Abstract
Many axonemal proteins enter cilia and flagella on intraflagellar transport (IFT) trains, which move bidirectionally along the axonemal microtubules. Certain axonemal substructures including the radial spokes and outer dynein arms are preassembled in the cell body and transported as multisubunit complexes into flagella by IFT. Here, we used in vivo imaging to analyze the transport and assembly of DRC2 and DRC4, two core subunits of the nexin–dynein regulatory complex (N-DRC). Tagged DRC2 moved by IFT in mutants lacking DRC4 and vice versa, showing that they do not depend on each other for IFT. Simultaneous imaging of tagged DRC2 and DRC4, expressed from transgenes that rescue a corresponding double mutant, mostly showed transport on separate IFT trains, but occasional cotransports were also observed. The results demonstrate that DRC2 and DRC4 are transported largely independently of each other into flagella. These studies suggest that the N-DRC assembles onto the axoneme by the stepwise addition of subunits.
INTRODUCTION
The assembly of most cilia and eukaryotic flagella depends on intraflagellar transport (IFT), the bidirectional motility of transport carriers or trains that move along the axonemal doublet microtubules (Kozminski et al., 1993). Imaging in several species has shown that IFT transports proteins of the ciliary membrane, matrix, and axoneme (Qin et al., 2005; Williams et al., 2014; Lechtreck, 2015; Ye et al., 2018). In Chlamydomonas, the transport frequency of the axonemal proteins is up-regulated during flagellar assembly, likely explaining the critical need of IFT to assemble cilia (Wren et al., 2013; Craft et al., 2015; Dai et al., 2018; Lechtreck et al., 2018, 2022). While tubulin dimers bind directly to the IFT-B proteins IFT74 and IFT81, the outer dynein arms, I1 inner dynein arms, and radial spoke precursors are preassembled in the cell body and attach to the IFT trains via specific adapter proteins (Ahmed et al., 2008; Kubo et al., 2016; Hunter et al., 2018; Van De Weghe et al., 2020; Lechtreck et al., 2022). These observations raise the question whether other axonemal substructures follow a similar pathway of preassembly in the cell body followed by the IFT-mediated transport of multisubunit complexes into cilia.
Here, we analyzed the transport of the nexin–dynein regulatory complex (N-DRC), which forms cross-bridges between adjacent doublet microtubules (DMTs; Heuser et al., 2009; Bower et al., 2013). The Chlamydomonas N-DRC consists of at least 11 distinct proteins (Lin et al., 2011; Bower et al., 2013). Mutations in several N-DRC subunits alter flagellar motility but also suppress flagellar paralysis seen in radial spoke mutants (Huang et al., 1982; Brokaw and Kamiya, 1987; Piperno et al., 1992; Piperno et al., 1994). Structural analysis further revealed that the N-DRC contacts the radial spokes, inner dynein arms, and outer dynein arms (Heuser et al., 2009). In a subset of Chlamydomonas N-DRC mutants, one of the two radial spokes (i.e., RS2), a subset of inner dynein arms, especially IDA e, and tektin are reduced or missing entirely (Gardner et al., 1994; Yanagisawa and Kamiya, 2004; Bui et al., 2012; Heuser et al., 2012; Austin-Tse et al., 2013; Wirschell et al., 2013; Bower et al., 2018). In humans, pathogenic mutations in N-DRC protein orthologues cause primary ciliary dyskinesia (Austin-Tse et al., 2013; Wirschell et al., 2013; Olbrich et al., 2015; Lewis et al., 2016). Despite its central role as an axonemal organizer and regulator of ciliary motility, the assembly of the N-DRC remains incompletely understood.
In mutants axonemes lacking certain N-DRC proteins, other N-DRC subunits are reduced or lost, establishing a partial hierarchy of N-DRC assembly. For example, numerous N-DRC subunits (i.e., DRC3 and DRC5-11) are reduced in mutants lacking DRC1, DRC2, or DRC4 indicating a central role of the latter subunits in forming the scaffold of the N-DRC base plate on the A-tubules of the DMTs, to which the more peripheral subunits of the linker region are attached (Gui et al., 2019). Recent ultrastructural analysis established that this base plate is formed by a heterodimer of the related DRC1 and DRC2 subunits and a homodimer of DRC4 (Gui et al., 2021). The N-DRC base plate interacts with the “axonemal ruler” composed of the coiled-coil proteins CCDC39/40 (Chlamydomonas FAP59/172) bound along the A-tubule wall to establish a 96-nm repeat (Oda et al., 2014; Gui et al., 2021). DRC1 is absent in axonemes of the DRC2-deficient mutant ida6, and DRC2 is missing in pf3/drc1 mutant axonemes, revealing a codependence between these two subunits for assembly onto the axoneme (Lin et al., 2011; Bower et al., 2013, 2018). In the ida6/drc2 mutant, DRC4 is present at wild-type levels in cell extracts and still assembled into axonemes, albeit in reduced amounts, indicating that it can enter cilia and bind to DMTs in the absence of DRC1/2 heterodimers (Bower et al., 2018). This raises the question whether the N-DRC, like some other axonemal substructures, is preassembled or partially preassembled in the cell body and transported into cilia as a holo complex or whether the N-DRC is assembled in a step-by-step manner from individual subunits or small subcomplexes that enter cilia on their own.
Here, we used in vivo imaging of tagged DRC2 and DRC4 subunits to investigate the characteristics and mechanism of N-DRC transport and assembly. Previously, we showed that DRC2 and DRC4 move by IFT and that DRC4 transport, like that of several other axonemal proteins, is up-regulated in short growing flagella (Wren et al., 2013; Craft et al., 2015; Dai et al., 2018). Here, we show that IFT of DRC2 continued in mutants lacking DRC4 and vice versa. Simultaneous two-color imaging revealed that these two proteins move mostly independently of each other on separate IFT trains. Together with other data, these observations support a model of stepwise N-DRC assembly by the transport and binding of individual proteins or smaller subcomplexes onto the DMTs.
