Abstract
Leishmaniasis remains a serious public health problem in many tropical regions of the world. Among neglected tropical diseases, the mortality rate of leishmaniasis is second only to malaria. All currently approved therapeutics have toxic side effects and face rapidly increasing resistance. To identify existing drugs with antileishmanial activity and predict the mechanism of action, we designed a drug-discovery pipeline utilizing both in-silico and in-vitro methods. First, we screened compounds from the Selleckchem Bio-Active Compound Library containing ~1622 FDA-approved drugs and narrowed these down to 96 candidates based on data mining for possible anti-parasitic properties. Next, we completed preliminary in-vitro testing of compounds against Leishmania amastigotes and selected the most promising active compounds, Lansoprazole and Posaconazole. We identified possible Leishmania drug targets of Lansoprazole and Posaconazole using several available servers. Our in-silico screen identified likely Lansoprazole targets as the closely related calcium-transporting ATPases (LdBPK_352080.1, LdBPK_040010.1, and LdBPK_170660.1), and the Posaconazole target as lanosterol 14-alpha-demethylase (LdBPK_111100.1). Further validation showed LdBPK_352080.1 to be the most plausible target based on induced-fit docking followed by long (100ns) MD simulations to confirm the stability of the docked complexes. We present a likely ion channel-based mechanism of action of Lansoprazole against Leishmania calcium-transporting ATPases, which are essential for parasite metabolism and infectivity. The LdBPK_111100.1 interaction with Posaconazole is very similar to the known fungal orthologue. Herein, we present two novel anti-leishmanial agents, Posaconazole and Lansoprazole, already approved by the FDA for different indications and propose plausible mechanisms of action for their antileishmanial activity.
Keywords: Drug repurposing, Lansoprazole, Leishmaniasis, Molecular dynamics, Posaconazole
1. Introduction
Among parasitic infections, leishmaniasis remains a leading cause of human morbidity and mortality in tropical regions around the world. Leishmaniasis is endemic in over 98 countries and affects 350 million people globally [1], inflicting great suffering in addition to a socioeconomic burden [2]. The disease is responsible for 57,000 deaths per year and 981,000 disability-adjusted life years (DALYs) [2]. Leishmaniasis is caused by the protozoan parasitic species (spp.) belonging to the phylum Kinetoplastida of the genus Leishmania and is transmitted by female sandflies [3]. The disease is divided into three clinical syndromes: visceral (VL), cutaneous (CL), and mucocutaneous (ML) leishmaniasis, and mainly affects populations that live in low and middle-income countries (LMIC) [4]. Furthermore, co-infection with HIV is concerning as observed in Brazil, India, Ethiopia, and Nigeria [5].
Liposomal amphotericin B, pentavalent antimonials, and miltefosine remain the primary drugs of choice for the management of leishmaniasis, however, the continued use of these medications is hampered by prohibitive costs, toxicities, and long treatment courses [8]. As steps have been taken to prevent the rise in the prevalence and to implement disease control measures, Leishmania spp. continue to develop resistance against these first-line medications [9]. Producing new chemotherapeutics would require significant cost and time, further delaying the development of new treatments. Therefore, new approaches are needed to identify existing drugs for combating drug resistance. As such, a promising approach to treating neglected tropical diseases (NTDs) including leishmaniasis is employing repurposed drugs [10,11]. One approach toward repurposing involves the application of both in-silico and in-vitro approaches. Through computeraided drug design (CADD) assisted screening, this integrated approach has identified several effective therapeutics against Leishmania [12]. By virtue of prior FDA approval, repurposed therapeutics have available ADME and toxicity data in humans, thereby reducing associated time and costs.
In the current study, we discuss the tandem insilico and in-vitro approaches used for discovering the repurposable drugs, Lansoprazole and Posaconazole, and their efficacy against Leishmania donovani amastigotes, the causal agents of VL. We also propose the plausible ion channel-based mechanism of action (MOA) of Lansoprazole targeting L. donovani calcium-transporting ATPases, and Posaconazole targeting the Lanosterol 14-α demethylase enzyme. Through the combination of in-silico target identification followed by in-vitro antileishmanial testing, we conclude that Lansoprazole and Posaconazole can likely be repurposed to effectively treat leishmaniasis.
2. Materials and methods
2.1. Cell line and parasite maintenance
THP-1 cells (human monocytic leukemia) were maintained in RPMI-1640 medium (pH 7.4) supplemented with 10% FBS and 1% streptomycin/penicillin (Gibco 15140) at 37 °C in a 5% CO2 incubator. The L. donovani (DsRed2 LV82) transgenic line expressing DsRed2 and LUC (SwaI fragment from plasmid pIR1SAT-LUC-DsRed2; strain B5947) [13,14] was kindly provided by Dr. Abhay Satoskar (The Wexner Medical Centre, The Ohio State University, USA). Promastigotes were maintained at 25 °C, 5% CO2, in M199 media supplemented with 4.62 mM NaHCO3, 5 mg/L hemin, and 10% heat-inactivated FBS (Gibco) and 1% streptomycin/penicillin (Gibco, 15140). Parasite density was maintained between 1 × 106 parasites/mL and 4 × 107 parasites/mL by dilution with complete media for less than 10 sub-culture cycles to maintain genetic variability. Fluorescence was monitored before each experiment to confirm the stability of the line.
2.2. Preliminary drug screening
The Selleckchem Bio-Active Compound Library (Catalog# Selleck_L1700, was comprised of a 4718-member Bioactive Compound Library-I as of May, 2019 which included 1622 FDA approved compounds and the rest were advanced investigational compounds at different stages in drug development and was subjected to curation based on known activity against different intracellular parasites. The sources for the data were gathered from all known available resources including journals, patents, and other published literature. To associate known activity against protozoan parasites within the FDA-approved compound library, data mining was performed using text-, structure-, and activity- etc. based queries on CHEMBL [15] and ChemSpider servers [16]. The FDA-approved library was thus shortlisted to 96 compounds. These compounds were subjected to initial exploratory screening against L. donovani amastigotes with two concentrations of 100 μM and 10 μM(from 10mM stocks) in triplicate for each compound using live-cell fluorescence imaging as described in detail under the drug susceptibility assay heading below.
2.3. Parasite infection and drug susceptibility assay
THP-1 cells at 5 × 105 cells/mL were differentiated with 50 ng/mL of phorbol 12-myristate 13-acetate (PMA, Sigma P1585) for 48 h at 37 °C, 5% CO2. Differentiated THP-1 cells were mixed with 6-dayold Leishmania promastigotes (enriched with metacyclic promastigotes) at a final density of 4 × 105 THP-1/mL and 2 × 107 parasites/mL in RPMI medium supplemented with 10% FBS. This homogeneous mixture of differentiated THP-1 cells and parasites was seeded in 96-well clear-bottom plates at 50 μL/well followed by 5 days incubation at 37 °C, 5% CO2. The wells were washed 2 times with incomplete media (RPMI-1640) and 1 time with complete media. Eight wells (11th column in the plate) were seeded with THP1 cells only and used as the control for 100% compound response. Eight wells (12th column in the plate) were seeded with promastigotes only, which were washed away as part of the washing step and hence were blanks. The first columns of the plates were flooded with an additional 50 μL complete media and 1 μL of drugs/test compounds from a 10 mM stock (100 μM final concentration). The media was gently mixed and 50 μL media was carried forward to an adjacent well, therefore diluting the original concentration by half. The same process was repeated up to the 10th well, generating a drug concentration gradient through serial dilution (100 μM; 50 μM; 25 μM; 12.5 μM; 6.25 μM; 3.12 μM; 1.56 μM; 0.78 μM; 0.39 μM; 0.19 μM). All the serial dilutions were carried out in triplicates for each compound. The antileishmanial reference drugs used were amphotericin B (Sigma, A9528), miltefosine (Merck, 475841), and pentamidine isethionate salt (Sigma, P0547). The reference drugs and tested compounds were added 24 h after infection and incubated at 37 °C and 5% CO2 for 4 days.
2.4. High-throughput image analysis
Following incubation with drug compounds, cells were washed twice to remove non-adherent cells and preincubated in the incubation/imaging chamber for imaging on the ImageXpress Pico Automated Cell Imaging System (#IX Pico; Molecular Devices, San Jose, CA). For each well, images were captured at 10x magnification in a stitched 4 × 4 grid to create an acquisition region that covered 75% of the well. The macrophage cell count and area of total red fluorescence area observed were quantified by CellReporterXpress (Version 2.5.1.0 Beta: Molecular Devices, San Jose, CA). The total red fluorescence area was directly proportional to the parasitemia observed and sigmoid curves were plotted for the same to calculate IC50 values. Values of IC50 were obtained using Prism® 8.0 software (Graph-Pad Software Inc.) with statistical significance set at P < 0.05 and then subjected to nonlinear regression analysis using normalized values as percentage parasitemia vs. Log 10 drug concentration.
2.5. Target protein prediction
The repurposable drugs that were found to be highly active in both preliminary screening as well as in the drug susceptibility assay were further analyzed for likely targets within the Leishmania proteome. The in-silico modeling was performed to predict target proteins in the parasites and understand the potential mechanism(s) of action. In this study, the identified drugs were Lansoprazole and Posaconazole. The Lansoprazole target in mycobacterium reported by Rybniker (qcrB, Rv2196) is absent in L. donovani [17] indicating that Lansoprazole has a different mechanistic target in leishmania. We data-mined several drug-target web servers including Stitch, Swiss Target Prediction, MolTar-Prep, Super Prediction, and Target Hunter. Through each of these drug-target prediction servers, Lansoprazole was repeatedly shown to bind to calcium-transporting ATPases (SERCA/P-type family). Posconazole’s anti-leishmaniasis activity is likely due to inhibition of a sterol 14α-demethylase LDBPK_11 1100 in leishmania, as this enzyme is a highly conserved orthologue of the enzyme responsible for the compound’s known antiparasitic activity.
2.6. Target sequence analysis
Sequences for each predicted target homolog and orthologues from different Leishmania species were downloaded from the VEuPathDB Bioinformatics Resource Center [18] previously known as EuPathDB [19]. This was carried out using the BLAST tool within one reference strain of each Leishmania spp. analyzed. The sequence set of P-type ATPases family for each Leishmania spp. reference strain was realigned using the ClustalΩ online server [20]. The alignments were highlighted by Boxshade 3.21 server [21] and trees were visualized using dendroscope-3 software [22].
