Abstract
NRF2/ARE signaling pathway is a principal regulator of cellular redox homoeostasis. The stress-induced transcription factor, NRF2, can shield cells from the oxidative damages via binding to the consensus antioxidant-responsive element (ARE) and driving several cyto-protective genes expression. Increasing evidence indicated that aberrant activation of NRF2 in malignant cells may support their survival through various pathways to detoxify chemotherapy drugs, attenuate drug-induced oxidative stress, or induce drug efflux, all of which are crucial in developing drug resistance. Accordingly, NRF2 is a potential drug target for improving the effectiveness of chemotherapy and to reverse drug resistance in cancer cells. A stable ARE-driven reporter human head and neck squamous cell carcinoma (HNSCC) cell line, HSC3-ARE9, was established and utilized to screen novel NRF2 inhibitors from a compound library. The cotton plant derived phenolic aldehyde-gossypol was selected for further analyses. The effects of gossypol in cancer cells were determined by western blotting, RT-qPCR, clonogenic assay, and cell viability assays. The gossypol-responsive gene expression levels were assessed in the Oncomine database. The effects of gossypol on conferring chemo-sensitization were evaluated in etoposide-resistant and cisplatin-resistant cancer cells. Our study is the first to identify that gossypol is effective to reduce both basal and NRF2 activator tert-butylhydroquinone (t-BHQ)-induced ARE-luciferase activity. Gossypol diminishes NRF2 protein stability and thereby leads to the suppression of NRF2/ARE pathway, which resulted in decreasing the expression levels of NRF2 downstream genes in both time- and dose-dependent manners. Inhibition of NRF2 by gossypol significantly decreases cell viabilities in human cancer cells. In addition, we find that gossypol re-sensitizes topoisomerase II poison treatment in etoposide-resistant cancer cells via suppression of NRF2/ABCC1 axis. Moreover, gossypol suppresses NRF2-mediated G6PD expression thereby leads to induce synthetic lethality with cisplatin not only in parental cancer cells but also in cisplatin-resistant cancer cells. These findings suggest that gossypol is a novel NRF2/ARE inhibitor, and can be a potential adjuvant chemotherapeutic agent for treatment of chemo-refractory tumor.
Keywords: Cancer therapy, Chemo-resistance, Gossypol, NRF2 inhibitor
1. Introduction
Nuclear factor erythroid 2-related factor 2 (NRF2) is a transcription factor, which belongs to basic leucine zipper (bZIP) family, and drives the antioxidant response elements (ARE)-containing genes [1]. Interaction with Kelch-like ECH-associated protein 1 (KEAP1), a substrate of Cullin 3-based E3 ubiquitin ligase complex, results in impairing transcriptional activity of NRF2 through inducing proteasomal degradation of NRF2 and then suppressing its nuclear translocation [2]. An uncontrolled over-activation of NRF2/ARE pathway has been viewed as a crucially tumor-promoting factor due to providing several advantages for cancel calls, such as maintaining excessive proliferative signaling, repressing the inhibitors of cell-cycle progression, modulating cancer cell metabolism, and increasing tolerance to excess reactive oxygen species (ROS) [3]. The crucial roles of persistent NRF2 activation in therapy resistance in cancers have been well established [4,5]. Over-activation of NRF2 induces the expressions of drug-metabolizing enzymes, drug efflux transporters and the proteins involved in redox homeostasis, subsequently offering protective effects to cancer cells against chemotherapy [4–6]. For instance, we previously identified that NRF2-induced over-expression of multidrug resistance-associated protein 1 (MRP1/ABCC1) and aldo-keto reductase family 1C (AKR1C) enzymes resulted in developing acquired resistance to topoisomerase II poisons and platinum drugs of cancer cells [7,8]. Hence, these findings provide strong evidence that inhibition of NRF2 is a potent therapeutic strategy for effective eradication of cancer [6].
Although no NRF2-targeted agents have been approved by FDA for cancer therapy so far, several promising strategies have been reported for inhibition of NRF2/ARE pathway; for instance, reducing expression, promoting degradation, impairing transcriptional activity, and suppressing nuclear translocation of NRF2 [9]. Several naturally occurring substances, such as brusatol, luteolin, wogonin, chrysin, convallatoxin, and procyanidins, have been reported to inhibit NRF2 signaling pathway through decreasing the mRNA or protein levels of NRF2, or promoting the degradation of NRF2 protein [9,10].
Through a high-throughput screening system, Bollong et al. identified a small molecule, AEM1, which has the ability to reduce transcriptional activity of NRF2, thereby suppressing the expression of NRF2-regulated genes [11]. Similarly, Singh et al., screened approximately 400,000 small molecules from the Molecular Libraries Small Molecule Repository (MLSMR) by using a cell-based quantitative high-throughput screening system [12]. They discovered a small molecule, ML385, which directly interacts with the Neh1 domain of NRF2 and then blocks the binding of the NRF2-MAFG complex to the ARE sequence. These NRF2 inhibitors have obvious cytotoxic activity against different types of cancer cells. However, the problems of solubility, specificity, and anti-cancer efficacy in vivo have led to the fact that currently available anti-NRF2 agents are far from successfully translated into clinical applications. Therefore, there is still a need to develop new NRF2 inhibitors.
In order to effectively and specifically identify novel NRF2/ARE inhibitors, we used the ARE-driven reporter cell system to screen 84 compounds from the SCREEN-WELL® REDOX Library, and identified gossypol as a new and effective NRF2/ARE inhibitor. The anti-cancer cytotoxic and chemo-sensitization effect of gossypol were observed through targeting NRF2/ARE pathway, suggesting that development of effective and specific NRF2 inhibitors provides a new opportunity for cancer therapy, particularly in chemo-refractory tumors.
2. Materials and methods
2.1. Cell culture
Human head and neck squamous cell carcinoma (HNSCC) cell lines, including TW 2.6, SCC-15, and HSC-3, were obtained and cultured as described previously [13]. The human lung carcinoma cell line, NCI–H460, was purchased from the American Type Culture Collection (ATCC, VA, USA) and followed the recommendations of the ATCC official website for cell culture. KB-derived etoposide-resistant cell line (KB-7D) [8] and HONE-1-derived cisplatin resistant cell lines (cis6 and cis15) were established from parental cancer cells by exposure to increasing concentrations of chemotherapeutic agents, such as etoposide (for establishment of KB-7D from KB cells) or cisplatin (for establishment of cis6 and cis15 from HONE-1 cells). When these drug-resistant cells grow stably in high concentrations of chemotherapy drugs, the individual clones were isolated and characterized. All cell lines were kept in the humidified 37 °C incubator with atmosphere of 95% air and 5% CO2.
