Abstract
Background
GLI-similar 1 (GLIS1) is one of of Krüppel-like zinc finger proteins, which are either stimulators or inhibitors of genetic transcription. Nevertheless, its effects on T cell were elusive.
Methods
In this study, we intend to explore the effects of GLIS1 on modulating the anticancer potency of CD8+ T cells in hepatocellular carcinoma (HCC). The expression of GLIS1 in CD8 peripheral blood mononuclear cell and CD8 tumor-infiltrating lymphocytes of HCC tissues was validated by quantificational real-time-PCR and flow cytometry. The anticancer potency of CD8+ T cells with GLIS1 knock down was confirmed in C57BL/6 mouse model and HCC patient-derived xenograft mice model. GLIS1–/– C57BL/6 mice was applied to explore the effects GLIS1 on tumor immune microenvironment. Chromatin immunoprecipitation and RNA transcriptome sequencing analysis were both performed in GLIS1-knock down of CD8+ T cells.
Results
GLIS1 was upregulated in exhausted CD8+ T cells in HCC. GLIS1 downregulation in CD8+ T cells repressed cancer development, elevated the infiltrate ability of CD8+ T cells, mitigated CD8+ T cell exhaustion and ameliorated the anti-PD1 reaction of CD8+ T cells in HCC. The causal link beneath this included transcriptional regulation of SGK1-STAT3-PD1 pathway by GLIS1, thereby maintaining the abundant PD1 expression on the surface of CD8+ T cells.
Conclusion
Our study revealed that GLIS1 promoted CD8+ T cell exhaustion in HCC through transcriptional regulating SGK1-STAT3-PD1 pathway. Downregulating the expression of GLIS1 in CD8+ T cells exerted an effect with anti-PD1 treatment synergistically, revealing a prospective method for HCC immune therapy.
Keywords: biomarkers, tumor; drug therapy, combination; immunity, cellular
WHAT IS ALREADY KNOWN ON THIS TOPIC
GLIS1 is one of of Krüppel-like zinc finger proteins which are either stimulators or inhibitors of genetic transcription.
WHAT THIS STUDY ADDS
This study revealed that GLIS1 promoted CD8+ T cell exhaustion in HCC through transcriptional regulating SGK1-STAT3-PD1 pathway.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Down-regulating the expression of GLIS1 in CD8+ T cells exerted an effect with anti-PD1 treatment synergistically, revealing a prospective method for HCC immune therapy.
Introduction
Liver cancer is a primary cause of tumor-associated mortality across the globe.1–3 Hepatocellular carcinoma (HCC) makes up most of primary liver cancer. HCC mainly impacts males, comprizing 75%–85% of primary liver cancer and ranking as the 5th most commonly seen malignant cancer.1 2 HCC usually derives from cirrhosis and is tightly associated with persistent hepatic illnesses.2 4 Virus hepatitis especially hepatitis B virus (HBV) in China and overmuch alcohol consumptions are primary risky factors for HCC. Surgical resection and liver transplant may cure early HCC; nevertheless, the majority of HCC sufferers exhibit non-resectable illness and in general, they respond unsatisfactorily to the existing therapeutic regimens.1 2 Even with the development of tumor therapies and elevated survival, HCC is among the few tumors with an increasing death rate.
Cancer cells are capable of evading immune surveillance, facilitating cancer development via activating diverse immuniey checkpoint signal paths. Monoclone antibody that target immunity checkpoints have witnessed a huge break through in tumor treatment, promoting the immunity-mediated eradication of oncocytes. Among those, programmed death-ligand 1(PD-L1)/PD1 and cytotoxic T-lymphocyte antigen (CTLA)-4 suppressors have already been used in some malignant cancers, but molecules that are capable of disrupting other cosuppressive signal paths are under exploration, like T cell immunoglobulins and ITIM domain proteins (TIGIT), lymphocyte activation gene-3 (LAG3), and T cell immunoglobulin containing the mucin domain 3 (TIM-3). In the era of immune therapy, immune checkpoint inhibitors (ICIs) have been examined for HCC sufferers as well, especially, nivolumab and pembrolizumab have been accepted for secondline treatment.5 Nevertheless, resembling other gastric intestinal malignant cancers,6 the HCC responsive rate of ICIs as a mono therapy is low, hence novel combined methods involving tyrosine kinase inhibitors, the supplement of diverse ICIs, antiangiogenic treatment, local treatment, other kinase suppressors, chemo, and other medicines are being investigated intensively at present. However, what causes HCC immunoescape is worth investigating.
Here, we found that GLIS1, a component of the Krüppel-like zinc finger transcription factor (TF) family,7 acted as an important regulator of the antitumor activity of CD8+ T cells in HCC. GLIS1 is vital for regulating various physiology process and has participated in various pathologies, such as neonatal diabetic diseases, glaucoma, cystic renal illness, neurology disorder, congenital hypothyrosis, and tumor.8–10 Nevertheless, the effects of GLIS1 on anticancer immunity remain elusive. This research revealed that GLIS1 was regulated upward in the process of CD8+ T cell exhaustion and exerted a vital effect on the establishment of the dysfunction status of CD8+ T cells in HCC.
Materials and methods
Geo data analysis
We downloaded data from GEO database (GSE84820 and GSE155192) and studied the difference of GLIS1 mRNA expressing in CD8+ T cells after 8 days and 30 days chronic infection and acute infection. Single-cell RNA-sequence data including HBV positive patients and HBV negative HCC patient were analyzed from GSE140228. Chromatin immunoprecipitation (CHIP) data of mouse STAT3-PD1 binding sites were obtained from GSE21669 and CHIP data of human STAT3-PD1 binding sites were obtained from GSE67183.
Samples collection
The research was completed strictly as per the Declaration of Helsinki and relevant government research requirements. Fresh cancer specimens and peripheral blood from HCC patients were harvested from Hepatobiliary Center, the First Affiliated Hospital of Nanjing Medical University. The tumor tissues were excised intraoperatively and were confirmed as HCC by pathology.
Acquisition and culture of CD8+ tumor-infiltrating lymphocytes from HCC patients
CD8+ tumor infiltrating lymphocytes (TILs) were extracted from the fresh cancer tissues of HCC patients (HBV infection) immediately. The tissue was cleaned twice with RPMI 1640, and connective tissue and necrotic tissue on the tumor tissue were removed with scissors. The tumor tissue was cut into 1–2 mm pieces with scissors, and the pieces were transferred into a 15 mL centrifuge tube. The DNA enzyme with final concentration of 30 µg/ mL, 0.1 mg/ mL hyaluronidase and 1 mg/ mL collagenase were added, and the samples were shaken slightly at room temperature (RT) for 2 hours. The digested tumor tissue was prepared into a single cell suspension using a 70 µm cell screen. Then, 10 mL ficoll-Paque (Sigma-Aldrich, USA) upper layer was added at 400 g rotation speed for 30 min at 20℃. Posterior to centrifugal treatment, the cloud lymph cell tier in the middle was separated by CD8+magnet beads(Miltenyi Biotec, Germany). CD8+ T cells were selected and cultured in vitro with 1 µg/mL CD3 mAb (Miltenyi Biotec, Germany), 5 µg/mL CD28 mAb (Miltenyi Biotec, Germany), 20 ng/mL recombination human IL-2 (PeproTech, USA).
Acquisition of peripheral blood mononuclear cell from HCC patients
A 10 mL ficoll-Paque (Sigma-Aldrich, USA) upper layer was added into the blood at 400 g rotation speed for 30 min at 20℃. Posterior to centrifugal treatment, the cloud lymph cell tier in the middle was separated by CD8+ magnet beads(Miltenyi Biotec, Germany).
RNA extraction and quantificational real-time PCR
Overall RNA was separated from samples and cells via TRIzol reagent (Invitrogen, USA) as per the supplier’s specification. Through reverse transcription kit (Takara, Japan), we reverse transcribed RNA into cDNA. Every primer sequence is presented by online supplemental table S1. GAPDH was used to standardize mRNA expressing levels.
jitc-2022-005126supp001.pdf (9.7MB, pdf)
Cell transfection
Day 1: CD8+ TILs were sorted out and cells were spread. A 300 μL of suspension cell specific lentivirus (sh-NC, sh-GLIS1 from Genechem, China) was added over the course of 3 days. Day 2: The virus was implanted, part of the supernatant was sucked out, then 300 μL of complete medium was added, and the virus was added until the well was full, 8–10 hours later, a new batch of virus and complete medium was replaced. Day 3: After 8–10 hours, the virus was replaced again. Day 4: A new 24-well plate was used to avoid long-term irritation to the cells. Day 7: Infection efficiency was measured.
Flow cytometry detection and analysis
The total amount and level of CD8+ T cells were computed by cellular counter technique. For cellular surface dyeing, cells were resuspended with PBS containing 2% serum, added with flow cytometry detection antibodies against PD1, TIM-3, LAG-3, CTLA-4 (1:100; BioLegend, USA) incubated on ice for 30 min, then cleaned in 2% PBS for two times, and then flow identification was performed.
For intracell dyeing, cells were first treated with immobilization, membrane rupture and perforation, and incubated with antibodies against IL-2, IFN-γ, TNF-α (1:100; eBioscience, USA) for 30 min at 4℃, and then detected by flow cytometry. For cells that need to be tested for cytokines, Phorbol Myristate acetate (Enzo, USA) was isolated from T cells at 37℃.1 µg/mL ionomycin (Enzo, USA) and 10 µg/mL Brefeldin A (Enzo, USA) were stimulated for 6 hours, and then corresponding staining treatments were given. Dead cells were marked with Fixable Dye 620 (BD Biosciences, USA) reagent and removed in the process flow cell technique and were not included in the analysis. FACS Aria II Cell Sorter (BD Biosciences, USA) was used for flow cell technique and FlowJo program (TreeStar, USA) was used to analyze the data. For the grouping of GLIS1 expression, we sorted its expression from low to high, and then divided it into two halves according to the median value, one half was a relatively high expression group, and half was a relatively low expression group.
