Rocha et al. show that C. elegans protein phosphatase 4 ensures robust outer kinetochore assembly prior to nuclear envelope breakdown, which is critical for timely and proper engagement of mitotic chromosomes with spindle microtubules.
Abstract
A landmark event in the transition from interphase to mitosis in metazoans is nuclear envelope breakdown (NEBD). Important mitotic events occur prior to NEBD, including condensation of replicated chromosomes and assembly of kinetochores to rapidly engage spindle microtubules. Here, we show that nuclear-enriched protein phosphatase 4 (PP4) ensures robust assembly of the microtubule-coupling outer kinetochore prior to NEBD. In the absence of PP4, chromosomes exhibit extended monopolar orientation after NEBD and subsequently mis-segregate. A secondary consequence of diminished outer kinetochore assembly is defective sister chromatid resolution. After NEBD, a cytoplasmic activity compensates for PP4 loss, leading to outer kinetochore assembly and recovery of chromosomes from monopolar orientation to significant bi-orientation. The Ndc80-Ska microtubule-binding module of the outer kinetochore is required for this recovery. PP4 associates with the inner kinetochore protein CENP-C; however, disrupting the PP4–CENP-C interaction does not perturb chromosome segregation. These results establish that PP4-dependent outer kinetochore assembly prior to NEBD is critical for timely and proper engagement of chromosomes with spindle microtubules.
Introduction
Successful metazoan mitosis requires that replicated chromosomes are extensively re-organized prior to NEBD so that they can be segregated to daughter cells by the microtubule-based spindle. This entails physical compaction and resolution of sister chromatids into discrete rod-shaped structures (chromosome condensation) and assembly of a microtubule attachment site at the centromeric locus on each sister chromatid (the kinetochore; Batty and Gerlich, 2019; Musacchio and Desai, 2017; Navarro and Cheeseman, 2021; Paulson et al., 2021). In late prophase, the chromatin-proximal inner kinetochore recruits the microtubule-coupling outer kinetochore, allowing rapid engagement of spindle microtubules after NEBD. Prometaphase chromosomes typically establish initial attachments to one spindle pole (mono-orientation) before forming stable load-bearing attachments that correctly connect sister kinetochores to opposite poles (bi-orientation). Bi-orientation is accompanied by chromosome alignment at the spindle equator, and a mitotic checkpoint ensures all chromosomes have time to bi-orient before sister chromatids separate in anaphase (Lara-Gonzalez et al., 2021). Efficient bi-orientation and alignment of chromosomes are crucial in the rapidly dividing cells of the early embryo, which lack a robust checkpoint response (Duro and Nilsson, 2020).
Chromosome condensation, kinetochore assembly, and kinetochore–microtubule interactions are extensively regulated by kinase and phosphatase activities. Regulation by Ser/Thr protein phosphatases (PP) is best understood for PP1 and PP2A (Moura and Conde, 2019; Nilsson, 2019), whereas the PP2A-like PP4 has received comparatively little attention (Park and Lee, 2020). In its most common form, PP4 is a heterotrimer consisting of the catalytic subunit PP4c and two regulatory subunits, PP4R2 and PP4R3, the latter targeting PP4 to substrates in the nucleus and cytoplasm through its N-terminal EVH1 domain, which binds to FxxP or MxPP motifs (Ueki et al., 2019; Karman et al., 2020).
In dividing cells, PP4 acts on both chromosomes and the microtubule cytoskeleton. Binding of human PP4 to the wings apart-like (Wapl) protein promotes cohesin release during prophase (Ueki et al., 2019), and targeting of Drosophila melanogaster PP4 to the inner kinetochore protein CENP-C is required for mitotic centromere integrity (Lipinszki et al., 2015; Torras-Llort et al., 2020). The Caenorhabditis elegans PP4c homolog PPH-4.1 promotes chromosome pairing and synapsis during female meiotic prophase and microtubule severing activity in mature oocytes (Gomes et al., 2013; Guo et al., 2022; Han et al., 2009; Sato-Carlton et al., 2014). Human and D. melanogaster PP4 localizes to mitotic spindle poles, and PP4c inhibition produces aberrant spindles with decreased centrosomal γ-tubulin levels (Brewis et al., 1993; Hastie et al., 2000; Helps et al., 1998; Martin-Granados et al., 2008). Similarly, C. elegans PPH-4.1 was reported to be required for mitotic centrosome maturation in the early embryo (Sumiyoshi et al., 2002), while no function has been described for its paralog PPH-4.2.
Here we report that inhibition of the C. elegans PP4R3 homolog SMK-1, known so far for post-mitotic functions in longevity and stress resistance (Sen et al., 2020; Wolff et al., 2006), results in chromosome alignment and segregation defects in the early embryo, which are not attributable to mis-regulation of the microtubule cytoskeleton. Our results establish that the PP4c paralogs PPH-4.1 and PPH-4.2 function redundantly and together with SMK-1 as part of a nuclear-enriched PP4 complex that promotes outer kinetochore assembly prior to NEBD for rapid engagement of spindle microtubules. We show that this novel function of PP4 is crucial for timely and error-free chromosome bi-orientation.
Results and discussion
PP4 inhibition in the early C. elegans embryo results in chromosome congression and segregation defects
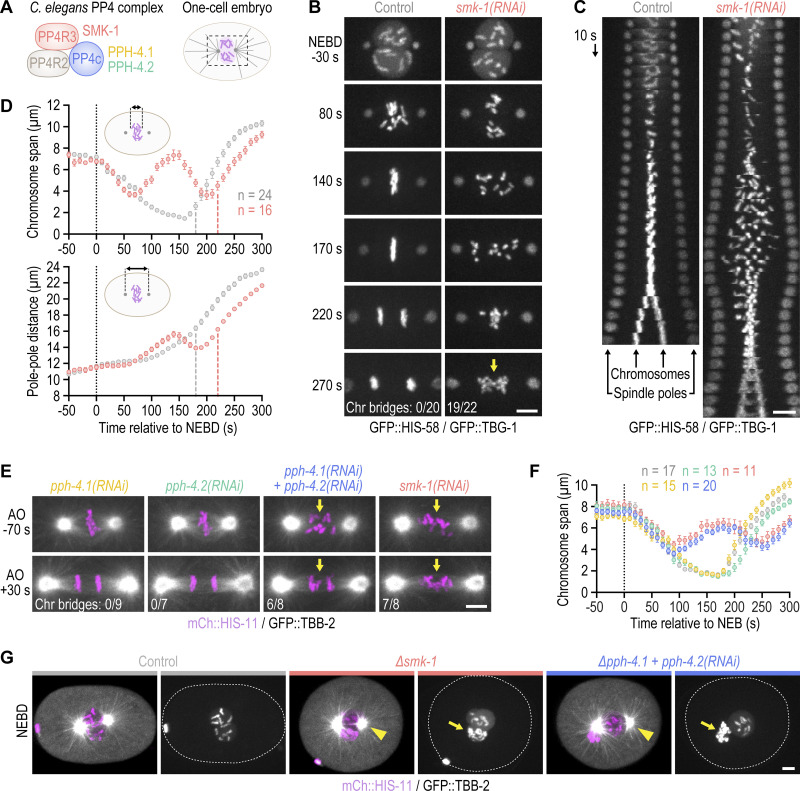
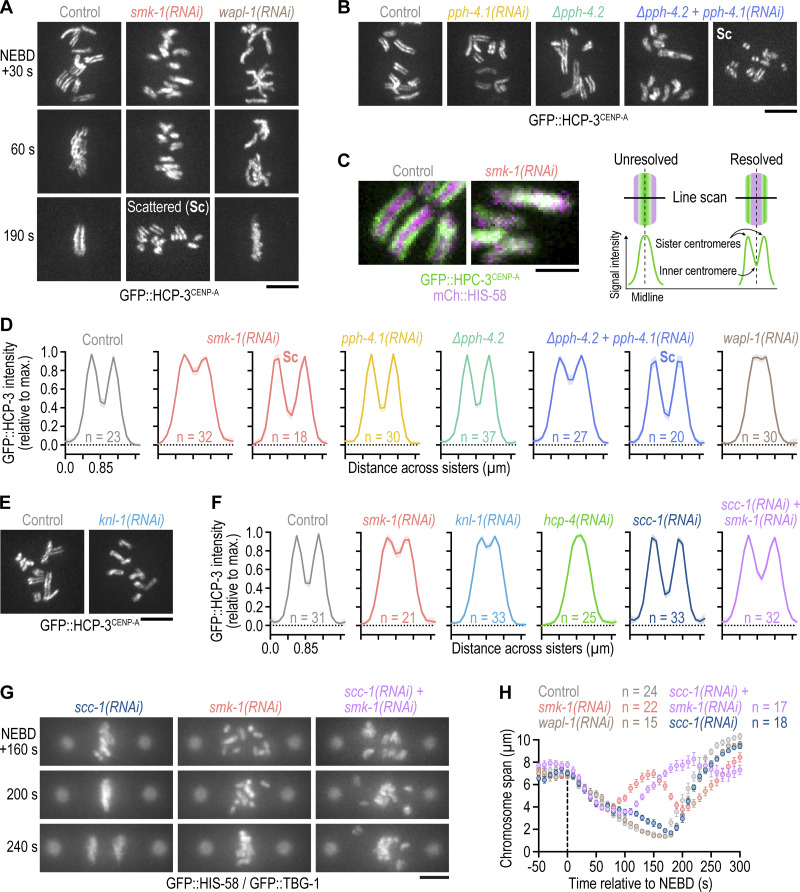
In a genome-wide RNAi screen whose hits were enriched for proteins with roles in embryonic mitosis, we identified SMK-1, the sole C. elegans PP4 targeting subunit (PP4R3) homolog (Fig. 1 A; and Fig. S1 A; Rocha et al., 2018). To assess whether SMK-1 plays a role in mitosis, we analyzed the effect of its depletion in one-cell embryos co-expressing fluorescent markers for chromosomes and spindle poles/microtubules. One-cell embryos depleted of SMK-1 exhibited a striking mitotic phenotype, with chromosomes scattering across the spindle after NEBD and then traversing back toward the spindle equator prior to anaphase onset (Fig. 1, B–F; and Video 1). This phenotype is particularly evident in time-aligned kymographs (Fig. 1 C and Fig. S1 B) and has not been observed in prior studies focused on the chromosome segregation machinery. Following anaphase onset, a high incidence of chromatin bridges was observed (Fig. 1, B and E), which in holocentric organisms is a hallmark of persistent chromosome-spindle attachment errors (Stear and Roth, 2002; Gassmann et al., 2008).
Figure 1.
PP4 inhibition in the early C. elegans embryo impairs chromosome congression and results in chromosome mis-segregation. (A) Cartoon of C. elegans PP4 (the catalytic subunit is represented by two paralogs) and of the dividing one-cell embryo with the spindle region imaged in this study outlined by dashed lines. (B) Selected images from time-lapse movies of one-cell embryos co-expressing GFP::histone H2B (HIS-58) and GFP::γ-tubulin (TBG-1). Time is relative to NEBD. The number of anaphases with chromatin (chr) bridges, highlighted by the arrow, relative to the total number of anaphases examined is indicated in the last frame. Scale bar, 5 µm. (C) Time-aligned kymographs generated from time-lapse movies such as those shown in B and Video 1. Scale bar, 5 µm. (D) Chromosome span (top) and pole–pole distance (bottom) versus time relative to NEBD, measured in one-cell embryos such as those shown in B, as indicated in the schematics. Values correspond to the mean of n embryos ± SEM, and vertical dashed lines mark the average time of anaphase onset. Conditions are color-coded as in B. (E) Selected images from time-lapse movies of one-cell embryos co-expressing mCherry::histone H2B (HIS-11) and GFP::β-tubulin (TBB-2). Time is relative to anaphase onset (AO). Arrows highlight defective chromosome congression and segregation in PP4-inhibited embryos. The frequency of anaphase chromatin (chr) bridges is indicated as described for B. Scale bar, 5 µm. (F) Chromosome span versus time, measured in one-cell embryos such as those shown in E and plotted as described for D. Conditions are color-coded as in B–E. (G) Selected images from time-lapse movies of one-cell embryos co-expressing mCherry::histone H2B (HIS-11) and GFP::β-tubulin (TBB-2), isolated from control mothers or homozygous mutant mothers before the onset of sterility. Time point corresponds to NEBD. Arrows point to structurally aberrant extra chromosomes in the female pronucleus, which are carried over into the first mitosis from defective meiotic divisions. This meiosis-derived defect is only observed in the two most penetrant PP4 inhibition conditions shown here. Centrosome maturation remains unaffected by penetrant PP4 inhibition (arrowheads). Scale bar, 5 µm.