2.2. Cell-based ARE-driven reporter and MTT assay
A stable ARE-driven reporter human HNSCC cell line, HSC3-ARE9, was generated previously by our group [14]. For compound screening, the HSC3-ARE9 cells were seeded at a density of 2.5 × 104 cells/well in 100 μL of culture medium in 96-well plates. After overnight incubation, the cells were treated with 25 μM of the various compounds from the SCREEN-WELL® REDOX Library (BML-2835, Enzo Life Sciences, Farminedale, USA) for 24 h. The luciferase activity was measured by Steady-Glo® Luciferase Assay System (E2520, Promega, Madison, USA) according to the manufacturer’s instructions. In order to exclude potential compounds that may interfere with luciferase activity due to cytotoxicity, we analyzed the cell viability under the same conditions as the ARE-driven reporter analysis. In brief, 2.5 × 104 cells were seeded in 96-well plates for overnight, and treated with 25 μM of the test compounds for 24 h. At the end of experiment, the cells were incubated with a serum-free medium containing MTT at a final concentration of 0.5 mg/ml for 4 h. The conversion of MTT to formazan by metabolically viable cells was assessed using 50% DMF solution containing 20% SDS at 37 °C overnight, and absorbance was measured at 570 nm.
2.3. Methylene blue assay
(1) Cell viability assay: cells were seeded in 24-well plates at a density of at a density of 8000 cells/well for overnight, then cells were treated with various concentrations of gossypol for the indicated times. Cell viability was examined by performing methylene blue assay [13]. (2) Clonogenic assay: cells were seeded at a density of 200 cells/well in 2 mL of culture medium in 6-well plates for overnight, then cells were treated with various concentrations of gossypol. After 24-h incubation, the gossypol-containing medium was replaced with fresh, drug-free culture medium. And then cells were incubated for 10 days to form colonies. The colonies were visualized by staining with 0.5% methylene blue for 2 h and the number of colonies in each group was counted.
2.4. Western blot analysis
The cells were lysed and the protein extracts were analyzed by performing western blotting as described previously [13]. All primary antibodies were used at dilutions of 1:500–2000 and listed in Table S1.
2.5. RNA extraction and real-time PCR
Total RNA was isolated from the cells and was analyzed by performing real-time RT-PCR (RTqPCR) by using gene-specific primers (Table S2), as described previously [13]. The changes of NRF2, NQO1, HO1, AKR1C1, AKR1C2, AKR1C3 and PRDX1 mRNA levels were expressed as fold to control after normalization to RPLP0.
2.6. Statistical analysis
Data was expressed as the mean ± SD. Student’s t-test was used to examine statistically significant difference between the control and test groups. *p < 0.05; **p < 0.01; ***p < 0.001.
3. Results
3.1. Screening of novel NRF2 inhibitors by using stable ARE-driven reporter cell line, HSC3-ARE9
The expression of NFE2L2 (NRF2 gene) in different tumor type of clinical cohorts was retrieved from the Oncomine (www.oncomine.org). The results demonstrated that the expression of NRF2 is significantly up-regulated in tumor tissues compared to normal tissues, especially in head and neck cancers (Table 1). Western blot analysis shows abundant expression of NRF2 in three different HNSCC cell lines, particularly in HSC-3 cells (Fig. S1A). Phosphorylation at serine 40 (Ser40) was an important mechanism of NRF2 stabilization and resulted in NRF2/ARE pathway activation [15]. We noted that the levels of serine 40 phosphorylated NRF2 was significantly increased in HSC-3 cells (Fig. S1A). Furthermore, knockdown of NRF2 significantly reduced cell growth in HSC-3 cells (Fig. S1B). These results indicated that NRF2 is unduly expressed and associated with cell growth in HNSCC.
Table 1.
Expression levels of NFE2L2 gene in cancer tissue versus normal in different cancer types.
| Cancer type | Cohorta | Fold Change | p-value | |
|---|---|---|---|---|
| Head-Neck | Talbot Head-Neck | Tongue SCC (n = 31) versus Tongue (n = 26) | 1.645 | 0.006 |
| Estilo Head-Neck | Tongue SCC (n = 31) versus Tongue (n = 26) | 1.644 | 0.005 | |
| Colorectal | Gaedcke Colorectal | Rectal Adenocarcinoma (n = 65) versus Rectum (n = 65) | 1.228 | 3.67E-6 |
| Leukemia | Haferlach Leukemia | Chronic Myelogenous Leukemia (n = 76) versus Peripheral Blood Mononuclear Cell (n = 74) | 1.150 | 7.38E-4 |
| Breast | Richardson Breast | Ductal Breast Carcinoma (n = 40) Versus Breast (n = 7) |
1.111 | 0.002 |
Data regarding NRF2 mRNA expression in normal and tumor specimen was retrieved from ONCOMINE database (www.oncomine.org).
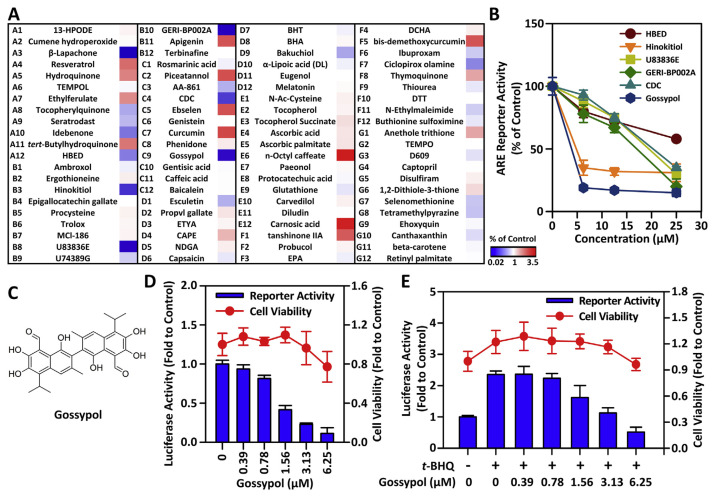
In order to discover novel and effective NRF2 inhibitors, we established a ARE-driven reporter cell line from HSC-3 cells, named HSC3-ARE9, which stably carried 9 repeats of consensus antioxidant response element [14]. Using this sensitive and efficient cell-based screening platform, we screened the SCREEN-WELL®REDOX Library, which contains 84 compounds with pro-oxidant or antioxidant properties. As the result, we found 12 compounds, including resveratrol (A4), hydroquinone (A5), ethylferulate (A7), t-BHQ (A11), apigenin (B11), piceatannol (C2), ebselen (C5), curcumin (C7), n-octyl caffeate (E6), carnosic acid (E12), tanshinone IIA (F1), and bis-demethoxycurcumin (F5), which exhibited NRF2/ARE-inducing properties with greater than 2-fold increases in luciferase reporter activities over control (Fig. 1A). We also identified 9 compounds, including β-lapachone (A3), idebenone (A10), HBED (A12), hinokitiol (B3), U83836E (B8), GERI-BP002A (B10), CDC (C4), gossypol (C9), ciclopirox olamine (F7), which displayed significant NRF2/ARE-inhibiting properties with around or greater than 2-fold reduction of luciferase reporter activities compared with control (Fig. 1A).
