Abstract
Long-lived proteins (LLPs), although less common than their short-lived counterparts, are increasingly recognized to play important roles in age-related diseases such as Alzheimer′s. In particular, spontaneous chemical modifications can accrue over time that serve as both indicators of and contributors to disrupted autophagy. For example, isomerization in LLPs is common and occurs in the absence of protein turnover while simultaneously interfering with protein turnover by impeding proteolysis. In addition to the biological implications this creates, isomerization may also interfere with its own analysis. To clarify, bottom-up proteomics experiments rely on protein digestion by proteases, most commonly trypsin, but the extent to which isomerization might interfere with trypsin digestion is unknown. Here, we use a combination of liquid chromatography and mass spectrometry to examine the effect of isomerization on proteolysis by trypsin and chymotrypsin. Isomerized aspartic acid and serine residues (which represent the most common sites of isomerization in LLPs) were placed at various locations relative to the preferred protease cleavage point to evaluate the influence on digestion efficiency. Trypsin was found to be relatively tolerant of isomerization, except when present at the residue immediately C-terminal to Arg/Lys. For chymotrypsin, the influence of isomerization on digestion was less predictable, resulting in long-range interference for some isomer/peptide combinations. Given the trypsin- and chymotrypsin-like behavior of the 20S proteasome, and to further establish the biological relevance of isomerization in LLPs, substrates with isomerized sites were also tested against proteasomal degradation. Significant disruption of 20S proteolysis was observed, suggesting that if LLPs persist long enough to isomerize, it will be difficult for cells to digest them.
Graphical Abstract

Introduction
Like a large movie with many supporting actors who appear only briefly, human biology is dominated by short-lived proteins that are synthesized and then degraded quickly. However, there also exist a number of long-lived proteins (LLPs), which, though their numbers may be few, can play important roles.1 For example, mature fiber cells in the eye lens contain extreme LLPs, where protein lifetime is roughly equal to the age of the organism itself. Despite their lengthy lifetimes, the crystallin proteins in the eye lens are vital for maintaining sight.2 An unavoidable consequence of increased lifespan is that LLPs have ample opportunity to accumulate post-translational modifications (PTMs), such as phosphorylation or glycosylation, or undergo spontaneous chemical modifications (SCMs), such as truncation, deamidation, oxidation, or isomerization.3 Of these, isomerization is the most difficult to study because there is no change in atomic composition or mass, and the structural differences are subtle. At the same time, isomerization can be abundant, especially in the case of aspartic acid where transformation is facilitated by a five-membered succinimide intermediate produced by loss of water from the side chain (as illustrated in Scheme 1). This ring preferentially reopens by addition of water to the opposite carbonyl, producing L-isoAsp where the natural backbone and side chain positions have inverted. In addition, the succinimide can switch chirality and reopen to produce D-Asp and D-isoAsp.
Scheme 1:

Aspartic acid isomerization
The structural changes imparted by isomerization in LLPs can influence subsequent interactions with enzymes. For example, isomerization of serine (which occurs second in frequency after aspartic acid) strongly interferes with kinase activity and prevents phosphorylation.4 It has also been recognized for some time that isomerization can inhibit protease action, which is relevant both for analytical analysis and biological function. In terms of analysis, modified protease behavior can be used to aid in the identification of peptides containing isomerized residues. For example, aminopeptidase M is unable to cleave D-amino acids from peptide N-termini, a shortcoming that has been used as an enrichment protocol to find D-amino acids in biological samples.5 Asp-N cleaves at the N-terminal side of L-Asp residues, but it is unable to cleave isomerized Asp residues, allowing for missed cleavages to pinpoint isomerization in LLPs.6 While examples are rare, certain enzymes recognize specific amino acid isomers and provide novel cleavage or modifications that can be used for identification of isomers. For example, protein isoaspartyl methyltransferase (PIMT) selectively binds L-isoAsp and methylates the side chain through S-adenosyl methionine, causing the succinimide ring to reform.7 PIMT has been used in conjunction with succinimide-reactive reagents such as tris and hydrazine to introduce mass shifts at Asp isomerization sites, allowing easy identification of Asp isomers.8,9 Paenidase is a novel protease which recognizes and cleaves at D-Aspartic acid and has been used to identify D-Asp in heavily isomerized proteins.6,10 In addition, cathepsin B recognizes the L-isoAsp side chain as a C-terminus analog, resulting in endopeptidase cleavage at L-isoAsp as well as ligase activity.11 For the majority of proteases however, isomerization causes structural changes that impede binding to the active site result and therefore reduce or prevent proteolysis.
Considering the biological perspective, substitution of D-amino acids in synthetic peptides used as antimicrobial drugs increases resistance to bacterial proteolysis thereby prolonging lifetime and improving effectiveness.12 Similarly, peptide neurotransmitters and toxins have been found to contain D-amino acids which increase lifetimes by conferring resistance to proteolysis.13,14 In terms of LLPs, isomerization can potentially disrupt proteostasis by preventing breakdown into constituent amino acids. For example, studies of the lysosomal cathepsins have shown that isomerized substrates cannot be fully digested, which could eventually lead to lysosomal storage observed in Alzheimer′s disease (AD).15,16
Although the lysosome appears to be unable to degrade isomerized substrates, the ubiquitin-proteasome system is another possible pathway for protein degradation. Within the mammalian cell, the 26S proteasome is a major complex composed of a core 20S unit responsible for enzymatic digestion and 1–2 outer 19S regulatory units that act together for digestion and recycling of protein/peptide substrates.17 Proteins tagged with ubiquitin are recognized by the 19S subunit and fed into the center of the complex for degradation. The core 20S proteasome unit can also operate independently without the attachment of 19S subunits and is an abundant form found in cells.18 The number of 20S substrates is quite substantial, comprising around 20% of cellular proteins, which are recognized and degraded by 20S without ubiquitination.19 Cleavage of substrates occurs through subunits β1, β2, and β5, which have caspase-, trypsin-, and chymotrypsin-like activity, respectively. In contrast to the 26S proteasome which recognizes ubiquitinated substrates via the 19S regulatory unit, the 20S proteasome recognizes and degrades damaged, unfolded, or otherwise disordered proteins.20 Interestingly, disordered proteins or disordered regions in proteins are more likely to undergo significant amounts of Asn deamidation and Asp isomerization due to increased backbone flexibility and solvent accessibility.21,22,23,24 In addition, isomerization itself can cause structural changes resulting in destabilization of ordered protein structure.25,26 However, the question remains unanswered as to whether the proteasome can digest peptide/protein substrates that have been isomerized.