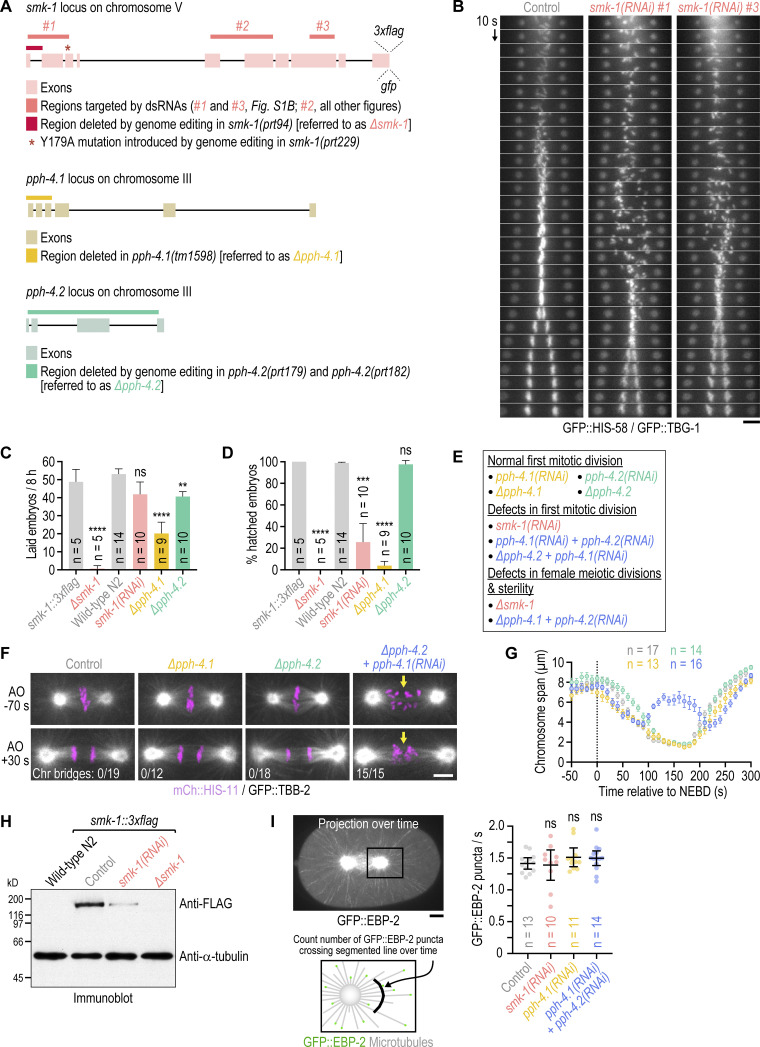
Figure S1.
Additional characterization of PP4 inhibition. (A) Genomic loci of PP4 subunits with annotation of existing mutations (∆pph-4.1) and modifications introduced by genome-editing for this study (smk-1::3xflag, smk-1::gfp, ∆smk-1, smk-1(Y179A), ∆pph-4.2). The ∆smk-1 mutation was introduced into the smk-1::3xflag background, and the smk-1(Y179A) mutation was introduced into the smk-1::gfp background. Regions in smk-1 targeted by dsRNAs are also indicated. (B) Time-aligned kymographs generated from time-lapse movies of one-cell embryos co-expressing GFP::histone H2B (HIS-58) and GFP::γ-tubulin (TBG-1). Of the three dsRNAs described in A, #1 and #3 were used here, and #2 was used for all other experiments. Scale bar, 5 µm. (C and D) Number of embryo progeny laid by a single mother in an 8-h interval (C), and embryonic viability (D), plotted as the percentage of hatched embryo progeny from a single mother (mean of n mothers ± 95% CI). Mothers were homozygous for the mutations analyzed. The smk-1::3xflag strain serves as the control for ∆smk-1, and the wild-type N2 strain serves as the control for the other conditions. Statistical significance (control versus perturbations) was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn's multiple comparison test. ****P < 0.0001; ***P < 0.001; **P < 0.01; ns = not significant, P > 0.05. (E) Summary of the effect of different PP4 inhibition conditions on chromosome segregation in embryonic mitosis and female meiosis. Homozygous ∆smk-1 animals and homozygous ∆pph-4.1 animals injected with dsRNA against pph-4.2 only produce a few embryos before becoming sterile and could therefore not be used to characterize mitotic PP4 function. (F) Selected images from time-lapse movies of one-cell embryos co-expressing mCherry::histone H2B (HIS-11) and GFP::β-tubulin (TBB-2). Time is relative to anaphase onset (AO). Arrows highlight defective chromosome congression and segregation in PP4-inhibited embryos. The number of anaphases with chromatin (chr) bridges relative to the total number of anaphases examined is indicated. Scale bar, 5 µm. (G) Chromosome span (mean of n embryos ± SEM) versus time relative to NEBD, measured in one-cell embryos such as those shown in F. Conditions are color-coded as in F. (H) Anti-FLAG immunoblot of adult animals, showing expression levels of endogenous 3xFLAG-tagged SMK-1, the absence of SMK-1::3xFLAG signal in the null mutant ∆smk-1, and residual SMK-1::3xFLAG signal after RNAi-mediated depletion. Anti-α-tubulin antibody serves as the loading control. Molecular weight is indicated in kilodaltons (kD). (I) Top left: Maximum intensity projection over successive frames from a time-lapse movie of a one-cell embryo expressing the microtubule plus-end marker GFP::EBP-2. Scale bar, 5 µm. Bottom left: Schematic illustrating the assay for quantification of microtubule nucleation rate. Right: Rate at which GFP::EBP-2 particles cross a segmented line near centrosomes (mean of n embryos ± 95% CI) as a measure of microtubule nucleation rate. Statistical significance (control versus perturbations) was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ns = not significant, P > 0.05.
Video 1.
One-cell embryos co-expressing GFP::TBG-1 (γ-tubulin) and GFP::HIS-58 (histone H2B). Control (left) and smk-1(RNAi) (right), aligned relative to NEBD. Time-lapse is 2 s and playback speed is 12 frames per second. Related to Fig. 1 B.
To determine whether SMK-1’s role in chromosome segregation reflects its molecular function as the PP4 targeting subunit, we inhibited the PP4 catalytic subunit. Individual inhibition of the PP4c paralogs PPH-4.1 and PPH-4.2 by RNAi or genetic knock-out (∆pph-4.1 and ∆pph-4.2, respectively; Fig. S1, A, C–E) had no adverse effect on the first embryonic division. By contrast, co-inhibition of PPH-4.1 and PPH-4.2 phenocopied SMK-1 depletion (Fig. 1, E and F; Fig. S1, F and G; and Video 2). Thus, PPH-4.1 and PPH-4.2 function redundantly in the early embryo, and SMK-1 contributes to mitotic chromosome segregation in the context of PP4 complexes.
Video 2.
One-cell embryos co-expressing GFP::TBB-2 (β-tubulin; white) and mCherry::HIS-11 (histone H2B; magenta). ∆pph-4.1 (top left), ∆pph-4.2 (top right), pph-4.1(RNAi) (bottom left), and ∆pph-4.2 + pph-4.1(RNAi) (bottom right), aligned relative to NEBD. Time-lapse is 10 s and playback speed is 6 frames per second. Related to Fig. 1 E.
Genetic knock-out of smk-1 (∆smk-1; Fig. S1, A and C–E) and penetrant inhibition of PP4c produced a variable number of structurally aberrant chromosomes in the maternal pronucleus (Fig. 1 G and Fig. S1 E), suggesting that PP4 is also required for chromosome segregation during oocyte meiosis. Immunoblotting suggested that this phenotype may not be observed in smk-1(RNAi) embryos because of residual protein (Fig. S1 H). Thus, PP4c acts during oocyte meiosis, in addition to ensuring proper chromosome segregation in the embryo.
Chromosomes in PP4-inhibited embryos go through an extended period of mono-orientation before recovering to a bi-oriented state
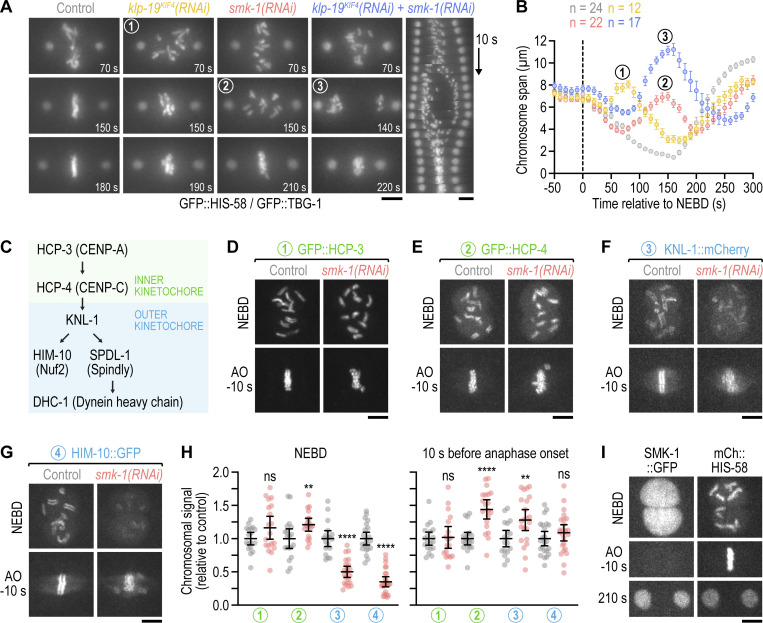
Inspection of the mitotic PP4 inhibition phenotype revealed unusual chromosome behavior during prometaphase: for the first ∼70 s following NEBD, chromosomes congressed toward the spindle equator before scattering back toward spindle poles between 70 and 150 s (Fig. 1, B and C). From 150 s until anaphase onset at 230 s (a ∼40-s delay relative to controls), chromosomes resumed congression, albeit without achieving full alignment at the spindle equator. The transient scattering of chromosomes manifests as a prominent bounce when the distance between outermost chromosomes along the spindle axis is plotted over time (“chromosome span”; Fig. 1, D and F; and Fig. S1 G). Plotting the distance between spindle poles over time showed that spindles in PP4-inhibited embryos elongated prematurely during chromosome scattering (Fig. 1 D). Spindle length subsequently recovered during chromosome re-congression, and by anaphase onset spindles in PP4-inhibited embryos were the same length as control spindles. The prometaphase bounce in spindle length indicates a delay in the formation of bi-oriented attachments capable of sustaining the tension generated by cortex-localized forces that position the spindle by pulling on astral microtubules (Grill et al., 2003; Nguyen-Ngoc et al., 2007; Oegema et al., 2001; Cheeseman et al., 2004). Taken together, these observations suggested that PP4 inhibition results in an extended period of mono-orientation before chromosomes recover to a bi-oriented state. This conclusion was reinforced by the striking consequence of co-inhibiting PP4 and polar ejection forces generated by the chromokinesin KLP-19Kif4 (Powers et al., 2004): in klp-19Kif4(RNAi);smk-1(RNAi) embryos, but not in klp-19Kif4(RNAi) or smk-1(RNAi) embryos, poleward chromosome scattering was so pronounced that the spindle equator became temporarily devoid of chromosomes; yet chromosomes eventually reversed direction and resumed congression (Fig. 2, A and B; and Video 3). We conclude that PP4 inhibition produces a defect at the time of NEBD that precludes rapid chromosome bi-orientation and instead leads to an extended mono-oriented state. The defect is not permanent, however, because significant bi-orientation is eventually achieved later in prometaphase, albeit at the cost of attachment errors that compromise the fidelity of chromosome segregation.
Figure 2.
PP4 inhibition diminishes outer but not inner kinetochore assembly prior to NEBD. (A) Selected images and kymograph from time-lapse movies of one-cell embryos co-expressing GFP::histone H2B (HIS-58) and GFP::γ-tubulin (TBG-1). Time is relative to NEBD. Scale bar, 5 µm. (B) Chromosome span versus time relative to NEBD (mean of n embryos ± SEM). Measurements were performed in time-lapse movies such as those shown in A. Numbers in A and B refer to corresponding time points in images and graph. (C) Assembly hierarchy of the kinetochore components analyzed in this figure and Fig. S2. (D–G) Selected images from time-lapse movies of one-cell embryos co-expressing fluorescently tagged kinetochore components (GFP::HCP-3, GFP::HCP-4, and HIM-10::GFP are endogenously tagged; KNL-1::mCherry is a functional transgene) and transgenic GFP- or mCherry-tagged histone H2B (HIS-58). Only the kinetochore component is shown. Time points correspond to NEBD and the last frame prior to anaphase onset (AO). Scale bars, 5 µm. (H) Intensity of the chromosomal signal at NEBD or just prior to AO in the one-cell embryo (mean ± 95% CI), normalized to the mean of the respective control, for the components shown in D–G. Statistical significance was determined by the Mann-Whitney test. ****P < 0.0001; **P < 0.01; ns = not significant, P > 0.05. (I) Selected images from a time-lapse movie of a one-cell embryo co-expressing endogenously tagged SMK-1::GFP and transgenic mCherry::histone H2B (HIS-58). Scale bar, 5 µm.
