Abstract
Organoids are widely used as a model system to study gut pathophysiology; however, they fail to fully reproduce the complex, multi-component structure of the intestinal wall. We present here a new gut on chip model that allows the co-culture of primary epithelial and stromal cells. The device has the topography and dimensions of the mouse gut and is based on a 3D collagen I scaffold. The scaffold is coated with a thin layer of laminin to mimic the basement membrane. To maintain the scaffold structure while preserving its cytocompatibility, the collagen scaffold was rigidified by threose-based post-polymerization treatment. This treatment being cytocompatible enabled the incorporation of primary intestinal fibroblasts inside the scaffold, reproducing the gut stromal compartment. We observed that mouse organoids, when deposited into crypts, opened up and epithelialized the scaffold, generating a polarized epithelial monolayer. Proper segregation of dividing and differentiated cells along the crypt–villus axis was achieved under these conditions. Finally, we show that the application of fluid shear stress allows the long-term culture of this intestinal epithelium. Our device represents a new biomimetic tool that captures key features of the gut complexity and could be used to study gut pathophysiology.
We report on a new gut on chip combining the co-culture of primary epithelial and stromal cells in 3D biomimetic scaffold. Proper segregation of dividing and differentiated cells along the crypt-villus axis was achieved in these unique conditions.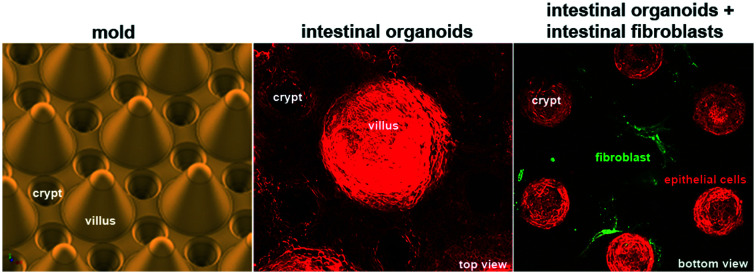
Introduction
Though still considered as the gold standard, animal testing faces several issues such as being time-consuming, costly, and often ineffective. The pharmaceutical industry has shifted towards testing toxicology and drug efficiency in vitro. However, results obtained with 2D in vitro models often fail to predict faithfully clinical response.1 Thus, there is a strong interest to develop more relevant alternative 3D in vitro models such as organoids2,3 or organs-on-chip4,5 that aim at recapitulating cellular, physical, biochemical and geometrical features of a specific organ or tissue in a microfluidic device. These approaches also enable the control of all system parameters independently, and allow real-time high-resolution imaging and extended functional analysis. Finally, because organ-on-chips are microfluidic-based approaches, they allow precise control of physical constraints or the implementation of chemical gradients.6 Organ-on-chip technology has a wide-range utility in all aspects of fundamental and pre-clinical research.7,8
The gut is an important organ as it acts as a barrier between the outside world and the body and has a role in food digestion and nutrient absorption. The small intestine is composed of villi that project into the lumen of the gut and crypts that descend into the stroma. They are overlaid with the monolayer of columnar epithelial cells. The integrity of this epithelium is maintained through its continuous renewal.9 Dividing stem cells are contained within the crypts and they give rise to specialized epithelial cells.9,10 Most of these differentiated cells travel upwards from the stem cell compartment towards the villus tip, where they are shed into the lumen.10 The basal surface of the epithelium is underlined by the basement membrane, a thin and dense sheet-like structure mainly composed of collagen IV and laminin.11 The basement membrane provides structural support for the epithelium, maintains cell polarity and has a role in the compartmentalization of the tissue by separating the epithelium from the stroma.11 The major components of the stroma are fibroblasts, immune cells, blood vessels and the ECM, which consists mostly of collagen I fibers. Among the different stromal components, fibroblasts play an important role in gut homeostasis in particular by producing growth factors, cytokines and ECM proteins.12–14
One of the most commonly used intestinal in vitro models is gut organoids,3,15 which are shown to be a more physiologically relevant model compared to 2D cell cultures.16 Organoids are derived from isolated intestinal crypts and they exhibit proper differentiation of intestinal epithelial cells. However, they fail to fully reproduce the crypt–villus architecture, as villi are represented, inadequately, by short, flat stretches of cells. Moreover, the organoids are closed structures, which prevent studies of the passage from the lumen through the epithelium. Microfluidics-based alternative gut-on-chip methods bring to this advanced 3D model a higher level of physiological relevance by integrating flow control, mechanical forces and chemical gradients. One of these approaches consists in growing cells on porous flat substrates while applying mechanical forces and biochemical gradients. In the pioneering and well-established device developed by Ingber's team, cells are grown on a PDMS membrane coated with ECM proteins.17,18 This platform is compelling, in particular, to study the interactions between microbes and the epithelium.17,19 The absence of crypts, however, limits the use of this device to investigate gut homeostasis. The second approach is based on growing cells on synthetic or bio-derived materials microstructured to recapitulate the gut topography. Developing such a 3D scaffold is a significant bioengineering challenge, as the scaffold should combine a high aspect ratio and biocompatibility. Synthetic polymers, like PDMS20 or biodegradable materials, such as poly(lactic-co-gycolic)acid,21,22 do not faithfully recapitulate the mechanical and chemical properties of the gut and its topography. The ability to photo-polymerize polyethyleneglycoldiacrylate (PEGDA) has been recently exploited to generate a 3D gut scaffold with high aspect ratio; however, PEGDA scaffolds are, so far, limited only to the use of epithelial cells.23,24 Natural materials such as collagen type I was also used to generate scaffolds.25 However, to restrict the scaffold deformation due to forces exerted by the cells, the chemical treatment of the scaffold was required. As chemical treatments are not cytocompatible, this precludes the integration of stromal cells within the scaffold. Thus, despite being a very active field that uses cutting edge technologies, none of the current gut-on-chip approaches provides cell substrates that support the 3D structure of the small intestine while allowing incorporation of stromal cells.
In this study, we report a new gut-on-chip device, which allows co-culture in a biomimetic 3D scaffold of both epithelial and stromal cells present in the small intestine. To achieve this, we first designed a mold with the typical topography of the mouse gut, which was used to cast the 3D scaffold entirely made of collagen I. The scaffold was then treated with threose to cross-link collagen, which was crucial for maintaining the topography of the device while retaining its cytocompatibility. This cross-linking approach enabled us to incorporate in the scaffold stromal cells, such as primary mouse intestinal fibroblasts. Then, in order to reconstitute the intestinal epithelium, we seeded mouse intestinal organoids on the scaffold and characterized the epithelialization process. Because intestinal cells experience forces in vivo, we subjected the device to shear stress, and investigated its influence on long-term cell culture. Finally, we evaluated how, in the presence of primary intestinal fibroblasts within the 3D collagen scaffold, the epithelial cell differentiation and spatial segregation can be achieved as found in vivo.
Materials and methods
Microstructuring collagen scaffolds
Collagen I was extracted from rat tails as described previously26 and labeled with tetramethyl rhodamine (TAMRA, Invitrogen) as described.27 TAMRA labelled collagen was mixed into non-labelled collagen in a weight ratio of 1 : 10. A neutralizing solution (1× PBS, 10% FBS, DMEM glutamax, MEM non-essential amino acids (Gibco), 1 mM sodium pyruvate (Gibco)) was prepared in another tube and the pH was adjusted to 7.4 with 1 M NaOH. The neutralizing reagent solution was mixed on ice and added to the top of the collagen solution. The gel was gently mixed on ice with a sterile spatula to avoid bubble formation. If bubbles formed, the mixture was centrifuged at 1000 g at 4 °C for 1 minute. The pH of the solution was controlled at 7.4 before cells were added. For fibroblast conditions, a fibroblast suspension is mixed with neutralized collagen, before being injected into the chip, to reach a final concentration of fibroblasts of 5 × 105 cells per mL.