Cell proliferation assay
CD8+ T cells were collected, removed from supernatant and added with 37 ℃ CFSE (carboxyfluorescein diacetate, Thermo Fisher, USA) working solution for incubation for 15 min. Serum was added to stop the reaction. PBS cleaned the CFSE molecules adhered to the surface. The complete medium was resuspended and inoculated in petri dishes. After 3 days, CD8+ T cells were removed, centrifuged, cleaned and rehung for detection.
Cell apoptosis assay
CD8+ T cells received the staining process by using FITC Apoptotic Identification Kit (BD Biosciences, USA). We employed FACScan (BD Biosciences, USA) for investigating cells stained. Furthermore, with the use of Flowjo V.10 software, a study was conducted on the apoptosis information pertaining to various cell lines (Tree Star, USA).
Western blot
Proteins were abstracted from cells via RIPA buffering solution, dissolved by SDS-PAGE and afterwards moved onto PVDF films. The first antisubstances (Thermo Fisher, USA) against GLIS1, SGK1, STAT3, p-STAT3, PD1 were used. Second antisubstance conjugated with peroxidase (CST) was used and the visualization of the antigen-antisubstance reactions were realized via enhanced chemiluminescence assay (ECL) (Thermo Fisher, USA).
RNA sequencing
Overall RNA was separated from CD8+T cells treated with sh-GLIS1 or sh-NC. Library establishment and sequence identification were completed at Beijing Novogene Company. The initial imaging data files acquired via high-flux sequence identification (illumina HiSeqTM2500) are transformed to sequenced reads via base calling analyses, and then mRNA sequencing are performed. KEGG pathway annotating and enriching assays were finished via DAVID. Function annotations were completed on the foundation of the GO data base. A Fisher’s exact test was used to screen important classifications, and the GO terms with calculated Q≤0.05 had significance.
CHIP sequencing
Cells were fixed with formaldehyde and DNA was cross-linked to proteins; cells were lysed, chromatin was separated, and chromatin was randomly cleaved by supersound or enzymetic treatment. Then, the specific recognition response of antigen antisubstance was used to realize the precipitation of the DNA fragments binding to the targeted protein GLIS1. DNA fragments were released and extracted by cross-linking. Library construction and high-throughput sequencing. Enough broken chromatin remains to be used directly for subsequent DNA extraction as a control, known as input. The effect of CHIP was verified by qPCR. After the library was constructed, quality inspection was carried out. After passing the quality inspection, subsequent machine sequencing was carried out.
Immunofluorescent and immunohistochemical assays
We sectioned the specimens embedded in paraffin to 4 mm slices. Antigen repair was completed via pressure cooking for 180 s in 0.01 mol/L citrate buffering solution (pH 6.0; Generalbiol, China). Immunofluorescent cells were subjected to fixation in 4% PFA (Paraformaldehyde, Generalbiol, China) for 20 min under RT and subjected to permeabilization via 0.05% Triton X-100 (Sigma-Aldrich, USA) in PBS for 600 s. The specimens were subjected to blockade in PBS containing 2% BSA for 60 min under RT and cultivated nightlong with antisubstances specific for GLIS1, CD8, PD1, SGK1, p-STAT3 (Thermo Fisher, USA) at 4°C, and subsequently with Alexa Fluor or HRP-conjugated second antisubstances (Abcam, UK) for 60 min under RT. Hoechst 33 342 (Sigma-Aldrich, USA) was used to counterstain the nucleus, and pictures were captured via laser scanning confocal microscopy (LSCM) (Zeiss, Germany).
For immunohistochemical assay, the samples were cultivated nightlong with antisubstances specific for Ki-67, CD8 (Abcam, UK) under 4℃, and immune detection was completed the nest day via 3’-diaminobenzidine (Generalbiol, China), as per the supplier’s specification.
Luciferase reporter assay
The pGL3‐SGK1 was constructed by inserting the human SGK1 promoter into the vector of pGL3‐basic (Genechem, China). To determine the minimal SGK1 promoter sequence required for constitutive and inducible activity, we constructed a series of promoter deletion fragments by qPCR. The plasmids of pGL3‐SGK1 mutant (AGATCTCAGTGATACT) were generated with the above-mentioned wild-type pGL3‐SGK1(AGATCTCTCACTATGT) as the template using a site‐directed mutagenesis kit (Genechem, China), according to the manufacturer’s instructions. The luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega, USA).
CHIP-qPCR assay
We performed the CHIP assay using the EZ-Magna CHIP Kit (Abcam, UK), according to the manufacturer’s instructions. In brief, we fixed CD8 T cells in 1% formaldehyde for 10 min at ambient temperature. The fixed cells were harvested, lysed, and sonicated for 10 cycles of 10 s ON/20 s OFF and 50% AMPL using Sonics VCX130 (Sonics & Materials, USA). Antibodies against GLIS1 and rabbit IgG (Thermo Fisher, USA) were used for immunoprecipitation. PCR amplification of the precipitated DNA was performed.
Mice model
The animal welfare board of our university accepted our animal tests (2018–672), and the entire experiment procedures and animal caring conformed to the institutional ethical guidelines for animals-associated experiment process.
Subcutaneous tumorbearing mouse model: 1×106 Hepa1-6 cells were inoculated into right groin of C57BL/6 mice. The groups were GLIS1+/+, GLIS1+/-, GLIS1-/- (n=5 for the respective group). GLIS1+/- and GLIS1-/- mice was purchased from biocytogen (China).
The tumor growth was observed. When the tumor grew to 2 cm in diameter, the mice were killed and the tumor tissue was taken out for weighing and further analysis. In addition, carcinoma transplanted tumor model mice fell to two groups, GLIS1-/- +PBS group and GLIS1-/-+αPD1 group (αPD1, bioxcell, USA). To be specific, GLIS1-/-+αPD1 group received 6.6 mg/kg intraperitoneal injection on the seventh day, and once per 7 days thereafter.
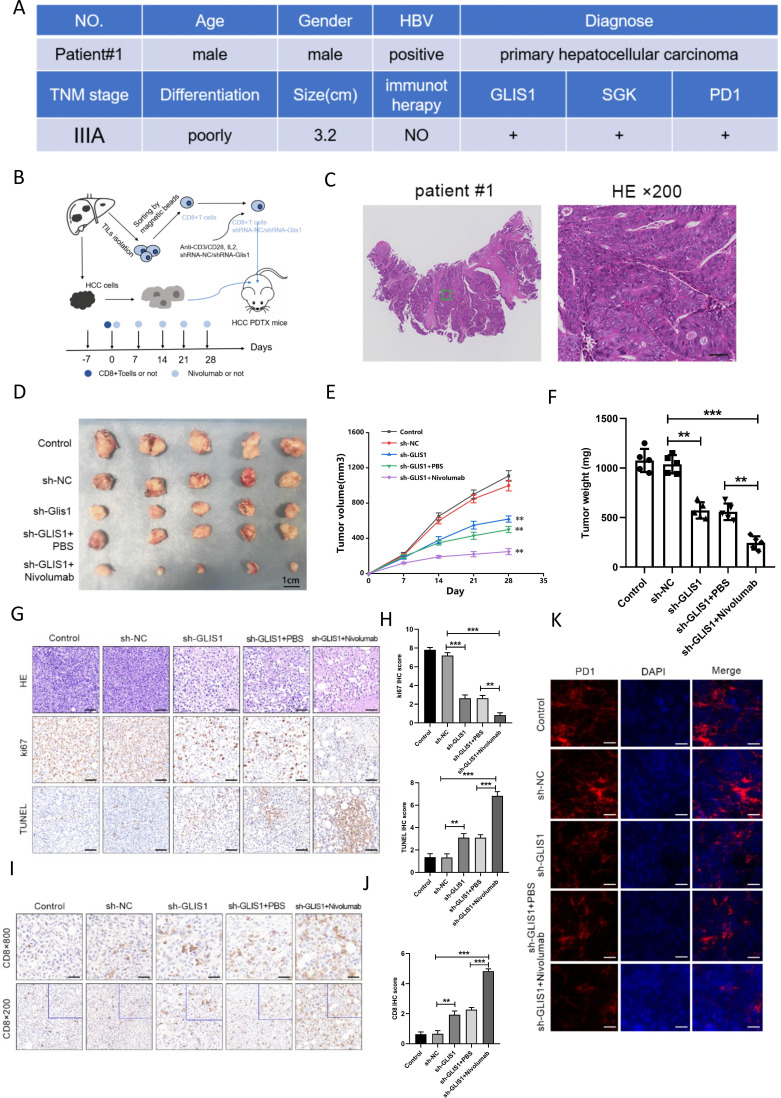
Patient-derived xenograft (PDX) mouse model: Fresh tissues from patients were collected to prepare single-cell suspension, and 2×106 HCC cells were obtained, mixed with matrix gel for 100 uL, and subcutaneously inoculated on the right spine of NOD/SCID mice. Tumor-infiltration CD8+ T cells separated from corresponding sufferers (infected with interfering sh-GLIS1 lentivirus or sh-NC lentivirus) were injected into mice by tail vein at a rate of 2×106 per mouse. The groups were sh-NC, sh-GLIS1, sh-GLIS1+PBS, sh-GLIS1+αPD1(Nivolumab, MCE, USA) (n=5 for the respective group). αPD1 group received 6.6 mg/kg intraperitoneal injection on the first day, and once per 7 days thereafter. On day 28, the animals were killed and their cancers were collected to verify the protein expression.