RESULTS AND DISCUSSION
Coexpression of tagged DRC2 and DRC4 transgenes rescues the corresponding ida6 pf2 double mutant
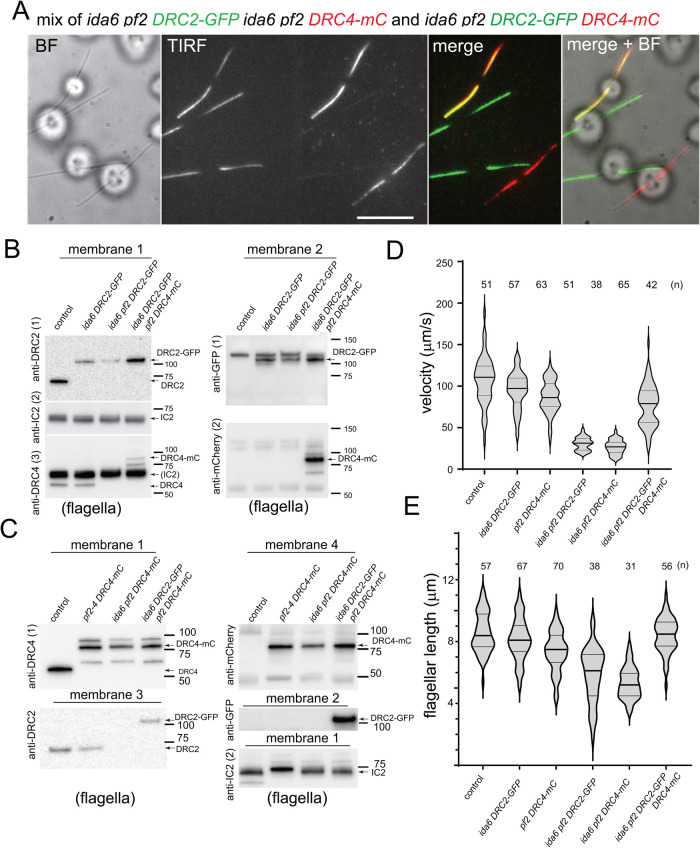
We previously reported that both DRC2-green fluorescent protein (GFP) and DRC4-GFP are cargoes of IFT (Wren et al., 2013). To address the question whether these two N-DRC subunits depend on each other for IFT and are transported as a single complex, we generated a pf2 DRC4-mCherry (mC) strain (Supplemental Figure S1A) and mated it to an ida6 DRC2-GFP strain. From the progeny, we isolated three different strains: a DRC2-GFP rescue (ida6 pf2 DRC2-GFP), a DRC4-mC rescue (ida6 pf2 DRC4-mC), and a double DRC2-GFP, DRC4-mC rescue (ida6 pf2-4 DRC2-GFP DRC4-mC). In a mixture of these strains, total internal reflection fluorescence (TIRF) microscopy of live cells revealed cells expressing DRC2-GFP only, DRC4-mC only, and both proteins (Figure 1A). Western blotting of isolated flagella confirmed both the absence of endogenous DRC2 and DRC4 subunits and the presence of either GFP-tagged DRC2, mC-tagged DRC4, or both subunits in these strains (Figure 1, B and C, and Supplemental Figure S1B). Flagella isolated from wild-type, ida6 DRC2-GFP, pf2 DRC4-mC rescue strains were loaded as controls. In the Western blots, DRC4-mC typically formed several immunoreactive bands (see Supplemental Figure S1B for details). The levels of DRC2-GFP and DRC4-mC were reduced in flagella of the respective double-mutant-single-rescue strains (Figure 1B, panels anti-DRC2 and anti-GFP, and Figure 1C, panels anti-DRC4 and anti-mC). These observations agree with the imaging data and previous mass spectrometry data showing reduced levels of DRC4 and other N-DRC proteins in ida6 axonemes (Bower et al., 2018), and reduced levels of DRC2 and other N-DRC subunits in pf2 axonemes (Lin et al., 2011; Bower et al., 2013).
FIGURE 1:
DRC2 and DRC4 mostly move independently of each other. (A) TIRF, bright field (BF), and merged images showing a mixture of double mutant–single rescued cells (ida6 pf2 DRC2-GFP and ida6 pf2 DRC4-mC) and double mutant–double rescued cells (ida6 pf2 DRC2-GFP DRC4-mC). TIRF, original frame from the TIRF recording with the GFP signal projected on the left side and the mC signal onto the right side of the chip. Bar = 10 μm. (B) Western blot analyzing flagella isolated from wild type (control), ida6 DRC2-GFP, ida6 pf2 DRC2-GFP, and ida6 pf2 DRC2-GFP DRC4-mC. Membranes 1 and 2 are replicate membranes with equal sample loading. Membrane 1 was first stained with anti-DRC2 (1), followed by anti-IC2 (2) as a loading control, and finally anti-DRC4 (3). Membrane 2 was stained first with anti-GFP (1) and then anti-mC (2). The positions of tagged and endogenous DRC2 and DRC4 and of the marker proteins are indicated. DRC4-mC ran as multiple bands, of which the middle one was the most prominent with both anti-mC and anti-DRC4. The mC tag tends to fragment upon sample heating, likely explaining the banding pattern (Gross et al., 2000). (C) Western blot analyzing flagella isolated from wild type (control), pf2-4 DRC4-mC, ida6 pf2 DRC4-mC, and ida6 pf2 DRC2-GFP DRC4-mC. Membranes 1–4 are replicate membranes with equal loading. Membrane 1 was first stained with anti-DRC4 (1), followed by anti-IC2 (2) as a loading control. Membrane 2 was stained with anti-GFP, membrane 3 with anti-DRC2, and membrane 4 with anti-mC. The positions of tagged and endogenous DRC2 and DRC4 and the marker proteins are indicated. (D) Violin plot of the swimming velocity of wild type, ida6 DRC2-GFP, pf2 DRC4-mC, ida6 pf2 DRC2-GFP, ida6 pf2 DRC4-mC, and ida6 pf2 DRC2-GFP DRC4-mC. The number of cells analyzed is indicated; error bars indicate SD. (E) Violin plot depicting the flagella length distribution in live cells from wild type, ida6 DRC2-GFP, pf2 DRC4-mC, ida6 pf2 DRC2-GFP, ida6 pf2 DRC4-mC, and ida6 pf2 DRC2-GFP DRC4-mC (see Materials and Methods for details).