Although perturbed proteolysis has been recognized as a potential aid for analysis of isomerization, it may also present problems for bottom-up proteomic analyses,27,28 which rely on digestion of whole proteins with proteases (most commonly trypsin), followed by tandem LC/MS analysis. Missed cleavages can result in significant problems for bottom-up proteomics such as fewer peptide identifications due to increased peptide size or inaccuracies in quantitative measurements due to inconsistent inclusion/exclusion of isomerized forms. Trypsin is the most abundant digestive enzyme present in the gut of all animals and is released to activate other digestive enzymes such as chymotrypsin as well as aid in the digestion of consumed protein.29 Trypsin dominates bottom-up approaches because it yields low missed cleavages and strongly prefers cleavage at Lys/Arg residues, which have the correct abundance in proteins to generate peptides at roughly the right length for mass-spectrometric analysis. Despite being robust and very specific, trypsin activity can be affected by the residues surrounding Lys/Arg, and certain sequence motifs are known to impair digestion. For example, proline or basic residues at P1′ can impair digestion.30 Acidic residues such as aspartic acid or glutamic acid in P2 or P1′ sites (see Scheme 2) are also known to cause moderate interference.31 If canonical residues such as these affect trypsin, isomerized residues may also interfere with proteolysis.
Scheme 2.
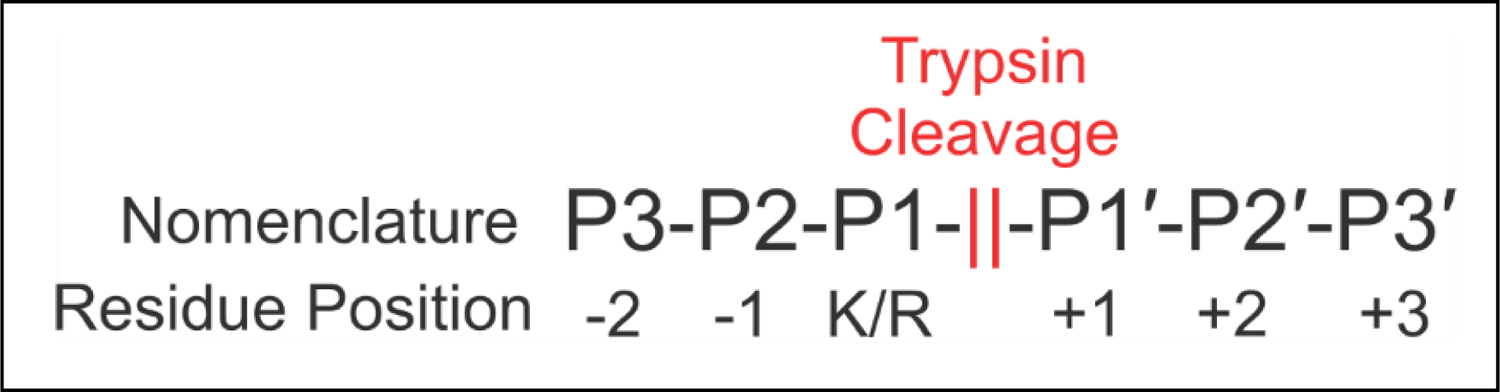
Standard proteolysis positioning
In this work we seek to answer two questions, (1) what is the effect of isomerization in proximity to trypsin and chymotrypsin cleavage sites on digestion efficiency, and (2) is the proteasome (which possesses trypsin and chymotrypsin-like activity) capable of degrading isomerized substrates? To this end, we test trypsin and chymotrypsin activity near isomerized residues in synthetic peptide substrates. Sequences were chosen with Asp residues near cleavage sites and tested by comparing trypsin/chymotrypsin cleavage of the all-L peptide against the sequences containing an isomerized Asp or epimerized Ser residue. Trypsin was found to primarily have difficulty processing isomerized substrates at the P1′ position, while for chymotrypsin, isomerization also caused substantial missed cleavages at sites further away. Similarly, the 20S proteasome was found to be unable to significantly degrade substrates with isomerized sites.
Experimental Methods
Materials.
Organic solvents and reagents were purchased from Fisher Scientific, Sigma-Aldrich, or Acros Organics and used without further purification. FMOC-protected amino acids and Wang resins were purchased from Anaspec, Inc or Chem-Impex International. Trypsin was purchased from Sigma Aldrich (T0303). α-chymotrypsin was also purchased from Sigma Aldrich (C-4129).
Peptide Synthesis.
Peptides were synthesized manually following a FMOC-protected solid-phase peptide synthesis protocol.32 Following synthesis, peptides were RP-HPLC purified using a Thermo UltiMate 3000 RS variable wavelength detector and pump connected to a Phenomenex Kinetex 5μm EVO C18 250 × 21.2 mm column (P/N 00G-4633-P0-AX) with Phenomenex SecurityGuard PREP Cartridge (P/N AJ0–9145). Samples were purified using water with 0.1% formic acid as mobile phase A, and acetonitrile with 0.1% formic acid as mobile phase B. Purified peptides were stored frozen in 50/50 v/v acetonitrile/water. Peptides were lyophilized and reconstituted in Fisher Optima water prior to determining concentrations and reaction with trypsin or chymotrypsin in buffer. Concentrations of peptides were obtained by measuring the absorbance at either 205nm or 280nm using an Agilent Cary 60 UV/Vis Spectrophotometer. Molar absorptivity constants were obtained from an online calculator made available by Marius Clore (https://spin.niddk.nih.gov/clore/).33
Digestion of Peptides with Trypsin/Chymotrypsin.
All-L sequences and sequences containing isomers were digested separately with trypsin in equal ratios of enzyme to substrate (1:100 w/w). Chymotrypsin digestions were carried out at a ratio of 1:20 w/w enzyme to substrate. Trypsin digestions were carried out in 100mM Tris pH 7.8 at 37C. Chymotrypsin digestions were carried out in 100mM Tris pH 7.8, 10mM CaCl2 at 37C. All time points were quenched with 1% TFA to halt reaction progress prior to LCMS analysis.
Analysis.
For LCMS analysis of synthetic peptides, an Agilent 1100 binary pump was used with a Thermo BetaBasic-18 3 μm C18 150 mm × 2.1 mm column interfaced to a Thermo Fisher Scientific LTQ mass spectrometer. Quenched reaction time points were diluted to a total concentration of ~5μM with water and 50μL of sample was loaded onto the sample loop for analysis of each time point. After sample loading but prior to analysis, peptides were desalted online by flushing the column at 1–5%B for 5 minutes with the inject valve directing column output to waste. After 5 minutes the valve was switched to the inject position to direct samples to the MS for analysis. Samples were eluted using water with 0.1% formic acid as mobile phase A, and acetonitrile with 0.1% formic acid as mobile phase B. Gradients were optimized for separation and elution of digestion products for synthetic sequences used. For example, trypsin cleavage products of SEMRLEKDRFSVNL were separated over a gradient of 1–65%B in 60 minutes.