Mass spectrometry
Tissue samples were obtained from samples of each group (GLIS1+/+, GLIS1+/-, GLIS1-/-) (treated with Hepa1-6 cells), respectively. The mouse tissue processing method referred to Miltenyi Mouse Tumor Dissociation Kit, Percoll removes impurities and split red. We totally got 3×106 cells. CyTOF staining steps consisted of 194Pt staining → Fc block → surface antibody staining → overnight DNA staining (191/193Ir) → intracellular antibody staining → collecting data on the computer. The data analysis steps comprised A. FlowJo pretreatment: circle and select single, live and complete CD45+ immune cells; B. Bioinformation analysis: X-shift algorithm performs cell subpopulation clustering, manual annotation, TSNE dimensionality reduction visual display, and statistical analysis. This experiment was performed in Puluopting Company (China).
Statistical analysis
This study carried out the analyzing processes largely based on Graphpad Prism V.8.0, and p<0.05 was reported with statistics-related significance. The study drew the comparing process of successive information on the foundation of a separate t-experiment in these groups, and categorically associated information received the analysis through the χ2 testing process.
Results
GLIS1 was upregulated in exhausted CD8+ T cells based on GEO database
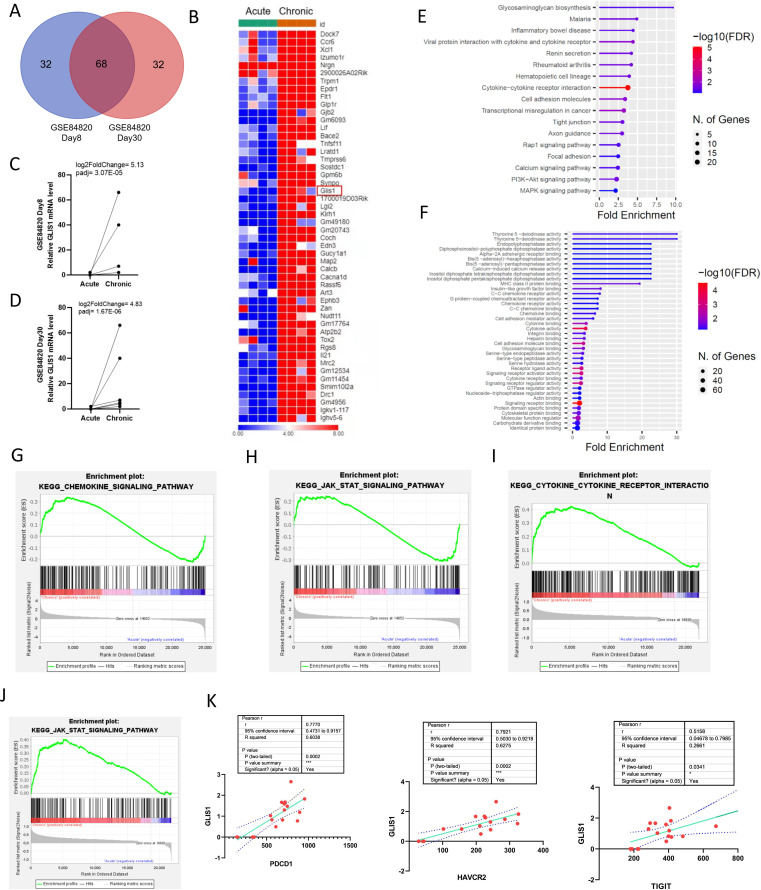
It is well known that chronic stimulation is one of the most important factors to induce T cell exhaustion in cancers. We downloaded two data from GEO database and studied the differential genes at day 8 and day 30 of acute and chronic viral infection respectively from GSE84820 data (online supplemental figure S1A, B). We cross-analyzed the results of the two data to search for common genes and the results showed that GLIS1 mRNA expression was higher in CD8+ T cells after persistent infections compared with acute infections at both day 8 and day 30 (figure 1A–D). Further GO, KEGG, and GESA analysis revealed that differential genes in chronic infections were closely related to cytokine secretion, T cell immunity, and the JAK/STAT signaling (figure 1E–J). Next, we analyzed the correlation between common markers of CD8+ T cell exhaustion (TIGIT, PDCD1, HAVCR2) and GLIS1 and found that the three markers were significantly positively correlated with GLIS1 expression in chronic infection based on GSE155192 and GSE84820 data (figure 1K). We also analyzed smart-seq based single cell RNA-sequence data including HBV positive patients and HBV negative patient in GSE140228 and the expression level of GLIS1 in exhausted CD8+ T cells has been compared between HCC tumor tissue and HBV positive normal tissue of HCC. The result showed that there was no difference in the expression level of GLIS1 between HCC tumor tissue and HBV positive normal tissue and GLIS1 was expressed in CD8+ T exhaustion cells in both HBV-positive and HBV non-positive patients (online supplemental figure S2). These outcomes revealed that GLIS1 might be associated with CD8+ T cell exhaustion in HCC. However, based on the fact that HCC patients in China are basically suffering from HBV, samples of HBV-infected HCC patients were selected in the following experiments.
Figure 1.
GLIS1 was upregulated in exhausted CD8+ T cells based on GEO database. (A) We cross-analyzed the differential genes at day 8 and day 30 of acute and chronic viral infection from GSE84820 data to find shared common genes. (B) Heatmap concretely showing shared common genes including GLIS1. (C, D) GLIS1 expression at day 8 and day 30 of acute and chronic viral infection respectively based on GSE84820 data.(E) KEGG pathway enrichment results based on GSE84820 data analysis. (F) GO analysis of GSE84820. (G, H) GSEA pathway enrichment results based on GSE155192 data analysis. (I, J) GSEA pathway enrichment results based on GSE84820 data analysis. (K) The relationship between GLIS1 and CD8 T cell exhausted markers expression based on based on GSE155192 and GSE 84820 data.
The GLIS1 expression in PBMC CD8+ T cells of HCC sufferers predicted the immune status in tissues
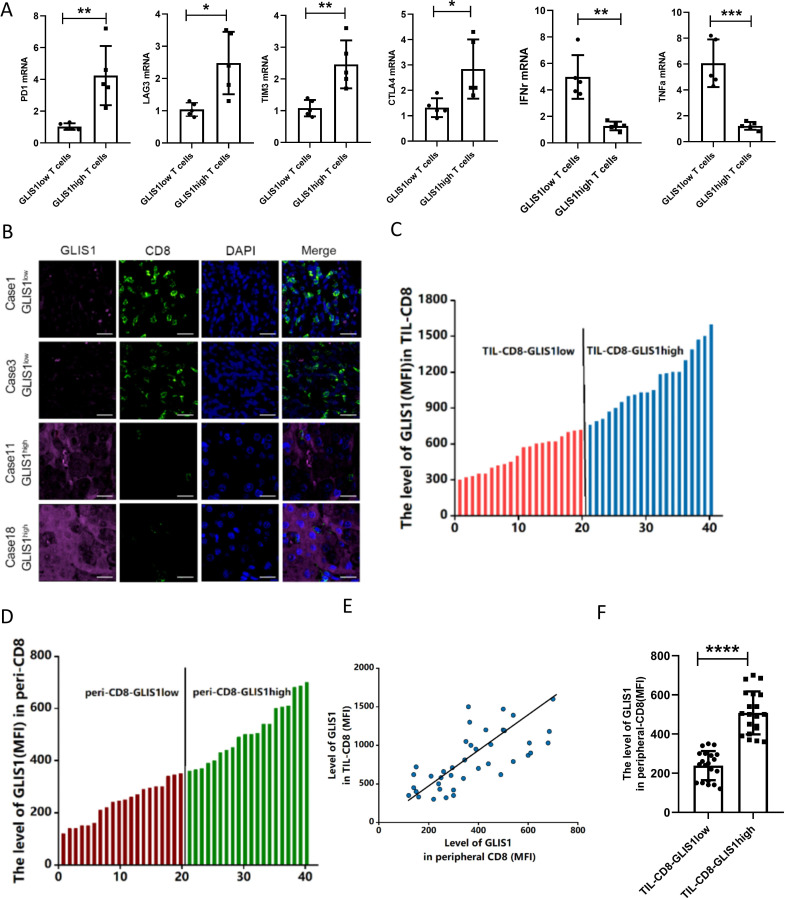
We analyzed GLIS1 expression in CD8+ TILs from 10 HCC patients (HBV infection) tissues by flow cytometry, and divided GLIS1 expressing into high GLIS1 expressing(n=5) and low GLIS1 expressing(n=5) groups according to the median value. The quantificational real-time PCR (qRT-PCR) outcomes revealed that the common markers of CD8+ T cell exhaustion (PD1, LAG3, TIM3 and CTLA4) were higher expressed in the GLIS1 high expression group and cytokines (IFN-γ, TNF-α) were lower expressed compared with the low GLIS1 expression group(figure 2A). After that, our team analyzed the expressing of GLIS1 in CD8+ TILs and corresponding peripheral blood mononuclear cell (PBMC) CD8+ T cells of 40 HCC (HBV infection) sufferers by flow cell technique, respectively and ranked them from low to high (figure 2C, D). The correlation analysis between the expression in CD8+ TILs and PBMC CD8+ T cells showed that they were correlated in a positive way (figure 2E). Patients with low GLIS1 expression in CD8+ TILs of cancer tissues also had relatively low GLIS1 expression in PBMC CD8+ T cells (figure 2F). According to the clinicopathological features, high expression of GLIS1 in PBMC was correlated with tumor size, TNM phase and vascular aggression in a positive way, whereas it was not significantly correlated with age, gender, and differentiation (online supplemental table S2). We selected samples from HCC patients CD8+ TILs for immunofluorescence detection and found that the high GLIS1 expression group had less CD8 fluorescence staining and more GLIS1 fluorescence staining, while the low GLIS1 expression group presented the contrary outcomes (figure 2B). Those studies revealed that GLIS1 expressing in peripheral blood CD8+ T cells of patients with HCC might be used to reflect the histological characteristics of patients’ tumors and the immune status in tissues.
