Abstract
Chikungunya virus (CHIKV) is a representative alphavirus causing debilitating arthritogenic disease in humans. Alphavirus particles assemble into two icosahedral layers: the glycoprotein spike shell embedded in a lipid envelope and the inner nucleocapsid (NC) core. In contrast to matrix-driven assembly of some enveloped viruses, the assembly/budding process of two-layered icosahedral particles remains poorly understood. Here we used cryogenic electron tomography (cryoET) to capture snapshots of the CHIKV assembly in infected human cells. Subvolume classification of the snapshots revealed 12 intermediates, representing different stages of assembly at the plasma membrane. Further subtomogram average structures ranging from subnanometer to nanometer resolutions show that immature, non-icosahedral NCs function as rough scaffolds to trigger icosahedral assembly of the spike lattice, which in turn progressively transforms the underlying NCs into icosahedral cores during budding. Further, analysis of CHIKV-infected cells treated with budding-inhibiting antibodies revealed wider spaces between spikes than in icosahedral spike lattice, suggesting that spacing spikes apart to prevent their lateral interactions prevents the plasma membrane bending around NC, thus blocking virus budding. These findings provide the molecular mechanisms for alphavirus assembly and antibody mediated budding inhibition that provide valuable insights in development of broad therapeutics targeting assembly of icosahedral enveloped viruses.
Keywords: Chikungunya virus, alphavirus, virus assembly and budding, enveloped virus, cryogenic electron tomography, subtomogram averaging, antibody
Introduction
Enveloped virus assembly is a highly coordinated process that requires budding the cell membrane and incorporating all necessary components into the viral particle for subsequent cell infection. This process is relatively well understood for viruses whose assembly and budding are driven solely by capsid or matrix proteins via recruiting host ESCRT machinery, such as retroviruses and filoviruses1. In contrast, assembly and budding of alphaviruses, a representative genus of enveloped spherical viruses containing two icosahedral structural protein layers, is less understood, with multiple contradictory models proposed in previous studies2. Chikungunya virus (CHIKV) is the most common alphavirus infecting humans and has caused global epidemics of debilitating, and often chronic polyarthritis3. No licensed vaccine or antivirals are available for treating any alphavirus infection. All alphaviruses share a spherical virion architecture ~70 nm in diameter, with concentric spike and NC layers separated by a lipid envelope. The outer glycoprotein (GP) shell of 240 copies of E1-E2·(E3) heterodimers are arranged as 80 prominent trimeric surface spikes embedded in the viral envelope, while an inner nucleocapsid (NC) core of 240 capsid proteins (Cps) encapsulates the 11.5kb plus-sense (+) genomic RNA4. At the late stage of virus infection, NC cores assembled in the cytosol and trimeric E1-E2·(E3) spikes, synthesized and processed through the membrane secretory system, co-assemble into virus particles at the plasma membrane. Vertical links across the lipid envelope between spikes and NC are mediated by insertion of the intracellular tail of E2 into the hydrophobic pocket of Cp5–7. Contradictory mechanisms were proposed for the assembly of two-layered icosahedral alphavirus particles, largely centered around whether Cps pre-assemble into icosahedral NCs5,6 that serve as structural templates or if spikes drive icosahedral co-assembly of outer-spike and inner-NC lattices2. Such assembly of spherical icosahedral shells during assembly/budding is relevant to other enveloped viruses, including the notable flaviviruses Dengue virus and Zika virus. Importantly, we previously demonstrated that the assembly of the two-layered icosahedral CHIKV particles can be blocked by antiviral antibodies8–10. Without direct visualization at molecular-resolution of both the assembly/budding process and spikes in a budding-blocked conformation in virus-infected cells, the mechanisms of icosahedral particle assembly and antibody-induced budding inhibition remain poorly defined.
Here we imaged vitrified, CHIKV-infected human cells and determined structures of viral assembly intermediates in situ using cryogenic electron tomography (cryo-ET). By analyzing both the full progression of CHIKV assembly in the cell and budding blocking by antibodies, we revealed the structural organization of spikes and NCs prior to budding, during their orchestrated co-assembly, and in a budding-blocked conformation. This resolved the structural mechanism of two-layered, icosahedral particle assembly, and importantly, that of budding-blocking molecules that can serve as anti-alphavirus therapeutics.
Results
Cell tomography captures CHIKV budding intermediates
To capture the dynamic CHIKV assembly/budding process in the near-native state, we imaged U2OS cells, a human bone osteosarcoma cell line, that were infected with CHIKV-181 vaccine strain on a cryo-EM grid and embedded in vitreous ice. Tomographic tilt series were collected at the infected cell peripheries at 8 hours post-infection (hpi) and revealed a variety of CHIKV assembly phenotypes (movie S1): budding at the PM of the cell body (Fig. 1A–D), budding on long intercellular extensions (>10 μm) and short extensions (typically 2–10 μm in length), and thin branching extensions composed solely of incomplete viral particles (Fig. 1A–B). Particles were observed at the PM at various stages of budding and as fully assembled virions released into the extracellular space (Fig. 1E–F), thus capturing snapshots of the entire CHIKV assembly/budding process. Interestingly, CHIKV replication spherules, where viral RNAs are synthesized, were occasionally observed near cytosolic nucleocapsid-like particles (NLPs) and budding viruses (Fig. 1C–D, Fig. S1). This is consistent with previous reports of CHIKV replication spherules located predominantly on the cell surface11. It is conceivable that viral RNAs can be synthesized and immediately packaged into NLPs that bud into virions, all near the PM, which is different from other old-world alphaviruses that have RNA replication and virus budding spatially separated12. We also identified thin extensions from the cell periphery, formed by incomplete particles often the width of a single virion (~70 nm diameter, <5 μm length) and linked by a continuous membrane (Fig. S2). These extensions displayed differences in the levels of particle completion and structural conformations (Fig. S2), but due to the lack of sufficient spikes to finish enwrapping the NC as an icosahedron, the linked particles are unlikely to complete the assembly of full virions. Therefore, such particles were excluded from analysis of two-layered icosahedral particle assembly.
Figure 1. CHIKV assembly and budding at the infected-cell periphery.

(A-B) Representative low magnification images of the cell periphery from 10+ repeated experiments reveal cell body (cyan arrow), long intercellular extension (white arrow), short extensions enriched in virus assembly features (green arrows), and thin extensions of viral particles (yellow arrows) emanating from the short extensions or cell body. Scale bars, 5 μm. (C) A representative tomographic slice image of cell periphery from 185 tomograms depicting virus budding events (red arrows) and RNA replication spherules (navy blue arrow) at the PM with (D) corresponding 3D segmentation of cellular features. (EX: extracellular space) Scale bar, 200 nm. (see also Fig. S1) (E) Tomogram slice of short extension with budding intermediate particles (red arrow), spikes (yellow arrow), and cell free virion (blue arrow). Scale bar 200 nm. (F) Enlarged view of the boxed region in (E) shows intermediate viral assembly complexes at the PM, composed of a spike budding shell and nucleocapsid (NC). Scale bar, 100 nm.
