Abstract
We previously identified the mechanistic target of rapamycin complex 2 (mTORC2) as an effector of Ras for the control of directed cell migration in Dictyostelium. Recently, the Ras-mediated regulation of mTORC2 was found to be conserved in mammalian cells, and mTORC2 was shown to be an effector of oncogenic Ras. Interestingly, mTORC2 has been linked to cancer cell migration, and particularly in breast cancer. Here, we investigated the role of Ras in promoting the migration and invasion of breast cancer cells through mTORC2. We observed that both Ras and mTORC2 promote the migration of different breast cancer cells and breast cancer cell models. Using HER2 and oncogenic Ras-transformed breast epithelial MCF10A cells, we found that both wild-type Ras and oncogenic Ras promote mTORC2 activation and an mTORC2-dependent migration and invasion in these breast cancer models. We further observed that, whereas oncogenic Ras-transformed MCF10A cells display uncontrolled cell proliferation and invasion, disruption of mTORC2 leads to loss of invasiveness only. Together, our findings suggest that, whereas the Ras-mediated activation of mTORC2 is expected to play a minor role in breast tumor formation, the Ras-mTORC2 pathway plays an important role in promoting the migration and invasion of breast cancer cells.
INTRODUCTION
The mechanistic target of rapamycin complex 2 (mTORC2) plays an evolutionarily conserved role in controlling eukaryotic cell migration, in addition to regulating cell survival and the metabolism (Oh and Jacinto, 2011; Xie et al., 2018). mTORC2 is one of two complexes formed by the kinase mTOR (Zhou and Huang, 2010). mTORC1 and mTORC2 have different substrates and functions in cells, which are determined by their mTORC-specific components that include raptor (mTORC1) and rictor (mTORC2). mTORC1 is a master regulator of cell growth, has been extensively studied, and much is understood about its biochemistry and regulation mechanisms (Liu and Sabatini, 2020). On the other hand, less is known about how mTORC2 is activated and regulated, but evidence suggests that different regulatory mechanisms are involved depending on the type of cell and stimulus (Smith et al., 2020). In the simple eukaryotic experimental model, Dictyostelium discoideum, others and we showed that mTORC2 is an effector of Ras proteins to promote directed cell migration toward cAMP (Sasaki and Firtel, 2006; Kamimura et al., 2008; Cai et al., 2010; Charest et al., 2010; Liao et al., 2013; Khanna et al., 2016). Importantly, the Ras-mediated regulation of mTORC2 was found to be conserved in mammalian cells, with evidence suggesting that mTORC2 is a direct effector of oncogenic H-, N-, and K-Ras4A and 4B, the four predominant human Ras isoforms (Kovalski et al., 2019; Lone et al., 2019).
Whereas a role for mTORC2 in tumorigenesis is often linked to its regulation of cell survival and metabolism, mTORC2 has also been shown to promote cancer cell migration, including downstream from human epidermal growth factor 2 (HER2) in breast cancer cells (Masri et al., 2007; Gulhati et al., 2011; Kim et al., 2011; Li et al., 2012; Gupta et al., 2013; Maru et al., 2013; Bera et al., 2014; Wang et al., 2014, 2016, 2017; Bian et al., 2015; Chen et al., 2015; Daulat et al., 2016; Joly et al., 2016; Zhang et al., 2016; Morrison Joly et al., 2017; Butt et al., 2019). Ras is rarely mutated in breast cancer, but it is often up-regulated due to its own overexpression, the overexpression of the epidermal growth factor receptor (EGFR) and HER2, activating EGFR mutations, or mutations affecting regulators of Ras (Clair et al., 1987; Galiè, 2019; von Lintig et al., 2000). Indeed, in addition to HER2-enriched breast cancer that displays amplification of HER2 (HER2+) including triple hormone receptor positive (estrogen receptor, progesterone receptor, and HER2; ER+PR+HER2+), ER and PR positive but HER2 negative breast cancer (ER+PR+), as well as triple negative breast cancer (TNBC) cells can display Ras hyperfunction (Galiè, 2019; von Lintig et al., 2000). Here, we investigated whether mTORC2 promotes cell migration and invasion downstream from Ras in transformed breast epithelial cells and breast cancer cells.
RESULTS
Ras and mTORC2 promote the migration of transformed breast epithelial cells
To investigate a role for Ras and mTORC2 in promoting the migration of breast cancer cells, we used MCF10A cells as an example of nontransformed breast epithelial cells, as well as HER2-expressing MCF10A cells as a model for HER2+ breast cancer cells. MCF10A cells have proven to be useful for investigating the specific effects of oncogenes and associated mechanisms implicated in epithelial tumor formation and progression (Debnath and Brugge, 2005). In addition, we used different breast cancer cell lines: MDAMB231 (TNBC, K-Ras oncogenic mutant), MDAMB453 (TNBC), MCF7 (ER+PR+), and SKBR3 (HER2+; Supplemental Table S1; Kao et al., 2009; Lehmann et al., 2011; Dai et al., 2017).
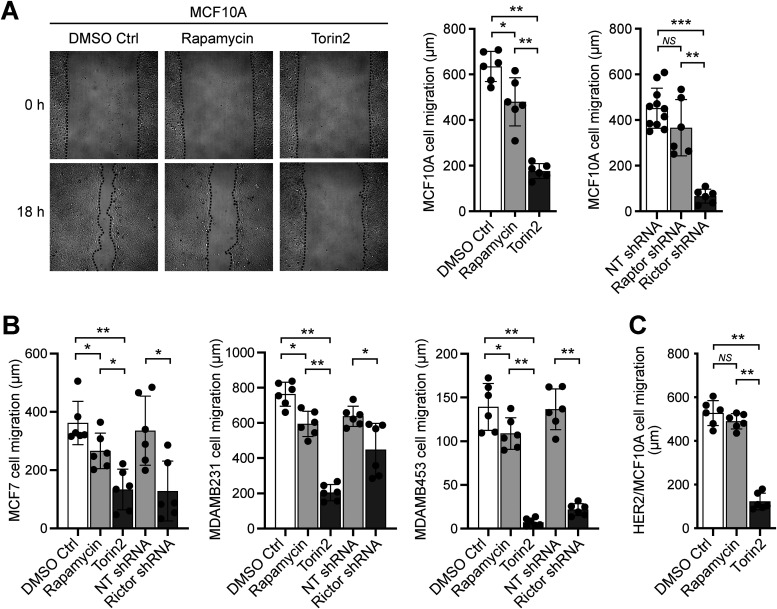
We first tested the role of mTORC2 in the migration of breast epithelial and cancer cells using a 2D migration wound closure assay, comparing the effect of the mTOR inhibitor Torin2 to that of the mTORC1-specific inhibitor rapamycin, as well as the effect of an established small hairpin RNA (shRNA) directed against the essential mTORC2 component rictor (Supplemental Figures S1 and S2). Because there are no mTORC2-selective inhibitors, comparing the effects of inhibiting only mTORC1 with rapamycin versus inhibiting both mTORC1 and mTORC2 with Torin2 can provide useful information regarding mTORC2-dependent cellular processes, which we further supported by also using previously validated rictor and raptor shRNAs to knock-down mTORC2 and mTORC1, respectively (Sarbassov et al., 2005; Liu et al., 2010). Moreover, because rapamycin inhibits cell growth (Tee, 2018), its use also allows one to evaluate the contribution of cell proliferation in wound closure (Supplemental Figure S2D). We observed that disruption of mTORC2 alone or both mTORCs significantly inhibits the wound closure of MCF10A cells, much more so than the inhibition of mTORC1 alone, suggesting that mTORC2 promotes cell migration and that cell proliferation has a significantly smaller contribution to wound closure (Figure 1A and Supplemental Figure S2D). Similarly, disruption of mTORC2 alone or both mTORCs also significantly suppressed the migration of breast cancer cells MCF7, MDAMB231, and MDAMB453, and significantly more than inhibiting mTORC1 and cell proliferation with rapamycin (Figure 1B). Unfortunately, assaying the migration of the HER2-overexpressing breast cancer cell line SKBR3 was unsuccessful because the cells do not spread uniformly on the substrate and, consequently, it is difficult to obtain a clean wound and properly analyze the cells’ migration. Instead, we used HER2-overexpressing MCF10A cells to model HER2+ breast cancer cell migration. Although our experimental conditions did not allow for effective silencing of rictor in HER2/MCF10A cells, we found that the migration of these cells is also significantly inhibited by the mTOR inhibitor Torin2 and not by the mTORC1-selective inhibitor rapamycin, suggesting that the observed effect of Torin2 on wound closure is due to inhibition of mTORC2-mediated cell migration (Figure 1C). Consequently, all together, these observations suggest that mTORC2 mediates the migration of nontransformed and transformed breast epithelial cells.
