Abstract
Nucleic acid amplification test (NAAT) is the gold standard for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection. However, genetic mutations in the virus can affect the result. Cycle threshold (Ct) values of N genes and their association with mutations using SARS-CoV-2 positive specimens diagnosed by the Xpert Xpress SARS-CoV-2 were examined in this study. In total, 196 nasopharyngeal swab specimens were tested for SARS-CoV-2 infection using the Xpert Xpress SARS-CoV-2, and 34 were positive. WGS was performed for four outlier samples with increased ΔCt identified by Scatterplot analysis as well as seven control samples without increased ΔCt in the Xpert Xpress SARS-CoV-2. The presence of the G29179T mutation was identified as a cause of increased ΔCt. PCR using the Allplex™ SARS-CoV-2 Assay did not show a similar increase in ΔCt. Previous reports focusing on N-gene mutations and their effects on SARS-CoV-2 testing including the Xpert Xpress SARS-CoV-2 were also summarized. While a single mutation that impacts one target of a multiplex NAAT is not a true detection failure, mutation compromising NAAT target region can cause confusion of the results and render the assay susceptible to diagnostic failure.
Keywords: SARS-CoV-2, G29179T, Single nucleotide polymorphism, Cycle threshold, Whole genome sequencing
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is spreading and re-emerging worldwide with multiple genetic mutations. It is an approximately 30 kb RNA virus comprising genes of open reading frame (ORF) 1a, ORF-1b, surface glycoprotein (S), envelope protein (E), matrix protein (M), other proteins such as nucleocapsid protein (N), and some nonstructural proteins (NSP) (Lu et al., 2020). ORF1b encodes 16 NSPs among which NSP12 and NSP15 encode RNA-dependent RNA polymerase (RdRp) and RNA helicase, respectively (Alanagreh et al., 2020). Because ORF1b contains genes to repair transcriptional errors, mutations in SARS-CoV-2 are less frequent compared with other RNA viruses. Although the mutation rate in SARS-CoV-2 is different by variants (Alpha; 28.3/year, Delta; 25.8/year, Omicron; 30.5/year), mutations in 29.5 base pairs have been estimated to occur annually overall (Nextstrain,, Wolf et al., 2023). Mutation has gained attention in terms of infectivity, pathogenicity, and vaccine effectiveness, but its impact on diagnostic tests is also important (Drain, 2022).
Nucleic acid amplification tests (NAATs), such as polymerase chain reaction (PCR), are gold standard methods for the diagnosis of coronavirus disease 2019 (COVID-19) (Hansen et al., 2021). Recently, many reagents and systems for SARS-CoV-2 detection have been developed, and several regions of its genome have been targeted. The number and region of target genes varied according to each protocol. Among several target regions, the sequence of the N gene is highly conserved among coronaviruses, with a low probability of point mutation and it is generally considered to be the most suitable target for in vitro diagnostic detection (Álvarez-Díaz et al., 2020, Dutta et al., 2020, Zóka and Bekő, 2020, Oliveira et al., 2020, Rahman et al., 2021, Wang et al., 2022). However, mutations in the N gene can reportedly affect NAAT results, such as give false negatives and delay cycle threshold (Ct) values. Ziegler et al. reported a C29200T single nucleotide polymorphism (SNP) compromising the N gene target of the Xpert Xpress (Cepheid, Sunnyvale, United States) (Ziegler et al., 2020). In this study, Ct values of N genes and their association with mutations using SARS-CoV-2 positive specimens diagnosed by the Xpert Xpress SARS-CoV-2 were investigated.
