Abstract
Drug substances are at risk of contamination with N-nitrosamines (NAs), well-known carcinogenic agents, during synthesis processes and/or long-term storage. Therefore, in this study, we developed an efficient data-based screening approach to systemically assess marketed products and investigated its scalability for benefiting both regulatory agencies and pharmaceutical industries. A substructure-based screening method employing DataWarrior, an open-source software, was established to evaluate the risks of NA impurities in drug substances. Eight NA substructures containing susceptible amino sources for N-nitrosation have been identified as screening targets: dimethylamine (DMA), diethylamine, isopropylethylamine, diisopropylamine, N-methyl-2-pyrrolidone, dibutylamine, methylphenylamine, and tetrazoles. Our method detected 192 drug substances with a theoretical possibility of NA impurity, 141 of which had not been reported previously. In addition, the DMA moiety was significantly dominant among the eight NA substructures. The results were validated using data from the literature, and a high detection sensitivity of 0.944 was demonstrated. Furthermore, our approach has the advantage of scalability, owing to which 31 additional drugs with suspected NA-contaminated substructures were identified using the substructures of 1-methyl-4-piperazine in rifampin and 1-cyclopentyl-4-piperazine in rifapentine. In conclusion, the reported substructure-based approach provides an effective and scalable method for the screening and investigation of NA impurities in various pharmaceuticals and might be used as an ancillary technique in the field of pharmaceutical quality control for risk assessments of potential NA impurities.
Keywords: Cheminformatics, Data mining, Nitrosamines, Risk assessment, Total quality management
1. Introduction
N-nitrosamines (NAs) are potent genotoxic agents that can affect several animal species and are classified as potentially carcinogenic to humans by the International Agency for Research on Cancer (IARC) [1]. They are referred to as “cohort of concern” compounds in the International Conference on Harmonisation (ICH) guidance for industry M7 (R1) Assessment and Control of DNA Reactive (Mutagenic) Impurities in Pharmaceuticals to Limit Potential Carcinogenic Risk [2].
In recent years, the presence of unexpected NA impurities in various pharmaceuticals has been reported. In May 2018, valsartan was reported as the first drug to be contaminated with the NA impurities N-nitrosodimethylamine (NDMA) and N-nitrosodiethylamine (NDEA), and this event triggered a series of drug recall actions worldwide [3]. Valsartan, like the other angiotensin II receptor blockers (ARBs), has a tetrazole ring. The specific set of reactants and reaction conditions used in the chemical synthesis of this ring has been linked to the formation of NA impurities [4]. Hence, the structural characteristics of tetrazole rings in pharmaceuticals may be utilized to reveal the possible existence of NA impurities owing to the firm relationship between the reactants and the synthesized product. In addition to ARBs, other drugs have been reported to be contaminated with NAs resulting from different routes of chemical synthesis. In September 2019, the FDA announced that NDMA was found in ranitidine, a common antihistamine antacid, and its structural sibling, nizatidine [5,6]. Taken together, these findings provide a rationale to search for a method to assess the risk of NA contamination based on the substructures of pharmaceuticals.
In April 2020, the FDA requested all ranitidine products to be immediately removed from the market. This recall was made following the discovery that ranitidine products ramped up NDMA impurities during storage, especially at higher temperatures [7,8]. Other antacids (e.g., famotidine and lansoprazole) seemed unaffected by the storage conditions. In May 2020, extended-release metformin products were found to be contaminated by NDMA at levels above the acceptable limit, and a voluntary recall was posted on these products in theUnited States [6]. Both metformin and ranitidine contain a dimethylamine moiety in their structures, suggesting that a specific amine-containing substructure can be utilized to pinpoint the possible existence of NA impurities in pharmaceuticals.
As recommended in the ICH Guideline M7 [2], potentially carcinogenic compounds should be controlled if “the existence of such substances is reasonably expected in the final drug substance or product.” From the pharmacovigilance point of view, the development of an effective method to detect NA impurities in pharmaceuticals for risk assessment and control is indeed required in the post-marketing surveillance of the drug products. Recent information gathered by the FDA regarding the root causes of NA impurities present in drug products suggests that the structural characteristics of pharmaceutical components, including active pharmaceutical ingredients (APIs), excipients, packing materials, and/or synthesis processes of APIs, are the prime factors delineated for NA formation [9]. For example, one of the root causes of the formation of NA impurities indicated in the FDA reports is that the APIs containing secondary or tertiary amine functional groups are susceptible to degradation under certain reaction conditions because the trace impurities from degraded APIs may react with nitrous acid or other nitrosating agents to form NAs under acidic conditions [8]. Thus, the presence of substructures of secondary or tertiary amine groups in APIs, excipients, or other materials might serve as an indicator to predict the potential risks of residual NA impurities in pharmaceuticals.