Similar to the original pf2 and ida6 mutants, the double-mutant-single-rescue strains swam with strongly reduced velocities in comparison to the wild-type control (Figure 1, D and E, and Supplemental Figure S1C). The swimming velocity of the double mutant–double rescue cells was well above that of the ida6 or pf2 mutant strains but was approximately 25% lower than that of control wild-type cells (Supplemental Figure S1C). Because the parental ida6 DRC2-GFP and pf2 DRC4-mC rescued strains also swam somewhat slower than wild-type cells, the combination of both tagged subunits in the double mutant–double rescue cells could lead to a more pronounced reduction of velocity. High-speed video microscopy of the double mutant–double rescue strain revealed a normal flagellar beat cycle similar to wild type (Supplemental Figure S1D and Supplemental Video 1). In contrast, flagella of the double mutant–single rescue strains beat with a reduced amplitude, consistent with previous observations (Brokaw and Kamiya, 1987). The motion analysis also indicated that the loss of DRC4 affects flagellar beating more profoundly than the loss of DRC2 (Supplemental Video 1). Like several other Chlamydomonas mutants with axonemal defects, the double mutant–single rescue strains typically had shorter than normal flagella (Figure 1E; Oda et al., 2014). The time course of flagellar regeneration was also significantly slower in both strains and, in a subset of cells, regeneration failed entirely (Figure 1 and unpublished data). Therefore, we did not attempt to analyze the transport of DRC subunits in the double-mutant-single-rescue strains during regeneration.
Movie S1.
Flagellar motion analysis of DRC mutant and rescue strains The video shows cells of the ida6 pf2 DRC2-GFP, ida6 pf2 DRC4-mC, and the ida6 pf2-4 DRC2-GFP DRC4-mC strains recorded at 1,000 fps. The timer counts seconds.milliseconds
DRC2 and DRC4 do not require each other for IFT
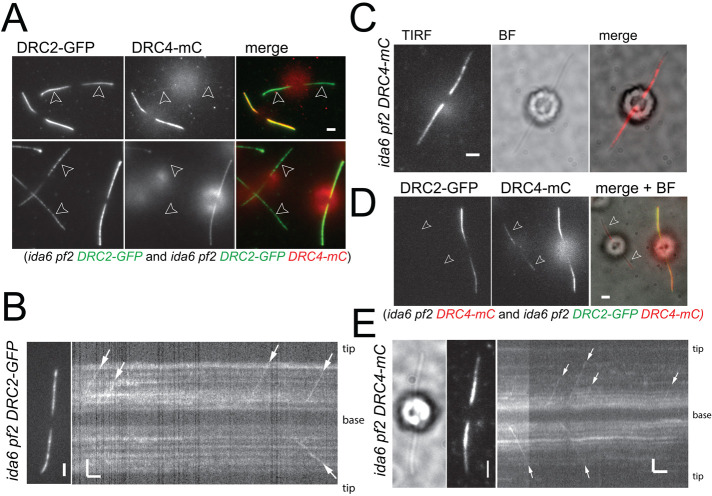
TIRF imaging confirmed the reduction of DRC2-GFP in the ida6 pf2 DRC2-GFP strain (Figure 2A), consistent with the Western blots (Figure 1B). The strength of the DRC2-GFP signal varied between individual clonal cells, as often observed for the expression of transgenes in Chlamydomonas (Koblenz et al., 2003; Schroda, 2019), but DRC2-GFP was present along the length of flagella (Figure 1A). In partially photobleached, full-length flagella, IFT of DRC2-GFP was apparent (Figure 2B). Thus, IFT of DRC2-GFP occurs in the absence of DRC4. In the ida6 pf2 DRC4-mC strain, the DRC4-mC signal was more prominent in the proximal region of the flagella and tapered off toward the tip; the signal strength varied between cells and was particularly weak in a subset of cells (Figure 1A and Figure 2, C and D). IFT transport of DRC4-mC was regularly observed (Figure 2E). Many of these transport events progressed to the distal region of the flagella or the tip. Thus, the reduction of the DRC4-mC signal in the distal region appears to be the result of preferred binding of DRC4-mC to the proximal region of ida6 axonemes rather than a failure to reach the distal region by IFT, as has been proposed for the pf27/armc2 mutant, which fails to transport the radial spoke precursor by IFT limiting spoke assembly to the proximal region of the axoneme (Lechtreck et al., 2022). A similar proximodistal decrease in signal strength along the axoneme was observed for polyglutamylated tubulin (Lechtreck and Geimer, 2000), and polyglutamylated tubulin appears to interact with DRC4 (Kubo and Oda, 2017), offering a possible explanation for the more stable binding of DRC4-mC to the proximal region of ida6 cilia. In conclusion, DRC2-GFP and DRC4-mC do not require each other for transport by IFT.
FIGURE 2:
DRC2-GFP moves by IFT in the absence of DRC4 and vice versa. (A) Still images of ida6 pf2 DRC2-GFP and ida6 pf2 DRC2-GFP DRC4-mC cells. A mixture of the two strains was used to highlight the reduction of DRC2-GFP in the double mutant–single rescue strain with the pf2 mutation. Flagella are marked with arrowheads. Bar = 2 µm. (B) Still image and kymogram showing anterograde IFT of DRC2-GFP (white arrows) in the single rescue strain (ida6 pf2 DRC2-GFP). Bars = 2 s and 2 μm. (C) TIRF, bright field (BF), and merged image of an ida6 pf2 DRC4-mC cell. Bar = 2 μm. Note that the DRC4-mC signal tappers off toward the flagellar tip in the ida6 background. Bar = 2 µm. (D) Still images of ida6 pf2 DRC4-mC and ida6 pf2 DRC2-GFP DRC4-mC cells. Shown are the GFP image, the mC image, and a merge of the two channels with the bright field (BF) image. Note the reduction of the DRC4-mC signal in the double mutant–single rescue strain. Flagella are marked with arrowheads. Bar = 2 μm. (E) Bright field (BF) and TIRF images and the corresponding kymogram showing IFT of DRC4-mC transport in ida6 pf2 DRC4-mC (white arrows). Bars = 2 s and 2 μm.