Proteasome Purification.
Rat 20S proteasomes were purified from rat livers according to a previously-established protocol.34 In brief, rat livers were homogenized on ice in lysis buffer (Tris-HCl 50 mM, pH 7.5, Sucrose 250 mM, EDTA 1 mM, DTT 1 mM, Benzonase endonuclease) and centrifuged at 4 °C for 15 min, at 1,000 g. The supernatant was mixed with NaCl to reach a final concentration of 0.5M and ultracentrifuged at 4 °C for 2 h, at 152,000 g. The resulting supernatant was ultracentrifuged again at 4 °C for 5 h, at 200,000 g. The resulting pellet was re-suspended in buffer containing Tris-HCl 25 mM, pH 7.5, EDTA 1 mM, DTT 1 mM and loaded onto a XK50/100 gel filtration column (•XK 50/100 column (GE Healthcare) packed with 1.8 L 4% Rapid Run™ Agarose Bead Standard 50–150 μm for gel filtration (GF) (Agarose Bead Technologies cat. # 4RRS-1000). Eluted fractions were tested by peptidase activity assay. Positive proteasome-containing fractions were pooled, loaded onto a Source 15Q strong anion exchange column (GE Healthcare cat. # 17-0947-01), and eluted over a gradient of 10–30% Buffer B (Tris-HCl 25 mM, pH 7.5, NaCl 1M, EDTA 1 mM, DTT 1 mM). Positive proteasome-containing fractions were pooled, loaded onto a HiTrap® DEAE Fast Flow weak anion exchange column (GE Healthcare cat. # 17-5055-01) and eluted over a step gradient of 10%, 30%, 40%, 50%, 60% and 70% buffer B. Positive proteasome-containing fractions were pooled, buffer exchanged to 10 mM phosphate buffer, pH 7.4 and loaded onto a CHT column •Tricorn 10/10 column packed with 8 ml of type I ceramic hydroxiapatite (CHT) (Bio-Rad cat. # 1582000). Proteins were eluted over a step gradient of 10%, 20%, 40%, 50% and 60% buffer (400 mM phosphate buffer, at pH 7.4). Positive proteasome-containing fractions were pooled and concentrated before storage.
Proteasome Digestion of Peptide Substrate.
Peptides were dissolved in 150mM ammonium acetate to a concentration of 20μM, and frozen in small aliquots. On the day of the measurement, a peptide aliquot was thawed and diluted 10x with ammonium acetate and kept on ice. A sample of purified rat 20S proteasomes, at a concentration of 4μM was buffer exchanged into 150 mM ammonium acetate. For each measurement, 1μl of the 20S proteasome was mixed with 1μl of the peptide. The sample was immediately loaded into a nano-electrospray needle and measured continuously over a period of 20 minutes. In control reactions, each peptide was diluted to 1μM and measured without the 20S proteasome over the same period of time (20 minutes). Spectra were recorded on a modified Q Exactive Plus at a resolution of 70,000, at 250 millisecond injection time and with low averaging −5. Capillary voltage was set to 1.7 kV, at a temperature of 180°C. Fore-vacuum pressure was set to 1.5 mbar, and the trapping gas pressure to 1, corresponding to HV pressure of 3.4×10–5 mbar, and UHV pressure of 9.2×10–11 mbar. The source was operated at a constant energy of 2V in the flatapole bias and interflatapole lens. Bent flatapole DC bias and gradient were set to 2 and 10V, respectively, and the HCD cell was operated at an energy of 1V. In each experiment, 20 time points were taken (averaged over minute, each). The results represent an average of three experiments, and error bars represent standard deviation. Peak intensities were extracted using the Qual Browser software, part of the Thermo Xcalibur 4.1.50 software package.
Results and Discussion
Isomerization near trypsin cleavage sites.
To test the effect of different Asp isomers on trypsin cleavage in a substrate with multiple cleavage sites, the crystallin-derived synthetic sequence SEMRLEKDRFSVNL with either L-/L-iso/D-/D-isoAsp at Asp-8 was incubated with trypsin. After 24 hours, the digestions were halted with acid and the products analyzed via LCMS. The chromatogram for the all-L sequence (Fig. 1a) reveals cleavage products at each of the three trypsin cleavage sites in the peptide. The inset bar plot in Fig. 1a quantifies the fractional abundance of peptides resulting from hydrolysis at each site (blue) relative to the fractional abundance of peptides containing a missed cleavage at the same site (orange). For example, the blue bar for all-L Arg-4 (Fig. 1a) represents the sum of the fractional abundances for SEMR, LEKDRFSVNL, LEKDR, and LEK as these fragments are all products resulting from cleavage at Arg-4. The orange bar represents the fractional abundances for SEMRLEK and SEMRLEKDR, which both contain a missed cleavage at Arg-4.
Figure 1:

Chromatograms showing major products after 24 hours of trypsin digestion of SEMRLEKDRFSVNL for the (a) All-L, (b) L-isoAsp-8, (c) D-Asp-8, and (d) D-isoAsp-8 sequences. Bar plots are shown inside each chromatogram summarizing the propensity for cleavage at each of the three possible sites. Blue bars represent the sum of the fractional abundance of all peptides terminating at the cleavage site (i.e. fragments with C-terminal Lys/Arg or the corresponding complement fragments). Orange bars represent the sum of fractional abundance of peptides containing missed cleavages (e.g. any peptide containing RL for Arg-4). P denotes the fractional abundance of the precursor. # corresponds to precursor peptide
The propensity for digestion as indicated by the size of the blue bars relative to the orange bars at each site reveals preferential digestion of canonical SEMRLEKDRFSVNL at Lys-7, followed by Arg-9 and Arg-4 (Fig. 1a). This result suggests that canonical Glu-6 and Asp-8 do not interfere with digestion at Lys-7 despite the known aversion of trypsin to acidic residues in these positions. The propensities for digestion at each site change dramatically for the L-isoAsp peptide, particularly at Arg-4, which becomes the preferred site, and at Lys-7, which shifts from the most cleaved to least cleaved site (Fig. 1b). For the D-Asp peptide, Arg-4 is again preferred, but very little cleavage occurs at Lys-7 or Arg-9 (Fig. 1c). The D-isoAsp isomer shifts proteolysis from Lys-7 to either Arg-4 or Arg-9 (Fig. 1d). In summary, isomerization at Asp-8 largely impedes hydrolysis at Lys-7 regardless of the Asp isomer. Importantly isomers only reside in the P1′ position relative to Lys-7, while for Arg-4 and Arg-9, the isomers occupy sites P4′ and P2, respectively. Although Arg-4 is not a preferred cleavage site for the canonical peptide, it becomes the preferred cleavage site for all Asp isomers. This most likely occurs because Arg-4 is most distant from the site of isomerization. Although both Lys-7 and Arg-9 are immediately adjacent to the site of isomerization, significantly greater interference is observed for Lys-7. In fact, for D-isoAsp, cleavage at Arg-9 actually increases relative to the canonical peptide. This suggests that the P1′ position for trypsin allows the least flexibility in accommodating isomerized substrates, which may be analogous to the similar trend observed previously for proline. Overall, the results clearly indicate that isomerization can interfere with trypsin digestion and increase the likelihood for missed cleavages to occur.