The regulatory agencies acted quickly to recall medications and pre-emptively test them to remove contaminated products fromthe market, continued to investigate other drug products, and advised pharmaceutical companies to take appropriate actions. However, the issue of a sudden lack of medicine still urges both regulatory agencies and pharmaceutical industries to develop a rapid screening approach to comprehensively evaluate marketed products. Hitherto, limited strategies have been available to efficiently identify medications that tend to produce NAs from various drug substances. Text mining, the most widely used approach for collecting historical results and conducting retrospective analysis, is an expert-oriented but labor-intensive methodology. Thus, a structure-based (i.e., substructure moiety) searching method capable of deriving the possible presence of NA impurities in drug products with superior efficiency would be highly regarded.
In this study, a structure-based method was established to determine the risks of NA impurities via mining eight types of substructures (i.e., seven identified amine sources of NAs and a tetrazole ring) in pharmaceuticals. Using DataWarrior, an open-source chemoinformatic software [10], the candidate compounds among all commercial drug substances were quickly identified, while their structural mark matched the defined NA substructures. In theory, NAs produced in drug products are related to the nitrogen-containing functional groups of reagents, solvents, catalysts, and/or APIs. The remnant of a NA-related structural mark in API can be first-hand evidence to differentiate whether these susceptible chemicals (i.e., as reagents) have been used in the synthesis of API. Since any newly identified NA root substructure can be added into the screening template, this computer-aided technique provides an effective and expandable methodology for risk assessments of potential NA impurities in pharmaceuticals [11].
2. Methods
2.1. Dataset preparation
To employ the screening feature of the method, a data file of approved small-molecule drugs with canonical simplified molecular-input line-entry system (SMILES) codes was prepared from DrugBank Release Version 5.1.7 (https://go.drugbank.com) for text encoding their chemical structures. This version contains 10,767 drug entries, including 2,471 approved small-molecule drugs (the collection of the FDA, Canada, and EMA approvals, but also including some withdrawn drug products), 107 nutraceuticals, and over 6,041 experimental (discovery-phase) drugs. However, only the data subset of approved small-molecule drugs was used in the study [12].
2.2. NA-related substructures’ definitions
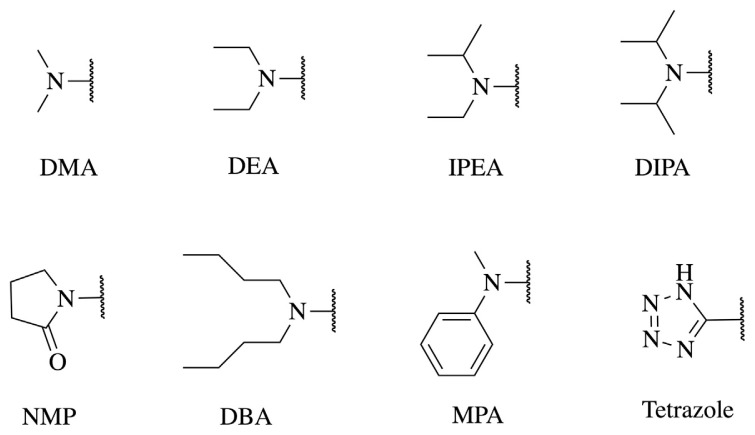
As mentioned previously, the FDA has identified seven NA impurities that can exist in drug products. Besides NDMA, NAs include N-nitrosodiethylamine (NDEA), N-nitroso-N-methyl-4-aminobutanoic acid (NMBA), N-nitrosoisopropylethylamine (NIPEA), N-nitrosodiisopropylamine (NDIPA), N-nitrosodibutylamine (NDBA), and N-nitrosomethylphenylamine (NMPA) [9]. Theoretically, these NAs are formed from precursors such as dimethylamine (DMA), diethylamine (DEA), isopropylethylamine (IPEA), diisopropylamine (DIPA), N-methyl-4-aminobutanoic acid (MBA), dibutylamine (DBA), and methylphenylamine (MPA) to react with nitrate under acidic conditions. Except for MBA, the six precursors were defined as NA-related substructures used in the query process. According to the European Medicines Agency (EMA) report in 2019, N-methyl-2-pyrrolidone (NMP) is converted to MBA through hydrolytic degradation; MBA can further undergo nitrosation into NMBA, a known animal and potential human carcinogen [13]. Therefore, we defined NMP as the NA-related substructure for MBA formation. Furthermore, previous reports have indicated that NDMA may be formed during the synthesis of tetrazole rings with the use of azides and dimethylformamide (DMF) as solvent [14], and several tetrazole drugs such as cephalosporins and cilostazol have been reported to be contaminated with NDMA. Consequently, tetrazole was set as the NA-related substructure for screening the possibility of NDMA impurities. Altogether, eight NA-related substructures, shown in Fig. 1, were selected for in-depth investigation in this study.