FIGURE 1:
mTORC2 promotes the migration of MCF10A and breast cancer cells. Wound closure cell migration assays were performed in the presence of rapamycin, Torin2, or 0.1% DMSO control (Ctrl); and with cells previously treated with shRNAs to silence raptor or rictor, or a nontargeting (NT) shRNA as control, as described in the Materials and Methods section. (A) MCF10A cell migration. Representative images of MCF10A cells in the wound closure assay are shown on the left, taken immediately after wounding and addition of the indicated treatments (0 h) and after 18 h. (B) Cell migration data for MCF7, MDAMB231, and MDAMB453 cells. (C) HER2/MCF10A cell migration. Graphs show the measured migration distances ±SD of at least six wounds from at least three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
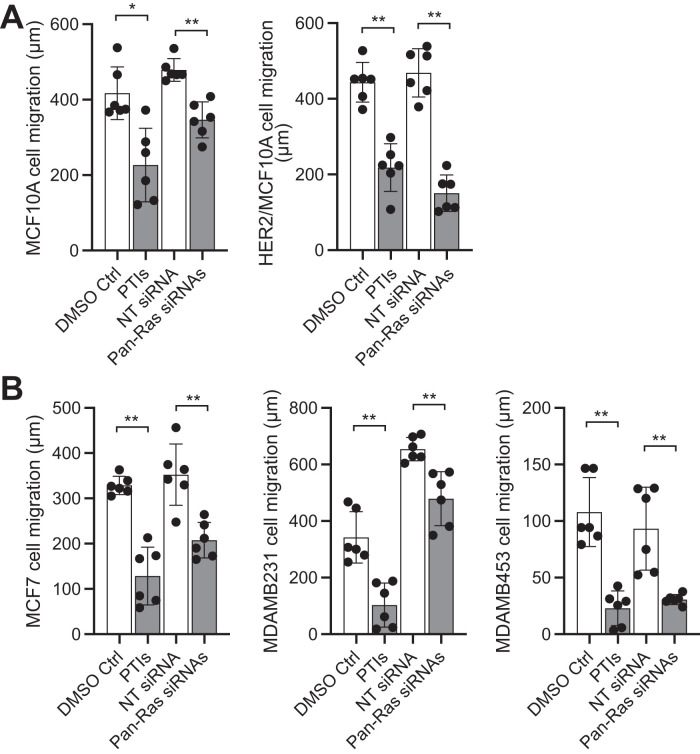
We then assessed the role of Ras in breast epithelial and cancer cell migration using prenylation inhibitors (prenyltransferase inhibitors [PTIs]), which effectively inhibit Ras function in cells (Supplemental Figure S3A; Philips and Cox, 2007; Fiordalisi and Cox, 2010), as well as the Pan-Ras small interfering RNA (siRNA)-mediated silencing of H-, N-, and K-Ras (Supplemental Figure S3B). We observed that disrupting Ras function significantly inhibits the migration of MCF10A, HER2/MCF10A, MCF7, MDAMB231, and MDAMB453 cells (Figure 2). Whereas loss of Ras function undoubtably also inhibits cell proliferation, which can contribute to the measured wound closures, this effect is expected to be less important given the minor effect of the mTORC1 and growth inhibitor rapamycin in similar experiments (Figure 1). Therefore, these observations suggest that Ras, in addition to mTORC2, promotes the migration of nontransformed and transformed breast epithelial cells, and independently of the type of breast cancer cells. This is further supported by the observation that the exogenous expression of Ras increases the migration and invasion of MCF10A cells (see below, Figures 3 and 4).
FIGURE 2:
Ras promotes the migration of MCF10A and breast cancer cells. Wound closure cell migration assays were performed in the presence of prenyltransferase inhibitors (PTIs), or 0.1% DMSO control; and with cells previously treated with siRNAs to silence H-, N-, and K-Ras (Pan-Ras), or nontargeting (NT) siRNAs as control, as described in the Materials and Methods section. (A) MCF10A cell migration. (B) HER2/MCF10A cell migration. (C) MCF7, MDAMB231, and MDAMB453 cell migration. Graphs show the measured migration distances ±SD of at least six wounds from at least three independent experiments. *, p < 0.05; **, p < 0.01.
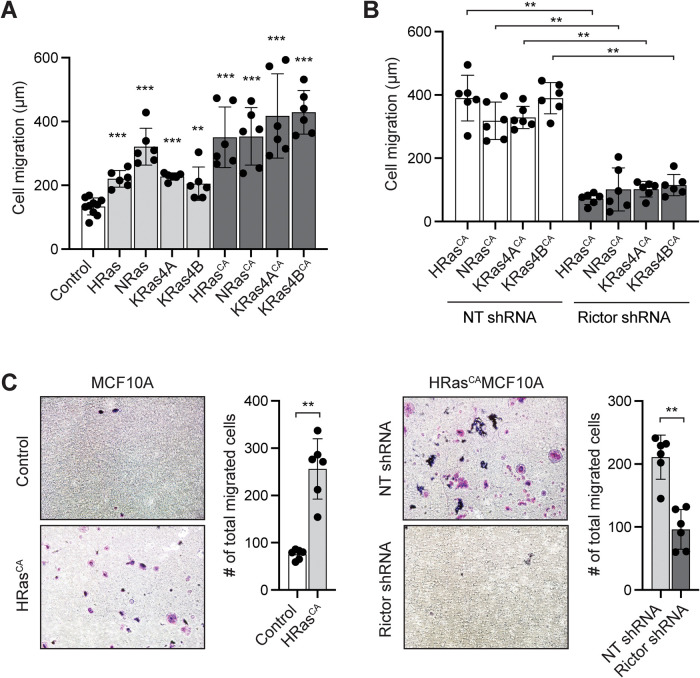
FIGURE 3:
Ras promotes the migration of MCF10A cells through mTORC2. (A) Wound closure cell migration assays of MCF10A cells expressing the indicated wild-type or constitutively active (CA; G12V mutation) Ras proteins, compared with control MCF10A cells transduced with empty vector. Data represent the measured migration distance ±SD of at least six wounds from at least three independent experiments. The statistics evaluate the difference in migration of Ras-expressing cells vs. the control. (B) Wound closure cell migration assays of MC10A cells expressing the indicated RasCA and treated with rictor shRNA or nontargeting (NT) shRNA control. Data represent the measured migration distance ±SD of at least six wounds from at least three independent experiments. (C) 3D cell migration transwell assay of HRasCA-expressing MCF10A cells compared with vector-transduced MCF10A cells, and HRasCA/MCF10A cells treated with rictor shRNA compared with NT shRNA. Representative images of crystal violet-stained cells that have migrated through the wells are shown (1 field of view of 13 areas analyzed and quantified). Graphs represent the total number of cells that have migrated through the entire well ±SD of six replicates performed in at least three independent experiments. **, p < 0.01; ***, p < 0.001.