Figure 2.
The expression of GLIS1 in PBMC CD8+ T cells of HCC patients predicted the immune status in tissues. (A) GLIS1 expression in CD8+ TILs from 10 HCC patients tissues was analyzed by flow cytometry, and were divided into high GLIS1 expression and low GLIS1 expression groups. The qRT-PCR results of the common markers of CD8+ T cell exhaustion (PD1, LAG3, TIM3 and CTLA4) and cytokines (IFN-γ, TNF-α) expression in each group. (B) Samples from HCC patients CD8+ TILs were used for immunofluorescence detection of CD8 and GLIS1 expression in each group. (C, D) The expression of GLIS1 in CD8+ TILs and corresponding peripheral blood CD8+ T cells of 40 HCC patients were analyzed by flow cytometry, respectively, and ranked them from low to high. (E, F) The correlation analysis between the GLIS1 expression in CD8+ TILs and PBMC CD8+ T cells. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. HCC, hepatocellular carcinoma; PBMC, peripheral blood mononuclear cell; qRT-PCR, quantificational real-time PCR; TILs, tumor infiltrating lymphocytes.
GLIS1 knockdown alleviated CD8+ T cell exhaustion in vitro
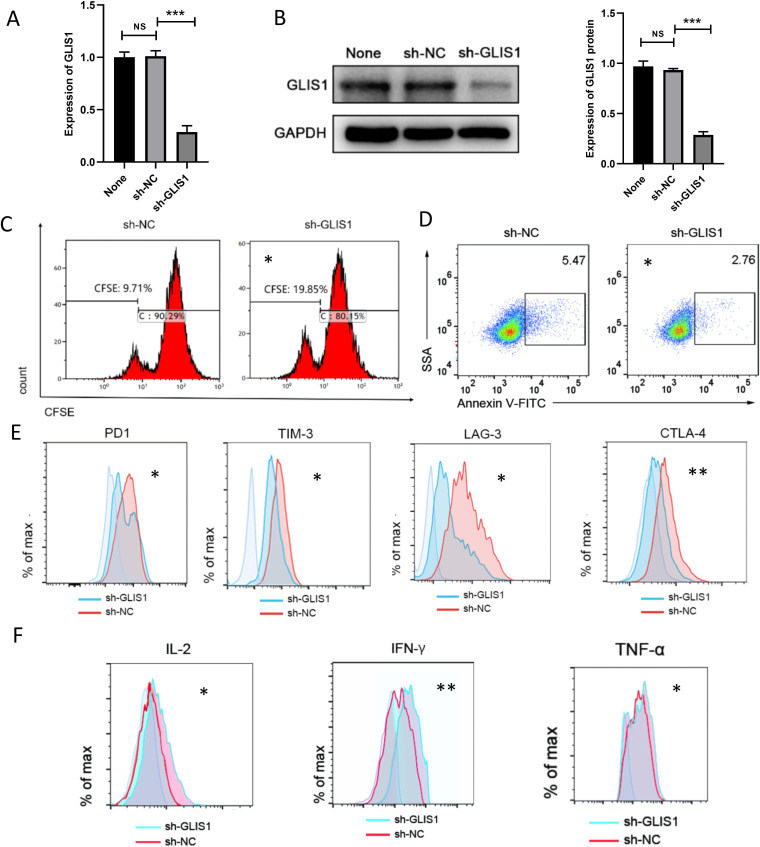
We developed shRNA targeting GLIS1 (sh-GLIS1) to silence GLIS1 in CD8+ TILs from 3 HCC tissues, and confirmed the success of this model by qRT-PCR and western blot (figure 3A, B). Downregulation of GLIS1 was capable of enhancing CD8+ T cell proliferation ability through CFSE experiment (figure 3C). Flow cell technique outcomes revealed that apoptosis of CD8+ T cells was reduced after GLIS1 reduction (figure 3D). As revealed from flow cytometry, results showed that when GLIS1was downregulated in CD8+ TILs, the expressions of common exhausted markers of CD8+ T cells (eg, PD1, TIM-3, LAG3, CTLA4) noticeably decreased, while the expression of cell factors (IL2, IFN-γ, TNF-α) increased significantly (figure 3E, F). Those outcomes revealed that GLIS1 knockdown alleviated CD8+ T cell exhaustion in HCC.
Figure 3.
GLIS1 knockdown alleviated CD8+ T cell exhaustion in vitro. (A, B) shRNA against GLIS1 was designed to silence GLIS1 level in CD8+ T cells, which was validated by qRT-PCR (A) and western blotting(B). (C) CFSE experiment showing CD8+ T cell proliferation ability in sh-NC and sh-GLIS1 group, respectively. (D) Flow cytometry was used to assess CD8+ T cell apoptosis in sh-NC and sh-GLIS1 group. (E) We measured the expressions of common exhausted markers of CD8+ T cells (PD1, TIM3, LAG3, CTLA4) by flow cytometry. (F) We measured the expressions of cytokines of CD8+ T cells (IL-2, IFN-γ, TNF-α) by flow cytometry. *p<0.05, **p<0.01, ***p<0.001. NS indicates no significant difference. CFSE, carboxyfluorescein diacetate; qRT-PCR, quantificational real-time PCR.
GLIS1-/- mice attenuated HCC growth and reinforced the anticancer ability of CD8+ T cells
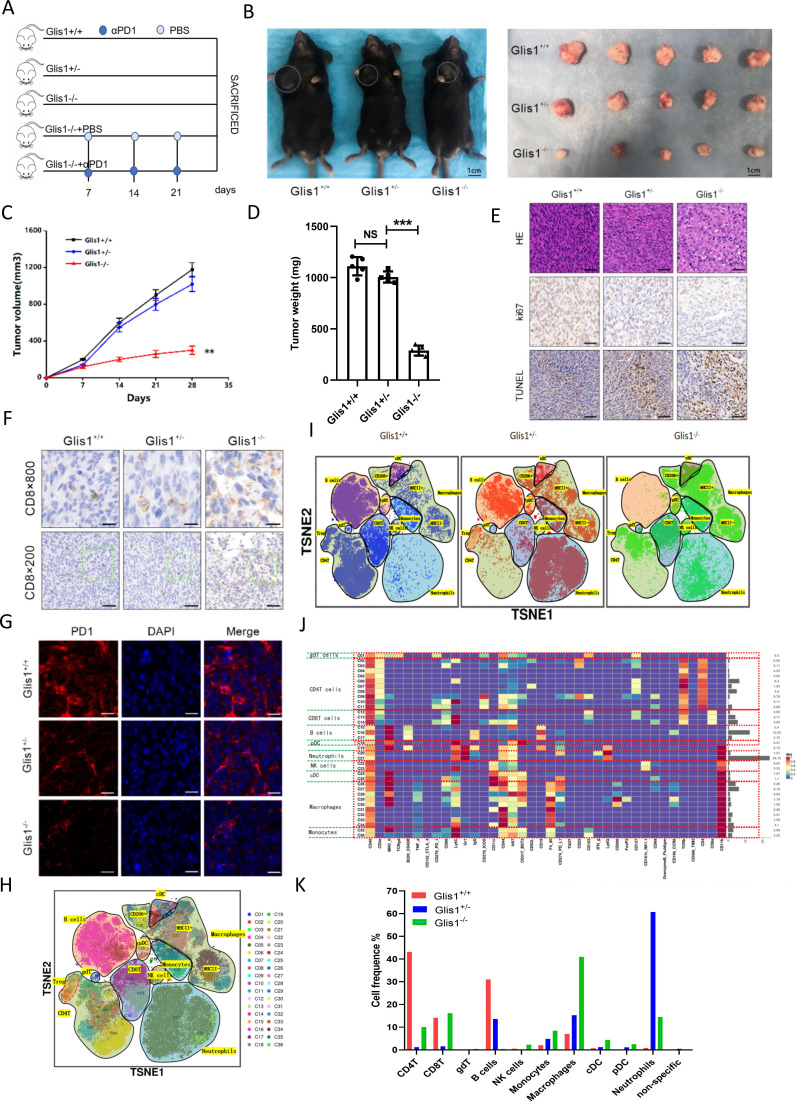
For addressing the possible effect exerted by GLIS1 on immunization responses to HCC, we established GLIS1-/- and GLIS1+/- C57BL/6 mice (online supplemental figure S3A), which was confirmed by agarose gel electrophoresis (online supplemental figure S3B, C). We injected cells (Hepa1-6) in GLIS1+/+, GLIS1+/-, and GLIS1-/- mice, respectively (figure 4A). Results showed that Hepa1-6 cells tended to grow in GLIS1+/+ mice and became slightly smaller in GLIS1+/- mice, but the difference was not significant, while they showed consistent attenuation in GLIS1-/- mice (figure 4B, C). The tumor weight in GLIS1-/- mice group was remarkably lower in contrast to the GLIS1+/+mice (figure 4D). Given the immunohistochemical results, Ki67 expression significantly decreased, and apoptosis as well as CD8+T cell expression was noticeably facilitated in GLIS1-/- mice group (figure 4E, F). Immunofluorescence results showed that PD1 expression in GLIS1-/- group was significantly reduced compared with GLIS1+/- or GLIS1+/+ group (figure 4G). In addition, when αPD1 was used simultaneously, the inhibition ratio of tumor in the GLIS1-/- mice group was remarkably increased compared with that of the PBS one (online supplemental figure S4A–D). Immunohistochemistry outcomes revealed that Ki67 expressing was noticeably reduced, and apoptosis and CD8 infiltration was significantly increased (online supplemental figure S4E, F). Immunofluorescence results showed that PD1 expression in GLIS1-/-+αPD1 was significantly reduced compared with GLIS1-/-+PSB group (online supplemental figure S4G). Collectively, this study revealed that inhibition of GLIS1 could attenuate HCC growth and improve the efficiency of αPD-1 treatment in HCC.