Fig. 1.
Eight substructures present in the drug substances of interest. DBA, dibutylamine; DEA, diethylamine; DIPA, diisopropylamine; DMA, dimethylamine; IPEA, isopropylethylamine; MPA, methylphenylamine; NMP, N-methyl-2-pyrrolidone.
2.3. Virtual screening based on NA-related substructures
The data of 2,471 small-molecule compounds were imported into Osiris DataWarrior 5.2.1 [10] and subjected to substructure screening. The virtual screening of the compounds suspected of having NA impurities was performed with the function “Add Substructure Count.” The search algorithm was then automatically run for each “query” substructure.
2.4. Method validation
Since the outcome of the virtual screening defined above indicated the possibility of NA impurities produced in pharmaceuticals, we used the index of sensitivity to validate this qualitative method. The APIs reported in the literature [14] with proven NA contamination were used to check the sensitivity index and were defined as the proportion of real positive cases that had been correctly identified [15]. For calculation, sensitivity was expressed as follows:
2.5. Scalability
One latest warning letter regarding NA impurities in market drugs was issued by FDA in August 2020, concerning about the presence of 1-methyl-4-nitrosopiperazine (MNP) in rifampin and 1-cyclopentyl-4-nitrosopiperazine (CPNP) in rifapentine [16]. Therefore, the root NA-precursors of these impurities - 1-methyl-piperazine (MP) and 1-cyclopentylpiperazine (CPP) moieties were set as the additional substructures for testing the scalability of this screening method.
3. Results
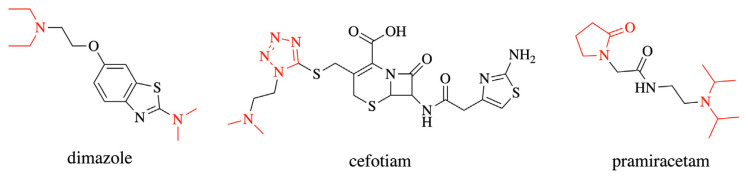
The results from the DataWarrior showed that 195 drugs met the screening criteria. Among these drugs, dimazole, cefotiam, and pramiracetam were found to contain two types of NA-related substructures, resulting in double counting (Fig. 2). Thus, a total of 192 drugs with the theoretical possibility of NA impurity production were detected using this method.
Fig. 2.
Double counting compounds: dimazole, cefotiam, and pramiracetam.
In the subgroup analysis, 124 compounds were found with a DMA moiety, 38 compounds with a DEA moiety, and 22 compounds with a tetrazole (Table 1).
Table 1.
Numbers of screening results for eight substructures.
| Substructures | Numbers (Double counting) | Double counting drugs |
|---|---|---|
| DMA | 122 (2) | Dimazole, Cefotiam |
| DEA | 38 | Dimazole |
| DIPA | 3 (1) | Pramiracetam |
| IPEA | 0 | |
| NMPa | 3 | Pramiracetam |
| MPA | 0 | |
| DBA | 4 | |
| Tetrazole | 22 | Cefotiam |
|
| ||
| Total | 192 | |
NMP is a root cause of MBA according to the EMA report 2019.
The structural formulae of all approved drugs meeting the screening criteria mentioned above are shown in the Supplemental data.
