Summary
Specific, regulated modification of RNAs is important for proper gene expression1,2. Transfer RNAs (tRNAs) are rich with various chemical modifications that affect their stability and function3,4. 7-methylguanosine (m7G) at tRNA position 46 is a conserved modification that modulates steady-state tRNA levels to affect cell growth5,6. The METTL1-WDR4 complex is responsible for m7G46 in humans, and dysregulation of METTL1-WDR4 has been linked to brain malformation and multiple cancers7–22. Here we show how METTL1 and WDR4 cooperate to recognize RNA substrates and catalyze methylation. A crystal structure of METTL1-WDR4 and cryogenic electron microscopy (cryo-EM) structures of METTL1-WDR4-tRNA show that the composite protein surface recognizes the tRNA Elbow through shape complementarity. The cryo-EM structures of METTL1-WDR4-tRNA with S-adenosylmethionine (SAM) or S-adenosylhomocysteine (SAH) along with METTL1 crystal structures provide additional insights into the catalytic mechanism by revealing the active site in multiple states. The METTL1 N-terminus couples cofactor binding with conformational changes in the tRNA, the catalytic loop, and WDR4 C-terminus, acting as the activation switch for m7G methylation. Thus, our structural models explain how post-translational modifications of the METTL1 N-terminus can regulate methylation. Together, our work elucidates the core and regulatory mechanisms underlying m7G modification by METTL1, providing the framework to understand its contribution to biology and disease.
INTRODUCTION
RNA sequence complexity expands vastly with > 150 different post-transcriptional modifications, with large implications for gene regulation1,2. tRNAs are especially rich with diverse chemical modifications3,4. Anticodon loop modifications can regulate wobble pairing to maintain the correct reading frame during translation23. Other tRNA modifications contribute to proper charging with cognate amino acids or other interactions with the ribosome. Certain tRNA modifications also promote structural stability24–26 and hypomodified tRNAs can be susceptible to decay via rapid tRNA decay6. As tRNAs play an integral role in the central dogma of gene expression, dysregulated tRNA modification has been linked to many human diseases, including neurological and metabolic disorders and cancers4,27,28.
m7G is one of the most conserved modifications from bacteria to humans at tRNA position 46 (m7G46)29,30. m7G46 makes triple base interactions with C13 and G22; the tertiary interactions increase the thermal stability of the tRNAs and protect from decay6. Although loss of m7G methyltransferase is tolerated in yeast, it leads to temperature-sensitive growth defects, especially when combined with ablation of other nonessential tRNA modifications5,6,31. m7G46 may have an even more important role in higher eukaryotes. Loss of the m7G methyltransferase is detrimental to spermatogenesis in Drosophila32 and mouse embryonic stem cell self-renewal and differentiation capacity33,34. The mammalian m7G writer contains two proteins, METTL1 and WDR4. Missense mutations of WDR4 in humans are associated with microcephalic primordial dwarfism, brain malformation, severe encephalopathy with seizure, and learning disabilities7–10. Overexpression of METTL1 has also been linked to oncogenesis in numerous contexts11–22. How METTL1 and WDR4 cooperate is important to understanding how m7G contributes to normal physiology and pathophysiology in mammals.
Molecular mechanisms underlying the m7G46 modification including how it is regulated are unclear. In bacteria and archaea, the methyltransferase is monomeric or homodimeric35,36. However, the eukaryotic m7G writer complex is heterodimeric; the catalytic domain of Trm8/METTL1 interacts with Trm82/WDR4 containing WD40 repeats5,37. The required role of the WD-repeat protein in m7G-writing is unclear. Only a subset of human tRNAs is known to be modified with m7G46, and hypermethylation correlates with higher tRNA levels11,12,33. The mechanism underlying the tRNA substrate specificity of m7G46 modification is also unknown. Furthermore, AKT/protein kinase B (PKB)-ɑ phosphorylates METTL1 is near the N-terminus (S27) to inactivate methylation in vitro and in vivo38,39. Without a mechanistic understanding of how METTL1 and WDR4 work together to recognize the substrates and catalyze the methylation, deciphering how m7G46 is regulated has been challenging.
Here we elucidate the molecular mechanisms underlying the tRNA-specific methyltransferase activity of the human m7G46 writer complex, METTL1-WDR4. Through a cryo-EM structure of the METTL1-WDR4-tRNA complex (3.72 Å) and a crystal structure of the METTL1-WDR4 complex (2.45 Å), we reveal how the composite surface of the heterodimeric complex of METTL1-WDR4 recognizes the tRNA via shape complementarity. To gain insight into the catalytic mechanism, we determined crystal structures of the METTL1 methyltransferase domain bound with SAM (2.25 Å) and SAH (1.93 Å), as well as cryo-EM structures of the METTL1-WDR4-tRNA complex with SAH (3.53 Å) and SAM (4.05 Å). The quaternary complexes reveal how the active sites change with cofactor binding, including a view of the precatalytic state where G46 is poised for methylation in the catalytic pocket. Furthermore, we show that the N-terminus of METTL1 plays an important role in coordinating cofactor binding with key conformational changes in the tRNA, the catalytic loop, and the C-terminal tail of WDR4, thereby orchestrating catalysis. Thus, our molecular model provides a structural explanation for how METTL1 N-terminus phosphorylation regulates methylation. Together, our work provides three-dimensional models to explain the core and regulatory mechanisms of METTL1 and opens new avenues to investigate how METTL1 dysregulation might affect biology and diseases.
RESULTS
Architecture of the METTL1-WDR4 complex
To examine the mechanisms underlying the human METTL1 methyltransferase, we first reconstituted the enzymatic activity using purified recombinant proteins. We were able to isolate METTL1 and the METTL1-WDR4 complex (Fig. 1a, Extended Data Fig. 1a), but we could not purify WDR4 alone, presumably because WDR4 requires METTL1 for proper folding when expressed in bacteria. For the RNA substrate, we chose tRNALys (tRNA-Lys-TTT-3) because of its stable structure with and without modifications or bound proteins40,41. Furthermore, tRNALys is consistently modified with m7G46 in multiple mammalian cell lines, and the methylation depends on the presence of METTL111,12,33. In an in vitro methylation assay, purified METTL1 without WDR4 does not support any detectable methylation of the tRNA, but the METTL1-WDR4 complex can robustly catalyze the modification of tRNALys. Electrophoretic mobility shift assays (EMSAs) show that METTL1 by itself does not bind the tRNA well, but in the presence of WDR4 the ternary complex is readily observed with an affinity in the low nanomolar range (Extended Data Fig. 1b, c). Thus, we successfully reconstituted the methylation activity of METTL1-WDR4 with tRNALys.
Fig. 1: Architecture of the human METTL1-WDR4 complex.
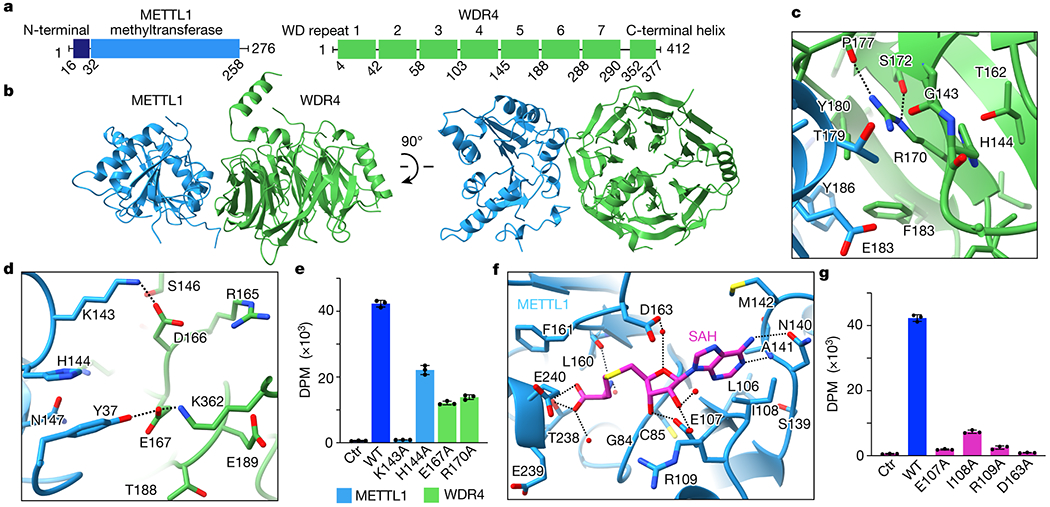
a, Domain organization of human METTL1 and WDR4. b, Crystal structure of the METTL1-WDR4 complex. c-d, Close-up views of the METTL1-WDR4 interface. R170 interactions with nearby backbone carbonyls build a scaffold to enable nearby hydrophobic intermolecular interactions. Side chains within 4 Å of the other protein or R170 of WDR4 are shown. Hydrogen bonds are indicated with dashed lines. e, In vitro methylation activity of full-length METTL1-WDR4 with indicated point mutations, shown as mean ± SD from 3 replicates. f, Close-up view of the SAH-binding pocket in the crystal structure of METTL1. Side chains are shown as sticks for the residues within 4 Å of SAH and waters as red spheres. Dashed lines indicate hydrogen bonds. g, In vitro methylation activity of the full-length METTL1-WDR4 complex with indicated point mutations in METTL1, shown as mean ± SD from 3 replicates.
To derive a three-dimensional model of the human METTL1-WDR4 complex, we obtained crystals using truncated constructs of each polypeptide (METTL1 20-265 and WDR4 1-389) because longer constructs did not crystallize (Fig. 1b, Extended Data Table 1). The overall architecture of the protein complex is similar to that of the yeast complex (Trm8-Trm82), but more differences are observed between WDR4 and Trm82 possibly due to the relatively low sequence identity (~23%) (Extended Data Fig. 1d)37. The C-terminal helix of WDR4 is newly resolved in our structure; since the helix engages in crystal contacts with symmetry molecules, its position relative to the beta-propeller domain is likely flexible. Upon complex formation, METTL1 and WDR4 bury a total of ~1746 Å2 of surface area containing many hydrophobic interactions. Polar residues such as R170 and E167 of WDR4 and K143 of METTL1 stabilize the interdomain contacts (Fig. 1c,d). Substitution of these residues reduces enzymatic activity, indicating that the observed intermolecular interactions are important for the optimal activity of METTL1-WDR4 (Fig. 1e). Homozygous mutations of WDR4 have been observed at R170 in patients with primordial dwarfism, highlighting that the observed decrease in m7G46 modification activity can manifest as a major brain abnormality8,9. Therefore, METTL1 and WDR4 form a stable complex with intricate intermolecular interactions that are important for optimal methyltransferase activity.
We were only able to crystallize the METTL1-WDR4 complex in the absence of SAM or SAH. To understand the catalytic function of METTL1 better, we determined crystal structures of the methyltransferase domain in complex with SAM (2.25 Å) and SAH (1.93 Å) (Extended Data Fig. 1e, Extended Data Table 1). The two structures align well with each other (RMSD, 0.15 Å), as well as with previously deposited structures in the protein data bank and our METTL1-WDR4 complex structure (Extended Data Fig. 1f,g). The adenine base of SAM/SAH fits into a hydrophobic pocket lined with the side chains of I108, M142, and A141, and it makes hydrogen bonds with N140 and the nearby peptide backbone (Fig. 1f). More H-bonds stabilize the interaction between the ribose hydroxyl groups and E107. The amino group of SAM/SAH makes polar contacts with the backbone carbonyls of G84 and L160, and the carboxy group makes H-bonds with the hydroxyl group of T238 and the backbone amino group of E240. To probe the catalytic pocket biochemically, we introduced single alanine substitutions of key interacting residues and assayed for in vitro methylation activity. Losing the observed side-chain interactions was detrimental to methylation (Fig. 1g). Therefore, through high-resolution crystal structures of METTL1-SAM, METTL1-SAH, and METTL1-WDR4 complexes and site-directed mutagenesis studies we gained mechanistic insight into how the human m7G writer complex forms and interacts with the methyl donor.