Fig. 1.
Screening of NRF2 modulators by using the stable ARE-driven reporter cell line, HSC3-ARE9.
(A) The heat map shows the change of luciferase activity after 24 h of incubation with 25 μM test compound in HSC3-ARE9 cells. (B) Dose–response effects of test compounds on ARE-driven luciferase activity after 24 h of incubation. (C) The schematic illustration of the chemical structure of gossypol. (D) The comparison of basal ARE-driven luciferase activity and cell viability of gossypol. HSC3-ARE9 cells were treated with the indicated concentration of gossypol. After 24 h, the ARE-driven luciferase activity and cell viability were measured. (E) The effects of gossypol on t-BHQ-induced luciferase activity and cell viability. HSC3-ARE9 cell were treated with the indicated concentration of gossypol in presence of 50 μM t-BHQ. After 24 h, ARE-driven luciferase activity was measured and cell viability was evaluated by MTT assay.
In order to exclude potential NRF2 inhibitors that may interfere with luciferase activity due to cytotoxicity, we analyzed cell viability under the same conditions as the ARE-driven reporter analysis to further confirm whether the effects of these compounds are actually due to inhibit the NRF2/ARE pathway, not due to impaired cell viability. After the test compound was treated at 25 μM for 24 h, the selectivity index was calculated by dividing the fold change of luciferase activity by the fold change of cell viability. A lower selectivity index indicates that the compound’s inhibitory activity on the NRF2/ARE pathway is higher than the inhibitory effect on cytotoxicity. Among the 9 compounds selected in primary screening, the selectivity indexes of idebenone and ciclopirox olamine are 0.86 and 0.77, respectively. This result indicates that the effects of these two compounds on NRF2 inhibition and viability suppression could not be distinguished (Table 2). In addition, the selectivity indexes of lapachone, HBED, hinokitiol, U83836E, GERIBP002A, CDC, and gossypol are all less than 0.6. Although lapachone showed the lowest selectivity index, however, the cell viability dropped sharply to 11%, suggesting that the inhibitory effect of lapachone on NRF2/ARE reporter activity may be caused by rapid cytotoxicity (Table 2). Therefore, we excluded idebenone, ciclopirox olamine, and lapachone, and selected 6 compounds (HBED, hinokitiol, U83836E, GERI-BP002A, CDC, gossypol) for further validation of the dose effect. Among them, we noticed that gossypol showed the most effective effect in inhibiting ARE-driven luciferase activity compared with other compounds (Fig. 1B).
Table 2.
The ARE-driven luciferase activity and cell viability of 25 μM test compounds in HSC3-ARE9 cells.
| Well | Group | Luciferase Activity (% of control) | Cell Viability (% of control) | Selectivity Indexa |
|---|---|---|---|---|
| – | Control | 100 | 100 | 1.00 |
| A3 | Lapachone | 2 | 11 | 0.18 |
| A10 | Idebenone | 55 | 64 | 0.86 |
| A12 | HBED | 58 | 98 | 0.59 |
| B3 | Hinokitiol | 31 | 83 | 0.37 |
| B8 | U83836E | 30 | 59 | 0.51 |
| B10 | GERI-BP002A | 16 | 47 | 0.34 |
| C4 | CDC | 35 | 90 | 0.39 |
| C9 | Gossypol | 19 | 48 | 0.40 |
| F7 | Ciclopirox olamine | 61 | 79 | 0.77 |
The Selectivity Index was calculated by diving luciferase activity by cell viability.
3.2. Gossypol effectively inhibits NRF2/ARE signaling pathway
Based on the above research, we noticed that gossypol (Fig. 1C), a natural polyphenolic compound isolated from cotton plant [16], is the most effective NRF2 inhibitor in SCREEN-WELL® REDOX Library. To accurately determine the range of efficacious doses of gossypol, we preformed luciferase reporter assay paralleled with cell viability assay by using a two-fold serial dilution. As shown in Fig. 1D, gossypol decreased ARE-driven luciferase activity in a concentration–dependent manner. The IC50 value of gossypol in suppressing ARE-driven luciferase activity was approximately 1.56 μM. At this concentration, gossypol did not affect cell viability. This result indicated that gossypol inhibited luciferase activity certainly owing to disturbing NRF2/ARE pathway, not because of impairing cell viability. Next, we examined the effect of gossypol on t-BHQ (a typical NRF2-activator)–induced ARE-driven luciferase activity. As shown in Fig. 1E, a 2.5-fold increase in ARE-driven luciferase activity was observed following treatment with 50 μMt-BHQ for 24 h. As consistent with basal ARE-luciferase activity, we noted that gossypol showed a concentration–dependent effect to suppress t-BHQ–induced ARE-driven luciferase activity in HSC3-ARE9 cells.