Microstructuring of collagen to replicate the 3D gut of the mouse gut consisted of several steps (Fig. 1A). The first step consisted of creating a brass mold. This was achieved by micromilling (Minitech). To maintain sterility the collagen was not molded directly onto the brass but instead onto a PDMS replica prepared with PDMS (Sylgard 184, Dow Corning) with a prepolymer/curing agent at a ratio of 10 : 1 (w/w). The PDMS mold was made of two parts. One part (mold 1) consisted of a chamber (1 cm × 2 cm × 3 mm) that permits both collagen injection and the handling of the collagen structure as the collagen structure was covalently grafted to the bottom part of this chamber. The second part (mold 2) consisted of a replica of the gut 3D structure and was used to mold the collagen. To allow collagen covalent bonding to the bottom of the PDMS chamber, the surface of mold 1 was plasma activated for 30 s (plasma cleaner, Harrick), the edges being protected by scotch tape to avoid further covalent bonding. This was done by filling the microfluidic chamber with 2% v/v of 3-aminopropyl-triethoxysilane (APTES) for 30 min at RT. After removing the APTES solution, the chamber was washed in water for 5 min, followed by incubation in 0.5% v/v glutaraldehyde for 30 min. Finally, the microfluidic chamber was washed twice in water and incubated in water overnight at 4 °C. To prepare mold 1 for collagen injection, its lower and upper parts were punched with a 4 mm puncher. The structured area of mold 2 was plasma activated, with the area around the microstructures screened with scotch tape. This activation improved the structure wettability and decreased the likelihood of trapped air bubbles.
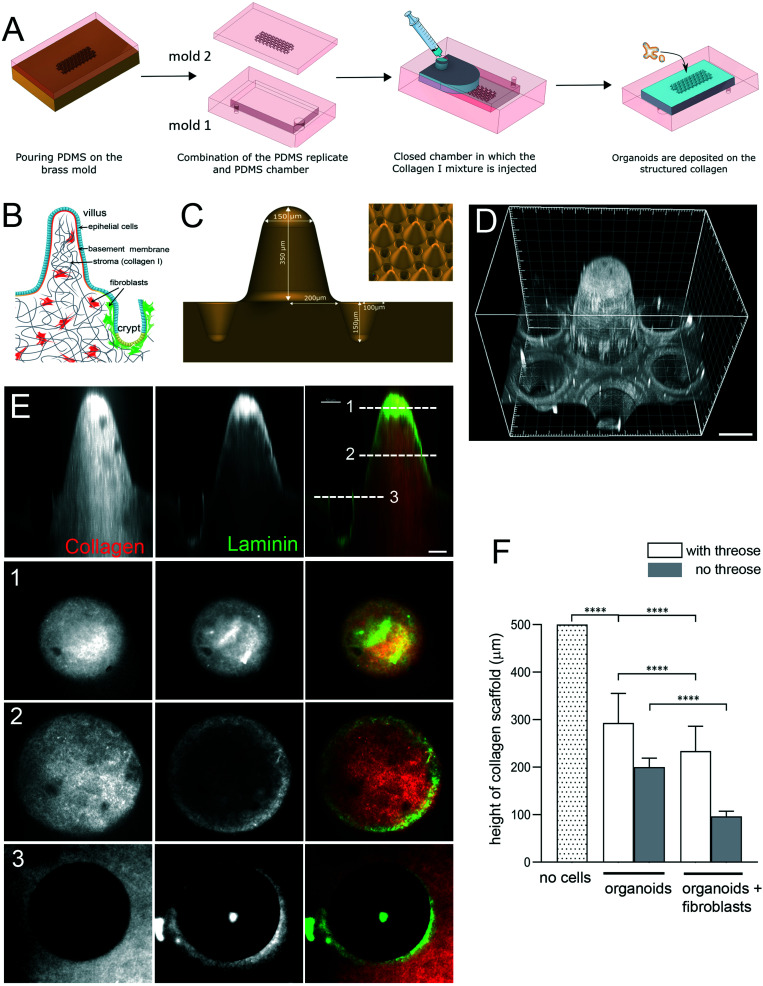
Fig. 1. Engineering microstructured scaffold. A. Scaffold microfabrication: PDMS (mold 2) replica of the micromilled brass mold is microfabricated. This PDMS replica is then non-covalently sealed with a second PDMS mold (mold 1) that consists in an open chamber (previously treated with APTES and glutaraldehyde). Collagen I solution is then injected within this closed device using a syringe. After collagen polymerization at 37 °C, the top part of the device is removed and organoids are seeded on top of the collagen 3D scaffold. B. Schematic organization of the small intestine. Simple columnar epithelium (blue) covers finger-like projections – villi and invaginations – crypts. Epithelial cells are attached to the basement membrane. Stroma, mostly made of collagen type I (gray), contains fibroblasts (red and green). C. Schematics of the brass mold used to prepare the PDMS intermediate replica. Villus height 350 μm, crypt depth 150 μm. Inset, arrays of villi and crypts. D. PDMS mold coated with laminin showing one unit consisting of one villus surrounded by six crypts. Scale bar, 150 μm. E. A scaffold made of TAMRA-labelled collagen type I (red) coated with Cy3-labeled laminin (green). Side view. Cross-sections at the top (1) and middle (2) of the villi and plateau/opening of the crypt (3). Scale bar, 50 μm. F. Collagen cross-linking preserves the scaffold structure. Quantification of the height of the scaffold (villus plus crypt) after 14 days. The initial height of the scaffold made of polymerized 10 mg ml−1 collagen I. Scaffold was either treated with threose or left untreated, and then populated with epithelial cells (organoids) or organoids and primary intestinal fibroblasts. n = 20 units, N = 3. Mean ± SEM, t-test **, p < 0.001; *, p < 0.05.
Molds 1 and 2 were then assembled and sealed with two clips. The collagen solution (300 μL/chip) was injected into the mold assembly through a Tygon tube using a conventional 1 mL syringe topped with a cut pipette tip as a needle. Once the collagen had polymerized for 1 h at 37 °C in a cell culture incubator, the clips were removed and the collagen structure was unmolded in PBS with antibiotics and finally each chip was placed in a Petri dish with culture medium. One day after unmolding, the collagen microstructures were incubated at 37 °C with 10 mM threose (Sigma) diluted in the fibroblast medium (see below) for 1 day. The collagen scaffolds were then rinsed two times for 30 min in DMEM followed by a third wash with DMEM or the fibroblast culture medium if they were embedded into the chip.
Laminin coating
Laminin LN1 (Sigma) was labelled with Cy3.28 Collagen scaffolds were incubated with 0.02 mg mL−1 laminin in DMEM for 30 min at 37 °C 5% CO2. Finally, the laminin solution was removed and the scaffolds were washed with cell culture medium for 5 min.
Rheology measurements
PDMS rings of 1 cm inner diameter and 0.5 cm height were cut out. Two holes (Ø 0.75 mm, Harris Uni Core) were punched in the wall of the ring and a small PTFE tube (O.D. 1.07 mm, Cole Parmer) was inserted in one of them. Since the hole of the tube was too thin to directly inject the collagen with the syringe, a larger tygon tube (I.D. 0.031′′, O.D. 0.093′′, Cole Parmer Instrument Company) was adapted at the end of the thin tube to avoid any leakage during collagen injection. Two pieces of PDMS (1 cm × 1cm) were adjusted on the top and bottom of the ring to form an enclosed chamber, hermetically sealed with two clips.
The collagen solution was prepared as described above and drawn into a 1 mL syringe. A 1 mL cone was cut and placed at the tip of the syringe. The solution was injected through the tubing in the PDMS enclosed chamber. The collagen was polymerized as previously described. Before collagen unmolding, the chamber was filled up with PBS. Immediately after the PDMS ring containing the collagen was transferred into the PBS bath. The collagen was gently detached from the PDMS ring wall. The PBS level was adjusted to reach the limit of the upper surface of the collagen gel to prevent the collagen from drying. The collagen was not completely immersed to avoid the recording of the fluctuations of the PBS solution instead of those of the collagen gel surface. Measurements of the viscoelastic modulus of the collagen at various frequencies were performed using the technique of surface fluctuation specular reflection spectroscopy (SFSRS). The laser beam (diameter of laser beam: 41.3 μm, 27.5 μm or 3.5 μm) was reflected by a prism and focused on the surface of the collagen gels. The surface deviated the laser beam, which was reflected a second time by the prism. The reflected laser beam was collected at the center of the detector composed of a two-quadrant photodiode. The difference in voltages between the two quadrants directly quantifies the fluctuations in the height of the surface of the sample. The duration of the acquisition varied from 1 to 5 minutes to investigate a larger range of frequencies. Subsequently, the diagram of viscoelastic modulus vs. frequency was plotted.