Cryo-EM structures of METTL1-WDR4-tRNA
To investigate how the METTL1-WDR4 complex engages with substrate tRNAs, we determined cryo-EM structures of the METTL1-WDR4-tRNALys complex without cofactor (apo, 3.72 Å), with SAM (4.05 Å), and with SAH (3.53 Å) (Fig. 2a–c, Extended Data Fig. 2–4, and Extended Data Table 2). We could use a full-length construct for METTL1 unlike in the crystallization experiments, but we had to truncate the C-terminal 23 residues of WDR4 due to aggregation. To stabilize the SAM-bound complex, we used METTL1D163A instead of the wild type to prevent catalysis (Fig. 1g, 2b). The final cryo-EM maps show clearly defined conformations of the protein domains as well as the bound tRNA, revealing how the intermolecular interactions are formed and change with the cofactor-bound state.
Fig. 2: Architecture of the METTL1-WDR4-tRNA complex in three states.
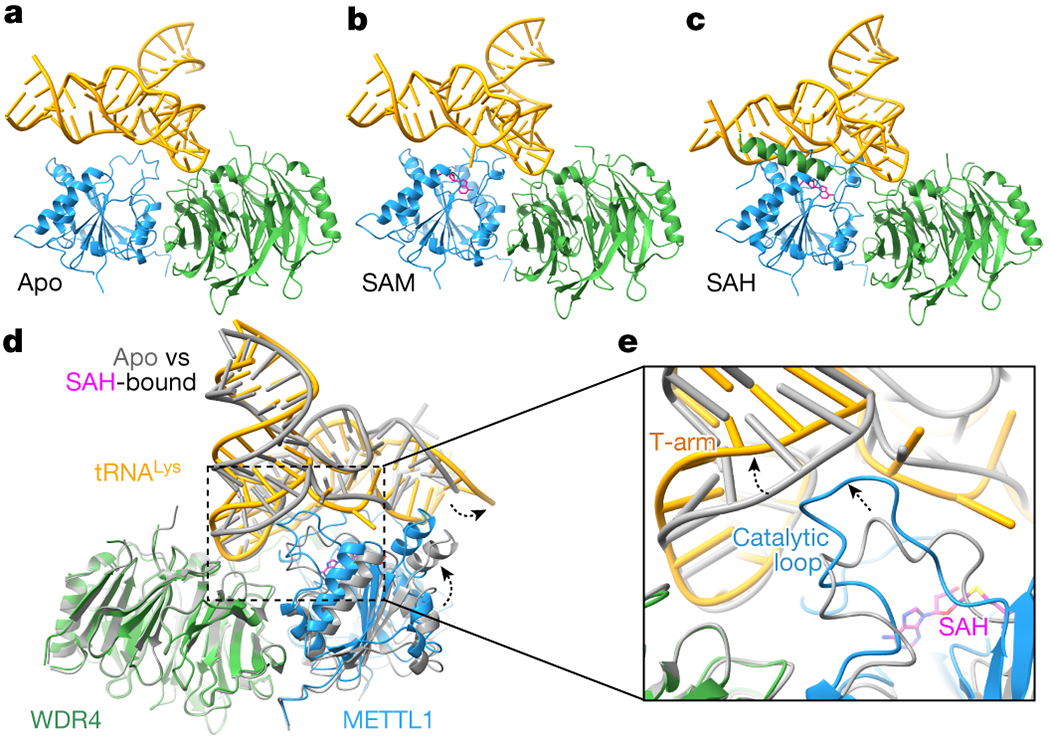
a-c, Models of the cryo-EM structures in the indicated cofactor binding states, in cartoon representation. METTL1, WDR4, tRNA and SAM/SAH are colored blue, green, gold, and magenta, respectively. d, Superimposition of the apo and SAH-bound cryo-EM structures of METTL1-WDR4-tRNA, aligning by WDR4. METTL1 moves closer to tRNA in the presence of SAH (black dashed arrow). e, Zoomed-in view of (d) showing the tRNA and the catalytic loop protruding toward the tRNA (black dashed arrow).
The composite surface of the METTL1-WDR4 complex accomplishes specific RNA binding through shape and charge complementarity. A stable apo METTL1-WDR4-tRNA complex maintains the same interdomain orientation as in the crystal structure of the METT1-WDR4 complex (RMSD, 0.83 Å) (Extended Data Fig. 5a). We could visualize more of the long loop (“catalytic loop”) next to the SAM binding site in the presence of RNA, but the structural changes are local and limited (Extended Data Fig. 5b). The conformation of tRNALys bound to METTL1-WDR4 is also similar to that of the unbound tRNALys (same sequence with full modification)40 and another structure of tRNALys in an unrelated protein-RNA complex with no modifications41 (RMSD of 1.2 and 1.0 Å, respectively, Extended Data Fig. 5c). Thus, to form the apo METTL1-WDR4-tRNA complex, the proteins and the RNA substrate undergo little induced folding upon binding, indicating that no significant conformational change is required to form a stable and specific protein-RNA complex.
While the overall domain architecture seems similar, local conformational changes occur when SAM or SAH binds METTL1-WDR4. An obvious change is in the C-terminal helix of WDR4 which becomes more ordered in the SAH-bound state (Fig. 2c and Extended Data Fig. 5d). The helix tends to be flexible and only a weak helical density is observed in the apo and SAM-bound state. However, in the presence of RNA and SAH, the C-terminal helix of WDR4 becomes significantly more stabilized to contact the tRNA and METTL1, further reinforcing the protein-RNA complex. When all three cryo-EM structures are superimposed, the most extreme conformational differences are observed between the apo and SAH-bound states, and the SAM-bound structure generally shows an intermediate state (Extended Data Fig. 5e). Anchoring on WDR4, METTL1 moves closer to the tRNA and the tRNA undergoes a slight twist when the cofactor is bound (Fig. 2d); and the catalytic loop adjacent extrudes into the tRNA in the SAM/SAH-bound structures (Fig. 2e). Therefore, even though the three states are largely similar with the tRNA docking on the preformed METTL1-WDR4 complex, cofactor binding induces local conformational changes.
tRNA shape recognition by METTL1-WDR4
Among the three cryo-EM structures, the most extensive RNA-protein interaction is observed in the SAH-bound state (Fig. 3a). Most cofactor-dependent contacts are made by METTL1, and WDR4 maintains similar interactions with the tRNA in all three structures, even with the rigidified C-terminal helix. Accordingly, the area of the WDR4-RNA binding interface in the three states is more constant (apo: 333 Å2, SAM: 337.5 Å2, and SAH: 527.5 Å2), compared with the METTL1-RNA interface (apo: 234 A2, SAM: 650 Å2, and SAH: 1422 Å2). The RNA-binding surfaces on METTL1-WDR4 overlap well with the basic patches, as expected for robust nucleic acid binding proteins (Fig. 3b). The residues in these RNA-binding surfaces are also relatively more conserved across different species (Extended Data Fig. 5f).
Fig. 3: Structural recognition of tRNA shape by METTL1-WDR4.
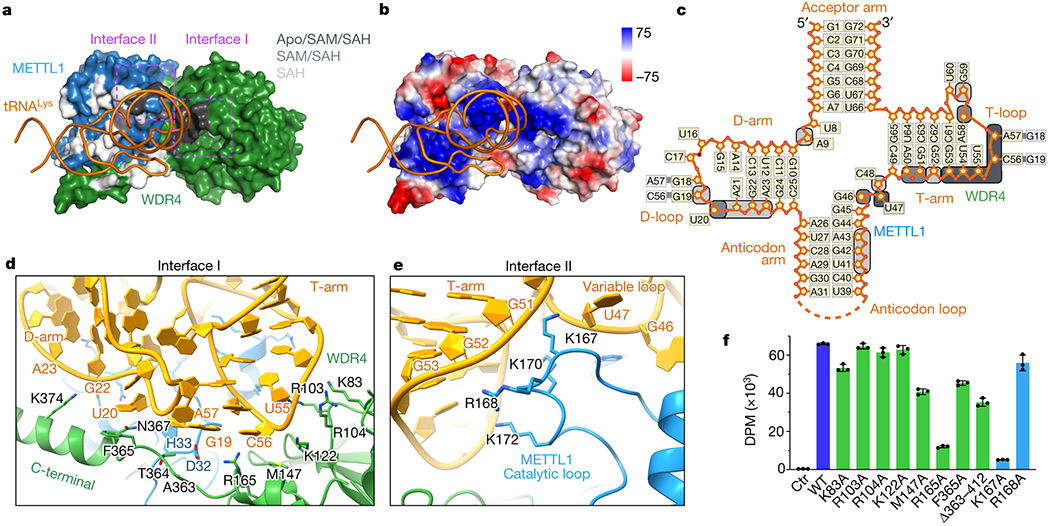
a, Surface representation of the METTL1-WDR4-tRNA-SAH structure, with only tRNA in ribbon representation. Residues within 4 Å of RNA are colored by conservation of the protein-RNA contact in the three states: black (Apo/SAM/SAH), grey (SAM/SAH), or white (SAH only). Two interface areas with conserved contacts in all three states are marked with dotted rectangles in magenta. b, Vacuum electrostatics surface representation of the protein complex in the same orientation as (a). c, Secondary structure diagram of tRNALys. Nucleotides within 4 Å of protein are indicated with rounded squares and colored using the same color key as in (a). d-e, Close-up views of METTL1-WDR4-tRNA-SAH where the residues within 4 Å of RNA are shown with side chain sticks for the interfaces marked in (a). f, In vitro methylation activity of METTL1-WDR4 with point mutations in WDR4 (green) or METTL1 (blue), shown as mean ± SD from 3 replicates.
The protein-RNA contacts conserved in all three structures involve the tRNA core “elbow” region—where the D- and T-loops meet (Fig. 3c). The kissing loop interaction creates a distinct tertiary structure of the tRNA, and the RNA fold is recognized by the composite surface formed by WDR4 (interface I) and METTL1 (interface II). The tRNA elbow is cupped by hydrophobic and pi-stacking interactions provided by M147, R165, and T364 of WDR4 as well as D32 of METTL1 (Fig. 3d). Additional contact with the D-arm is observed for the C-terminal helix of WDR4, including ring stacking through F365. Furthermore, the T-arm phosphate backbone makes several favorable contacts with two clusters of basic side chains from WDR4 and METTL1 (Fig. 3d,e). Disrupting the phosphate interactions with single alanine substitutions was not enough to cause a significant change in the enzymatic activity in most cases, but the R165A and M147A mutations near the elbow tip cause a significant decrease in methylation, indicating that detecting the unique structural features of the tRNA elbow is critical for proper substrate recognition (Fig. 3f). Truncating the WDR4 C-terminal helix or introducing an F365A mutation in the middle of it reduced methylation, suggesting that the helix-RNA contacts also contribute to productive binding. The cluster of basic residues in METTL1 belongs to the catalytic loop that changes conformation in SAM/SAH-bound states to maintain contact with the tRNA despite some twist in the RNA conformation (Fig. 2g,h). K167 of METTL1 seems to be critical for productive catalysis—the amino group is sandwiched between the two phosphates of C48 and A50 in the SAH-bound state (Fig. 3e,f). Almost all of the RNA-protein contacts are electrostatic interactions with the phosphate backbone, pi, or hydrophobic, rather than nucleobase-specific contacts, likely because the methyltransferase has to recognize many different tRNA sequences through their tertiary fold. Therefore, METTL1-WDR4 and tRNA can minimally form stable complexes through shape and charge complementarity, but local rearrangements that accompany cofactor binding are critical for optimal enzymatic activity.
Insight into the catalytic mechanism
In the apo structure of METTL1-WDR4-tRNA, the RNA structure is remarkably similar to the structure of the isolated tRNA41 (Extended Data Fig. 5c). Maintaining the free conformation of tRNA is unlike what happens with some methyltransferases that need to pry apart the D- and T-loop interactions, resulting in the partial unfolding of the tRNA42. Indeed, the variable loop is only partially interacting with the protein in the apo state, with an obvious gap at the interface (Fig. 4a). To support G46 methylation, the guanine base would still need to disengage from the helical stack where it is buried between other bases and stabilized with a favorable hydrogen bonding network. We observe the largest amount of conformational heterogeneity in the tRNA in the apo state, especially near the anticodon arm adjacent to the variable loop. To isolate the most stable conformation of the tRNA, we had to use a mask near the anticodon arm to improve the map after initial 3D classification and refinement (Extended Data Fig. 2). The observed heterogeneity suggests that the tRNA bound to METTL1-WDR4 breathes and forms alternative conformations, most of which are not prevalent enough to be reconstructed. Therefore, although METTL1, WDR4, and tRNA make stable complexes in all three states, the tRNA conformation is more flexible in the apo state.