2. Materials and methods
2.1. Clinical specimens and ethics
Nasopharyngeal swabs collected from patients tested with the Xpert Xpress SARS-CoV-2 from August 7, 2020, to June 2, 2021, at the University of the Ryukyus Hospital, were used in this study. For the Xpert Xpress SARS-CoV-2 positive samples, the Ct values of the E gene (CtE) and N2 gene (CtN2) of each positive sample were checked and the numerical difference between these two Ct values, ΔCt (CtN2 − CtE), was calculated. The need for informed consent from each patient for inclusion in this study was waived because this study was retrospective in approach, which caused no additional adverse events in any subject. Patients were given the opportunity for opt-out via website of the Division of Infectious, Respiratory, and Digestive Medicine, University of the Ryukyus Graduate School of Medicine. This study was approved by the Institutional Review Board of the University of Ryukyus (approval number: 1862).
2.2. PCR
Viral RNA was extracted from the nasal swabs stored in a deep freezer using magLEAD 12gC (Precision System Science, Chiba, Japan). To ensure N gene detection in another PCR assay, additionally extracted RNA was subjected to real-time PCR with primer sets developed by the Allplex™ SARS-CoV-2 Assay (Seegene, Seoul, South Korea) which targets the E, N, and RdRp/S genes, according to the manufacturer’s instructions using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The results of the Xpert Xpress SARS-CoV-2 and the Allplex™ SARS-CoV-2 Assay for these samples were compared. Additionally, Novaplex™ SARS-CoV-2 Variants I Assay (Seegene), which can test N501Y, E484K mutation and H69/V70 deletion, was used for identification of SARS-CoV-2 variants.
2.3. Whole genome sequences (WGS)
To determine the genome sequences of SARS-CoV-2, next-generation sequencing libraries of the collected samples were prepared using the hybridization capture-based target enrichment method. Extracted RNA was transformed into double-stranded cDNA using SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) and NEBNext Ultra II Non-Directional RNA Second Strand Synthesis Module (New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s protocols. Sequencing libraries were generated using the SureSelectXT Low Input (Agilent Technologies, Santa Clara, CA, USA) with a custom panel targeting SARS-CoV-2 sequences. Paired-end sequencing with a read length of 2 × 151 bp was performed on a MiSeq platform (Illumina, San Diego, CA, USA). After trimming the adapter sequences using Cutadapt version 3.2, sequence reads were aligned to the reference genome of SARS-CoV-2 (GenBank accession number: NC_045512.2) using BWA version 0.7.17. After marking duplicate reads in BAM files using Samtools version 1.11 and Picard in GATK 4.2.0.0, variant calling was executed using Mutect2 in GATK 4.2.0.0. Consensus sequences were obtained using bcftools version 1.9.
2.4. Statistical analysis
Continuous variables were compared using Wilcoxon/Kruskal–Wallis test. Statistical significance was set at p < 0.05. Statistical analysis was performed using JMP Pro 15 (SAS Institute Inc., Cary, NC, USA).
3. Results
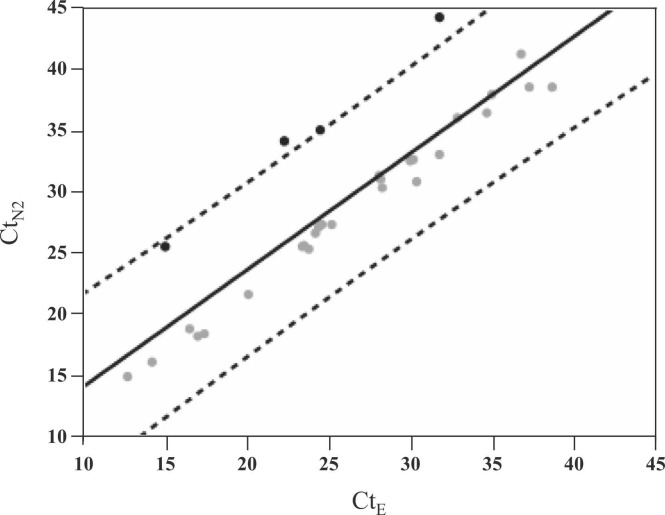
During the study period, 196 nasopharyngeal swab specimens were tested for SARS-CoV-2 using the Xpert Xpress SARS-CoV-2 and 34 were positive. Supplementary Table 1 shows the details of the positive samples with Ct values. CtE values were not available in 4 samples (Sample No. 11, 12, 22, 28) due to the low viral loads below the detection limit. For the remaining 30 samples where both E and N2 genes were detectable, the average of CtE, CtN2, and ΔCt (CtN2 − CtE) were 26.2, 29.4, and 3.2, respectively. Scatter plots of the 30 samples generated by CtE and CtN2 identified 4 outlier samples (Sample No. 30, 32, 33, 34) with an increase in ΔCt plotted around the dashed line indicating + 2-standard deviation (SD) ( Fig. 1). These 4 samples with increased ΔCt values were all collected between May and June 2021.