2.7. Target protein modeling and preliminary docking
Four L. donovani target proteins, LdBPK_352080.1, LdBPK_040010.1, and LdBPK_170660.1, were found to be closely related to the SERCA family of calcium transporting ATPases as likely Lansoprazole targets, while Posaconazole’s predicted target LDBPK_11 1100 (CYP51) involved in sterol synthesis belongs to the Cytochrome P450 superfamily and is membrane-associated with a single transmembrane domain at the N-terminus. I-TASSER was used to predict the target protein structure of the four targets and the active site map through COACH analysis [23]. Protein structures were validated by online servers accredited by CASP-13 [24]. Preliminary target protein prediction for Lansoprazole was tested with molecular docking analysis using PatchDock followed by FireDock refinement without selecting a target site, which showed a significant interaction of Lansoprazole with the ATP binding site of all three ATPase proteins, whereas Posaconazole showed a highly specific binding interaction with CYP51 with heme involvement in the sterol 14ɑ-demethylase [25]. For the in-silico work, both (S)- & (R)-enantiomers of Lansoprazole were docked and simulated separately, as while Lansoprazole is marketed as a racemate, the two enantiomers differ in their pharmacodynamics and pharmacokinetics. As these four proteins are transmembrane-associated, the complete analysis of Lansoprazole and Posaconazole interactions was carried out to further confirm the target protein for each drug.
2.8. Induced fit docking
The Lansoprazole (Pubchem Compound ID: 3883) & Posaconazole (Pubchem Compound ID: 468595) 2D and 3D structures were retrieved from the NCBI PubChem database including the enantiomers (S)-Lansoprazole: 138530-95-7, and (R)-Lansoprazole: 138530-94-6 [26]. These structures were then prepared using the LigPrep application in Schrödinger [27] with energy minimizations and energy optimizations. The Induced Fit Docking (IFD) application/workflow in Schrödinger was used to perform flexible protein-ligand docking in the presence of an implicit membrane position predicted by the OPM server [28]. The ATP binding site predicted by the COACH server was used to generate a receptor grid of the protein for the Lansoprazole targets. For the Posaconazole target, the heme and Posaconazole binding grids were generated based on the solved X-ray structure of the fungal homolog (PDB-ID; 6E8Q). The “trim-side chains” and “protein preparation constrained refinement” options were not selected as preliminary docking with automated servers (Patchdock/Firedock) did not indicate a ligand entry block. Follow-up re-docking and scoring were performed by extra precision (XP) within 30.0 kcal/mol of the best structures, comprising 20 structures overall utilizing the generated Glide score (kcal/mol) and IFD score (kcal/mol). While the Glide score is based on various energies involved in ligand and binding site interactions, the IFD score is calculated by the addition of Prime energy calculations. The top conformer was selected by re-ranking using Prime-MMGBSA, which calculates additional ligand strain in the predicted docked pose [29]. For Posaconazole the docking was performed in two sequential steps by first docking the heme, then docking Posaconazole.
2.9. Molecular dynamic simulations
Although IFD considers the ligand-induced receptor flexibility, the channels studied had transmembrane regions very close to the ATP binding site. Also, Posaconazole had a very typical binding with a slot canyon-like binding site stabilized by the heme. Therefore, a long 100ns MD simulation was additionally performed using the Desmond module of the Schrödinger suite to evaluate the binding stability of the ligand in the presence of water, physiological ions, and transmembrane surface tension [30]. The boundary conditions were defined by forming a 10 Å × 10 Å x 10 Å orthorhombic box around the protein-ligand complex. A POPC membrane system was set up around the transmembrane regions predicted by the Orientations of Proteins in Membranes (OPM) server [28]. The systems were then solvated in a predefined TIP3P water arrangement. The protein model was neutralized by access to Na+/Cl− ions with an excess 0.15M NaCl solution mimicking physiological conditions. Relaxations of model systems were performed before simulations in NPγT ensemble class at constant temperature (300.0 K), pressure (1.013 bar), and surface tension (0.0 bar Å). The energy was recorded at regular intervals of 1.2ps for a total 100ns simulation time with a 20ps trajectory recording (5K frames). Trajectory analysis was performed by generating a simulation interaction diagram (SID).
3. Results
3.1. Drug susceptibility in parasite infection assay: IC50 values for FDA approved therapeutics and cytotoxicity assay results
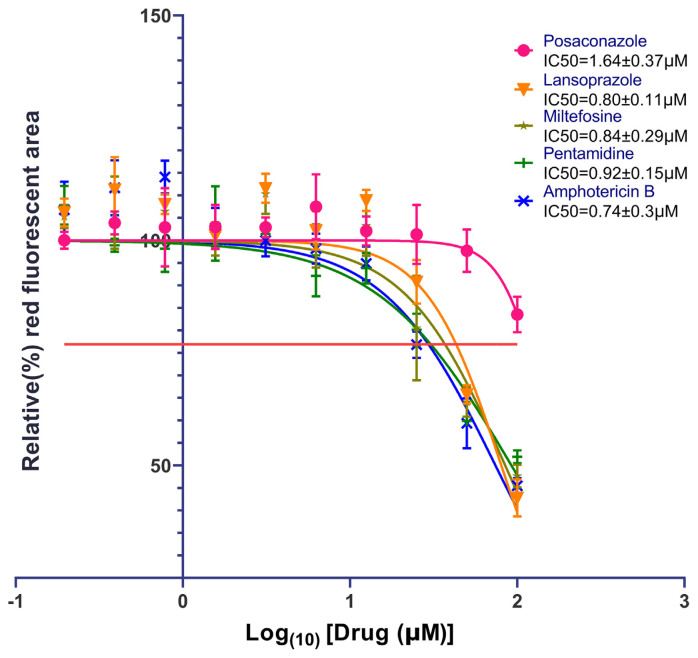
The IC50 values from the dose–response experiments for Lansoprazole, Posaconazole, and the standard antileishmanial chemotherapeutics are shown in Fig. 1. The IC50 values of Lansoprazole and Posaconazole were 0.80 μm and 1.64 μm, respectively. These values compare favorably to the IC50 values of the approved antileishmanial agents’ Miltefosine (hexadecylphosphocholine) (0.84 μm), Pentamidine (Pentamidine diisethionate) (0.92 μm), and Amphotericin B (0.74 μm). Compared to these standard drugs, Lansoprazole is as effective in inhibiting L. donovani as Miltefosine and pentamidine. Amphotericin B appears to be slightly more potent, but that difference is not statistically significant. Another important factor is that the inherent red fluorescence of Lansoprazole has been shown to possibly give a false negative reading, consistent with a potentially higher potency of Lansoprazole in killing the parasites than indicated by the measured IC50. Posaconazole is shown to have a higher (less potent) IC50 than Lansoprazole and the standard anti-leishmanial drugs, indicating that Lansoprazole is more effective against Leishmania amastigotes. Nevertheless, the approved dosage of Posaconazole is 20 times that of Lansoprazole given its tolerable toxicity profile making it also a prime candidate for a repurposable drug to treat leishmaniasis. Neither compound showed any toxicity up to 100 μM in HEK293 cells in an Alamar blue assay, consistent with their known breakpoints. The pharmacologic profiles of Lansoprazole & Posaconazole are summarized in Table S2.
Fig. 1.
IC50 values for Lansoprazole and Posaconazole were tested and obtained against standard anti-leishmanial drug therapeutics. Figures were created and IC50 values were calculated using GraphPad Prism version 8 software (GraphPad Software, Inc., La Jolla, CA, USA). The dose–response curve function of this software was used for IC50 calculations for standard as well as experimentally tested drugs. The horizontal straight orange line on the Y-axis is the average concentration of drugs tested that give half-maximal effects which is higher than 50% (~77%) because the maximum inhibition is at 40% due to residual background fluorescence from lysed pathogens. The Parasiticidal activity was calculated with the live fluorescent imaging (Fig. S3.) after 24h treatment with different concentrations of control and experimental drugs (50, 25, 12.5, 6.2, 3.1, 1.6, 0.8, 0.4, 0.2, and 0.1 μM). The relative infection rate in live imaging was determined by red fluorescent area (parasite expressing reporter gene DsRed2) in the captured well image of a 96 well plate. Inset is the calculated IC50s of all the drugs. The relative infection rate of untreated infected cells was the starting point of the curve.
3.2. Putative Lansoprazole and Posaconazole targets and their homology among Leishmania spp
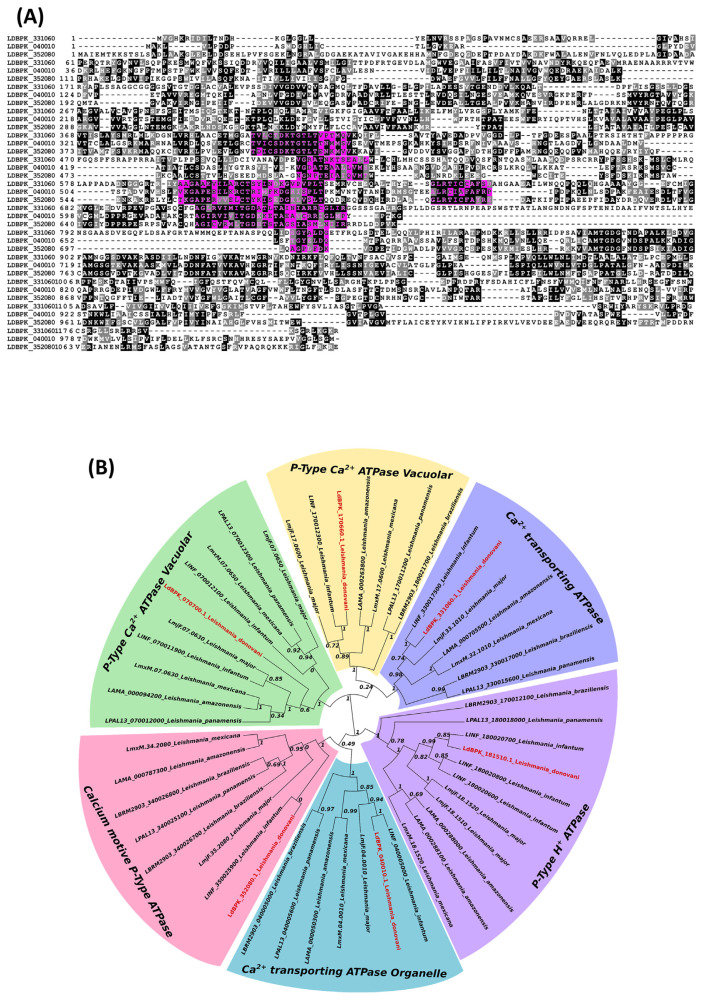
The putative Ca2+ transporting ATPase protein targets in L. donovani share high homology, indicating a potential for broad-spectrum antileishmanial (VL and CL) activities for compounds that target these proteins. The amino acid sequences of the respective protein targets of L. donovani are illustrated in Fig. 2A. As shown in Fig. 2B., the genetic origins of Ca2+ ATPase channels are shared among different species of leishmania. In addition, the genetic origins of calcium-transporting ATPases can be traced back to the origins of P-type H+ ATPases, calcium-transporting ATPase organelles, calcium motive P-type ATPases, P-type calcium ATPase vacuolar, and P-type calcium ATPase vacuolar antileishmanial targets. Proteins LdBPK_040010.1, LdBPK_352080.1, and LdBPK_170660.1 are located in the calcium-transporting ATPase organelle, in the calcium motive Ptype ATPase, and the P-Type calcium ATPase vacuolar, respectively. The genetic map provides further insight into potential protein targets that are essential to the lifecycle of L. donovani and other Leishmania spp. It is important to note that the similarities between protein targets create opportunities for broad-spectrum application of chemotherapeutics targeting these proteins in all strains sharing these homologous proteins. Table S1. shows the essential nature of orthologs of calcium ion transporting ATPase enzymes through the results of knock-out studies and for lanosterol 14α demethylase for Posaconazole. Phylogenetic analysis of both targets (LdBPK_352080.1 for Lansoprazole and LdBPK_111100.1 for Posaconazole) with the orthologues revealed high conservancy of these targets among different Leishmania spp. infecting humans. Figure S4. shows more than 92% conservancy of LdBPK_352080.1 among human leishmaniasis orthologs with no gaps in the Lansoprazole or ATP binding regions. For the Posaconazole target LdBPK_111100.1 (Fig. S5.) there was 85% identity, while the rest of the polymorphisms showed biochemical conservancy including the posaconazole and heme-binding sites which were 100% conserved.