Fig. 4: Detailed view of the active site.
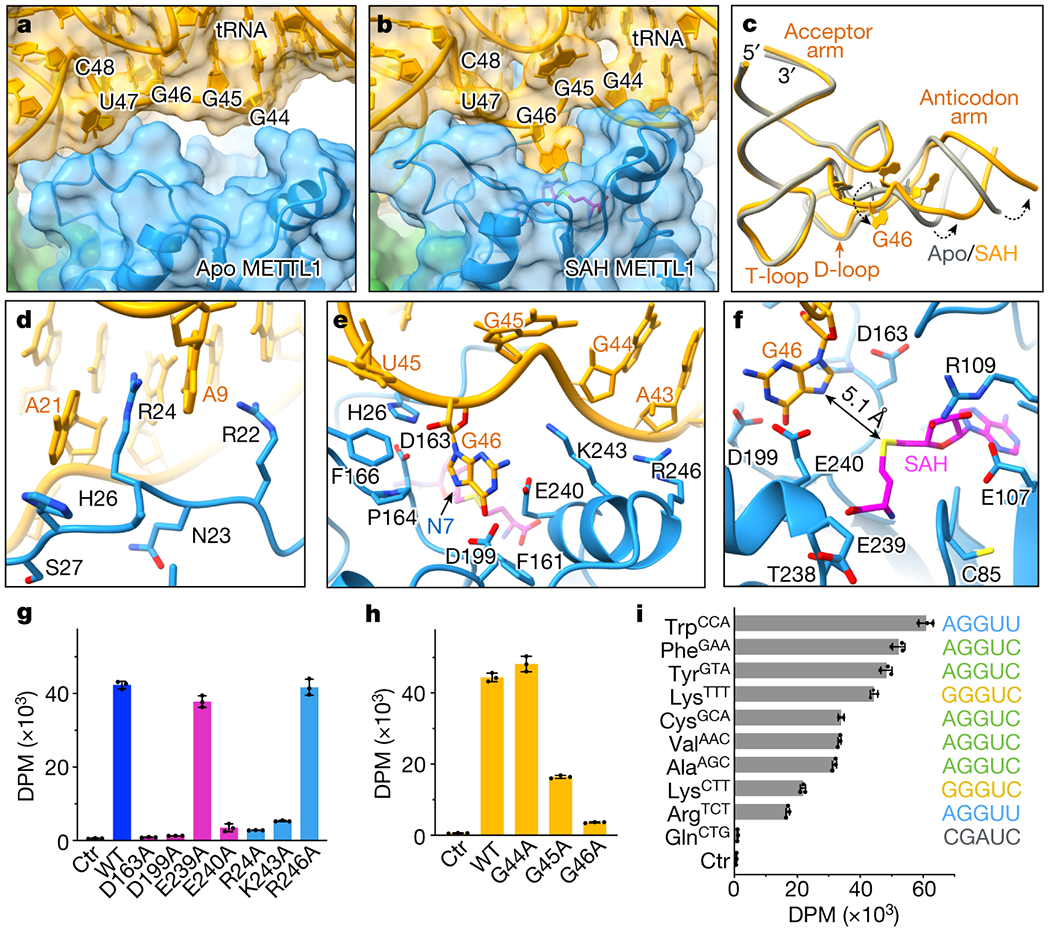
a-b, View of the tRNA variable near METTL1. RNA (yellow) and METTL1 (blue) are shown with transparent surface representation and cartoon, and SAH is shown in magenta. The variable loop is indicated with nucleotide numbers. The gap in apo state is filled as G46 is released from the helical stack and buried in the catalytic pocket after binding SAH. The orientation is the same as in (a). c. Twisting near the variable loop exposes G46 and rotates the anticodon arm relative to the rest of the tRNA. d, R24 of METTL1 occupies the space left behind by G46 to stack between A21 and A9. e, View of G46 base surrounded by a triad of acidic residues next to SAH. N7 position is indicated with an arrow, and side chains are shown only for the residues within 4 Å of RNA. f, Distance between the sulfur in SAH and N7 in G46 is indicated with an arrow. g-i, In vitro methylation activity of METTL1-WDR4 for tRNALys with point mutations in METTL1 (g) or tRNALys (h), or with wild-type proteins and different human tRNAs (i). All bar graphs are shown as mean ± SD from 3 replicates.
In the SAH-bound structure, the variable loop unfurls to release the G46 base into the catalytic pocket, causing a more intimate protein-RNA contact (Fig. 4b, Extended Data Fig. 6a,b). With the flipped G46, the anticodon arm also twists relative to the rest of the tRNA (Fig. 4c). To make the release of the G46 base energetically favorable, the void left behind between the rings of A21 and A9 is filled by the R24 side chain of METTL1. The distorted RNA conformation is supported by additional nearby basic residues, such as R22 (pi-pi and pi-cation with rings) and K243 (with the phosphate backbone) (Fig. 4d,e). The resulting RNA conformation in the SAH-bound state seems to be more uniform across the particles (unlike in the apo state), as no masks were necessary to derive a cryo-EM map with a unique tRNA structure (Extended Data Fig. 4).
The N7 position of G46 is ~5 Å away from the sulfur that would donate the methyl group in SAM, suggesting that the SAH-bound structure is closer to the active conformation (Fig. 4f). The guanine ring has three acidic residues (D163, D199, and E240) with the side chains facing the base within 4 Å distance (Fig. 4e). D163 is the closest to activate N7, but D199 and E240 side chains near the larger ring may also be important to deprotonate the guanine to dissipate the positive charge gained by methylation. Substituting any of the three acidic residues with alanine is detrimental to the methylation activity (while a nearby E239A mutation has a negligible effect), suggesting that they are all likely to contribute significantly to catalysis (Fig. 4g). Furthermore, R24 that substitutes for G46 in the RNA helix is also critical to the enzymatic activity. Therefore, we revealed key molecular elements for productive m7G methylation: R24 intercalates in the tRNA stem to help the G46 base to flip out, the methyl acceptor is closest to D163 for activation, and more acidic residues—D199 and E240—surrounding the guanine support catalysis.
We investigated how the sequence of the variable loop contributes to m7G modification due to the large contact it makes with the protein upon changing the conformation. We introduced transition mutations for the purines in the variable loop of tRNALys and found that the methylation was inhibited when G45 or G46 were mutated, although mutating G44 did not affect methylation significantly (Fig. 4h). For a broader search of mutations without breaking the tRNA fold, we measured methylation activity using other human tRNAs as substrates (Fig. 4i, Extended Data Fig. 6c). Most methylated tRNAs contain the sequence RGGUY in the variable loop33. However, reconstituting the RGGUY motif in tRNAGln—normally refractive to m7G modification—can only partially rescue the catalytic activity, suggesting that there are additional features involved in substrate recognition than the variable loop sequence (Extended Data Fig 6d). Only a few nucleobases contact METTL1-WDR4 directly in our cryo-EM structures, and their variance across different tRNAs does not seem to correlate with changes in methylation efficiency (Extended Data Fig. 6c). Furthermore, methylation efficiency does not correlate with RNA affinity measured by EMSA, suggesting that an event after complex formation might be determining the methylation level (Extended Data Fig 6e). From these experiments, we conclude that the sequence of the variable loop and the rest of the tRNA scaffold are both important to determine the propensity for m7G modification.
METTL1 N-terminus organizes catalysis
When we compare the structures of METTL1-WDR4-tRNA in apo and SAH-bound states, the cryo-EM map near the N-terminal end of METTL1 (N-term) becomes more prominent with cofactor, allowing us to extend the model for amino acids P16-D32 (Fig. 5a,b). This region is disordered in the other structures we report here. In the METTL1-WDR4-tRNA-SAH complex, the METTL1 N-term passes through a narrow tunnel between SAH and the D-arm, which may explain why the peptide becomes ordered only in the presence of both RNA and cofactor (Fig. 5c).
Fig. 5: METTL1 N-term coordinates SAH binding with RNA and protein conformational changes.
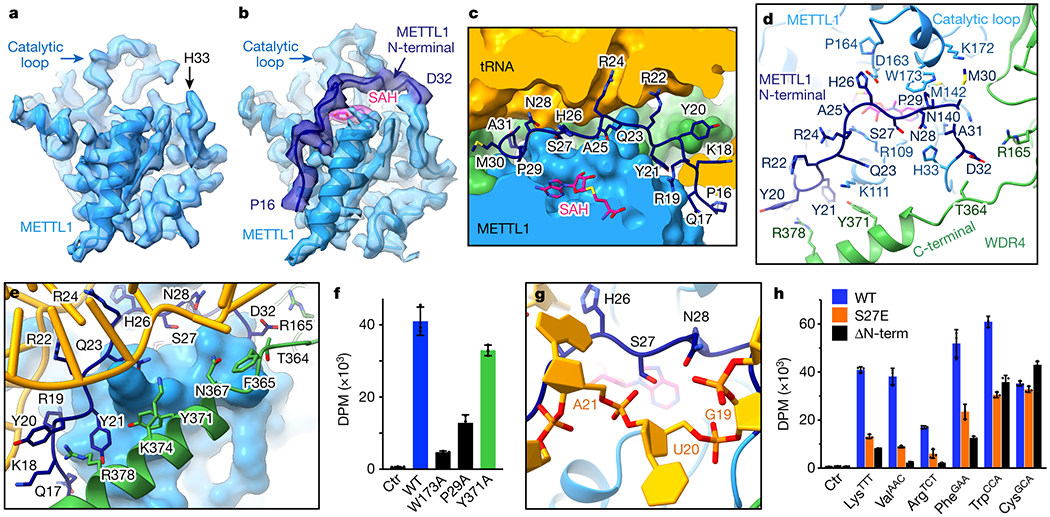
a-b, Sharpened cryo-EM maps of METTL1-WDR4-tRNA ± SAH structures reveal that the METTL1 N-terminal peptide (dark blue) becomes ordered upon binding SAH. c, METTL1 N-term and SAH shown in stick representation with surrounding proteins and RNA shown as surfaces. N-term seals the gap in the protein-RNA interface. d, METTL1 N-term supports conformational changes in the catalytic loop and the WDR4 C-terminal helix. Side chains within 4 Å of the N-term are shown in stick representation. e, METTL1 N-term and WDR4 C-terminal helix support each other to bind RNA together. Side chains within 4 Å of RNA or N-term are shown with sticks. f, In vitro methylation activity of full-length METTL1-WDR4 constructs with point mutations in WDR4 (green) or METTL1 (dark blue), shown as mean ± SD from 3 replicates. g, Close-up view of METTL1 S27 lying on top of SAH and under the RNA backbone. h, In vitro methylation activity of full-length METTL1-WDR4, using WT, S27E, or N-terminally truncated (Δ1-19) METTL1. Data are shown as mean ± SD from 3 replicates.
METTL1 N-term makes important specific contacts with the tRNA, the catalytic loop, and the C-terminal helix of WDR4 to coordinate the quaternary complex assembly near the catalytic pocket. R22 and R24 within the METTL1 N-term have already been discussed for their ability to release G46 from the helical stack (Fig. 4f–g and 5c). The SAM/SAH binding pocket is further stabilized by P29 of the N-term packing against M142 in the hydrophobic binding pocket for the adenyl ring. The METTL1 N-term also supports the extruded catalytic loop conformation—P29 and M30 make hydrophobic interactions with W173 and the aliphatic portion of K172, respectively (Fig 3e and 5d). Moreover, H26 of N-term is positioned close to the catalytic pocket, within hydrogen-bonding distance from D163 and the backbone of the methylated guanine. To obtain the SAM-bound structure, we used the catalytically inactive METTL1D163A. Mutating D163 likely also perturbs its ability to stabilize the ordered N-term, which may be why we were only able to partially resolve the N-term, to H26. Since the catalytic loop extrusion is similar in both SAM and SAH-bound states, the presence of the cofactor seems to suffice in triggering certain conformational changes. In summary, the METTL1 N-term couples cofactor binding with more extensive RNA-protein contact where G46 is properly bound in the catalytic pocket.
