W., Schwartz T. (2012). The CIPRES science gateway: Enabling high-impact science for phylogenetics researchers with limited resources. Assoc. Comput. Mach. 39, 1–8. doi: 10.1145/2335755.2335836 [DOI] [Google Scholar]
- Nylander J. A. A. (2004). MrModeltest v2. program distributed by the author (Uppsala: Evolutionary Biology Centre; ). [Google Scholar]
- Petersen J. H. (1996). Farvekort. the Danish mycological society´s colour-chart. Foreningen til Svampekundskabens Fremme Greve., 1–6. [Google Scholar]
- Piepenbring M., Maciá-Vicente J. G., Codjia J. E. I., Glatthorn C., Kirk P., Meswaet Y., et al. (2020). Mapping mycological ignorance checklists and diversity patterns of fungi known for West Africa. IMA Fungus 11, 13. doi: 10.1186/s43008-020-00034-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu M. H., Wang D. Q., Zhao C. L. (2022). A phylogenetic and taxonomic study on Xylodon (Hymenochaetales): Focusing on three new Xylodon species from southern China. J. Fungi 8, 35. doi: 10.3390/jof8010035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Hohna S., et al. (2012). Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryvarden L. (1978). A study of Hydnum subcrinale and Odontia laxa. Nord. J. Bot. 25, 293–296. [Google Scholar]
- Snell W. H., Dick E. A. (1958). Notes on the pileate Hydnum. IV. Lloydia 21, 34–37. [Google Scholar]
- Spirin W. A., Zmitrovitch I., Malysheva V. (2007). Steccherinum tenuispinum (Polyporales, basidiomycota), a new species from Russia, and notes on three other species. Ann. Bot. Fenn. 44, 298–302. [Google Scholar]
- Swofford D. L. (2002). PAUP*: phylogenetic analysis using parsimony (*and other methods). version 4.0b10 (Sunderland, Massachusetts: Sinauer Associates; ). [Google Scholar]
- Vu D., Groenewald M., de Vries M., Gehrmann T., Stielow B., Eberhardt U., et al. (2019). Large-Scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 92, 135–154. doi: 10.1016/j.simyco.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. Q., Zhao C. L. (2021). Morphological and phylogenetic evidence for recognition of two new species of Phanerochaete from East Asia. J. Fungi 7, 1063. doi: 10.3390/jof7121063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphalen M. C., Motato-Vásquez V., Tomšovský M., Gugliotta A. M. (2021). Additions to the knowledge of hydnoid steccherinaceae: Cabalodontia, Etheirodon, Metuloidea, and Steccherinum . Mycologia 113, 791–806. doi: 10.1080/00275514.2021.1894536 [DOI] [PubMed] [Google Scholar]
- Westphalen M. C., Rajchenberg M., Tomšovský M., Gugliotta A. M. (2018). A re-evaluation of neotropical Junghuhnia s.lat. (Polyporales, basidiomycota) based on morphological and multigene analyses. Persoonia 41, 130–141. doi: 10.3767/persoonia.2018.41.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols: A guide to methods and applications. Eds. Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (San Diego: Academic Press; ), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Wijayawardene N. N., Hyde K. D., Fu L. (2017). Towards incorporating asexual fungi in a natural classification: Checklist and notes 2012–2016. Mycosphere 8 (9), 1457–1555. doi: 10.5943/mycosphere/8/9/10 [DOI] [Google Scholar]
- Wijayawardene N. N., Hyde K. D., Phookamsak R. (2018. a). Outline of ascomycota: 2017. Fungal Divers. 88 (1), 167–263. doi: 10.1007/s13225-018-0394-8 [DOI] [Google Scholar]