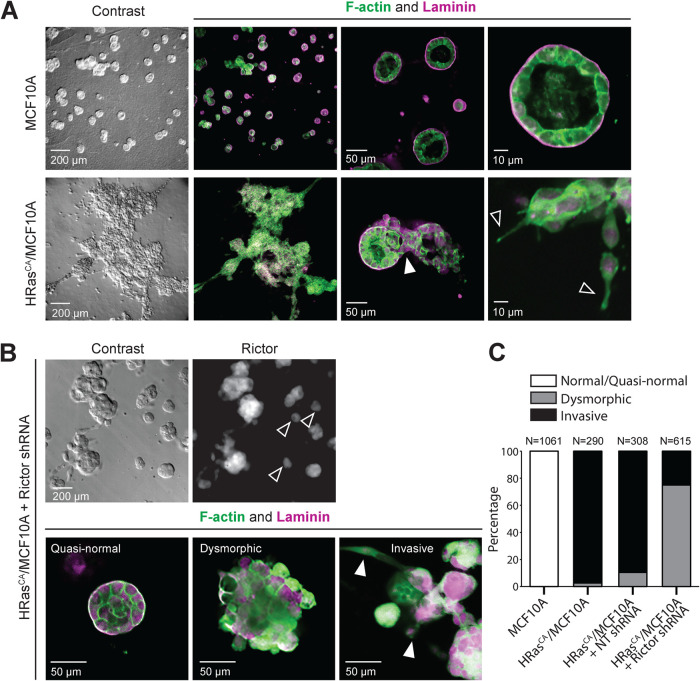
FIGURE 4:
mTORC2 mediates the invasion of HRasCA-transformed MCF10A cells. MCF10A and HRasCA/MCF10A cells (A), and rictor shRNA-treated HRasCA/MCF10A cells (B) were grown for 12 d in 3D cell cultures on reconstituted basement membrane. The acini were stained for F-actin and laminin or rictor and imaged as described in the Materials and Methods section. (A) Filled arrow indicates the area where cells have invaded through the basal membrane; empty arrows indicate invasive cellular projections. (B) Images are representative of the three general acini phenotypes in rictor shRNA-treated HRasCA/MCF10A cells. To visualize otherwise very low rictor labeling signal, the rictor-stained cells were imaged using a long exposure time and high gain. Empty arrows indicate quasinormal acini with very low rictor expression; filled arrows indicate cells invading the surrounding matrix, migrating away from the cell clusters. (C) Classification and quantification of the acini observed in the indicated cell lines according to the observed phenotypes. The representative images and quantified data were obtained from at least three independent experiments.
mTORC2 promotes the migration and invasion of Ras CA-transformed MCF10A cells
All four main Ras isoforms have been implicated in breast oncogenesis (Galiè, 2019). To assess whether Ras promotes breast epithelial cell migration through mTORC2, we individually overexpressed H-, N-, and K-Ras 4A and 4B, as well as their respective constitutively active (CA), oncogenic G12V mutant forms in MCF10A cells (Supplemental Figure S4A). We observed that exogenous expression of either wild-type or RasCA mutant of each isoform significantly increases the 2D migration of MCF10A cells, with a greater effect from the RasCA mutants, further indicating that Ras plays an important role in promoting cell migration of breast epithelial cells (Figure 3A). To then assess the role of mTORC2 in mediating the Ras-induced cell migration, we inhibited mTORC2 using shRNA-mediated rictor silencing in RasCA-expressing MCF10A cells (Supplemental Figure S4B). We observed that loss of mTORC2 function significantly suppresses the RasCA-induced migration of cells (Figure 3B). Similarly, using HRasCA/MCF10A cells in a transwell assay to assess their migration in 3D, we observed increased migration compared with control MCF10A cells, as previously reported (Basolo et al., 1991), and that this migration was significantly inhibited by loss of mTORC2 function (Figure 3C). Together, these observations indicate that Ras promotes breast epithelial cell migration through mTORC2.
HRasCA was previously shown to induce an invasive phenotype in MCF10A cells grown in 3D cultures (Moon et al., 2000). When cultured on a reconstituted basement membrane, MCF10A cells grow as spheroids of ∼70 µm in diameter with a hollow lumen, forming acinar structures that recapitulate the mammary gland epithelial architecture (Figure 4A; Debnath et al., 2003; Shaw et al., 2004). However, the spheroids formed by HRasCA/MCF10A cells are much bigger (up to 300 µm) and display a filled lumen, indicative of oncogenic growth, as well as extensive cellular projections (visualized by F-actin staining) and invasion of cells through the basal membrane (visualized by laminin staining) and surrounding matrix (as seen by the presence of single cells; Figure 4A). We then assessed the role of mTORC2 in mediating the HRasCA-induced invasion by silencing rictor in these cells. We found that loss of mTORC2 function in HRasCA/MCF10A cells resulted in a mixed phenotype of invasive, dysmorphic, and quasinormal (size of ∼70 μm with intact basal lamina but filled lumen), presumably due to a heterogenous knock-down of rictor in the cell population (Figure 4B). In fact, the smaller, noninvasive spheroids displayed the lowest rictor expression levels (Figure 4B). Interestingly, whereas 90% of spheroids of HRasCA/MCF10A cells treated with a nontargeting shRNA control displayed invasiveness and 10% were dysmorphic, the silencing of rictor led to a significant loss of invasion, with 25% invasive, 74.8% dysmorphic, and ∼0.2% quasinormal spheroids (Figure 4C). Taken together, these observations suggest that the HRasCA-induced invasiveness of MCF10A cells is mediated by mTORC2, whereas HRasCA-induced cell transformation and proliferation are not.