Fig. 1.
Scatterplot of Ct values for N2 gene and E gene in the Xpert Xpress positive samples. Regression line was drawn as solid line, and confidence intervals for individual values [α = 0.05, ± 2 SD] were drawn as dashed lines. Four samples were located near the dashed line on the + 2 SD side, indicating an increase in ΔCt.
To genetically investigate the cause of the increased ΔCt, WGS was performed in these 4 samples, as well as 7 control samples (Sample No. 20, 23, 24, 25, 27, 29, 31) with low Ct values available for WGS collected near the above 4 samples. Among the N-gene region SNPs, G29179T alone was identified as a cause of increased ΔCt (Supplementary Table 2).
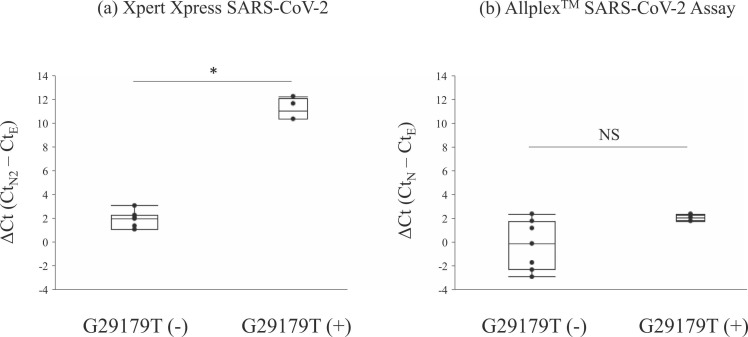
To investigate whether G29179T mutation affected another PCR kit, Allplex™ SARS-CoV-2 Assay was performed on the above 11 samples and found no significant increase in ΔCt (CtN − CtE) values ( Table 1). Comparing the ΔCt results of the Xpert Xpress and the Allplex™ SARS-CoV-2 Assay in these 11 samples, the group with the G29179T mutation showed a significant increase in ΔCt in the Xpert Xpress SARS-CoV-2, but no similar finding in the Allplex™ SARS-CoV-2 Assay ( Fig. 2). SARS-CoV-2 variant typing was further performed using the Novaplex™ Variants I Assay and all specimens with the G29179T mutation were alpha variants (PANGO lineage B.1.1.7).
Table 1.
Impact of G29179T mutation on ΔCt in two PCR assays.