Video 3.
One-cell embryos co-expressing GFP::TBG-1 (γ-tubulin) and GFP::HIS-58 (histone H2B). klp-19(RNAi) (left) and klp-19(RNAi);smk-1(RNAi) (right), aligned relative to NEBD. Time-lapse is 10 s and playback speed is 6 frames per second. Related to Fig. 2 A.
PP4 inhibition diminishes outer but not inner kinetochore assembly prior to NEBD
We next set out to understand why PP4 inhibition precludes rapid bi-orientation after NEBD. Live imaging in one-cell embryos showed that PP4 inhibition had no discernable effect on the size of centrosomes or their capacity to nucleate microtubules (Fig. 1, B, C, E, and G; and Fig. S1, F and I), which contrasts with the conclusion of a prior study (Sumiyoshi et al., 2002). Consistent with this, PP4 inhibition affected neither pronuclear migration nor assembly or positioning of the mitotic spindle. The chromosome congression and segregation defects observed following PP4 inhibition are therefore not attributable to mis-regulation of the microtubule cytoskeleton.
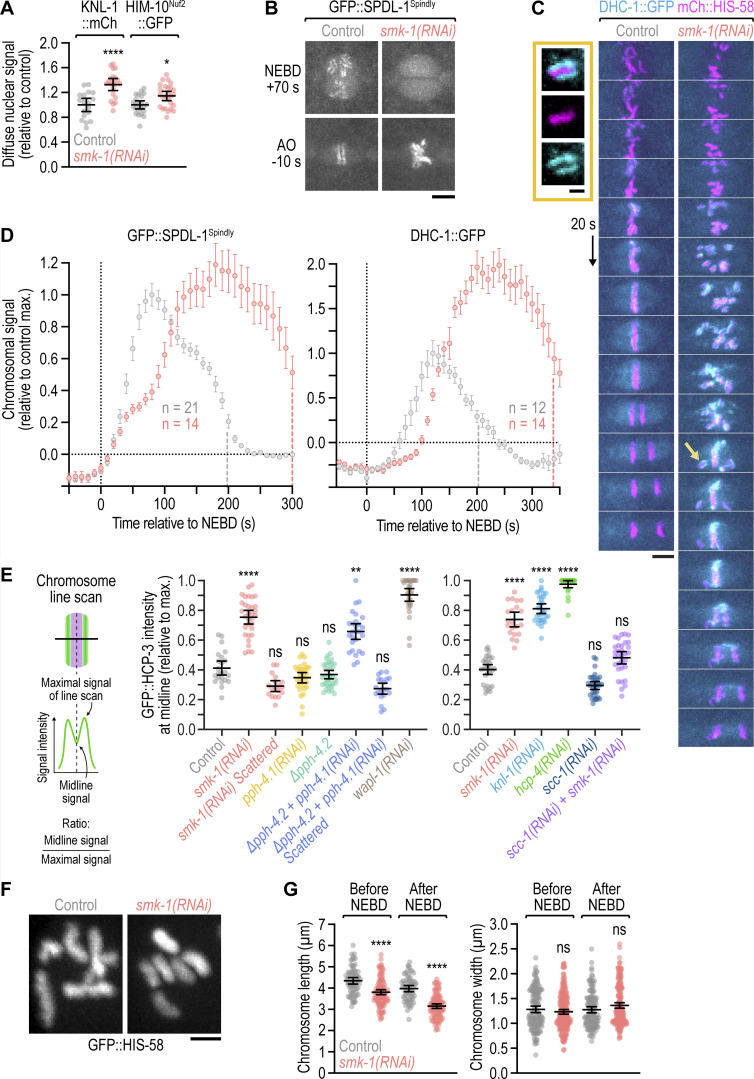
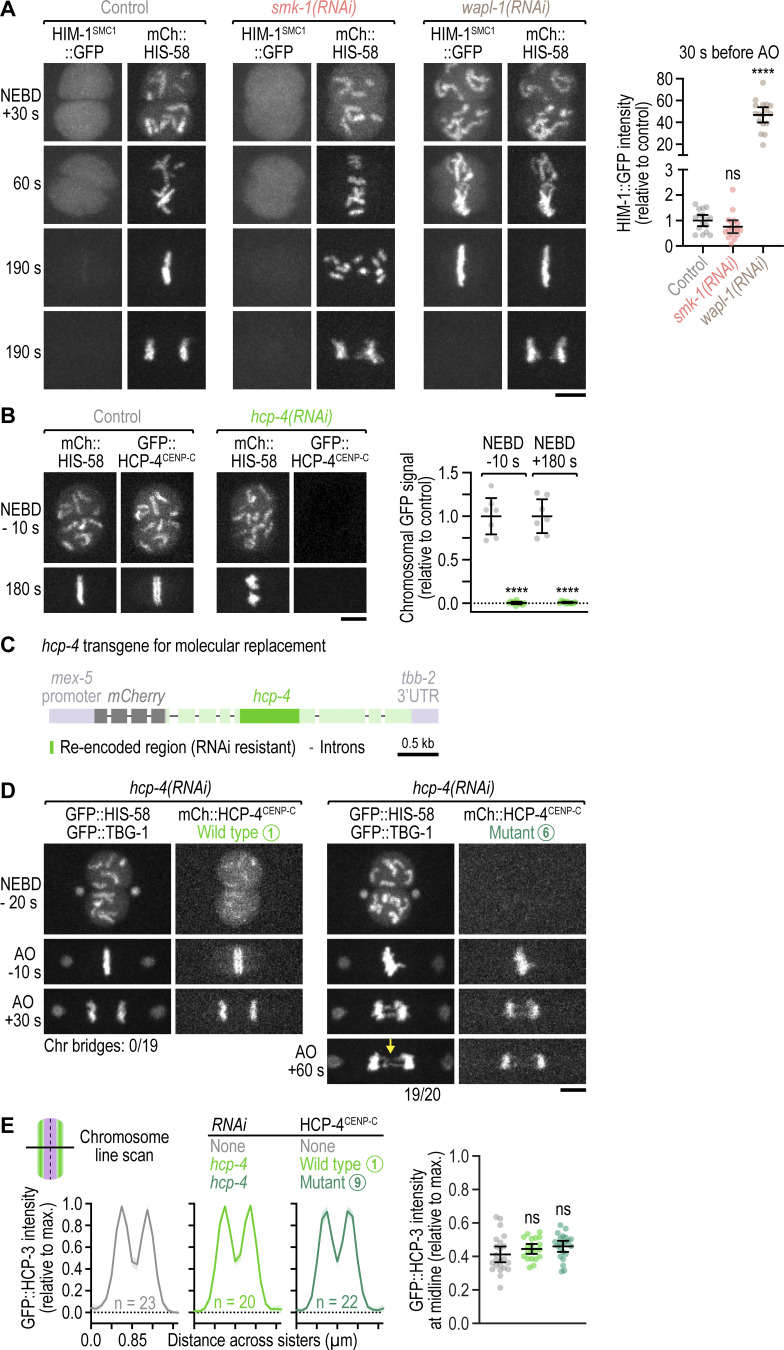
We noticed that PP4 inhibition in KLP-19Kif4-depleted embryos suppressed the kinetochore-powered poleward chromosome movements in early prometaphase that are typically observed after KLP-19Kif4 loss (Powers et al., 2004; Fig. 2, A and B; and Video 3). This suggested that PP4 inhibition prevented kinetochores from promptly engaging spindle microtubules at NEBD. We therefore examined the effect of PP4 inhibition on kinetochore assembly (Fig. 2 C). Kinetochores are built in a hierarchical manner starting with the inner kinetochore components HCP-3CENP-A and HCP-4CENP-C, which through HCP-4CENP-C recruit components of the outer kinetochore, including the scaffolding protein KNL-1, the Ndc80 complex (represented here by the subunit HIM-10Nuf2), and the dynein module (represented here by SPDL-1Spindly and dynein heavy chain DHC-1; Cheeseman et al., 2004; Desai et al., 2003; Gassmann et al., 2008; Oegema et al., 2001). Using functional fluorescent versions of these components (Barbosa et al., 2017; Cheerambathur et al., 2019; Espeut et al., 2012; Yamamoto et al., 2008), we confirmed that control embryos assemble a robust outer kinetochore prior to NEBD (Fig. 2, D–G). In smk-1(RNAi) embryos, chromosomal levels of GFP::HCP-3CENP-A and GFP::HCP-4CENP-C were similar and higher at NEBD than in controls, respectively (Fig. 2, D, E, and H; and Video 4). By contrast, chromosomal levels of outer kinetochore components were significantly lower at NEBD than in controls but subsequently recovered after NEBD, approximating control levels by anaphase onset (Fig. 2, F–H and Video 5). Thus, PP4 inhibition diminishes outer kinetochore assembly downstream of GFP::HCP-4CENP-C and does so specifically prior to NEBD. Importantly, non-chromosomal levels of outer kinetochore components in prophase nuclei were increased in smk-1(RNAi) relative to controls (Fig. S2 A), ruling out defective nuclear import as the cause for diminished outer kinetochore assembly. Chromosomal levels of SPDL-1Spindly and dynein, which assemble on outer kinetochores after NEBD, eventually surpassed control levels in smk-1(RNAi) embryos (Fig. S2, B–D), underscoring that PP4 inhibition delays outer kinetochore assembly rather than preventing it.
Video 4.
One-cell embryos expressing GFP::HCP-4 (CENP-C). Control (left) and smk-1(RNAi) (right), aligned relative to NEBD. Time-lapse is 10 s and playback speed is 6 frames per second. Related to Fig. 2 E.
Video 5.
One-cell embryos co-expressing HIM-10::GFP (Nuf2) and mCherry::HIS-58 (histone H2B). Control (left) and smk-1(RNAi) (right), aligned relative to NEBD. Time-lapse is 10 s and playback speed is 6 frames per second. Related to Fig. 2 G.
Figure S2.
Nuclear import of outer kinetochore components is unaffected by PP4 inhibition; the outermost kinetochore component dynein hyperaccumulates in late prometaphase following PP4 inhibition; quantification of the sister centromere resolution defects shown in Fig. 3; mitotic chromosomes have a compact morphology following PP4 inhibition. (A) Intensity of the non-chromosomal (diffuse) nuclear signal of fluorescently tagged outer kinetochore components at NEBD in the one-cell embryo (mean ± 95% CI), normalized to the mean of the respective control. Statistical significance (control versus smk-1(RNAi)) was determined by the Mann-Whitney test. ****P < 0.0001; *P < 0.05. (B) Selected images from time-lapse movies of one-cell embryos co-expressing transgenic GFP::SPDL-1 in a spdl-1 knock-out background and transgenic mCherry::histone H2B (HIS-58). Only the GFP::SPDL-1 signal is shown. AO, anaphase onset. Scale bar, 5 µm. (C) Selected images from time-lapse movies of one-cell embryos co-expressing endogenously GFP-tagged dynein heavy chain (DHC-1) and transgenic mCherry::histone H2B (HIS-58). Image sequences start at the same time point after NEBD. Arrow points to the unaligned chromosome shown at greater magnification. Scale bar, 5 µm; magnified chromosome, 2 µm. (D) Intensity of chromosomal GFP::SPDL-1 (left) and DHC-1::GFP (right) signal (mean of n embryos ± SEM) versus time relative to NEBD, determined by averaging the signal of the top 15 local maxima detected on chromosomes in time-lapse movies of one-cell embryos. Traces are normalized to the peak signal in the control. Vertical dashed lines mark the average time of anaphase onset. (E) Quantification of the line scans shown in Fig. 3, D and F (left and right graph, respectively). GFP::CENP-A (HCP-3) signal intensity at the chromosome midline relative to the maximal signal intensity in the line scan (mean of n chromosomes from at least 10 embryos ± 95% CI) was determined as illustrated in the schematic. Statistical significance (control versus perturbations) was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn's multiple comparison test. ****P < 0.0001; **P < 0.01; ns = not significant, P > 0.05. (F) Morphology of early prometaphase chromosomes in one-cell embryos, marked by transgenic GFP::histone H2B (HIS-58). Scale bar, 2 µm. (G) Length and width of mitotic chromosomes in late prophase and early prometaphase (mean ± 95% CI), derived from 3D measurements in z-stacks acquired of GFP-marked chromosomes as shown in F. Data points correspond to values of individual chromosomes from at least 5 embryos per condition, normalized to the mean of the control. Statistical significance (control versus smk-1(RNAi)) was determined by the Mann-Whitney test. ****P < 0.0001; ns = not significant, P > 0.05.