Ras promotes mTORC2 activation in transformed breast epithelial cells
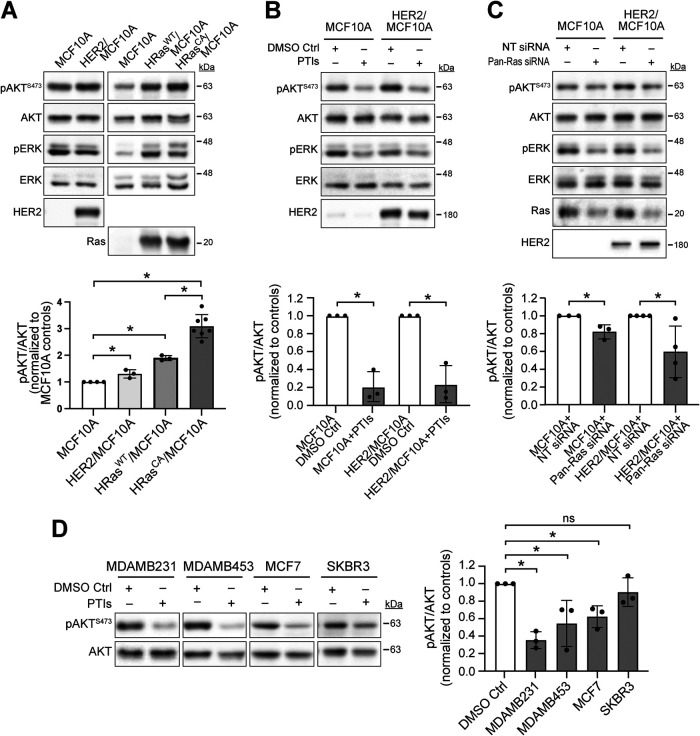
We then evaluated whether Ras regulates the cellular activity of mTORC2 in breast epithelial and cancer cells, using mTORC2’s substrate AKT and its mTORC2-dependent phosphorylation at serine residue 473 (pAKTS473) as read out. First, we assessed the activity of mTORC2 in MCF10A cells exogenously expressing HER2, HRas, or HRasCA, compared to that in control MCF10A cells. We observed that the pAKTS473 levels in the HER2-transformed cells is significantly more elevated (∼1.3-fold average increase) than those in MCF10A cells, as was reported previously (Yang et al., 2016), and that cells expressing HRas and HRasCA at similar levels also display significant increases of 1.8- and 3-fold average in pAKTS473, respectively (Figure 5A). Interestingly, the effects of HRas and HRasCA on mTORC2 activity strongly correlate with their effect on MCF10A cell migration, which is increased on average by 1.6- and 2.6-fold, respectively (Figure 3A). Therefore, this further supports that Ras promotes cell migration through mTORC2 in breast epithelial cells. On the other hand, we observed that the exogenous expressions of HRas and HRasCA induce similar increases in ERK phosphorylation (pERK) in resting MCF10A cells, whereas we did not detect any effect of HER2 expression on pERK levels, similar to a previous report (Yang et al., 2016). Furthermore, we observed that PTI treatment or Pan-Ras siRNAs significantly reduce pAKTS473 levels in MCF10A (PTI to ∼20% and Pan-Ras siRNAs to ∼80% average inhibition) and HER2/MCF10A cells (PTI to ∼20% and Pan-Ras siRNAs to ∼60% average inhibition), suggesting that at least part of the mTORC2 activity in these cells is dependent on Ras (Figure 5, B and C). We then investigated whether Ras-dependent mTORC2 activity is conserved in breast cancer cells. We observed that the PTI treatment decreased pAKTS473 levels in the cell lines tested (MDAMB231, ∼65%; MDAMB453, ∼40%; MCF7, ∼40%; and SKBR3, ∼10% average inhibition; Figure 5D). Interestingly, the greatest effect was observed on MDAMB231 cells, a TNBC cell line expressing oncogenic K-Ras (American Type Culture Collection). Altogether, these results strongly suggest that mTORC2 activity in transformed MCF10A and breast cancer cells is, at least in part, dependent on Ras signaling.
FIGURE 5:
Ras promotes mTORC2 activation in transformed MCF10A and breast cancer cells. AKT phosphorylation at the mTORC2-dependent site (S473; pAKTS473) and total AKT proteins were detected by immunoblot as described in the Materials and Methods section. Immunoblots of pERK, ERK, HER2, and Ras were used as controls and performed as described in the Materials and Methods section. (A) pAKTS473 levels in HER2/MCF10A and HRasCA/MCF10A and cells compared with their respective MCF10A control cells. (B) MCF10A and HER2/MCF10A cells were pretreated with PTIs or 0.2% DMSO control. (C) MCF10A and HER2/MCF10A cells were pretreated with Pan-Ras siRNAs or nontargeting (NT) siRNA control. (D) The indicated breast cancer cells were treated with PTIs or 0.1% DMSO control. Graphs represent pAKTS473 over total AKT levels quantified by densitometry and normalized to the control conditions ±SD of at least three independent experiments. *, p < 0.05.
DISCUSSION
Altogether, our study suggests that the Ras-mediated regulation of mTORC2 promotes the migration of breast epithelial and cancer cells, and that this mechanism is likely involved independently of Ras’s mutational status and the type of breast cancer. Indeed, our observations suggest that this pathway regulates the migration of nontransformed breast epithelial cells (MCF10A), HER2-transformed breast epithelial cells (HER2/MCF10A), as well as different breast cancer cells; including luminal A MCF7 and triple negative MDAMB453 cells that are more epithelial-like and less migratory, and triple negative Ras mutant-positive MDAMB231 that are mesenchymal-like and more migratory. Furthermore, because the epithelial-like breast cancer cells have been reported to migrate collectively whereas mesenchymal-like cells can often also migrate as single cells (Lu and Lu, 2021), our data suggest that Ras-mTORC2 is important independently of the mode of migration used by different breast cancer cells. Finally, we found that mTORC2 promotes the invasiveness of HRasCA-transformed breast epithelial cells in an in vitro tumor model, indicating that the Ras-mediated activation of mTORC2 may play a central role in the early stages of breast cancer metastasis.
Our observation that PTI treatment in the breast cancer cells tested has the strongest effect on mTORC2 activity in the oncogenic Ras-expressing MDAMB231 cells could suggest that mutant Ras is the main mechanism promoting mTORC2 activation in these cells, whereas Ras-mediated mTORC2 activation is likely one of several mechanisms regulating mTORC2 in cells expressing wild-type Ras proteins. However, more studies need to be performed to investigate this possibility. Previous evidence supports our observations that both wild-type and oncogenic mutant Ras can promote mTORC2 activation in different cell types (Smith et al., 2020). Our own previous findings in Dictyostelium suggests that the human Ras protein homologue, RasC, can directly activate mTORC2 in response to chemoattractant stimulation by interacting with mTOR (Khanna et al., 2016). This Ras interaction with mTOR and mTORC2 activation was later found to be conserved for both wild-type and the oncogenic forms of the main human Ras isoforms, H-, N-, and K-Ras, with a much greater effect with the RasCA mutants (Kovalski et al., 2019). Additional evidence suggests that both wild-type and oncogenic Ras can also regulate mTORC2 activity by interacting with its component SIN1 (Kovalski et al., 2019; Lone et al., 2019). However, a role for the interaction of Ras with SIN1 in the regulation of mTORC2 activity is debated, and evidence suggests that the Ras-SIN1 interaction could have cellular functions independently of mTORC2 (Castel et al., 2021).