| Mutation | Sample No. | Xpert Xpress SARS-CoV-2 |
Allplex™ SARS-CoV-2 Assaya |
Novaplex™ SARS-CoV-2 Variant Ⅰ Assay | ||||
|---|---|---|---|---|---|---|---|---|
| E (Ct) | N2 (Ct) | ΔCt | E (Ct) | N (Ct) | ΔCt | |||
| G29179T (−) | 20 | 28.2 | 31.3 | 3.1 | 29.6 | 28.0 | −1.7 | (R.1) |
| 23 | 12.8 | 14.9 | 2.1 | 9.7 | 9.6 | −0.1 | (R.1) | |
| 24 | 30.3 | 32.6 | 2.3 | 31.3 | 28.9 | −2.3 | (R.1) | |
| 25 | 25.3 | 27.3 | 2.0 | 26.8 | 23.9 | −2.9 | alpha variant (B.1.1.7) | |
| 27 | 23.9 | 25.3 | 1.4 | 25.0 | 23.7 | 1.2 | alpha variant (B.1.1.7) | |
| 29 | 17.1 | 18.2 | 1.1 | 18.1 | 16.3 | 1.8 | alpha variant (B.1.1.7) | |
| 31 | 31.9 | 33.0 | 1.1 | 30.7 | 28.3 | 2.4 | alpha variant (B.1.1.7) | |
| G29179T (+) | 30 | 24.6 | 35.0 | 10.4 | 25.9 | 23.8 | 2.1 | alpha variant (B.1.1.7) |
| 32 | 31.9 | 44.2 | 12.3 | 33.0 | 30.9 | 2.1 | alpha variant (B.1.1.7) | |
| 33 | 22.4 | 34.1 | 11.7 | 22.9 | 21.1 | 1.8 | alpha variant (B.1.1.7) | |
| 34 | 15.1 | 25.5 | 10.4 | 15.2 | 12.8 | 2.4 | alpha variant (B.1.1.7) | |
Allplex™ SARS-CoV-2 Assay detects the E, N, and RdRp/S genes, but the results for the RdRp/S gene were omitted (the Ct values were similar to those for the E and N genes).
Fig. 2.
Comparison of ΔCt values between samples with and without G29179T in the two assays. * p-value < 0.01. NS, not significant.
4. Discussion
In this study, the Xpert Xpress SARS-CoV-2 was performed on 196 clinical specimens during the study period and detected SARS-CoV-2 in 34 specimens. Among these, 4 samples with increased ΔCt (CtN2 − CtE) values were identified by scatterplot analysis. WGS identified G29179T mutation as a cause of increased ΔCt. Vanaerschot et al. have described the influence of G29140U mutation, although it is not the target for Xpert Xpress SARS-CoV-2, in the N gene on PCR testing for SARS-CoV-2 using scatterplot analysis of ΔCt and WGS (Vanaerschot et al., 2020), as performed in this study. The primers and probes used in the Xpert Xpress SARS-CoV-2 are proprietary and this cannot be confirmed. However, the G29179T mutation falls within a genomic region consistent with the forward primer of the CDC protocol N gene target (Miller et al., 2021, Foster et al., 2022). As a result, CtN2 was increased compared to CtE, leading to an increase in ΔCt. The Allplex™ SARS-CoV-2 Assay did not show the same difference in ΔCt values as the Xpert Xpress SARS-CoV-2.
Hong et al. investigated the ΔCt (CtN2 − CtE) in the Xpert Xpress SARS-CoV-2 positive specimens collected in Korea in September 2020 and reported that three samples showed delayed CtN2 (ΔCt: 11.8, 10.8, 12.1), whereas two samples were negative for the N2 gene (Hong et al., 2022). Sanger sequencing was performed on these five samples and the G29179T mutation was identified. In addition, the authors searched the GISAID database for Korean samples and found that most of the G29179T mutations belonged to PANGO lineage B.1.497, which has been prevalent in Korea since May 2020. The G29179T mutation has been reported in several countries, including Peru, Congo, and the United States, but the detection rate was much lower than that in Korea. Subsequently, Foster et al. investigated samples with altered profile defined as > 3 Ct value difference between the E and N gene (Foster et al., 2022). As a result, the median ΔCt was 10 and G29179T mutation was identified as a cause of increased ΔCt. In addition to the G29179T mutation, several reports have shown that mutations in the N gene region adversely affect SARS-CoV-2 testing targeting this region. Previous reports focusing on N gene mutations and their effects on SARS-CoV-2 tests have been summarized in Supplementary Table 3 (Ziegler et al., 2020, Vanaerschot et al., 2020, Miller et al., 2021, Foster et al., 2022, Hong et al., 2022, Jian et al., 2022, Bourassa et al., 2021, Sánchez-Calvo et al., 2021, Hasan et al., 2021a, Amato et al., 2021, Leelawong et al., 2021, Fox-Lewis et al., 2021, Hasan et al., 2021b). In all reports, the mutations caused false-negative N gene detection or delayed Ct.