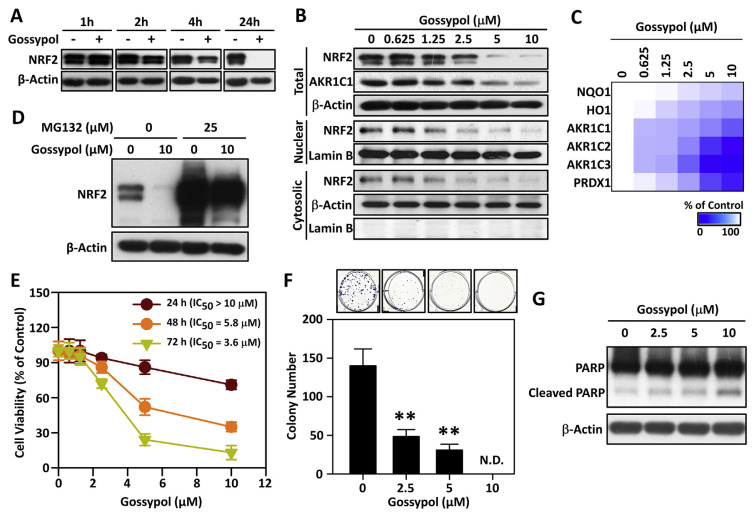
We further investigated the effects of gossypol on the protein levels of NRF2 in HSC-3 cells by Western blot analysis. As shown in Fig. 2A, gossypol reduced the levels of NRF2 protein in a time–dependent manner. Furthermore, treatment with gossypol for 24 h, we found that as the concentration of gossypol increases, the total, cytoplasmic and nuclear NRF2 protein gradually decreases (Fig. 2B). In order to confirm the inhibitory effect of gossypol on the NRF2/ARE pathway, we evaluated the expression of NRF2 regulatory genes by RT-qPCR. After 24 h of treatment with different concentrations of gossypol, the mRNA levels of NRF2 downstream genes, such as NQO1, HO1, AKR1C1/2/3, and PRDX1, decreased in a concentration-dependent manner (Fig. 2C). These results indicated that gossypol can indeed disrupt the NRF2/ARE pathway, thereby blocking the transcription of NRF2 downstream genes.
Fig. 2.
Gossypol effectively inhibits NRF2/ARE signaling pathway through impairing NRF2 protein stability and leads to reducing the viability of HNSCC cells.
(A) Time-dependent effects of 10 μM gossypol on the expression levels of NRF2 protein. (B) Dose-dependent effects of gossypol on the expression levels of NRF2 protein after 24 h of treatment. β-Actin was used as the internal control. (C) The heat-map of NRF2-regulated genes. HSC-3 cells were treated with various concentrations of gossypol for 24 h, and the mRNA levels of NRF2-regulated genes were analyzed by real-time RT-PCR. (D) Proteasome inhibitor, MG132, restored NRF2 protein level after gossypol treatment. HSC-3 cells were pre-treated with 25 μM MG132 for 30 min and then treated with or without 10 μM gossypol for additional 8 h. The level of NRF2 protein was determined by Western blot assay. β-Actin was used as the internal control. (E) HSC-3 cells were treated with various concentrations of gossypol at different time points. The percentage of viable cells was calculated compared to the control cells, and the IC50 values were calculated. (F) Gossypol significantly reduces colony forming ability in HSC-3 cells. Cells were seeded in 6-well plates with the indicated concentrations of gossypol for 24 h, then cells were washed twice with PBS and continued to culture in gossypol-free medium for 10 days. The methylene blue dye assay has been used to measure the number of colonies, and manual interpretation counts the colonies. Upper panel: the stained colonies were photographed; Bottom panel: the total number of colonies in each group was counted. Data are presented as the mean ± S.D. from three independent experiments (**p < 0.01 vs. Control). (G) Gossypol treatment induced cleaved PARP in HSC-3 cells. Cells were treated with 2.5, 5, 10 μM of gossypol for 24 h, the full length and cleaved PARP were detected by Western blot analysis. β-Actin was used as the internal control.
3.3. Gossypol exhibits antitumor activity by reducing the stability of NRF2 protein
Since gossypol reduces the expression level of NRF2 protein in a time– and dose–dependent manner (Fig. 2A–B), we further clarified whether the reduction of NRF2 protein level by gossypol is related to changes in NRF2 mRNA. The result demonstrated that treatment with different concentrations of gossypol for 24 h had no significant effect on the level of NFE2L2 (NRF2) mRNA (Fig. S2). Therefore, we hypothesized that gossypol may reduce the level of NRF2 protein by impairing the stability of NRF2 protein. As shown in Fig. 2D, pre-treatment of the proteasome inhibitor MG132 can effectively restore the NRF2 protein level caused by gossypol, indicating that gossypol promotes the proteasome degradation of NRF2 protein.
To further evaluate the effects of gossypol on anticancer effect of HNSCC cells, HSC-3 cells were exposed to various concentrations of gossypol for 24, 48 or 72 h and the viable cells were determined by staining with methylene blue. As shown in Fig. 2E, the effect of gossypol on cell viability is not obvious after 24 h of treatment. However, cell growth was markedly inhibited by gossypol in a time– and concentration–dependent manner after 48- and 72- h treatment. The IC50 values of gossypol at 24, 48, and 72 h were >10 μM, 5.8 μM and 3.6 μM, respectively.
Clonogenic assay is an in vitro cell survival assay based on the ability of a single cell to grow into a colony, thus this assay could exactly distinguish whether gossypol causes cell death or growth inhibition. As shown in (Fig. 2F), we found that a gradual reduction of colony forming ability was obviously observed with increasing concentrations of gossypol, indicating that gossypol treatment resulted in impaired HNSCC cell viability. Furthermore, we detected the level of cleaved PARP, a defining feature of apoptosis, in HSC-3 cells after treatment with gossypol for 24 h by Western blot analysis (Fig. 2G). Even though a basal level of cleaved PARP was observed in the control group, gossypol treatment significantly induced the cleavage of PARP in a concentration–dependent manner. These findings suggested that gossypol induced anti-cancer effect in part through apoptosis induction in HNSCC cells.
Over-activation of NRF2 has been considered to play an important driver role for promoting tumor progression and therapeutic resistance in non-small cell lung cancers (NSCLC) [17]. Therefore, in addition to HNSCC cells, we also evaluated the anti-cancer effect of gossypol on the non-small cell lung cancer (NSCLC) cell line NCI–H460. The results showed that gossypol exhibited the dose-related effect on cell viability of NCI–H460 cells with the IC50 value of 2.6 μM (Fig. S3A). In addition, we also observed that gossypol significantly reduced NRF2/ARE downstream proteins in a concentration-dependent manner (Fig. S3B), suggesting that gossypol is also an effective NRF2 inhibitor in NSCLC cells.