Figure 4.
GLIS1-/- mice attenuated HCC growth and enhanced the anti-tumor activity of CD8+ T cells. (A) Schematic diagram of drug administration in mice. (B) Picture display of the respective group (GLIS1+/+, GLIS1+/- and GLIS1-/- mice) of subcutaneous tumors. (C) The volume and (D) weight statistics of subcutaneous tumors in the respective group (GLIS1+/+, GLIS1+/- and GLIS1-/- mice). (E) The tumors were confirmed by HE staining. Immunohistochemical results of Ki67 and TUNEL expression in the respective group. (F) Immunohistochemical results of CD8 expression in the respective group. (G) Immunofluorescence results of PD1 expression in the respective group. (H) TSNE plot showing distributions of 36 cell clusters. (I) The TSNE diagram showing the distribution of cell clusters in the respective sample. (J) A total of 36 cell clusters were divided, and we defined the respective group of the mass cytometry results. (K) The histogram showing the number of the respective cell cluster in different groups by mass cytometry. **p<0.01,***p<0.001. NS indicates no significant difference. HCC, hepatocellular carcinoma.
To further assess the overall immune microenvironment variations of the tumor in each group, we used mass cytometry to measure the expression of the respective immune cell clusters. We cycled single, live and intact CD45+ immune cells from the selected cells in the respective tumor tissues (online supplemental figure S5A). Clustering and subgroup annotation of CD45+immune cells were displayed in all samples (figure 4H, I). There were 36 cell clusters in total, and we defined the respective cell cluster based on the specific markers of the respective cell type (figure 4J, online supplemental figure S5B). The results showed that the expressing of CD8+T cells were upregulated after GLIS1 was knocked out (figure 4I, K). In addiction, NK cells, DC, monocytes and macrophages were all increased when GLIS1 was knocked out compared with wild type (figure 4I, K). The mentioned results suggested that GLIS1-/- mice attenuated HCC growth and enhanced antitumor activity of CD8+ T cells.
GLIS1 knockout alleviated CD8+ T cell exhaustion and improved the reaction to αPD1 treatment in PDX model of HCC
We investigated if GLIS1 knockout affected the anti-cancer potency of CD8+ TIL cells. CD8+ TIL cells from patients with HCC were pretreated with a control interfering slow virus (sh-NC) or an interfering lentivirus that targeted the GLIS1 gene (sh-GLIS1) and the cells were afterwards adoptively introduced into NOD/SCID mice with HCC PDX in the presence or absence of αPD1 treatment (figure 5A–C). We discovered that GLIS1 knockout in CD8+ TILs restricted the development and weight of HCC cancers, with αPD1 treatment limiting cancer development and weight in a synergistic way (figure 5D–F). The outcomes of immune histochemistry revealed that in contrast to the sh-NC group, the expression of Ki67 in the sh-GLIS1 group was significantly reduced, and apoptosis and CD8+ T cell expression were significantly promoted, while the combined use of αPD1 enhanced the anti-tumor effect (figure 5G–J). Immunofluorescence outcomes revealed that in contrast to the sh-NC group, the expression of PD1 in the sh-GLIS1 group was decreased and further inhibited while the combined use of αPD1 (figure 5K). These results indicated that GLIS1 knockdown alleviated CD8+ T cell exhaustion and improved the reaction to αPD1 treatment in PDX model of HCC.
Figure 5.
GLIS1 knockdown alleviated CD8+ T cell exhaustion and improved the response to αPD1 therapy in PDX model of HCC. (A) Basic clinical information of the HCC patient used for PDX model. (B) Schematic diagram of the mouse experiment. CD8+ TIL cells from patients with HCC were pretreated with a control interfering lentivirus (shRNA-NC) or an interfering lentivirus targeting the GLIS1 gene (sh-GLIS1) and the cells were subsequently adoptively transferred into NOD/SCID mice-harboring HCC PDX with or without αPD1 therapy. (C) The tumors were confirmed by HE staining in HCC patient used for PDX model. (D) Picture display of the respective group (Control, sh-NC, sh-GLIS1, sh-GLIS1+PBS, sh-GLIS1+αPD1) of subcutaneous tumors. (E) The volume and (F) weight statistics of subcutaneous tumors in the respective group (Control, sh-NC, sh-GLIS1, sh-GLIS1+PBS, sh-GLIS1+αPD1). (G, H) The tumors were confirmed by HE staining. Immunohistochemical results of Ki67 and TUNEL expression in the respective group. (I, J) Immunohistochemical results of CD8 expression in the respective group. (K) Immunofluorescence results of PD1 expression in the respective group. **p<0.01, ***p<0.001. HCC, hepatocellular carcinoma; PDX, patient-derived xenograft.
CHIP and RNA sequence analysis when GLIS1 was knockdown in CD8+ TIL from HCC patients
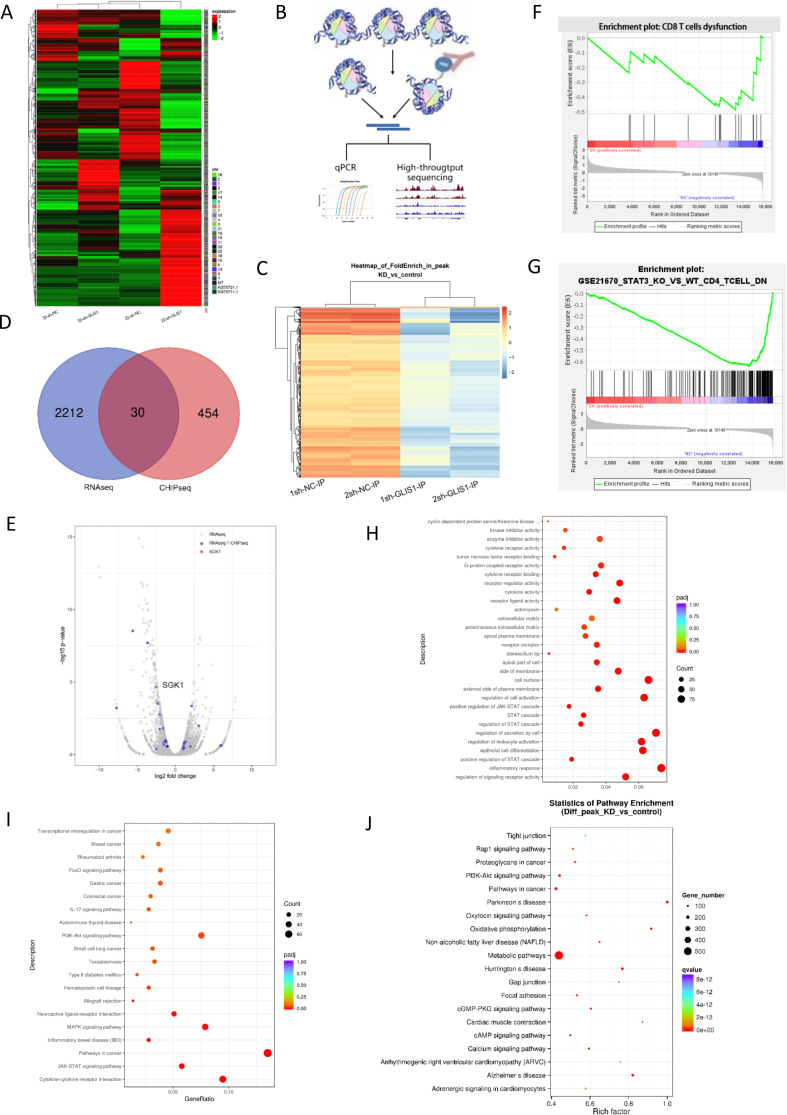
For addressing the potential mechanism of GLIS1 regulating CD8+ T cell exhaustion, we use RNA-seq analysis to explore the genes and enrichment pathways that change after GLIS1 was knocked down in CD8+ TILs isolated from two HCC patients (22sh-GLIS1, 32 sh-GLIS1, respectively) (figure 6A). CHIP-seq results showed that GLIS1 might positively transcript some genes in two groups with four samples, respectively (figure 6B, C). We cross-analyzed the results of RNA-seq and CHIP-seq to search for common genes and found that 30 genes including SGK1 were the genes shared by the two sequencing results (figure 6D, E).
Figure 6.