The effect of smk-1(RNAi) on prophase kinetochore assembly implies that PP4 acts in the nucleus. In agreement with this, live imaging of one-cell embryos expressing endogenously tagged SMK-1::GFP revealed that SMK-1 is strongly enriched in the male and female pronucleus (Fig. 2 I and Video 6). The nuclear signal remained diffusive throughout prophase with no detectable enrichment on condensing chromosomes. After NEBD, SMK-1::GFP gradually dissipated from the spindle region and rapidly concentrated in nascent daughter nuclei at mitotic exit. We also added tags to PPH-4.1, but homozygous animals were either very sick (C-terminal 3xFLAG or GFP) or inviable (N-terminal 3xFLAG), precluding conclusive localization analysis.
Video 6.
One-cell embryo co-expressing SMK-1::GFP and mCherry::HIS-58 (histone H2B). Time-lapse is 10 s and playback speed is 6 frames per second. Related to Fig. 2 I.
Taken together, these results suggest that nuclear-enriched PP4 acts on prophase chromosomes to ensure timely outer kinetochore assembly prior to NEBD. In PP4-inhibited embryos, suppression of outer kinetochore assembly prior to NEBD, and subsequent relief of this suppression after nuclear dissolution, presumably by a cytoplasmic phosphatase activity, likely accounts for the unusual chromosome scattering and recovery phenotype.
Delayed outer kinetochore assembly following PP4 inhibition also delays sister centromere resolution
Our analysis of kinetochore assembly showed that PP4 inhibition affects prophase kinetochore assembly downstream of HCP-4CENP-C. Since HCP-4CENP-C is not only required for kinetochore assembly but also for resolution of sister centromeres (Moore and Roth, 2001; Moore et al., 2005), we analyzed sister centromere resolution following PP4 inhibition. We monitored sister centromere resolution using GFP::HCP-3CENP-A, which localizes to centromeres upstream of HCP-4CENP-C, and developed a live-imaging assay to quantify the extent of resolution. This revealed that sister centromeres in PP4-inhibited embryos are poorly resolved when examined within the first minute after NEBD but are fully resolved by the time chromosomes have scattered later in prometaphase (Fig. 3, A–D; Fig. S2 E, and Video 7). Importantly, mitotic chromosomes in smk-1(RNAi) embryos had a compact morphology and were in fact slightly shorter than in controls (Fig. S2, F and G), demonstrating that the resolution defect is not a byproduct of defective chromosome condensation. Moreover, in contrast to wapl-1(RNAi), smk-1(RNAi) did not increase chromosomal levels of the cohesin subunit HIM-1SMC1::GFP (Fig. S3 A), suggesting that the resolution defect is not caused by impaired cohesin removal. Instead, KNL-1 depletion revealed that outer kinetochore assembly is a prerequisite for sister centromere resolution (Fig. 3, E and F; and Fig. S2 E). We therefore conclude that the failure to assemble an outer kinetochore contributes to the sister centromere resolution defect that was previously reported for HCP-4CENP-C depletion and that we confirmed in this study (Fig. 3 F and Fig. S2 E). Taken together, these results suggest that the sister centromere resolution defect in PP4-inhibited embryos is a consequence of diminished outer kinetochore assembly during prophase and that recovery of outer kinetochore assembly after NEBD also restores sister centromere resolution.
Figure 3.
Delayed outer kinetochore assembly following PP4 inhibition also delays sister centromere resolution. (A and B) Selected images from time-lapse movies of one-cell embryos expressing endogenously tagged GFP::CENP-A (HCP-3) to mark sister centromeres. Time in A is relative to NEBD. Time point in B corresponds to 30 s after NEBD. For ∆pph-4.2 + pph-4.1(RNAi), the time of chromosome scattering (Sc) is also shown. Scale bars, 5 µm. (C) Left: Selected images from time-lapse movies of one-cell embryos co-expressing endogenously tagged GFP::HCP-3 and transgenic mCherry::histone H2B (HIS-58). Time point corresponds to 30 s after NEBD. Scale bar, 2 µm. Right: Schematic illustrating how line scans of the GFP::HCP-3 signal across the short axis of condensed mitotic chromosomes report on the extent of sister centromere resolution. (D and F) Line scan profiles (mean of n chromosomes from at least 10 embryos ± 95% CI), generated as described in C. Profiles in D are of endogenously tagged GFP::HCP-3, and profiles in F are of transgenic GFP::HCP-3 in an hcp-3 knock-out background. Also see Fig. S2 E. (E) Selected images from time-lapse movies of one-cell embryos expressing transgenic GFP::HCP-3. Time point corresponds to 30 s after NEBD. Scale bar, 5 µm. (G) Selected images from time-lapse movies of one-cell embryos co-expressing GFP::histone H2B (HIS-58) and GFP::γ-tubulin (TBG-1). Time is relative to NEBD. Scale bar, 5 µm. (H) Chromosome span (mean of n embryos ± SEM) versus time relative to NEBD. Measurements were performed in embryos such as those shown in G.
Video 7.
One-cell embryos co-expressing GFP::HCP-3 (CENP-A) and mCherry::HIS-58 (histone H2B). Control (left) and smk-1(RNAi) (right), aligned relative to NEBD. Time-lapse is 10 s and playback speed is 6 frames per second. Related to Fig. 3 A.
Figure S3.
Mitotic chromosomes do not exhibit defects in cohesin removal following PP4 inhibition; hcp-4(RNAi) for molecular replacement is penetrant; the HCP-4(F438A) mutation prevents nuclear import of HCP-4; the SMK-1–HCP-4 interaction is dispensable for sister centromere resolution. (A) Left: Selected images from time-lapse movies of one-cell embryos co-expressing endogenously tagged HIM-1::GFP (the SMC1 subunit of the cohesin complex) and transgenic mCherry::histone H2B (HIS-58). Time is relative to NEBD. Scale bar, 5 µm. Right: Integrated intensity of chromosomal HIM-1::GFP signal (mean ± 95% CI) three frames before anaphase onset (AO) in the one-cell embryo, normalized to the mean of the control. Signal was measured in embryos such as those shown in A. Statistical significance (control versus perturbations) was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ****P < 0.0001; ns = not significant, P > 0.05. (B) Left: Selected images from time-lapse movies of one-cell embryos co-expressing endogenously tagged GFP::CENP-C (HCP-4) and transgenic mCherry::histone H2B (HIS-58). Time is relative to NEBD. Scale bar, 5 µm. Right: Integrated intensity of chromosomal GFP::HCP-4 signal (mean ± 95% CI) before NEBD and at metaphase in the one-cell embryo, normalized to the mean of the control. Signal was measured in embryos such as those shown on the left. Statistical significance (control versus smk-1(RNAi)) was determined by the Mann-Whitney test. ****P <0.0001. The dsRNA targets the region of hcp-4 that is re-encoded in the RNAi-resistant transgene. (C) Schematic of the mCherry-tagged RNAi-resistant hcp-4 transgene used for molecular replacement experiments in one-cell embryos. (D) Selected images from time-lapse movies of one-cell embryos co-expressing RNAi-resistant transgenic mCherry::HCP-4, transgenic GFP::histone H2B (HIS-58), and transgenic GFP::γ-tubulin (TBG-1). Time is relative to NEBD or anaphase onset (AO). The HCP-4 mutant corresponds to mutant number 6 in Fig. 4 B, which fails to localize to the prophase nucleus but is recruited to chromosomes after NEBD. A comparison with other FxxP motif mutants shows that the effect is attributable to the F438A mutation. Arrow points to defective chromosome segregation in the mutant. The number of anaphases with chromatin (chr) bridges relative to the total number of anaphases examined is indicated. Scale bar, 5 µm. (E) Left: Line scan profiles (mean of n chromosomes from at least 10 embryos ± 95% CI), generated as described in Fig. 3 C. Right: Ratio of GFP::CENP-A (HCP-3) signal intensity at the midline of condensed chromosomes to the maximal signal intensity in the line scan (mean of n chromosomes from at least 10 embryos ± 95% CI), determined as described in Fig. S2 E. Statistical significance (control versus perturbations) was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn's multiple comparison test. ns = not significant, P > 0.05. The HCP-4 mutant corresponds to mutant number 9 in Fig. 4 B.
To address whether delayed sister centromere resolution contributes to poleward chromosome scattering in PP4-inhibited embryos, we asked whether inhibiting sister centromere resolution by preventing cohesin removal also results in chromosome scattering and whether rescuing prophase sister centromere resolution in PP4-inhibited embryos by depleting cohesin prevents chromosome scattering. Despite the failure to resolve sister centromeres due to defective cohesin removal (Fig. 3, A and D; Fig. S2 E; and Fig. S3 A), chromosomes in wapl-1(RNAi) embryos congressed normally and formed a tight metaphase plate (Fig. 3, A and H). Thus, chromosomes can congress in a timely manner even when sister centromere resolution is substantially impaired. Moreover, depletion of the cohesin subunit SCC-1 rescued sister centromere resolution in smk-1(RNAi) embryos but failed to rescue chromosome congression (Fig. 3, F–H and Fig. S2 E). Taken together, these results argue against the idea that delayed sister centromere resolution is causative for the chromosome congression defects in PP4-inhibited embryos. However, delayed sister centromere resolution likely contributes to chromosome mis-segregation in PP4-inhibited embryos by elevating merotelic attachments, in which a kinetochore is simultaneously connected to both spindle poles (Stear and Roth, 2002).
Substrate recognition by SMK-1, but not its interaction with HCP-4CENP-C, is required for timely congression and error-free segregation of chromosomes
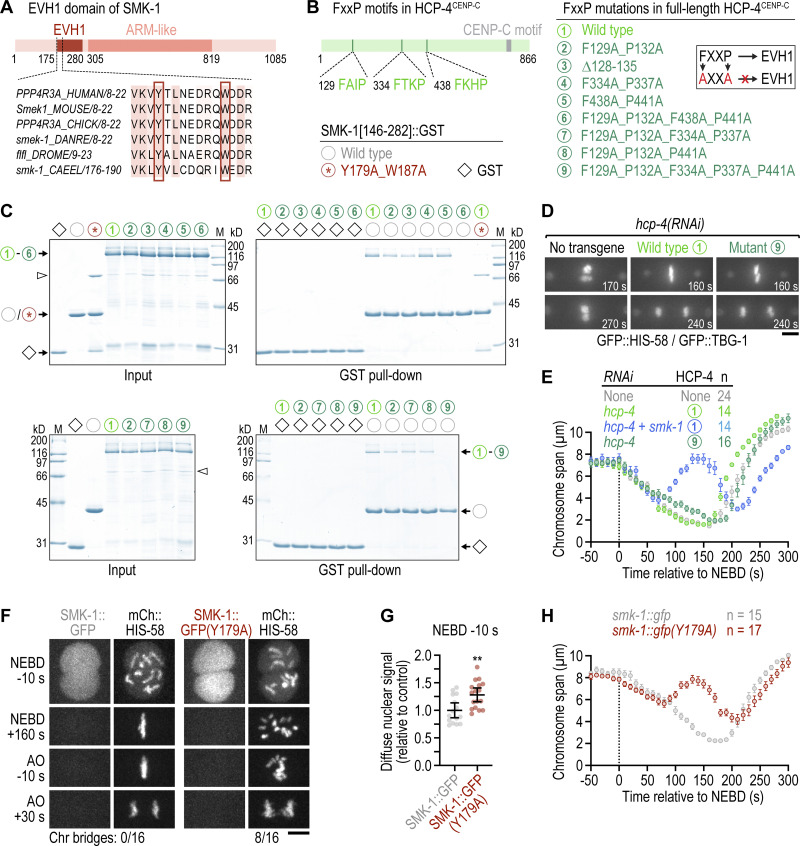
Our results show that PP4 inhibition delays outer kinetochore assembly and sister centromere resolution, both of which require HCP-4CENP-C. Prior work in D. melanogaster has shown that CENP-C binds the SMK-1 homolog Falafel and becomes hyperphosphorylated following PP4 inhibition (Lipinszki et al., 2015). Thus, an attractive hypothesis is that HCP-4CENP-C recruits PP4 through an interaction with SMK-1 and that the outer kinetochore assembly defect in PP4-inhibited embryos reflects defective PP4 targeting of HCP-4CENP-C. To test this idea, we first asked whether HCP-4CENP-C binds SMK-1. Like Falafel, SMK-1 contains an EVH1 domain near its N-terminus (Fig. 4 A). Using purified recombinant proteins, we performed GST pull-downs with full-length HCP-4CENP-C and the GST-tagged EVH1 domain (residues 146–282). Full-length HCP-4CENP-C bound to GST::EVH1, and mutating the conserved EVH1 residues Y179 and W187 to alanine abrogated the interaction (Fig. 4, B and C). Thus, the EVH1 domain of SMK-1 binds FxxP motifs in the same manner as the EVH1 domains of the fly and human homologs (Lipinszki et al., 2015; Ueki et al., 2019).