Next, we studied how icosahedral alphavirus particles form at the PM by analyzing snapshot images of 1,918 individual budding states picked from the 3D tomograms. These snapshots were subjected to subtomogram-alignment and classification through an unbiased and iterative multi-reference refinement protocol based on structural similarity (Fig. S3A, see “Methods” section). This resulted in 12 class average maps with distinct stages of the virus assembly (Fig. 2) that displayed two-layered particles with different budding levels. We defined the budding level of an intermediate assembly state as the percentage completion of the outer glycoprotein shell relative to the final spherical virion. Budding level was estimated for each class average map by using the mature icosahedral virion with 80 glycoprotein spikes as a reference and counting the number of spikes covering its partially-formed budding shell (Fig. 2D). The 12 class average maps were ranked by increasing budding level and the particle numbers of individual states within each class were determined (Fig. 2D). This classification scheme resulted in large numbers of particles (868 particles, 63% of total) grouped into classes (9–12) at the very late stages of assembly/budding with 94–100% complete glycoprotein shells. In this analysis, the 100% assembled particles are still tethered to the PM. The proportion of particles within early budding classes (1–3, 12–50% particle completion) is less (288 particles, 21% of total), and even fewer particles (191, 14% of total) were grouped in the remaining classes (4–8, 50–88% completion). In our analysis, we assumed that a snapshot of any intermediate state can be captured and that at the time we imaged the cells (8 hpi), virus assembly and supply of Cp and spikes were at a steady state across cells on the grid. The discrete classes with a larger number of particles could be interpreted as the states having a low free energy and would thus transition to the next assembly state at a slower rate. On the contrary, the classes with fewer particles could have a faster rate of transition to the next assembly state. Our results suggest the kinetic progression of CHIKV budding is non-uniform, with formation of the first half of the glycoprotein shell a minor rate-limiting step, and final pinch-off of the complete virions the major rate-limiting step. Further work is warranted to support this proposed model of assembly kinetics, while alternative interpretations related to bursts of virus assembly in the cells cannot be excluded from the current study.
Figure 2. Classification and enumeration of CHIKV budding intermediates.
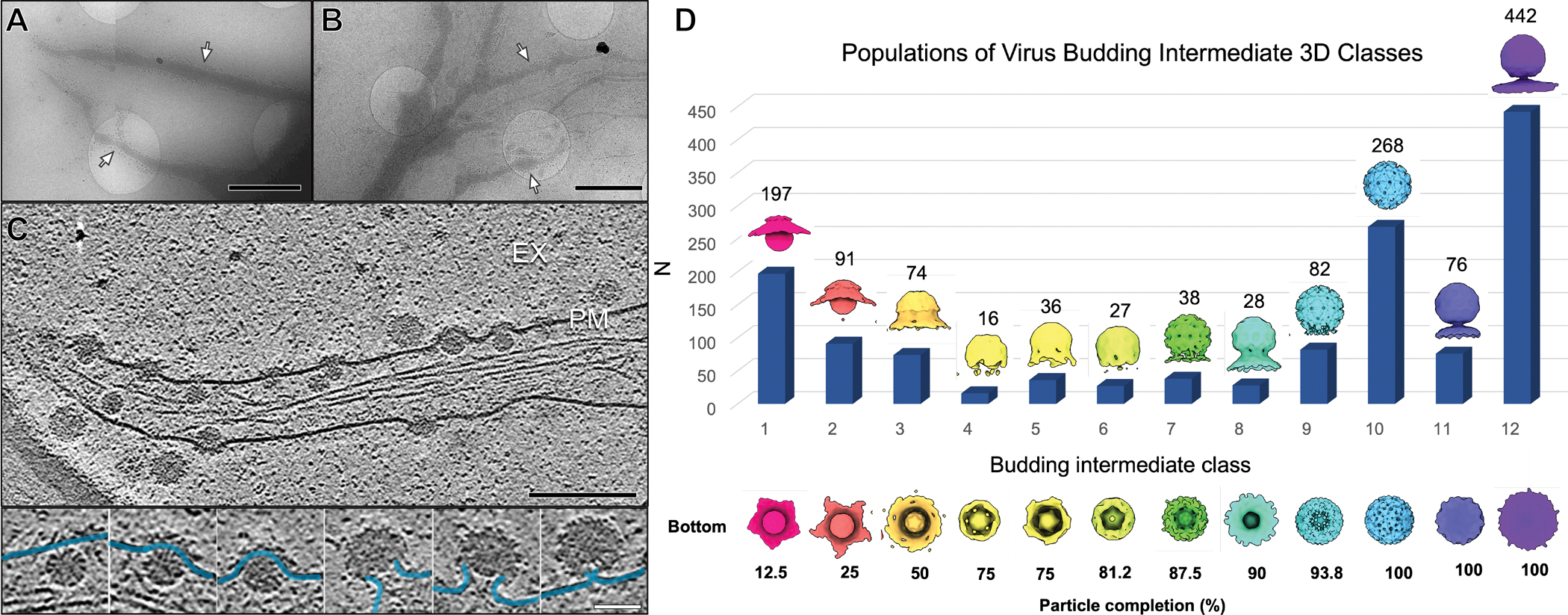
(A-B) Representative images of virus-infected cells with extensions enriched in virus assembly (white arrows) from 185 tomograms. Scale bars, 2 μm. (see also Fig. S2) (C) Tomogram slice image depicts snapshots of the virus budding process. (EX-extracellular space, PM-plasma membrane). Scale bar, 200 nm. Selected particle images (insets below) reveal heterogeneity based on conformations of the bending PM (blue). Scale bar, 50 nm. (D) CHIKV-budding intermediate 3D classes determined by subvolume classification. Density maps of each class average (1–12) are colored uniquely and displayed with side-view and bottom-view (viewed from below the PM). The number of particles (N) assigned to each class displayed as a bar graph with respective N listed above. (see also Fig. S3)
In light of previous studies that described significant inter- and intra-particle heterogeneity in alphavirus particles following in vitro purification, we then evaluated images of released virus particles in the 3D tomograms6,13. Concerns about the fragility of enveloped viruses to purification have called into question the relevance of observed structural heterogeneity in single particle cryoEM analysis of biochemically purified virus to alphavirus assembly in situ13,14. Therefore, our direct imaging of CHIKV-infected cells eliminates potential structural perturbations to the biochemically purified virus particles.
Using a Volta phase plate (VPP) to collect images for increased contrast, released virions typically displayed relatively weak or absent density at one side of the particle between spike and NC core layers (Fig. S4). In some rare extreme events, multiple NC cores bud into one released particle and similarly at the un-enveloped side of each core an absent density was observed (Fig. S4E). Interestingly, the trailing-end of late-stage budding particles still tethered to the PM also displayed relatively absent density with similar non-icosahedral geometry of the viral envelope (Fig. S4B). In some tomograms, unidentified molecular complexes were observed at the base of the viral envelope in the relatively absent density region (Fig. S4D). Therefore, released CHIKV particles possess imperfect icosahedral symmetry that likely results from the final membrane scission step of virus, reminiscent of imperfect symmetry of flaviviruses15.