During validation based on literature data [14], 4 out of 58 APIs with known or suspected precursors of NAs, including methapyrilene, methyl orange, methylene blue, and oleandomycin, were not detected because they were not collected in Drugbank as approved drugs (Table 2). Among the 54 real positive APIs, only three, namely aminopyramine, benzalkonium chloride, and chloramphenicol, were not detected with this method, yielding a detection sensitivity of 0.944. This high sensitivity suggests that our approach could efficiently detect suspicious APIs from Drugbank’s API pool with high accuracy.
Table 2.
Active pharmaceutical ingredients reported as known or suspected precursors of N-nitrosamines.
| Order | Compounds | Order | Compounds | Order | Compounds |
|---|---|---|---|---|---|
| 1 | Aminopyraminea | 21 | Chlortetracycline | 41 | Methylene Blueb |
| 2 | Alfentanil | 22 | Cilostazol | 42 | Mifepristone |
| 3 | Aminophenazone | 23 | Citalopram | 43 | Minocycline |
| 4 | Amitriptyline | 24 | Clarithromycin | 44 | Nizatidine |
| 5 | Azithromycin | 25 | Clomipramine | 45 | Oleandomycind |
| 6 | Benzalkonium Chloridea | 26 | Diethyltoluamide | 46 | Olmesartan |
| 7 | Candesartan | 27 | Diltiazem | 47 | Oxytetracycline |
| 8 | Carbinoxamine | 28 | Dimenhydrinate | 48 | Promazine |
| 9 | Cefamandole | 29 | Diphenhydramine | 49 | Propoxyphene |
| 10 | Cefazolin | 30 | Doxepin | 50 | Quinupristin |
| 11 | Cefmenoxime | 31 | Doxylamine | 51 | Ranitidine |
| 12 | Cefonicid | 32 | Erythromycin | 52 | Roxithromycin |
| 13 | Cefoperazone | 33 | Escitalopram | 53 | Spiramycin |
| 14 | Cefotetan | 34 | Imipramine | 54 | Sumatriptan |
| 15 | Cefotiam | 35 | Irbesartan | 55 | Tetracycline |
| 16 | Cefpiramide | 36 | Losartan | 56 | Tramadol |
| 17 | Chloramphenicola | 37 | Meropenem | 57 | Trimipramine |
| 18 | Chlorpheniramine | 38 | Metformin | 58 | Venlafaxine |
| 19 | Chlorpromazine | 39 | Methapyrileneb | ||
| 20 | Chlorprothixene | 40 | Methyl Orangec |
Drugbank dataset covers drug products approved by the FDA, Health Canada and EMA; including some withdrawn drug products as well.
Aminopyramine, benzalkonium chloride, and chloramphenicol were not detected.
Methapyrilene was withdrawn from the market in 1979.
Methyl orange and methylene blue are not collected in Drugbank as approved drugs.
Oleandomycin, a synthesized macrolide antibiotic, is an approved vet medicine.
In the scalability testing of this method, 33 out of 2,471 approved drugs were quickly identified as the suspected NA-contaminated compounds by using MP and CPP moieties as the filtering substructures. A detailed list of APIs containing the MP and CPP moieties can be found in Table 3.
Table 3.
Active pharmaceutical ingredients identified containing the R-piperazine moieties (MP&CPP).
| Substructure moiety | Root case | Other compounds |
|---|---|---|
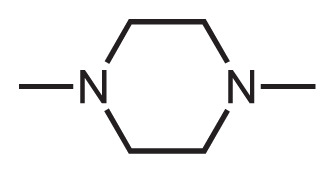 1-methyl-piperazine (MP) |
Rifampicin | Bosutinib, Brigatinib, Butaperazine, Chlorcyclizine, Clothiapine, Clozapine, Cyclizine, Diethylcarbamazine, Entrectinib, Eszinib, Levofloxacin, Loxapine, Netupitant, Nintedanib, Ofloxacin, Olanzapine, Pefloxacin, Perazine, Pipecuronium, Pirenzepine, Ponatinib, Prochlorperazine, Sildenafil, Thiethylperazine, Thiothixene, Trifluoperazine, Zopiclone |
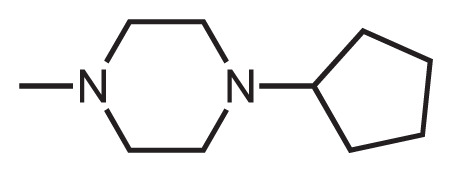 1-cyclopentyl-piperazine (CPP) |
Rifapentine | None |
4. Discussion
The present study is the first to demonstrate an effective and scalable screening methodology for the identification of possible NA impurities in drug substances by matching the substructure of APIs with NA precursors using open-source software. This method effectively indicates suspicious drug substances from a comprehensive API library pool using the artificial intelligence technique of cheminformatics.