Animals
All studies and procedures involving animals were in strict accordance with the European and National Regulation for the Protection of Vertebrate Animals used for Experimental and other Scientific Purposes (facility license #C75-05-18). We have used the following mice strains: C57/B6 mice (Charles Rivers), villin:Cre-ERT,29 fluorescent reporter mouse CAG:Tomato/GFP (mT/mG),30 αSMA:Cre-ERT2.31
Crypt isolation and organoid culture
Organoid cultures were prepared as described in ref. 32. Briefly, the small intestine was isolated from 3 to 6 month-old mice and cleared of its inner materials with cleaning solution (2% (v/v) antibiotic–antimycotic (Gibco), 1% (v/v) gentamicin (Gibco) in cold sterile PBS). The intestine was cut longitudinally, divided into 3 cm-long pieces and incubated in the cleaning solution with constant shaking at 4 °C twice for 15 min. The clean tissue was chopped and incubated in 2 mM EDTA diluted in PBS for 30 min at 4 °C. The EDTA supernatant was then replaced by a cleaning solution and pipetted ten times with a 10 mL pipette to mechanically dissociate the intestinal tissue. This step was repeated four times, and each time the supernatant was replaced by a fresh cleaning solution. The last supernatant was centrifuged at 200 g for 3 min at 4 °C. The pellet was suspended in a cleaning solution, filtered through a 70 μm filter and centrifuged at 400 g for 6 min. The pellet was suspended in Matrigel (Corning) half diluted in cold PBS and plated in 48 well plates. The Matrigel was allowed to polymerize for 30 min at 37 °C. After polymerization, the organoid ENR culture medium (DMEM F12, Glutamax (ThermoFisher) supplemented with 20 ng mL−1 EGF (Peprotech), 10 ng mL−1 FGF (Peprotech), 100 ng mL−1 Noggin (Peprotech), 2,5% Glutamax (v/v) (Gibco), 500 ng mL−1 R-spondin (R&D System), 1× B27 (Gibco), 1× N2 (Gibco)) was added.
Organoid seeding and culture on the 3D scaffold
The organoids were extracted from Matrigel after one day by flushing the matrix in cold PBS several times in order to break them and obtain smaller cystic like organoids of about 100 μm in diameter. Then, the organoids were centrifuged at 200 g for 5 min at 4 °C. The pellet made of organoids was resuspended in ENR medium. 300 μl of organoid solution were seeded on laminin-coated collagen scaffolds and gently shaken to make the organoids fall into the crypts. The organoids were left to adhere for 3 h at 37 °C 5% CO2. For culture under shear stress, the samples were placed on a rocking plate (PMR-30 from Grant-Bioone) one day after seeding. Rocking parameters were fixed to apply a shear stress of 0.06 dyne per cm2.33 The cell culture medium is changed every two days.
Isolation of intestinal fibroblasts
Mouse intestinal fibroblasts were isolated from αSMA–CreERT2/R26mT/mG transgenic mice, which were obtained by crossing a R26mT/mG reporter mouse30 with a mouse expressing CreERT2 recombinase under the αSMA promoter.31 Fibroblasts were isolated following the procedure described in ref. 34 with some modifications. The intestine was isolated from 12 day-old mice and incubated for 15 min in HBSS solution without calcium or magnesium, supplemented with 20 mg mL−1 of d-glucose (ThermoFisher), 1% Glutamax (v/v) and 2% anti–anti (v/v). The tissue was diced into 0.5 mm2 pieces and transferred into the dissociation solution (0.31 mg mL−1 dispase and 0.25 mg mL−1 collagenase XI diluted in HBSS with calcium and magnesium supplemented with 1% Glutamax (v/v) and 2% anti–anti (v/v)). After 30 min at 37 °C under slow agitation, 25 mL of DMEM Glutamax supplemented with 5% FBS (v/v), 1% Glutamax (v/v), d-sorbitol (20 mg ml−1), 2% anti–anti (v/v) were added. The solution was spun down at 100 g for 2 min at 4 °C and the pellet obtained was suspended into HBSS with calcium and magnesium and centrifuged at 200 g for 3 min. The tissue pieces were resuspended into 10 mL culture medium (DMEM-Glutamax) supplemented with 10% FBS (v/v), 2% anti–anti (v/v), 1× ITS (v/v), 50 μg mL−1 EGF and placed in a 10 cm diameter Petri dish to adhere. The medium was changed every 4 days. After 7 days cells that escaped the tissue and adhered to the Petri dish were collected. Upon treatment with 1 μM 4-hydroxytamoxifen, about 85% of isolated fibroblasts were αSMA positive: they expressed green fluorescent protein (GFP) (and so could be classified as myofibroblasts, based on αSMA expression), while about 15% were αSMA negative and expressed red fluorescent protein Tomato.
Immunostaining
For EdU staining, the samples were pre-incubated with 10 μM EdU (ThermoFisher) for 30 min at 37 °C. Prior to immunostaining, the culture medium was removed and the chip was incubated in 4% PFA in PBS (v/v) for 30 min at RT to perform cell fixation. The chip was washed 3 times for 15 min with PBS. The cells were permeabilized with 0.5% Triton X-100 (v/v) for 5 min at RT. The chip was rinsed 2 times for 15 min with PBS at RT. 400 μL of staining solution was injected and the chips were incubated for 30 min at RT for EdU detection or at 4 °C overnight for protein staining. The chip was rinsed 3 times for 30 min with PBS at RT. The chips were incubated with 400 μL of secondary antibody solution for 2 h at RT, in the dark and then rinsed 3 times with PBS for 30 min at RT. The chips were stored at 4 °C until imaged. The following antibodies and dyes were used: Alexa Fluor 488 phalloidin (ThermoFisher, A12379, 1 unit per mL), Click-iT® EdU Alexa Fluor® 647 imaging kit (ThermoFisher, C10340), rabbit anti-mucin 2 (Santa Cruz, sc-15334, 10 μg mL−1), rabbit anti-lysosyme (DAKO, A0099), and rabbit anti-L-FABP (Santa Cruz, sc-50380), and secondary antibodies were purchased from ThermoFisher as used as 25 μg mL−1.
Imaging
Collagen scaffolds were imaged using an upright spinning disk (Roper Zeiss) microscope equipped with 10×/0.3NA/WD520 (working distance 5.2 mm) or 40×/1.3NA/WD210 objective, and 405 nm, 491 nm, 561 nm and 634 nm lasers and controlled using Metamorph imaging software (Universal Imaging). Z stacks were acquired every 1 μm (40×) to 5 μm (10×). Images were processed using Image J and Imaris software.
Quantifications and statistics
For the quantification of the epithelialization process, the percentage of the surface covered by epithelial cells has been quantified using 2D projection of the Z stacks acquired with a 10× objective. All experiments were performed in triplicate in 2–3 independent experiments. Statistical analysis and graphic representations were performed using Graph Pad Prism software. Data were represented as mean ± SEM. The statistical significance was determined with a t-test as indicated; ***p < 0.0001; **p < 0.001; *p < 0.05.