Fig. 2.
(A) Multiple sequence alignment of P-type calcium transporting ATPases of L. donovani. These isotypes have high homology in the ATP binding site, which is highlighted in pink. Proteins LdBPK_170660.1, LdBPK_040010.1, and LdBPK_352080.1 are the accession numbers of amino acid sequences used for analysis. (B) Phylogenetic tree of P-type calcium transporting ATPases from different species of Leishmania commonly pathogenic to humans. Gene accession prefixes represent reference strains of different species: LbrM (L.{Viannia}braziliensis {MHOM/BR/75/M2904}), LmjF (Leishmania major{ strain Friedlin}), LmxM (Leishmania mexicana{MHOM/GT/2001/U1103}), LdBPK (L. donovani {BPK282A1}), LinJ (Leishmania infantum{JPCM5}) LpaL13(Leishmania panamensis {MHOM/COL/81/L13}) and LAMA (Leishmania amazonensis{ MHOM/BR/71973/M2269 }).
The calcium-transporting ATPase in L. donovani is shown to have homology with a similar isotype pattern among different Leishmania species, indicating highly specialized and energy-driven Calcium regulation and sequestration in all the species responsible for the majority of human infections. We know there are unique calcium storing organelles called calciosomes in kinetoplexans as well as apicomplexans [31]. The putative protein targets LdBPK_040010.1, LdBPK_352080.1, and LdBPK_170660.1 are shown in calcium-transporting ATPase organelle, calcium motive P-type ATPase, and P-type calcium ATPase vacuolar, respectively.
3.3. Induced fit docking and validation by MD simulations of drug-bound Lansoprazole and Posaconazole targets
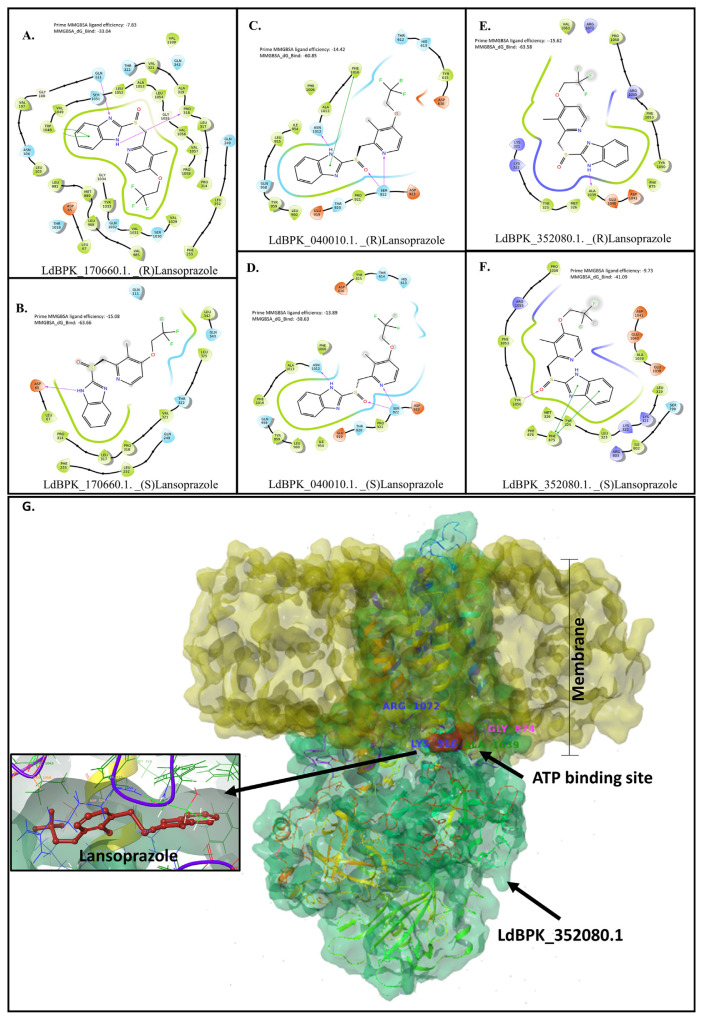
Induced fit docking of Lansoprazole yielded very stable complexes according to follow-up MM-GBSA analysis (Fig. 3). All the isoforms of P-type Ca2+ ATPase had very strong predicted interactions with all three targets for both Lansoprazole enantiomers. Docking of Posaconazole was comparable to reported interactions with the fungal homolog [32]. The heme moiety played an essential role in stabilizing the complex participating in π-stacking interactions with the 1,2,4-triazole ring. LdBPK_352080.1 seems to have similar and energetically favorable interactions with both Lansoprazole enantiomers, but (R)-Lansoprazole bound with LdBPK_352080.1 is the most stable complex in terms of binding energy and ligand strain.
Fig. 3.
(A, B, C, D, E, and F) Schematic representation of the 2D interaction maps of Lansoprazole enantiomers bound to the ATP binding site of three plausible targets belonging to P-type calcium channels. The MMGBSA energy scores (ΔG) and ligand efficiencies denote ligand strain and unfavorable steric interactions in a particular docking pose. (G) Schematic representation of (S)-Lansoprazole with LdBPK_352080.1. The yellow surface represents the membrane topology, a protein embedded in the membrane is green, and the ATP binding site with the docked (S)-Lansoprazole molecule (red) is close to the transmembrane region. Inset is the (S)-Lansoprazole molecule (red) within the binding pocket. Based on docking energies and stability during MD simulations this protein is the likely target for Lansoprazole antileishmanial activity.
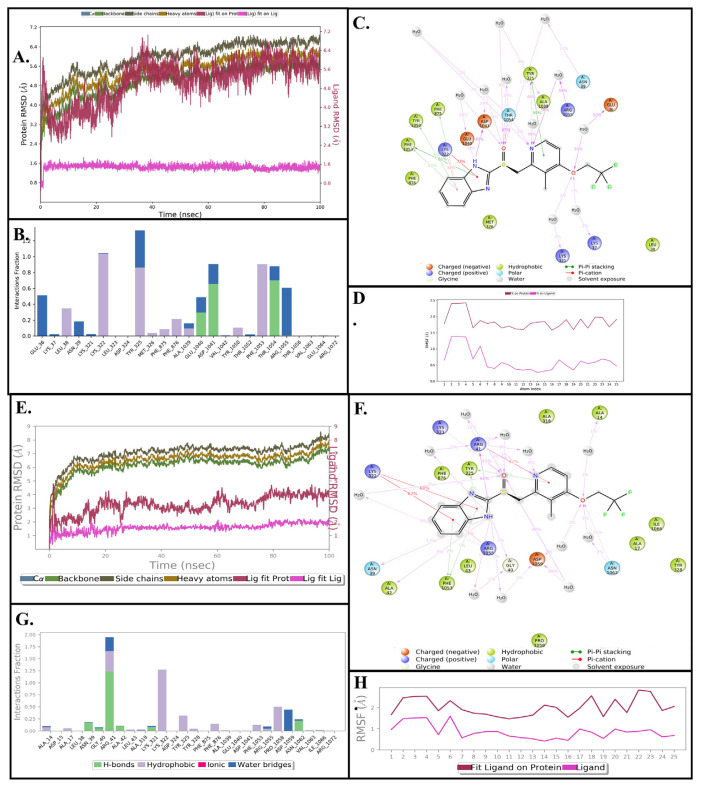
To validate the stability and dynamics of the three putative leishmanial protein targets (LdBPK_35 2080.1, LdBPK_040010.1, and LdBPK_170660.1) and the Posaconazole target (LdBPK_111100.1), we completed a 100ns MD simulation of the induced fit docked ligand-protein complexes that showed high affinity and low relative binding energy. MD simulation allows for analysis of root means square deviation (RMSD) and root mean square fluctuation (RMSF). RMSD measures the average displacement of the protein during the simulation, thus it correlates with the stability of the ligand-protein interaction, while a lower RMSD correlates with greater protein stability. Meanwhile, the RMSF measures local changes along the protein chain or the deviation between the position of the ligand and a set reference position within the protein [33]. As shown in Figs. 4 & 5, the data demonstrate the molecular binding for Lansoprazole with the three target proteins and Posaconazole with its target. Figs. 4 & 5 show the schematic of ligand atom interactions with protein residues, demonstrating the percentage of time that a particular interaction occurs. A higher percentage for an interaction correlates with a more significant interaction with the binding site.
Fig. 4.
(A, B, C, & D) Results of a 100 nanosecond (ns) molecular dynamics simulation of (S)-Lansoprazole bound to LdBPK_352080.1. (A) Root mean square deviations between the calcium pump ATP binding site and bound ligand. The graph obtained for the RMSF value of the ligand (purple line) from the protein backbone (green line) revealing that there was no major conformational change and the ligand stayed in the binding site throughout the simulation. (B) Critical protein-ligand contacts of amino acid side chain residues with their interaction types designated by color. (C) Schematic 2D representation of bound ligand interactions of (S)-Lansoprazole with the side chains of amino acid residues lining the binding pocket throughout the simulation. (D) Atomic index showing RMSF of different components of the ligand showing individual movements. The trifluoromethoxy has a wide rotational range of motion but it does not influence the binding affinity of the ligand as a whole. (E, F, G, & H) Results of a 100 nanosecond (ns) molecular dynamics of (R)-Lansoprazole bound to LdBPK_352080.1. (E). Root mean square difference between the calcium pump ATP binding site and bound ligand. The graph obtained for the RMSF value of ligand (purple line) from the protein backbone (green line) revealing that there was no major conformational change and that the ligand stayed in the binding site throughout the simulation. The total RMSD of ligand post 10ns stabilization was less than 2 Å compared to the maximum allowed value of 4 Å. (F) Critical protein-ligand contacts of amino acid side chain residues with the interaction types designated by color. (G) Schematic 2D representation of bound ligand interactions of (S)-Lansoprazole with sidechains of amino acid residues of the binding pocket throughout the simulation. (H) Atomic index showing RMSF of different components of ligands showing individual movements. The trifluoromethoxy has a wide rotational range of motion but it does not influence the binding affinity of the ligand as a whole.