DRC2 and DRC4 largely move independently of each other by IFT
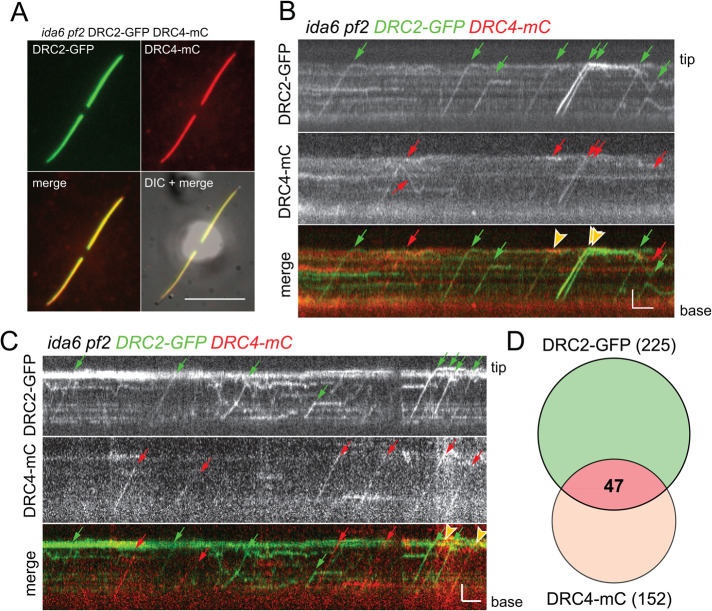
The observation that DRC2 and DRC4 can be transported by IFT into flagella independently of each other does not exclude the possibility that, when both are present, they are typically cotransported as a single complex, potentially including other N-DRC subunits. To address this question, we analyzed the transport of both subunits in regrowing, partially photobleached flagella of the ida6 pf2-4 DRC2-GFP DRC4-mC strain by simultaneous two-color TIRF microscopy. Because the double mutant–double rescue strain contains both proteins prominently along the length of flagella (Figure 3A), the flagella were partially photobleached first. Likely, photobleaching will reduce the signal of unbound subunits, and residual fluorescence from unbleached subunits could obscure some of the trajectories of the traveling DRC2-GFP and DRC4-mC subunits, which could affect the particle counts of N-DRC transport events. However, DRC2-GFP and DRC4-mC particles were mostly observed to be traveling independently of each other, although cotransport events with both proteins present on the same IFT train were occasionally observed (Figure 3, B and C). In detail, we analyzed 52 flagella for 3158 s and observed a total of 225 DRC2-GFP transports, of which 212 were anterograde transports, and 152 DRC4-mC transports, of which 142 were anterograde transports (Table 1). Only 47 of these events were cotransports, corresponding to 21% of the DRC2-GFP particles and 31% of the DRC4-mC particles (Figure 3D). While relatively low, the number of DRC2/DRC4 cotransports observed in regrowing flagella was approximately four times higher than expected by chance. Occasionally, cotransport events were also observed in full-length double mutant–double rescue (ida6 pf2-4 DRC2-GFP DRC4-mC) flagella, when IFT of DRC subunits was infrequent and the probability of cotransports occurring by chance is even lower. Thus, DRC2, DRC4, and likely other components of the N-DRC appear to occasionally move into flagella as a larger complex, suggesting prior association in the cell body.
FIGURE 3:
DRC2 and DRC4 mostly move independently of each other in regrowing flagella. (A) TIRF still images generated by averaging 10 individual frames of an ida6 pf2 DRC2-GFP DRC4-mC cell. Shown are GFP signal, mC signal, the merged signals, and an overlay of the fluorescence and DIC image (DIC + merge). Bar = 10 μm. (B, C) Kymograms showing DRC2-GFP and DRC4-mC transport in regenerating ida6 pf2 DRC2-GFP DRC4-mC flagella. Anterograde transport is indicated by trajectories from the bottom left (flagella base) to the top right (flagella tip), whereas retrograde IFT results are indicated in the top left to bottom right trajectories. DRC2-GFP transports are indicated with green arrows, and red arrows mark DRC4-mC transports. Cotransports are marked by yellow arrowheads in the merge kymogram (bottom). Bars = 2 s and 2 μm. (D) Venn diagram depicting the number of observed DRC2 and DRC4 transports in ida6 pf2 DRC2-GFP DRC4-mC flagella. The number of DRC2-GFP (green) and DRC4-mC (red) IFT events is indicated. The overlapping section depicts the share of DRC2-GFP DRC4-mC cotransport.
TABLE 1:
Frequency of cargo cotransport.
| Strain | DRC2-GFP (N) (ante/retro) | DRC4-mC (N) (ante/retro) | Cotransports (N) (ante/retro) | Cilia (N) | Time (s) |
|---|---|---|---|---|---|
| ida6 pf2 DRC2-GFP DRC4-mC (regenerating flagella) | 225(212/13) | 152(142/10) | 47(44/3) | 52 | 3158 |
| CC-5325 × ida6 pf2 DRC2-GFP DRC4-mC (mutant-derived flagella) | 137(130/7) | 117(107/10) | 11(9/2) | 73 | 5165 |
The table lists the observed IFT transports for DRC2-GFP and DRC4-mC in flagella of the double mutant–double rescue strain and in wild-type–derived zygotic cilia obtained by mating CC-5325 and ida6 pf2 DRC2-GFP DRC4-mC.
For the interdependent RSP3-NG/ARMC2-mS cargo/adapter pair, we previously observed that 80% of the RSP3 transport events in regenerating flagella were accompanied by the radial spoke adapter ARMC2. Also, tagged RSP3 and RSP4, two proteins of the radial spoke, comigrate on IFT trains in approximately 90% of the transport events observed in regenerating flagella of a pf1 pf14 RSP4-GFP mS-RSP3 double mutant–double rescue strain (Lechtreck et al., 2018). The cotransport rate for DRC2/DRC4 is well below those observed for the transport of the interlinked pairs RSP3/RSP4 and RSP3/ARMC2 (Lechtreck et al., 2018, 2022). In conclusion, in vivo imaging indicates that independent IFT transport is the preferred mode of flagellar delivery for DRC2-GFP and DRC4-mC. This conclusion is further supported by the apparent normal IFT of DRC2-GFP and DRC4-mC in the double mutant–single rescue strains.