- Wijayawardene N. N., Pawlowska J., Hyde K. D. (2018. b). Notes for genera: Basal clades of fungi (including aphelidiomycota, basidiobolomycota, blastocladiomycota, calcarisporiellomycota, caulochytriomycota, chytridiomycota, entomophthoromycota, glomeromycota, kickxellomycota, monoblepharomycota, mortierellomycota, mucoromycota, neocallimastigomycota, olpidiomycota, rozellomycota and zoopagomycota). Fungal Divers. 92 (1), 43–129. doi: 10.1007/s13225-018-0409-5 [DOI] [Google Scholar]
- Wu Y. X., Dong J. H., Zhao C. L. (2021. a). Steccherinum puerense and s. rubigimaculatum spp. nov. (Steccherinaceae, polyporales), two new species from southern China. Nova Hedwigia 113, 243–258. doi: 10.1127/nova_hedwigia/2021/0636 [DOI] [Google Scholar]
- Wu F., Man X. W., Tohtirjap A., Dai Y. C. (2022. a). A comparison of polypore funga and species composition in forest ecosystems of China, north America, and Europe. For. Ecosyst. 9, 1–7. doi: 10.1016/j.fecs.2022.100051 [DOI] [Google Scholar]
- Wu Z. Q., Shen S., Luo K. Y., Wang Z. H., Zhao C. L. (2017). Morphological and molecular identification of a new species of Atraporiella (Polyporales, basidiomycota) in China. Phytotaxa 332, 31–40. doi: 10.11646/phytotaxa.332.1.3 [DOI] [Google Scholar]
- Wu Y. X., Wu J. R., Zhao C. L. (2021. b). Steccherinum tenuissimum and s. xanthum spp. nov. (Polyporales, basidiomycota): New species from China. PloS One 16, e0244520. doi: 10.1371/journal.pone.0244520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. Q., Xu T. M., Shen S., Liu X., Luo K. Y., Zhao C. L. (2018). Elaphroporia ailaoshanensis gen. et sp. nov. in polyporales (Basidiomycota). MycoKeys 29, 81–95. doi: 10.3897/mycokeys.29.22086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhou L. W., Vlasák J., Dai Y. C. (2022. b). Global diversity and systematics of hymenochaetaceae with poroid hymenophore. Fungal Divers. 113, 1–192. doi: 10.1007/s13225-021-00496-4 [DOI] [Google Scholar]
- Yuan H. S. (2013). Antrodiella chinensis sp. nov., a Chinese representative of the antrodiella americana complex. Mycol. Prog. 12, 437–443. doi: 10.1007/s11557-012-0852-8 [DOI] [Google Scholar]
- Yuan H. S. (2014). Molecular phylogenetic evaluation of Antrodiella and morphologically allied genera in China. Mycol. Prog. 13, 353–364. doi: 10.1007/s11557-013-0921-7 [DOI] [Google Scholar]
- Yuan H. S., Dai Y. C. (2005. a). Two new species of Steccherinum (Basidiomycota) from China. Mycotaxon 93, 173–178. [Google Scholar]
- Yuan H. S., Dai Y. C. (2005. b). Two species of Steccherinum (Basidiomycota, aphyllophorales) new to China. Fung. Sci. 20(1 2), 35–39. [Google Scholar]
- Yuan H. S., Lu X., Qin W. M. (2019). Molecular and morphological analyses separate Junghuhnia pseudocrustacea sp. nov. (Basidiomycota) from Junghuhnia crustacea complex. Nova Hedwigia 108, 255–264. doi: 10.1127/nova_hedwigia/2018/0497 [DOI] [Google Scholar]
- Yuan H. S., Wu S. H. (2012). Two new species of Steccherinum (Basidiomycota, polyporales) from Taiwan. Mycoscience 53, 133–138. doi: 10.1007/S10267-011-0139-Y [DOI] [Google Scholar]
- Zhao C. L., Wu Z. Q. (2017). Ceriporiopsis kunmingensis sp. nov. (Polyporales, basidiomycota) evidenced by morphological characters and phylogenetic analysis. Mycol. Prog. 16, 93–100. doi: 10.1007/s11557-016-1259-8 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.