Fig. 5.
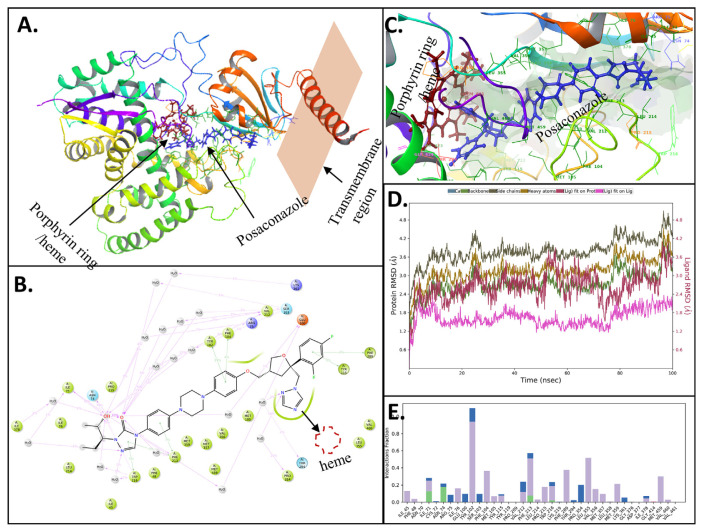
Induced fit docking and Molecular dynamics (100ns) of Posaconazole with LDBPK_111100 (CYP51): (A) Schematic representation of Posaconazole bound to LDBPK_111100. The protein is represented in ribbon form, and the ligand-binding site with the docked (S)-heme molecule (red) is close to the Posaconazole (blue) binding site. (B) Schematic 2D representation of bound ligand interactions of Posaconazole with sidechains of amino acid residues lining the binding pocket throughout the simulation. (C) Schematic representation of the binding topology of Posaconazole (blue) and heme (red). (D) Root mean square deviations between the calcium pump ATP binding site and bound ligand. The graph obtained for the RMSF value of ligand (purple line) from the protein backbone (green line) revealing that there was no major conformational change and that the ligand stayed in the binding site throughout the simulation total RMSD of ligand post 10ns stabilization was less than 2 Å compared the maximum allowed value of 4 Å (E). Critical protein-ligand contacts of amino acid side chain residues with the interaction properties.
3.3.1. Molecular dynamics simulations of lansoprazole enantiomers docked to the ATP binding domain in the target proteins
Figs. 4A, & E, and 5D illustrate the protein RMSD (left Y-axis), which provides insight into the structural conformation of the protein and ligand during the simulation. A stable value in Å (<4Å fluctuations) indicates that the interaction has equilibrated. The ligand RMSD (right Y-axis) indicates how stable the ligand is within the protein’s binding pocket. Figures 4B, C, F and G, and 5 B and E show the protein interactions at the amino acid side chain residue level throughout the simulation that is categorized into four types: hydrogen bonds, hydrophobic, ionic, and water bridges. Specific amino acid residue side chains within the binding pocket are found on the X-axis. The fraction of interactions noted on the Y-axis correlates to the proportion of time that a specific interaction is maintained. Generally, the nitrogen and carbon protein termini fluctuate the most, while secondary structures are usually more rigid than loop regions, as expected.
As lansoprazole is marketed as a racemate, both enantiomers were investigated in-silico, and the LdBPK_352080.1 ATP binding site has a strong binding affinity to both Lansoprazole enantiomers. The overall RMSD for this complex was in the desirable range (<4 Å) after an initial stabilization phase of 10ns, and the ligand strain was minimal as the ligand-binding further improved and water molecules enhanced the stability of the complex rather than displacing the ligand. The RMSD of the LdBPK_352080.1 protein was found to increase initially and reached 6.4 Å at 40ns and remained constant for the remaining simulation. Protein-ligand interactions correlated well and demonstrated the conformational stability of ligand-protein interaction. For the (S)-Lansoprazole/LdBPK_352080.1 complex (Figs. 3G & 4A–D), Asp-1041 and Thr-1054 played critical roles in ligand binding. Asp-1041 accepted hydrogen bonds 65% of the time from (S)-Lansoprazole, while Thr-1054 donated hydrogen bonds to the oxygen of (S)-Lansoprazole 67% of the time. Asp-1041 is in the same site that ATP binds to on the LdBPK_3 52080.1 protein (Fig. 4). For (R)-Lansoprazole LdBPK_352080.1 (Fig. 4 E-H) the complex seems to be more stable, while initial docking has quite a similar interaction profile, both enantiomers interacted differently in the MD phase. (R)-Lansoprazole was more stable throughout the simulation with less than 2 Å deviation post initial 10ns stabilization. Arg-41 and Lys-322 played critical roles in stabilizing pyridine and benzimidazole substructures respectively. Asp-1059 formed a water bridge with the trifluoromethoxy oxygen intermittently from (R)-Lansoprazole. The simulation with the docked molecule was most stable not only concerning retained binding characteristics throughout the simulation, but in addition, the water molecules further strengthened the interactions as the energy of the complex decreased and were hence stabilized in the course of the long simulation.
LdBPK_111100.1 was also subjected to in silico simulation and was initially docked with the heme, forming a complex very similar to fungal orthologues [32]. The binding site for Posaconazole is a slot canyon-like linear fissure. The piperazine and phenyl rings in the middle regions bind to the hydrophobic trench formed by amino acid residues: multiple Met residues, two Phe, and one Val (Fig. 5 B and E). The triazole stacks with heme moiety. The tetrahydrofuran and alkoxy phenyl substructures sit in a stable pocket comprised of Phe-289 and Glu-100. Throughout MD simulations the hydrophobic region was most stable and was nearly immobile. Following the initial stabilization of 10ns, the total RMSD of the ligand was less than 2 Å. This was surprising given the large size and minimum intramolecular folding of the ligand suggesting a strong and highly specific binding.
4. Discussion
We report the first known activity of Lansoprazole and Posaconazole against L. donovani amastigotes and extended the study to present a plausible MOA of these FDA-approved compounds in the parasites. Based on our studies, lansoprazole targets the Leishmania calcium-transporting ATPases, and Posaconazole targets lanosterol 14-alpha-demethylase (LDBPK_111100). In the current study, three of these closely related ATPase targets were found to have high affinity and low binding energy with Lansoprazole: LdBPK_352080.1, LdBPK_040010.1, and LdBPK_170660.1. These proteins act as a calcium motive P-type ATPase, a calcium-transporting ATPase, and a P-type ATPase, respectively. Through induced-fit docking followed by longer MD simulations of 100ns to assess the long-term stability of binding, we have demonstrated that Lansoprazole binds to the ATP binding site of these proteins. Furthermore, we show that LdBPK_352080.1 is the most plausible Lansoprazole target based on the greater stability of the drug-target interactions (Fig. 3G). As the docking with the other two homologs was favorable, and though the simulation was not optimal as described in the respective result section, the ligand remained in the binding site throughout the molecular dynamics run. Therefore, efficacy may be a cumulative effect derived from somewhat weaker binding to each of these three homologs. As these proteins are ATPases, they are performing energetically essential functions, and blocking these functions would have a detrimental effect on parasite calcium homeostasis. Calcium homeostasis has been shown to play a role in critical functions of leishmania, including the process of cell invasion in Leishmania amazonensis [34]. Furthermore, none of these proteins have any significant homology with human host sequences. They contain highly conserved sites at the pore region, ATP binding site, and calcium-sensing/binding region, and have only 0–6% similarity with the closest human orthologues [31].
Compared to Amphotericin B, Pentamidine, and Miltefosine, Lansoprazole has a comparable IC50. While the approved dosage of Lansoprazole is 20–50 mg daily which is much lower than a normal antibiotic dose with similar activity. Oral doses up to 5000 mg/kg in rats (approximately 1300 times the 30 mg human dose based on body surface area) and mice (about 675.7 times the 30 mg human dose based on body surface area) did not produce deaths or any clinical signs; also, there is a case report with a patient consuming 600 mg of Lansoprazole (Prevacid) without any adverse side effects [35]. This further indicates that compared to the standard drug therapeutics, Lansoprazole is both safe and equally effective in inhibiting L. donovani’s life cycle, as measured by a decrease in relative red fluorescence in the assay. Posaconazole is shown to have a higher IC50 than Lansoprazole and the standard anti-leishmanial drugs, indicating it is not as potent. However, the approved dosage of Posaconazole is 20X that of Lansoprazole suggesting a strong case for repurposing for both drugs.
Lanosterol C-14 demethylase (LDBPK_111100.1) is a lipid metabolism enzyme and a member of the ergosterol biosynthesis pathway. Many members of this pathway, such as HMG-CoA reductase, squalene epoxidase, lanosterol C-14 demethylase, and sterol C-24 methyltransferase, are proven drug targets [36]. Lanosterol-C14-demethylase is a critical P450 family enzyme for trypanosomatids, including Leishmania, with its inhibition resulting in membrane permeability alterations, reduced infectivity, and impaired mitochondrial functions [37]. In leishmania, another member of the cytochrome P450 family, CYP5122A1 (LdBPK_270090.1) [38], also modulates ergosterol levels and probably supports extracellular survival of Leishmania upon inhibition of lanosterol C-14 demethylase [39]. CYP51 (sterol 14α-demethylase) is highly conserved across eukaryotes at the structural level [40] but there is negligible homology of the same with human orthologues. The essentiality and drug targetability of CYP51 have been well documented with examples of successful drug targeting [41]. Through induced-fit docking followed by longer MD simulations of 100ns to assess the stability of binding, we have demonstrated that Posaconazole binding to the known binding site of Lanosterol C-14 demethylase (LDBPK_111100.1) is comparable to fungal counterparts and has the heme (porphyrin ring) as an interacting partner.