DRC2 and DRC4 are transported and assembled mostly independently of each other into zygotic wild-type flagella
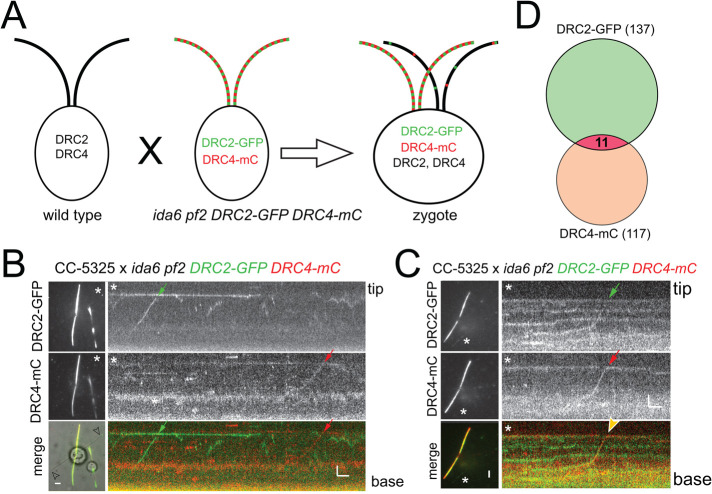
The extremely short flagella observed in the ida6 pf2 double mutant prevented us from performing a dikaryon rescue experiment using ida6 pf2 × ida6 pf2 DRC2-GFP DRC4-mC zygotes to analyze the entry of tagged DRC2 and DRC4 subunits in the absence of the untagged endogenous proteins. Instead, we mated the ida6 pf2 DRC2-GFP DRC4-mC strain to a wild-type strain (CC-5325) and analyzed the wild-type–derived zygotic flagella by TIRF. Because the two flagella derived from the wild-type parent are initially devoid of fluorescent proteins, this experiment allowed us to image transport events without prior photobleaching of the flagella and the associated loss of signal (Figure 4A). We reasoned that if DRC2 and DRC4 form a stable complex in the cell body, one would expect that labeled complexes provided by the double mutant–double rescue parent would enter the wild-type–derived flagella. We analyzed 44 zygotic wild-type–derived full-length flagella for 5165 s and observed a total 137 IFT events for DRC2-GFP and 117 for DRC4-mC. Of these, only 11 events showed cotransport of DRC2-GFP and DRC4-mC subunits, corresponding to ∼8% and 10% of all observed DRC2-GFP and DRC4-mC particles, respectively (Figure 4, B–D, and Supplemental Figure S2B). Thus, cotransport of these two N-DRC subunits was rare in both photobleached regenerating flagella and unbleached zygotic flagella. However, occasionally DRC2-GFP and DRC4-mC diffused together inside flagella, indicative of the presence of a mobile complex (Supplemental Figure S2A). Incorporation of tagged DRC proteins into the wild-type–derived flagella was frequently observed near the tip, where both proteins typically overlapped (Supplemental Figure S2, A and C). This can be explained by axonemal elongation of zygotic flagella and preferred exchange of subunits near the axonemal tip (Mesland et al., 1980). The incorporation of fluorescent DRC proteins along the length of the wild-type–derived flagella, which likely occurs by exchange of endogenous DRC proteins with tagged ones, was much less common. Such stationary DRC2-GFP and DRC4-mC signals typically formed distinct patterns, indicating that they incorporated into the axoneme independently of each other (Supplemental Figure S2A).
FIGURE 4:
DRC2 and DRC4 mostly move independently of each other in zygotic flagella. (A) Schematic of a mating experiment using wild-type (CC-5325) and ida6 pf2 DRC2-GFP DRC4-mC parental strains. The zygotes possess both tagged and endogenous versions of DRC2 and DRC4. (B) TIRF and merged still images of a zygote and the corresponding kymogram of the cilium marked by an arrowhead. IFT of DRC2-GFP and DRC4-mC is marked by green and red arrows, respectively. The star marks the flagellum used for kymographic analysis. Bars = 2 s and 2 µm. (C) as in B, but showing cotransport of DRC4-mC and DRC2-GFP. Bars = 2 s and 2 µm. (D) Venn diagram depicting the number of observed DRC2-GFP and DRC4-mC transports in the wild-type–derived flagella of CC-5325 × ida6 pf2 DRC2-GFP DRC4-mC zygotes. The number of IFT events of DRC2-GFP (green) and DRC4-mC (red) are indicated. The overlapping section depicts the share of DRC2-GFP DRC4-mC cotransport.
We also mated the pf2 ida6 DRC2-GFP and ida6 pf2 DRC4-mC strains to the double mutant–double rescue strain to analyze how these subunits are added during the repair of flagella initially lacking them. As previously described, DRC2-GFP and DRC4-mC were incorporated into full-length flagella lacking them starting from the tip and progressing toward the base (Supplemental Figure S2, D and E). In ida6 pf2 DRC2-GFP–derived zygotic cilia, the DRC4-mC signal typically exceeded the DRC2-GFP signal in the distal region of the cilia (Supplemental Figure S2D). In ida6 pf2 DRC4-mC–derived zygotic cilia, DRC2-GFP assembled also from the tip toward the base rather than being added first to the proximal flagellar region already occupied by DRC4-mC (Supplemental Figure S2E). To summarize, the in vivo imaging data indicate that DRC2 and DRC4 are typically transported into flagella and assembled into the axonemes separately of each other.
The independent transport and assembly of DRC2 and DRC4 shown here are consistent with previous quantitative mass spectrometry data showing a reduction of DRC4 and DRC2 associated with ida6 and pf2 mutations, respectively (Huang et al., 1982; Piperno et al., 1994; Bower et al., 2018). Recent structural studies have also shown that the DRC1/DRC2 heterodimer and the DRC4 homodimer play distinct but important roles in binding the N-DRC to the A-tubule and scaffolding the assembly of other N-DRC subunits, radial spokes, and inner dynein arms (Gui et al., 2019, 2021). Yet other work has demonstrated that mutations in the more peripheral N-DRC subunits (e.g., DRC3, DRC5, DRC7, and DRC11) have minimal impact on the assembly of the N-DRC (Bower et al., 2013; Awata et al., 2014; Song et al., 2015; Gui et al., 2019). Early dikaryon rescue experiments further suggest that DRC5 can be transported and assembled into drc5 (=spf4) mutant flagella independently of other N-DRC subunits (Huang et al., 1982). Although additional studies are needed to further define how each subunit is transported and incorporated into the axoneme, the work shown here suggests that the N-DRC assembles via a complex step-by-step process rather than by delivery and docking of a preformed precursor as described for radial spokes and dynein arms (Fowkes and Mitchell, 1998; Qin et al., 2004; Diener et al., 2011).
MATERIALS AND METHODS
Strains, culture conditions, and genotyping
The Chlamydomonas strains ida6-1 (CC-3090), pf2-4 (CC-4483), and ida6 IDA6-GFP (CC-4495) have been previously described (Kato et al., 1993; Rupp and Porter, 2003; Bower et al., 2013, 2018). To make a DRC4 transgene that is tagged with mC, the DRC4-GFP construct (Bower et al., 2013) was digested with BstEII to release the GFP tag. We then amplified a Chlamydomonas codon optimized version of the mC tag (Rasala et al., 2013) using two primers containing BstEII sites (forward primer 5′-GGCTTCCGGCCACTGATGGTGAGCAAGGGCGAG-3′ and reverse primer 5′-CCGGTGCTGGTGTTGACCTTGTACAGCTCGTCCATGC-3′). The digested vector and PCR product were purified by agarose gels and assembled using NEBuilder (New England Biolabs, Ipswich, MA). The resulting DRC4-mC construct, encoding a DRC4 polypeptide with an mC tag located between amino acids 457 and 458 of the original DRC4 sequence, was verified by sequencing (Azenta, Chelmsford, MA). The plasmid was linearized with EcoRI and cotransformed with the selectable marker plasmid pSI103, encoding the AphVIII resistance gene (Sizova et al., 2001), into pf2-4. Transformants were selected by growth on solid TAP medium containing 10 µg/ml paromomycin, picked into a 96-well plate containing liquid TAP medium, and screened by phase contrast microscopy for rescue of the pf2 motility defect. Rescued strains were verified by Western blotting of isolated axonemes using anti-DRC4 (Bower et al., 2013) and polyclonal anti-mCherry (600-401-P16; Rockland Immunochemicals, Limerick, PA) and by fluorescence microscopy for the presence of an mC signal in flagella.