NAAT is the gold standard for the detection of SARS-CoV-2 and diagnosis of COVID-19. However, the constantly mutating nature of this virus can affect test results. Therefore, multiplex NAAT, which targets multiple gene regions, is more reliable than monoplex NAAT, which targets a single gene region (Petrillo et al., 2020, Peñarrubia et al., 2020, LeBlanc et al., 2020). In general, retesting is recommended when the test result is negative in patients suspected of COVID-19 to reduce the incidence of false negatives (Wikramaratna et al., 2020). In a multiplex NAAT, negativity of one target gene is not necessarily interpreted as a negative test result. In the case of Xpert Xpress SARS-CoV-2 for example, if the E gene is positive and the N2 gene is negative, the result is interpreted as a presumptive positive and requires retesting. However, repeating the same test does not address the problem of presumptive positives or false negatives due to mutations in the target gene; hence, it is preferable to use a different test targeting other genes or gene sequences, if available. Although the multi-target approach is of value, it should be understood that mutation can affect the result even in a multiplex NAAT.
The present study has several limitations. First, it was a retrospective study conducted in a single institution, and the sample size was small. Second, a review of the patient information to determine the clinical impact of delayed N gene expression caused by the G29179T mutation was not performed. Third, viral nucleic acids extracted from nasopharyngeal samples were analyzed directly by viral WGS, resulting in some sequences derived from primary specimens being of poor quality, thereby limiting genomic analyses. Although there are previous studies focusing on this topic, this study adds to the literature by characterizing the G29179T in Japan.
NAAT-based detection of SARS-CoV-2 is being performed worldwide. However, the target regions are different for each assay; thus, the mutations occurring during the evolution of SARS-CoV-2 may affect various assays. This study demonstrates that a mutation affecting NAAT target region can render the assay susceptible to diagnostic failure, therefore ongoing surveillance for such mutations is important.
CRediT authorship contribution statement
Kami W: Conceptualization, Investigation, Data curation, Writing – original draft, Validation, Visualization, Writing – review & editing, and final approval of submitted version. Kinjo T: Conceptualization, Writing – original draft, Visualization, Writing – review & editing, final approval of the submitted version. Hashioka H: Investigation, Data curation, final approval of the submitted version. Arakaki W: Investigation, Data curation, final approval of the submitted version. Uechi K: Conceptualization, Investigation, Data curation, Resources, Supervision, final approval of the submitted version. Takahashi A: Investigation, Data curation, Resources, final approval of the submitted version. Oki H: Software, Data curation, Formal analysis, final approval of the submitted version. Tanaka K: Software, Data curation, Formal analysis, final approval of the submitted version. Motooka D: Software, Formal analysis, Visualization, Writing – review & editing, final approval of the submitted version. Nakamura S: Software, Formal analysis, Visualization, Supervision, final approval of the submitted version. Nakamatsu M: Supervision, final approval of the submitted version. Maeda S: Investigation, Supervision, final approval of the submitted version. Yamamoto K: Investigation, Supervision, Project administration, final approval of the submitted version. Fujita J: Investigation, Supervision, Project administration, final approval of the submitted version.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank Editage (www.editage.com) for English language editing.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jviromet.2023.114692.
Appendix A. Supplementary material
Supplementary material
.
Data availability
Data will be made available on request.