Results
Engineering a cytocompatible 3D scaffold for gut-on-chip
To produce a biomimetic gut scaffold that will enable the co-culture of epithelial and stromal cells on a chip, we first replicated the composition and architecture of the mucosal extracellular matrix (Fig. 1A and B). Because collagen type I is the main constituent of the intestinal stroma, we developed a microfabrication strategy compatible with type I collagen molding (Fig. 1A). To produce structures of high aspect ratio mimicking the dimensions of the murine small intestine, we first micro-milled an array of 80 structural units on a brass mold, each unit being composed of one villus surrounded by six crypts (Fig. 1C). Villi were 350 μm in height with 400 μm in diameter at their base and 150 μm at their top, with a slope of 68°, while crypts were 150 μm deep and 200 μm wide, the distance between villi and crypts were 50 μm. Then, by employing soft lithography techniques, we produced an intermediate silicon (PDMS) replica of the brass mold (Fig. 1D) and used this replica to cast collagen I into structural units (Fig. 1E). Because collagen hydrogels have low stiffness, which could be increased by increasing the collagen concentration,35 to generate a scaffold more resistant to traction forces exerted by cells, we selected the highest collagen concentration that was still compatible with reproducible microstructuring. We used 10 mg ml−1 of native type I collagen. We characterized the linear viscoelastic modulus of these collagen gels. Their structure being heterogeneous at the microscale, the measurements of their viscoelasticity at a larger scale have been required. We thus used a method based on the thermal fluctuations of the free surface of collagen gels called surface fluctuation specular reflection spectroscopy (SFSRS) and obtained a modulus of 1.21 ± 0.1 kPa for collagen I at 10 mg mL−1 in its polymerized state.35,36 The obtained collagen scaffold replicated the mold dimensions with high fidelity (Fig. 1E and F). However, we observed that, despite being made of highly concentrated collagen, this scaffold only partially resisted the traction forces exerted by epithelial and stromal cells. Indeed, after 14 days of co-culture, the scaffold flattened, retaining only 19 ± 10% of the original height of the scaffold (Fig. 1F). We thus further increased its stiffness by using post-polymerization treatment with threose, a carbohydrate reacting with lysine and arginine residues, in order to cross-link collagen fibers via glycation. We selected glycation over other stiffening methods because as a post-polymerization treatment, it preserves the fibrillary structure of the collagen.37 Also, as glycation is a physiological process that occurs in vivo with aging,38 threose treatment is cytocompatible with cells embedded into collagen scaffolds,39 in contrast to collagen scaffolds treated with chemical cross-linkers.25,40 To test if cells can survive threose treatment, we mixed primary intestinal fibroblasts with non-polymerized collagen, and once polymerized in the mold, the collagen was treated with threose. We found that after this treatment, fibroblasts retained their characteristic spindle shape and were highly dynamic, demonstrating that cross-linked collagen is indeed cytocompatible for primary fibroblasts (ESI† Movie S1), as already shown for cancer cells.39 To evaluate the effect of threose treatment on the mechanical properties of collagen we measured its shear modulus by SFSRS and showed that threose treatment increased the shear modulus of the collagen scaffold 1.6 fold, which decreased scaffold flattening: scaffold retained 49 ± 20% of its initial height after 14 days in culture (Fig. 1F). This property is pivotal to allow the co-culture of stromal and epithelia cells within and on the 3D scaffold, respectively.
In vivo, intestinal epithelial cells do not interact directly with type I collagen, they are anchored at their basal side to the basement membrane, which overlays the collagen-rich stromal compartment. To refine the 3D scaffold, we coated the surface of the collagen structures with laminin, the main component of the basement membrane (Fig. 1E). Altogether, we have developed a biomimetic scaffold that replicates both the architecture and molecular composition of the mouse intestinal mucosa while being compatible with stromal and epithelial cells co-culture.
Epithelialization of 3D scaffolds
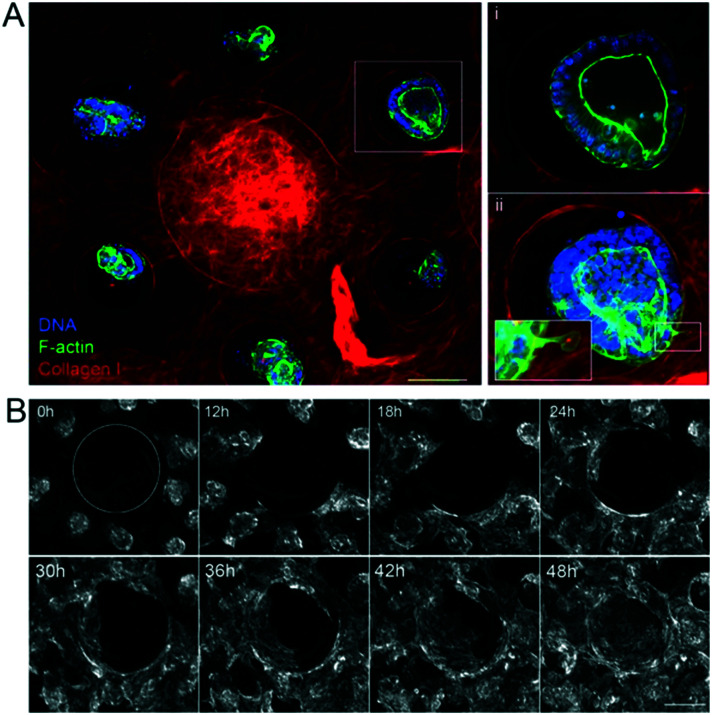
Once the biomimetic scaffold has been developed, we characterize the epithelialization process. To reconstitute the intestinal epithelium, we seeded mouse intestinal organoids of around 100 μm in diameter into the crypt regions of the scaffold containing primary mouse intestinal fibroblasts (Fig. 2A and ESI† Movie S2). The organoids first attached to the collagen scaffold via actin-rich protrusions (Fig. 2Aii and ESI† Movie S3) and about six hours after seeding, they started spreading over the scaffold (Fig. 2B and ESI† Movies S4 and S5). The cells migrated mostly collectively; the leaders of the clusters exhibited an elongated mesenchymal phenotype with broad lamellipodia-like protrusions at the migration front. Frequent exchanges of cells between adjacent organoids were also observed. Upon collision, the organoids fused (ESI† Movies S4–S6). At earlier time points, however, fusion was unstable and resulted in rapid cell segregation, whereas at later time points, when the cell density was increased, the fusion becomes stabilized resulting in a cohesive monolayer (Fig. 2B and ESI† Movie S7). This transition from cystic organoids to a flat monolayer resembles the wetting transitions of cellular aggregates on 2D surfaces,41 suggesting that the newly formed cell-matrix adhesions were stronger than existing cell–cell adhesions.
Fig. 2. Epithelialization of the scaffolds. A. The unit consists of one villus and six crypts. Three hours after seeding the organoids on the collagen scaffold in the presence of mouse intestinal fibroblasts. Collagen type I (TAMRA-labeled, red); F-actin (phalloidin, green), DNA (DAPI, blue). Top view, maximal projection of 343 μm. Scale bar, 150 μm. Insets, higher magnification of one crypt containing an organoid. Single plane (i), maximal projection of 152 μm (ii). Scale bar, 20 μm. B. Organoids expressing LifeAct-GFP (F-actin label) spreading over collagen scaffolds. Time in hours. Top view. The dashed line outlines the base of the villus. Scale bar, 200 μm.