Isomerization-site proximity effect.
Having established that trypsin is affected by isomerization near cleavage sites, we further examined a series of four synthetic sequences derived from highly isomerized LLPs. These peptides were chosen to test the effects of L-isoAsp in the P3, P2, P1′, and P2′ positions on trypsin digestion as shown in Fig. 2a. Each L-isoAsp-containing peptide and the corresponding all-L peptide were incubated separately with trypsin and the progress of cleavage compared at a series of time points over the course of the reaction via LCMS. One example of raw data is shown in Fig. 2b, where it is clear that the canonical peptide is digested much more than the isomerized version. To quantitatively assess the relative extent of trypsin digestion for each isomerization site, rate constants for degradation of each peptide were calculated by fitting a first-order rate equation (Fig. S1). The ratios of the all-L rate constants to the L-isoAsp rate constants are shown in Fig. 2c. Referencing the extent of digestion to the canonical peptide in each case allows examination of the influence of the L-isoAsp substitution independent of other sequence effects that may vary between peptides. Isomerization at P1′ (GDYKDSSDF) leads to a clear preference for digestion of the all-L form, as indicated by the large ratio shown in Fig. 2c. Digestion of the all-L peptide is also favored relative to L-isoAsp at the P3, P2, and P2′ positions, though to much lesser extent. The results again suggest that isomerization at P1′ is particularly problematic for trypsin recognition. Further investigation of this position revealed that L-isoAsp is not the only Asp isomer found to inhibit cleavage when present at P1′, D-Asp and D-isoAsp also inhibit trypsin cleavage, and D-Serine also yields similar effects (Fig. S2). This suggests that while isomerization at other nearby locations causes a reduction in trypsin cleavage efficiency, isomerization at P1′ is particularly likely to slow proteolysis. These findings also agree with the data in Fig. 1 for SEMRLEKDRFSVNL, in which cleavage at Lys-7 was strongly perturbed by isomerization at Asp-8, which also occupies the P1′ position.
Figure 2:
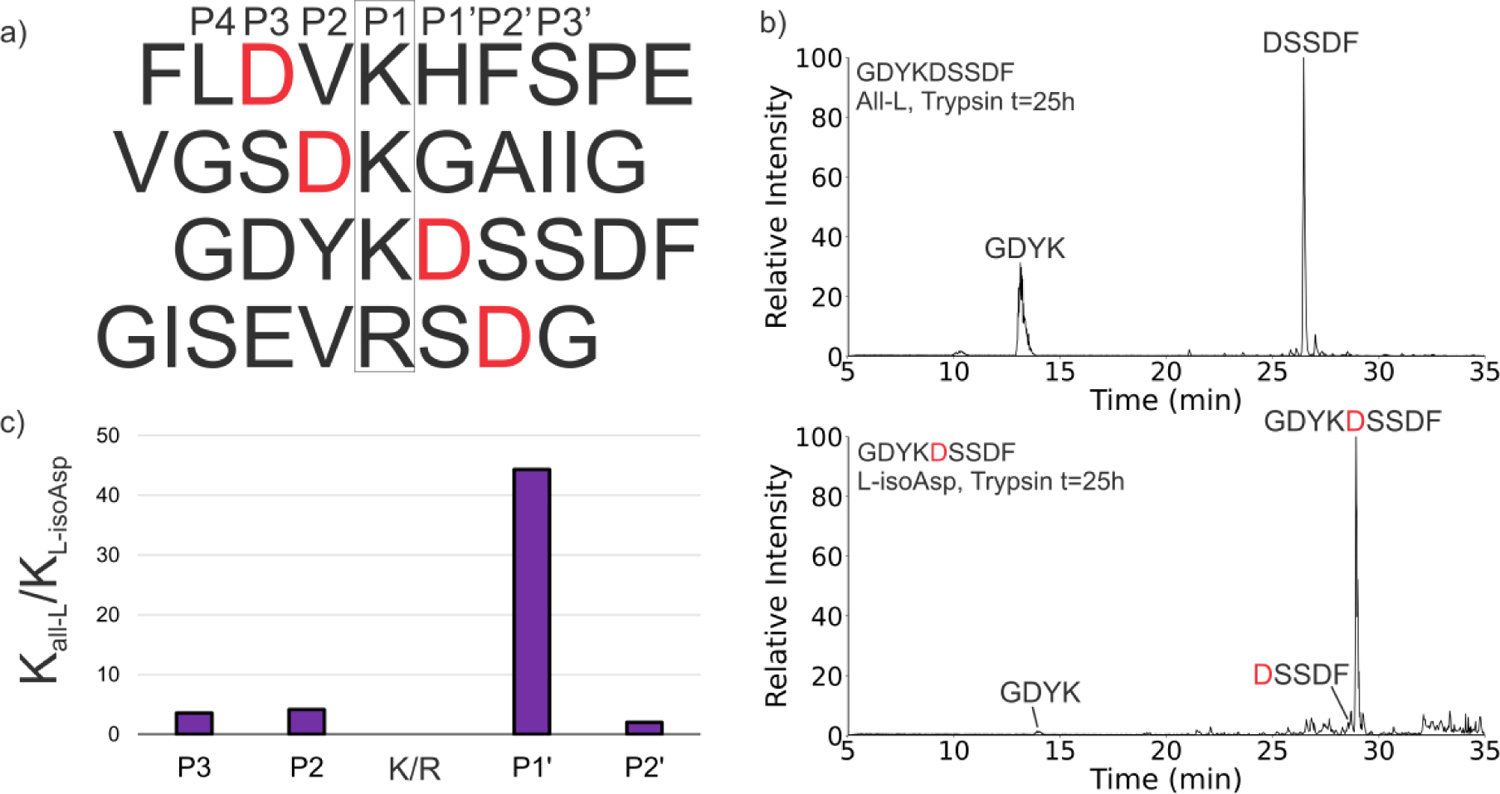
(a) Four sequences with an Asp residue at either P3, P2, P1′, or P2′ (labeled in red) were tested with trypsin. (b) Representative chromatogram showing trypsin cleavage of GDYKDSSDF All-L at t=25h and cleavage of GDYKDSSDF L-isoAsp-5 at t=25h. (c) Bar plot showing the comparison of the ratios of first-order rate constants for all-L precursor (Kall-L) and L-isoAsp precursor (KL-isoAsp). Larger bars indicate a higher preference for all-L sequence cleavage relative to the L-isoAsp containing sequence.