The strains generated in this study, that is, pf2-4 DRC4-mC (CC-5510), ida6-1 DRC2-GFP pf2-4 (CC-5514), ida6-1 pf2-4 DRC4-mC (CC-5515), and pf2-4 ida6-1 DRC2-GFP DRC4-mC (CC-5516 for mt− and CC-5517 for mt+), are available from the Chlamydomonas stock center (Supplemental Table S1; https://www.chlamycollection.org/). The cells were maintained in minimal (M) medium at ∼24°C with a light/dark cycle of 14:10 h. (https://www.chlamycollection.org/methods/media-recipes/minimal-or-m-medium-and-derivatives-sager-granick/). Large cultures of M medium (∼1 l) aerated with air supplemented with 0.5% CO2 were used to isolate flagella. Cells maintained in unaerated flasks were used for imaging and phenotypical analysis.
Swimming velocity and flagella length measurement
To measure swimming velocity, cells were resuspended in fresh M medium, placed in a chambered plastic slide (Fisherbrand; 14-377-259), and observed under an inverted light microscope (TMS; Nikon). The images were recorded using a MU500 camera (Amscope) with the associated Topview software at a fixed exposure time of 1 s. The length of the swimming trajectories was analyzed in ImageJ (National Institutes of Health).
To measure flagella length, observation chambers were constructed by applying a ring of vacuum grease or petroleum jelly to a 24 × 60 mm2 no. 1.5 coverslip, to which ∼15 μl of cell suspension was added and allowed to settle for ∼1 min. The chamber was sealed by placing a 22 × 22 mm2 no. 1.5 cover glass with approximately a 5–10 μl drop of 5 mM N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid) (HEPES), pH 7.3, supplemented with 3–5 mM ethylene glycol tetraacetic acid (EGTA) onto the larger cover glass to immobilize the cells. Live adhered cells were then imaged through the large cover glass at room temperature. Because the base of the flagella bends toward the cell body, the measured flagellar length is shorter than that of the actual flagella. For DIC imaging, we used an Eclipse Ti-U microscope (Nikon) and an Andor iXon EMCCD camera. Swimming velocity and flagella lengths were measured using FIJI (National Institutes of Health), and Prism (GraphPad) was used to generate violin plots.
Flagellar regeneration
Cells with regenerating flagella were obtained as follows: Cells in fresh M medium were deflagellated by a pH shock, sedimented, resuspended in a small volume of M medium, and stored on ice for 15 min or until needed. Then, cells were diluted with M medium at room temperature and allowed to regenerate flagella in bright light on a shaker at room temperature. Aliquots were mounted for TIRF microscopy at various time points.
High-speed video analysis
For high-speed video analysis at 1000 fps, we used an inverted Eclipse Ti2 microscope (Nikon) equipped with a long-distance DIC condenser and a 40× 0.95 PlanApo objective. Images were recorded using an EoSens 3CL camera (Mikrotron) and a CORE2 DVR Express rapid storage device (IO Industries). Cells were concentrated and placed in an observation chamber. Recordings were exported in AVI format and analyzed using ImageJ.
Flagellar isolation and Western blotting
For Western blot analyses, cells were concentrated by centrifugation, washed in 10 mM HEPES, resuspended in 10 mM HEPES, 5 mM MgSO4, 4% sucrose (wt/vol), and immediately deflagellated by the addition of dibucaine and vigorous pipetting. The cell bodies were removed by two differential centrifugations and flagella were sedimented from the supernatant by centrifugation at 40,000 × g, 20 min, 4°C as previously described (Witman, 1986). Flagella were dissolved in Laemmli SDS sample buffer, separated on Mini-Protean TGX gradient gels (BioRad), and transferred electrophoretically to PVDF membrane (Millipore). After blocking in TBS-T supplemented with bovine serum albumin, powdered dry milk, or fish gelatin (3–5% final concentration) for at least 30 min, the blots were incubated overnight in the rabbit polyclonal primary antibodies anti-DRC2 (1:100; Bower et al., 2018), anti-DRC4 (1:750; Bower et al., 2013), anti-GFP (1:2000; Invitrogen A11122), and anti-mCherry (1:1000; BioVision 5993), and mouse monoclonal anti-IC2 (1:4,000; King and Witman, 1990). Secondary antibodies (Invitrogen anti-rabbit and anti-mouse IgG conjugated to horseradish peroxidase) were applied for 90–120 min at room temperature with agitation. After several washes in TBS-T, substrate (Femtoglow by Michigan Diagnostics or ECL Prime Western Blotting Detection Reagent by GE Healthcare) was added, and the chemiluminescent signals were documented using a BioRad ChemiDoc imaging system, exported in tif format, adjusted for contrast and brightness in Photoshop, and assembled in Illustrator.
Mating experiments
Cells grown in M medium were pelleted and incubated overnight in M-N medium with aeration in constant light. The next morning, cells were transferred to 1/5th M-N medium supplemented with 10 mM HEPES and incubated for ∼2–4 h in bright light with agitation. For in vivo imaging, plus and minus gametes were mixed and imaged at different time points. To generate progeny, mixed gametes were incubated in light for 3–5 h without agitation, plated on 4% agar or 1.8% phytogel M-medium plates, air-dried, incubated overnight in constant light, wrapped in aluminum foil and incubated for at least 10 d in the dark, transferred to −20°C for 2 d, defrosted, dried, and incubated in constant light until colonies appeared. Individual colonies were streaked for single cells and clonal progeny colonies were transferred to 96-well plates for further analysis.