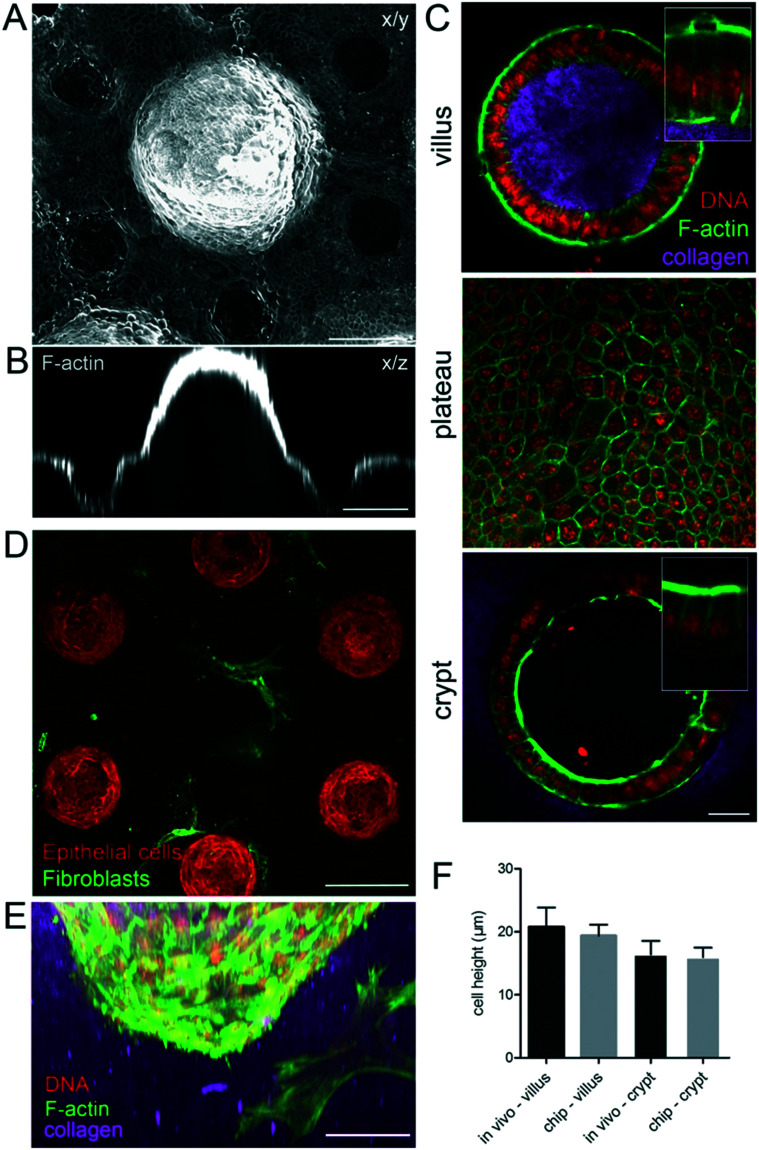
After four days, the epithelialization process was achieved, giving rise to a locally confluent monolayer of epithelial cells (Fig. 3A–E). Both in the villi and crypt domains, this epithelial monolayer was tightly packed with columnar cells with a hexagonal shape at their apical side (Fig. 3C) characteristic of in vivo intestinal epithelium. In these confluent regions, the cells were polarized, having well-defined apical domains rich in actin and with nuclei oriented perpendicular to the cell base (Fig. 3C). The cells had an average width of 6.1 ± 0.2 μm and height of 19.4 ± 0.6 μm similar to the epithelial cells in vivo (Fig. 3F). Altogether, these data show that the morphology of the epithelial cells in the microdevice faithfully resembles those in the mouse intestinal epithelium in vivo.
Fig. 3. Cell morphology on the microdevice resembles the in vivo epithelium. A. A continuous monolayer of epithelial cells over the collagen scaffold four days after seeding the organoids, in the presence of mouse intestinal fibroblasts. x/y, top view, maximal projection of 433 μm. B. Orthogonal section through one villus and two crypts. Scale bar, 150 μm. C. Cross-section through the villus and crypt and top view of the region between villi and crypts (plateau) on collagen scaffolds. Collagen type I (TAMRA-labeled, pink); F-actin (phalloidin, green), DNA (DAPI, red). Scale bar, 20 μm. Insets, higher magnification of boxed regions. D. A monolayer of epithelial cells (F-actin labeled, red) four days after seeding the organoids over the collagen scaffold containing fibroblasts (expressing αSMA, green). A cross-section through the crypt region, 348 μm from the top of the scaffold showing fibroblasts in contact with crypts. Scale bar, 150 μm. E. Side view of the crypt. Epithelial cells and mouse intestinal fibroblasts (F-actin, green; DNA, DAPI, red), collagen (TAMRA-labelled, pink). Scale bar, 50 μm. F. Comparison of the epithelial cell height on the chip and in vivo in villi and crypts, n = 10–60 cells, from N = chips or mice.
Fluid shear stress preserves epithelium integrity
Next, we examined if the generated epithelial monolayer could be cultured for extended periods. We first observed that under these initial conditions the survival of the epithelium was limited to short-term culture even in the presence of fibroblasts within the scaffold (Fig. 4A and B). After five days, patches of epithelial cells detached from the collagen scaffold, indicating that homeostasis was not maintained.
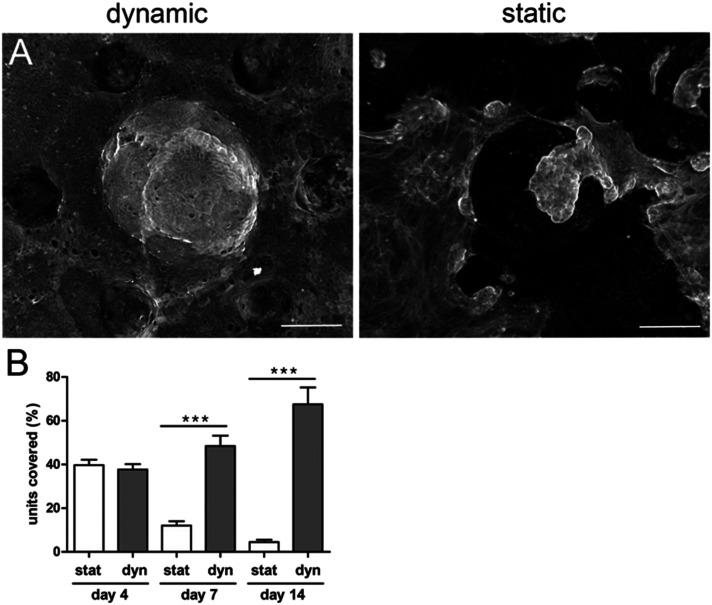
Fig. 4. Shear stress improves epithelial integrity. A. Epithelial cells (F-actin, phalloidin) 14 days after seeding the organoids over the collagen scaffold containing mouse intestinal fibroblasts with fluid shear stress (right panel, dynamic conditions) and without shear stress (right panel, static conditions). Top view, maximal projection. Scale bar, 150 μm. B. Percentage of units covered with epithelial cells after 14 days after seeding the organoids on collagen scaffolds containing mouse intestinal fibroblasts under static conditions (stat) and with fluid shear stress (dynamic conditions, Dyn). n = 128 units, N = 3 independent experiments. Mean ± SEM, t-test ***, p < 0.0001.
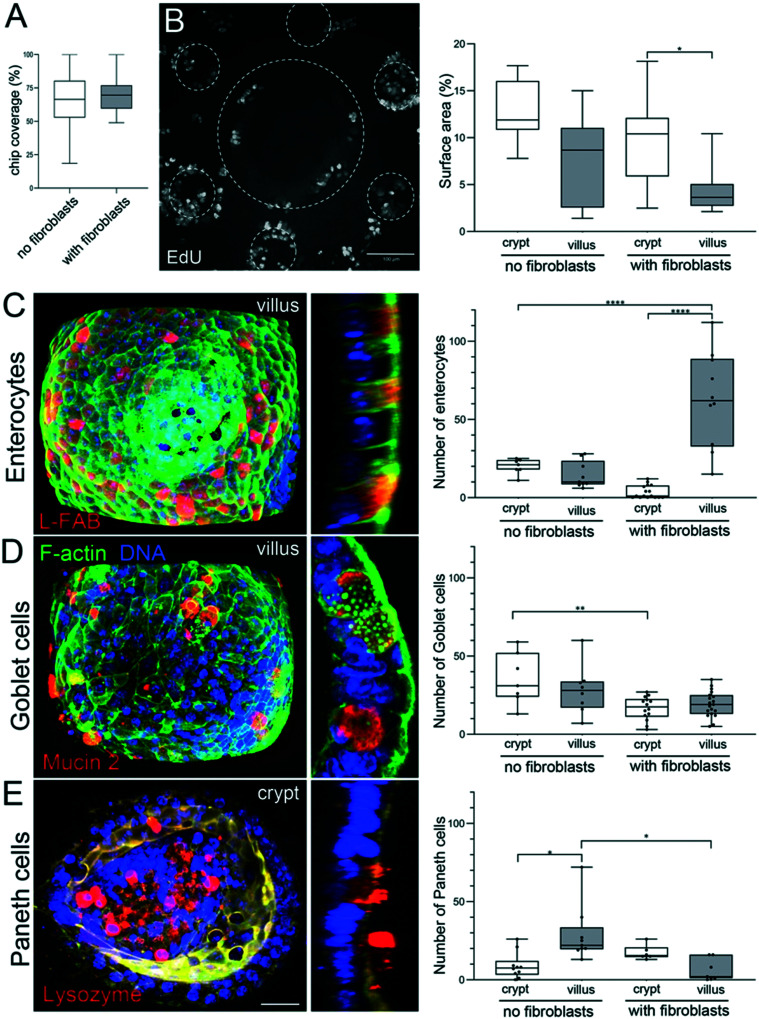
In vivo, the intestinal mucosa is exposed to shear stress caused by nutrients progressing throughout the gut lumen.42 Recently, it has also been shown that intestinal cell lines, cultured within a V-shape microchamber, are highly sensitive to fine changes in the fluid shear stress with an impact on the cell morphology, phenotype and function.43 We thus studied how fluid shear stress might favor cell adhesion and survival in our chip. One day after organoid seeding, when the cells successfully adhered to the scaffold, we applied to the microdevices a fluid shear stress of about 0.06 dyne per cm.33 This value is a rough estimate as it has been calculated for a flat surface and is of the same order of magnitude compared to the shear stress applied to other gut on chip models17 and the one observed in vivo.17 For short-term culture (up to four days), no significant difference was observed between the static and dynamic (with fluid shear stress) culture (Fig. 4B). After five days, massive patches of cells detached from the scaffolds cultured under static conditions, while no detachment of cells was observed on microdevices subjected to shear stress. This effect was striking after seven days of culture. The percentage of fully covered units on static scaffolds was reduced. The differences were even more pronounced after long-term culture. While around 70% of the units remained covered with cohesive and densely packed epithelial cells after 14 days in dynamic culture (Fig. 4B), only 5% of units remained covered with the epithelium under static conditions. The remaining epithelial cells were less cohesive, more elongated and flat (Fig. 4A). It is also worth noting that, under dynamic culture conditions, the percentage of the surface covered by epithelial cells was similar in the presence or absence of fibroblasts (Fig. 5A).