When confronting the risk assessments and surveillances for the massive amount of marketed pharmaceutical products, implementing such a cheminformatics tool to detect the possibility of NA impurities in pharmaceutical substances would be much more efficient and comprehensive than analyzing those substances item by item in the laboratory. DataWarrior is an open-source chemical software that facilitates the visual and quantitative analysis of information relevant to drug discovery. The user can input a chemical structure using the text coding of the SMILES string, and DataWarrior can automatically convert this code into a chemical structure with the corresponding atom coordinates. The built-in chemistry intelligence allows working with chemical structures as easily as with alphanumeric data. In this study, the filter function we utilized was a substructure filter, which possesses the features of a flexible query and real-time filtering. By running this function, the search for a given substructure in a molecule’s structure is readily achieved. In addition, this function allows the manual definition of the atom and/or bond features of the query fragment to narrow down the search results.
Compared to the conventional methodologies, the developed cheminformatics-based methodology has tremendous advantages to carry out NA risk assessments, with profound effectiveness and scalability [11]. This study utilized the structure-mining methodology, which enabled comprehensive screening of all commercial APIs by merely setting suspicious NA-related substructures. In contrast, conventional methods based on text-mining methodology have a low throughput because these kinds of analyses are all based on retrospective literature analysis, which is very time consuming. Nevertheless, this study provides an effective and prospective way to perform NA risk assessment for both pharmaceutical regulatory agencies and industries.
Another advantage of our approach is that it can be scaled for other NA impurities such as 1-methyl-4-nitrosopiperazine (MNP) and 1-cyclopentyl-4-nitrosopiperazine (CPNP), as long as the theoretical basis for the formation of these NA impurities can be established. Among 33 drugs with the theoretical possibility of NA impurity production, only rifapentine contained a CPP moiety, and the remaining suspected compounds all contained the MP moiety, implying that the cases of MNP contamination in pharmaceuticals might be more severe.
During validation of our screening results by analyzing known cases in the literature [14], we found that our method failed to detect quinupristin, irbesartan, and losartan. Modifying the query of the DMA substructure, the appended sub-query for the moiety fragment coordinated to double-bond structures including azo linkages, imine linkages, and aromatic rings, resulting in 10 new hits in the API screening, including dacarbazine (azo), altretamine (imine), iopodic acid (imine), gentian violet cation (aromatic), methylene blue (aromatic), quinupristin (aromatic), pyrvinium (aromatic), ulipristal (aromatic), pamidate O (aromatic), and mifepristone (aromatic). Among these substances, methylene blue was previously reported as a precursor of NA impurity.
While quinupristin could be detected through our approach with the appendix of sub-query, the antihypertensive drugs irbesartan and losartan exhibiting known issues of NA impurities omitted in the first screening run, still needed further investigation. In this regard, we found that the tetrazole ring can exist in two tautomeric forms [17,18], which mutually convert between 1H-tetrazole and 2H-tetrazole. Considering our previous query setting including only 1H-tetrazole, some tetrazole compounds might be omitted. The screening query was revised with the addition of tautomeric 2H-tetrazole, and the results covered all the aforementioned sartans, including irbesartan, losartan, valsartan, and pemirolast, as well as tedizolid and tedizolid phosphate. These results showed that our approach could provide a very accurate screening via well and precisely defined query settings.
As indicated in the Results section, aminopyramine, benzalkonium chloride, and chloramphenicol were not detected using our approach, although they are documented in the literature to contain NAs. Therefore, we analyzed the possible causes of this omittance and found that aminopyramine and chloramphenicol do not have a structural fragment matching the eight NA-related substructures used in this study. According to studies on the variation of NDMA content in aminopyramine-containing products, it was found that tablet and suppository formulations have higher levels of NDMA, suggesting that the production of NDMA impurities may be associated with other pharmaceutical components, such as excipients and packaging materials, which have been used in the entire lifecycle of drug products, and their amounts vary among different formulations [19]. On the contrary, in addition to our approach, it appeared that universal NA analysis was still needed as a second-line method to reduce the risk of NA impurities for some nitrogen-containing compounds. In accordance, chloramphenicol was not detected through our approach. Thus, the proposal of new NA-related substructures based on the organic chemistry theorem will help greatly improve the comprehensiveness and effectiveness of our approach [20].