Live cell microscopy
The TIRF microscope has been described previously (Lechtreck, 2013, 2016). In brief, we used an Eclipse Ti-U microscope (Nikon) equipped with 60× NA 1.49 TIRF objective and through-the-objective TIRF illumination provided by a 40-mW, 488-nm and a 75-mW, 561-nm diode laser (Spectraphysics). The excitation lasers were cleaned up with a Nikon GFP/mCherry TIRF filter cube, and the emission was separated using an image splitting device (Photometrics DualView2 with filter cube 11-EM) supplemented with a meniscus lens to adjust for focus and an et595/33m filter to reduce chlorophyll autofluorescence. Cells were mixed 1:1 with 5 mM HEPES, pH 7.3, supplemented with 3–5 mM EGTA and placed in an observation chamber consisting of a 24 × 60 mm no. 1.5 and a 22 × 22 mm no. 1.5 cover glass separated by a ring of petroleum jelly. An iXON3 (Andor) and the NIS-Elements Advanced Research software (Nikon) were used to record images at 10 fps. FIJI (National Institutes of Health) was used to generate kymograms using the KymoResliceWide plugin (https://imagej.net/KymoResliceWide). Adobe Photoshop was used to adjust image contrast and brightness, and figures were prepared in Adobe Illustrator.
To analyze the cotransport probability, we assumed an IFT frequency of ∼60 trains/min as repeatedly reported (Dentler, 2005; Engel et al., 2012; Reck et al., 2016). The observed probability of a train carrying both tagged DRC4 and DRC2 subunits was ∼0.015 (i.e., 47 cotransports observed over a period of 3158 s or 1 in ∼67 trains carrying both proteins). This value is approximately four times higher than the theoretical value of 0.003 for the cotransport of DRC2-GFP and DRC4-mC subunits occurring by chance (i.e., observed probability of DRC4 transport [0.071] × observed probability of DRC2 transport [0.05] = expected probability of cotransport = 0.0034).
Supplementary Material
Acknowledgments
We thank Jared Rieck (University of Minnesota) for analyzing the swimming velocity of the pf2 DRC4-mC strain. This study was supported by grants by the National Institutes of Health (R01GM-110413 to K.L. and 5R01GM-055667 to M.P.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations used:
- DMT
doublet microtubule
- GFP
green fluorescent protein
- mC
mCherry
- N-DRC
nexin–dynein regulatory complex
- IFT
intraflagellar transport.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-11-0524) on January 4, 2023.
REFERENCES
- Ahmed NT, Gao C, Lucker BF, Cole DG, Mitchell DR (2008). ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J Cell Biol 183, 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin-Tse C, Halbritter J, Zariwala MA, Gilberti RM, Gee HY, Hellman N, Pathak N,, Liu Y, Panizzi JR, Patel-King RS, et al. (2013). Zebrafish ciliopathy screen plus human mutational analysis identifies C21orf59 and CCDC65 defects as causing primary ciliary dyskinesia. Am J Hum Genet 93, 672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awata J, Takada S, Standley C, Lechtreck KF, Bellve KD, Pazour GJ, Fogarty KE, Witman GB (2014). NPHP4 controls ciliary trafficking of membrane proteins and large soluble proteins at the transition zone. J Cell Sci 127, 4714–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower R, Tritschler D, Mills KV, Heuser T, Nicastro D, Porter ME (2018). DRC2/CCDC65 is a central hub for assembly of the nexin–dynein regulatory complex and other regulators of ciliary and flagellar motility. Mol Biol Cell 29, 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower R, Tritschler D, Vanderwaal K, Perrone CA, Mueller J, Fox L, Sale WS, Porter ME (2013). The N-DRC forms a conserved biochemical complex that maintains outer doublet alignment and limits microtubule sliding in motile axonemes. Mol Biol Cell 24, 1134–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw CJ, Kamiya R (1987). Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskeleton 8, 68–75. [DOI] [PubMed] [Google Scholar]
- Bui KH, Yagi T, Yamamoto R, Kamiya R, Ishikawa T (2012). Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J Cell Biol 198, 913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft JM, Harris JA, Hyman S, Kner P, Lechtreck KF (2015). Tubulin transport by IFT is upregulated during ciliary growth by a cilium-autonomous mechanism. J Cell Biol 208, 223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Barbieri F, Mitchell DR, Lechtreck KF (2018). In vivo analysis of outer arm dynein transport reveals cargo-specific intraflagellar transport properties. Mol Biol Cell 29, 2553–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentler W (2005). Intraflagellar transport (IFT) during assembly and disassembly of Chlamydomonas flagella J Cell Biol 170, 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener DR, Yang P, Geimer S, Cole DG, Sale WS, Rosenbaum JL (2011). Sequential assembly of flagellar radial spokes. Cytoskeleton (Hoboken) 68, 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel BD, Ishikawa H, Wemmer KA, Geimer S, Wakabayashi K, Hirono M, Craige B, Pazour GJ, Witman GB, Kamiya R, Marshall WF (2012). The role of retrograde intraflagellar transport in flagellar assembly, maintenance, and function. J Cell Biol 199, 151–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowkes ME, Mitchell DR (1998). The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol Biol Cell 9, 2337–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner LC, O’Toole E, Perrone CA, Giddings T, Porter ME (1994). Components of a “dynein regulatory complex” are located at the junction between the radial spokes and the dynein arms in Chlamydomonas flagella. J Cell Biol 127, 1311–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY (2000). The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA 97, 11990–11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M, Ma M, Sze-Tu E, Wang X, Koh F, Zhong ED, Berger B, Davis JH, Dutcher SK, Zhang R, Brown A (2021). Structures of radial spokes and associated complexes important for ciliary motility. Nat Struct Mol Biol 28, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui L, Song K, Tritschler D, Bower R, Yan S, Dai A, Augspurger K, Sakizadeh J, Grzemska M, Ni T, et al. (2019). Scaffold subunits support associated subunit assembly in the Chlamydomonas ciliary nexin-dynein regulatory complex. Proc Natl Acad Sci USA 116, 23152–23162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Dymek EE, Lin J, Smith EF, Nicastro D (2012). The CSC connects three major axonemal complexes involved in dynein regulation. Mol Biol Cell 23, 3143–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D (2009). The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol 187, 921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Ramanis Z, Luck DJ (1982). Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell 28, 115–124. [DOI] [PubMed] [Google Scholar]
- Hunter EL, Lechtreck K, Fu G, Hwang J, Lin H, Gokhale A, Alford LM, Lewis B, Yamamoto R, Kamiya R, et al. (2018). The IDA3 adapter, required for intraflagellar transport of I1 dynein, is regulated by ciliary length. Mol Biol Cell 29, 886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Kagami O, Yagi T, Kamiya R (1993). Isolation of two species of Chlamydomonasreinhardtii flagellar mutants, ida5 and ida6, that lack a newly identified heavy chain of the inner dynein arm. Cell Struct Funct 18, 371–377. [DOI] [PubMed] [Google Scholar]