3.4. Gossypol enhances the anticancer effect of etoposide in topoisomerase II poison-resistant KB-7D cancer cells by inhibiting the NRF2/MRP1 axis
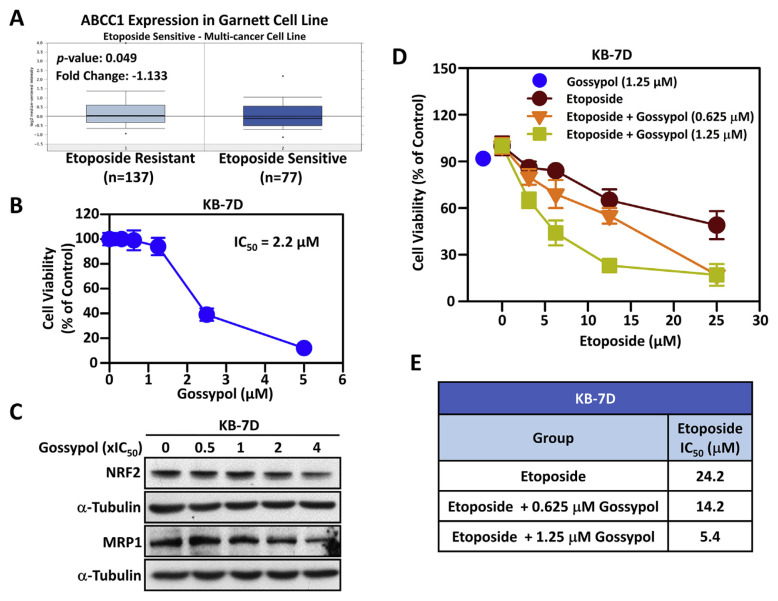
We have previously demonstrated that enhanced B-RAF-mediated NRF2 gene transcription and HATs-mediated NRF2 protein acetylation contributes to multidrug resistance-associated protein 1 (MRP1/ABCC1)-mediated chemoresistance in acquired topoisomerase II poison-resistant KB-7D cancer cells [8]. In addition, gene expression analysis of hundreds of cancer cell lines demonstrated that etoposide sensitive cells showed higher ABCC1 (MRP1) mRNA level than etoposide resistant cells (Fig. 3A). Therefore, we wondered whether gossypol can reverse the resistance of topoisomerase II poison–resistant KB-7D cells to etoposide. As shown in Fig. 3B, gossypol decreases cell viability of KB-7D cells in a dose–dependent manner and the IC50 value is 2.2 μM. Furthermore, we confirmed that gossypol can dose-dependently reduce NRF2 and MRP1 protein levels in KB-7D cells (Fig. 3C). Compared with the group treated with etoposide alone, the non-toxic concentration (0.625 and 1.25 μM) of gossypol significantly increases the sensitivity of KB-7D cells to etopoiside (Fig. 3D). Markedly, the IC50 values of etoposide in KB-7D cells decreased significantly from 24.2 μM to 14.2 (in combination with 0.625 μM gossypol) and 5.4 μM (in combination with 1.25 μM gossypol), respectively (Fig. 3D). These results suggested that gossypol effectually re-sensitizes etoposide-resistant cells to topoisomerase II poisons by reducing the protein expression of NRF2 and MRP1.
Fig. 3.
The effects of gossypol on the NRF2/MRP1 axis and etoposide sensitivity in etoposide-resistant KB-7D cells.
(A) The expression levels of MRP1 gene (ABCC1) in etoposide-resistant (left plot) and etoposide-sensitive (right plot) cancer cells were retrieved from ONCOMINE database. (B) Drug sensitivity of gossypol in etoposide-resistant HNSCC KB-7D cells. Cells were treated with the indicated concentrations of gossypol for 72 h and IC50 value was calculated. (C) Dose-dependent effect of gossypol on protein levels of NRF2 and MRP1. After treatment with the indicated concentrations of gossypol for 24 h, the NRF2 and MRP1 protein levels were detected by Western blot analysis. β-Actin was used as the internal control. (D) The combined effect of gossypol and etoposide on viability of KB-7D cells. Cells were co-treated with non-toxic concentrations of gossypol (0.625 or 1.25 μM) and etoposide for 72 h. Cell viability was determined by methylene blue assays. (E) IC50 values was calculated when combination of etoposide with non-toxic gossypol in KB-7D cells.
3.5. Gossypol may attenuate cisplatin resistance through inhibiting NRF2/G6PD axis in head and neck squamous cell carcinoma
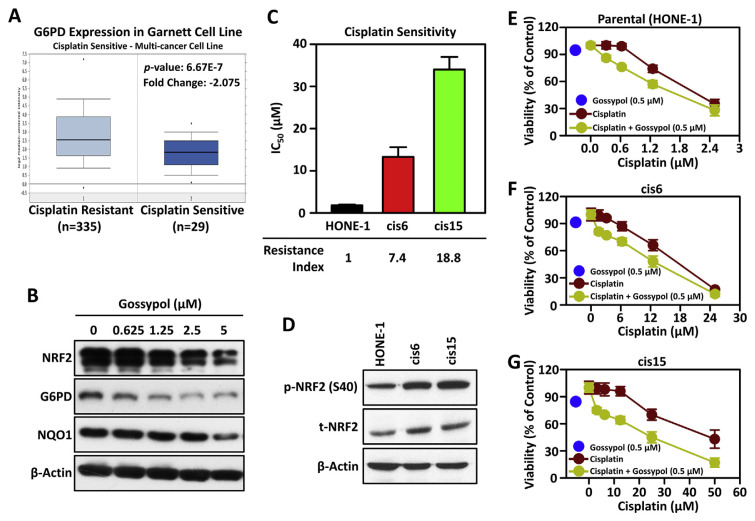
Anomalistic activation of the NRF2/ARE signaling pathway is believed to be the key mechanism that causes cancer cells to develop resistance to platinum drugs [7,17,18]. The discovery of effective anti-NRF2 therapeutics is a promising strategy for sensitizing cancer cells to platinums [6,10,19]. We recently identified that NRF2-mediated up-regulation of glucose-6-phosphate dehydrogenase (G6PD) played an important role to promote tumor progression in HNSCC. Blocking the NRF2/G6PD axis can effectively induce the synthetic lethality of HNSCC cells to cisplatin [13]. According to data retrieved from ONCOMINE, we observed that the expression level of G6PD mRNA is significantly increased in cisplatin-resistant cancer cells compared with cisplatin-sensitive cancer cells (Fig. 4A). Therefore, the inhibitory potential of gossypol in NRF2/G6PD axis was examined. The results demonstrated that HNSCC cells treated with gossypol effectively reduce the expression level of NRF2 and G6PD protein in a concentration-dependent manner (Fig. 4B). Therefore, we wondered whether gossypol can re-sensitize the anti-cancer effect of cisplatin in platinum resistant HNSCC cells.