Several drugs are currently used to treat leishmaniasis [42]. The first-line treatment for Leishmania infection is pentavalent antimony sodium stibogluconate, an organometallic prodrug that works by inhibiting trypanothione reductase [43]. Sodium stibogluconate is very toxic to the veins at the site of injection, and pancreatitis is a common side effect of the drug. Second-line medications are pentamidine (PTM) and amphotericin B, which work by inhibiting DNA and sterol biosynthetic pathways, respectively [44]. PTM is no longer used due to toxic side effects in humans and drug resistance in parasites, while amphotericin B treatment requires hospitalization and high cost due to the requirement of lipid formulation [45,46]. Other medications used to treat Leishmania include miltefosine and paromomycin. Miltefosine is the only orally available anti-leishmaniasis drug and exhibits its effect by inhibiting phosphatidylcholine, but it is teratogenic.
In this study, we have also explored the importance of calcium channels (CCs) and P-type ATPases as novel therapeutic targets. Several studies have shown that calcium concentrations in L. donovani are critical for parasite metabolism and invasion [31]. Calcium is tightly regulated through transporters in the plasma membrane, ER, mitochondria, and acidocalcisomes [47]. Drugs that modulate CCs have been used for chemosensitization, which improves the efficacy of antiparasitic drugs [31]. CCs are a promising antiparasitic drug target to combat growing drug resistance [31]. In addition, P-type ATPases have an integral function in maintaining lipid membrane asymmetry and cellular ion homeostasis by moving phospholipids and ions against their concentration gradients [48]. ATPases are important for Leishmania spp. to withstand changes in the external environment they encounter throughout the parasitic life cycle. This study demonstrates that Lansoprazole and Posaconazole should be investigated clinically as repurposed antileishmanial drugs.
Lansoprazole is an FDA-approved PPI drug, extensively used to reduce gastric acid production via inhibition of H+/K+ ATPase pumps in humans. It is also used to treat gastric Helicobacter pylori infections in combination with antibiotics. Like all PPIs, Lansoprazole is labile in strong acid and forms an active sulfenamide derivative below pH 4. The reactive sulfenamide intermediate leads to the formation of a covalent disulfide bond between cysteine residues of the gastric proton pump αsubunit, thus deactivating the enzyme [49]. Our studies demonstrate the utility of the parent molecule, independent of acid-mediated sulfonamide formation. Importantly for its use as a repurposable drug, Lansoprazole is available as a generic oral drug at low cost from multiple international pharmaceutical manufacturers. Furthermore, we have shown Lansoprazole to be effective against multidrug-resistant Plasmodium falciparum, the parasite responsible for causing severe malaria, with an IC50 range of 7–11 μM [50]. However, the underlying antimalarial mechanism of action for Lansoprazole against P. falciparum is unknown. Jiang et al. demonstrated that another PPI, omeprazole, is effective against Leishmania by targeting P-type K+/H+-ATPase on the surface membrane [51]. As noted by Riel et al., it is unlikely that the internal environment of a parasite could produce a pH low enough to form the active Lansoprazole intermediate, suggesting an alternate antiparasitic mechanism of action. Lansoprazole may be effective against parasites through an intracellular sulfoxide reduction (a similar pathway is reported in mycobacterium) [17].
Herein, we present Lansoprazole and Posaconazole as potentially repurposable chemotherapeutics for treating leishmaniasis, targeting proteins specific in L. donovani with negligible similarity to host orthologues. Our study employed in-silico approaches combined with in-vitro studies to demonstrate that Lansoprazole and Posaconazole are effective in combating L. donovani. The low cost and limited side effects of these compounds make them promising candidates for drug repurposing while maintaining potency comparable to currently used therapeutics for treating leishmaniasis. This is of critical importance as L. donovani and other Leishmania spp. have developed multidrug resistance to available therapeutics. The utilization of Lansoprazole as a repurposed therapeutic may eliminate the need to create new therapeutics, which would require significant time and economic cost. As these compounds are already FDA-approved, they should be immediately tested in animal models for in-vivo efficacies and subsequently assessed in human trials in endemic regions. Furthermore, the drug discovery pipeline utilized in the current study can be used to further discover new treatments utilizing new targets in other infectious diseases via high-throughput virtual screening and integrated drug-target identification.
5. Conclusion
Our results from integrated drug discovery methodologies demonstrate that FDA-approved drugs and other well-characterized drug libraries can be utilized to facilitate the screening of target-specific inhibitors of L. donovani and other parasitic species. In the current study, we have discovered two widely used FDA-approved drugs, Lansoprazole and Posaconazole, as promising chemotherapeutics to combat leishmaniasis. Our results also predicted a conserved mechanism for Posaconazole (sterol 14-alpha-demethylase) and a novel mechanism of action for Lansoprazole pointing to calcium-transporting ATPases, which act in the ER membrane of Leishmania. These target proteins have been deemed essential and critical to L. donovani survival. Stability in MD simulations further supports LdBPK_352080.1 as the most plausible target for Lansoprazole. Further experiments are needed to confirm this likely mechanism of action as well as demonstrate a possible synergy of Lansoprazole and/or Posaconazole with other approved therapeutics as seen with various calcium modulators. The current study complements parallel studies that have demonstrated Lansoprazole and Posaconazole to be effective antiparasitic drugs. The already-marketed drugs Lansoprazole and Posaconazole are inexpensive and carry minimal side effects compared to currently used anti-leishmaniasis drugs and may prove to be powerful tools in combatting drug resistance in L. donovani observed with current therapeutics.
Acknowledgement
We thank Dr. Abhy Satoskar (Ohio State University) for graciously providing the L. donovani strain expressing the reporter gene. Authors sincerely thank the Department of Medicine, Loyola University Chicago Stritch School of Medicine for providing the initial funding support for the Drug Discovery Program and Software acquisition.
ABBREVIATIONS
- ADME
Absorption, distribution, metabolism, and excretion
- CC
Calcium channel
- CADD
computer-aided drug design
- CL
cutaneous leishmania
- VL
visceral leishmania
- DALY
disability-adjusted life year
- ER
endoplasmic reticulum
- FDA
Food and Drug Administration
- IC50
50% inhibitory concentration
- IFD
induced-fit docking
- LMIC
low and middle income countries
- MOA
mechanism of action
- OPM
Orientations of Proteins in Membranes
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- PPI
proton-pump inhibitor
- PTM
pentamidine
- RMSD
root means square deviation
- RMSF
root mean square fluctuation
- spp.
several species
- SID
simulation interaction diagram
- WHO
World Health Organization
Appendix
Molecular dynamics of (S)-Lansoprazole with LdBPK_040010.1. (i) Gln 111 donated hydrogen bonds to oxygen on Lansoprazole 71% of the time. Gln 259 donated hydrogen bonds to nitrogen on Lansoprazole 40% of the time. (ii) The ligand Å values are similar to the RMSD of LdBPK_040010.1, indicating that Lansoprazole is stable within the binding site. The RMSD of LdBPK_040010.1 was found to increase initially and reached 8.0 Å at 20ns and remained constant until 60ns when it decreased to 7.0 Å, where it remained stable for the remainder of the simulation. (iii) Protein-ligand interactions are consistent with conformational stability and correlated well. Gln 111 and Gln 249 were integral to ligand binding. (iv, v) There was no significant deviation in the RMSF of each amino acid residue measured with respect to its Cα carbon atom. The RMSD of LdBPK_040010.1 increased initially and reached 8.0 Å at 20ns and remained steady until 60ns when it decreased to 7.0 Å, thereafter it continued stably for the remainder of the simulation. Like LdBPK_352080.1, there was no significant deviation in the RMSF of each amino acid residue measured with respect to the Cα atom. Protein-ligand interactions demonstrated conformational stability and correlated well. Gln 111 and Gln 249 were integral to ligand binding. Gln donated hydrogen bonds to oxygen on Lansoprazole 71% of the time. Gln 259 donated hydrogen bonds to nitrogen on Lansoprazole 40% of the time (Fig. 5). While the ligand stayed in the docked site throughout simulation a significant wobble was observed with the trifluoride region completely dislodging from the predicted binding site causing an increase in ‘wobble’ and the energy decrease during simulation can be attributed to the protein component of the complex rather than the intermolecular interactions.
Molecular dynamics of (S)-Lansoprazole with LdBPK_170660.1. (i) Ser922 donated hydrogen bonds to oxygen on Lansoprazole 99% of the time (ii) The RMSD of LdBPK_170660.1 was found to increase initially and reached 10.0 Å from 5 to 40ns and then increased again to 15 Å at 50ns. The RMSD of Lansoprazole remained constant at 7 Å throughout the simulation. (iii) Protein-ligand interactions correlated well and are consistent with the conformational stability of ligand-protein interaction. For LdBPK_170660.1, Ser 1041 played an integral role in ligand binding. (iv, v) There was a moderate deviation in the RMSF of each amino acid residue measured with respect to its Cα carbon atom. The RMSD of LdBPK_170660.1 was found to increase during initial simulations and reached 10.0 Å from 5 to 40ns and then increased again to 15 Å at 50ns. The RMSD of Lansoprazole remained constant at 7 Å throughout the simulation. There was a moderate deviation in the RMSF of each amino acid residue measured with respect to the Cα atom. Furthermore, protein-ligand interactions demonstrated conformational stability and correlated well. Ser 922 was critical to ligand binding and donated hydrogen bonds to oxygen on Lansoprazole 99% of the time (Fig. 4). The interactions were mostly electrostatic which was weakened by water solvation during the simulation and the ligand interactions became destabilized during simulation without much change in the ATP binding site architecture. These findings support low docking scores by poor performance in simulation in terms of complex stability.
An example image set (each panel is 1/8th area of one well from a triplicate set) of live imaging (ImageExpress Pico) at 10X with L. donovani amastigotes infected macrophages post-treatment with different drugs. Here we have shown a three-drug i.e. Amphotericin B (A, B, C & D), Lansoprazole (E, F, G, & H), and Posaconazole (I, J, K, & L). The dose–response area with red fluorescence i.e. parasites expressing reporter gene DsRed2 is shown. Macrophages infected with leishmania attract more macrophages forming infection masses making the total fluorescence area a reliable parasite load estimation tool as compared to counting individual parasites. Latter requires much higher resolution screening but is less affected by background fluorescence from lysed parasites. Panels A, E, & I are negative controls with untreated and uninfected macrophages. Panels B, F, J (50 μM) and C, G, K (3.1 μM) are infected macrophages treated with different drug concentrations. Panels D, H, J are positive control panels with untreated and infected macrophages. All pannels were seeded with the same number of THP-1 cells and except for uninfected controls, the rest were introduced with the same number of promastigotes.