Chymotrypsin cleavage of isomerized substrates.
Chymotrypsin shares many similarities with trypsin in terms of sequence, structure, and catalytic site while simultaneously exhibiting quite different substrate specificity.35 Consequently, chymotrypsin is frequently used in bottom-up proteomics to produce digestion at aliphatic amino acids. To evaluate the effect of isomers on Chymotrypsin digestion, six variants of APSWFDTGLSEMR (all-L, L-isoAsp-6, D-Asp-6, DisoAsp-6, D-Ser-3, D-Ser-10) were examined. After incubation with chymotrypsin for 21 hours, all digestion products were determined by LCMS and the results are shown in Fig. 3 as a function of cleavage propensity. The all-L sequence is preferentially cut at Phe-5 with minor activity at Trp-4 and Leu-9 (Fig. 3a). Digestion of the L-isoAsp-containing peptide reveals a shift in behavior with the Leu-9 site being targeted, while no activity was observed at Trp-4 and only minimal cleavage at Phe-5 (Fig. 3b). This behavior can be most easily rationalized by the proximity of Trp-4 and Phe-5 to L-isoAsp. In contrast, the D-Asp variant yields virtually no chymotrypsin digestion at any of the three sites (Fig. 3c). Presumably the stereochemical inversion of the L-Asp side chain to D-Asp results in steric clash that largely prevents access to the chymotrypsin active site. Interestingly, D-isoAsp behaves similarly to L-isoAsp (Fig. 3d). Changing L-Ser-3 to D-Ser-3 does not completely impair digestion, but instead allows significant cleavage at Phe-5 while slightly increasing digestion at Leu-9 relative to the all-L sequence (Fig. 3e). In contrast, D-Ser-10 essentially only permits cleavage at Phe-5 while shutting down digestion at Leu-9 and Trp-4 (Fig. 3f). The reduced recognition at Leu-9 is easily explained by proximity, but the high digestion at Phe-5 coupled with reduced digestion at Trp-4, which is further away from the isomerized residue, suggests that longer range effects can sometimes influence behavior in unexpected ways. While D-Ser-3 and D-Ser-10 were both found to affect chymotrypsin cleavage, a direct comparison of Ser isomerization to Asp isomerization is difficult to make due to the positional differences relative to the chymotrypsin cleavage sites. To more directly compare Asp and Ser isomerization, we prepared the all-L serine analog APSWFSTGLSEMR as well as the D-Ser-6 epimer and digested with chymotrypsin (Fig. 3g and 3h). The all-L peptide yields results very similar to the original sequence (compare Fig. 3a/3g). Surprisingly, the D-Ser-6 epimer was almost untouched by chymotrypsin, yielding results similar to the D-Asp epimer for APSWFDTGLSEMR. This suggests that the sidechain stereochemistry may be more important than side chain identity for impairing active site fit in some cases. In sum, these results show that chymotrypsin is sensitive to isomerization at a variety of positions relative to the cleavage site, which could make chymotrypsin a less predictable choice for the digestion of potentially isomerized LLPs.
Figure 3:
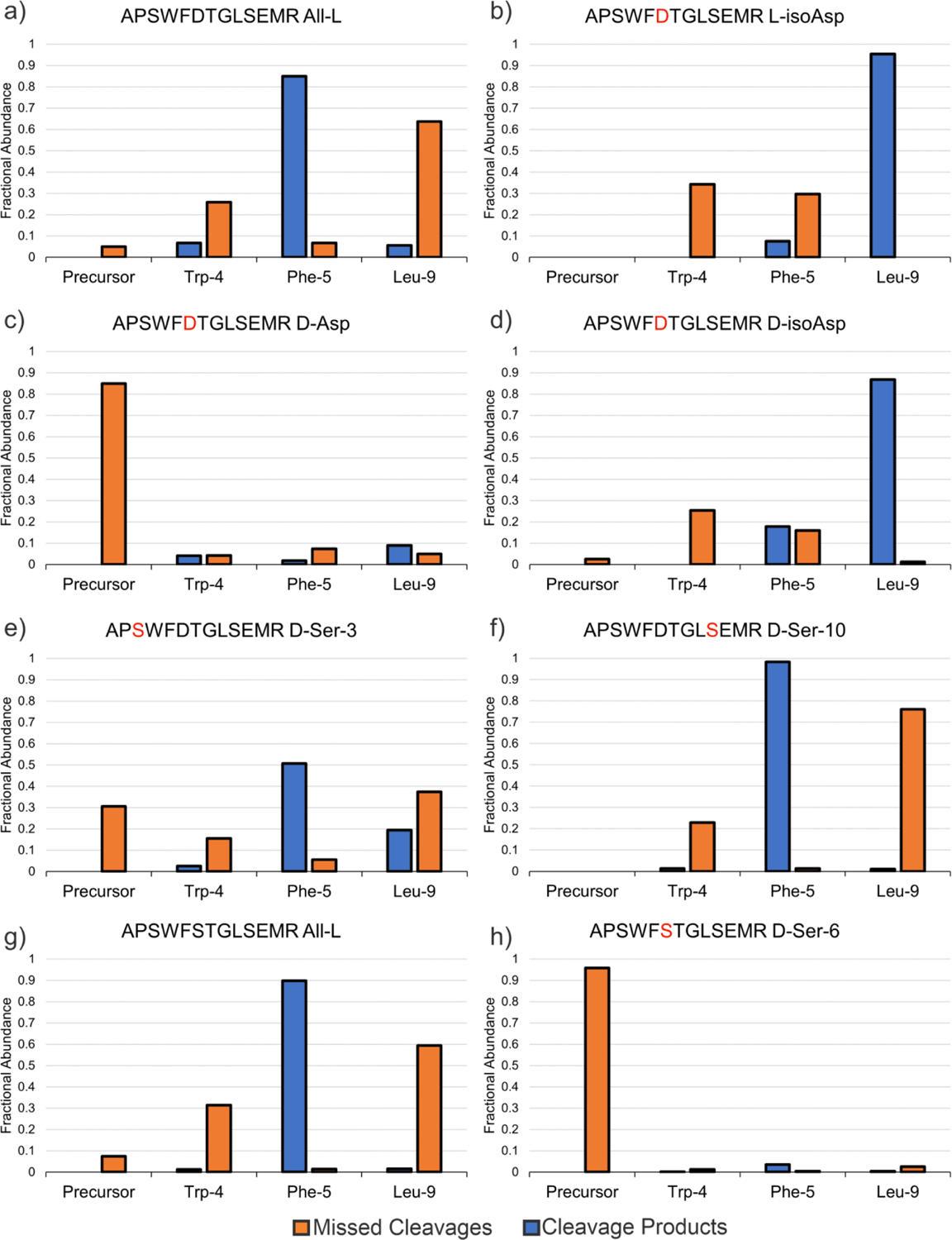
Chymotrypsin cleavage results at 21h of (a) APSWFDTGLSEMR All-L, (b) APSWFDTGLSEMR L-isoAsp, (c) APSWFDTGLSEMR D-Asp, (d) APSWFDTGLSEMR D-isoAsp, (e) APSWFDTGLSEMR D-Ser-3, (f) APSWFDTGLSEMR D-Ser-10, (g) APSWFSTGLSEMR All-L, and (h) APSWFSTGLSEMR D-Ser-6. Blue bars represent the sum of fractional abundance of sequences produced from cleavage at a particular site (e.g. APSW and FDTGLSEMR for Trp-4 cleavage), while orange bars represent the fractional abundance of missed cleavage products at a specific site.