Detailed CHIKV budding structures
Two-layered, icosahedral (5-3-2) symmetry is a feature of purified and mature CHIKV particles4, but the role of spike and NC layers in assembly/budding has not been established. We next explored how the glycoprotein spike and NC layers were organized at different stages of budding to decipher the mechanism of alphavirus assembly. We performed further subtomogram-refinement of released virions and four budding intermediate classes that displayed weak icosahedral features during previous 3D classification (Fig. 2), applying five-fold symmetry (see Methods). Among the resulting five maps, the earliest budding structure displays a 5-fold pentagon of spikes only at the leading end of budding, while the other four structures displayed excellent pentagon and hexagon assemblies with 5-fold, 3-fold, and 2-fold symmetry axes (Fig. 3B). The maps ranged in resolution from 8.3 Å (released) to ~44 Å (“docking”) (0.143 FSC criterion) (Fig. 3B, Fig. S3B). Subnanometer resolution in the released virion average is validated by the visualization of E1/E2 transmembrane helices spanning the viral envelope (Fig. S3C–D). The resolution progressively decreases in maps of less-complete particles and is likely caused by increased conformational flexibility due to lack of icosahedral symmetry constraints. A striking correlation between icosahedrally-symmetric regions of the spike budding shells and Cps of the underlying NC core was observed at the “leading end” of budding (Fig. 3B). In contrast, there is a lack of detectable icosahedral symmetry in the “trailing end” of each intermediate NC’s structure, where no spikes are present (Fig. 3B, Fig. S4A–C). The NC of the earliest budding map (“docking”) only shows five-fold symmetry at the “leading end” with a small cluster of spikes above, and is significantly smaller (long axis: 37 nm, short axis: 31 nm) than the icosahedral NC of a released virion (41 nm). Overall, as the growing icosahedral spike shell enwraps the NC it reorganizes Cps from asymmetry into matching icosahedral symmetry. This result explains the origin of two-layered icosahedral symmetry evident in mature alphavirus virions7.
Figure 3. In situ refined structures of CHIKV assembly intermediates after extensive subtomogram classification and averaging.

(A) Tomogram slice image displays budding intermediate particles at the PM (white arrows) and an apparently-cytosolic NLP (red arrow). Scale bar, 200 nm. (B) Radially-colored density maps of five CHIKV ensemble subtomogram averages arranged in accordance with budding progression from earliest budding level (“I.Docking”) to latest (“V.Released” virion) with half-cut representations (middle row) and NC-zoned densities (bottom row). PM and NC density (“Half-Domed”, bottom row) from class averages prior to additional 3D refinement depicted as white surfaces. (see also Fig. S3 & S4) (C) Subtomogram average structures of two apparently-cytosolic NLP classes. Class I displays no interpretable 5-fold symmetry while class II shows weak five-fold symmetry at one pole that indicates it is at an early stage of budding. (see also Fig. S4 & S5)
To assemble a two-layered icosahedral particle, the previously proposed NC-centric model of alphavirus budding16 suggested that preformed icosahedral NCs provide a symmetric template for spike incorporation. To test this model and our previous interpretation of spike-driven reorganization of NCs during budding, we performed subtomogram-classification and refinement of apparently cytosolic NLPs present in the 3D tomograms. 545 apparently cytosolic NLP subvolumes were picked from the 3D tomograms on condition there was no clear budding or attachment to spikes at the PM (Fig. 3A, Fig. S5A). Due to the orientation of the membrane in the tomograms, clear budding structures at the plasma membrane were only visible in the side view, while the top and bottom membrane surfaces were invisible. Subtomogram-classification of the apparently cytosolic NLP particles was performed with five-fold symmetry applied and resulted in two classes with NC-like structures (class I&II). Further subtomogram-refinement of those two sets of particles, again with applied five-fold symmetry, produced low-resolution structures shown in Fig 3C at resolutions of 47.6Å (class I) and 43.5Å (class II). However, both maps lacked icosahedral symmetry, while only class II displayed local five-fold symmetry features at one pole (Fig. S4A). The diameters of the two maps were both ~37 nm, with one structure (class I) largely spherical and the other (class II) an oblate spheroid with short axis 33 nm (Fig. 3C, Fig. S4A). Due to overall similarity between the class II map with the NC of the earliest budding intermediate, we next asked whether the local five-fold symmetry at one pole of the class II map was a result of interactions with spikes at the PM in orientations that were not clear in the 3D tomograms.
We proceeded by utilizing the refined subvolume orientations of each particle in class II to map the particles back into the original 3D tomograms for cellular context. Most particles within class II were positioned in the cell with the five-fold symmetric pole oriented towards the PM. This indicates that five-fold structure arises from interactions with membrane-embedded spikes (Fig. S5), and different from cytosolic NLPs, these particles are early stage budding NCs docking at the plasma membrane. Patches of spikes were observed in tomogram slices above or below many of these particles, adding further support to this conclusion (Fig S5). At the current resolution, the lack of symmetry in class I cytosolic NLP particles cannot rule out that the Cp-RNA mixture is organized into assemblies with alternate, non-icosahedral and non-five-fold symmetry. Taken together, the results indicate the spike lattice must reorganize cytosolic NLPs from asymmetric assemblies to icosahedral viral cores during assembly/budding.
Assembly of spike lattices
Our structures of budding-intermediate states (Fig. 2) revealed a progressive spike-driven NC morphogenesis, demonstrating the mechanistic role of the spike lattice in alphavirus assembly. However, how the spike layer acquires the proper icosahedral symmetry is not well understood. Detailed spike organization at the PM has never been observed in virus-infected cells due to technical challenges in resolving small macromolecules in different orientations without chemical stains. Here we applied contrast-enhancing VPP cryo-ET imaging to investigate how spikes organize outside virus budding sites. From 3D tomograms of infected cells, we observed rare non-icosahedral spike assembly products that provided unexpected insights into icosahedral spike assembly. First, we identified rare instances of near-planar sheets of hexagonal spike lattices without underlying NCs (Fig. S6). Secondly, we observed highly-curved tubular spike lattices in filopodia-like structures: on short extensions with budding particles at the tip (Fig. 4A), on long extensions entirely devoid of NCs (Fig. 4C, Fig. S7), and on thin extensions loaded with budding particles (Fig. 4E, Fig. S6). We determined subvolume averages of three different tubular spike lattices by applying helical rotations to compensate for the tomographic missing wedge. The results revealed helical organization of trimeric spikes arranged as hexagons with uniform spacing and conformation, but no internal NCs or helically-arranged Cps were found within any of the membrane tubes (Fig. 4). Interestingly, the average diameter of these spike coated membrane tubes varies from 55 to 65 nm, close to the diameter of the icosahedral, enveloped CHIKV virion (~70 nm) (Table S2). Of the different spike assemblies we observed, pentagons of trimeric spikes were only observed on the surface of budding-intermediates and released particles (Fig 3, Fig. S6). Self-assembled hexagonal spike lattices can cover flat and highly curved membrane surfaces (Fig. 4, S6 & S7) but do not form spherical icosahedrons. Taken together with the heterogeneous non-icosahedral feature of cytosolic NLPs (Fig. 3), these data suggest NCs serve as rough spherical scaffolds that initiate assembly of the proper icosahedral spike lattice that then drives assembly/budding.
Figure 4. CHIKV envelope spikes arranged in hexagonal lattices form helical tubes in situ.

(A,C,E) Tomogram 2D slice images of cell extensions with membrane-embedded spike arrays (white dashed boxes) with enlarged inset views. Arrays form (A) at the base of a budding intermediate particle with NC at the leading end (white arrow), (C) on a cell extension without nearby NCs (see also Fig. S7), and as a (E) segment within a thin extension containing nearby budding viral particles (see also Fig. S6). Scale bars, 200 nm. Inset images show a different tomogram slice of each respective lattice (scale bars 75 nm). (B,D,F) 2D slice view of envelope lattice 3D subtomogram average (left) and density map (middle) (corresponding to A,C,E dashed white boxes) reveal helical arrays of trimeric spikes arranged as hexagon lattices. Density maps of each tube, rotated to view down the helical axis (right), revealed no underlying NCs below the spike lattice and membrane bilayer. Scale bars, 20 nm.
Besides the hexagonal spike lattices that were observed on thin membrane extensions of CHIKV-infected cells, no organized spike assemblies were observed at the PM near virus budding sites. Some spike-like proteins were often identified around the base of the budding particles, but were not organized into detectable higher-order assemblies (Fig. S8), in marked contrast to organized spikes in the hexagonal and icosahedral lattices (Fig. 3, 4, S6 & S7). These results suggest that the icosahedral spike lattice on budding particles is unlikely to be reorganized from any pre-assembled higher-order spike assembly on the plasma membrane.