Fig. 4.
Gossypol enhances cisplatin sensitivity through inhibition of NRF2/G6PD axis in cisplatin resistant HNSCC cells.
(A) The box plot of G6PD expression in different cancer cell lines. The expression levels of G6PD in cisplatin-resistant (left plot) and cisplatin-sensitive cancer cells (right plot) were retrieved from the Oncomine database. (B) Dose-dependent effect of gossypol on protein levels of NRF2, G6PD and NQO1. After treatment with 0.625, 1.25, 2.5, and 5 μM of gossypol for 24 h, the expression levels of NRF2, G6PD and NQO1 proteins were detected by Western blot analysis. β-Actin was used as the internal control. (C) The IC50 values of cisplatin in parental HONE-1 cell and the two resistant sub-lines, cis6 and cis15. The Resistance Index was calculated by dividing the IC50 value of cisplatin in resistant sub-line by the IC50 value of cisplatin in parental HONE-1 cell. (D) Total and phospho-NRF2 levels in parental and cisplatin-resistant HONE-1 cells. β-Actin was used as the internal control. (E ~ G) Cytotoxicity effects of gossypol combined with cisplatin in HONE-1-derieved cells. The cells were co-treated with non-toxic concentration of gossypol (0.5 μM) with cisplatin for 72 h, and cell viability was determined by methylene blue assays (E: parental HONE-1 cells; F: cisplatin-resistant cis6 cells; G: cisplatin-resistant cis15 cells).
To this end, cisplatin-resistant cells were established from HONE-1 cells (a representative HNSCC cell line derived from nasopharyngeal carcinoma) by exposure to increasing concentrations of cisplatin, and named cis6 and cis15. Compared with the parental HONE-1 cells, the IC50 values of the two drug-resistant sublines cis6 and cis15 to cisplatin were 7.4 and 18.8 times higher, respectively (Fig. 4C). Moreover, the expression levels of total and phospho-NRF2 proteins were also significantly higher in cis6 and cis15 cells than in parental HONE-1 cell (Fig. 4D). The anti-cancer cytotoxic effect of gossypol is equal potent in HONE-1, cis6, and cis15 with IC50 values of 0.89–1.32 μM (Table 3). Compared with the group treated with cisplatin alone, the non-toxic concentration of gossypol (0.5 μM) can significantly increase the sensitivity of HONE-1, cis6, and cis15 cells to cisplatin (Fig. 4E–G). Importantly, with the increase of cisplatin resistance, the chemo-sensitization ability of gossypol has been significantly improved. As shown in Table 3, the chemo-sensitization multiples of gossypol were 1.3-, 1.5-, and 1.8-fold in HONE-1, cis6 and cis15, respectively. These data indicated that gossypol effectively enhances the anti-cancer effect of cisplatin, especially in chemo-refractory cancer cells.
Table 3.
IC50 values for gossypol and cisplatin in HONE-1 and HONE-1 derived cisplatin-resistant cells.
| Group | HONE-1 (parental) | cisplatin-resistant | ||
|---|---|---|---|---|
|
| ||||
| cis6 | cis15 | |||
| IC50 for Gossypol (μM) | 1.32 | 1.13 | 0.89 | |
| IC50 for Cisplatin (μM) | Cisplatin alone | 2.02 | 18.1 | 42.5 |
| Cisplatin + Gossypolb | 1.55 | 12.2 | 23.4 | |
| Fold of chemo-sensitizationa | 1.3 | 1.5 | 1.8 | |
The Fold of chemo-sensitization was calculated by diving the IC50 value of cisplatin alone group by cisplatin combined with gossypol group.
The effects of gossypol on the cisplatin re-sensitization were assessed by combining the non-toxic concentration of gossypol (0.5 μM) with various concentrations of cisplatin.
4. Discussion
There is abundant evidence that the activation of NRF2 can reduce carcinogens and oxidative stress by inducing detoxification and antioxidant proteins, thereby inhibiting carcinogenesis, especially in its early stages [20]. Chemically diverse chemopreventive agents are known to exhibit cytoprotective effects by activating the NRF2 signaling pathway, many of which are dietary phytochemicals [21]. Based on our ARE reporter screening platform [14], we found that resveratrol, hydroquinone, ethylferulate, t-BHQ, apigenin, piceatannol, ebselen, curcumin, n-octyl caffeate, carnosic acid, tanshinone IIA, and bis-demethoxycurcumin exhibited significant NRF2/ARE-activating capacity with more than a two-fold increase of luciferase activity (Fig. 1). Indeed, all of these compounds were reported to activate NRF2/ARE pathway [22–24]. Among them, t-BHQ, resveratrol, and curcumin were deemed as the typical NRF2 activators [24]. These results indicated that our ARE reporter screening platform can fully verify previous publications, showing that this platform has excellent accuracy and credibility for discovery of NRF2 modulators.
Considering the role of NRF2 in regulating a battery of genes that act to detoxify anti-cancer drugs, attenuate drug-induced oxidative stress, or induce drug efflux, it is not surprise to note that NRF2/ARE pathway also plays an important role in developing cancer chemo-resistance [4,5]. Therefore, we aimed to identify novel NRF2 inhibitors as adjuvant chemotherapeutic agents. As the results, we found that HBED, hinokitiol, U83836E, GERIBP002A, CDC, and gossypol were potential NRF2 inhibitors. Among them, gossypol exhibited the highest ARE-driven luciferase inhibitory activity, and the second most effective compound was hinokitiol (Fig. 1). Hinokitiol is a known NRF2 inhibitor [25]. Of note, HBED, U83836E, GERIBP002A, CDC and gossypol have never been reported to have NRF2 inhibitory activity, and they were first clarified in this study.