Proteasome digestion of isomerized substrates.
Although not typically used for proteomics experiments, the proteasome houses both trypsin-like and chymotrypsin-like proteolytic sites as well as caspase-like activity. Furthermore, given that previous work has established that the lysosomal system cannot fully degrade isomerized peptide substrates, the potential digestion of isomerized peptides by the proteasome remains an important biological question. In order to test proteasome activity, larger peptide targets are required to make suitable substrates. We synthesized VKKDQLGKNE EGAPQEGILE DMPVDPDNEA YEMPSEEG, a sequence derived from alpha-synuclein, in both the all-L-Asp and all-L-isoAsp forms (L-isoAsp/Asp sites indicated in bold). The individual peptides were mixed with the 20S proteasome and monitored over 20 minutes via nano-electrospray. The signal for the intact all-L peptide decreased rapidly to zero over 10–15 minutes digestion time, while digestion product peaks appeared (Fig. 4a). In contrast, the all-(L-isoAsp) peptide persisted when incubated in the presence of the 20S proteasome, being virtually untouched by 20S digestion after 20 minutes and producing only minor cleavage products (Fig. 4b). Interestingly, the two main degradation products for the all-(L-Asp) sequence were identified as all-(L-Asp) 5–38, resulting from a single caspase-like cleavage at Asp-4, and all-(LAsp) 5–25, resulting from sequential caspase cleavage at Asp-4 and Asp-25 (Fig. 4c). Changing Asp-4 and Asp-25 to L-isoAsp seems to prevent any caspase-like cleavage of the substrate, as these products are not observed in significant amounts for the all-(L-isoAsp) sequence. Furthermore, digestion is not shifted to other locations as was observed for trypsin/chymotrypsin. To quantify the 20S digestion results, the fractional abundance for each peptide precursor and observed cleavage products are plotted versus reaction time in Figure 4d. The all-L substrate decreases in fractional abundance quickly (Fig. 4d, purple trace), while the all-(L-isoAsp) substrate changes very little in fractional abundance at any time point (Fig. 4d, yellow trace). Given that a full 16-residue portion of the peptide contains no isomerized sites, and multiple potential cleavage sites are located several residues from any isomers, it appears that the proteasome may have difficulty processing isomerized substrates. This may be related to reduced conformational flexibility available to substrates within the proteasome. Although this proximity may be efficient for promoting rapid, successive digestion events, it may also make it more difficult for modified substrates to be accommodated in the active sites.
Figure 4:
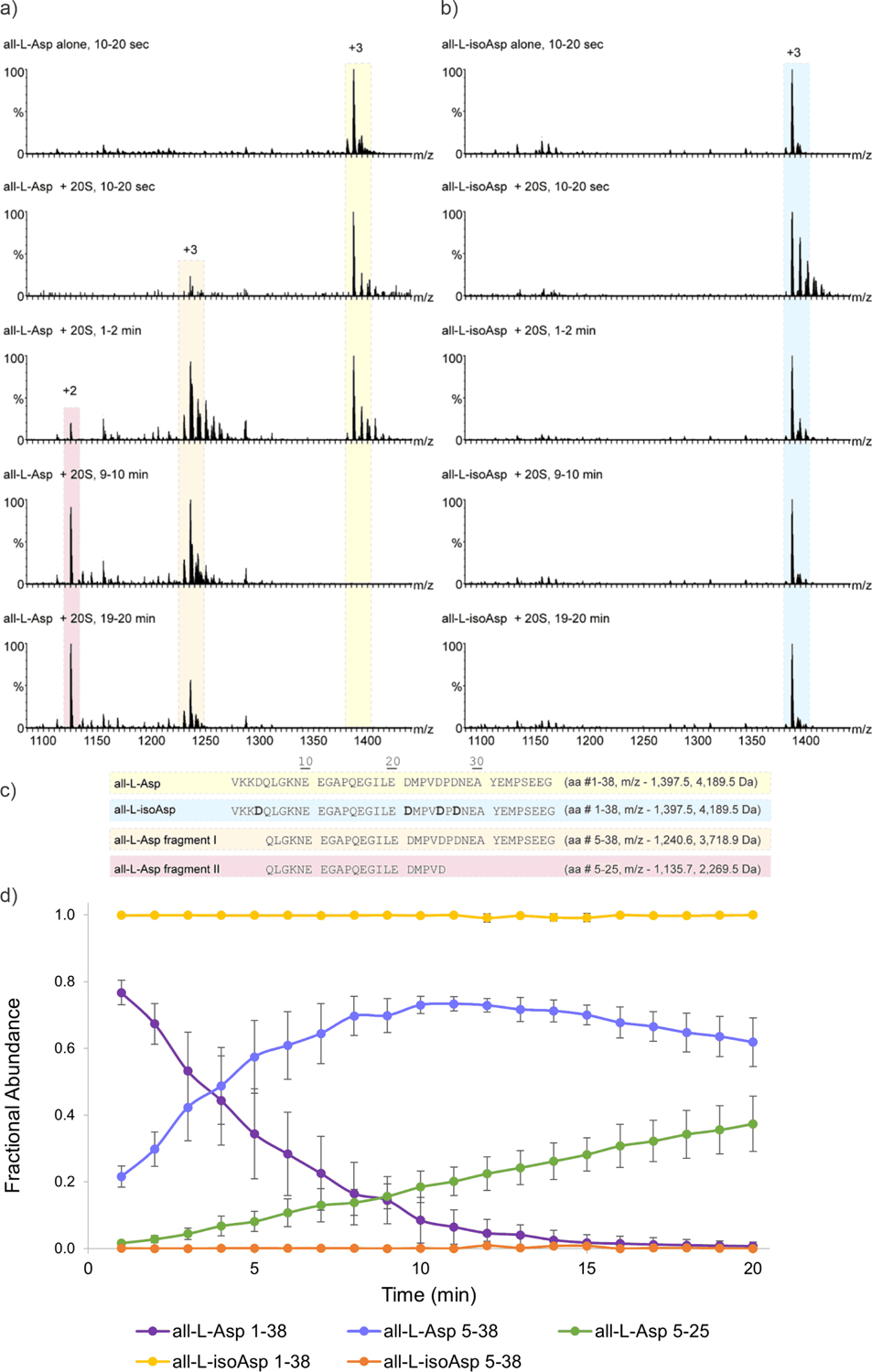
20S Proteasome digestion of alpha-synuclein 1–38. (a) Averaged mass spectra for 20S digestion of all-(L-Asp) substrate at specific time ranges. Precursor is highlighted in yellow, major digestion products are highlighted in orange and red. (b) 20S digestion of all-(L-isoAsp) substrate. Precursor is highlighted in blue. (c) Sequences of precursor and cleavage products. Isomerized residues are indicated with bold letters. (d) Fractional abundance plot of precursors and cleavage products. Each point represents the average of three experiments, and error bars represent the standard deviation.
Conclusion
The inherent longevity of LLPs enables the possibility for Asp isomerization, which presents nearly universal problems for proteolysis. In the context of LLP analysis, trypsin exhibits the least sensitivity to isomerization and therefore remains the preferred protease for proteomic experiments. However, missed cleavages should be enabled during data analysis to identify those sites where isomerization may have blocked digestion, and special care will need to be taken in quantitative experiments if dealing with isomerized LLPs. In particular, for LCMS experiments where isomers elute with partial or complete separation, the quantitation should include all isomers, or (ideally) each isomer should be quantified individually. If isomerization is less pervasive, proteases other than trypsin will likely be suitable, although the same caveats for missed cleavages would still apply. In terms of cell biology, we demonstrated that the 20S proteasome, although containing trypsin-like sites, is not able to efficiently digest isomerized peptides. When considered in light of previous work showing that the lysosome is also incapable of digesting isomerized LLPs, it is not clear how cells (particularly post-mitotic cells) would be able to clear isomerized LLPs. Recent work has shown that Asp isomerization in tau is higher in AD versus control samples and is well-correlated with markers indicative of impaired autophagic flux.36 Given that the burden of LLP isomerization is also unlikely to be reduced by the proteasome, the presence of isomerization should be an excellent indicator of disrupted proteostasis.