Fig. 5. Segregation of proliferative and differentiated cells. A. Percentage of the chip covered with epithelial cells 14 days after seeding the organoids on collagen scaffolds, in the absence or presence of fibroblasts. n = 128 units, N = 3 independent experiments. Mean ± SEM, t-test **, p < 0.001; *, p < 0.05. B. Left: Dividing (EdU positive cells) in the monolayer of epithelial cells stained 14 days after seeding the organoids over the collagen scaffold containing fibroblasts with fluid shear stress. Top view, maximal projection of 493 μm. Scale bar, 100 μm. Right: Percentage of the surface area (2D maximal projection) occupied with dividing, EdU positive cells, in crypts and villi, in the absence or presence of fibroblasts. n = 10 units (unit: 1 villi surrounded by 6 crypts), N = 3. Mean ± SEM, t-test **, p < 0.001; *, p < 0.05. C. Left: Fully differentiated enterocytes labeled with anti-L-FABP antibodies (red) in the epithelial monolayer 14 days after seeding the organoids over the scaffold containing primary mouse intestinal fibroblasts with continuous fluid shear stress. F-Actin (green), DNA (DAPI, blue). Left panel, maximal projection of 95 μm from the top of the villus. Scale bar 20 μm. Right panel, a cross-section through the villus. Right: The number of enterocytes in crypts and villi, in the absence or presence of fibroblasts. n = 12 units, N = 2. Mean ± SEM, t-test **, p < 0.001; *, p < 0.05. D. Left: Fully differentiated goblet cells labeled with anti-mucin2 antibodies (red) in the epithelial monolayer 14 days after seeding the organoids over the scaffold containing primary mouse intestinal fibroblasts with continuous fluid shear stress. F-Actin (green), DNA (DAPI, blue). Left panel, maximal projection of 95 μm from the top of the villus. Scale bar 20 μm. Right panel, a cross-section through the villus. Right: The number of goblet cells in crypts and villi, in the absence or presence of fibroblasts. n = 15 units, N = 2. Mean ± SEM, t-test **, p < 0.001; *, p < 0.05. E. Left: Fully differentiated Paneth cells labeled with anti-lysozyme antibodies (red) in the epithelial monolayer 14 days after seeding the organoids over the scaffold containing primary mouse intestinal fibroblasts with continuous fluid shear stress. F-Actin (green), DNA (DAPI, blue). Left panel, maximal projection of 55 μm from the bottom of the crypt. Scale bar 20 μm. Right panel, a cross-section through the crypt. Right: The number of Paneth cells in crypts and villi, in the absence or presence of fibroblasts. n = 10 units, N = 2. Mean ± SEM, t-test **, p < 0.001; *, p < 0.05.
Together these data show that fluid shear stress improves the maintenance of the epithelial monolayer under long-term culture conditions. This response to fluid shear stress could be related to epithelium mechanosensitivity and/or to improved convection at the apical side of the epithelium.
Recapitulating on-chip epithelial cell differentiation and spatial segregation
Next, we explored if the reconstitution of epithelial and stromal cell co-culture within the collagen 3D scaffold allows capturing in vivo gut features in particular, segregation of proliferative and differentiated cells.
To evaluate if the proliferation of epithelial cells is maintained in the device, we performed EdU labeling that marks cells in the S phase of the cell cycle. We found that cell proliferation was maintained even after 14 days of culture in the presence or absence of fibroblasts in the scaffold. In vivo, proliferative cells are located exclusively in the crypts. To assess if spatial segregation of dividing cells is also achieved in our device, we quantified the percentage of crypt and villus surfaces occupied by proliferative cells. We found that, in the presence of fibroblasts, proliferative cells are mostly restricted to the crypts (Fig. 5B) whereas in the absence of fibroblasts, the surface occupied by proliferative cells was only slightly higher in the crypts compared to villi (Fig. 5B).
Next, we assessed if our gut on chip model reproduces epithelial cell differentiation and their spatial partitioning. To identify and quantify the different cell types, epithelial cells were stained with specific markers of three main specialized cell types. The absorptive cells, enterocytes, are the predominant cell type in the small intestine and were identified based on the expression of liver fatty acid binding protein (L-FABP). Goblet cells, secretory cells specialized in the production of the mucus blanket, were identified using mucin 2 staining. Finally, lysozyme staining was used to identify Paneth cells, secretory cells that play a role in host defense and regulation of microbiota. We found that after 14 days of culture, all the three main types of differentiated cells (Fig. 5C–E) were observed on the scaffold, illustrating that cell differentiation takes place in our gut-on-chip in the presence or absence or fibroblasts within the scaffold. In vivo, enterocytes and goblet cells are present all along the villus, while Paneth cells are restricted to the bottom of the crypts. To assess if the differentiated cells are properly positioned along the crypt–villus axis in our device, we quantified the number of differentiated cells on villus and crypt domains. We found that in the absence of fibroblasts, the proper segregation of differentiated cells was not achieved. The number of enterocytes was almost the same in crypts and villi, and in addition, the total number of enterocytes was very low (e.g. less than 20 enterocytes per villus) (Fig. 5C). Furthermore, goblet and Paneth cells showed the opposite segregation in comparison with in vivo conditions. Goblet cells were found in higher numbers in crypts, while Paneth cells mostly localized in villi (Fig. 5D and E).
In contrast, in the presence of fibroblasts, we observed that segregation of differentiated cells in our chip occurs with a remarkable resemblance to that in the in vivo intestine. Only a few enterocytes were found in the crypts44 while more than 30 times more enterocytes were located on the villi (Fig. 5C). Although goblet cells were present in a higher number in the villi (Fig. 5D), this difference was not as pronounced as in vivo. Finally, Paneth cells were properly segregated in the crypts, as in vivo (Fig. 5E). Our results show that in the presence of primary intestinal fibroblasts in the 3D collagen scaffold, in vivo features of the epithelium, such as spatial segregation of proliferative and differentiated cells were recapitulated on the chip.