CHIP and RNA sequence analysis when GLIS1 was knockdown in CD8+ TIL from HCC patients. (A) RNA transcriptome sequencing was used to assess the different mRNAs of sh-NC and sh-GLIS1 group. 22 and 32 represented two groups.Heatmap was presented. (B) Schematic diagram of the experimental steps of CHIP-seq. (C) CHIP sequencing was used to assess genes regulated by GLIS1 transcription. 1 and 2 represented two groups.Volcano map was presented. (D) We cross-analyzed the results of RNA sequencing and CHIP to search for common genes shared by the two sequencing. (E) Volcano plot showing cross-analyzed genes including SGK1. (F) GSEA analysis of total RNA-seq, indicating the different genes were enriched in CD8+ T exhaustion. (G) Pathway analysis of mRNAs in sh-NC and sh-GLIS1 group. (H) GO analysis of RNA-seq22. (I) KEGG analysis of RNA-seq22. (J) KEGG analysis of CHIP-seq. CHIP, chromatin immunoprecipitation; HCC, hepatocellular carcinoma.
Overall or separate analysis of RNA sequencing pathways showed that these different genes were mainly enriched in T cell exhaustion and STAT3 pathways (figure 6F, G). For the pathway analysis of CHIP-seq data, we found that it is enriched in pathways in cancer, PI3K, and cAMP signals (figure 6H–J, online supplemental figure S6A, B). In addition, the overall distribution ratio of CHIP-site and the distribution of CHIP-site from transcription start site (TSS) were shown in online supplemental figure S6C, D, respectively. We used GEPIA software to analyze and fortunately found that GLIS1 was related to the expression of SGK1 in a positive way in HCC samples and then chosen for further study (online supplemental figure S7A).
GLIS1 promoted the exhaustion of CD8+ T cells by targeting SGK1-STAT3-PD1 pathway in HCC
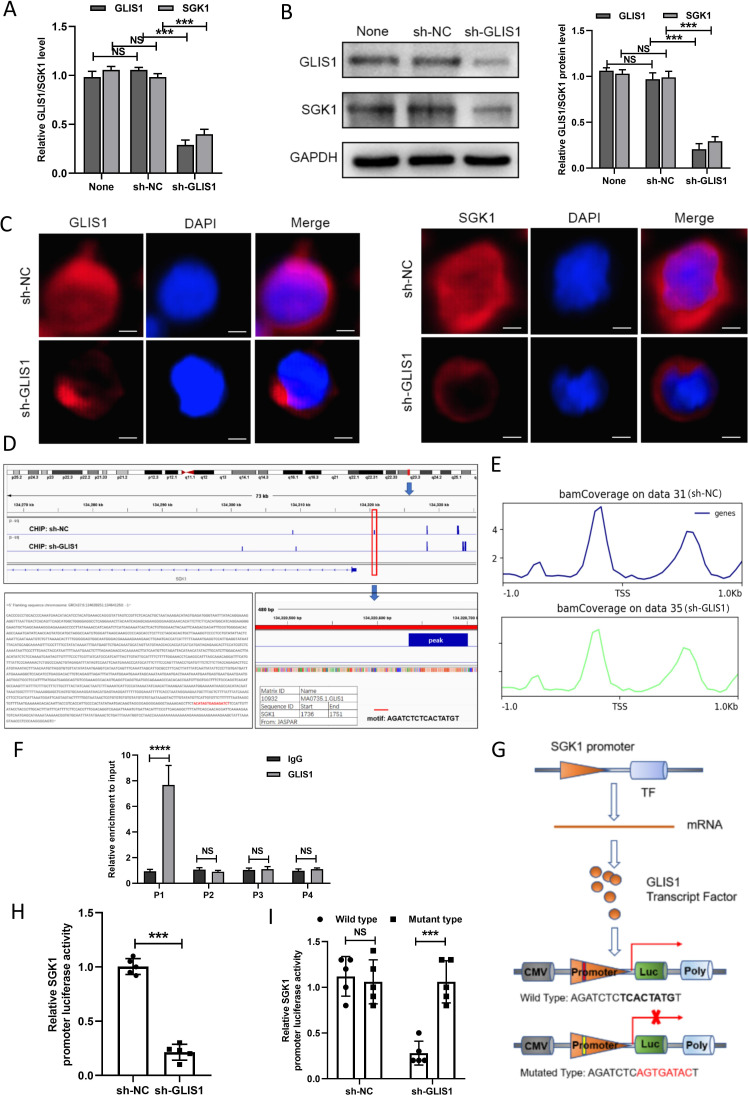
To further study the mechanism of GLIS1 and SGK1 causing CD8+ T cell exhaustion in HCC, we discovered that the mRNA and protein expression levels of GLIS1 and SGK1 were significantly reduced after knocked down GLIS1 in CD8+ T cells (figure 7A, B). Immunofluorescence results showed that in contrast to the sh-NC group, the expressing of GLIS1 and SGK1 were both decreased in sh-GLIS1 group (figure 7C). Our CHIP sequencing results showed that the peak existing in sh-NC group disappeared in sh-GLIS1 group, suggesting that GLIS1 had a binding relationship at the upstream peak peak of SGK1 (figure 7D). In addition, we analyzed the possible binding sites of GLIS1 and SGK1 in TSS, center sites and Transcription end site (TES), respectively, and results revealed that there was peaks in TSS (figure 7E) and center sites (online supplemental figure S8A) while no peaks in TES (online supplemental figure S8B), which indicated that GLIS1 might regulate SGK1 transcription in TSS sites. As demonstrated by the CHIP assay, GLIS1 was directly correlated with the SGK1 promoter within P1 (figure 7F). Furthermore, GLIS1 downregulation noticeably decreased reporter activity driven by the SGK1 promoter (figure 7G, H) and the mutated sequence did not affect the SGK1 promoter-driven reporter activity, while the wild type had a significant effect (figure 7I).
Figure 7.
GLIS1 promoted the exhaustion of CD8+ T cells by targeting SGK1 in HCC. (A) qRT-PCR revealed the expressions of GLIS1 and SGK1 under GLIS1 knockdown in CD8+ T cells. (B) Western blotting revealed the expressions of GLIS1 and SGK1 under GLIS1 knockdown in CD8+T cells. (C) Immunofluorescence results of GLIS1 and SGK1 expression under GLIS1 knockdown in CD8+ T cells. (D) Our CHIP sequencing results of the peak in sh-NC group and sh-GLIS1 group. (E) Possible binding sites of GLIS1 and SGK1 in transcription start site (TSS). (F) CHIP assay confirmed that GLIS1 can be directly correlated with the SGK1 promoters within P1, while it has no obvious significance in other sites (P2, P3, P4). (G) Schematic diagram of the dual luciferase assay. (H) SGK1 promoter-driven reporter activity was measured under GLIS1 downexpression. (I) The luciferase reporter gene experiment revealed the result of the mutated and wild type sequence affecting the promoter-driven reporter activity of SGK1 under GLIS1 down-expression. ***p<0.001, ****p<0.0001. NS indicates no significant difference. CHIP, chromatin immunoprecipitation; HCC, hepatocellular carcinoma; qRT-PCR, quantificational real-time PCR.
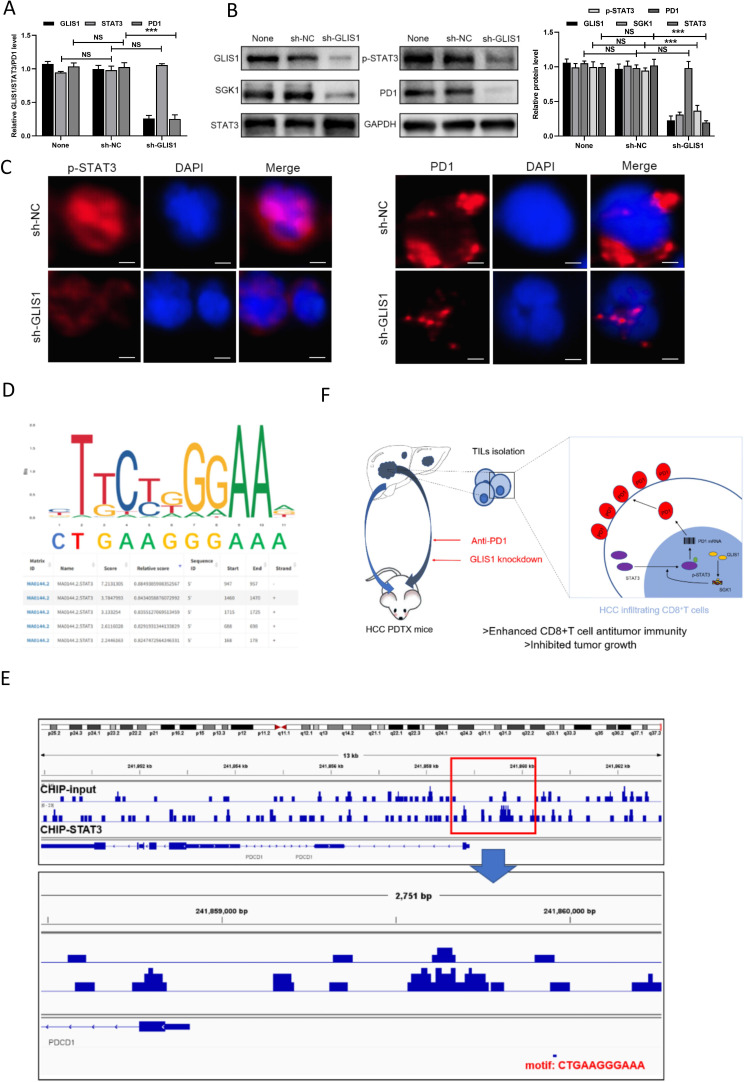
We used GEPIA software to analyze and found that SGK1 was related to the expressing of PD1 in a positive way as well (online supplemental figure S7B). Moreover, the expression of SGK1 was significantly related to exhaustion of CD8+T cells (online supplemental figure S7C). Previous study has reported that SGK1 deficiency decreased STAT3 in macrophage at M2 or M1 macrophage-priming status and STAT3/PD1 transcriptional axes elicited an immune suppressive pulmonary cancer micro environment.11 12 Therefore, we hypothesized that GLIS1 might promote CD8+ T cell exhaustion by targeting the SGK1-STAT3-PD1 pathway. To confirm this, the mRNA of STAT3 and PD1 were found significantly decreased after knocked down GLIS1 in CD8+ T cells via qRT-PCR (figure 8A). The protein of p-STAT3 and PD1 were significantly decreased after knocked down GLIS1 in CD8+ T cells via western blot (figure 8B). Immunofluorescence results showed that in contrast to the sh-NC group, the expressing of p-STAT3 and PD1 in the sh-GLIS1 group was reduced (figure 8C). Moreover, the binding sites and motif of human TF STAT3 in PD1 were predicted by JASPAR database (GSE21669) (figure 8D). The human CHIP data showed that STAT3 had a significant peak in the upstream of PD1, suggesting that there was an obvious binding site (figure 8E). Moreover, JASPAR database (GSE67183) predicted the possible binding sites and motifs of mouse TF STAT3 on PD1 (online supplemental figure S8C). The mouse CHIP data showed that STAT3 had a significant peak in the upstream of PD1, suggesting that there was an obvious binding site (online supplemental figure S8D). All of these results demonstrated that GLIS1 promoted the exhaustion of CD8+ T cells by targeting SGK1-STAT3-PD1 pathway in HCC (figure 8F).