Orthologs of Posaconazole targeted Lanosterol C-14 demethylase (LDBPK_111100.1) from reference strains of different species of leishmania were obtained from plasmoDB (VEUpathDB) [52] using BLAST similarity searches. Orthologs further analyzed were: Leishmania infantum (LINJ_111100), Leishmania Mexicana (LMXM_111100), Leishmania major (LMJF_111100), Leishmania braziliensis (LBRM_110880), and Leishmania panamensis (LPMP111090). The phylogenetic analyses of the orthologs were performed on the Phylogeny.fr server [53]. A. The phylogenetic trees were constructed using the PhyML program (v3.1/3.0 aLRT) with the maximum-likelihood method with default settings. B. The multiple sequence alignment (MSA) was obtained with MUSCLE (v3.8.31) and gaps and/or poorly aligned regions were removed by Gblocks (v0.91b) with default settings. Publication-quality outputs of the sequence alignments were generated with the BOXSHADE 3.21.5 server [21].
Supplementary Video. Molecular dynamics of (S)-Lansoprazole with LdBPK_352080.1. The membrane is shown as a surface translucent cloud and protein is represented as ribbons. The interacting aminoacids side-chain residues and drug molecules are represented as line models of their chemical structure. The transmembrane topology remains stable throughout the 100ns simulation. However, the protein had a lot of swinging but stable movement showing it to be a good model. (S)-Lansoprazole remained in the binding site, and though had movements throughout the simulation they seem to be partly due to whole protein movements and not due to weakness in interaction or ligand strain. While the molecule remains within the ATP binding site and does not fly off an alternate stable pose is formed at 80ns time point with less than 4 Å deviation and retaining 85% of interacting amino-acid side chain residues. This transition doesn’t affect the binding energy but having multiple binding poses increase the engagement time favoring the competitive inhibitor over the natural substrate. The Supplementary video can be found at https://www.jfda-online.com/cgi/viewcontent.cgi?filename=12&article=3394&context=journal&type=additional&preview_mode=1.
Orthologs of Lansoprazole targeted calcium channel (LdBPK_352080.1) from reference strains of different species of leishmania were obtained from plasmoDB (VEUpathDB) [52] using BLAST similarity searches. Orthologs further analyzed were: L. infantum (LINJ_352080), L. major (LMJF_352080), L. Mexicana (LMXM_342080), L. braziliensis (LBRM_341990), and L. panamensis (LPMP_341910). The phylogenetic analyses of the orthologs were performed on the Phylogeny.fr server [53]. A. The phylogenetic trees were constructed using the PhyML program (v3.1/3.0 aLRT) with the maximum-likelihood method with default settings. B. The multiple sequence alignment (MSA) was obtained with MUSCLE (v3.8.31) and gaps and/or poorly aligned regions were removed by Gblocks (v0.91b) with default settings. Publication-quality outputs of the sequence alignments were generated with the BOXSHADE 3.21.5 server [21].
Table S1.
Gene significance analysis based on experimental evidence from knock-out studies with orthologs of calcium ion transporting ATPase channels and Sterol 14α demethylase in related organisms.
| Gene/Orthologs | Organism | Essentiality/Phenotype | Source Study (TDRtargets.org) | |
|---|---|---|---|---|
| Lansoprazole | Tb927.5.3400 | Trypanosoma brucei | significant loss of fitness in the bloodstream (3 days) | [54] |
| Tb927.5.3400 | Trypanosoma brucei | significant loss of fitness in the bloodstream (6 days) | ||
| Tb927.5.3400 | Trypanosoma brucei | A significant gain of fitness in procyclic | ||
| Tb927.5.3400 | Trypanosoma brucei | no significant loss or gain of fitness in the differentiation of procyclic to bloodstream forms | ||
| CELE_K11D9.2 | Caenorhabditis elegans | embryonic lethal | [55] | |
| CELE_K11D9.2 | Caenorhabditis elegans | larval arrest | ||
| CELE_K11D9.2 | Caenorhabditis elegans | larval lethal | ||
| CELE_K11D9.2 | Caenorhabditis elegans | slow growth | ||
| CELE_K11D9.2 | Caenorhabditis elegans | sterile | ||
| PBANKA_0207000 | Plasmodium berghei | essential | [56] | |
| PBANKA_0610400 | Plasmodium berghei | slow | ||
| TGME49_230420 | Toxoplasma gondii | probably essential | [57] | |
| Posaconazole | PF3D7_1355300 | Plasmodium falciparum | Highly essential | [58] |
| Tb11.02.4080 | Trypanosoma brucei | significant loss of fitness in the bloodstream (6 days) | [59] | |
| Tb11.02.4080 | Trypanosoma brucei | significant loss of fitness in the differentiation of procyclic to bloodstream forms | ||
| YHR007C | Saccharomyces cerevisiae | inviable | [60] |
Table S2.
Lansoprazole & Posaconazole: human pharmacological profile and completed trials.
| Category | aLansoprazole | aPosaconazole |
|---|---|---|
| General Use | Reduce gastric acid secretion to treat gastric ulcers, duodenal ulcers, esophagitis, and gastroesophageal reflux disease. | Triazole antifungal drug to treat invasive infections by Candida species and Aspergillus species in severely immunocompromised patients |
| Pharmaco-dynamics | Targets H+/K + ATPase on gastric parietal cells. | Inhibits the fungal enzyme lanosterol 14α-demethylase |
| Mechanism of Action | Lansoprazole requires protonation in a strongly acidic environment to become activated as a PPI. Once activated, lansoprazole interacts with cysteine residues on the H+/K+ ATPase enzyme on parietal cells to form a stable disulfide bond. This covalent bond provides prolonged inhibition of gastric acid secretion. | Posaconazole exerts its antifungal activity through blockage of the cytochrome P-450 dependent enzyme, sterol 14α-demethylase in fungi by binding to the heme cofactor located on the enzyme. This leads to the inhibition of the synthesis of ergosterol, a key component of the fungal cell membrane, with an accumulation of methylated sterol precursors |
| Side Effects and Toxicity | Manifestations include abdominal pain, constipation, diarrhea, and nausea. Classified for pregnancy as category B indicating that no risk was observed in animals and that there are not adequate and well-controlled studies in pregnant women. | No related adverse events were noted up to 1,600 mg/day |
| US Clinical Trials | Safety and Efficacy of Lansoprazole in Patients with Reflux Disease. An Open, Single-Arm, Long-term Study (2002–2008), ClinicalTrials.gov Identifier: NCT01135368. Therapeutic Response to Lansoprazole Among Different Subgroups of Functional Dyspepsia: a Multicenter, Randomized, Double-blind, Placebo-controlled Trial (2009–2013), ClinicalTrials.gov Identifier: NCT01040455. Long-term Use of Takepron on the Prevention of Recurrence of Gastric/Duodenal Ulcer in Patients Receiving Non-Steroidal Anti-inflammatory (2010–2014), ClinicalTrials.gov Identifier: NCT02099708. |
aPosaconazole Pharmacokinetics in Patients Receiving Chemotherapy or Stem Cell Transplants (POPULAR) (ClinicalTrials.gov Identifier: NCT03717623) (Currently recruiting). A New Posaconazole Dosing Regimen for Pediatric Patients with Cystic Fibrosis and Aspergillus Infection (cASPerCF) (NCT04966234), (Recruiting now). Pharmacokinetics and Safety of Intravenous Posaconazole (MK-5592) in Chinese Participants at High Risk for Invasive Fungal Infections (MK-5592-120) (NCT03336502, just completed). Posaconazole for Pulmonary Fungal Infection Prophylaxis in Hematopoietic Stem Cell Transplantation Patients (Recruitment not started) (NCT04725942). |
Multiple sources: https://go.drugbank.com/drugs/DB01263 and https://clinicaltrials.gov.
Funding Statement
Authors sincerely thank the Department of Medicine, Loyola University Chicago Stritch School of Medicine for providing the initial funding support for the Drug Discovery Program and Software acquisition.
Footnotes
Author contributions
YG and PK conceived and designed the HTVS study. YG, SG, and RM performed in-vitro antileishmanial testing and in-silico target analysis. TRC helped with advanced in-silico analysis and JGR participated in analyzing calcium pump isotypes, structural features, and drug interactions inhibitory potential. RD covered the clinical aspect of the leishmaniasis and DPB helped to interpret the target–ligand interactions of the in-silico analysis. All authors contributed to writing and reviewing the manuscript.
Conflicts of interests
The authors declare no conflict of interest.
References
- 1.Jain K, Jain N. Vaccines for visceral leishmaniasis: a review. J Immunol Methods. 2015;422:1–12. doi: 10.1016/j.jim.2015.03.017. [DOI] [PubMed] [Google Scholar]
- 2.W. World Health Organization. WHO Factsheet; leishmaniasis. 2019. [Accessed 12 February 2020]. https://www.who.int/en/news-room/fact-sheets/detail/leishmaniasis .