References
- Alanagreh L., Alzoughool F., Atoum M. The human coronavirus disease COVID-19: its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens. 2020;9(5):331. doi: 10.3390/pathogens9050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Díaz D.A., Franco-Muñoz C., Laiton-Donato K., Usme-Ciro J., Franco-Sierra N.D., Flórez-Sánchez A.C., et al. Molecular analysis of several in-house rRT-PCR protocols for SARS-CoV-2 detection in the context of genetic variability of the virus in Colombia. Infect. Genet. Evol. 2020;84 doi: 10.1016/j.meegid.2020.104390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato L., Jurisic L., Puglia I., Lollo V.D., Curini V., Torzi G., et al. Multiple detection and spread of novel strains of the SARS-CoV-2 B.1.177 (B.1.177.75) lineage that test negative by a commercially available nucleocapsid gene real-time RT-PCR. Emerg. Microbes Infect. 2021;10(1):1148–1155. doi: 10.1080/22221751.2021.1933609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourassa L., Perchetti G.A., Phung Q., Lin M.J., Mills M.G., Roychoudhury P., et al. A SARS-CoV-2 nucleocapsid variant that affects antigen test performance. J. Clin. Virol. 2021;141 doi: 10.1016/j.jcv.2021.104900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drain P.K. Rapid diagnostic testing for SARS-CoV-2. N. Engl. J. Med. 2022;386(3):264–272. doi: 10.1056/NEJMcp2117115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta N.K., Mazumdar K., Gordy J.T. The nucleocapsid protein of SARS–CoV-2: a target for vaccine development. J. Virol. 2020;94(13) doi: 10.1128/JVI.00647-20. e00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster C.S.P., Madden M., Chan R., Agapiou D., Bull R.A., Rawlinson W.D., et al. SARS-CoV-2 N-gene mutation leading to Xpert Xpress SARS-CoV-2 assay instability. Pathology. 2022;54(4):499–501. doi: 10.1016/j.pathol.2022.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox-Lewis S., Fox-Lewis A., Harrower J., Chen R., Wang J., de Ligt J., et al. Lack of N2-gene amplification on the Cepheid Xpert Xpress SARS-CoV-2 assay and potential novel causative mutations: a case series from Auckland, New Zealand. IDCases. 2021;25 doi: 10.1016/j.idcr.2021.e01233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K.E., Caliendo A.M., Arias C.A., Hayden M.K., Englund J.A., Lee M.J., et al. The Infectious Diseases Society of America Guidelines of the diagnosis of COVID-19: molecular diagnostic testing. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab048. Jan 22; ciab048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan R., Hossain M.E., Miah M., Hasan M.M., Rahman M., Rahman M.Z. Identification of novel mutations in the N Gene of SARS-CoV-2 that adversely affect the detection of the virus by reverse transcription-quantitative PCR. Microbiol. Spectr. 2021;9(1) doi: 10.1128/spectrum.00545-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M.R., Sundararaju S., Manickam C., Mirza F., Al-Hail H., Lorenz S., et al. A novel point mutation in the N Gene of SARS-CoV-2 may affect the detection of the virus by reverse transcription-quantitative PCR. J. Clin. Microbiol. 2021;59(4):e03278–20. doi: 10.1128/JCM.03278-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K.H., In J.W., Lee J., Kim S.Y., Lee K.A., Kim S., et al. Prevalence of a single-nucleotide variant of SARS-CoV-2 in Korea and its impact on the diagnostic sensitivity of the Xpert Xpress SARS-CoV-2 assay. Ann. Lab Med. 2022;42(1):96–99. doi: 10.3343/alm.2022.42.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian M.J., Chung H.Y., Chang C.K., Lin J.C., Yeh K.M., Chen C.W., et al. SARS-CoV-2 variants with T135I nucleocapsid mutations may affect antigen test performance. Int. J. Infect. 2022;114:112–114. doi: 10.1016/j.ijid.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc J.J., Gubbay J.B., Li Y., Needle R., Arneson S.R., Marcino D., et al. Real-time PCR-based SARS-CoV-2 detection in Canadian laboratories. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelawong M., Mitchell S.L., Fowler R.C., Gonzalez E., Hughes S., Griffith M.P., et al. SARS-CoV-2 N gene mutations impact detection by clinical molecular diagnostics: reports in two cities in the United States. Diagn. Microbiol. Infect. Dis. 2021;101(3) doi: 10.1016/j.diagmicrobio.2021.115468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S., Lee T., Merritt A., Pryce T., Levy A., Speers D. Single-point mutations in the N gene of SARS-CoV-2 adversely impact detection by a commercial dual target diagnostic assay. Microbiol. Spectr. 2021;9(3) doi: 10.1128/Spectrum.01494-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nextstrain website: 〈https://nextstrain.org/ncov/gisaid/global/all-time?d=tree&dmax=2023–01-24&l=clock&p=full〉. (Accessed 24 January).