Furthermore, it was found that benzalkonium chloride, a class of quaternary ammonium antimicrobial agents, could not be detected with our approach; however, this result was misleading. Quaternary ammonium salts may contain residual secondary and tertiary amines as impurities or degradants, and the presence of quaternary amines and trace nitrite salts under acidic reaction conditions are known to generate NA impurities. After carefully examining our dataset, it was found that benzalkonium chloride could not be classified as an approved drug in Drugbank; therefore, it was not detected using our approach. To solve this issue, other chemical databases such as ChEMBL can be integrated to expand the compound coverage of the dataset and enrich the sensitivity of our approach.
Notably, among eight structural fragments used in this study to detect NA impurities, DMA, DEA moieties, and tetrazole ring were found to be the primary substructures with the highest hit rates in detecting suspicious APIs. Using the known cases cited in the literature [14] for subtraction, this study showed that our approach disclosed 141 drug substances with possible NA impurities (Table 4).
Table 4.
The numbers of new active pharmaceutical ingredients in each sub-group.
| Substructures | Hit numbers | Confirmed numbers [14] | Undiscovered numbers |
|---|---|---|---|
| DMA | 122 | 37 | 85 |
| DEA | 38 | 1 | 37 |
| DIPA | 3 | 0 | 3 |
| IPEA | 0 | 0 | 0 |
| NMP | 3 | 0 | 3 |
| MPA | 0 | 0 | 0 |
| DBA | 4 | 0 | 4 |
| Tetrazole | 22 | 13 | 9 |
|
| |||
| Total | 192 | 51 | 141 |
DBA, dibutylamine; DEA, diethylamine; DIPA, diisopropylamine; DMA, dimethylamine; IPEA, isopropylethylamine; MPA, methylphenylamine; NMP, N-methyl-2-pyrrolidone.
In this study, DMA was found to be the most sensitive substructure for detecting suspicious drug substances. This moiety detected 122 suspicious drug substances, with 37 proven cases having the risk of NA impurities. Several of the remaining 85 compounds were also evident in the literature. For example, triptans are used for the treatment of severe migraine attacks or patients who do not respond to NSAIDs or other over-the-counter drugs. Several triptans, including sumatriptan, zolmitriptan, almotriptan, and rizatriptan, with DMA moieties were detected with our approach. Structurally, sumatriptan’s DMA fragment is bound to an electron-donating indole ring, as demonstrated in a recent study [21], and could form NDMA in NA-formation potential tests with a molar conversion rate of more than 6%[22]. Therefore, both pharmaceutical manufacturers and regulatory authorities were aware of the risk of NDMA formation in sumatriptan. Nevertheless, according to our screening results, a warning for NDMA risk in other triptans such as zolmitriptan, almotriptan, and rizatriptan should be also be given [14].
On the other hand, it is known that amine-based pharmaceuticals and personal care products (PPCPs) containing DMA or DEA in their structures that could contribute to NA formation during chloramine disinfection of water [21]. While our method could detect all twenty PPCPs including two DEA compounds (i.e., diethyltoluamide (DEET) and lidocaine) tested in the study, it is worthy to note that these DEA-related compounds with the susceptible NA problem can be liberally underestimated.
In this study, the screening library was limited to a pool of APIs. Therefore, our approach ran with no hits for two substructure groups: isopropylethylamine (IPEA) and methylphenylamine (MPA) [16]. After further examination, the root causes of no hits were rationalized as follows: 1) either IPEA or MPA is the side product of the nitrogen-containing reagent; 2) IPEA or MPA is present in small amounts in the reagent or solvent; 3) IPEA or MPA is not involved in the synthesis of API and therefore does not integrate into API’s structure as a structural mark. In fact, the route of IPEA formation was identified owing to the use of tertiary diisopropylethylamine (DIPEA) as a reagent in API synthesis. DIPEA consists of a tertiary hindered nitrogen that is bonded to an ethyl group and two isopropyl groups, commonly employed as a proton scavenger in several reactions, such as alkylation and amide coupling [23]. The DIPEA reagent might undergo de-alkylation to give the tertiary amine IPEA, following the occurrence of N-nitrosation with nitrite sources, yielding ethylisopropyl-N-NA (EIPNA). Similarly, N-dimethylaniline is expected to be a potential source of N-methylaniline (another name of MPA).