Ras is most often associated with the transduction of mitogenic signals and promoting cell proliferation through activation of the ERK cascade (Gimple and Wang, 2019; Lavoie et al., 2020). However, multiple evidence indicate that Ras can also promote cell migration and invasion (Gimple and Wang, 2019). Previous work implicated phosphatidylinositol 3-kinase (PI3K), ERK, and p38 downstream from Ras in mediating cancer cell migration and/or invasion, as well as the Ras-mediated regulation of the expression of the matrix metallopeptidase MMP2 and of the transcription factor p63, both implicated in epithelial to mesenchymal transition (EMT; Moon et al., 2000; Kim et al., 2003; Lee et al., 2016; Yoh et al., 2016; Gimple and Wang, 2019; Soriano et al., 2021). We do not know whether and how mTORC2 is linked with these Ras downstream effectors in breast epithelial cells, although PI3K could be involved in regulating mTORC2 activity as was previously found in other cell contexts (Smith et al., 2020). mTORC2 could also be mediating EMT downstream from Ras and/or PI3K by promoting the expression of key genes, similar to its previously suggested role in transforming growth factor β (TGF β)-induced EMT (Lamouille et al., 2012). As for a role for ERK, although we have not studied this in detail, our results suggest it is unlikely that ERK is involved in promoting cell migration downstream from Ras in MCF10A cells because, contrary to the detected mTORC2 activity, we did not observe any correlation between the Ras and RasCA-mediated increase in cell migration and ERK activity. Finally, previous studies performed with breast cancer cells demonstrated that mTORC2 promotes cell motility, invasion, and metastasis through its activation of AKT and protein kinase C (PKC), and their downstream activation of Rac1 (Morrison et al., 2015; Morrison Joly et al., 2017). Thus, it is likely that these pathways play a role in promoting the mTORC2-mediated cell migration and invasion downstream from Ras. In the future, it will also be interesting to define how these pathways regulate the motility machinery, including actin polymerization, myosin assembly, and focal adhesion dynamics, as well as how they affect cell migration speed, persistence, and directionality.
Interestingly, we observed that whereas rictor silencing significantly inhibits the invasion of HRasCA-transformed MCF10A cells, it has little effect on the oncogenic Ras-induced uncontrolled cell proliferation. This result suggests that the Ras-mTORC2 pathway functions in parallel to a mitogenic Ras pathway (e.g., ERK cascade) promoting cell proliferation. The premalignant proliferation of transformed breast epithelial cells in the lumen of mammary ducts, termed ductal carcinoma in situ (DCIS), is a nonobligate precursor of breast cancer with 40% of cases progressing to invasive disease (Cowell et al., 2013). While the mechanisms involved in promoting the transition from DCIS to invasive breast cancer remain elusive, our results suggest that the Ras-mediated activation of mTORC2 could play a role. Therefore, our study supports a rationale for using mTOR inhibitors to prevent invasive breast cancer, and for the usefulness of developing selective inhibitors targeting the Ras-mTORC2 pathway in breast cancer.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Materials
Reagents.
Farnesyltransferase inhibitor was obtained from Enzo Life Sciences (Farmingdale, NY) and geranylgeranyltransferase I inhibitor was from Tocris Biosciences (Bristol, UK). Horse serum and penicillin-streptomycin were from Life Technologies, Alexa Fluor 488 phalloidin and lipofectamine RNAiMAX transfection reagent were from Invitrogen, and EGF was from PeproTech, at Thermo Fisher Scientific (Waltham, MA). Hygromycin B and ProBlock gold protease inhibitor cocktail were obtained from Gold Biotechnology (St-Louis, MI). DMEM, DMEM/F12, RPMI-1640, and McCoy’s 5A media were from Corning (Corning, NY). Rapamycin was from Research Products International (Mount Prospect, IL); Torin2 was from ApexBio (Houston, TX); fetal bovine serum (FBS) was from Avantor (Radnor Township, PA); hydrocortisone was from United States Pharmacopeia (Rockville, MD); cholera toxin was from List Biological Laboratories (Campbell, CA); puromycin was from Mirus Bio LLC (Madison, WI); and insulin was from Sigma-Aldrich (St. Louis, MO).
Antibodies.
Phospho AKT (Ser473, D9E) XP rabbit mAb, phospho-p44/42 MAPK (ERK1/2) (Thr202/Tyr204) antibody, HER2/ErbB2 antibody, and AKT (pan; 40D4) mouse mAb were purchased from Cell Signaling Technology (Danvers, MA). ERK 1 (C-19) antibody was from Santa Cruz Biotechnology, Inc. (Dallas, TX), and Anti-Pan-Ras (Ab-3) mouse mAb was from Sigma-Aldrich (St. Louis, MO). Anti-laminin 5 antibody and Alexa Fluor 568 goat anti-rabbit IgG were from Abcam (Cambridge, UK). Rictor mouse mAb 1G3P2C9 was obtained from Bethyl Laboratories (Montgomery, TX). Peroxidase AffiniPure goat anti-mouse IgG, peroxidase AffiniPure goat anti-rabbit IgG, and AMCA AffiniPure goat anti-mouse IgG were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
RNA and DNA constructs.
Dharmacon ON-TARGETplus nontargeting siRNA Control Pool and SMARTPool of human HRas (3265) siRNA, human NRas (4893) siRNA, and human KRas (3845) siRNA were obtained from Horizon Discovery (Waterbeach, UK). pLKO.1 GFP shRNA (Addgene: plasmid #30323) and Raptor_1 shRNA (Addgene; plasmid #1857) were gifts from David Sabatini (Whitehead Institute, Cambridge, MA) and are described elsewhere (Sarbassov et al., 2005; Sancak et al., 2008). Rictor shRNA in pGIPZ lentiviral vector was a gift from Carole Parent (University of Michigan, Ann Arbor, MI) and was described previously (Liu et al., 2010). Pax2 packaging plasmid (Addgene; plasmid #35002) was a gift from Malin Parmar (Lund University, Lund, Sweden) and was described elsewhere (Pfisterer et al., 2011). pMD2.G envelope plasmid (Addgene; plasmid #12259) was a gift from Didier Trono (Swiss Federal Institute of Technology, Lausanne, Switzerland). HRas, NRas, KRas4A, KRas4B, HRas(G12V), NRas(G12V), Kras4A(G12V), and KRas4B(G12V) in pLenti were purchased from GenScript Biotech (Piscataway, NJ), using pLenti-PGK-KRAS4B(G12V) (Addgene; plasmid #35633) as the vector template, which was a gift from Daniel Haber (Harvard Medical School, Boston, MA) and was described elsewhere (Singh et al., 2012).