The folded METTL1 N-term also promotes the ordering of the WDR4 C-terminal helix. Two tyrosine rings of Y20 and Y21 are well-positioned to make pi-pi interactions with R378 and Y371, making the C-terminal helix location more favorable (Fig. 5e). Since the C-terminal helix of WDR4 also binds the RNA (Fig. 3d), rigidifying its conformation via the METTL1 N-term would result in a more stable protein-RNA complex. As a result, when the METTL1 N-term and the WDR4 C-term become ordered concomitantly, the two together provide a more expanded RNA-binding site.
Given the multiple roles that the METTL1 N-term plays in stabilizing the complex structure, we tested how the contacts affect the enzymatic activity. We found that mutations that would perturb favorable interactions with the catalytic loop or the WDR4 C-terminal helix both reduced the methylation activity significantly, highlighting the importance of the METTL1 N-term in organizing the observed conformational changes for optimal methylation efficiency (Fig. 5f). Therefore, our structural and biochemical data show that the METTL1 N-term acts as a switch to coordinate multiple molecular events important for productive m7G writing. Located at the center of the catalytic center, METTL1 N-term supports the protruding catalytic loop conformation, stabilizes the SAM binding pocket, reinforces WDR4-RNA interactions, and activates the release of the G46 base.
Previous studies have shown that S27 in the METTL1 N-term is phosphorylated by key signaling kinases such as AKT to render the enzyme inactive38,39. In our METTL1-WDR4-tRNA-SAH structure, S27 is located between SAH and the RNA backbone, closest (~ 5 Å) to the phosphate group of U20. Phosphorylation of S27 is likely to create a clash, due to steric clash and charge repulsion (Fig. 5g). Indeed, when we used METTL1S27E to mimic phosphorylation, we observe a significant decrease in in vitro methylation activity (Fig. 5h). When we truncate the first 19 amino acids of METTL1, there is a similar loss of enzymatic activity highlighting the positive contribution of METTL1 N-term to productive methylation. Most tRNAs are sensitive to the truncation or mutation of the METTL1 N-term. However, the degree of sensitivity varies, and certain tRNAs such as tRNACys and tRNATrp were not affected to the same degree, whereas others such as tRNAVal were more responsive to changes in the N-term. Together, our data show that the N-terminal end of METTL1 regulates m7G46 methylation activity, and it works by orchestrating SAM binding, RNA binding, and conformational changes in the tRNA and METTL1-WDR4.
DISCUSSION
Modification of tRNAs with m7G46 is important for normal cellular processes, and dysregulated methylation is linked to diseases. Our structural and biochemical data allow us to present a comprehensive mechanistic model for how METTL1 and WDR4 cooperate to recognize tRNAs and catalyze m7G modification at position 46 (Fig. 6). The METTL1-WDR4 heterodimer forms a composite docking site that recognizes the elbow region of tRNAs by shape and charge complementarity. Upon binding SAM, the METTL1 N-term becomes ordered between the bound cofactor and the tRNA. Concomitantly, the catalytic loop extends toward the tRNA, and the WDR4 C-terminal helix rigidifies against the METTL1 N-term to strengthen the hold around the outer tRNA Elbow. The same METTL1 N-term also activates the tRNA by replacing G46 (via R24) in the helical core, unleashing the guanine to flip out and insert into the active site containing SAM and three acidic residues critical for catalysis, poised for methylation at the N7 position. As the variable loop unfurls, the sequence RGG46UY likely helps to position the modified base optimally. Truncations or modifications of the METTL1 N-term—such as phosphorylation of S27—inhibit proper organization of the catalytic pocket, providing a structural explanation for how m7G modification can be regulated post-translationally. Interestingly, the METTL1 N-term regulates only a subset of tRNAs. Therefore, through molecular snapshots of METTL1 in various contexts, we elucidate the mechanisms underlying the specific methyltransferase function and regulation of METTL1.
Fig. 6: Mechanistic model for tRNA m7G46 methylation by METTL1-WDR4.
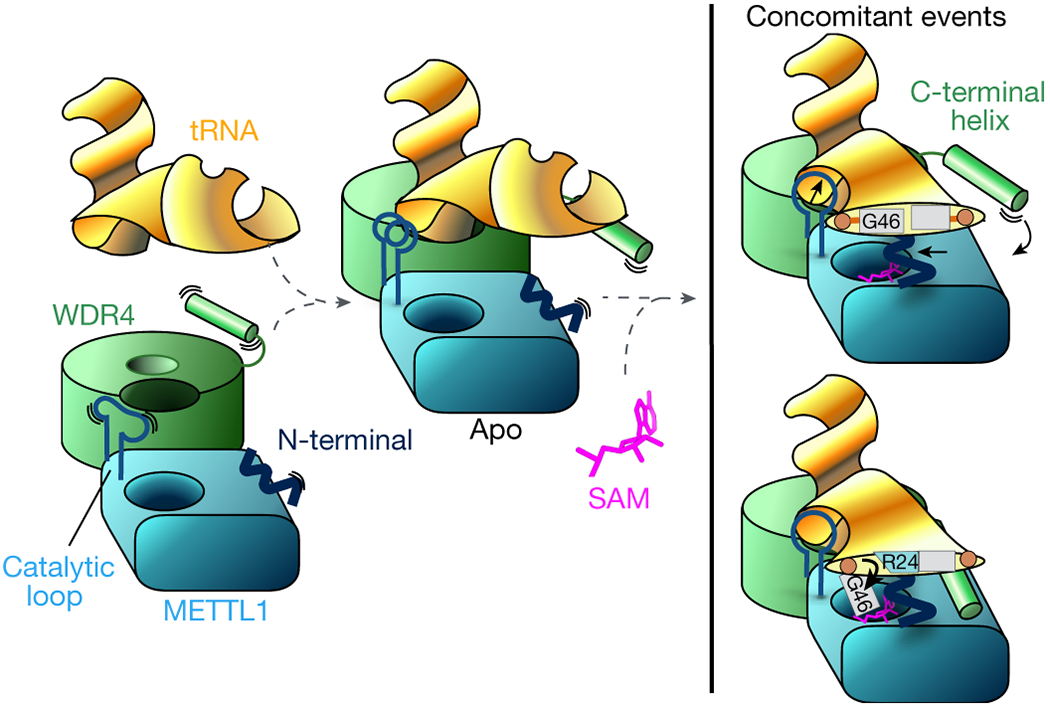
The stable heterodimeric METTL1-WDR4 protein complex provides a docking site for the tRNA Elbow. The protein-RNA docking can occur without cofactor, and METTL1 can bind SAM without tRNA. In the apo state without SAM or SAH, the catalytic loop becomes more ordered with bound RNA. WDR4 C-terminal helix moves close to RNA but remains flexible and METTL1 N-term is also disordered. When both tRNA and SAM are bound, METTL1 shifts even closer to the tRNA—the catalytic loop protrudes toward the tRNA, METTL1 N-term becomes ordered sandwiched between RNA and SAM, and WDR4 C-term attaches to the METTL1 N-term to stabilize the bound RNA together. The same N-term also supports the twisting of the tRNA to release G46 by replacing the stacking interactions with R24.
The arrangement of the human METTL1-WDR4-tRNA complex we observe in the cryo-EM structures is distinct from the binding mode that has been proposed for the yeast homologs (Trm8-Trm82) or the B. subtilis m7G45 methyltransferase (TrmB)37,43. Although there may be species-specific differences (Extended Data Fig. 7,8), it may be more likely that small-angle X-ray scattering does not provide enough high-resolution information to pinpoint the accurate orientation of tRNA relative to the protein complex. Our cryo-EM structure nevertheless is congruent with the previous biochemical findings that the D- and T-arm structures are important for recognition by the m7G46 methyltransferase44. We used unmodified nascent tRNAs for our structural and biochemical experiments. Our tRNA structures align well with previously reported crystal structures of fully modified or unmodified tRNALys (Extended Data Fig. 5c), in agreement with the findings that the m7G46 modification is inert to other modifications for tRNAPhe45. However, given that minor structural changes could affect tRNA dynamics to result in different methylation kinetics, our work will help identify whether other modifications or factors could modulate m7G modification.
Recent findings suggest that the METTL1-WDR4 complex promotes cancer in multiple contexts. Our comprehensive model for the mechanism of function and regulation of METTL1-WDR4 provides the necessary groundwork to dissect how pathogenesis would occur. Not all tRNAs are affected equally when METTL1-WDR4 is suppressed or overexpressed11,12,33. Our results also show that the intrinsic methylation efficiency is different for distinct tRNA sequences. Our structural explanation for the importance of the N-terminus provides the framework to understand how METTL1 is regulated, by phosphorylation38 or yet other modifications (eg. ubiquitination) in the N-terminus46. Our finding that regulation via the N-terminus does not equally affect all tRNA sequences will also provide new insight for future research on how METTL1 affects individual tRNAs and its implications on disease.
Methods
Protein expression and purification
Genes coding for human WDR4 and METTL1 were cloned into pETDuet (Novagen) and expressed in E.coli Rosetta (DE3) cells, grown in ZYM-5052 media47. Cells were harvested and lysed in buffer containing 50 mM Tris-HCl pH 7.5, 150 mM NaCl, and 1 mM DTT, supplemented with protease inhibitors using a sonicator. Clarified cell extract was used for Nickel affinity chromatography using Ni-NTA resin and eluted with imidazole. Eluted protein was then used for cation exchange chromatography and eluted using a linear gradient of NaCl. Fractions containing both polypeptides were pooled together and purified further using size-exclusion chromatography previously equilibrated with 50 mM Tris-HCl pH 7.5, 150 mM NaCl and 2 mM DTT. All protein constructs were purified similarly. Crystallization constructs contained residues 1–389 of WDR4 and 20–265 of METTL1. Cryo-EM constructs contained 1-389 of WDR4 and full-length METTL1.
Protein crystallization
The purified WDR4-METTL1 complex (residues 1-389 and 20-265 respectively) was concentrated to 20 mg/mL and used to set up crystallization experiments. Crystals were obtained using hanging drop vapor diffusion by mixing 1 μl of protein solution with 1 μL of 50 mM MES pH 6.0, 6 mM MgCl2, 10% Isopropanol, 2 mM reduced L-Glutathione, 2 mM oxidized L-Glutathione at room temperature. Crystals were washed three times in mother liquor and cryocooled in liquid nitrogen. METTL1 crystals were grown using hanging drop vapor diffusion by mixing 1 μL of purified METTL1 (20-265) in the presence of 2 mM SAM or 2 mM SAH with 1 μL 50 mM Sodium Cacodylate pH 5.7, 1.5 M Lithium Sulfate, 10 mM Magnesium Sulfate. METTL1 crystals were equilibrated on mother liquor supplemented with 20% glycerol and cryocooled in liquid nitrogen.
Crystallography
Crystal diffraction data was collected at the 19-ID beamline of the Advanced Photon Source (APS). Diffraction data was indexed, integrated and scaled using HKL3000 v.72248. The phase for WDR4-METTL1 was determined by molecular replacement, using PHASER49 with the S. cerevisiae Trm8-Trm82 structure (PDB: 2VDU) as a search model37. The phase information for METTL1-SAH and METTL1-SAM was obtained using molecular replacement of human METTL1 structure (PDB: 3CKK). The models for all three structures were further built using COOT and refined using Phenix50,51.
RNA in-vitro transcription
Human tRNA sequences were assembled using primer extension PCR (Supplementary Information Figure 1), and the DNA fragments were purified after agarose gel electrophoresis. Using the DNA templates, RNA was transcribed in vitro using recombinant T7 RNA polymerase. RNA transcripts were extracted using phenol-chloroform and precipitated with sodium acetate and ethanol, and then purified using denaturing polyacrylamide gel electrophoresis, using the crush and soak method as previously described52.