The NMP moiety was detected in three suspicious drug substances: piracetam, levetiracetam, and pramiracetam. However, one study showed that the NMP moiety was degraded under alkaline stress conditions, instead of acidic conditions [24,25], which are essential for NA impurity formation. Although we would infer that those piracetam-like drugs bear a lower risk of NA formation compared to DMA moiety-containing drugs, appropriate measures are recommended as these solvents/reagents are commonly used and unavoidable.
In summary, the effectiveness or impotence of our approach utilizing a substructure-matching technique to detect the risks of NA impurities mainly depends on the compound library used. The potential presence of tertiary amine impurities in solvents, reagents, and/or catalysts requires the addition of a solvent/reagent/catalyst library for the detection of side products. While awareness and detection of NAs have increased in recent years, our approach could provide a rapid and effective method to estimate the risk of NA impurities in pharmaceuticals.
5. Conclusion
Our study shows that numerous of drug substances are still undiscovered yet bear a potential to produce NAs, DMA-containing drug substances taking the lead. The screening process used could provide a practical risk assessment strategy involving an effective and scalable method for screening and investigation of NA impurities in various pharmaceuticals. Additionally, the validation data reflected the method’s compliance in line with the regulatory expectations and requirements and published literature. Our approach has successfully demonstrated broad applicability not only for drug substances but also possibly for nutraceuticals and investigational products.
Acknowledgments
This work was supported by a grant from the Taiwan Food and Drug Administration (Grant No. 109TFDA-DF-029).
Appendix. Supplemental data
The screening results for DMA candidates.
The screening results for DEA candidates.
The screening results for DIPA candidates.
The screening results for NMP candidates.
The screening results for DBA candidates.
The screening results for tetrazoles candidates.
The screening results for MP candidates.
Funding Statement
This work was supported by a grant from the Taiwan Food and Drug Administration (Grant No. 109TFDA-DF-029).
Footnotes
Conflict of interest
The authors declare no conflicts of interest with respect to the authorship and publication of this article.
References
- 1.International Agent for Research on Cancer, World Health Organization. Smokeless tobacco and some tobacco-specific N-nitrosamines. 2007.
- 2.Teasdale A, Ich M. ICH quality guidelines. John Wiley & Sons, Ltd; 2017. pp. 667–99. [DOI] [Google Scholar]
- 3.Charoo NA, Ali AA, Buha SK, Rahman Z. Lesson learnt from recall of valsartan and other angiotensin II receptor blocker drugs containing NDMA and NDEA impurities. AAPS PharmSciTech. 2019;20:66. doi: 10.1208/s12249-019-1376-1. [DOI] [PubMed] [Google Scholar]
- 4.Snodin DJ, Elder DP. Short commentary on NDMA (N-Nitrosodimethylamine) contamination of valsartan products. Regul Toxicol Pharmacol. 2019;103:325–9. doi: 10.1016/j.yrtph.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Aldawsari FS, Alshehri YM, Alghamdi TS. N-nitrosodimethylamine (NDMA) contamination of ranitidine products: a review of recent findings. J Food Drug Anal. 2021;29:39–45. doi: 10.38212/2224-6614.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaik KM, Sarmah B, Wadekar GS, Kumar P. Regulatory updates and analytical methodologies for nitrosamine impurities detection in sartans, ranitidine, nizatidine, and metformin along with sample preparation techniques. Crit Rev Anal Chem. 2020:1–19. doi: 10.1080/10408347.2020.1788375. [DOI] [PubMed] [Google Scholar]
- 7.Zhao B, Zhou J, Nakada N. N-nitrosodimethylamine formation potential (NDMA-FP) of ranitidine remains after chlorination and/or photo-irradiation: identification of transformation products in combination with NDMA-FP test. Chemosphere. 2021;267:29200. doi: 10.1016/j.chemosphere.2020.129200. [DOI] [PubMed] [Google Scholar]
- 8.Ashworth IW, Dirat O, Teasdale A, Whiting M. Potential for the formation of N-nitrosamines during the manufacture of active pharmaceutical ingredients: an assessment of the risk posed by trace nitrite in water. Org Process Res Dev. 2020;24:1629–46. doi: 10.1021/acs.oprd.0c00224. [DOI] [Google Scholar]
- 9.Research, C. for D.E. and. Control of Nitrosamine Impurities in Human Drugs. 2021. [Accessed 23 June 2021]. https://www.fda.gov/regulatoryinformation/search-fda-guidance-documents/controlnitrosamine-impurities-human-drugs .