- Oliveira S.C., de Magalhães M.T., Homan E.J. Immunoinformatic analysis of SARS-CoV-2 nucleocapsid protein and identification of COVID-19 vaccine targets. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.587615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñarrubia L., Ruiz M., Porco R., Rao S.N., Juanola-Falgarona M., Manissero D., et al. Multiple assays in a real-time RT-PCR SARS-CoV-2 panel can mitigate the risk of loss of sensitivity by new genomic variants during the COVID-19 outbreak. Int. J. Infect. Dis. 2020;97:225–229. doi: 10.1016/j.ijid.2020.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrillo S., Carrà G., Bottino P., Zanotto E., Santis M.C.D., Margaria J.P., et al. A novel multiplex qRT-PCR assay to detect SARS-CoV-2 infection: high sensitivity and increased testing capacity. Microorganisms. 2020;8(7):1064. doi: 10.3390/microorganisms8071064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M.S., Islam M.R., Alam A.R.U., Islam I., Hoque M.N., Akter S., et al. Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences. J. Med. Virol. 2021;93:2177–2195. doi: 10.1002/jmv.26626. [DOI] [PubMed] [Google Scholar]
- Sánchez-Calvo J.M., Arboledas J.C.A., Vidal L.R., Francisco J.L., Prieto M.D.L. Diagnostic pre-screening method based on N-gene dropout or delay to increase feasibility of SARS-CoV-2 VOC B.1.1.7 detection. Diagn. Microbiol. Infect. 2021;101(4) doi: 10.1016/j.diagmicrobio.2021.115491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanaerschot M., Mann S.A., Webber J.T., Kamm J., Bell S.M., Bell J., et al. Identification of a polymorphism in the N Gene of SARS-CoV-2 that adversely impacts detection by reverse transcription-PCR. J. Clin. Microbiol. 2020;59(1) doi: 10.1128/JCM.02369-20. e02369-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Jean S., Wilson S.A., Lucyshyn J.M., McGrath S., Wilson R.K., et al. A deletion in the N gene of SARS-CoV-2 may reduce test sensitivity for detection of SARS-CoV-2. Diagn. Microbiol. Infect. Dis. 2022;102 doi: 10.1016/j.diagmicrobio.2021.115631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramaratna P.S., Paton R.S., Ghafari M., Lourenço J. Estimating the false-negative test probability of SARS-CoV-2 by RT-PCR. Eur. Surveill. 2020;25(50) doi: 10.2807/1560-7917.ES.2020.25.50.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J.M., Wolf L.M., Bello G.L., Maccari J.G., Nasi L.A. Molecular evolution of SARS-CoV-2 from December 2019 to August 2022. J. Med. Virol. 2023;95(1) doi: 10.1002/jmv.28366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler K., Steininger P., Ziegler R., Steinmann J., Korn K., Ensser A. SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Eur. Surveill. 2020;25(39) doi: 10.2807/1560-7917.ES.2020.25.39.2001650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zóka A., Bekő G. Distinct changes in the real-time PCR detectability of certain SARS-CoV-2 target sequences. Clin. Chim. Acta. 2020;507:248–249. doi: 10.1016/j.cca.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.