Cell culture
MDAMB453 cells were a gift from Joyce Schroeder (University of Arizona, Tucson, AZ); and HEK293T, MCF10A, MCF7, MDAMB231, and SKBR3 cells were obtained from the University of Arizona Cancer Center Experimental Mouse Shared Resources. HER2-expressing MCF10A and their puromycin-resistant MCF10A control cell line (transduced with empty vector) were gifts from Cheuk Leung (University of Minnesota, Minneapolis, MN). To generate these cells, retroviruses were produced by cotransfecting pBABE-puro-ErbB2 or pBABE-puro (Debnath et al., 2002) in HEK293 cells, with the packaging vector pCL-Ampho (Novus Biologicals, Littleton, CO). To generate the stable cell lines MCF10A/pBABE-puro-ErbB2 and MCF10A/pBABE-puro, MCF10A cells were transduced with the corresponding viral vectors overnight and selected in 2 µg/ml puromycin. HEK293T and MDAMB231 cells were cultured in DMEM with 10% FBS; MCF7 cells were cultured in DMEM with 10% FBS supplemented with 10 µg/ml insulin; MDAMB453 cells were cultured in RPMI-1640 medium with 10% FBS; SKBR3 cells were cultured in McCoy’s 5A medium with 10% FBS; MCF10A cells were cultured in DMEM/F12 with 5% horse serum supplemented with 20 ng/ml EGF, 0.5 mg/ml hydrocortisone, 100 ng/ml cholera toxin, and 10 µg/ml insulin. All cell culture media were supplemented with 100 U/ml penicillin/streptomycin. Cells were transiently transfected with siRNAs using lipofectamine RNAiMAX transfection reagent according to the manufacturer’s protocol. HEK293T cells were transiently transfected with Pax2 (packaging) and pMD2.G (envelope) plasmids together with shRNA-containing lentiviral vectors using the calcium phosphate precipitation method to produce shRNA-containing lentiviruses, which were then used to transduce cells for shRNA expression. MCF10A cells were transduced in a similar manner for the expression of wild-type Ras proteins and RasCA mutants. Cells were selected for stable shRNA or gene expression in 200 µg/ml hygromycin-containing media or 2.5 µg/ml puromycin as appropriate. After the selection was completed, the stable cell lines were maintained in regular MCF10A culture media.
Migration and invasion assays
Wound closure 2D migration.
Cells were grown in six-well plates to confluency. Scratches were made manually using flat tip pipette tips, removing a uniform, thin line of cells from one side of the well to the other. Removed cells were washed off the plates and contrast images were captured to measure the initial wound sizes. The wounded cells were then incubated in media at 37°C for 18 h (MCF10A, HER2/MCF10A), 32 h (MDAMB231), 48 h (MCF7), or 72 h (MDAMB453). Images of the wounds were captured before and after the incubation using a Motic Stereo Zoom microscope. Cell migration distances were determined by analyzing the difference in wound sizes at the end of the incubation by calculating the average of six measurements of wound width from one side of the well to the other using the Motic Images Plus software. Where indicated, 0.1% dimethyl sulfoxide (DMSO; control), 100 nM rapamycin, 100 nM Torin2, or 50 μM prenyltransferase inhibitors (PTIs; 25 μM farnesyltransferase inhibitor [FTI] and 25 μM geranylgeranyltransferase inhibitor [GGTI]) was added after wounding of the cells. For the PTI treatments of HER2/MCF10A cells, twice as much PTIs and DMSO (control) were used due to FTI and GGTI lots that were less potent.
Transwell 3D migration.
A Corning BioCoat Matrigel Invasion Chamber with 8.0 μm PET membrane in six-well plates from Corning (Corning, NY) were used following the manufacturer’s protocol. Briefly, 2.5 × 105 cells were seeded in the upper chambers while growth media was added to both chambers and then incubated for 24 h at 37°C. The cells that did not migrate were then removed from the upper surface of the membrane, and the cells that migrated to the lower chamber and attached to the lower surface of the membrane were fixed with 3.7% formaldehyde, permeabilized with methanol, and stained with 0.5% crystal violet in 20% ethanol. Using a light microscope at 200× magnification, the stained cells were then counted across 13 different areas encompassing the center of the chamber and avoiding the edges.
MCF10A acini invasion.
The 3D cell culture of MCF10A cells in chamber slides on reconstituted basement membrane was performed as previously described (Debnath et al., 2003). After 12 d in culture, cells formed matured acini that were then fixed with 4% paraformaldehyde and 1% glutaraldehyde, and the cells permeabilized with 0.5% Triton X-100. To visualize the cells and acini structures, F-actin and laminin were stained for fluorescence microscopy. F-actin was stained using 2.5% Alexa Fluor 488 phalloidin, and laminin was revealed by immunofluorescence (IF) using sequential incubations with anti-laminin 5 antibody and Alexa Fluor 568 goat anti-rabbit IgG in IF buffer (130 mM NaCl, 7.7 mM NaN3, 7 mM Na2HPO4, 3.5 mM NaH2PO4, 0.2% bovine serum albumin, 0.2% Triton X-100, 0.01% Tween-20). For imaging, slides were mounted on glass coverslips, imaged by confocal microscopy using a 3i Marianna spinning-disk confocal system, and analyzed using the Slidebook 6.0 software. Blinded visual scoring of the acini morphological phenotypes by a distinct researcher was performed to classify them in one of three categories: 1) normal/quasinormal—spherical architecture with intact basal lamina and no cellular projections into the extracellular matrix; 2) dysmorphic—misshapen acinar structures, with filled lumen and with or without a disrupted lamina, and no cellular projections into the extracellular matrix; and 3) invasive—misshapen and complex structures, with filled lumen and disrupted or absent basal lamina, and displaying cellular projections and invasion of cells into the extracellular matrix. Assessment of rictor expression in the rictor shRNA-transduced HRasCA/MCF10A acini was performed by IF using mouse anti-rictor antibody and AMCA AffiniPure goat anti-mouse IgG and imaged by epifluorescence using the 3i Marianna imaging system.
Immunoblot
Log phase growing cells were collected and lysed on ice in RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS, 10 mM Na-pyrophosphate, 10 mM beta-glycerophosphate, 0.5 mM Na-orthovanadate, 50 mM NaF, supplemented with protease inhibitors). Lysates were cleared by centrifugation, and protein concentration in lysates was determined and normalized between conditions. Samples were mixed 1:6 with 6X Laemmli sample buffer leading to 100 mM dithiothreitol final, resolved on SDS–PAGE and analyzed by immunoblotting using phospho-AKT (Ser473) antibody to detect phosphorylated AKT at the mTORC2-dependent site. Pan-AKT antibody was used to detect total AKT protein expression, phospho-ERK and ERK antibodies were used to detect phosphorylated and total ERK, respectively, and HER2 antibody was used to detect HER2 expression. Where indicated, cells were pretreated with 0.1% DMSO (control) or 50 μM PTIs (25 μM FTI, 25 μM GGTI) for 24 h at 37°C. For the PTI treatments of HER2/MCF10A cells, twice as much PTIs and DMSO were used due to FTI and GGTI lots that were less potent. Protein bands from the immunoblots were quantified by densitometry using ImageJ.
Statistical analyses
Quantified cell migration data were graphed and analyzed in Prism using the unpaired nonparametric two-tailed Mann-Whitney test, whereas the quantified immunoblots were analyzed using a one-tailed Mann-Whitney test. p values below 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by a Research Scholar Grant from the American Cancer Society, Grant no. RSG-15-024-01-CSM, and National Institutes of Health (NIH) R01 Grant no. GM-131200 to P.G.C. S.E.C. and A.N.W. were also supported by NIH T32 Grant no. GM-008804, and M.E.W. by NIH T32 Grant no. GM-136536. G.M. also acknowledges support from NIH R01 Grant no. CA-196885. We are grateful to David Sabatini, Carole Parent, Malin Parmar, Didier Trono, Daniel Haber, Cheuk Leung, and Joyce Schroeder for providing material. All confocal images were collected in the W.M. Keck Center for Nano-Scale Imaging in the Department of Chemistry and Biochemistry at the University of Arizona. This instrument purchase was partially supported by Arizona Technology and Research Initiative Fund (A.R.S.§15-1648).