- 10.Sander T, Freyss J, von Korff M, Rufener C. DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model. 2015;55:460–73. doi: 10.1021/ci500588j. [DOI] [PubMed] [Google Scholar]
- 11.Lagarde N, Rey J, Gyulkhandanyan A, Tufféry P, Miteva MA, Villoutreix BO. Online structure-based screening of purchasable approved drugs and natural compounds: retrospective examples of drug repositioning on cancer targets. Oncotarget. 2018;9:32346–61. doi: 10.18632/oncotarget.25966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ion GND, Nitulescu GM. Drugs and PAINs: a DrugBank analysis of pan-assay interference compounds. Proceedings of 5th International Electronic Conference on Medicinal Chemistry. sciforum.net. MDPI; 2019; p. 6378. [DOI] [Google Scholar]
- 13.Fitt H. Nitrosamine impurities. 2021. [Accessed 23 June 2021]. https://www.ema.europa.eu/en/human-regulatory/post-authorisation/referral-procedures/nitrosamine-impurities .
- 14.Parr MK, Joseph JF. NDMA impurity in valsartan and other pharmaceutical products: analytical methods for the determination of N-nitrosamines. J Pharm Biomed Anal. 2019;164:536–49. doi: 10.1016/j.jpba.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Powers DMW. Evaluation: from precision, recall and Fmeasure to ROC, informedness, markedness and correlation. arXiv 2020. preprint arXiv:2010.16061; Ar.Xiv201016061. [Google Scholar]
- 16.Updates and press announcements on nitrosamines in rifampin and rifapentine. FDA; 2021. [Google Scholar]
- 17.Mazurek AP, Osman R. Molecular orbital studies of tautomerlsm in tetrazole. J Phys Chem. 1985;89:460–3. doi: 10.1021/j100249a018. [DOI] [Google Scholar]
- 18.Kiselev VG, Cheblakov PB, Gritsan NP. Tautomerism and thermal decomposition of tetrazole: high-level ab initio study. J Phys Chem. 2011;115:1743–53. doi: 10.1021/jp112374t. [DOI] [PubMed] [Google Scholar]
- 19.Castegnaro M, Pignatelli B, Walker EA. Analysis of volatile N-nitrosamines in commercial drugs. Food Chem Toxicol. 1981;19:489–91. doi: 10.1016/0015-6264(81)90455-7. [DOI] [PubMed] [Google Scholar]
- 20.Xu L, Zhang W-X, Xi Z. Mechanistic considerations of the catalytic guanylation reaction of amines with carbodiimides for guanidine synthesis. Organometallics. 2015;34:1787–801. doi: 10.1021/acs.organomet.5b00251. [DOI] [Google Scholar]
- 21.Shen R, Andrews SA. Demonstration of 20 pharmaceuticals and personal care products (PPCPs) as nitrosamine precursors during chloramine disinfection. Water Res. 2011;45:944–52. doi: 10.1016/j.watres.2010.09.036. [DOI] [PubMed] [Google Scholar]
- 22.Brambilla G, Martelli A. Update on genotoxicity and carcinogenicity testing of 472 marketed pharmaceuticals. Mutat Res. 2009;681:209–29. doi: 10.1016/j.mrrev.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Sorgi KL. Encyclopedia of Reagents for Organic Synthesis. American Cancer Society; 2001. Diisopropylethylamine; pp. 1–4. [DOI] [Google Scholar]
- 24.Arayne MS, Sultana N, Siddiqui FA, Mirza AZ, Qureshi F, Zuberi MH. Simultaneous determination of piracetam and its four impurities by RP-HPLC with UV detection. J Chromatogr Sci. 2010;48:589–94. doi: 10.1093/chromsci/48.7.589. [DOI] [PubMed] [Google Scholar]
- 25.Sahu K, Shaharyar M, Siddiqui AA, Sahu S. Isolation, identification and characterization of degradation product of piracetam using analytical techniques. Int J Adv Res Chem Sci. 2014;1:8–16. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The screening results for DMA candidates.
The screening results for DEA candidates.
The screening results for DIPA candidates.
The screening results for NMP candidates.
The screening results for DBA candidates.
The screening results for tetrazoles candidates.
The screening results for MP candidates.