In vitro methylation assay
Methyltransferase activity reactions contained 150 nM tRNA substrate and 100 nM enzyme in 20 mM Tris pH 7.5, 0.01% Triton-X, 1 mM DTT, 1% glycerol, and 230 nM [3H]-SAM. Reactions were incubated at 37 °C for 1 hour. Reactions were blotted on Biodyne-B nylon membrane and the blotted membranes were washed with buffer containing 20 mM Tris-HCl pH 7.5, 0.01% Triton X-100. Incorporated tritium was measured using TriCarb 2010 TR Scintillation Counter (Perkin Elmer).
Electrophoretic mobility shift assay (EMSA)
Each purified tRNA was radiolabeled at the 5’ end with T4 polynucleotide kinase and [γ-32P] ATP. The concentrations of proteins are indicated in the figure legends, and the RNA concentration was 1 nM in all experiments. RNA-protein complexes were assembled in buffer containing 20 mM Tris-HCl pH 7.5, 1 mM DTT, 0.01% Triton X-100, 150 mM NaCl, 5% Glycerol, 2 mM MgCl2 and nonspecific DNA oligonucleotides. The reactions were incubated at RT for an hour and resolved using Tris-borate native PAGE. Phosphor screens were imaged using a Typhoon phosphorimager (GE) after overnight exposure. Bound and unbound bands were quantified by densitometry (ImageJ v.1.53) and curve-fitting was completed by Prism 9. Representative gels are shown and raw images are included in Supplementary Information Figure 2.
Cryo-EM sample preparation and data collection
The METTL1-WDR4-tRNA complex was assembled in a buffer containing 50 mM Tris-HCl pH 7.5, 100 mM NaCl, 2 mM DTT, and 1 mM MgCl2. Protein and RNA were mixed in a 1:1.2 molar ratio (protein:RNA) and incubated for 15 min on ice before applying to grids. For the SAM-bound complex, METTL1D163A was used instead of wild type, and SAM or SAH was added to the complex at 250 μM. Cryo-EM grids were prepared by applying 3 μL of the complex to a Quantifoil Au 1.2/1.3 200 mesh grid using Vitrobot previously equilibrated at 4 °C and 100% relative humidity. Data collection was performed on a Titan Krios equipped with a K3 bioquantum detector and an energy filter using SerialEM v4.0.853. Collected movies were preprocessed using Cryosparc v3.3.154 and the particles were picked using TOPAZ v0.2.455. 3D analysis was performed using Cryosparc. The METTL1-WDR4 crystal structure and a previously reported tRNALys structure were first docked into the cryo-EM map using ChimeraX56. The model was further built and refined using COOT v1.0.06 and Phenix v1.20.150,51,57. Sequence alignments were performed using Clustal Omega58.
Statistics and Reproducibility
All protein and RNA reagents were verified for purity to ensure >95% purity and homogeneity by using SDS-PAGE or Urea-PAGE, respectively. Using these purified reagents, all quantified biochemical assays were performed at least 3 times (3 technical replicates). For micrographs and gel pictures, only representative images are shown. For micrographs similar motion-corrected images were obtained hundreds of times as indicated in Extended Data Fig. 2–4. For SDS-PAGE or EMSA gels, at least 3 similar results were obtained. All attempts at replication were successful.
Extended Data
Extended Data Fig. 1: METTL1-WDR4 protein purification and tRNA complex reconstitution.
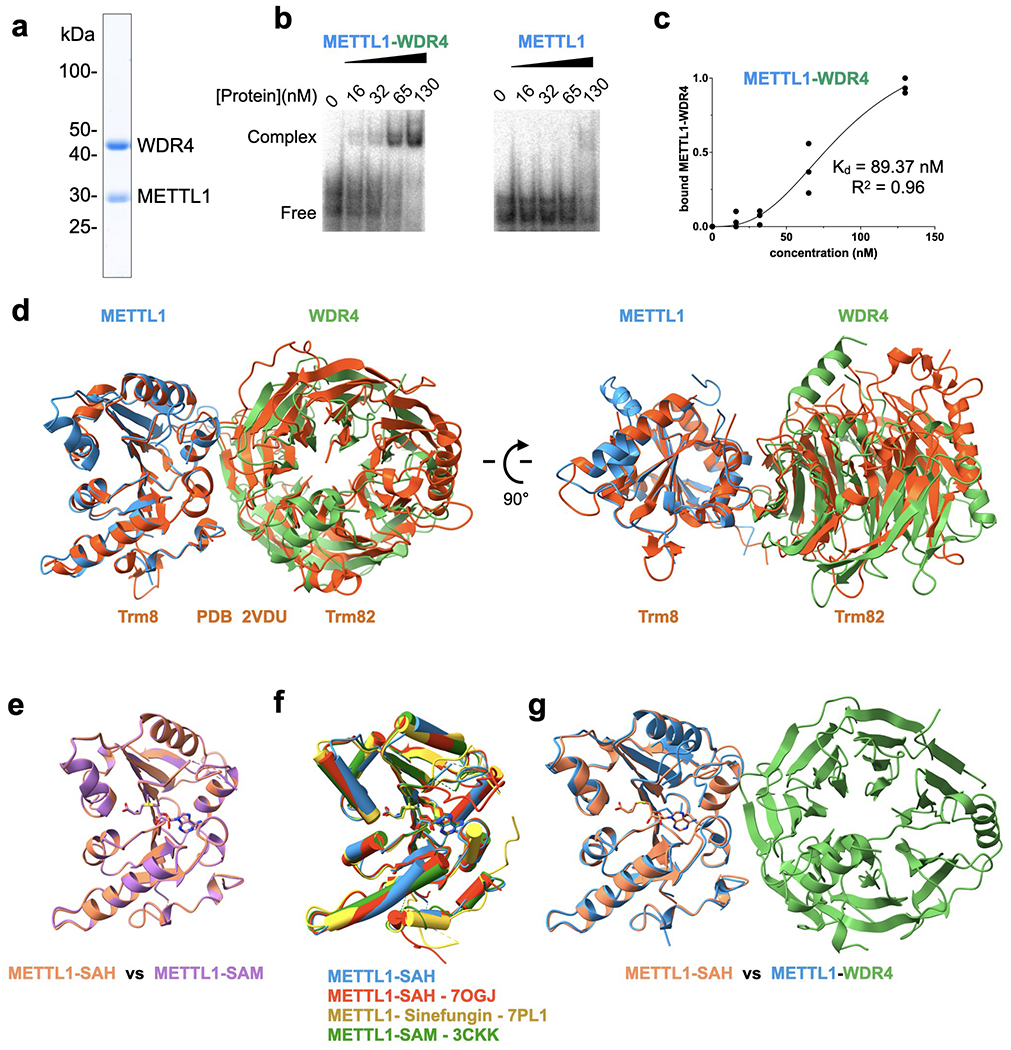
a, SDS-PAGE of purified full-length wild-type METTL1-WDR4 complex. A representative gel among 3 replicates is shown. b, EMSA to measure the affinity for tRNALys. For each gel, protein concentrations are 0, 16, 32, 65, and 130 nM, left to right. Representative gel among 3 replicates is shown. c, Quantification of EMSA for METTL1-WDR4 from 3 replicate experiments. d, Superimposition of yeast Trm8-Trm82 (PDB 2VDU, orange) onto the crystal structure of human METTL1-WDR4. The complex structures were superimposed by aligning METTL1 with Trm8. e-g, Superimposition of METTL1 structures as indicated. The structures missing PDB codes are from this study.
Extended Data Fig. 2: Cryo-EM data processing for the METTL1-WDR4-tRNALys structure.
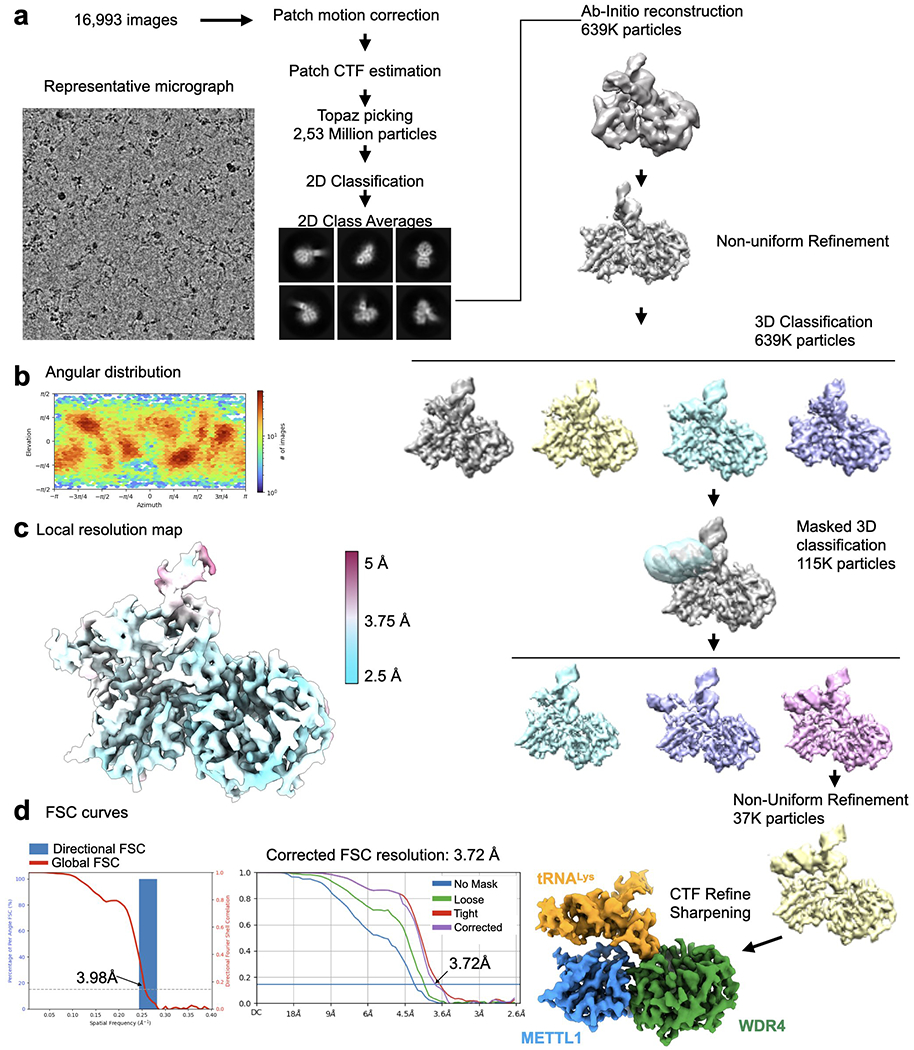
a, Cryo-EM data processing workflow. Left, a representative micrograph of the METTL1-WDR4-tRNA complex particles. A total of 16,993 images were used for picking and 2D classification. 2D class averages that showed high-resolution features were used for further 3D analysis using Cryosparc. 3D classification identified different populations that showed partial or no density around the anticodon arm. 3D classification applying the indicated mask revealed a homogeneous population of particles showing contiguous density in the tRNA region. b, Angular distribution plot. c, Local resolution map shown with colors on the sharpened map. d, Directional FSC plot and FSC curves showing the resolution at 0.143 cutoff.
Extended Data Fig. 3: Cryo-EM data processing for the METTL1-WDR4-tRNALys-SAM structure.