Disrupting lateral spike interactions blocks CHIKV budding
Building on our results showing icosahedral spike lattice formation is essential to CHIKV assembly/budding, we investigated the mechanism for budding inhibition by anti-CHIKV antibodies that we previously reported8,10. We utilized anti-CHIKV NAb C9 that bivalently binds to spikes at the PM and blocks virus budding, to investigate the organization of spikes in a budding-blocked state8,10. CHIKV-181-infected U2OS cells were treated with C9 at 2 hrs. post-infection and we collected tomographic tilt series of the cells 6 hrs. later using VPP. 3D cryoET tomograms revealed large numbers of cytosolic NLPs docked to the inner leaflet of the PM without virus budding (Fig. 5, movie S2), consistent with our previous report10. Computational slices of budding-arrested NLPs clearly resolved a flattened docking end below the near-planar PM. 3D classification of 1,727 budding-arrested NLPs revealed six conformations, two of which contained membrane density, and four without any membrane feature that resembled the cytosolic NLP class I described above (Fig. 3C, Fig. S3A). Budding-arrested NLPs were predominantly docked to the plasma membrane (~82% of particles) with flattened ends that directly interact with the cytoplasmic tail of spikes (Fig. 5B, I). The lack of icosahedral or five-fold structure in cytosolic budding-arrested NLPs, and a transformation in structure upon docking to the PM, further supports the model of spike-driven NC reorganization.
Figure 5. Neutralizing antibody C9 crosslinks spikes at the cell surface and induces coalescence of spike-C9 complexes.
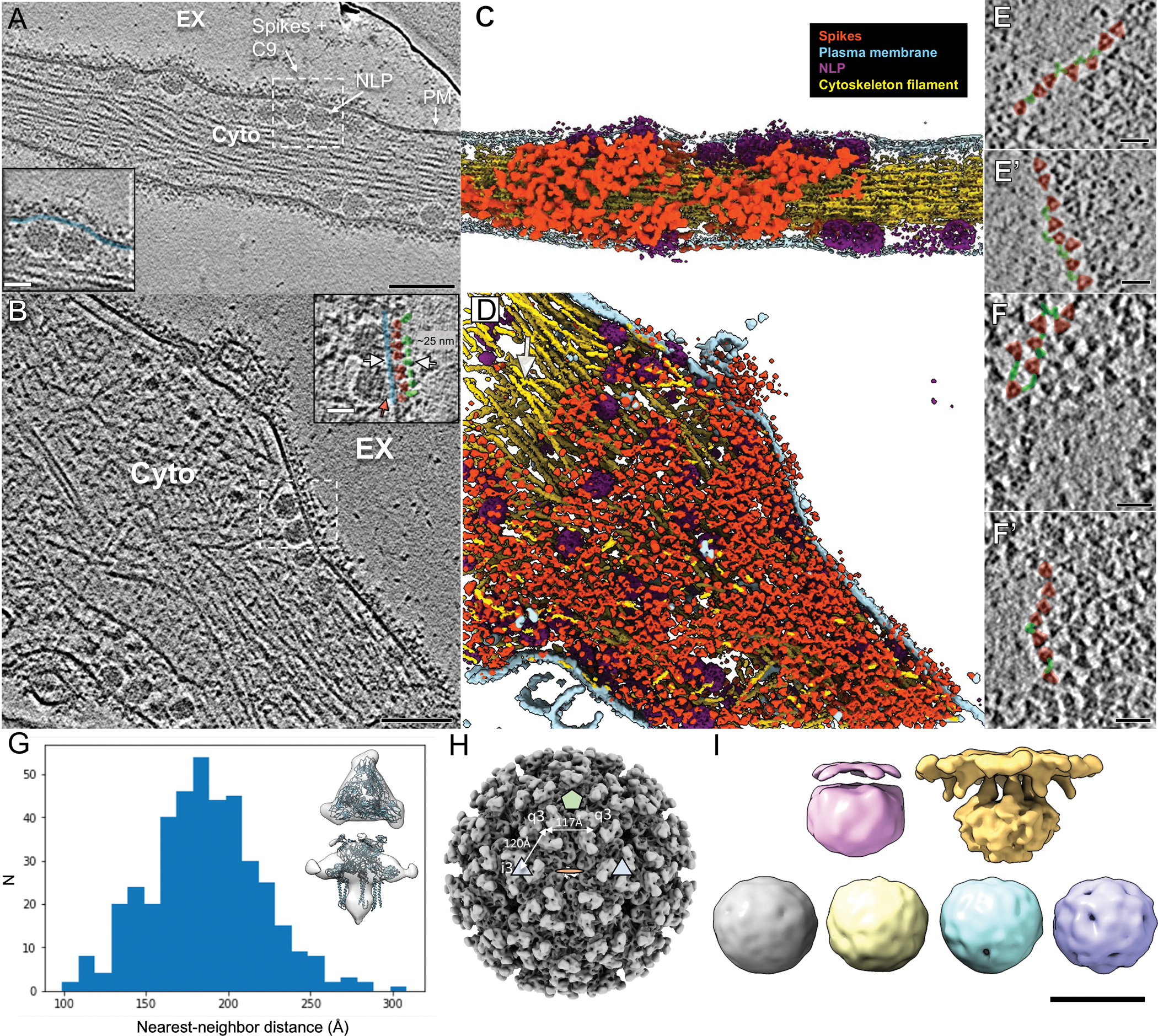
(A-B) Individual slices of Volta phase plate cryoET tomograms displaying CHIKV-infected cells treated with NAb C9 revealed arrested NLPs at the PM inner leaflet and dense, coalesced spike-C9 complexes on the PM outer leaflet (inset images: docked NLPs interacting with spike intracellular tails (red arrow) at the PM inner leaflet (blue), with spike ectodomains (pink) bound to NAb C9 (green) outside the cell. Scale bars, 100nm. Inset images scale bars, 25nm. EX-extracellular, Cyto-cytosol. (C-D) Corresponding 3D cellular annotations of tomograms (A-B), with trimeric spikes (orange-red), PM (light blue), NLPs (purple) and cytoskeleton filaments (gold) colored. (E, E’, F, F’) Zoomed-in top views of envelope spikes (orange-red) embedded at the PM with C9 (green) intercalating trimeric spikes. Spikes with bridging C9 density (green) often arranged with clear, near-linear boundaries. Scale bars 25 nm. (G) Subvolume average of C9-linked trimeric spikes and plot of distance between spike and nearest neighbor after refinement of orientation for each extracted spike subvolume in representative tomogram. Low-resolution subvolume spike average shows general agreement with the CHIKV spike atomic model (PDB:3J0C). (H) Distances on the virus particle between icosahedral-3-fold (i3) spikes and quasi-3-fold spikes (q3), as well as q3-q3 spikes, displayed on the virus particle with icosahedral 2-fold (orange disc), 3-fold (blue triangle) and 5-fold (green penton) for reference. (I) Subtomogram class averages of NLPs within the cytoplasm of C9-treated cells. Two classes (top) display reorganization at the membrane-proximal end while other four class averages (bottom) are roughly spherical and lack icosahedral symmetry. Docking end of egress-blocked NLPs is flattened in comparison to cytosolic NLP.