Supplementary Material
Acknowledgements.
The authors are grateful for funding from the NIH (R01 AG066626). MS is grateful for the support of the Sagol Institute for Longevity Research grant and a Moross Proof-of-Concept grant. MS is the incumbent of the Aharon and Ephraim Katzir Memorial Professorial Chair.
Footnotes
Supporting Information
First order rate equation plots for trypsin sequences, plots of precursor vs time for trypsin sequences
References
- 1.Truscott RJW; Schey KL; Friedrich MG Old Proteins in Man: A Field in Its Infancy. Trends Biochem. Sci 2016, 41 −8, 654–664.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samuel Zigler J; Goosey J Aging of Protein Molecules: Lens Crystallins as a Model System. Trends Biochem. Sci 1981, 6 (1), 133–136. [Google Scholar]
- 3.Toyama BH; Hetzer MW Protein Homeostasis : Live Long, Won’t Prosper. 2013, 14 (January), 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyon YA; Collier MP; Riggs DL; Degiacomi MT; Benesch JLP; Julian RR Structural and Functional Consequences of Age-Related Isomerization in α-Crystallins. J. Biol. Chem 2019, 294 (19), 7546–7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Livnat I; Tai HC; Jansson ET; Bai L; Romanova EV; Chen TT; Yu K; Chen SA; Zhang Y; Wang ZY; et al. A D-Amino Acid-Containing Neuropeptide Discovery Funnel. Anal. Chem 2016, 88 (23), 11868–11876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujii N; Maeda H; Takata T; Fujii N; Sakaue H; Nirasawa S; Takahashi S; Sasaki H Rapid Survey of Four Asp Isomers in Disease-Related Proteins by LC-MS Combined with Commercial Enzymes. Anal. Chem 2015, 87 (1), 561–568. [DOI] [PubMed] [Google Scholar]
- 7.Lowenson JD; Clarke S Structural Elements Affecting the Recognition of L-Isoaspartyl Residues by the L-Isoaspartyl/D-Aspartyl Protein Methyltransferase. Implications for the Repair Hypothesis. J. Biol. Chem 1991, 266 (29), 19396–19406. [PubMed] [Google Scholar]
- 8.Silzel JW; Lambeth TR; Julian RR PIMT-Mediated Labeling of L-Isoaspartic Acid with Tris Facilitates Identification of Isomerization Sites in Long-Lived Proteins. J. Am. Soc. Mass Spectrom 2022, 33 −3, 548–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alfaro JF; Gillies LA; Sun HG; Dai S; Zang T; Klaene JJ; Byung JK; Lowenson JD; Clarke SG; Karger BL; et al. Chemo-Enzymatic Detection of Protein Isoaspartate Using Protein Isoaspartate Methyltransferase and Hydrazine Trapping. Anal. Chem 2008, 80 10, 3882–3889. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi S; Ogasawara H; Hiwatashi K; Hori K; Hata K; Tachibana T; Itoh Y; Sugiyama T Paenidase, a Novel d-Aspartyl Endopeptidase from Paenibacillus Sp. B38: Purification and Substrate Specificity. J. Biochem 2006, 139 (2), 197–202. [DOI] [PubMed] [Google Scholar]
- 11.Lambeth TR; Dai Z; Zhang Y; Julian RR A Two-Trick Pony: Lysosomal Protease Cathepsin B Possesses Surprising Ligase Activity. RSC Chem. Biol 2021, 2 (2), 606–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamamoto K; Kida Y; Zhang Y; Shimizu T; Kuwano K Antimicrobial Activity and Stability to Proteolysis of Small Linear Cationic Peptides with D-Amino Acid Substitutions. Microbiol. Immunol 2002, 46 (11), 741–749. [DOI] [PubMed] [Google Scholar]
- 13.Checco JW; Zhang G; Yuan W; Yu K; Yin S; Roberts-Galbraith RH; Yau PM; Romanova EV; Jing J; Sweedler JV Molecular and Physiological Characterization of a Receptor for D -Amino Acid-Containing Neuropeptides. ACS Chem. Biol 2018, 13, 1343–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genchi G An Overview on D-Amino Acids. Amino Acids 2017, 49 (9), 1521–1533. [DOI] [PubMed] [Google Scholar]
- 15.Lambeth TR; Riggs DL; Talbert LE; Tang J; Coburn E; Kang AS; Noll J; Augello C; Ford BD; Julian RR Spontaneous Isomerization of Long-Lived Proteins Provides a Molecular Mechanism for the Lysosomal Failure Observed in Alzheimer’s Disease. ACS Cent. Sci 2019, 5 −8, 1387–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandey G; Julian RR LC-MS Reveals Isomeric Inhibition of Proteolysis by Lysosomal Cathepsins. Anal. Sens 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glickman MH; Ciechanover A The Ubiquitin-Proteasome Proteolytic Pathway: Destruction for the Sake of Construction. Physiol. Rev 2002, 82 (2), 373–428. [DOI] [PubMed] [Google Scholar]