- King SM, Witman GB (1990). Localization of an intermediate chain of outer arm dynein by immunoelectron microscopy. J Biol Chem 265, 19807–19811. [PubMed] [Google Scholar]
- Koblenz B, Schoppmeier J, Grunow A, Lechtreck KF (2003). Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation and maturation. J Cell Sci 116, 2635–2646. [DOI] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL (1993). A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA 90, 5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Brown JM, Bellve K, Craige B, Craft JM, Fogarty K, Lechtreck KF, Witman GB (2016). The IFT81 and IFT74 N-termini together form the major module for intraflagellar transport of tubulin. J Cell Sci 129, 2106–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Oda T (2017). Electrostatic interaction between polyglutamylated tubulin and the nexin-dynein regulatory complex regulates flagellar motility. Mol Biol Cell 28, 2260–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF (2013). In vivo Imaging of IFT in Chlamydomonas flagella. Methods Enzymol 524, 265–284. [DOI] [PubMed] [Google Scholar]
- Lechtreck KF (2015). IFT-cargo interactions and protein transport in cilia. Trends Biochem Sci 40, 765–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF (2016). Methods for studying movement of molecules within cilia. Methods Mol Biol 1454, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Geimer S (2000). Distribution of polyglutamylated tubulin in the flagellar apparatus of green flagellates. Cell Motil Cytoskeleton 47, 219–235. [DOI] [PubMed] [Google Scholar]
- Lechtreck KF, Liu Y, Dai J, Alkhofash RA, Butler J, Alford L, Yang P (2022). Chlamydomonas ARMC2/PF27 is an obligate cargo adapter for intraflagellar transport of radial spokes. Elife 11, e74993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Mengoni I, Okivie B, Hilderhoff KB (2018). In vivo analyses of radial spoke transport, assembly, repair and maintenance. Cytoskeleton (Hoboken) 75, 352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis WR, Malarkey EB, Tritschler D, Bower R, Pasek RC, Porath JD, Birket SE, Saunier S, Antignac C, Knowles MR, et al. (2016). Mutation of growth arrest specific 8 reveals a role in motile cilia function and human disease. PLoS Genet 12, e1006220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Tritschler D, Song K, Barber CF, Cobb JS, Porter ME, Nicastro D (2011). Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella. J Biol Chem 286, 29175–29191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesland DA, Hoffman JL, Caligor E, Goodenough UW (1980). Flagellar tip activation stimulated by membrane adhesions in Chlamydomonas gametes. J Cell Biol 84, 599–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Kamiya R, Kikkawa M (2014). Cilia and flagella. A molecular ruler determines the repeat length in eukaryotic cilia and flagella. Science 346, 857–860. [DOI] [PubMed] [Google Scholar]
- Olbrich H, Cremers C, Loges NT, Werner C, Nielsen KG, Marthin JK, Philipsen M, Wallmeier J, Pennekamp P, Menchen T, et al. (2015). Loss-of-function GAS8 mutations cause primary ciliary dyskinesia and disrupt the nexin–dynein regulatory complex. Am J Hum Genet 97, 546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Mead K, LeDizet M, Moscatelli A (1994). Mutations in the “dynein regulatory complex” alter the ATP-insensitive binding sites for inner arm dyneins in Chlamydomonas axonemes. J Cell Biol 125, 1109–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Mead K, Shestak W (1992). The inner dynein arms I2 interact with a “dynein regulatory complex” in Chlamydomonas flagella. J Cell Biol 118, 1455–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Burnette DT, Bae YK, Forscher P, Barr MM, Rosenbaum JL (2005). Intraflagellar transport is required for the vectorial movement of TRPV channels in the ciliary membrane. Curr Biol 15, 1695–1699. [DOI] [PubMed] [Google Scholar]
- Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL (2004). Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J Cell Biol 164, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Barrera DJ, Ng J, Plucinak TM, Rosenberg JN, Weeks DP, Oyler GA, Peterson TC, Haerizadeh F, Mayfield SP (2013). Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonasreinhardtii. Plant J 74, 545–556. [DOI] [PubMed] [Google Scholar]
- Reck J, Schauer AM, VanderWaal Mills K, Bower R, Tritschler D, Perrone CA, Porter ME (2016). The role of the dynein light intermediate chain in retrograde IFT and flagellar function in Chlamydomonas. Mol Biol Cell 27, 2404–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp G, Porter ME (2003). A subunit of the dynein regulatory complex in Chlamydomonas is a homologue of a growth arrest-specific gene product. J Cell Biol 162, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M (2019). Good news for nuclear transgene expression in Chlamydomonas. Cells 8, 1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova I, Fuhrmann M, Hegemann P (2001). A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonasreinhardtii. Gene 277, 221–229. [DOI] [PubMed] [Google Scholar]
- Song K, Awata J, Tritschler D, Bower R, Witman GB, Porter ME, Nicastro D (2015). In situ localization of N and C termini of subunits of the flagellar nexin-dynein regulatory complex (N-DRC) using SNAP tag and cryo-electron tomography. J Biol Chem 290, 5341–5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Weghe JC, Harris JA, Kubo T, Witman GB, Lechtreck KF (2020). Diffusion rather than IFT likely provides most of the tubulin required for axonemal assembly. J Cell Sci 133, jcs249805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CL, McIntyre JC, Norris SR, Jenkins PM, Zhang L, Pei Q, Verhey K, Martens JR (2014). Direct evidence for BBSome-associated intraflagellar transport reveals distinct properties of native mammalian cilia. Nat Commun 5, 5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M, Olbrich H, Werner C, Tritschler D, Bower R, Sale WS, Loges NT, Pennekamp P, Lindberg S, Stenram U, et al. (2013). The nexin-dynein regulatory complex subunit DRC1 is essential for motile cilia function in algae and humans. Nat Genet 45, 262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman GB (1986). Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol 134, 280–290. [DOI] [PubMed] [Google Scholar]
- Wren KN, Craft JM, Tritschler D, Schauer A, Patel DK, Smith EF, Porter ME, Kner P, Lechtreck KF (2013). A differential cargo-loading model of ciliary length regulation by IFT. Curr Biol 23, 2463–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa HA, Kamiya R (2004). A tektin homologue is decreased in Chlamydomonas mutants lacking an axonemal inner-arm dynein. Mol Biol Cell 15, 2105–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Nager AR, Nachury MV (2018). BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J Cell Biol 217, 1847–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.