Figure 4.
Substrate recognition by SMK-1, but not its interaction with HCP-4CENP-C, is required for timely congression and error-free segregation of chromosomes. (A) Schematic of SMK-1 and sequence alignment in the highly conserved EVH1 domain. The tyrosine and tryptophan residues required for FxxP motif binding are highlighted. (B) Schematic of HCP-4CENP-C showing the location of FxxP motifs and overview of HCP-4 and SMK-1 mutations evaluated in pull-down assays. Mutating FxxP motifs to AxxA prevents their interaction with the EVH1 domain, as indicated in the cartoon. (C) Coomassie Blue-stained protein gels showing purified recombinant proteins (Input) and proteins eluted from glutathione agarose resin after GST pull-down. Molecular weight of size markers (M) is indicated in kilodaltons (kD). (D) Selected images from time-lapse movies of one-cell embryos co-expressing GFP::histone H2B (HIS-58) and GFP::γ-tubulin (TBG-1). Time is relative to NEBD. Numbers refer to transgene-encoded wild-type and mutant HCP-4, as described in B. Scale bar, 5 µm. (E) Chromosome span (mean of n embryos ± SEM) versus time relative to NEBD. Measurements were performed in time-lapse movies such as those shown in D. (F) Selected images from time-lapse movies of one-cell embryos co-expressing endogenously tagged wild-type or mutant SMK-1::GFP together with transgenic mCherry::histone H2B (HIS-58). The number of anaphases with chromatin (chr) bridges relative to the total number of anaphases examined is indicated. Scale bar, 5 µm. (G) Intensity of the chromosomal SMK-1::GFP signal just prior to NEBD in the one-cell embryo (mean ± 95% CI), normalized to the mean of wild-type SMK-1::GFP, measured in embryos such as those shown in F. Statistical significance was determined by the Mann-Whitney test. **P < 0.01. (H) Chromosome span (mean of n embryos ± SEM) versus time relative to NEBD. Measurements were performed in time-lapse movies such as those shown in F. Source data are available for this figure: SourceData F4.
HCP-4CENP-C contains three FxxP motifs in its N-terminal half (Fig. 4 B), an arrangement that contrasts with the single C-terminally located Falafel-interacting FxxP motif of D. melanogaster CENP-C. HCP-4CENP-C in which any of the three FxxP motifs was mutated to AxxA (or in which the first FxxP motif was deleted) still bound GST::EVH1, as did HCP-4CENP-C in which the first and second FxxP motifs were mutated to AxxA (Fig. 4, B and C). By contrast, the combination of first and third AxxA abrogated the interaction (mutant number 6; Fig. 4 B), as did the combination of first AxxA, second AxxA, and third FxxA (mutant number 9). Furthermore, in vitro translated HCP-4CENP-C fragments containing individual FxxP motifs bound to GST::EVH1 (data not shown). We conclude from these binding experiments that all three FxxP motifs in HCP-4CENP-C participate in the interaction with SMK-1.
To test whether SMK-1 binding to HCP-4CENP-C is important for outer kinetochore assembly prior to NEBD, we set up a molecular replacement system based on transgenic expression of RNAi-resistant mCherry::HCP-4CENP-C from a defined chromosomal locus after single-copy integration (Fig. S3, B and C). Depletion of endogenous HCP-4CENP-C resulted in the characteristic kinetochore-null phenotype, as expected (Cheerambathur et al., 2017; Cheeseman et al., 2004; Desai et al., 2003), and was rescued by RNAi-resistant wild-type mCherry::HCP-4CENP-C (Fig. 4 D). For the SMK-1 binding-defective HCP-4CENP-C mutant, we chose mutant number 9 rather than number 6 (Fig. 4 B), because mutating the phenylalanine in the third FxxP motif to alanine unexpectedly interfered with nuclear localization of mCherry::HCP-4CENP-C (Fig. S3 D). The SMK-1 binding-defective HCP-4CENP-C mutant number 9 supported near-normal chromosome congression, timely sister centromere resolution, and error-free chromosome segregation (Fig. 4, D and E; and Fig. S3 E). By contrast, nuclear localization-defective mutant number 6 exhibited penetrant chromosome segregation defects (Fig. S3 D). These results imply that the interaction of PP4 with HCP-4CENP-C mediated by the EVH1 domain of SMK-1 is dispensable for normal mitosis.
To test whether substrate recognition by SMK-1 per se is required for PP4 function in chromosome segregation, we introduced the EVH1 domain mutation (Y179A) into endogenously tagged SMK-1::GFP. SMK-1(Y179A)::GFP induced chromosome congression and segregation defects that were analogous to those observed after SMK-1 depletion, albeit less severe (Fig. 4, F–H and Video 8). Taken together, these results show that PP4 function in mitosis requires substrate recognition by the EVH1 domain of SMK-1 and suggest that HCP-4CENP-C is not the relevant PP4 target.
Video 8.
One-cell embryos expressing SMK-1::GFP (left) or SMK-1::GFP(Y179A) (right) together with mCherry::HIS-58 (histone H2B), aligned relative to NEBD. Time-lapse is 10 s and playback speed is 6 frames per second. Related to Fig. 4 F.
The Ndc80-Ska module is essential for recovery from mono-orientation induced by kinetochore dynein following PP4 inhibition
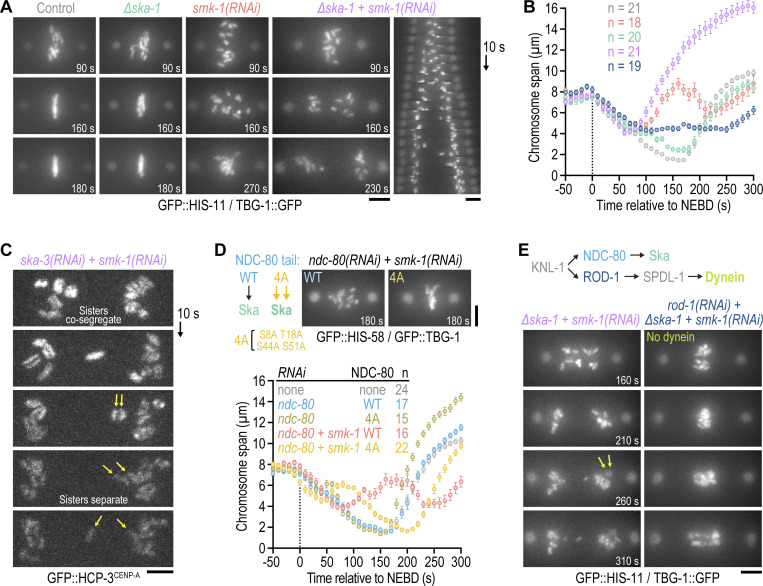
Scattered chromosomes in PP4-inhibited one-cell embryos eventually resume congression, even when initial mono-orientation is exacerbated by co-depletion of KLP-19Kif4 (Fig. 2, A and B; and Video 3), and form bi-oriented attachments capable of sustaining tension (a fraction of these being merotelic). We therefore sought to address the mechanism responsible for this striking recovery. The microtubule-binding Ska complex (a trimer that in C. elegans consists of two SKA-1 subunits and one SKA-3 subunit) has emerged as a conserved attachment maturation factor at kinetochores (Cheerambathur et al., 2017; Hanisch et al., 2006; Janczyk et al., 2017; Schmidt et al., 2012; Welburn et al., 2009). To test whether Ska promotes bi-orientation of scattered chromosomes, we depleted SMK-1 in ∆ska-1 embryos. Loss of SKA-1 on its own caused a very mild chromosome congression defect. By contrast, ∆ska-1;smk-1(RNAi) completely blocked re-congression of scattered chromosomes, which remained close to spindle poles until the end of mitosis (Fig. 5, A and B; and Video 9). Consistent with a persistent mono-oriented state of ∆ska-1;smk-1(RNAi) chromosomes, spindle poles in the co-inhibition separated prematurely without a subsequent recovery of spindle length. The same phenotype was observed in ska-3(RNAi);smk-1(RNAi) embryos, and examination of GFP::HCP-3CENP-A confirmed that sister chromatids co-segregate to the same spindle pole prior to their separation (Fig. 5 C). Consistent with Ska being important for recovery from initial mono-orientation, promoting Ska recruitment to kinetochores by mutation of phosphorylation sites in NDC-80 (Cheerambathur et al., 2017) ameliorated chromosome scattering in smk-1(RNAi) embryos (Fig. 5 D).
Figure 5.
The Ndc80-Ska module is essential for recovery from mono-orientation induced by kinetochore dynein following PP4 inhibition. (A and E) Selected images from time-lapse movies of one-cell embryos co-expressing GFP::histone H2B (HIS-11) and γ-tubulin::GFP (TBG-1). Time is relative to NEBD. A kymograph for ∆ska-1 + smk-1(RNAi) is also shown in A. Cartoon in E shows recruitment hierarchy at the outer kinetochore relevant for this figure. Arrows in E point to separating sister chromatids. Scale bars, 5 µm. (B) Chromosome span (mean of n embryos ± SEM) versus time relative to NEBD. Measurements were performed in embryos such as those shown in A. Conditions are color-coded as in A and E. (C) Successive frames from a time-lapse movie of a one-cell embryo expressing endogenous GFP::CENP-A (HCP-3) to mark centromeres. Top frame corresponds to late prometaphase. Arrows point to a pair of co-segregated sister chromatids which later separate. Scale bar, 2 µm. (D) Top left: Cartoon illustrating that phosphorylation site mutations in NDC-80's N-terminal tail promote Ska recruitment to kinetochores. Top right: Selected images from time-lapse movies of one-cell embryos expressing transgene-encoded WT or mutant (4A) NDC-80 in the background of GFP::histone H2B (HIS-58) and GFP::γ-tubulin (TBG-1). Time is relative to NEBD. Scale bar, 5 µm. Bottom: Chromosome span (mean of n embryos ± SEM) versus time relative to NEBD, measured in embryos such as those shown above the graph.
Video 9.
One-cell embryos co-expressing TBG-1::GFP (γ-tubulin) and GFP::HIS-11 (histone H2B). smk-1(RNAi) (left) and ∆ska-1;smk-1(RNAi) (right), aligned relative to NEBD. Time-lapse is 10 s and playback speed is 6 frames per second. Related to Fig. 5 A.
We next investigated the mechanism leading to mono-orientation of chromosomes in PP4-inhibited embryos that persists when Ska is inhibited. A potential candidate driver of this mono-orientation is kinetochore dynein, whose recruitment requires the RZZ complex (comprised of ROD-1, ZWL-1Zwilch, and CZW-1Zw10) and SPDL-1Spindly (Gassmann et al., 2008; Fig. 5 E). Consistent with this idea, the persistent mono-orientation and sister chromatid co-segregation in ∆ska-1;smk-1(RNAi) embryos was suppressed by depleting ROD-1 to prevent kinetochore dynein recruitment: chromosomes in ∆ska-1;smk-1(RNAi);rod-1(RNAi) embryos remained near the spindle equator and became stretched out along the spindle axis in anaphase (Fig. 5 E and Video 10). Thus, kinetochore dynein is required for chromosome scattering following PP4 inhibition. This likely reflects dynein’s ability to power poleward chromosome movement and/or promote mono-orientation by limiting the formation of bipolar merotelic attachments (Gassmann et al., 2008; Edwards et al., 2018). The prominent stretching of chromosomes in anaphase indeed suggests that most kinetochores are merotelically attached in ∆ska-1;smk-1(RNAi);rod-1(RNAi) embryos (Fig. 5 E).
Video 10.
One-cell embryos co-expressing TBG-1::GFP (γ-tubulin) and GFP::HIS-11 (histone H2B). ∆ska-1;smk-1(RNAi);C09B9.4(RNAi) (left) and ∆ska-1;smk-1(RNAi);rod-1(RNAi) (right), aligned relative to NEBD. C09B9.4(RNAi) serves as the control for double RNAi. Time-lapse is 10 s and playback speed is 6 frames per second. Related to Fig. 5 E.
Collectively, these data establish that the outer kinetochore assembly defect prior to NEBD in PP4-inhibited embryos results in an imbalance of kinetochore-based microtubule-interfacing activities after NEBD, with kinetochore dynein being responsible for aberrant mono-orientation and poleward scattering followed by subsequent bi-orientation mediated by the Ndc80-Ska module. Notably, this imbalance results in extensive mis-segregation, likely due to merotely, highlighting the critical importance of properly timed outer kinetochore assembly for faithful mitosis.