- 3.Chapman LA, Spencer SE, Pollington TM, Jewell CP, Mondal D, Alvar J, et al. Inferring transmission trees to guide targeting of interventions against visceral leishmaniasis and post–kala-azar dermal leishmaniasis. Proc Natl Acad Sci Unit States Am. 2020;117:25742–50. doi: 10.1073/pnas.2002731117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey F, Mondragon-Shem K, Haines LR, Olabi A, Alorfi A, Ruiz-Postigo JA, et al. others, Cutaneous leishmaniasis and co-morbid major depressive disorder: a systematic review with burden estimates. PLoS Neglected Trop Dis. 2019;13:e0007092. doi: 10.1371/journal.pntd.0007092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kone AK, Niaré DS, Piarroux M, Izri A, Marty P, Laurens MB, et al. Visceral leishmaniasis in West Africa: clinical characteristics, vectors, and reservoirs. Journal of Parasitology Research. 2019:2019. doi: 10.1155/2019/9282690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghorbani M, Farhoudi R. Leishmaniasis in humans: drug or vaccine therapy? Drug Des Dev Ther. 2018;12:25. doi: 10.2147/DDDT.S146521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponte-Sucre A, Gamarro F, Dujardin JC, Barrett MP, Lopez-Velez R, Garcia-Hernandez R, et al. Drug resistance and treatment failure in leishmaniasis: a 21st century challenge. PLoS Neglected Trop Dis. 2017;11:e0006052. doi: 10.1371/journal.pntd.0006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konreddy AK, Rani GU, Lee K, Choi Y. Recent drug-repurposing-driven advances in the discovery of novel antibiotics. Curr Med Chem. 2019;26:5363–88. doi: 10.2174/0929867325666180706101404. [DOI] [PubMed] [Google Scholar]
- 11.Pazhayam NM, Chhibber-Goel J, Sharma A. New leads for drug repurposing against malaria. Drug Discov Today. 2019;24:263–71. doi: 10.1016/j.drudis.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Kwofie SK, Broni E, Dankwa B, Enninful KS, Kwarko GB, Darko L, et al. Others, outwitting an old neglected nemesis: a review on leveraging integrated data-driven approaches to aid in unraveling of leishmanicides of therapeutic potential. Curr Top Med Chem. 2020;20:349–66. doi: 10.2174/1568026620666200128160454. [DOI] [PubMed] [Google Scholar]
- 13.Lezama-Dávila CM, Isaac-Márquez AP, Kapadia G, Owens K, Oghumu S, Beverley S, et al. Leishmanicidal activity of two naphthoquinones against L. donovani. Biol Pharm Bull. 2012;35:761. doi: 10.1248/bpb.b12-00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terrazas C, Varikuti S, Oghumu S, Steinkamp HM, Ardic N, Kimble J, et al. Ly6C(hi) inflammatory monocytes promote susceptibility to Leishmania donovani infection. Sci Rep. 2017;7:4693. doi: 10.1038/s41598-017-14935-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Awale M, Reymond JL. The polypharmacology browser: a web-based multi-fingerprint target prediction tool using ChEMBL bioactivity data. J Cheminf. 2017;9:1. doi: 10.1186/s13321-017-0199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pence HE, Williams A. ChemSpider: an online chemical information resource. ACS Publications; 2010. [Google Scholar]
- 17.Rybniker J, Vocat A, Sala C, Busso P, Pojer F, Benjak A, et al. Lansoprazole is an antituberculous prodrug targeting cytochrome bc 1. Nat Commun. 2015;6:1–8. doi: 10.1038/ncomms8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warrenfeltz S, Basenko EY, Crouch K, Harb OS, Kissinger JC, Roos DS, et al. Eukaryotic genomic databases. Springer; 2018. EuPathDB: the eukaryotic pathogen genomics database resource; pp. 69–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aurrecoechea C, Barreto A, Basenko EY, Brestelli J, Brunk BP, Cade S, et al. others, EuPathDB: the eukaryotic pathogen genomics database resource. Nucleic Acids Res. 2017;45:D581–91. doi: 10.1093/nar/gkw1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sievers F, Higgins DG. Clustal omega. Curr Prot Bioinform. 2014;48:3–13. doi: 10.1002/0471250953.bi0313s48. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann K, Baron M. Boxshade 3.21, pretty printing and shading of multiple-alignment files. Lausanne, Switzerland: Kay Hofmann ISREC Bioinformatics Group; 1996. [Google Scholar]
- 22.Huson DH, Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol. 2012;61:1061–7. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 23.Wu Q, Peng Z, Zhang Y, Yang J. COACH-D: improved protein–ligand binding sites prediction with refined ligand-binding poses through molecular docking. Nucleic Acids Res. 2018;46:W438–42. doi: 10.1093/nar/gky439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Liang Y, Zhang Y. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure. 2011;19:1784–95. doi: 10.1016/j.str.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrusier N, Nussinov R, Wolfson HJ. FireDock: fast interaction refinement in molecular docking. Proteins. 2007;69:139–59. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 26.PubChem. PubChem. n.d. [Accessed 20 July 2020]. https://pubchem.ncbi.nlm.nih.gov/
- 27.Schrodinger P. Release 2020-2. New York, NY: Maestro, Schrodinger, LLC; 2020. [Google Scholar]
- 28.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–6. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrodinger L. Small-molecule drug discovery suite 2020-1. 2020:2020. [Google Scholar]
- 30.DE Shaw research. New York, NY: Maestro-Desmond Interoperability Tools, Schrödinger; 2017. Release S 3: Desmond molecular dynamics system. [Google Scholar]
- 31.Gupta Y, Goicoechea S, Pearce CM, Mathur R, Romero JG, Kwofie SK, et al. others, the emerging paradigm of calcium homeostasis as a new therapeutic target for protozoan parasites. Medicinal Research Reviews. 2021 doi: 10.1002/med.21804. [DOI] [PubMed] [Google Scholar]
- 32.Monk BC, Keniya MV, Sabherwal M, Wilson RK, Graham DO, Hassan HF, et al. Azole resistance reduces susceptibility to the tetrazole antifungal VT-1161. Antimicrob Agents Chemother. 2019;63:e02114–8. doi: 10.1128/AAC.02114-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aier I, Varadwaj PK, Raj U. Structural insights into conformational stability of both wild-type and mutant EZH2 receptor. Sci Rep. 2016;6:1–10. doi: 10.1038/srep34984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coelho AC, Gentil LG, da Silveira JF, Cotrim PC. Characterization of Leishmania (Leishmania) amazonensis promastigotes resistant to pentamidine. Exp Parasitol. 2008;120:98–102. doi: 10.1016/j.exppara.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 35.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. others, DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–82. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts C, McLeod R, Rice D, Ginger M, Chance M, Goad L. Fatty acid and sterol metabolism: potential antimicrobial targets in apicomplexan and trypanosomatid parasitic protozoa. Mol Biochem Parasitol. 2003;126:129–42. doi: 10.1016/s0166-6851(02)00280-3. [DOI] [PubMed] [Google Scholar]
- 37.Mukherjee S, Moitra S, Xu W, Hernandez V, Zhang K. Sterol 14-α-demethylase is vital for mitochondrial functions and stress tolerance in Leishmania major. PLoS Pathog. 2020;16:e1008810. doi: 10.1371/journal.ppat.1008810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verma S, Mehta A, Shaha C. CYP5122A1, a novel cytochrome P450 is essential for survival of Leishmania donovani. PLoS One. 2011;6:e25273. doi: 10.1371/journal.pone.0025273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu W, Hsu F-F, Baykal E, Huang J, Zhang K. Sterol biosynthesis is required for heat resistance but not extracellular survival in Leishmania. PLoS Pathog. 2014;10:e1004427. doi: 10.1371/journal.ppat.1004427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lepesheva GI, Waterman MR. Structural basis for conservation in the CYP51 family. Biochim Biophys Acta Protein Proteonomics. 2011;1814:88–93. doi: 10.1016/j.bbapap.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCall L-I, El Aroussi A, Choi JY, Vieira DF, De Muylder G, Johnston JB, et al. others, Targeting ergosterol biosynthesis in Leishmania donovani: essentiality of sterol 14alpha-demethylase. PLoS Neglected Trop Dis. 2015;9:e0003588. doi: 10.1371/journal.pntd.0003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhir N, Jain A, Mahendru D, Prakash A, Medhi B. Drug repurposing. IntechOpen; 2020. Drug repurposing and orphan disease therapeutics. [Google Scholar]
- 43.Nagle AS, Khare S, Kumar AB, Supek F, Buchynskyy A, Mathison CJ, et al. Recent developments in drug discovery for leishmaniasis and human African trypanosomiasis. Chem Rev. 2014;114:11305–47. doi: 10.1021/cr500365f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raj S, Sasidharan S, Balaji S, Saudagar P. An overview of biochemically characterized drug targets in metabolic pathways of Leishmania parasite. Parasitol Res. 2020;119:2025–37. doi: 10.1007/s00436-020-06736-x. [DOI] [PubMed] [Google Scholar]
- 45.Carvalheiro M, Vieira J, Faria-Silva C, Marto J, Simões S. Amphotericin B-loaded deformable lipid vesicles for topical treatment of cutaneous leishmaniasis skin lesions. Drug Deliv Translat Res. 2021;11:717–28. doi: 10.1007/s13346-021-00910-z. [DOI] [PubMed] [Google Scholar]
- 46.Jafari M, Abolmaali SS, Tamaddon AM, Zomorodian K, Shahriarirad B. Nanotechnology approaches for delivery and targeting of Amphotericin B in fungal and parasitic diseases. Nanomedicine. 2021;16:857–77. doi: 10.2217/nnm-2020-0482. [DOI] [PubMed] [Google Scholar]
- 47.Moreno SN, Docampo R. Calcium regulation in protozoan parasites. Curr Opin Microbiol. 2003;6:359–64. doi: 10.1016/S1369-5274(03)00091-2. [DOI] [PubMed] [Google Scholar]
- 48.Meade JC. P-type transport ATPases in leishmania and trypanosoma. Parasite. 2019:26. doi: 10.1051/parasite/2019069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin JM, Sachs G. Pharmacology of proton pump inhibitors. Curr Gastroenterol Rep. 2008;10:528–34. doi: 10.1007/s11894-008-0098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riel MA, Kyle DE, Bhattacharjee AK, Milhous WK. Efficacy of proton pump inhibitor drugs against Plasmodium falciparum in vitro and their probable pharmacophores. Antimicrob Agents Chemother. 2002;46:2627–32. doi: 10.1128/AAC.46.8.2627-2632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang S, Anderson SA, Winget GD, Mukkada AJ. Plasma membrane K+/H+-ATPase from leishmania donovani. J Cell Physiol. 1994;159:60–6. doi: 10.1002/jcp.1041590109. [DOI] [PubMed] [Google Scholar]
- 52.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, et al. A functional genomic database for malaria parasites. Nucleic Acids Res. 2008;37:D539–43. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–9. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glover L, Alsford S, Baker N, Turner DJ, Sanchez-Flores A, Hutchinson S, et al. Genome-scale RNAi screens for highthroughput phenotyping in bloodstream-form African trypanosomes. Nat Protoc. 2015;10:106–33. doi: 10.1038/nprot.2015.005. [DOI] [PubMed] [Google Scholar]
- 55.Simmer F, Moorman C, Van Der Linden AM, Kuijk E, Van Den Berghe PV, Kamath RS, et al. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:e12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stanway RR, Bushell E, Chiappino-Pepe A, Roques M, Sanderson T, Franke-Fayard B, et al. Genome-Scale identification of essential metabolic processes for targeting the Plasmodium liver stage. Cell. 2019;179:1112–28. doi: 10.1016/j.cell.2019.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sidik SM, Huet D, Ganesan SM, Huynh M-H, Wang T, Nasamu AS, et al. A genome-wide CRISPR screen in Toxoplasma identifies essential apicomplexan genes. Cell. 2016;166:1423–35. doi: 10.1016/j.cell.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas P, Sedillo J, Oberstaller J, Li S, Zhang M, Singh N, et al. Phenotypic screens identify parasite genetic factors associated with malarial fever response in Plasmodium falciparum piggyBac mutants. mSphere. 2016;1:e00273–316. doi: 10.1128/mSphere.00273-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, et al. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011;21:915–24. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sousa M, Duarte AM, Fernandes TR, Chaves SR, Pacheco A, Leão C, et al. Genome-wide identification of genes involved in the positive and negative regulation of acetic acid-induced programmed cell death in Saccharomyces cerevisiae. BMC Genom. 2013;14:1–15. doi: 10.1186/1471-2164-14-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Molecular dynamics of (S)-Lansoprazole with LdBPK_040010.1. (i) Gln 111 donated hydrogen bonds to oxygen on Lansoprazole 71% of the time. Gln 259 donated hydrogen bonds to nitrogen on Lansoprazole 40% of the time. (ii) The ligand Å values are similar to the RMSD of LdBPK_040010.1, indicating that Lansoprazole is stable within the binding site. The RMSD of LdBPK_040010.1 was found to increase initially and reached 8.0 Å at 20ns and remained constant until 60ns when it decreased to 7.0 Å, where it remained stable for the remainder of the simulation. (iii) Protein-ligand interactions are consistent with conformational stability and correlated well. Gln 111 and Gln 249 were integral to ligand binding. (iv, v) There was no significant deviation in the RMSF of each amino acid residue measured with respect to its Cα carbon atom. The RMSD of LdBPK_040010.1 increased initially and reached 8.0 Å at 20ns and remained steady until 60ns when it decreased to 7.0 Å, thereafter it continued stably for the remainder of the simulation. Like LdBPK_352080.1, there was no significant deviation in the RMSF of each amino acid residue measured with respect to the Cα atom. Protein-ligand interactions demonstrated conformational stability and correlated well. Gln 111 and Gln 249 were integral to ligand binding. Gln donated hydrogen bonds to oxygen on Lansoprazole 71% of the time. Gln 259 donated hydrogen bonds to nitrogen on Lansoprazole 40% of the time (Fig. 5). While the ligand stayed in the docked site throughout simulation a significant wobble was observed with the trifluoride region completely dislodging from the predicted binding site causing an increase in ‘wobble’ and the energy decrease during simulation can be attributed to the protein component of the complex rather than the intermolecular interactions.