- 18.Kumar Deshmukh F; Yaffe D; Olshina M; Ben-Nissan G; Sharon M The Contribution of the 20S Proteasome to Proteostasis. Biomolecules 2019, 9 −5, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baugh JM; Viktorova EG; Pilipenko EV Proteasomes Can Degrade a Significant Proportion of Cellular Proteins Independent of Ubiquitination. J. Mol. Biol 2009, 386 −3, 814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickering AM; Davies KJA Degradation of Damaged Proteins. Prog. Mol. Biol. Transl. Sci 2012, 109, 227–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie M; Schowen RL Secondary Structure and Protein Deamidation. J. Pharm. Sci 1999, 88 (1), 8–13. [DOI] [PubMed] [Google Scholar]
- 22.Stevenson CL; Friedman AR; Kubiak TM; Donlan ME; Borchardt RT Effect of Secondary Structure on the Rate of Deamidation of Several Growth Hormone Releasing Factor Analogs. Int. J. Pept. Protein Res 2009, 42 −6, 497–503. [DOI] [PubMed] [Google Scholar]
- 23.Lyon YA; Sabbah GM; Julian RR Identification of Sequence Similarities among Isomerization Hotspots in Crystallin Proteins. J. Proteome Res 2017, 16 −4, 1797–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wakankar AA; Borchardt RT; Eigenbrot C; Shia S; Wang YJ; Shire SJ; Liu JL Aspartate Isomerization in the Complementarity-Determining Regions of Two Closely Related Monoclonal Antibodies. Biochemistry 2007, 46 −6, 1534–1544. [DOI] [PubMed] [Google Scholar]
- 25.Noguchi S; Miyawaki K; Satow Y Succinimide and Isoaspartate Residues in the Crystal Structures of Hen Egg-White Lysozyme Complexed with Tri-N-Acetylchitotriose. J. Mol. Biol 1998, 278 (1), 231–238. [DOI] [PubMed] [Google Scholar]
- 26.Noguchi S Structural Changes Induced by the Deamidation and Isomerization of Asparagine Revealed by the Crystal Structure of Ustilago Sphaerogena Ribonuclease U2B. Biopolymers 2010, 93 (11), 1003–1010. [DOI] [PubMed] [Google Scholar]
- 27.Fujii N; Sakaue H; Sasaki H; Fujii N A Rapid, Comprehensive Liquid Chromatography-Mass Spectrometry (LC-MS)-Based Survey of the Asp Isomers in Crystallins from Human Cataract Lenses. J. Biol. Chem 2012, 287 (47), 39992–40002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukherjee S; Perez KA; Lago LC; Klatt S; McLean CA; Birchall IE; Barnham KJ; Masters CL; Roberts BR Quantification of N-Terminal Amyloid-β Isoforms Reveals Isomers Are the Most Abundant Form of the Amyloid-β Peptide in Sporadic Alzheimer’s Disease. Brain Commun. 2021, 3 (2), 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitcomb DC; Lowe ME Human Pancreatic Digestive Enzymes. Dig. Dis. Sci 2007, 52 (1), 1–17. [DOI] [PubMed] [Google Scholar]
- 30.Siepen JA; Keevil E-J; Knight D; Hubbard SJ Prediction of Missed Cleavage Sites in Tryptic Peptides Aids Protein Identification in Proteomics. J. Proteome Res 2007, 6 (1), 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Šlechtová T; Gilar M; Kalíková K; Tesařová E Insight into Trypsin Miscleavage: Comparison of Kinetic Constants of Problematic Peptide Sequences. Anal. Chem 2015, 87 (15), 7636–7643. [DOI] [PubMed] [Google Scholar]
- 32.Hood CA, Fuentes G, Patel H, Page K, Menakuru M, Park JH Fast Conventional Fmoc Solid-Phase Peptide Synthesis With HCTU. J. Pept. Sci 2008, 14, 97–101. [DOI] [PubMed] [Google Scholar]
- 33.Anthis NJ & Clore GM Sequence-Specific Determination of Protein and Peptide Concentrations By Absorbance at 205 nm. Protein Sci. 2013, 22, 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben-Nissan G, Vimer S, Tarnavsky M, Sharon M. Structural Mass Spectrometry Approaches To Study The 20S Proteasome. Methods Enzymol. 2019, 619, 179–223. [DOI] [PubMed] [Google Scholar]
- 35.Ma W; Tang C; Lai L Specificity of Trypsin and Chymotrypsin: Loop-Motion-Controlled Dynamic Correlation as a Determinant Biophys. J 2005, 89, 1183–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hubbard EE; Heil LR; Merrihew GE; Chhatwal JP; Farlow MR; McLean CA; Ghetti B; Newell KL; Frosch MP; Bateman RJ; et al. Does Data-Independent Acquisition Data Contain Hidden Gems? A Case Study Related to Alzheimer’s Disease. J. Proteome Res 2022, 21 (1), 118–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