Figure 8.
GLIS1 promoted the exhaustion of CD8+ T cells by targeting SGK1-STAT3-PD1 pathway in HCC. (A) qRT-PCR revealed the expressions of GLIS1, STAT3, and PD1 under GLIS1 knockdown in CD8+ T cells. (B) Western blotting revealed the expressions of GLIS1, STAT3, p-STAT3 and PD1 under GLIS1 knockdown in CD8+ T cells. (C) Immunofluorescence results of p-STAT3 and PD1 expression under GLIS1 knockdown in CD8+ T cells. (D) The binding sites and motif of human transcription factor STAT3 in PD1 were predicted by JASPAR database (GSE21669). (E) The human CHIP data results of the peak between STAT3 and PD1. (F) Pattern diagram reveals that GLIS1 promoted the exhaustion of antitumor CD8+ T cells by targeting SGK1-STAT3-PD1 pathway in HCC. GLIS1 promotes CD8+ T cell exhaustion in HCC through transcription regulating SGK-STAT3-PD1 pathway. Downregulating GLIS1 expression in CD8+ T cells exerts synergistic effects with anti-PD1 therapy, highlighting a promising strategy for HCC immunotherapy. ***p<0.001. NS indicates no significant difference. CHIP, chromatin immunoprecipitation; HCC, hepatocellular carcinoma; qRT-PCR, quantificational real-time PCR.
Discussion
T cell exhaustion is a gradual and kinetic process.13 14 The exhausted T-cell population is inhomogeneous and involves precursor-like exhausted or terminally exhausted cells with different features and reactions to ICIs. The expressing of immune checkpoint receptors (ICRs) and TFs participating in T-cell exhaustion, like TF T-cell factor 1 (TCF-1), T-box TF TBX21 (T-bet), Eomesodermin (Eomes), thymocyte selection-associated high mobility group box protein (TOX) determines the differentiative status and inhomogeneity of exhausted T cells.15–20 In this research, we revealed that exhausted HCC-infiltration CD8+ T cells expressed substantial GLIS1. The GLIS1 knockout in CD8+ T cells reinforced the anticancer potency of CD8+ T cells, which displayed certain effects with PD1 blocking treatment synergistically. Hence, GLIS1 might be a prospective target for the reversal of T cell exhaustion and facilitating anticancer immunity.
Some researches have revealed the effects of GLIS1 on reprogramming. Body cells can be converted to induced pluripotent stem cells (iPSCs) via reprogramming through ectopic expressing of some reprogramming factors, OCT3/4 (POU5F1), SOX2, KLF4, and c-MYC (OSKM).21 GLIS1 remarkably facilitates the reprogramming of mankind and murine fibroblasts into iPSCs when coexpressed with OSK (called OSKG), but downregulating GLIS1 via shRNAs decreased the efficiency of reprogramming.22 23 Recently, a research has discovered a positive effect of GLIS1 on the differentiative ability of brown adipose cells.24 This was revealed that exogenetic expressing of GLIS1 in murine myoblast C2C12 cells reduced the expressing of myogenesis biomarkers and reinforced adipogenesis biomarker expressing, but downregulating GLIS1 via siRNAs exhibited a reverse effect. Li et al highlighted GLIS1 as a potent reprogramming factor, and discovered an epigenome-metabolome-epigenome signal transmission cascade involving the glycolysis coordination of histone acetylization and lactylation pertaining to determining cellular fate.25
However, the effects of GLIS1 on cancers are still elusive. Tao et al found that increasing miRNA-1–3 p carried by ttumor-related fibroblasts-originated exocellular vesicles suppressed mammal carcinoma development and metastases via suppressing GLIS1.26 Shimamoto et al reported that hypoxia-triggered GLIS1 was vital for mammal carcinoma cells through regulating genetic expression associated with cellular motility and aggression abilities, causing unsatisfactory prognoses in sufferers with late period mammal carcinoma.27 Kim Joung et al suggested that cancer-associated fibroblasts (CAFs) support migration and metastasis of ovarian cancer cells by GLIS1 overexpression and GLIS1 in CAFs might be a potential therapeutic target to inhibit ovarian cancer metastasis.28 Ren et al reported that SGK1 deficiency reduced STAT3 whereas elevated FoxO1 expressing in macrophagus at M2 or M1 macrophagus-priming status, separately.11 Previous study has also revealed that STAT3/PD1 transcriptional axes elicited an immune suppressive pulmonary carcinoma micro environment.12 Our study is the first to report the role of GLIS1 in HCC and investigate its effects and causal link in exhausted CD8+ T cells. GLIS1 downregulation in CD8+ T cells suppressed cancer development, elevated CD8+ T cell infiltrative ability, mitigated CD8+ T cell exhaustion and uplifted the anti-PD1 reaction of CD8+ T cells. In addition, our research showed that the mechanism involves transcriptional regulation of SGK1-STAT3-PD1 pathway by GLIS1, thereby maintaining the abundant PD1 expression on the surfaces of CD8+ T cells in HCC.
Of course, this is not the first study to report that transcriptional regulation of PD1 expression leads to CD8 T exhaustion. In the transcriptional activity and expressing of PD1, the gene encoding PD1, two conservative areas (CR-B and CR-C) upstream of the TSS of the PDCD1 gene are pivotal.29 Scholars have discovered that modulators exist in transcription modulation and epigenesis modulation of PD1 expressing tightly related to those two conservative areas. Xiao et al discovered substantial activator protein 1 (AP-1) subtype protein c-Fos in complex with JunB on cancer-infiltration T cells, and discovered that the latter was capable of binding to the AP-1 binding spot in the CR-B area, causing upregulation of PD1 expressing. T-bet was the first modulatory factor determined to exhibit the capability of repressing PD1 transcript ability.30 Bally et al discovered that in the stimulation of macrophagus, PD1 transcript was stimulated via NF-κB, and the p65 sub-type of NF-κB was capable of binding to the relevant element in the CR-C area and upregulating PD1 expressing.31 In our study, we found that GLIS1 facilitated CD8+ T cell exhaustion in HCC through transcription modulating SGK1-STAT3-PD1 pathway. We look forward to more studies to report on the specific mechanisms regulating PD1, which will contribute to the immunotherapy of tumors especially HCC.
For addressing the possible effect exerted by GLIS1 on immunization responses to HCC, we used mass cytometry to measure the expression of the respective immune cell clusters. With its advantages of ultra-high flux, sensitivity and stability, mass cytometry has been widely used in many research fields such as immunity, tumor, blood, medicine, and genetics. Its main technical characteristics include a very large number of channels; There is no interference between adjacent channels, no need to calculate compensation; Low background interference and high signal-to-noise ratio; in addition, its ultra-high sensitivity ensures the detection of low abundance proteins, and its diverse data processing methods allow for in-depth analysis of samples.32–34 In this study, we applied mass cytometry and resuts showed that the expressing of CD8+T cells were up-egulated after GLIS1 was knocked out. In addiction, NK cells, DC, monocytes and macrophages were all increased when GLIS1 was knocked out compared with wild type. Previous studies have shown that dendritic cells are the most powerful professional antigen presenting cells (APCs) in the body’s immune system. It plays a key role in the initiation and regulation of immune response. Using the biological characteristics of DC to load tumor antigen to make tumor therapeutic vaccine is currently a relatively hot tumor therapeutic method.35 36 The results of this study suggest that the increase of DC caused by GLIS1 knockout may provide potential help for subsequent research in related fields to a certain extent. NK cells are important members of the innate immune system, which can play a rapid and direct antitumor role. At present, adoptive immunotherapy of NK cells targeting tumors is attracting more and more attention.37 Our study showed that knockout of GLIS1 can directly activate NK cells and increase their number. These results all confirmed that GLIS1 knockout reactivated the antitumor immunity of the body, suggesting that GLIS1 is a potential immunotherapeutic target for HCC.