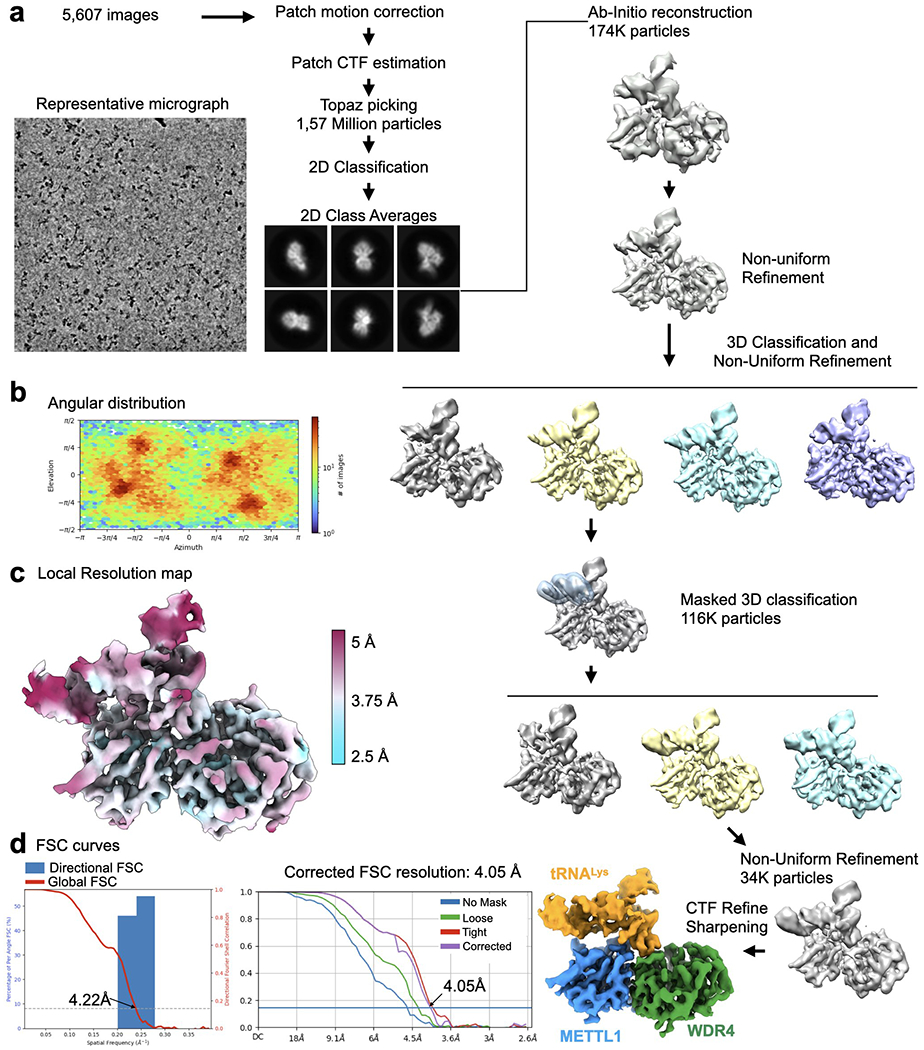
a, Cryo-EM data processing workflow. Left, a representative micrograph of the METTL1-WDR4-tRNA-SAM complex particles. A total of 5,607 images were used for picking and 2D classification. 2D class averages that showed high-resolution features were used for further 3D analysis using Cryosparc. 3D classification identified different populations that showed partial density around the anticodon arm. 3D classification applying the indicated mask revealed a homogeneous population of particles showing contiguous density in the tRNA region. b, Angular distribution plot. c, Local resolution map shown with colors on the sharpened map. d, Directional FSC plot and FSC curves showing the resolution at 0.143 cutoff.
Extended Data Fig. 4: Cryo-EM data processing for the METTL1-WDR4-tRNALys-SAH structure.
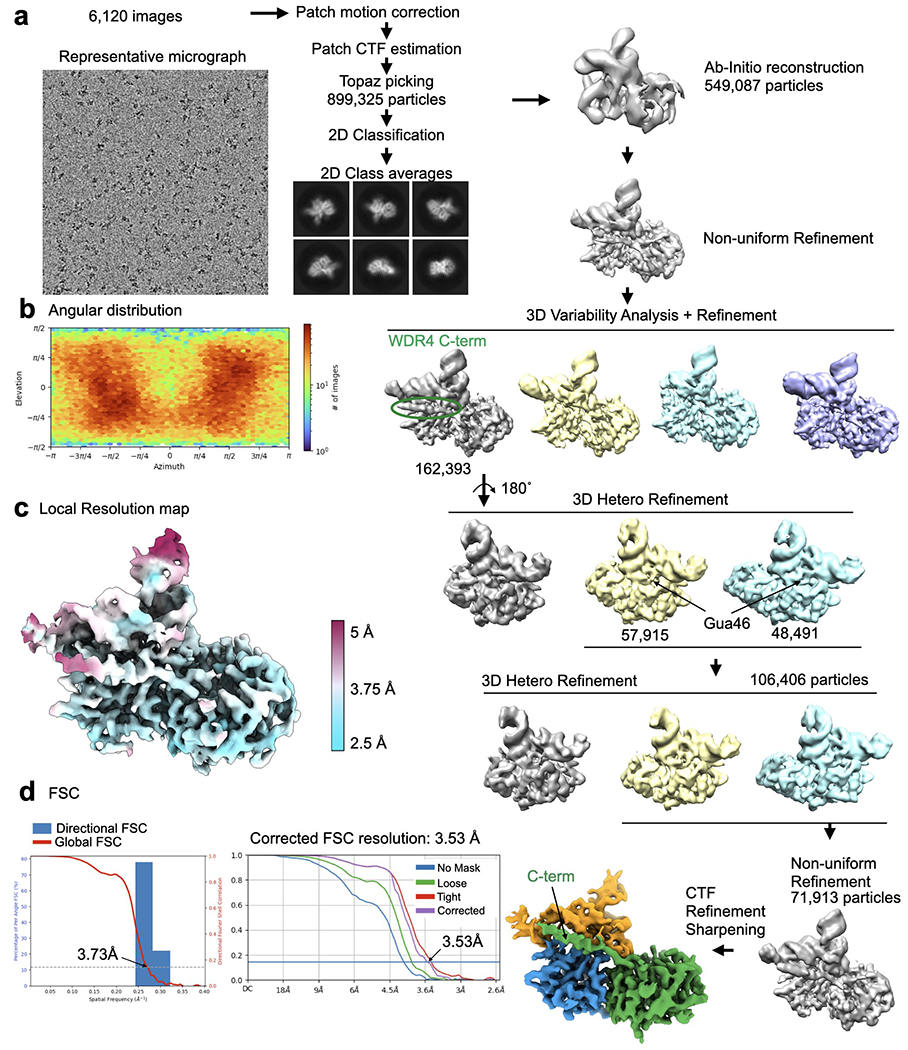
a, Cryo-EM data processing workflow. Left, a representative micrograph of the METTL1-WDR4-tRNA-SAH complex particles. A total of 6,120 images were used for picking and 2D classification. 2D class averages that showed high-resolution features were used for further 3D analysis using Cryosparc. 3D variability analysis identified a population of particles that showed consistent density for the WDR4 C-terminal helix and several rounds of 3D Heterogeneous Refinement identified particles showing prominent density for G46. b, Angular distribution plot. c, Local resolution map shown with colors on the sharpened map. d, Directional FSC plot and FSC curves showing the resolution at 0.143 cutoff.
Extended Data Fig. 5: Conformational changes in structures containing the METTL1-WDR4-tRNA complex.
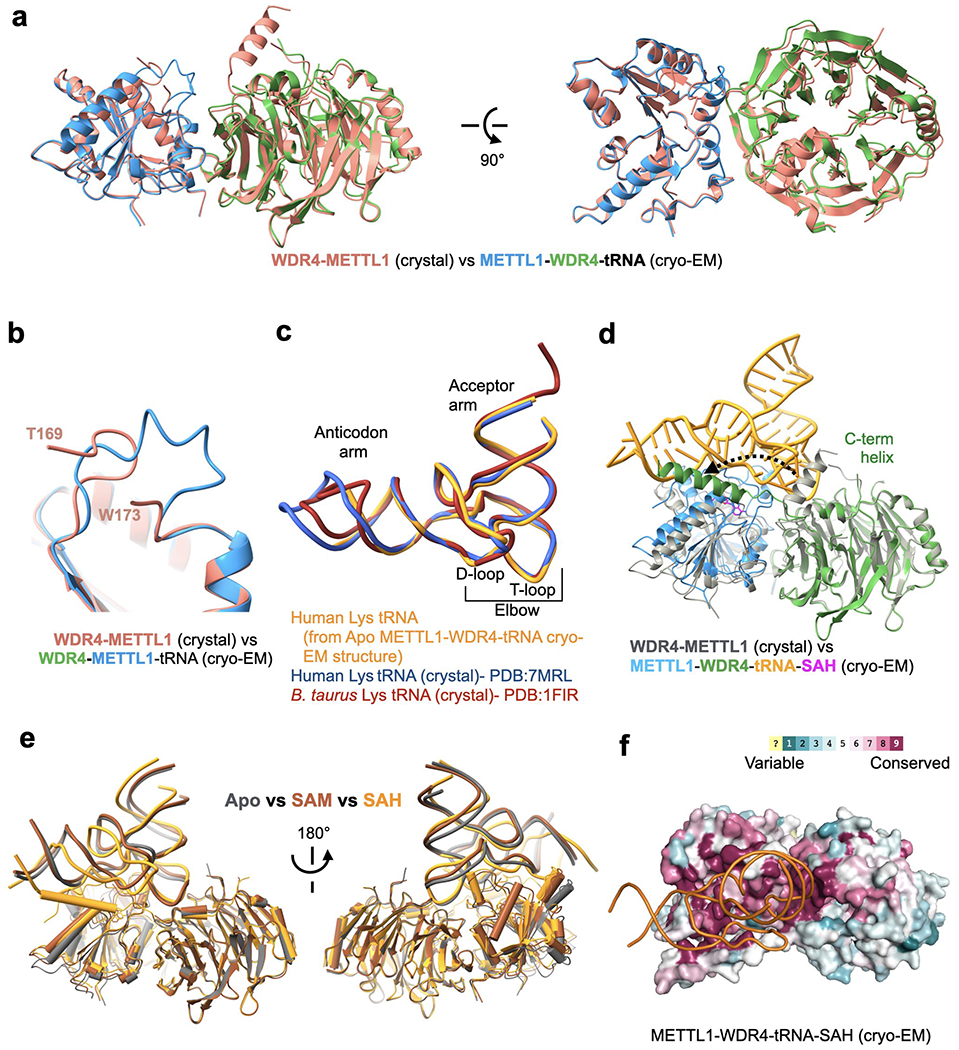
a, Superimposition of the METTL1-WDR4 crystal structure on the METTL1-WDR4-tRNA cryo-EM structure, aligned by METTL1. b, Flexible loop of METTL1 (161-175, catalytic loop) becomes more ordered with RNA. c, Superimposition of different tRNALys structures. All three have the same sequence except the tips of the anticodon arm and the acceptor arm. The B. Taurus structure (PDB:1FIR) was with a fully modified tRNA and the other two structures were obtained for unmodified RNA after in vitro transcription. d, Superimposition of the METTL1-WDR4 crystal structure (gray) onto the SAH-bound quaternary complex cryo-EM structure (multiple colors), aligned by WDR4. Movement of the WDR4 C-terminal helix upon binding RNA is shown with a dashed arrow. e, Superimposition of all three cryo-EM structures (colored by state) presented in this study, aligned by WDR4. f. Surface representation of the SAH-bound cryo-EM structure colored by evolutionary sequence conservation (Consurf server). The orientation is identical to Fig. 3a and 3b.
Extended Data Fig. 6: Structural and sequence organization of tRNA.