Conclusion
Here we show that nuclear-enriched PP4 is critical for properly timed outer kinetochore assembly and that this in turn is important for the proper order and fidelity of chromosome–microtubule interactions. It is interesting that a phosphatase is important for this function, as mitotic assembly reactions are typically promoted by phosphorylation and reversed by dephosphorylation. While the regulatory EVH1 domain-containing subunit of PP4 complexes is known to interact with the inner kinetochore protein CENP-C, this interaction seems dispensable for timely outer kinetochore assembly. Thus, a major future goal will be to address the key substrate(s) of PP4 whose dephosphorylation underlies this critical event ensuring orderly and accurate execution of mitosis. Our analysis of PP4 has also uncovered the importance of outer kinetochore assembly for sister centromere resolution, a critical event in ensuring properly bi-oriented attachments. Thus, timely outer kinetochore assembly also restrains the frequency of merotelic attachments that are responsible for chromosome mis-segregation. Finally, our results suggest that the Ndc80-Ska module promotes the conversion from mono- to bi-orientation. We propose that this function of Ndc80-Ska is ordinarily masked by mechanisms that ensure rapid bi-orientation in early prometaphase but are suppressed in PP4-inhibited embryos because outer kinetochore assembly is diminished in prophase. For example, polar ejection forces mediated by the chromokinesin KLP-19Kif4 are strongest at NEBD, i.e., when chromosomes are closest to spindle poles (Ke et al., 2009), and are proposed to promote bi-orientation by generating torque on mono-oriented chromosomes such that sister kinetochores come to face opposite poles (Powers et al., 2004). Although the Ndc80-Ska module can compensate for the failure of kinetochores to rapidly engage microtubules at NEBD by promoting bi-orientation later in prometaphase, overreliance on this pathway produces an unacceptable number of attachment errors and is incompatible with proper chromosome segregation. PP4 inhibition therefore demonstrates that outer kinetochore assembly prior to NEBD is crucial for error-free mitosis.
Materials and methods
C. elegans strains
Worm strains (Table S1) were maintained at 20°C on standard nematode growth media (NGM) plates seeded with OP50 bacteria. A Mos1 transposon-based strategy (MosSCI; Frøkjær-Jensen et al., 2012) was used to generate strains stably expressing mCherry::HCP-4 under the control of the mex-5 promoter and tbb-2 3′ UTR for expression in germline cells. Transgenes were cloned into pCFJ352 for insertion on chromosome 1 (ttTi4348 locus) or pCFJ151 for insertion on chromosome 2 (ttTi5605 locus), and transgene integration was confirmed by PCR and sequencing. Other fluorescent markers were subsequently introduced by mating.
The alleles smk-1::3xflag, ∆smk-1, smk-1(Y179A), and ∆pph-4.2 were generated by CRISPR/Cas9-mediated genome editing using single-stranded repair templates (Integrated DNA Technologies) with 50-bp homology regions (Paix et al., 2014). Genomic sequences targeted by guide RNAs are listed in Table S2. The repair template and a ribonucleoprotein (RNP) mixture consisting of crRNA/tracrRNA (Integrated DNA Technologies) and purified recombinant Cas9 enzyme were injected into gonads of young N2 adults. To identify potentially successful edits, animals were co-injected with RNPs to generate the R92C mutation in dpy-10, which causes a dominant roller phenotype (Arribere et al., 2014). Screening was performed by PCR and edits were confirmed by sequencing. The self-excising cassette method (Dickinson et al., 2015) was used to generate smk-1::gfp.
Strains were outcrossed 4–6 times against the wild-type N2 strain to remove potential background mutations, and fluorescent markers were subsequently introduced by mating. Homozygous ∆pph-4.2 animals are fully viable, and smk-1(Y179A) animals exhibit some embryonic lethality but can be propagated in a homozygous state. By contrast, homozygous ∆smk-1 and ∆pph-4.1 animals (F1) grow to adulthood and produce embryos that are inviable (Fig. S1, C and D). ∆smk-1 animals additionally become sterile soon after they start to produce embryos, which are very fragile and remain inside the mother. ∆smk-1 and ∆pph-4.1 were therefore maintained in a heterozygous state using a GFP-marked genetic balancer, and homozygous F1 progeny from heterozygous mothers were identified by the absence of GFP fluorescence in the pharynx.
RNA interference
For production of double-stranded RNA (dsRNA), oligonucleotides with tails containing T3 and T7 promoters (Table S3) were used to amplify regions from wild-type N2 cDNA (ndc-80 and pph-4.1) or genomic DNA (all other genes). PCR products were purified (NucleoSpin Gel and PCR Clean-up, Macherey-Nagel) and used as templates for T3 and T7 transcription reactions (MEGAscript, Invitrogen). Transcription reactions were purified (NucleoSpin RNA Clean-up, Macherey-Nagel) and annealed in 3× soaking buffer (32.7 mM Na2HPO4, 16.5 mM KH2PO4, 6.3 mM NaCl, 14.1 mM NH4Cl). dsRNAs were delivered by injecting L4 hermaphrodites. After injection, animals were kept on NGM plates seeded with OP50 bacteria for 48 h at 20°C before embryo progeny was isolated for analysis. For strain TH66 (Fig. S1 I), injected animals were kept at 25°C for 24–30 h to prevent silencing of the gfp::ebp-2 transgene. As a control for the smk-1(RNAi);rod-1(RNAi) and smk-1(RNAi);ndc-80(RNAi) double depletions (Fig. 5, D and E), smk-1(RNAi) alone was performed with a 1:1 dilution of dsRNA targeting smk-1 with dsRNA targeting C09B9.4, which does not have any obvious function in the one-cell embryo.
Live imaging
Gravid hermaphrodites were dissected in a watch glass filled with a 0.7 × dilution of Egg Salts medium (1 × medium is 118 mM NaCl, 40 mM KCl, 3.4 mM MgCl2, 3.4 mM CaCl2, 5 mM HEPES, pH 7.4). Embryos were mounted on a 2% agarose pad and covered with an 18 × 18 mm coverslip (No. 1.5H; Marienfeld). All imaging was performed in temperature-controlled rooms kept at 20°C. Two microscopes were used: a Zeiss Axio Observer microscope, equipped with an Orca Flash 4.0 camera (Hamamatsu) and a Colibri 2 light source (Zeiss), controlled by ZEN software (Zeiss); and a Nikon Eclipse Ti microscope coupled to an Andor Revolution XD spinning disk confocal system, composed of an iXon Ultra 897 CCD camera (Andor Technology), a solid-state laser combiner (ALC-UVP 350i; Andor Technology), and a CSU-X1 confocal scanner (Yokogawa Electric Corporation), controlled by Andor IQ3 software (Andor Technology).
Image acquisition and analysis
Chromosome span and pole–pole distance
Time-lapse movies of one-cell embryos co-expressing GFP::HIS-58/GFP::TBG-1 (Fig. 1 D, Fig. 2 B, Fig. 3 H, Fig. 4 E, and Fig. 5 D), GFP::HIS-11/TBG-1::GFP (Fig. 5 B), or mCherry::HIS-11/GFP::TBB-2 (Fig. 1 F and Fig. S1 G) were acquired on the Axio Observer microscope with a 63× NA 1.4 Plan-Apochromat objective (Zeiss) at 2 × 2 binning. A 9 × 1.5 µm z-stack was captured every 10 s from just prior to NEBD until chromosome decondensation. Time-lapse movies of one-cell embryos co-expressing SMK-1::GFP/mCherry::HIS-58 (Fig. 4 H) were acquired on the spinning disk confocal microscope with a 60× NA 1.4 Plan-Apochromat objective (Nikon) at 1 × 1 binning. A 9 × 1 µm z-stack was captured every 10 s from just prior to NEBD until chromosome decondensation. Maximum intensity projections of z-stacks were used for measurements in Fiji (Image J version 2.0.0-rc-56/1.53 c).
For chromosome span and pole–pole distance plots, the x and y coordinates of centrosomes, and of the chromosomes closest to each centrosome, were recorded for each frame using the MTrackJ plugin by manually clicking in the center of centrosomes and on the outer edge of chromosomes. Chromosome span was defined as the distance between the two outermost chromosomes measured along the spindle axis. For movies of SMK-1::GFP/mCherry::HIS-58 (Fig. 4 H), which did not have markers for spindle poles, the spindle axis was assumed to be perpendicular to the forming metaphase plate. Tracks from individual embryos were aligned relative to NEBD, defined as the first frame with a decrease in the diffusely nuclear fluorescent histone signal due to equilibration with the cytoplasmic pool. Anaphase onset was defined as the first frame with discernible sister chromatid separation. Replicate profiles were aligned relative to the time of NEBD and plotted as mean ± SEM.
Anaphase chromatin bridges
Time-lapse movies of one-cell embryos co-expressing GFP::HIS-58/GFP::TBG-1 (Fig. 1 B), mCherry::HIS-11/GFP::TBB-2 (Fig. 1 E; Fig. S1 F), or mCherry::HIS-58/SMK-1::GFP (Fig. 4 F) were acquired on the spinning disk confocal microscope with a 60× NA 1.4 Plan-Apochromat objective (Nikon) at 1 × 1 binning. A 9 × 1 µm z-stack was captured every 10 s from just prior to NEBD until chromosome decondensation, and maximum intensity projections were analyzed in Fiji. Anaphases were scored as having chromatin bridges if fluorescent histone signal connecting the two masses of segregating chromosomes was visible three frames after the onset of sister chromatid separation.
Chromosomal and nuclear levels
Time-lapse movies of one-cell embryos expressing GFP::HCP-3/mCherry::HIS-58, GFP::HCP-4/mCherry::HIS-58, KNL-1::mCherry/GFP::HIS-58, HIM-10::GFP/mCherry::HIS-58, GFP::SPDL-1/mCherry::HIS-58, HIM-1::GFP/mCherry::HIS-58, or SMK-1::GFP/mCherry::HIS-58 were acquired on the spinning disk confocal microscope with a 60× NA 1.4 Plan-Apochromat objective (Nikon) at 1 × 1 binning. A 9 × 1 µm z-stack was captured every 10 s from just prior to NEBD until chromosome decondensation, and maximum intensity projections were analyzed in Fiji.
For analysis of chromosomal levels at NEBD (Fig. 2 H, left; Fig. S3 B), the integrated intensity was measured within a region corresponding to the area occupied by the nucleus, as well as within three smaller intranuclear regions that cumulatively covered a large fraction of the nuclear area not occupied by chromosomes (nuclear background). The chromosomal signal was calculated by subtracting the integrated nuclear background intensity (multiplied by the ratio of nuclear region area to background region area) from the integrated intensity of the nuclear region. The diffuse non-chromosomal nuclear signal (Fig. 4 G and Fig. S2 A) was calculated by subtracting the mean background intensity measured in cytoplasmic regions adjacent to the nucleus from the mean intensity of the three intranuclear regions not occupied by chromosomes. Signal intensities of replicates were averaged, normalized to the average of the respective control, and plotted as mean ± 95% CI.
For analysis of chromosomal levels just prior to anaphase onset (Fig. 2 H, right; Fig. S3, A and B), defined as one to three frames before sister chromatid separation, the integrated intensity was determined in the region encompassing the chromosomes. The chromosomal region was expanded by 2 pixels, and the difference in integrated intensity between the expanded and chromosomal region was used to define the background intensity. The chromosomal signal was calculated by subtracting the integrated background intensity (multiplied by the ratio of chromosomal region area to background region area) from the integrated intensity of the chromosomal region. Signal intensities of replicates were averaged, normalized to the average of the respective control, and plotted as mean ± 95% CI.
For analysis of chromosomal signal over time (Fig. S2 D), the “Find Maxima” function in Fiji was used to identify the top 15 local maxima in the region containing the chromosomes, and the values were averaged. The averaged intensity of the top 15 local maxima of an adjacent cytoplasmic region was subtracted as background. Signal intensities of replicate time-lapse series were averaged, normalized to the peak signal within the time-lapse series of the respective control, aligned relative to the time of NEBD, and plotted as mean ± SEM.
Sister centromere resolution
Time-lapse movies of one-cell embryos expressing endogenous GFP::HCP-3 (Fig. 3 D; Fig. S2 E, left; Fig. S3 E) or transgenic GFP::HCP-3 in an hcp-3 knock-out background (Fig. 3 F; Fig. S2 E, right) were acquired on the spinning disk confocal microscope with a 100× NA 1.45 Plan-Apochromat objective (Nikon) at 1 × 1 binning. A 9 × 1 µm z-stack was captured every 10 s from just prior to NEBD until chromosome decondensation, and maximum intensity projections were analyzed in Fiji. Measurements were made in the first six frames following NEBD, and, in PP4 inhibitions, also later in prometaphase when chromosomes had scattered.