Gossypol is a lipid-soluble and non-steroidal polyphenolic compound that naturally occurs in the cotton plant (genus Gossypium) and the portia tree (Thespesia populnea (L.) Sol. ex Corrêa) [26]. Gossypol and its derivatives was known to have potential anti-tumor effects [26]. Among them, the anti-apoptotic protein Bcl-2 and Bcl-xL is the most striking anti-tumor mechanism of gossypol and its derivatives [26,27]. Among them, AT101, the R-(−) enantiomer of gossypol acetic acid, has been studied in clinical for treatment of gastroesophageal carcinoma, advanced adrenal cortical carcinoma, prostate cancer, refractory metastatic breast cancer, and small cell lung cancer [26]. The combination of gossypol with other anti-cancer agents, such as topotecan [28], androgen deprivation therapy [29], docetaxel [30], and trastuzumab [31], have been also studied in clinical. AT101 has been applied to clinical use in the treatment of cancer in China [26]. In addition to induce cell death, gossypol has been also reported to diminish the epithelial-to-mesenchymal transition and metastatic potential of cancer cells by suppression of FAK signaling pathway and key molecules involved in EMT, such as N-cadherin, fibronectin and vimentin [32,33]. Gossypol also significantly inhibited the expression level or enzymatic activity of extracellular matrix-degrading proteases, such as Urokinase-type plasminogen activator (uPA) and matrix metallopeptidase 2 (MMP-2), thereby reducing the ability of cancer cells to metastasize [32,33].
We first time revealed that gossypol was a novel and potential NRF2 inhibitor. It not only suppressed basal ARE-driven luciferase activity but also inhibited t-BHQ-induced ARE-driven luciferase activity in HNSCC cells. Further studies demonstrated that gossypol significantly suppressed the expression levels of NRF2 protein and NRF2-regulated cytoprotective genes in a concentration- and time-dependent manners (Fig. 2A–C). We explored the potential mechanism of gossypol inhibiting the level of NRF2 protein, and found that gossypol reduced the expression levels of NRF2 protein by inducing protein degradation without affecting the level of NRF2 mRNA (Fig. 2D & S2). Interestingly, we noted that under short-term treatment (24 h), gossypol significantly inhibited NRF2 activity without affecting cell survival. Once the treatment time was extended (48 or 72 h), gossypol significantly reduced the viability of cancer cells (Fig. 2E–F & S3A). Gossypol has been reported to induce both apoptosis and autophagic cell death in cancer cells [34–36]. In the present study, we observed that treatment with gossypol significantly induced apoptosis (Fig. 2G). In addition, we also found that gossypol caused the conversion of LC3-I to LC3-II, an autophagy marker, in HSC-3 cells (Fig. S4), suggesting that gossypol may also trigger autophagy in HNSCC cells. However, the role of gossypol-induced autophagy in HNSCC cells whether triggering autophagic cell death remains to be elucidated. Furthermore, the cell cycle analysis demonstrated that gossypol strikingly induced S and G2/M phases arrest in the time- and dose-dependent manners (Fig. S5A–B). The obtained results adumbrated that gossypol-induced suppression of NRF2/ARE pathway may act on the S/G2/M phases and then reduces the viability of cancer cells. A previous study demonstrated that apogossypolone (ApoG2), a derivate of gossypol, stimulated cell cycle arrest at S phase through downregulation of proto-oncogene c-MYC and G1/S checkpoints cyclin D1/cyclin E and induction of tumor suppressor p21 proteins [37]. The molecular mechanism underlying the cell cycle perturbation and NRF2 inhibition by gossypol in HNSCC cells remains further clarifications.
It has been reported that excessive activation of NRF2/ARE pathway has brought great benefits for tumor progression [3], and prolonged activation of NRF2/ARE signal pathway confers therapeutic resistance of cancer cells through inductions of antioxidant proteins, metabolizing/detoxifying enzymes, and drug-efflux transporters, etc. [4,5,7,8]. Therefore, inhibition of NRF2 is not only a potential treatment to eradicate tumors but also a chemosensitization strategy to reverse chemoresistance [6–8,10,13]. For instance, ERK-stimulated hyperactivation of the NRF2/ARE pathway played an important role in acquired resistance against tamoxifen in human breast cancer cells [38]. Epicatechin gallate (EGCG), the major catechin found in green tea, which was found that exhibited anti-proliferative activity in patient-derived triple-negative breast cancer xenograft mouse model [39], strikingly suppressed NRF2/ARE pathway and efficiently re-sensitized tamoxifen-resistant breast cancer cells [40]. Furthermore, etoposide, a topoisomerase II poison, is a widely used chemotherapeutic agent. We previously established an etoposide-resistant cell line KB-7D and found that activation of NRF2/multidrug resistance protein 1 (MRP1/ABCC1) axis caused acquired resistance to topoisomerase II poisons [8]. It was worth noting that the combination of etoposide and gossypol can significantly improve the therapeutic efficacy of etoposide and reverse drug resistance by reduction of the expression levels of NRF2 and MRP1 proteins in etoposide-resistant KB-7D cells (Fig. 3). These results indicated that gossypol may have potential to enhance therapeutic efficacy of topoisomerase II poison-refractory tumors clinically.
Platinum-based chemotherapeutic drugs, such as cisplatin, carboplatin, and oxaliplatin, are the most powerful and widely used anti-cancer agents [41]. Although platinum-based chemotherapy remains the cornerstone for cancer treatment, but most patients experienced relapse and further developed platinum resistance [41]. We and others previous studies demonstrated that NRF2 promotes the resistance of cancer cells to platinum-based chemotherapy by inducing the antioxidant proteins and metabolizing/detoxifying enzymes [5,7,18]. More recently, we revealed that inhibiting NRF2-mediated glucose-6-phosphate dehydrogenase (G6PD) expression stimulated the synthetic lethality with cisplatin in HNSCC cells [13], indicating that NRF2-mediated metabolic reprogramming also contributes to cisplatin resistance. In this study, we first identified that gossypol strikingly suppressed both NRF2 and G6PD protein levels in a concentration-dependent manner. Similar to the inhibitory effect on the protein level, we further verified that gossypol significantly reduced the expression of G6PD mRNA (Fig.S6). These results implied that gossypol may suppress G6PD through inhibiting NRF2-mediated G6PD gene transactivation. Therefore, we inferred that gossypol inhibits the NRF2/G6PD axis as one of the mechanisms to alleviate cisplatin resistance in HNSCC cells. To further simulate the clinical treatment conditions, we established two cisplatin resistant cell lines cis6 and cis15 from HONE-1 cells for further study. As compared to HONE-1 cells, cis6 and cis15 cells showed 7.4- and 18.8-fold resistance to cisplatin, respectively, and resistant cells also expressed higher level of total NRF2 and phospho-NRF2 than parental cells (Fig. 4A–D). Unlike with cisplatin, gossypol was equal potent between parental and resistant cells with IC50 values approximately 1 μM. Notably, combination of non-toxic concentration of gossypol markedly increased cisplatin sensitivity thereby reduced the IC50 values of cisplatin in HONE-1, cis6, and cis15 cells (Fig. 4E–G & Table 3). Importantly, as the resistance of cells to cisplatin increases, gossypol had a better ability to sensitize cisplatin (Table 3). Previous studies have shown that gossypol enhanced the sensitivity of cancer cells to cisplatin through inhibiting anti-apoptotic Bcl-2 family proteins, blocking APE1-mediated IL-6/STAT3 signaling, and up-regulating pro-apoptotic molecule Smac [26,27,42]. Nevertheless, our research clearly demonstrated that in addition to the aforementioned regulatory mechanisms, the inhibition of NRF2 by gossypol was a novel and important mechanism for chemo-sensitization and drug resistance reversal in cisplatin resistant cells.