Tomogram slices displaying side-views of the PM revealed three characteristic layers: the PM, spike ectodomains above the PM, and a second protein layer above the spikes, approximately 150–250 Å from the inner leaflet of the PM (Fig. 5A–B). The dense protein layer above spike ectodomains, not seen in regular CHIKV-infected cells (Fig. 1F), indicated binding of C9 IgGs to the previously reported epitope at the crest of spikes8. Computational slices corresponding to top- and bottom-views of the cells revealed clear trimeric spikes in large patches at micrometer scale (Fig. 5E–F, Movie S2). Densities were observed bridging across spikes with Y-shaped features, consistent with IgG structure (Fig. 5E–F). Taken together, these observations indicated spikes were bridged by C9 and blocked from the structural rearrangements necessary for icosahedral assembly. The flat feature of spike patches and the docking end of underlying NLPs further supports our model of outside-in, spike-driven reorganization of Cps in those interacting NLPs.
After confirming the lack of virus budding and NAb crosslinking spikes in the cell tomograms, we asked how the NAbs blocked budding at the molecular level. We first determined the precise location of individual spikes in the 3D tomograms. 7,678 trimer spikes were manually picked, followed by subtomogram refinement that yielded a density map at ~24 Å resolution (0.143 FSC criterion). The map approximately matches the atomic model of the CHIKV trimeric spike (Fig. 5G). Using the refined orientations of each spike in a tomogram, we computed the distance between a spike and its nearest neighbor spike at the PM. The median distance between centers of nearest-neighbor spikes in the C9-coalesced spike patches was 185.2 Å (range from 98 Å to 309 Å) (Fig. 5G). This distance is significantly greater than that between neighboring spikes in pentagons and hexagons of icosahedral CHIKV particles (117 Å and 120 Å respectively) (Fig. 5H). This data revealed the molecular mechanism of budding inhibition by anti-alphavirus antibodies: crosslinking spikes to space them apart and therefore preventing lateral spike-spike interactions required to drive the icosahedral shell assembly. This further supports the spike-driven alphavirus assembly/budding model. The absence of ordered spike assemblies in the spike-C9 coalescence also suggests that spikes are again unlikely to be delivered to the PM as pre-assembled lattices, arguing against what was proposed from the observation of hexagonal spike lattice tubes inside cytopathic vacuole type-II in Semliki Forest virus-infected cells17.
Discussion
CryoEM has been a revolutionary tool capable of resolving near atomic resolution structures of purified macromolecules and macromolecular assemblies both with and without global symmetry. Advances in image reconstruction have further allowed the possibility of resolving multiple structures from compositionally or conformationally mixed assemblies within a single biochemical preparation18. This work presents the technical feasibility of imaging an entire virus assembly/budding process in situ and computationally classifying the snapshots into discrete intermediate state structures at subnanometer- to nanometer-resolution. Importantly, such an approach is scalable with large datasets, where more particles per class result in more interpretable intermediate structures that give further insights into the assembly path.
Here, we captured cytosolic NLPs prior to budding and the entire progressive co-assembly of two-layered icosahedral shells upon NC interaction with spikes in CHIKV-infected human cells. These structures revealed the molecular mechanism of alphavirus assembly/budding as illustrated by our model (Fig. 6). Briefly, cytosolic NLPs lack observable symmetry and serve as a rough scaffold to trigger icosahedral assembly of the envelope spike lattice. The spike lattice likely grows through lateral self-interactions with additional spikes and progressively reorganizes the underlying, initially asymmetric NCs into expanded, icosahedral viral cores through direct spike:Cp contacts. Without underlying NCs, spikes can self-assemble into rare hexagonal lattices covering flat sheets or thin membrane tubes. These spike lattices can potentially shed from the cell as capsid-less subviral particles like reported for many virus families19–25, or form long extensions to promote virus cell-to-cell transmission although different from other reported virus-induced cytoskeleton containing cell extensions26–29. Our spike-driven assembly/budding model for alphaviruses can also be applicable to other two-layer icosahedral enveloped viruses like flaviviruses, which similarly assemble concentric icosahedral spike and NC lattices but apparently without utilizing a preformed NC. In the cell we show that asymmetric, preformed NCs are not required for assembly of highly-curved tubular spike lattices but actually initiate formation of a less-curved, icosahedral spike lattice of T=4 symmetry around them. This mechanistic function of NCs in serving as rough scaffolds to promote the less-curved icosahedral architecture of the infectious particle is consistent with the role of flavivirus capsid protein in promoting assembly of T=3 virions rather than T=1 subviral particles formed by spikes alone. Further, this NC scaffold function in RNA viruses is reminiscent of dsDNA bacteriophage scaffolding proteins that control the hexamer:pentamer ratio during icosahedral capsid assembly and lead to production of infectious, larger icosahedral particles30. Our work finally reconciles previous contradictory NC- or spike-centric budding models derived from structures of purified fully-assembled virions7, in vitro assembled nucleocapsids31,32 and various mutations disrupting virus assembly and budding33–35. Similar to this study, we expect further mechanistic discoveries will be made using this experimental framework to image assembly processes and functional cycles of other biological assemblies in the native cellular context.
Figure 6. Mechanistic model of alphavirus budding and assembly.
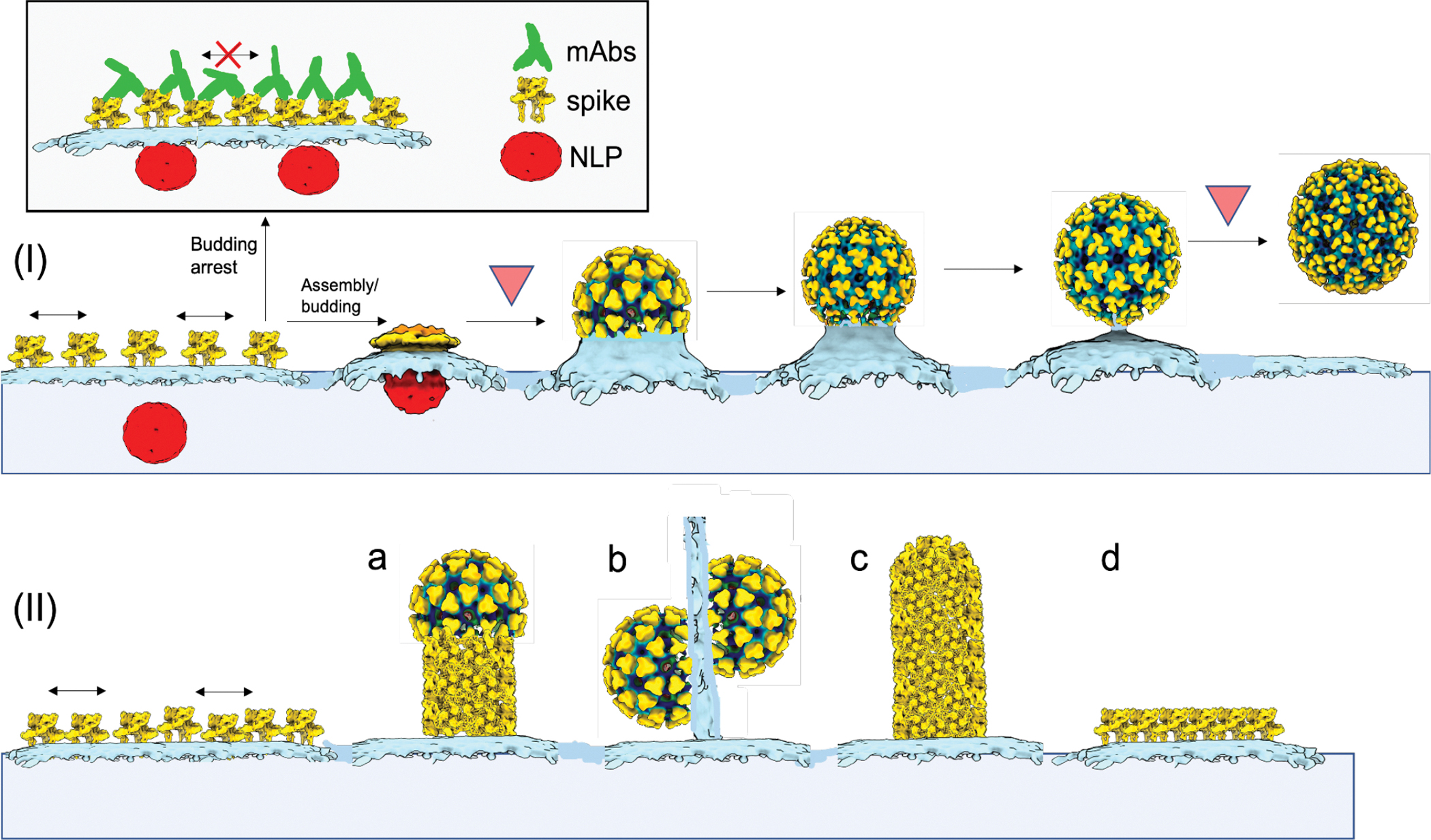