Discussion
In this work, we reported a new and advanced gut chip device in which we have cultured an intestinal epithelium monolayer derived from mouse organoids on a laminin-coated microstructured collagen I scaffold containing primary mouse intestinal fibroblasts. Our device possesses several unique features. 1) It has the typical topography of the mouse gut, consisting of an array of villi surrounded by crypts. 2) The collagen scaffold is covered with a thin layer of the ECM protein laminin, mimicking the basement membrane, which is necessary for the survival of the epithelial cells. 3) Our 3D scaffold is generated entirely from collagen I, the major constituent of the mouse gut stroma. To maintain the topography of the scaffold, we have cross-linked and thus rigidified the collagen scaffold while maintaining its cytocompatibility. 4) This permits, for the first time, the incorporation of stromal cells into the collagen 3D scaffold. 5) The seeding of intestinal organoids on these scaffolds results in the formation of a columnar epithelium monolayer with a morphology closely resembling the intestinal epithelium in vivo. 6) The culture of the epithelium over a long period is achieved thanks to the application of fluid shear stress. 7) Finally, we showed that under these conditions epithelial cell differentiation and spatial segregation take place in vivo.
Compared to existing devices,17,23,25 our device is made of collagen I and cross-linked by a cytocompatible reagent, which allowed the incorporation of primary intestinal fibroblasts in the 3D scaffold and thus a better mimic of epithelial–stromal cell interactions on a chip. Incorporation of stromal cells in gut on chip devices has not been achieved so far; the devices reported in the literature are limited either by the nature of the scaffold material that requires photopolymerization23 or by the cytotoxic chemical treatment used to crosslink collagen.25 The ability to uncouple the contribution of different components of this device opens up numerous possibilities to decipher the role of stromal components in intestine physiology or pathophysiology. By adapting the dimensions of the device to match the size of the human crypts and villi, this gut-on-chip could be used, in the near future, to study the pathophysiology of the human gut. By using epithelial cells and/or fibroblasts extracted from colorectal cancer tissues, it would be also possible to study the interplay between the stroma and cancer cells during tumor growth or invasion. Beyond tumors, the intestine is susceptible to a range of chronic pathologies,45 such as Crohn's and celiac diseases. This device may be a good model of inflammatory bowel disease because the role of microbiota composition,46 mucus defects,47 overexpression of basement membrane components,48 activation of fibroblasts,49 remodeling of the extracellular matrix and reduced peristalsis50 could be addressed independently.
Author contributions
JLV, SD and DMV conceived the study. MV, AS, MBD, RGV and LG performed the majority of the experiments. DF and LG microfabricated the brass molds. LT performed rheology measurements and analysis. DK performed quantification of cell dimensions. MV, SD and DMV wrote the manuscript with input from all the authors. DMV and SD prepared the figures and supervised the project.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
We thank all members of the Descroix (UMR 168) and Vignjevic (UMR144) labs for helpful discussions. We acknowledge imaging facilities PICT-IBiSA@Lhomond and PICT@BDD and in particular O. Leroy and O. Renaud. We also thank all members of the animal house facility. We thank Remy Fert and Eric Nicolau (UMR168) for their support in micro-milling. We thank D. Metzger and P. Chambon (Strasbourg) for their generous gift of the αSMA:Cre mouse line, and C. Rosse (Institut Curie) for providing collagen labeled Cy5. D. F. thanks the ARC foundation for young researcher fellowship. This work has received the support of Institut Pierre-Gilles de Gennes (équipement d'excellence and LABEX, “Investissements d'avenir”, program ANR-10-EQPX-34, ANR-10-IDEX-0001-02 PSL and ANR-10-LABX-31). This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 Research and Innovation Programme (grant agreement no. 772487) (D. M. V.) and ERCadg CellO FP7-IDEAS-ERC-321107 (J. L. V.), ANR HOMEOGUT (D. M. V. and S. D.) and financial support from Inserm Cancer (D. M. V. and S. D.).
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0lc00672f
References
- Sontheimer-Phelps A. Hassell B. A. Ingber D. E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer. 2019;19:65. doi: 10.1038/s41568-018-0104-6. [DOI] [PubMed] [Google Scholar]
- Sato T. Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- Sato T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Shin Y. Han S. Jeon J. S. Yamamoto K. Zervantonakis I. K. Sudo R. Kamm R. D. Chung S. Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels. Nat. Protoc. 2012;7:1247–1259. doi: 10.1038/nprot.2012.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S. N. Ingber D. E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- Verhulsel M. Vignes M. Descroix S. Vignjevic D. Viovy J. L. A review of microfabrication and hydrogel engineering for micro-organs on chips. Biomaterials. 2014;35:1816–1832. doi: 10.1016/j.biomaterials.2013.11.021. [DOI] [PubMed] [Google Scholar]
- Sobrino A. Phan D. Datta R. et al. 3D microtumors in vitro supported by perfused vascular networks. Sci. Rep. 2016;6:31589. doi: 10.1038/srep31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M. De Ninno A. Mencattini A. Mermet-Meillon F. Fornabaio G. Evans S. S. Cossutta M. Khira Y. Han W. Sirven P. Pelon F. Di Giuseppe D. Bertani F. R. Gerardino A. Yamada A. Descroix S. Soumelis V. Mechta-Grigoriou F. Zalcman G. Camonis J. Martinelli E. Businaro L. Parrini M. C. Dissecting effects of anti-cancer drugs and of cancer-associated fibroblasts by on-chip reconstitution of immunocompetent tumor microenvironments. Cell Rep. 2018;25:3884–3893. doi: 10.1016/j.celrep.2018.12.015. [DOI] [PubMed] [Google Scholar]
- Van der Flier L. G. Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- Krndija D. El Marjou F. Guirao B. Richon S. Leroy O. Bellaiche Y. Hannezo E. Matic Vignjevic D. Active cell migration is critical for steady-state epithelial turnover in the gut. Science. 2019;365(6454):705–710. doi: 10.1126/science.aau3429. [DOI] [PubMed] [Google Scholar]
- Glentis A. Gurchenkov V. Matic Vignjevic D. Assembly, heterogeneity, and breaching of the basement membranes. Cell Adhes. Migr. 2014;8:236–245. doi: 10.4161/cam.28733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stzepourginski I. Nigro G. Jacob J. M. Dulauroy S. Sansonetti P. J. Eberl G. Peduto L. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E506–E513. doi: 10.1073/pnas.1620059114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell D. W. Mifflin R. C. Valentich J. D. Crowe S. E. Saada J. I. West A. B. Myofibroblasts. II. Intestinal subepithelial myofibroblasts. Am. J. Physiol. 1999;277:C183–C201. doi: 10.1152/ajpcell.1999.277.2.C183. [DOI] [PubMed] [Google Scholar]
- McCarthy N. Manieri E. Strom E. E. Saadatpour A. Luoma A. M. Kapoor V. N. Madha S. Gaynor L. T. Cox C. Keerthivasan S. Wucherpfenning K. Yuan G. C. de Sauvage F. J. Turley S. J. Shivdasani R. A. Distinct Mesenchymal Cell Populations Generate the Essential Intestinal BMP Signaling Gradient. Cell Stem Cell. 2020;26:391–402. doi: 10.1016/j.stem.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; , e395
- Sato T. Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- Lehmann R. et al. Human organoids: a new dimension in cell biology. Mol. Biol. Cell. 2019;30:1129–1137. doi: 10.1091/mbc.E19-03-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J. Huh D. Hamilton G. Ingber D. E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–2174. doi: 10.1039/C2LC40074J. [DOI] [PubMed] [Google Scholar]