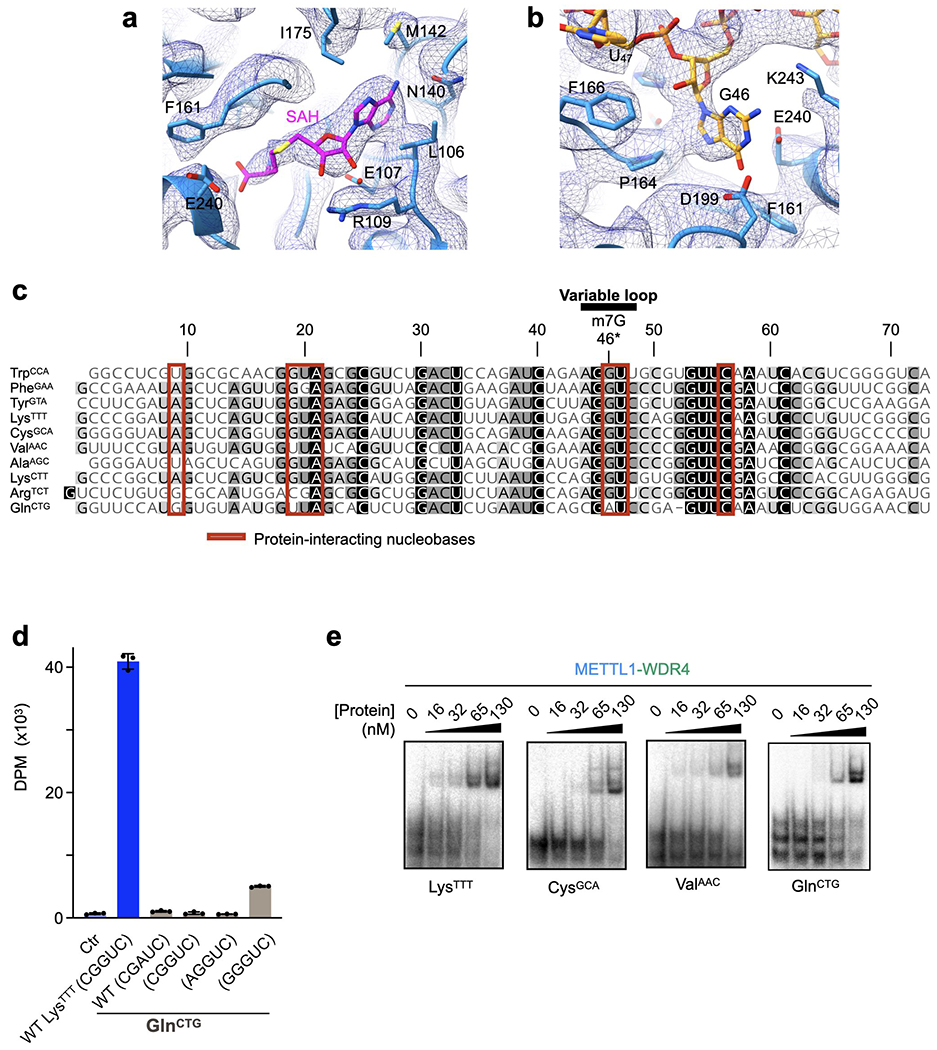
a-b, Sharpened cryo-EM map (mesh) near the SAH-binding site (a) and the G46 binding pocket (b). c, Sequence alignment of the human tRNAs used in this study shaded by conservation. Red boxes indicate nucleobases within 4 Å of protein. Sequences were aligned using Clustal Omega and visualized by Geneious Prime. d, In vitro methylation activity of full-length METTL1-WDR4 for the indicated tRNAs with the specified variable loop sequences, shown as mean ± SD from 3 replicates. e, EMSA using METTL1-WDR4 with different tRNAs shows no dramatic differences in affinities. Representative images from 3 replicate experiments are shown. For each gel, protein concentrations are 0, 16, 32, 65, and 130 nM, left to right.
Extended Data Fig. 7: Sequence alignment of METTL1 protein homologs.
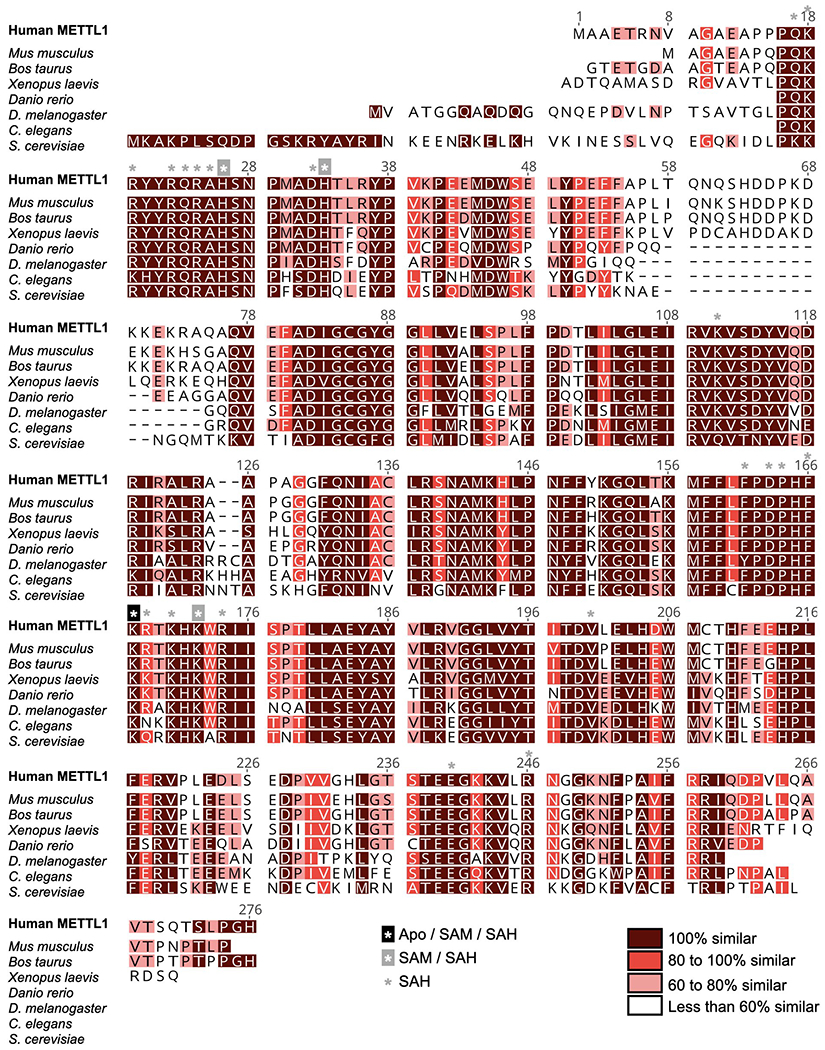
Sequences used are from human (Q9UBP6), Mus musculus (Q9Z120), Bos taurus (Q2YDF1), Xenopus laevis (Q6NU94), Danio rerio (Q5XJ57), Drosophila melanogaster (O77263), Caenorhabditis elegans (Q23126) and Saccharomyces cerevisiae (Q12009) METTL1. Sequences were aligned using Clustal Omega and visualized by Geneious Prime. Residues within 4 Å of RNA in different states are indicated with asterisks.
Extended Data Fig. 8: Sequence alignment of WDR4 protein homologs.
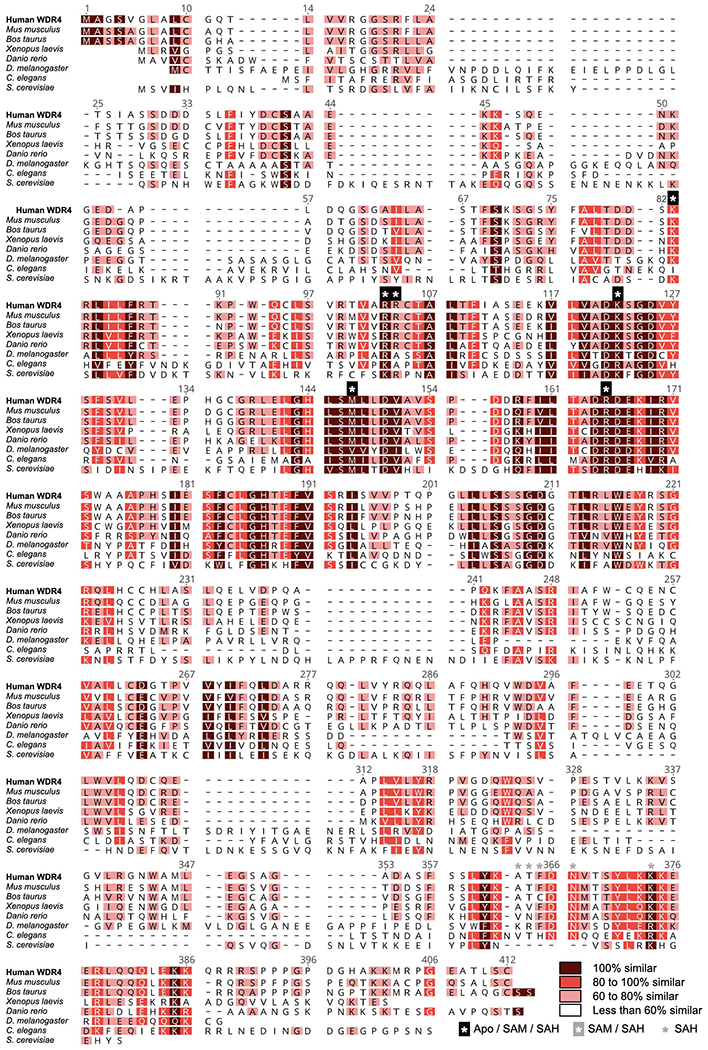
Sequences used are from human (P57081), Mus musculus (Q9EP82), Bos taurus (A7E3S5), Xenopus laevis (Q7ZY78), Danio rerio (A4IGH4), Drosophila melanogaster (Q9W415), Caenorhabditis elegans (Q23232) and Saccharomyces cerevisiae (A6ZYC3) WDR4. Sequences were aligned using Clustal Omega and visualized by Geneious Prime. Residues within 4 Å of RNA in different states are indicated with asterisks.
Extended Data Table 1:
Crystallography Data Collection and Refinement Statistics.
| METTL1-WDR4 (PDB-ID: 8D58) |
METTL1-SAM (PDB-ID: 8D59) |
METTL1-SAH (PDB-ID: 8D5B) |
|
|---|---|---|---|
| Data collection | |||
| Space group | P21 21 21 | P61 | P61 |
| Cell dimensions | |||
| a, b, c (Å) | 79.49, 91.11, 122.83 | 86.02, 86.02, 65.57 | 86.03, 86.03, 65.48 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Wavelength (Å) | 0.9791 | 0.9789 | 0.9789 |
| Resolution (Å) | 42.71- 2.45 (2.49- 2.45) * | 37.25- 2.25 (2.29- 2.25) * | 37.25- 1.93 (1.96- 1.93) * |
| R merge | 0.242 (3.76) | 0.292 (1.89) | 0.242 (3.22) |
| I/σI | 17.48 (0.8) | 13.5 (1.2) | 16.24 (1.3) |
| CC1/2 | 0.998 (0.319) | 0.983 (0.414) | 0.996 (0.450) |
| Completeness (%) | 99.9 (100.0) | 99.2 (91.6) | 100.0 (99.9) |
| Redundancy | 11.9 (8.2) | 15.5 (5.7) | 18.1 (10.5) |
| Refinement | |||
| Resolution (Å) | 42.71- 2.47 | 37.25- 2.26 | 37.25- 1.93 |
| No. reflections | 32035 | 12757 | 20756 |
| Rwork / Rfree | 0.203 / 0.229 | 0.207 / 0.229 | 0.180 / 0.212 |
| No. atoms | |||
| Protein | 4320 | 1601 | 1620 |
| Ligand/ion | 20 | 49 | 28 |
| Water | 60 | 53 | 144 |
| B-factors | |||
| Protein | 64.87 | 53.05 | 35.80 |
| Ligand/ion | 78.27 | 68.69 | 63.84 |
| Water | 60.05 | 52.39 | 44.00 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.002 | 0.002 | 0.006 |
| Bond angles (°) | 0.436 | 0.546 | 0.951 |
| Ramachandran agreement | |||
| favored (%) | 97.79 | 98.97 | 99.48 |
| allowed (%) | 2.21 | 1.03 | 0.52 |
| outliers (%) | 0.00 | 0.00 | 0.00 |
Values in parentheses are for the highest resolution shell.