At the CHIKV-infected cell surface, immature non-icosahedral NLPs and membrane-embedded spikes converge. Subsequent virus budding (I) is predicated on assembly of the icosahedral spike lattice that enwraps NLPs and reorganizes them into icosahedral NCs through sequential spike:Cp interactions. Rate-limiting steps to particle formation likely occur at early- and late-stages associated with assembly of the first half of the virions and membrane scission following completion of full virions, respectively (upside-down red triangles). Released virions contain near-icosahedral spike and NC layers, with local disruptions in the lattices likely related to membrane scission and virus release from the PM. Binding of mAbs to exposed spike surfaces at the PM (boxed) inhibits virus biogenesis by preventing formation of the curved, icosahedral spike shell. (II) Spikes can self-assemble into non-icosahedral structures, giving rise to rare alternative assembly products, including (a,c) helical tubes formed by spike hexagons, (b) thin extensions of linked, incomplete particles and (d) planar hexagonal sheets of spikes.
Our data also resolved the structural organization of NAb crosslinked spikes on the CHIKV-infected cell surface and revealed the molecular mechanism for antibody-mediated budding inhibition of alphaviruses8,9,36. Crosslinking antibodies serve as molecular wedges between neighboring spikes and prevent spike-spike lateral interactions required for spike lattice assembly that is shown to drive budding through bending of the PM around NCs. Based on this mechanism, targeting conserved regions of spikes with antibodies36,37 or other cross-linking molecules that multivalently bind spikes can serve as pan-alphavirus antivirals without the need to neutralize virus entry. Our study thus provides a mechanistic basis for the development of novel therapeutics against alphaviruses and other enveloped viruses with similar spike-driven assembly mechanisms.
Methods
Cells, virus and antibody
Human bone epithelial cell line U2OS cells (Cat. #HTB-96), hamster fibroblast cell line BHK21 cells (Cat. #CCL-10) and adenovirus 5 DNA transformed human kidney epithelial cell line 293 c18 (Cat. #CRL-10852) were purchased from American Type Culture Collection (ATCC). Cells were maintained at 37°C with 5% humidified CO2 in DMEM (Invitrogen) supplemented with penicillin and streptomycin, 10 mM HEPES, non-essential amino acids, and 10% FBS (Hyclone). For 293 c18, 0.25 mg/ml Geneticin (ThermoFisher, Cat. #11811023) was added to the medium. CHIKV vaccine strain 181/clone 25 (CHIKV-181) (Cat. #NR-13222) was obtained through BEI Resources, NIAID, NIH and amplified in BHK21 cells. NAb C98 was produced in 293 c18 cells by transfecting cells with plasmids expressing heavy chain and light chain of C938 using TransIT 2020 transfection reagent (Mirus, Cat. #MIR 5406) and antibodies were purified from the supernatant of the transfected cells with HiTrap protein G HP (MilliporeSigma, Cat. #GE29-0485-81).
Cell infection and vitrification
U2OS cells grown on fibronectin (Sigma-Aldrich, Cat. #F1141)-coated gold 200 mesh R2/2 grids (Quantifoil, Cat. #Q2100AR2) were infected with CHIKV-181 at an MOI of 50 for an incubation period of 8 hrs. In the case of NAb C9-treated cells, after 3 hrs of infection, the grids were washed extensively and incubated with 5 ug/mL NAb C9 for an additional 5 hrs. The grids were then washed with PBS and a solution of 10 nm BSA gold tracer (EMS, Cat. #25486) was added directly prior to vitrification. Grids were blotted and plunged into liquid ethane using the LEICA EMGP plunge freezer device. Grids were stored under liquid nitrogen conditions until required for data collection.
Acquisition and processing of cryo-ET tilt series
Grids of vitrified virus-infected cells were imaged on two instruments: (1) a Titan Krios microscope (ThermoFisher) operated at 300kV with post-column energy filter (20eV) and K2 Summit detector (Gatan) with a calibrated pixel size of 2.72Å and (2) a Talos Arctica (ThermoFisher) operated at 200kV with post-column energy filter (20eV) and K2 Summit detector with calibrated pixel size of 3.54Å. Single-axis, bi-directional tilt series were collected using Tomography Software 4.0 (ThermoFisher, https://www.thermofisher.com/us/en/home/electron-microscopy/products/software-em-3d-vis/tomography-software.html) or SerialEM software v3.739,40 (http://bio3d.colorado.edu/SerialEM/) with low-dose settings and defocus range of −3 to −5.5 μm. For data of CHIKV-181-infected cells collected with the Titan Krios, a total cumulative dose of 110e−/A2 was applied to the specimen, while for data collected with Talos Arctica, the total average dose at the specimen was 90e−/A2. In both cases the electron dose was distributed over 51 tilt images, covering an angular range of −50° to +50°, with an angular increment of 2°. Additional data collection on both electron microscopes was collected using a Volta phase plate, whereby the objective aperture was removed, phase plate inserted and activated, and tilt series collected under the above conditions. The activated Volta phase plate was operated at phase shift 0.3–0.6𝝅 radians as measured by AutoCTF software (ThermoFisher). The motion between frames of each tilt image in the tilt series was corrected using MotionCor2 software v1.3.042 (https://emcore.ucsf.edu/ucsf-software). Tilt images were compiled, fiducial-based aligned and reconstructed using IMOD software v4.741 (https://bio3d.colorado.edu/imod/), or automatically aligned and reconstructed using EMAN2 software v2.343 (https://blake.bcm.edu/emanwiki/EMAN2). In total, 144 tomograms were judged as sufficient for further analysis from the Titan Krios data collections and 20 tomograms from the Talos Arctica data collections. A summary of the Cryo-ET data collection can be found in Supplementary Table 1.
For analysis of CHIKV-181-infected cells treated with NAb C9, 61 single-axis, bi-directional tilt series were collected on the Titan Krios microscope operated at 300kV with post-column energy filter and K2 Summit detector and calibrated pixel size of 2.72Å. Data was acquired using SerialEM software with low-dose settings and defocus range of −3 to −5.5 μm. Tilt series were collected with a total cumulative electron dose of 120e−/A2 distributed over 51 tilt images, again covering an angular range of −50° to +50°, with an angular increment of 2°. Data was exclusively collected using an activated Volta phase plate, with phase shift targeted in the range 0.3–0.65 radians. 51 tomograms were judged as sufficient for further analysis, based on achieved phase shift and tomogram reconstruction quality, and were used for subvolume analysis.