For line scan plots (Fig. 3, D and F; Fig. S3 E), a 2-µm-long and 3-pixel-wide line, drawn perpendicularly to the long axis of an individual chromosome, was centered on the chromosome midline and the fluorescence profile was determined using the “Plot Profile” function in Fiji. Signal intensities were normalized to the maximum intensity within each profile. Profiles of replicates were averaged and plotted as mean ± 95% CI.
For signal ratio plots (Fig. S2 E and Fig. S3 E), the intensity at the chromosome midline was divided by the maximum intensity within the line scan. Ratios of replicates were averaged, normalized to the average ratio of the respective control, and plotted as mean ± 95% CI.
Chromosome size
Z-stacks with step size 0.1 µm were acquired in one-cell embryos co-expressing GFP::HIS-58/GFP::TBG-1 on the spinning disk confocal microscope with a 100× NA 1.45 Plan-Apochromat objective (Nikon) at 1 × 1 binning in late prophase or early prometaphase. Z-stacks were visualized in 3D using Imaris 9.5.0 (Bitplane). Chromosome length and width measurements (Fig. S2 G) were made by visually delimiting chromosomes and assigning points at the extremities and in the middle of individual chromosomes. Replicate measurements were averaged and plotted as mean ± 95% CI.
Microtubule nucleation
Time-lapse movies of one-cell embryos expressing GFP::EBP-2 were acquired on the spinning disk confocal microscope with a 100× NA 1.45 Plan-Apochromat objective (Nikon) at 1 × 1 binning. A single central z-section that included both centrosomes was captured every 200 ms at metaphase for a total of 1 min.
For analysis of microtubule nucleation rates (Fig. S1 I), an arc positioned 30 pixels away from the centrosome was drawn using the segmented line tool in Fiji, and a kymograph of at least 200 frames was generated using the Fiji Multi kymograph plugin. GFP::EBP-2 puncta on the kymograph were manually counted and the number was divided by the duration of imaging. Replicate measurements were averaged and plotted as mean ± 95% CI.
Embryonic viability and brood size
Brood size (Fig. S1 C) and embryonic viability assays (Fig. S1 D) were performed at 20°C. Wild-type N2 L4 hermaphrodites, injected with dsRNA, or untreated L4 hermaphrodites of homozygous mutants, were grown for 40 h on NGM plates containing OP50 bacteria. Single adults were then placed on new plates with a small amount of OP50, removed 8 h later, and the plates were incubated for another 16–24 h to give viable embryos time to hatch. Embryonic viability was calculated by dividing the number of hatched larvae by the total number of progeny on the plate. Replicate counts were averaged and plotted as mean percentage ± 95% CI.
Plasmids for recombinant protein expression
cDNA encoding for full-length HCP-4 (UniProt entry G5EDA3) was inserted into a 2CT vector (N-terminal 6xHis::maltose binding protein [MBP] followed by a TEV protease cleavage site and C-terminal StrepTagII), and cDNA encoding for SMK-1 (UniProt entry H2KYN6; residues 146–282) was inserted into pGEX-6P-1 (N-terminal glutathione S-transferase [GST] followed by a Prescission protease cleavage site) containing C-terminal 6xHis.
Recombinant protein expression and purification
Plasmids were transformed into E. coli strains BL21 or Rosetta. Expression was induced at an OD600 of 0.9 with 0.1 mM IPTG. After expression overnight at 18°C, cells were harvested by centrifugation at 4,000 × g for 20 min.
For purification of 6xHis::MBP::HCP-4::StrepTagII, bacterial pellets were resuspended in lysis buffer A (50 mM HEPES, 250 mM NaCl, 10 mM imidazole, 1 mM DTT, 1 mM PMSF, 2 mM benzamidine-HCl, pH 8.0), lysed by sonication, and cleared by centrifugation at 40,000 × g for 45 min at 4°C. Proteins were purified by tandem affinity chromatography using HisPur Ni-NTA resin (Thermo Fisher Scientific) followed by strep-tactin sepharose resin (IBA). Ni-NTA resin was incubated in batch with the cleared lysate in the presence of 0.1% (v/v) Tween 20 for 1 h at 4°C and washed with wash buffer A (25 mM HEPES, 250 mM NaCl, 25 mM imidazole, 0.1% Tween 20, 1 mM DTT, 2 mM benzamidine-HCl, pH 8.0). Proteins were eluted on a gravity column with elution buffer A (50 mM HEPES, 150 mM NaCl, 250 mM imidazole, 1 mM DTT, 2 mM benzamidine-HCl, pH 8.0). Fractions containing the recombinant protein were pooled, incubated in batch with strep-tactin sepharose resin for 1 h at 4°C, and washed with wash buffer B (25 mM HEPES, 250 mM NaCl, 0.1% Tween 20, 1 mM DTT, 2 mM benzamidine-HCl, pH 8.0). Proteins were eluted on a gravity column with elution buffer B (100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 10 mM desthiobiotin, pH 8.0), and the eluate was dialyzed against storage buffer (25 mM HEPES, 150 mM NaCl, pH 7.5). Glycerol and DTT were added to a final concentration of 10% (v/v) and 1 mM, respectively, and aliquots were flash frozen in liquid nitrogen and stored at −80°C.
For purification of GST::SMK-1(146–282)::6xHis, bacterial pellets were resuspended in lysis buffer B (50 mM HEPES, 250 mM NaCl, 10 mM EDTA, 10 mM EGTA, 1 mM DTT, 1 mM PMSF, 2 mM benzamidine-HCl, pH 8.0), lysed by sonication, and cleared by centrifugation at 40,000 × g for 45 min at 4°C. Proteins were purified by tandem affinity chromatography using glutathione agarose resin (Thermo Fisher Scientific) followed by Ni-NTA resin. Glutathione agarose resin was incubated in batch with the cleared lysate in the presence of 0.1% (v/v) Tween 20 for 1 h at 4°C, washed with wash buffer C (25 mM HEPES, 250 mM NaCl, 0.1% Tween 20, 1 mM DTT, 2 mM benzamidine-HCl, pH 8.0), and proteins were eluted on a gravity column with elution buffer C (50 mM HEPES, 150 mM NaCl, 10 mM reduced L-glutathione, 1 mM DTT, 2 mM benzamidine-HCl, pH 8.0). Fractions containing the proteins were pooled and further purified by size exclusion chromatography using a Superose 6 10/300 column (GE Healthcare) equilibrated with storage buffer. Glycerol and DTT were added to a final concentration of 10% (v/v) and 1 mM, respectively, and aliquots were flash-frozen in liquid nitrogen and stored at −80°C.
GST pull-down
50 pmol of purified GST::SMK-1(146–282)::6xHis were incubated with 50 pmol of purified 6xHis::MBP::HCP-4::StrepTagII for 1 h at 4°C in 150 μl pull-down buffer (50 mM HEPES, 100 mM NaCl, 5 mM DTT, pH 7.5) containing 0.1% Tween 20 and supplemented with 15 μl of glutathione agarose resin. After washing the resin with 3 × 500 μl pull-down buffer, proteins were eluted with 50 µl pull-down buffer containing 15 mM reduced L-glutathione. Eluted proteins were separated by SDS-PAGE and visualized by Coomassie Blue staining.
Immunoblotting
For immunoblots of C. elegans lysate, ∼60 adult hermaphrodites were collected into 500 μl M9 buffer and washed 2 × with M9 buffer and 2 × with M9 buffer containing 0.05% Triton X-100. To 100 μl of worm suspension, 33 μl 4× SDS-PAGE sample buffer (250 mM Tris-HCl, pH 6.8, 30% [v/v] glycerol, 8% [w/v] SDS, 200 mM DTT and 0.04% [w/v] bromophenol blue) and ∼20 μl of glass beads were added. Samples were incubated for 3 min at 95°C and vortexed for 2 × 5 min with intermittent heating. After centrifugation at 20,000 × g for 1 min at room temperature, proteins in the supernatant were resolved by 10% SDS-PAGE and transferred to a 0.2-μm nitrocellulose membrane (Hybond ECL, Amersham Pharmacia Biotech). Membranes were rinsed 3 × with TBS (50 mM Tris-HCl, 145 mM NaCl, pH 7.6), blocked with 5% non-fat dry milk in TBST (TBS containing 0.1% Tween 20), and probed at 4°C overnight with mouse monoclonal anti-FLAG M2 (F3165, 1:1,000; Sigma-Aldrich) and mouse monoclonal anti-α-tubulin B512 (T5168, 1:5,000; Sigma-Aldrich). The membrane was washed 3× with TBST, incubated with goat polyclonal anti-mouse IgG antibody coupled to HRP (115-035-044, 1:10,000; Jackson ImmunoResearch) for 1 h at room temperature, and washed again 3× with TBST. Proteins were detected by chemiluminescence using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) and x-ray film (GE Healthcare).
Statistical analysis
Statistical analysis was performed with Prism 9.0 software (GraphPad). Statistical significance was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test, or by a two-sided Mann-Whitney test, where ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, and ns = not significant, P > 0.05. The analytical method used is specified in the figure legends.
Online supplemental material
Fig. S1 shows results related to Fig. 1 (description of PP4 mutants). Fig. S2 shows results related to Fig. 2 (nuclear levels of kinetochore components and kinetochore dynein localization) and Fig. 3. (quantification of sister centromere resolution and mitotic chromosome morphology). Fig. S3 shows results related to Fig. 3 (cohesin levels on mitotic chromosomes) and Fig. 4 (validation of RNAi penetrance for molecular replacement and further characterization of hcp-4 mutants). Table S1 lists C. elegans genotypes. Table S2 lists sequences targeted by CRISPR/Cas9. Table S3 list oligonucleotides for dsRNA production. Video 1 shows embryos co-expressing GFP::TBG-1 and GFP::HIS-58 related to Fig. 1 B. Video 2 shows embryos co-expressing GFP::TBB-2 and mCherry::HIS-11 related to Fig. 1 E. Video 3 shows embryos co-expressing GFP::TBG-1 and GFP::HIS-58 related to Fig. 2 A. Video 4 shows embryos expressing GFP::HCP-4 related to Fig. 2 E. Video 5 shows embryos expressing HIM-10::GFP related to Fig. 2 G. Video 6 shows an embryo co-expressing SMK-1::GFP and mCherry::HIS-58 related to Fig. 2 I. Video 7 shows embryos co-expressing GFP::HCP-3 and mCherry::HIS-58 related to Fig. 3 A. Video 8 shows embryos co-expressing SMK-1::GFP or SMK-1::GFP(Y179A) together with mCherry::HIS-58 related to Fig. 4 F. Video 9 shows embryos co-expressing TBG-1::GFP and GFP::HIS-11 related to Fig. 5 A. Video 10 shows embryos co-expressing TBG-1::GFP and GFP::HIS-11 related to Fig. 5 E.
Supplementary Material
lists C. elegans strains.
lists genomic sequences targeted by gRNAs for CRISPR/Cas9-mediated genome editing.
lists oligonucleotides for dsRNA production.
is the source file for Fig. 4.
Acknowledgments
The authors thank the Biochemical and Biophysical Technologies Scientific Platform at i3S for support. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440).
This study was funded by the European Research Council under the European Union’s Seventh Framework Programme (ERC grant agreement no. ERC-2013-StG-338410-DYNEINOME), by the Fundação para a Ciência e a Tecnologia (FCT)/Ministério da Ciência, Tecnologia e Ensino Superior through project grant PTDC/BIA-CEL/30507/2017, and by National Funds through FCT under the project UIDB/04293/2020. R. Gassmann is supported by FCT Principal Investigator position CEECIND/00333/2017. H. Rocha was supported by FCT PhD fellowship SFRH/BD/103495/2014.
Author Contributions: Conceptualization: R. Gassmann, J. Dumont. Formal analysis: H. Rocha, R. Gassmann. Funding acquisition: R. Gassmann, A. Desai. Investigation: H. Rocha, P.A. Simões, J. Budrewicz. Project administration: R. Gassmann, A. Desai. Resources: R. Gassmann, P. Lara-Gonzalez, A.X. Carvalho, A. Desai. Supervision: R. Gassmann, A.X. Carvalho, A. Desai, P. Lara-Gonzalez. Validation: H. Rocha, P.A. Simões, A.X. Carvalho, A. Desai, R. Gassmann. Visualization: R. Gassmann, H. Rocha, P.A. Simões. Writing - original draft: R. Gassmann, A. Desai, H. Rocha, A.X. Carvalho. Writing - review & editing: R. Gassmann, A. Desai, H. Rocha, A.X. Carvalho, J. Dumont, P. Lara-Gonzalez, P.A. Simões.