Extended Data Table 2:
Cryo-EM Data Collection, Refinement, and Validation Statistics.
| METTL1-WDR4-tRNA (EMDB-27264) (PDB 8D9K) |
METTL1-WDR4-tRNA-SAM (EMDB-27265) (PDB 8D9L) |
METTL1-WDR4-tRNA-SAH (EMDB-28108) (PDB 8EG0) |
|
|---|---|---|---|
| Data collection and processing | |||
| Microscope | Titan Krios | Titan Krios | Titan Krios |
| Magnification | 105,000x | 105,000x | 105,000x |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e–/Å2) | 62 | 62 | 62 |
| Exposure time (s) | 5.5 | 5.5 | 5.5 |
| Defocus range (μm) | 0.7 – 2.2 | 0.7 – 2.2 | 0.7 – 2.2 |
| Pixel size (Å) | 0.415 | 0.415 | 0.415 |
| Number of frames | 62 | 62 | 62 |
| Energy filter slit width (keV) | 20 | 20 | 20 |
| Symmetry imposed | CI | CI | CI |
| Number of micrographs | 16,993 | 5,607 | 6,120 |
| Initial particle images (no.) | 2,531,779 | 395,556 | 899,325 |
| Particles used for 3DVA (no.) | 1,571,798 | 321,258 | 594,087 |
| Final particle images (no.) | 37,033 | 34,558 | 71,913 |
| Map resolution (Å) | 3.72 / 4.20 | 4.05/4.70 | 3.53/4.10 |
| 0.143 FSC (masked/unmasked) | |||
| Map resolution range (Å) | 2.5-4 | 3-5 | 2.5-4 |
| Refinement | |||
| Software | Phenix | Phenix | Phenix |
| Initial model used (PDB code) | 8D58 – 7MRL | 8D58 – 7MRL | 8D58 – 7MRL |
| Model resolution (Å) | 3.7 (0.143) | 4.1 (0.143) | 3.5 (0.143) |
| FSC threshold | |||
| Model resolution range (Å) | 2.5-4 | 3-5 | 2.5-4 |
| Map sharpening B factor (Å2) | −138 | −168 | −149 |
| Model composition | |||
| Non-hydrogen atoms | 5708 | 5757 | 6031 |
| Protein residues | 546 | 553 | 584 |
| Nucleotide residues | 65 | 65 | 65 |
| Ligands | None | SAM | SAH |
| B factors (Å2) | |||
| Protein | 160.96 | 245.13 | 182.88 |
| Nucleotide | 255.72 | 338.24 | 248.42 |
| Ligand | 255.74 | 182.8 | |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.001 | 0.001 | 0.002 |
| Bond angles (°) | 0.364 | 0.385 | 0.420 |
| Validation | |||
| MolProbity score | 1.35 | 1.40 | 1.43 |
| Clashscore | 6.27 | 7.25 | 7.87 |
| Poor rotamers (%) | 0.64 | 0.43 | 0.00 |
| Ramachandran plot | |||
| Favored (%) | 98.13 | 98.15 | 99.47 |
| Allowed (%) | 1.87 | 1.85 | 0.53 |
| Disallowed (%) | 0.00 | 0.00 | 0.00 |
Supplementary Material
Acknowledgements
We thank the Structural Biology Laboratory and the Cryo-Electron Microscopy Facility (supported by the Cancer Prevention Research Institute of Texas (CPRIT) RP170644) at UT Southwestern for support with synchrotron and cryo-EM data collection. The use of the SBC 19ID beamline at Advanced Photon Source is supported by the United States Department of Energy contract DE-AC02-06CH11357. This work was supported by the National Institutes of Health (R01GM122960 and R01CA258589), Cancer Prevention Research Institute of Texas (RP190259), and the Welch Foundation (I-2115-20220331). Y.N. is a Packard Fellow, Pew Scholar, and Southwestern Medical Foundation Scholar in Biomedical Research.
Footnotes
Competing Interests
The authors declare no competing interests.
Data Availability
The atomic models for METTL1-WDR4 (8D58), METTL1-WDR4-tRNA (8D9K), METTL1-SAM (8D59), METTL1-SAH (8D5B), METTL1-WDR4-tRNA-SAM (8D9L), METTL1-WDR4-tRNA-SAH (8EG0) are deposited in the Protein Data Bank (PDB: https://www.rcsb.org/). Cryo-EM maps and masks of METTL1-WDR4-tRNA (EMD-27264), METTL1-WDR4-tRNA-SAM (EMD-27265), and METTL1-WDR4-tRNA-SAH (EMD-28108) used to build the models are deposited in the Electron Microscopy Data Bank (EMDB: https://www.ebi.ac.uk/pdbe/emdb/).
References
- 1.Frye M, Harada BT, Behm M & He C RNA modifications modulate gene expression during development. Science 361, 1346–1349 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiener D & Schwartz S The epitranscriptome beyond m(6)A. Nat Rev Genet 22, 119–131 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Pan T Modifications and functional genomics of human transfer RNA. Cell Res 28, 395–404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki T The expanding world of tRNA modifications and their disease relevance. Nat Rev Mol Cell Biol 22, 375–392 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Alexandrov A, Martzen MR & Phizicky EM Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8, 1253–1266 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexandrov A et al. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21, 87–96 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Filonava L, Torres AG & Ribas de Pouplana L A novel cause for primordial dwarfism revealed: defective tRNA modification. Genome Biol 16, 216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaheen R et al. Mutation in WDR4 impairs tRNA m(7)G46 methylation and causes a distinct form of microcephalic primordial dwarfism. Genome Biol 16, 210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trimouille A et al. Further delineation of the phenotype caused by biallelic variants in the WDR4 gene. Clin Genet 93, 374–377 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Chen X et al. Speech and language delay in a patient with WDR4 mutations. Eur J Med Genet 61, 468–472 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Orellana EA et al. METTL1-mediated m(7)G modification of Arg-TCT tRNA drives oncogenic transformation. Mol Cell 81, 3323–3338 e3314 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai Z et al. N(7)-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol Cell 81, 3339–3355 e3338 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Ying X et al. METTL1-m(7) G-EGFR/EFEMP1 axis promotes the bladder cancer development. Clin Transl Med 11, e675 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han H et al. N(7)-methylguanosine tRNA modification promotes esophageal squamous cell carcinoma tumorigenesis via the RPTOR/ULK1/autophagy axis. Nat Commun 13, 1478 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z et al. METTL1 promotes hepatocarcinogenesis via m(7) G tRNA modification-dependent translation control. Clin Transl Med 11, e661 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J et al. Aberrant translation regulated by METTL1/WDR4-mediated tRNA N7-methylguanosine modification drives head and neck squamous cell carcinoma progression. Cancer Commun (Lond) 42, 223–244 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C et al. Methyltransferase-like 1 regulates lung adenocarcinoma A549 cell proliferation and autophagy via the AKT/mTORC1 signaling pathway. Oncol Lett 21, 330 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma J et al. METTL1/WDR4-mediated m(7)G tRNA modifications and m(7)G codon usage promote mRNA translation and lung cancer progression. Mol Ther 29, 3422–3435 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y et al. Overexpressed methyltransferase-like 1 (METTL1) increased chemosensitivity of colon cancer cells to cisplatin by regulating miR-149-3p/S100A4/p53 axis. Aging (Albany NY) 11, 12328–12344 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian QH et al. METTL1 overexpression is correlated with poor prognosis and promotes hepatocellular carcinoma via PTEN. J Mol Med (Berl) 97, 1535–1545 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Chen B et al. N(7)-methylguanosine tRNA modification promotes tumorigenesis and chemoresistance through WNT/beta-catenin pathway in nasopharyngeal carcinoma. Oncogene 41, 2239–2253 (2022). [DOI] [PubMed] [Google Scholar]
- 22.Luo Y et al. The potential role of N(7)-methylguanosine (m7G) in cancer. J Hematol Oncol 15, 63 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki T Fine-Tuning of RNA Functions by Modification and Editing. Topics in Current Genetics, 23–69 (2005). [Google Scholar]
- 24.Motorin Y & Helm M tRNA stabilization by modified nucleotides. Biochemistry 49, 4934–4944 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Lorenz C, Lunse CE & Morl M tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules 7, 35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohira T et al. Reversible RNA phosphorylation stabilizes tRNA for cellular thermotolerance. Nature 605, 372–379 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonkhout N et al. The RNA modification landscape in human disease. RNA 23, 1754–1769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirchner S & Ignatova Z Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet 16, 98–112 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Boccaletto P et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res 50, D231–D235 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Juhling F et al. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37, D159–162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexandrov A, Grayhack EJ & Phizicky EM tRNA m7G methyltransferase Trm8p/Trm82p: evidence linking activity to a growth phenotype and implicating Trm82p in maintaining levels of active Trm8p. RNA 11, 821–830 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J, Hou JH & Hsieh TS A new Drosophila gene wh (wuho) with WD40 repeats is essential for spermatogenesis and has maximal expression in hub cells. Dev Biol 296, 219–230 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin S et al. Mettl1/Wdr4-Mediated m(7)G tRNA Methylome Is Required for Normal mRNA Translation and Embryonic Stem Cell Self-Renewal and Differentiation. Mol Cell 71, 244–255 e245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng Y, Zhou Z, Ji W, Lin S & Wang M METTL1-mediated m(7)G methylation maintains pluripotency in human stem cells and limits mesoderm differentiation and vascular development. Stem Cell Res Ther 11, 306 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Bie LG et al. The yggH gene of Escherichia coli encodes a tRNA (m7G46) methyltransferase. J Bacteriol 185, 3238–3243 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou H et al. Monomeric tRNA (m(7)G46) methyltransferase from Escherichia coli presents a novel structure at the function-essential insertion. Proteins 76, 512–515 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Leulliot N et al. Structure of the yeast tRNA m7G methylation complex. Structure 16, 52–61 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Cartlidge RA et al. The tRNA methylase METTL1 is phosphorylated and inactivated by PKB and RSK in vitro and in cells. EMBO J 24, 1696–1705 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okamoto M et al. tRNA modifying enzymes, NSUN2 and METTL1, determine sensitivity to 5-fluorouracil in HeLa cells. PLoS Genet 10, e1004639 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benas P et al. The crystal structure of HIV reverse-transcription primer tRNA(Lys,3) shows a canonical anticodon loop. RNA 6, 1347–1355 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bou-Nader C et al. HIV-1 matrix-tRNA complex structure reveals basis for host control of Gag localization. Cell Host Microbe 29, 1421–1436 e1427 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finer-Moore J, Czudnochowski N, O’Connell JD 3rd, Wang AL & Stroud RM Crystal Structure of the Human tRNA m(1)A58 Methyltransferase-tRNA(3)(Lys) Complex: Refolding of Substrate tRNA Allows Access to the Methylation Target. J Mol Biol 427, 3862–3876 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blersch KF et al. Structural model of the M7G46 Methyltransferase TrmB in complex with tRNA. RNA Biol 18, 2466–2479 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsumoto K et al. RNA recognition mechanism of eukaryote tRNA (m7G46) methyltransferase (Trm8-Trm82 complex). FEBS Lett 581, 1599–1604 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Schultz SK & Kothe U tRNA elbow modifications affect the tRNA pseudouridine synthase TruB and the methyltransferase TrmA. RNA 26, 1131–1142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akimov V et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat Struct Mol Biol 25, 631–640 (2018). [DOI] [PubMed] [Google Scholar]
References for Methods
- 47.Studier FW Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Minor W, Cymborowski M, Otwinowski Z & Chruszcz M HKL-3000: the integration of data reduction and structure solution – from diffraction images to an initial model in minutes. Acta Crystallographica Section D: Biological Crystallography 62, 859–866 (2006). [DOI] [PubMed] [Google Scholar]
- 49.McCoy AJ et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petrov A, Wu T, Puglisi EV & Puglisi JD RNA purification by preparative polyacrylamide gel electrophoresis. Methods Enzymol 530, 315–330 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Bepler T et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat Methods 16, 1153–1160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pettersen EF et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murshudov GN et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sievers F et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic models for METTL1-WDR4 (8D58), METTL1-WDR4-tRNA (8D9K), METTL1-SAM (8D59), METTL1-SAH (8D5B), METTL1-WDR4-tRNA-SAM (8D9L), METTL1-WDR4-tRNA-SAH (8EG0) are deposited in the Protein Data Bank (PDB: https://www.rcsb.org/). Cryo-EM maps and masks of METTL1-WDR4-tRNA (EMD-27264), METTL1-WDR4-tRNA-SAM (EMD-27265), and METTL1-WDR4-tRNA-SAH (EMD-28108) used to build the models are deposited in the Electron Microscopy Data Bank (EMDB: https://www.ebi.ac.uk/pdbe/emdb/).