Abbreviations used:
- CA
constitutively active
- DCIS
ductal carcinoma in situ
- DMSO
dimethyl sulfoxide
- EGFR
epidermal growth factor receptor
- EMT
epithelial-mesenchymal transition
- ER
estrogen receptor
- ERK
extracellular signal-regulated kinase
- FTI
farnesyltransferase inhibitor
- GGTI
geranylgeranyltransferase inhibitor
- HER2
human epidermal growth factor receptor 2
- mTOR
mechanistic target of rapamycin
- mTORC2
mTOR complex 2
- NT
nontargeting
- PI3K
phosphoinositide 3-kinase
- PR
progesterone receptor
- PTIs
prenyltransferase inhibitors
- shRNA
small hairpin RNA
- siRNA
small interfering RNA
- TNBC
triple negative breast cancer.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-06-0236) on December 21, 2022.
REFERENCES
- American Type Culture Collection records, Center for Biological Sciences Archives, Collection 10, Special Collections, University of Maryland, Baltimore County (Baltimore, MD).
- Basolo F, Elliott J, Tait L, Chen XQ, Maloney T, Russo IH, Pauley R, Momiki S, Caamano J, Klein-Szanto AJP, et al. (1991). Transformation of human breast epithelial cells by c-Ha-ras oncogene. Mol Carcinog 4, 25–35. [DOI] [PubMed] [Google Scholar]
- Bera A, Das F, Ghosh-Choudhury N, Kasinath BS, Abboud HE, Choudhury GG (2014). MicroRNA-21-induced dissociation of PDCD4 from rictor contributes to Akt-IKKβ-mTORC1 axis to regulate renal cancer cell invasion. Exp Cell Res 328, 99–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Y, Wang Z, Xu J, Zhao W, Cao H, Zhang Z (2015). Elevated Rictor expression is associated with tumor progression and poor prognosis in patients with gastric cancer. Biochem Biophys Res Commun 464, 534–540. [DOI] [PubMed] [Google Scholar]
- Butt G, Shahwar D, Qureshi MZ, Attar R, Akram M, Birinci Y, Karatoprak GS, Gasparri ML, Farooqi AA (2019). Role of mTORC1 and mTORC2 in breast cancer: therapeutic targeting of mTOR and its partners to overcome metastasis and drug resistance. Adv Exp Med Biol 1152, 283–292. [DOI] [PubMed] [Google Scholar]
- Cai H, Das S, Kamimura Y, Long Y, Parent CA, Devreotes PN (2010). Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. J Cell Biol 190, 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel P, Dharmaiah S, Sale MJ, Messing S, Rizzuto G, Cuevas-Navarro A, Cheng A, Trnka MJ, Urisman A, Esposito D, et al. (2021). RAS interaction with Sin1 is dispensable for mTORC2 assembly and activity. Proc Natl Acad Sci USA 118, e2103261118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest PG, Shen Z, Lakoduk A, Sasaki AT, Briggs SP, Firtel RA (2010). A ras signaling complex controls the RasC-TORC2 pathway and directed cell migration. Dev Cell 18, 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Cheng H, Pan T, Liu Y, Su Y, Ren C, Huang D, Zha X, Liang C (2015). mTOR regulate EMT through RhoA and Rac1 pathway in prostate cancer. Mol Carcinog 54, 1086–1095. [DOI] [PubMed] [Google Scholar]
- Clair T, Miller W, Cho-Chung Y (1987). Prognostic significance of the expression of a ras protein with molecular weight of 21,000 by human breast cancer. Cancer Res 15, 5290–5293. [PubMed] [Google Scholar]
- Cowell CF, Weigelt B, Sakr RA, Ng CKY, Hicks J, King TA, Reis-Filho JS (2013). Progression from ductal carcinoma in situ to invasive breast cancer: revisited. Mol Oncol 7, 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Cheng H, Bai Z, Li J (2017). Breast cancer cell line classification and its relevance with breast tumor subtyping. J Cancer 8, 3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daulat AM, Bertucci F, Audebert S, Sergé A, Finetti P, Josselin E, Castellano R, Birnbaum D, Angers S, Borg JP (2016). PRICKLE1 contributes to cancer cell dissemination through its interaction with mTORC2. Dev Cell 37, 311–325. [DOI] [PubMed] [Google Scholar]
- Debnath J, Brugge JS (2005). Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer 5, 675–688. [DOI] [PubMed] [Google Scholar]
- Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS (2002). The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell 111, 29–40. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS (2003). Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268. [DOI] [PubMed] [Google Scholar]
- Fiordalisi JJ, Cox AD (2010). Chapter 222 - Farnesyltransferase inhibitors. Handbook of Cell Signaling, Part II, 2nd ed., New York: Academic Press, 1819–1826. [Google Scholar]
- Galiè M (2019). RAS as supporting actor in breast cancer. Front Oncol 9, 1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimple RC, Wang X (2019). RAS: Striking at the core of the oncogenic circuitry. Front Oncol 9, 965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulhati P, Bowen KA, Liu J, Stevens PD, Rychahou PG, Chen M, Lee EY, Weiss HL, O’Connor KL, Gao T, et al. (2011). mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res 71, 3246–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Hau AM, Beach JR, Harwalker J, Mantuano E, Gonias SL, Egelhoff TT, Hansel DE (2013). Mammalian target of rapamycin complex 2 (mTORC2) is a critical determinant of bladder cancer invasion. PLoS One 8, e81081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly MM, Hicks DJ, Jones B, Sanchez V, Estrada MV, Young C, Williams M, Rexer BN, Sarbassov DD, Muller WJ, et al. (2016). Rictor/mTORC2 drives progression and therapeutic resistance of HER2-amplified breast cancers. Cancer Res 76, 4752–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y, Xiong Y, Iglesias PA, Hoeller O, Bolourani P, Devreotes PN (2008). PIP3-independent activation of TorC2 and PKB at the cell’s leading edge mediates chemotaxis. Curr Biol 18, 1034–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao J, Salari K, Bocanegra M, Choi YL, Girard L, Gandhi J, Kwei KA, Hernandez-Boussard T, Wang P, Gazdar AF, et al. (2009). Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One 4, e6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna A, Lotfi P, Chavan AJ, Montaño NM, Bolourani P, Weeks G, Shen Z, Briggs SP, Pots H, Van Haastert PJM, et al. (2016). The small GTPases Ras and Rap1 bind to and control TORC2 activity. Sci Rep 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Lee E, Kim H, Moon A (2003). p38 kinase is a key signaling molecule for h-Ras-induced cell motility and invasive phenotype in human breast epithelial cells. Cancer Res 63, 5454–5461. [PubMed] [Google Scholar]
- Kim EK, Yun SJ, Ha JM, Kim YW, Jin IH, Yun J, Shin HK, Song SH, Kim JH, Lee JS, et al. (2011). Selective activation of Akt1 by mammalian target of rapamycin complex 2 regulates cancer cell migration, invasion, and metastasis. Oncogene 30, 2954–2963. [DOI] [PubMed] [Google Scholar]
- Kovalski JRR, Bhaduri A, Zehnder AMM, Neela PHH, Che Y, Wozniak GGG, Khavari PAA (2019). The functional proximal proteome of oncogenic Ras includes mTORC2. Mol Cell 73, 830–844.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Connolly E, Smyth JW, Akhurst RJ, Derynck R (2012). TGF-β-induced activation of mTOR complex 2 drives epithelial-mesenchymal transition and cell invasion. J Cell Sci 125, 1259–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie H, Gagnon J, Therrien M (2020). ERK signalling: a master regulator of cell behaviour, life and fate. Nat Rev Mol Cell Biol 21, 607–632. [DOI] [PubMed] [Google Scholar]