Statistics and Reproducibility
Of the 144 reconstructed tomograms from CHIKV-181-infected cells imaged with Talos Artica and selected for further analysis based on proper alignment, 117 contained features of the cell periphery of short extensions with virus budding events (Fig. 1A–B, Fig. 2A–B). 20 tomograms contained strands of incomplete particles extending from the cell surface (Fig. S2). 7 tomograms contained cell extensions with self-assembled spike lattice without nearby virus budding or NCs (Fig S7).
Budding intermediate subvolume classification
Subvolume analysis steps were performed using the previously published EMAN2 Tomo pipeline of version 2.343. CTF estimation for each tilt image was performed using the EMAN2 program e2spt_tomoctf.py. 1,918 budding intermediate particles were manually picked using the EMAN2 3D slice picker and extracted into subvolumes with x4, x2, x1 binning. 50 high-SNR particles (x4 binning) were picked from the dataset for each of three rough stages of budding (early-, mid-, and late-) for initial model generation. The initial model for each budding class was produced using the EMAN2 initial model generation program e2spt_sgd.py, first imposing c1 symmetry and running 5 iterations. After aligning the C1 initial models to the symmetry axes, 5 additional iterations were run with C5 symmetry imposed for each. These three maps were then used as initial models for subtomogram multi-reference refinement (e2spt_refinemulti.py).
The full dataset of 1,918 budding-intermediate subvolumes (x4 binning) was input into EMAN2 multi-reference refinement with 10 initial models (three early-, three mid-, four late-budding) and run for 12 iterations, imposing C5 symmetry and limiting resolution to 40Å for alignments. Due to poor convergence of the earliest-budding classes, all budding particles in the tomograms were re-picked with two points defining an initial budding orientation: one at the center of NC and one at the apex of the budding shell. Multi-reference refinement of the pre-oriented subvolumes was repeated as described above, with a refinement angular difference constraint of 30° to prevent particle “flipping” from the initial rough budding orientation. If a resulting class displayed budding virus structural features with sufficient particle count, those particles were subjected to further classification with either two or three low-passed versions of the class average as initial references. In this way, particles within five of the 10 3D classes were subjected to a second round of multi-reference refinement for further identification of budding conformations, with refinement parameters described above (Fig. S3). Between the two rounds of classification, 12 different 3D budding structures were determined in total. Subvolume particles within “junk” class averages lacking interpretable structure were viewed in the original 3D tomograms, revealing these particles covered a wide range of budding levels and were typically located near high density gold fiducials that biased the alignment.
Subtomogram averaging of budding intermediates, released virions and NLPs
For each of those budding intermediate 3D classes (5) displaying low-resolution icosahedral features, particles were re-extracted (x4, x2, x1 binning) for subtomogram refinement (e2spt_refine.py). For each class, 4–6 iterations of refinement were performed for each binned (x4, x2, x1) particle set, imposing C5 symmetry at each step and following gold-standard protocol: all particles were split into two independent subsets and resolution measured by Fourier shell correlation (0.143 FSC criterion) of the two density maps. Following subtomogram refinement of the least-binned particle set for each class, 2 iterations of sub-tilt refinement (e2spt_tiltrefine.py) with imposed C5 symmetry were performed to produce final budding-intermediate subvolume averages. A summary of the CryoET data collection and subtomogram analysis of viral intermediates can be found in Table S1.
Subtomogram averaging of released virions was performed by manually picking and extracting 521 released particles (x4, x2, x1 binning) into subvolumes, followed by EMAN2 3D refinement and sub-tilt refinement. An initial reference for 3D refinement was generated from 50 high SNR particles with different defocuses using EMAN2, with C5 symmetry imposed as described previously. 3D refinement was performed with C5 symmetry imposed, working from x4 to x2 to x1 binned subvolumes as resolution improved. After visual observation of icosahedral structure in the map, icos. symmetry was applied during final sub-tilt refinement of x1 binned subvolumes. This resulted in a converged map with pixel size 2.72 Å/pixel and resolution (0.143 FSC criterion) of 8.2 Å.
For subtomogram averaging of NLPs, 545 NLPs apparently within the cytosol of virus-infected cell tomograms were manually picked using the EMAN2 3D slice picker and extracted (x4 binning) into subvolumes. 50 high SNR particles with varying defocus were used to generate an initial reference with C5 symmetry as described above. Multi-reference refinement of the 545 NLPs (x4 binning) was performed with three classes and similar refinement parameters described above for budding intermediate classification, without the angular difference constraints. This resulted in two cytosolic NLP 3D classes (class I & II) with interpretable structure (Fig. S3). Additional 3D refinements of particles within those two respective classes, with imposed C5 symmetry, resulted in maps with resolutions of 47.6Å (class I) and 43.5Å (class II) (Gold-standard, 0.143 FSC criterion). The refined orientations of cytosolic NLPs within one class displaying local five-fold symmetry (class II, Fig. 3, S4) were mapped back in 3D to originating tomogram reconstructions using EMAN2 program e2spt_mapptclstotomo.py.
Subvolume analysis of NAb-crosslinked spikes and budding-arrested NLPs
For analysis of the C9-treated CHIKV-181-infected cells, 7,678 individual spikes were automatically picked from tomograms based on a low-resolution reference and judged individually for false positives. Any additional spikes in the tomogram were picked manually. This extensive manual picking protocol was meant to ensure all spikes were properly extracted for nearest-neighbor distance analysis. 3D subvolumes (x4, x2 binning) of each spike were then extracted and a c3-symmetric initial model was built from a subset of 500 (x4 binning) high SNR particles using the reference-free initial model program in EMAN2 (e2spt_sgd.py). The full set of 7,678 (x4, x2 binned) spike particles was then subjected to iterative 3D subtomogram refinement (e2spt_refine.py) with C3 symmetry imposed until no improvement in refined orientations was achieved. The final converged average map had resolution 24.4Å (Gold-standard, 0.143 FSC criterion) and pixel size 5.44Å/pixel. The Euclidean distance between each refined spike and its nearest neighbor in the dataset was determined using the refined center-of-mass orientations of spike subvolumes in each tomogram.
From the same tomograms, 1,727 budding-arrested NLPs were manually picked, extracted into subvolumes (x4 binned) and an initial model was generated from 50 high-contrast particles in the dataset as described above with C5 symmetry imposed. The initial model was used as input for 3D classification of the 1,727 NLP subvolumes, imposing C5 symmetry, performed using e2spt_refinemulti.py. The multi-reference refinement resulted in six class average structures ranging from ~37Å to 63Å resolution (Gold-standard, 0.143 FSC criterion) (Fig. S3A).
Visualization, figure generation and model docking were performed in UCSF Chimera v1.1644 (https://www.cgl.ucsf.edu/chimera/) or UCSF ChimeraX v1.3 and its built-in tools45 (https://www.rbvi.ucsf.edu/chimerax/).
Supplementary Material
Acknowledgements
We thank SLAC National Accelerator Laboratory for access and support of these studies, and all SLAC cryoEM staff for technical support and assistance. We also thank Dr. Muyuan Chen for helpful discussions and providing technical advice in data analysis. This research was supported by the NIH grants R01AI148382 and S10OD021600 (to W.C.) and R01AI119056 (to G.S.).