Conclusion
GLIS1 promotes CD8+ T cell exhaustion in HCC through transcription regulating SGK-STAT3-PD1 pathway. Downregulating GLIS1 expression in CD8+ T cells exerts synergistic effects with anti-PD1 therapy, highlighting a promising strategy for HCC immunotherapy.
Footnotes
DR, YW, LL and HC contributed equally.
Contributors: There are four first authors in this manuscript and they have equally contributed to this project. DR, YW, LL and HC were responsible for performing the experiments as well as drafting the manuscript. YW and LL analyzed the data. TH, HL, XH, GuaS, GuoS, ZZ and JK performed part of the experiments and interpreted the data. Furthermore, we have 4 corresponding authors in this manuscript. YX, ZC, WT and XW have contributed to designed the study and critical revised the manuscript. XW was responsible for the overall content as the guarantor and the work and/or the conduct of the study, had access to the data, and controlled the decision to publish. All authors read and approved the final manuscript.
Funding: This work was supported by grants from the National Natural Science Foundation of China (Grant No.31930020 belonging to XW; No.81902485 belonging to ZC; No.82070676 belonging to YX).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
No data are available.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved by the Ethical Board of the First Affiliated Hospital of Nanjing Medical University(2019-SRFA-238). Participants gave informed consent to participate in the study before taking part.
References
- 1.Bruix J, Han K-H, Gores G, et al. Liver cancer: approaching a personalized care. J Hepatol 2015;62:S144–56. 10.1016/j.jhep.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villanueva A. Hepatocellular carcinoma. N Engl J Med 2019;380:1450–62. 10.1056/NEJMra1713263 [DOI] [PubMed] [Google Scholar]
- 3.Banales JM, Cardinale V, Carpino G, et al. Expert consensus document: cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European network for the study of cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol 2016;13:261–80. 10.1038/nrgastro.2016.51 [DOI] [PubMed] [Google Scholar]
- 4.Colotta F, Allavena P, Sica A, et al. Cancer-Related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 2009;30:1073–81. 10.1093/carcin/bgp127 [DOI] [PubMed] [Google Scholar]
- 5.Longo V, Gnoni A, Casadei Gardini A, et al. Immunotherapeutic approaches for hepatocellular carcinoma. Oncotarget 2017;8:33897–910. 10.18632/oncotarget.15406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basile D, Garattini SK, Bonotto M, et al. Immunotherapy for colorectal cancer: where are we heading? Expert Opin Biol Ther 2017;17:709–21. 10.1080/14712598.2017.1315405 [DOI] [PubMed] [Google Scholar]
- 7.Kim YS, Lewandoski M, Perantoni AO, et al. Identification of Glis1, a novel Gli-related, Kruppel-like zinc finger protein containing transactivation and repressor functions. J Biol Chem 2002;277:30901–13. 10.1074/jbc.M203563200 [DOI] [PubMed] [Google Scholar]
- 8.Kang HS, ZeRuth G, Lichti-Kaiser K, et al. Gli-Similar (glis) Krüppel-like zinc finger proteins: insights into their physiological functions and critical roles in neonatal diabetes and cystic renal disease. Histol Histopathol 2010;25:1481–96. 10.14670/HH-25.1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lichti-Kaiser K, ZeRuth G, Kang HS, et al. Gli-Similar proteins: their mechanisms of action, physiological functions, and roles in disease. Vitam Horm 2012;88:141–71. 10.1016/B978-0-12-394622-5.00007-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scoville DW, Kang HS, Jetten AM. GLIS1-3: emerging roles in reprogramming, stem and progenitor cell differentiation and maintenance. Stem Cell Investig 2017;4:80. 10.21037/sci.2017.09.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren J, Han X, Lohner H, et al. Serum- and glucocorticoid-inducible kinase 1 promotes alternative macrophage polarization and restrains inflammation through FoxO1 and STAT3 signaling. J Immunol 2021;207:268–80. 10.4049/jimmunol.2001455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo IY, Yang YE, Yang PS, et al. Converged rab37/IL-6 trafficking and STAT3/PD-1 transcription axes elicit an immunosuppressive lung tumor microenvironment. Theranostics 2021;11:7029–44. 10.7150/thno.60040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLane LM, Abdel-Hakeem MS, Wherry EJ. Cd8 T cell exhaustion during chronic viral infection and cancer. Annu Rev Immunol 2019;37:457–95. 10.1146/annurev-immunol-041015-055318 [DOI] [PubMed] [Google Scholar]
- 14.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015;15:486–99. 10.1038/nri3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doering TA, Crawford A, Angelosanto JM, et al. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity 2012;37:1130–44. 10.1016/j.immuni.2012.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim H-D, Song G-W, Park S, et al. Association between expression level of PD1 by tumor-infiltrating CD8+ T cells and features of hepatocellular carcinoma. Gastroenterology 2018;155:1936–50. 10.1053/j.gastro.2018.08.030 [DOI] [PubMed] [Google Scholar]
- 17.Utzschneider DT, Charmoy M, Chennupati V, et al. T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity 2016;45:415–27. 10.1016/j.immuni.2016.07.021 [DOI] [PubMed] [Google Scholar]
- 18.Siddiqui I, Schaeuble K, Chennupati V, et al. Intratumoral tcf1+PD-1+CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 2019;50:195–211. 10.1016/j.immuni.2018.12.021 [DOI] [PubMed] [Google Scholar]
- 19.Han HS, Jeong S, Kim H, et al. TOX-expressing terminally exhausted tumor-infiltrating CD8+ T cells are reinvigorated by co-blockade of PD-1 and TIGIT in bladder cancer. Cancer Lett 2021;499:137–47. 10.1016/j.canlet.2020.11.035 [DOI] [PubMed] [Google Scholar]
- 20.Wang X, He Q, Shen H, et al. TOX promotes the exhaustion of antitumor CD8+ T cells by preventing PD1 degradation in hepatocellular carcinoma. J Hepatol 2019;71:731–41. 10.1016/j.jhep.2019.05.015 [DOI] [PubMed] [Google Scholar]
- 21.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature 2010;465:704–12. 10.1038/nature09229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maekawa M, Yamaguchi K, Nakamura T, et al. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature 2011;474:225–9. 10.1038/nature10106 [DOI] [PubMed] [Google Scholar]
- 23.Yoshioka N, Gros E, Li H-R, et al. Efficient generation of human ipscs by a synthetic self-replicative RNA. Cell Stem Cell 2013;13:246–54. 10.1016/j.stem.2013.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tosic M, Allen A, Willmann D, et al. Lsd1 regulates skeletal muscle regeneration and directs the fate of satellite cells. Nat Commun 2018;9:366. 10.1038/s41467-017-02740-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Chen K, Wang T, et al. Glis1 facilitates induction of pluripotency via an epigenome-metabolome-epigenome signalling cascade. Nat Metab 2020;2:882–92. 10.1038/s42255-020-0267-9 [DOI] [PubMed] [Google Scholar]
- 26.Tao S, Li H, Ma X, et al. Elevating microrna-1-3p shuttled by cancer-associated fibroblasts-derived extracellular vesicles suppresses breast cancer progression and metastasis by inhibiting GLIS1. Cancer Gene Ther 2021;28:634–48. 10.1038/s41417-020-00244-x [DOI] [PubMed] [Google Scholar]
- 27.Shimamoto K, Tanimoto K, Fukazawa T, et al. GLIS1, a novel hypoxia-inducible transcription factor, promotes breast cancer cell motility via activation of WNT5A. Carcinogenesis 2020;41:1184–94. 10.1093/carcin/bgaa010 [DOI] [PubMed] [Google Scholar]
- 28.Kim MJ, Jung D, Park JY, et al. GLIS1 in cancer-associated fibroblasts regulates the migration and invasion of ovarian cancer cells. Int J Mol Sci 2022;23:2218. 10.3390/ijms23042218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oestreich KJ, Yoon H, Ahmed R, et al. NFATc1 regulates PD-1 expression upon T cell activation. J Immunol 2008;181:4832–9. 10.4049/jimmunol.181.7.4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao G, Deng A, Liu H, et al. Activator protein 1 suppresses antitumor T-cell function via the induction of programmed death 1. Proc Natl Acad Sci U S A 2012;109:15419–24. 10.1073/pnas.1206370109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bally APR, Lu P, Tang Y, et al. Nf-Κb regulates PD-1 expression in macrophages. J Immunol 2015;194:4545–54. 10.4049/jimmunol.1402550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simoni Y, Chng MHY, Li S, et al. Mass cytometry: a powerful tool for dissecting the immune landscape. Curr Opin Immunol 2018;51:187–96. 10.1016/j.coi.2018.03.023 [DOI] [PubMed] [Google Scholar]
- 33.Tanner SD, Baranov VI, Ornatsky OI, et al. An introduction to mass cytometry: fundamentals and applications. Cancer Immunol Immunother 2013;62:955–65. 10.1007/s00262-013-1416-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartmann FJ, Bendall SC. Immune monitoring using mass cytometry and related high-dimensional imaging approaches. Nat Rev Rheumatol 2020;16:87–99. 10.1038/s41584-019-0338-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palucka K, Banchereau J. Dendritic-cell-based therapeutic cancer vaccines. Immunity 2013;39:38–48. 10.1016/j.immuni.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gardner A, Ruffell B. Dendritic cells and cancer immunity. Trends Immunol 2016;37:855–65. 10.1016/j.it.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat Immunol 2016;17:1025–36. 10.1038/ni.3518 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-005126supp001.pdf (9.7MB, pdf)
Data Availability Statement
No data are available.