- Lee KH, Koh M, Moon A (2016). Farnesyl transferase inhibitor FTI-277 inhibits breast cell invasion and migration by blocking H-Ras activation. Oncol Lett 12, 2222–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA (2011). Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121, 2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lin J, Wang X, Yao G, Wang L, Zheng H, Yang C, Jia C, Liu A, Bai X (2012). Targeting of mTORC2 prevents cell migration and promotes apoptosis in breast cancer. Breast Cancer Res Treat 134, 1057–1066. [DOI] [PubMed] [Google Scholar]
- Liao XH, Buggey J, Lee YK, Kimmel AR (2013). Chemoattractant stimulation of TORC2 is regulated by receptor/G protein targeted inhibitory mechanisms that function upstream and independently of an essential GEF/Ras activation pathway in dictyostelium. Mol Biol Cell 24, 2146–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GY, Sabatini DM (2020). mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol 21, 183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Das S, Losert W, Parent CA (2010). mTORC2 regulates neutrophil chemotaxis in a cAMP- and RhoA-dependent fashion. Dev Cell 19, 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone M-U-D, Miyan J, Asif M, Malik SA, Dubey P, Singh V, Singh K, Mitra K, Pandey D, Haq W, et al. (2019). Direct physical interaction of active Ras with mSIN1 regulates mTORC2 signaling. BMC Cancer 19, 1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Lu Y (2021). Born to run? Diverse modes of epithelial migration. Front Cell Dev Biol 9, 2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maru S, Ishigaki Y, Shinohara N, Takata T, Tomosugi N, Nonomura K (2013). Inhibition of mTORC2 but not mTORC1 up-regulates E-cadherin expression and inhibits cell motility by blocking HIF-2α expression in human renal cell carcinoma. J Urol 189, 1921–1929. [DOI] [PubMed] [Google Scholar]
- Masri J, Bernath A, Martin J, Jo OD, Vartanian R, Funk A, Gera J (2007). mTORC2 activity is elevated in gliomas and promotes growth and cell motility via overexpression of rictor. Cancer Res 67, 11712–11720. [DOI] [PubMed] [Google Scholar]
- Moon A, Kim M-S, Kim T-G, Kim S-H, Kim HE, Chen YQ, Choi KIM H-R (2000). H-Ras, but not N-Ras, induces an invasive phenotype in human breast epithelial cells: a role for MMP-2 in the H-Ras-induced invasive phenotype. J Cancer 85, 176–181. [DOI] [PubMed] [Google Scholar]
- Morrison Joly M, Williams MMM, Hicks DJJ, Jones B, Sanchez V, Young CDD, Sarbassov DDD, Muller WJJ, Brantley-Sieders D, Cook RSS (2017). Two distinct mTORC2-dependent pathways converge on Rac1 to drive breast cancer metastasis. Breast Cancer Res 19, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison MM, Young CD, Wang S, Sobolik T, Sanchez VM, Hicks DJ, Cook RS, Brantley-Sieders DM (2015). mTOR directs breast morphogenesis through the PKC-α-Rac1 signaling axis. PLoS Genet 11, e1005291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WJ, Jacinto E (2011). mTOR complex 2 signaling and functions. Cell Cycle 10, 2305–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Björklund A, Lindvall O, Jakobsson J, Parmar M (2011). Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci USA 108, 10343–10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips MR, Cox AD (2007). Geranylgeranyltransferase I as a target for anti-cancer drugs. J Clin Invest 117, 1223–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM (2008). The rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM (2005). Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101. [DOI] [PubMed] [Google Scholar]
- Sasaki AT, Firtel RA (2006). Regulation of chemotaxis by the orchestrated activation of Ras, PI3K, and TOR. Eur J Cell Biol 85, 873–895. [DOI] [PubMed] [Google Scholar]
- Shaw KR, Wrobel CN, Brugge JS (2004). Use of three-dimensional basement membrane cultures to model oncogene-induced changes in mammary epithelial morphogenesis. J Mammary Gland Biol Neoplasia 9, 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Sweeney MF, Yu M, Burger A, Greninger P, Benes C, Haber DA, Settleman J (2012). TAK1 inhibition promotes apoptosis in KRAS-dependent colon cancers. Cell 148, 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SF, Collins SE, Charest PG (2020). Ras, PI3K and mTORC2 – three’s a crowd? J Cell Sci 133, jcs234930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano O, Alcón-Pérez M, Vicente-Manzanares M, Castellano E (2021). The crossroads between RAS and RHO signaling pathways in cellular transformation, motility and contraction. Genes 12, 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee AR (2018). The target of rapamycin and nechanisms of cell growth. Int J Mol Sci 19, 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lintig FC, Dreilinger AD, Varki NM, Wallace AM, Casteel DE, Boss GR (2000). Ras activation in human breast cancer. Breast Cancer Res Treat 62, 51–62. [DOI] [PubMed] [Google Scholar]
- Wang L, Qi J, Yu J, Chen H, Zou Z, Lin X, Guo L (2017). Overexpression of Rictor protein in colorectal cancer is correlated with tumor progression and prognosis. Oncol Lett 14, 6198–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lai P, Zhang Z, Huang M, Wang L, Yin M, Jin D, Zhou R, Bai X (2014). Targeted inhibition of mTORC2 prevents osteosarcoma cell migration and promotes apoptosis. Oncol Rep 32, 382–388. [DOI] [PubMed] [Google Scholar]
- Wang D, Wu P, Wang H, Zhu L, Zhao W, Lu Y (2016). SIN1 promotes the proliferation and migration of breast cancer cells by Akt activation. Biosci Rep 6, e00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Wang X, Proud CG (2018). Who does TORC2 talk to? Biochem J 475, 1721–1738. [DOI] [PubMed] [Google Scholar]
- Yang X, Palasuberniam P, Myers KA, Wang C, Chen B, Yang X, Palasuberniam P, Myers KA, Wang C, Chen B (2016). Her2 oncogene transformation enhances 5-aminolevulinic acid-mediated protoporphyrin IX production and photodynamic therapy response. Oncotarget 7, 57798–57810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh KE, Regunath K, Guzman A, Lee S-M, Pfister NT, Akanni O, Kaufman LJ, Prives C, Prywes R (2016). Repression of p63 and induction of EMT by mutant Ras in mammary epithelial cells. Proc Natl Acad Sci USA 113, E6107–E6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang G, Kong C, Gong D (2016). PP242 suppresses bladder cancer cell proliferation and migration through deactivating the mammalian target of rapamycin complex 2/AKT1 signaling pathway. Mol Med Rep 13, 333–338. [DOI] [PubMed] [Google Scholar]
- Zhou H, Huang S (2010). The complexes of mammalian target of rapamycin. Curr Protein Pept Sci 11, 409–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.