Abbreviations
- GP
glycoprotein
- Cp
capsid protein
- NC
nucleocapsid
- Cryo-ET
cryogenic electron tomography
- NLP
nucleocapsid-like particle
- PM
plasma membrane
- CHIKV
chikungunya virus
- Nab
neutralizing antibody
Footnotes
Competing Interest
All authors declare no competing interest.
Data availability
Cryo-EM maps reported in this study have been deposited in the Electron Microscopy Data Bank (EMDB) under the following accession codes: EMDB-26446 (released virion), EMDB-26447, -26448, -26449, -26450 (budding intermediates), and EMDB-26451, -26452 (cytosolic NLPs). The publicly deposited atomic model of VEEV TC-83 (PDB:3J0C) was used for comparison to the subtomogram average structure of the CHIKV spike trimer determined in this study. All other data supporting the findings of this study are available within the article and its supplementary files.
REFERENCES
- 1.Rheinemann L & Sundquist WI Virus Budding. Encyclopedia of Virology 519–528 (2021) doi: 10.1016/b978-0-12-814515-9.00023-0. [DOI] [Google Scholar]
- 2.Brown RS, Wan JJ & Kielian M The Alphavirus Exit Pathway: What We Know and What We Wish We Knew. Viruses 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silva LA & Dermody TS Chikungunya virus: epidemiology, replication, disease mechanisms, and prospective intervention strategies. J. Clin. Invest. 127, 737–749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun S et al. Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. Elife 2, e00435 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang J et al. Molecular links between the E2 envelope glycoprotein and nucleocapsid core in Sindbis virus. J. Mol. Biol. 414, 442–459 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang R et al. 4.4 Å cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J. 30, 3854–3863 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng RH et al. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80, 621–630 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin J et al. Neutralizing Monoclonal Antibodies Block Chikungunya Virus Entry and Release by Targeting an Epitope Critical to Viral Pathogenesis. Cell Reports vol. 13 2553–2564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox JM et al. Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress. Cell 163, 1095–1107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin J et al. Neutralizing Antibodies Inhibit Chikungunya Virus Budding at the Plasma Membrane. Cell Host Microbe 24, 417–428.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thaa B et al. Differential Phosphatidylinositol-3-Kinase-Akt-mTOR Activation by Semliki Forest and Chikungunya Viruses Is Dependent on nsP3 and Connected to Replication Complex Internalization. J. Virol. 89, 11420–11437 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jose J, Taylor AB & Kuhn RJ Spatial and Temporal Analysis of Alphavirus Replication and Assembly in Mammalian and Mosquito Cells. MBio 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L et al. Implication for alphavirus host-cell entry and assembly indicated by a 3.5Å resolution cryo-EM structure. Nature Communications vol. 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukhopadhyay S, Chipman PR, Hong EM, Kuhn RJ & Rossmann MG In Vitro-Assembled Alphavirus Core-Like Particles Maintain a Structure Similar to That of Nucleocapsid Cores in Mature Virus. Journal of Virology vol. 76 11128–11132 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Therkelsen MD et al. Flaviviruses have imperfect icosahedral symmetry. Proc. Natl. Acad. Sci. U. S. A. 115, 11608–11612 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garoff H, Sjöberg M & Cheng RH Budding of alphaviruses. Virus Res. 106, 103–116 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Soonsawad P et al. Structural evidence of glycoprotein assembly in cellular membrane compartments prior to Alphavirus budding. J. Virol. 84, 11145–11151 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cogan DP et al. Mapping the catalytic conformations of an assembly-line polyketide synthase module. Science 374, 729–734 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruss V & Ganem D The role of envelope proteins in hepatitis B virus assembly. Proc. Natl. Acad. Sci. U. S. A. 88, 1059–1063 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferlenghi I et al. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7, 593–602 (2001). [DOI] [PubMed] [Google Scholar]
- 21.de Haan CA, Kuo L, Masters PS, Vennema H & Rottier PJ Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 72, 6838–6850 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allison SL et al. Two distinct size classes of immature and mature subviral particles from tick-borne encephalitis virus. J. Virol. 77, 11357–11366 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stange A, Lüftenegger D, Reh J, Weissenhorn W & Lindemann D Subviral particle release determinants of prototype foamy virus. J. Virol. 82, 9858–9869 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P-G et al. Efficient Assembly and Secretion of Recombinant Subviral Particles of the Four Dengue Serotypes Using Native prM and E Proteins. PLoS ONE vol. 4 e8325 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heilingloh CS & Krawczyk A Role of L-Particles during Herpes Simplex Virus Infection. Front. Microbiol. 8, 2565 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherer NM & Mothes W Cytonemes and tunneling nanotubules in cell–cell communication and viral pathogenesis. Trends in Cell Biology vol. 18 414–420 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikolic DS et al. HIV-1 activates Cdc42 and induces membrane extensions in immature dendritic cells to facilitate cell-to-cell virus propagation. Blood 118, 4841–4852 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sowinski S et al. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat. Cell Biol. 10, 211–219 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Martinez MG & Kielian M Intercellular Extensions Are Induced by the Alphavirus Structural Proteins and Mediate Virus Transmission. PLoS Pathog. 12, e1006061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen D-H et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc. Natl. Acad. Sci. U. S. A. 108, 1355–1360 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukhopadhyay S, Chipman PR, Hong EM, Kuhn RJ & Rossmann MG In vitro-assembled alphavirus core-like particles maintain a structure similar to that of nucleocapsid cores in mature virus. J. Virol. 76, 11128–11132 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snyder JE et al. Rescue of infectious particles from preassembled alphavirus nucleocapsid cores. J. Virol. 85, 5773–5781 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forsell K, Xing L, Kozlovska T, Cheng RH & Garoff H Membrane proteins organize a symmetrical virus. EMBO J. 19, 5081–5091 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skoging U, Vihinen M, Nilsson L & Liljeström P Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure 4, 519–529 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Jose J et al. Interactions of the cytoplasmic domain of Sindbis virus E2 with nucleocapsid cores promote alphavirus budding. J. Virol. 86, 2585–2599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williamson LE et al. Therapeutic alphavirus cross-reactive E1 human antibodies inhibit viral egress. Cell 184, 4430–4446.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim AS et al. Pan-protective anti-alphavirus human antibodies target a conserved E1 protein epitope. Cell 184, 4414–4429.e19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selvarajah S et al. A neutralizing monoclonal antibody targeting the acid-sensitive region in chikungunya virus E2 protects from disease. PLoS Negl. Trop. Dis. 7, e2423 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mastronarde DN SerialEM: A Program for Automated Tilt Series Acquisition on Tecnai Microscopes Using Prediction of Specimen Position. Microsc. Microanal. 9, 1182–1183 (2003). [Google Scholar]
- 40.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Kremer JR, Mastronarde DN & McIntosh JR Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen M et al. A complete data processing workflow for cryo-ET and subtomogram averaging. Nat. Methods 16, 1161–1168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettersen EF, Goddard TD & Huang CC UCSF Chimera—a visualization system for exploratory research and analysis. Journal of (2004). [DOI] [PubMed] [Google Scholar]
- 45.Pettersen EF et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps reported in this study have been deposited in the Electron Microscopy Data Bank (EMDB) under the following accession codes: EMDB-26446 (released virion), EMDB-26447, -26448, -26449, -26450 (budding intermediates), and EMDB-26451, -26452 (cytosolic NLPs). The publicly deposited atomic model of VEEV TC-83 (PDB:3J0C) was used for comparison to the subtomogram average structure of the CHIKV spike trimer determined in this study. All other data supporting the findings of this study are available within the article and its supplementary files.