References
- Arribere, J.A., Bell R.T., Fu B.X.H., Artiles K.L., Hartman P.S., and Fire A.Z.. 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 198:837–846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa, D.J., Duro J., Prevo B., Cheerambathur D.K., Carvalho A.X., and Gassmann R.. 2017. Dynactin binding to tyrosinated microtubules promotes centrosome centration in C. elegans by enhancing dynein-mediated organelle transport. PLoS Genet. 13:e1006941. 10.1371/journal.pgen.1006941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batty, P., and Gerlich D.W.. 2019. Mitotic chromosome mechanics: How cells segregate their genome. Trends Cell Biol. 29:717–726. 10.1016/j.tcb.2019.05.007 [DOI] [PubMed] [Google Scholar]
- Brewis, N.D., Street A.J., Prescott A.R., and Cohen P.T.. 1993. PPX, a novel protein serine/threonine phosphatase localized to centrosomes. EMBO J. 12:987–996. 10.1002/j.1460-2075.1993.tb05739.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur, D.K., Prevo B., Hattersley N., Lewellyn L., Corbett K.D., Oegema K., and Desai A.. 2017. Dephosphorylation of the Ndc80 tail stabilizes kinetochore-microtubule attachments via the Ska complex. Dev. Cell. 41:424–437.e4. 10.1016/j.devcel.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur, D.K., Prevo B., Chow T.-L., Hattersley N., Wang S., Zhao Z., Kim T., Gerson-Gurwitz A., Oegema K., Green R., and Desai A.. 2019. The kinetochore-microtubule coupling machinery is repurposed in sensory nervous system morphogenesis. Dev. Cell. 48:864–872.e7. 10.1016/j.devcel.2019.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., Niessen S., Anderson S., Hyndman F., Yates J.R. III, Oegema K., and Desai A.. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268. 10.1101/gad.1234104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., Rybina S., Müller-Reichert T., Shevchenko A., Shevchenko A., Hyman A., and Oegema K.. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435. 10.1101/gad.1126303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, D.J., Pani A.M., Heppert J.K., Higgins C.D., and Goldstein B.. 2015. Streamlined genome engineering with a self-excising drug selection cassette. Genetics. 200:1035–1049. 10.1534/genetics.115.178335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duro, J., and Nilsson J.. 2020. SAC during early cell divisions: Sacrificing fidelity over timely division, regulated differently across organisms. BioEssays. 29:2000174. 10.1002/bies.202000174 [DOI] [PubMed] [Google Scholar]
- Edwards, F., Maton G., Gareil N., Canman J.C., and Dumont J.. 2018. BUB-1 promotes amphitelic chromosome biorientation via multiple activities at the kinetochore. Elife. 7:e40690. 10.7554/eLife.40690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeut, J., Cheerambathur D.K., Krenning L., Oegema K., and Desai A.. 2012. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J. Cell Biol. 196:469–482. 10.1083/jcb.201111107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen, C., Davis M.W., Ailion M., and Jorgensen E.M.. 2012. Improved mos1-mediated transgenesis in C. elegans. Nat. Methods. 9:117–118. 10.1038/nmeth.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann, R., Essex A., Hu J.-S., Maddox P.S., Motegi F., Sugimoto A., O’Rourke S.M., Bowerman B., McLeod I., Yates J.R. III, et al. 2008. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the rod/Zwilch/Zw10 complex. Genes Dev. 22:2385–2399. 10.1101/gad.1687508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, J.-E., Tavernier N., Richaudeau B., Formstecher E., Boulin T., Mains P.E., Dumont J., and Pintard L.. 2013. Microtubule severing by the katanin complex is activated by PPFR-1-dependent MEI-1 dephosphorylation. J. Cell Biol. 202:431–439. 10.1083/jcb.201304174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill, S.W., Howard J., Schäffer E., Stelzer E.H.K., and Hyman A.A.. 2003. The distribution of active force generators controls mitotic spindle position. Science. 301:518–521. 10.1126/science.1086560 [DOI] [PubMed] [Google Scholar]
- Guo, H., Stamper E.L., Sato-Carlton A., Shimazoe M.A., Li X., Zhang L., Stevens L., Tam K.C.J., Dernburg A.F., and Carlton P.M.. 2022. Phosphoregulation of DSB-1 mediates control of meiotic double-strand break activity. Elife. 11:e77956. 10.7554/eLife.77956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, X., Gomes J.-E., Birmingham C.L., Pintard L., Sugimoto A., and Mains P.E.. 2009. The role of protein phosphatase 4 in regulating microtubule severing in the Caenorhabditis elegans embryo. Genetics. 181:933–943. 10.1534/genetics.108.096016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch, A., Silljé H.H.W., and Nigg E.A.. 2006. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 25:5504–5515. 10.1038/sj.emboj.7601426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie, C.J., Carnegie G.K., Morrice N., and Cohen P.T.W.. 2000. A novel 50 kDa protein forms complexes with protein phosphatase 4 and is located at centrosomal microtubule organizing centres. Biochem. J. 347:845–855. 10.1042/bj3470845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helps, N.R., Brewis N.D., Lineruth K., Davis T., Kaiser K., and Cohen P.T.. 1998. Protein phosphatase 4 is an essential enzyme required for organisation of microtubules at centrosomes in Drosophila embryos. J. Cell Sci. 111:1331–1340. 10.1242/jcs.111.10.1331 [DOI] [PubMed] [Google Scholar]
- Janczyk, P.Ł., Skorupka K.A., Tooley J.G., Matson D.R., Kestner C.A., West T., Pornillos O., and Stukenberg P.T.. 2017. Mechanism of Ska recruitment by Ndc80 complexes to kinetochores. Dev. Cell. 41:438–449.e4. 10.1016/j.devcel.2017.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karman, Z., Rethi-Nagy Z., Abraham E., Fabri-Ordogh L., Csonka A., Vilmos P., Debski J., Dadlez M., Glover D.M., and Lipinszki Z.. 2020. Novel perspectives of target-binding by the evolutionarily conserved PP4 phosphatase. Open Biol. 10:200343. 10.1098/rsob.200343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, K., Cheng J., and Hunt A.J.. 2009. The distribution of polar ejection forces determines the amplitude of chromosome directional instability. Curr. Biol. 19:807–815. 10.1016/j.cub.2009.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez, P., Pines J., and Desai A.. 2021. Spindle assembly checkpoint activation and silencing at kinetochores. Semin. Cell Dev. Biol. 117:86–98. 10.1016/j.semcdb.2021.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinszki, Z., Lefevre S., Savoian M.S., Singleton M.R., Glover D.M., and Przewloka M.R.. 2015. Centromeric binding and activity of protein phosphatase 4. Nat. Commun. 6:5894. 10.1038/ncomms6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Granados, C., Philp A., Oxenham S.K., Prescott A.R., and Cohen P.T.W.. 2008. Depletion of protein phosphatase 4 in human cells reveals essential roles in centrosome maturation, cell migration and the regulation of Rho GTPases. Int. J. Biochem. Cell Biol. 40:2315–2332. 10.1016/j.biocel.2008.03.021 [DOI] [PubMed] [Google Scholar]
- Moore, L.L., and Roth M.B.. 2001. HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J. Cell Biol. 153:1199–1208. 10.1083/jcb.153.6.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, L.L., Stanvitch G., Roth M.B., and Rosen D.. 2005. HCP-4/CENP-C promotes the prophase timing of centromere resolution by enabling the centromere association of HCP-6 in Caenorhabditis elegans. Mol. Cell. Biol. 25:2583–2592. 10.1128/MCB.25.7.2583-2592.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura, M., and Conde C.. 2019. Phosphatases in mitosis: Roles and regulation. Biomolecules. 9:55. 10.3390/biom9020055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio, A., and Desai A.. 2017. A Molecular view of kinetochore assembly and function. Biology (Basel). 6:5–47. 10.3390/biology6010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, A.P., and Cheeseman I.M.. 2021. Kinetochore assembly throughout the cell cycle. Semin. Cell Dev. Biol. 117:62–74. 10.1016/j.semcdb.2021.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ngoc, T., Afshar K., and Gönczy P.. 2007. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat. Cell Biol. 9:1294–1302. 10.1038/ncb1649 [DOI] [PubMed] [Google Scholar]
- Nilsson, J. 2019. Protein phosphatases in the regulation of mitosis. J. Cell Biol. 218:395–409. 10.1083/jcb.201809138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema, K., Desai A., Rybina S., Kirkham M., and Hyman A.A.. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. 10.1083/jcb.153.6.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix, A., Wang Y., Smith H.E., Lee C.-Y.S., Calidas D., Lu T., Smith J., Schmidt H., Krause M.W., and Seydoux G.. 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics. 198:1347–1356. 10.1534/genetics.114.170423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J., and Lee D.-H.. 2020. Functional roles of protein phosphatase 4 in multiple aspects of cellular physiology: A friend and a foe. BMB Rep. 53:181–190. 10.5483/BMBRep.2020.53.4.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson, J.R., Hudson D.F., Cisneros-Soberanis F., and Earnshaw W.C.. 2021. Mitotic chromosomes. Semin. Cell Dev. Biol. 117:7–29. 10.1016/j.semcdb.2021.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, J., Rose D.J., Saunders A., Dunkelbarger S., Strome S., and Saxton W.M.. 2004. Loss of KLP-19 polar ejection force causes misorientation and missegregation of holocentric chromosomes. J. Cell Biol. 166:991–1001. 10.1083/jcb.200403036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha, H., Maia A.F., and Gassmann R.. 2018. A genome-scale RNAi screen for genetic interactors of the dynein co-factor nud-2 in Caenorhabditis elegans. Sci. Data. 5:180047. 10.1038/sdata.2018.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato-Carlton, A., Li X., Crawley O., Testori S., Martinez-Perez E., Sugimoto A., and Carlton P.M.. 2014. Protein phosphatase 4 promotes chromosome pairing and synapsis, and contributes to maintaining crossover competence with increasing age. PLoS Genet. 10:e1004638. 10.1371/journal.pgen.1004638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, J.C., Arthanari H., Boeszoermenyi A., Dashkevich N.M., Wilson-Kubalek E.M., Monnier N., Markus M., Oberer M., Milligan R.A., Bathe M., et al. 2012. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell. 23:968–980. 10.1016/j.devcel.2012.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, I., Zhou X., Chernobrovkin A., Puerta-Cavanzo N., Kanno T., Salignon J., Stoehr A., Lin X.X., Baskaner B., Brandenburg S., et al. 2020. DAF-16/FOXO requires Protein Phosphatase 4 to initiate transcription of stress resistance and longevity promoting genes. Nat. Commun. 11:138. 10.1038/s41467-019-13931-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stear, J.H., and Roth M.B.. 2002. Characterization of HCP-6, a C. elegans protein required to prevent chromosome twisting and merotelic attachment. Genes Dev. 16:1498–1508. 10.1101/gad.989102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyoshi, E., Sugimoto A., and Yamamoto M.. 2002. Protein phosphatase 4 is required for centrosome maturation in mitosis and sperm meiosis in C. elegans. J. Cell Sci. 115:1403–1410. 10.1242/jcs.115.7.1403 [DOI] [PubMed] [Google Scholar]
- Torras-Llort, M., Medina-Giró S., Escudero-Ferruz P., Lipinszki Z., Moreno-Moreno O., Karman Z., Przewloka M.R., and Azorín F.. 2020. A fraction of barrier-to-autointegration factor (BAF) associates with centromeres and controls mitosis progression. Commun. Biol. 3:454. 10.1038/s42003-020-01182-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki, Y., Kruse T., Weisser M.B., Sundell G.N., Larsen M.S.Y., Mendez B.L., Jenkins N.P., Garvanska D.H., Cressey L., Zhang G., et al. 2019. A consensus binding motif for the PP4 protein phosphatase. Mol. Cell. 76:953–964.e6. 10.1016/j.molcel.2019.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn, J.P.I., Grishchuk E.L., Backer C.B., Wilson-Kubalek E.M., Yates J.R. III, and Cheeseman I.M.. 2009. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev. Cell. 16:374–385. 10.1016/j.devcel.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, S., Ma H., Burch D., Maciel G.A., Hunter T., and Dillin A.. 2006. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 124:1039–1053. 10.1016/j.cell.2005.12.042 [DOI] [PubMed] [Google Scholar]
- Yamamoto, T.G., Watanabe S., Essex A., and Kitagawa R.. 2008. SPDL-1 functions as a kinetochore receptor for MDF-1 in Caenorhabditis elegans. J. Cell Biol. 183:187–194. 10.1083/jcb.200805185 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists C. elegans strains.
lists genomic sequences targeted by gRNAs for CRISPR/Cas9-mediated genome editing.
lists oligonucleotides for dsRNA production.
is the source file for Fig. 4.