Molecular dynamics of (S)-Lansoprazole with LdBPK_170660.1. (i) Ser922 donated hydrogen bonds to oxygen on Lansoprazole 99% of the time (ii) The RMSD of LdBPK_170660.1 was found to increase initially and reached 10.0 Å from 5 to 40ns and then increased again to 15 Å at 50ns. The RMSD of Lansoprazole remained constant at 7 Å throughout the simulation. (iii) Protein-ligand interactions correlated well and are consistent with the conformational stability of ligand-protein interaction. For LdBPK_170660.1, Ser 1041 played an integral role in ligand binding. (iv, v) There was a moderate deviation in the RMSF of each amino acid residue measured with respect to its Cα carbon atom. The RMSD of LdBPK_170660.1 was found to increase during initial simulations and reached 10.0 Å from 5 to 40ns and then increased again to 15 Å at 50ns. The RMSD of Lansoprazole remained constant at 7 Å throughout the simulation. There was a moderate deviation in the RMSF of each amino acid residue measured with respect to the Cα atom. Furthermore, protein-ligand interactions demonstrated conformational stability and correlated well. Ser 922 was critical to ligand binding and donated hydrogen bonds to oxygen on Lansoprazole 99% of the time (Fig. 4). The interactions were mostly electrostatic which was weakened by water solvation during the simulation and the ligand interactions became destabilized during simulation without much change in the ATP binding site architecture. These findings support low docking scores by poor performance in simulation in terms of complex stability.
An example image set (each panel is 1/8th area of one well from a triplicate set) of live imaging (ImageExpress Pico) at 10X with L. donovani amastigotes infected macrophages post-treatment with different drugs. Here we have shown a three-drug i.e. Amphotericin B (A, B, C & D), Lansoprazole (E, F, G, & H), and Posaconazole (I, J, K, & L). The dose–response area with red fluorescence i.e. parasites expressing reporter gene DsRed2 is shown. Macrophages infected with leishmania attract more macrophages forming infection masses making the total fluorescence area a reliable parasite load estimation tool as compared to counting individual parasites. Latter requires much higher resolution screening but is less affected by background fluorescence from lysed parasites. Panels A, E, & I are negative controls with untreated and uninfected macrophages. Panels B, F, J (50 μM) and C, G, K (3.1 μM) are infected macrophages treated with different drug concentrations. Panels D, H, J are positive control panels with untreated and infected macrophages. All pannels were seeded with the same number of THP-1 cells and except for uninfected controls, the rest were introduced with the same number of promastigotes.
Orthologs of Posaconazole targeted Lanosterol C-14 demethylase (LDBPK_111100.1) from reference strains of different species of leishmania were obtained from plasmoDB (VEUpathDB) [52] using BLAST similarity searches. Orthologs further analyzed were: Leishmania infantum (LINJ_111100), Leishmania Mexicana (LMXM_111100), Leishmania major (LMJF_111100), Leishmania braziliensis (LBRM_110880), and Leishmania panamensis (LPMP111090). The phylogenetic analyses of the orthologs were performed on the Phylogeny.fr server [53]. A. The phylogenetic trees were constructed using the PhyML program (v3.1/3.0 aLRT) with the maximum-likelihood method with default settings. B. The multiple sequence alignment (MSA) was obtained with MUSCLE (v3.8.31) and gaps and/or poorly aligned regions were removed by Gblocks (v0.91b) with default settings. Publication-quality outputs of the sequence alignments were generated with the BOXSHADE 3.21.5 server [21].
Orthologs of Lansoprazole targeted calcium channel (LdBPK_352080.1) from reference strains of different species of leishmania were obtained from plasmoDB (VEUpathDB) [52] using BLAST similarity searches. Orthologs further analyzed were: L. infantum (LINJ_352080), L. major (LMJF_352080), L. Mexicana (LMXM_342080), L. braziliensis (LBRM_341990), and L. panamensis (LPMP_341910). The phylogenetic analyses of the orthologs were performed on the Phylogeny.fr server [53]. A. The phylogenetic trees were constructed using the PhyML program (v3.1/3.0 aLRT) with the maximum-likelihood method with default settings. B. The multiple sequence alignment (MSA) was obtained with MUSCLE (v3.8.31) and gaps and/or poorly aligned regions were removed by Gblocks (v0.91b) with default settings. Publication-quality outputs of the sequence alignments were generated with the BOXSHADE 3.21.5 server [21].
Table S1.
Gene significance analysis based on experimental evidence from knock-out studies with orthologs of calcium ion transporting ATPase channels and Sterol 14α demethylase in related organisms.
| Gene/Orthologs | Organism | Essentiality/Phenotype | Source Study (TDRtargets.org) | |
|---|---|---|---|---|
| Lansoprazole | Tb927.5.3400 | Trypanosoma brucei | significant loss of fitness in the bloodstream (3 days) | [54] |
| Tb927.5.3400 | Trypanosoma brucei | significant loss of fitness in the bloodstream (6 days) | ||
| Tb927.5.3400 | Trypanosoma brucei | A significant gain of fitness in procyclic | ||
| Tb927.5.3400 | Trypanosoma brucei | no significant loss or gain of fitness in the differentiation of procyclic to bloodstream forms | ||
| CELE_K11D9.2 | Caenorhabditis elegans | embryonic lethal | [55] | |
| CELE_K11D9.2 | Caenorhabditis elegans | larval arrest | ||
| CELE_K11D9.2 | Caenorhabditis elegans | larval lethal | ||
| CELE_K11D9.2 | Caenorhabditis elegans | slow growth | ||
| CELE_K11D9.2 | Caenorhabditis elegans | sterile | ||
| PBANKA_0207000 | Plasmodium berghei | essential | [56] | |
| PBANKA_0610400 | Plasmodium berghei | slow | ||
| TGME49_230420 | Toxoplasma gondii | probably essential | [57] | |
| Posaconazole | PF3D7_1355300 | Plasmodium falciparum | Highly essential | [58] |
| Tb11.02.4080 | Trypanosoma brucei | significant loss of fitness in the bloodstream (6 days) | [59] | |
| Tb11.02.4080 | Trypanosoma brucei | significant loss of fitness in the differentiation of procyclic to bloodstream forms | ||
| YHR007C | Saccharomyces cerevisiae | inviable | [60] |
Table S2.
Lansoprazole & Posaconazole: human pharmacological profile and completed trials.
| Category | aLansoprazole | aPosaconazole |
|---|---|---|
| General Use | Reduce gastric acid secretion to treat gastric ulcers, duodenal ulcers, esophagitis, and gastroesophageal reflux disease. | Triazole antifungal drug to treat invasive infections by Candida species and Aspergillus species in severely immunocompromised patients |
| Pharmaco-dynamics | Targets H+/K + ATPase on gastric parietal cells. | Inhibits the fungal enzyme lanosterol 14α-demethylase |
| Mechanism of Action | Lansoprazole requires protonation in a strongly acidic environment to become activated as a PPI. Once activated, lansoprazole interacts with cysteine residues on the H+/K+ ATPase enzyme on parietal cells to form a stable disulfide bond. This covalent bond provides prolonged inhibition of gastric acid secretion. | Posaconazole exerts its antifungal activity through blockage of the cytochrome P-450 dependent enzyme, sterol 14α-demethylase in fungi by binding to the heme cofactor located on the enzyme. This leads to the inhibition of the synthesis of ergosterol, a key component of the fungal cell membrane, with an accumulation of methylated sterol precursors |
| Side Effects and Toxicity | Manifestations include abdominal pain, constipation, diarrhea, and nausea. Classified for pregnancy as category B indicating that no risk was observed in animals and that there are not adequate and well-controlled studies in pregnant women. | No related adverse events were noted up to 1,600 mg/day |
| US Clinical Trials | Safety and Efficacy of Lansoprazole in Patients with Reflux Disease. An Open, Single-Arm, Long-term Study (2002–2008), ClinicalTrials.gov Identifier: NCT01135368. Therapeutic Response to Lansoprazole Among Different Subgroups of Functional Dyspepsia: a Multicenter, Randomized, Double-blind, Placebo-controlled Trial (2009–2013), ClinicalTrials.gov Identifier: NCT01040455. Long-term Use of Takepron on the Prevention of Recurrence of Gastric/Duodenal Ulcer in Patients Receiving Non-Steroidal Anti-inflammatory (2010–2014), ClinicalTrials.gov Identifier: NCT02099708. |
aPosaconazole Pharmacokinetics in Patients Receiving Chemotherapy or Stem Cell Transplants (POPULAR) (ClinicalTrials.gov Identifier: NCT03717623) (Currently recruiting). A New Posaconazole Dosing Regimen for Pediatric Patients with Cystic Fibrosis and Aspergillus Infection (cASPerCF) (NCT04966234), (Recruiting now). Pharmacokinetics and Safety of Intravenous Posaconazole (MK-5592) in Chinese Participants at High Risk for Invasive Fungal Infections (MK-5592-120) (NCT03336502, just completed). Posaconazole for Pulmonary Fungal Infection Prophylaxis in Hematopoietic Stem Cell Transplantation Patients (Recruitment not started) (NCT04725942). |
Multiple sources: https://go.drugbank.com/drugs/DB01263 and https://clinicaltrials.gov.