- Kasendra M. Tovaglieri A. Sontheimer-Phelps A. Jalili-Firoozinezhad S. Bein A. Chalkiadaki A. Scholl W. Zhang C. Rickner H. Li H. Breault D. T. Ingber D. E. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018;8:2871. doi: 10.1038/s41598-018-21201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassart A. Malardé V. Gobaa S. Sartori-Rupp A. Kerns J. Karalis K. Marteyn B. Sansonetti P. J. Sauvonnet N. Bioengineered Human Organ-on-Chip Reveals Intestinal Microenvironment and Mechanical Forces Impacting Shigella Infection. Cell Host Microbe. 2019;26:435–444. doi: 10.1016/j.chom.2019.08.007. [DOI] [PubMed] [Google Scholar]
- Salomon J. Gaston C. Magescas J. Duvauchelle J. Canioni D. Sengmanivong L. Mayeux A. Michaux G. Campeotto F. Lemale J. Viala J. Poirier F. Minc N. Schmitz J. Brousse N. Ladoux B. Goulet O. Delacour D. Contractile forces at tricellular contacts modulate epithelial organization and monolayer integrity. Nat. Commun. 2017:13998. doi: 10.1038/ncomms13998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello C. M. Sorna R. M. Goh Y. L. Cengic I. Jain N. K. March J. C. 3-D intestinal scaffolds for evaluating the therapeutic potential of probiotics. Mol. Pharmaceutics. 2014;11:2030–2039. doi: 10.1021/mp5001422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffiey S. A. Jia H. Keane T. Costello C. Wasserman D. Quidgley M. Dziki J. Badylak S. Sodhi C. P. March J. C. Hackam D. J. Instestinal stem cell growth and differentiation on a tubular scaffold with evaluation in small and large animals. Regener. Med. 2016;11:45–61. doi: 10.2217/rme.15.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creff J. Courson R. Mangeat T. Foncy J. Souileile S. Thibault C. Besson A. Malaquin L. Fabrication of 3D scaffolds reproducing intestinal epithelium topography by high-resolution 3D stereolithography. Biomaterials. 2019;221:119404. doi: 10.1016/j.biomaterials.2019.119404. [DOI] [PubMed] [Google Scholar]
- Castaño A. G. García-Díaz M. Torras N. Altay G. Comelles J. Martínez E. Dynamic photopolymerization produces complex microstructures on hydrogels in a moldless approach to generate a 3D intestinal tissue model. Biofabrication. 2019;11:025007. doi: 10.1088/1758-5090/ab0478. [DOI] [PubMed] [Google Scholar]
- Wang Y. Gunasekara D. B. Reed M. I. DiSalvo M. Butman S. J. Sims C. E. Magness S. T. Allbritton N. L. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials. 2017;128:44–55. doi: 10.1016/j.biomaterials.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo S. Simon A. Vignjevic D. Revealing the cytoskeletal organization of invasive cancer cells in 3D. J. Visualized Exp. 2013:e50763. doi: 10.3791/50763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo S. Simon A. Elkhatib N. Louvard D. Fteler L. Vignjevic D. M. Do cancer cells have distinct adhesions in 3D collagen matrices and in vivo? Eur. J. Cell Biol. 2012;91:930–937. doi: 10.1016/j.ejcb.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Schoumacher M. Glentis A. Gurchenkov V. V. Vignjevic D. M. Basement Membrane Invasion Assays: Native Basement Membrane and Chemoinvasion Assay. Methods Mol. Biol. 2013;1046:133–144. doi: 10.1007/978-1-62703-538-5_8. [DOI] [PubMed] [Google Scholar]
- el Marjou F. et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- Muzumdar M. D. Tasic B. Miyamichi K. Li L. Luo L. A. Global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Wendling O. Bornert J. M. Chambon P. Metzger D. Efficient temporally-controlled targeted mutagenesis in smooth muscle cells of the adult mouse. Genesis. 2009;47:14–18. doi: 10.1002/dvg.20448. [DOI] [PubMed] [Google Scholar]
- Sato T. Clevers H. Primary mouse small intestinal epithelial cell cultures. Methods Mol. Biol. 2013;945:319–328. doi: 10.1007/978-1-62703-125-7_19. [DOI] [PubMed] [Google Scholar]
- Zhou X. Liu D. You L. Wang L. J. Biomech. 2010;43(8):1598–1602. doi: 10.1016/j.jbiomech.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahar N. Lei N. Y. Wang J. Jabaji Z. Tung S. C. Joshi V. Lewis M. Stelzner M. Marting M. G. Dunn J. C. Intestinal subepithelial myofibroblasts support in vitro and in vivo growth of human small intestinal epithelium. PLoS One. 2011;6:e26898. doi: 10.1371/journal.pone.0026898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulsel M. Shivokhin M. Simon A. Descroix S. Fretigny C. Talini L. High bandwidth non invasive measurements of the linear viscoelasticity of collagen gels. J. Rheol. 2016;60:1269–1278. doi: 10.1122/1.4965039. [DOI] [Google Scholar]
- Pottier B. Ducouret G. Frétigny F. Lequeux F. Talini L. High bandwith linear viscoelastic properties of complex fluids from the measurement of their free surface fluctuations. Soft Matter. 2011;7:7843. doi: 10.1039/C1SM05258F. [DOI] [Google Scholar]
- Mason B. N. Starchenko A. Williams R. M. Bonassar L. J. Reinhart-King C. A. Tuning three-dimensional collagen matrix stiffness independently of collagen concentration modulates endothelial cell behavior. Acta Biomater. 2013;9:4635–4644. doi: 10.1016/j.actbio.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- Staneva R. Burla F. Koenderink G. H. Descroix S. Vignjevic D. M. Attieh Y. Verhulsel M. A new biomimetic assay reveals the temporal role of matrix stiffening in cancer cell invasion. Mol. Biol. Cell. 2018;29:2979–2988. doi: 10.1091/mbc.E18-01-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer D. P. Chvapil M. Eskelson C. D. Ulreich J. Biological effects of residual glutaraldehyde in glutaraldehyde-tanned collagen biomaterials. J. Biomed. Mater. Res. 1980;14:753–764. doi: 10.1002/jbm.820140607. [DOI] [PubMed] [Google Scholar]
- Douezan S. Dumond J. Brochard-Wyart F. Wetting transitions of cellular aggregates induced by substrate rigidity. Soft Matter. 2012;8:9. doi: 10.2324/ejsm.8.9. [DOI] [Google Scholar]
- Lentle R. G. Janssen P. W. M. Physical Characteristics of Digesta and Their Influence on Flow and Mixing in the Mammalian Intestine: A Review. J. Comp. Physiol., B. 2008;178:673–690. doi: 10.1007/s00360-008-0264-x. [DOI] [PubMed] [Google Scholar]
- Delona G. Oszmianab C. Gibsonc L. C. Zhaobin A. Chia-Chi R. Prestidge T. Clive B. A systematic investigation of the effect of the fluid shear stress on Caco-2 cells towards the optimization of epithelial organ-on-chip models. Biomaterials. 2019;225:119521. doi: 10.1016/j.biomaterials.2019.119521. [DOI] [PubMed] [Google Scholar]
- Moor A. E. Harnik Y. Ben-Moshe S. Massasa E. E. Rozenberg M. Eilam R. Bahar Halpern K. Itzkovitz S. Spatial Reconstruction of Single Enterocytes Uncovers Broad Zonation along the Intestinal Villus Axis. Cell. 2018;175:1156–1167. doi: 10.1016/j.cell.2018.08.063. [DOI] [PubMed] [Google Scholar]
- Kaplan G. G. Ng S. C. Globalisation of inflammatory bowel disease: perspectives from the evolution of inflammatory bowel disease in the UK and Chin. Lancet Gastroenterol. Hepatol. 2016;1:307–316. doi: 10.1016/S2468-1253(16)30077-2. [DOI] [PubMed] [Google Scholar]
- Manichanh C. Borruel N. Casellas F. Guarner F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- Johansson M. E. Mucus layers in inflammatory bowel disease. Inflammatory Bowel Dis. 2014;20:2124–2131. doi: 10.1097/MIB.0000000000000117. [DOI] [PubMed] [Google Scholar]
- Spenlé C. Lefebvre O. Lacroute J. Méchine-Neuville A. Barreau F. Blottière H. M. Duclos B. Arnold C. Hussenet T. Hemmerlé J. Gullberg D. Kedinger M. Sorokin L. Orend G. Simon-Assmann P. The laminin response in inflammatory bowel disease: protection or malignancy? PLoS One. 2014;9:e111336. doi: 10.1371/journal.pone.0111336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrance I. C. Maxwell L. Doe W. Altered response of intestinal mucosal fibroblasts to profibrogenic cytokines in inflammatory bowel disease. Inflammatory Bowel Dis. 2001;7:226–236. doi: 10.1097/00054725-200108000-00008. [DOI] [PubMed] [Google Scholar]
- Bassotti G. Antonelli E. Villanacci V. Salemme M. Coppola M. Annese V. Gastrointestinal motility disorders in inflammatory bowel diseases. World J. Gastroenterol. 2014;20:37–44. doi: 10.3748/wjg.v20.i1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.