In conclusion, we identified that gossypol was a novel NRF2/ARE inhibitor. This was the first study to corroborate that gossypol reduces the stability of NRF2 protein and leads to inhibition of NRF2- mediated transcriptional signature. Importantly, gossypol not only inhibited growth of cancer cells, but also enhanced the therapeutic effects of etoposide and cisplatin in drug-resistant cancer cells by inhibiting the NRF2/MRP1 and NRF2/G6PD axis, respectively. These results suggested that blockade of NRF2 signal by gossypol may have the potential to enhance therapeutic efficacy of chemo-refractory tumors in the clinic.
Availability of data and materials
The analysis of NFE2L2 (NRF2), ABCC1 (MRP1) and G6PD mRNA expression levels (Figs. 1A, 5A and 6A and S1) were obtained from the Oncomine (https://www.oncomine.org/). All data generated or analyzed during this study were included in this published article and its supplementary information file. All data in this study is available from the corresponding author upon reasonable request.
Acknowledgements
Thanks to the Taiwan Bioinformatics Institute Core Facility for assistances on using Oncomine database (National Core Facility Program for Biotechnology, MOST 109-2740-B-400-002). Ya-Chu Tang carried out her thesis research under the auspices of the Graduate Program of Biotechnology in Medicine, National Tsing Hua University and National Health Research Institutes.
List of abbreviations
- AKR1C
aldo-keto reductase family 1C
- ARE
antioxidant response elements
- t-BHQ
tert-butylhydroquinone
- G6PD
glucose-6-phosphate dehydrogenase
- HNSCC
head and neck squamous cell carcinoma
- KEAP1
Kelch-like ECH-associated protein 1
- MRP1/ABCC1
multidrug resistance-associated protein 1
- NRF2
Nuclear factor erythroid 2-related factor 2
- NSCLC
non-small cell lung cancer
- ROS
reactive oxygen species
Appendix. Supplementary materials and methods
Chemical reagents
The SCREEN-WELL® REDOX Library (BML-2835, Version 1.0) was obtained from Enzo Life Sciences (NY., USA). Other chemicals were purchased from Sigma–Aldrich (Saint Louis, MO., USA) unless otherwise stated. Dulbecco’s Modified Eagle Medium (DMEM), Minimum Essential Media (MEM), Ham’s F-12, 100 – Penicillin-Streptomycin-Glutamine solution, fetal bovine serum (FBS), trypsin, and G418 were purchased from Gibco-Thermo Fisher Scientific (MA., USA). The primary antibodies used in this study were listed in Table S1.
RNA extraction and real-time PCR
Total RNA were obtained from cells by using the NucleoSpin RNA purification kit (740955, Macherey–Nagel, Düren, Germany), and then the cDNA was synthesized from equal amounts (2 μg) of total RNA using SuperScriptTM III Reverse Transcriptase (12574026, Invitrogen, MA, USA). Real-time PCR was performed under the following conditions: an initial denaturing cycle at 95 °C for 3 min, followed by 40 cycles of amplification consisting of denaturation at 95 °C for 3 s, annealing at 60 °C for 30 s. An extend program as following: 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s was added after the last cycle for melting curve stage. The forward and reverse primer sequences used to amplify each target gene were listed in Table S2. The changes of NRF2, NQO1, HO1, AKR1C1, AKR1C2, AKR1C3 and PRDX1 mRNA levels were expressed as fold to control after normalization to RPLP0.
Western blot analysis
Western blot analysis was performed on total protein isolated from cells. Total protein was extracted, and protein concentration was determined by the Bradford assay (#5000006, Bio-Rad Laboratories, CA, USA). Equal amounts (40 μg) of total protein were separated by 10% SDS-PAGE with a loading buffer (100 mMTris–HCl, pH 6.8, 4% SDS, 20% glycerol, 0.2% bromophenol blue, and 10% β-mercaptoethanol). The separated protein in SDS-PAGE was transferred to PVDF membrane (Millipore Co., Bedford, MA). After blocking with 5% skim milk in 1 – PBST for 1 h at room temperature, the membrane was incubated with primary antibody at 4 °C for overnight. After washing three times for 10 min each with TBST, the membrane was incubated with the secondary antibody for 1 h at room temperature. The immunoreactive bands were visualized by using WesternBright ECL (R-03031, Advansta, CA, USA). All primary antibodies were used at dilutions of 1:500–2000 and listed in Table S1. Donkey anti-rabbit IgG and donkey anti-mouse IgG conjugated with horseradish peroxidase were used as the secondary antibody at a dilution of 1:5000.
Cell cycle analysis
For analysis of the cell cycle distribution, HSC-3 cells were incubated with various concentrations of gossypol (0, 2.5, and 5 μM) for 24 and 48 h, and then cells were collected by centrifugation. The harvested cell pellets were fixed in 80% ethanol (dissolved in 1 – PBS, pH 7.4) and stored at −20 °C at least 2 h. For analysis, cell pellets were washed with cold 1 – PBS (pH 7.4), incubated at 4 °C for 30 min with 50 μg/ml RNase A (Sigma), and then stained with 50 μg/ml propidium iodide (Sigma) in 1 – PBS. Cell cycle profiles and distributions were determined by flow cytometry analysis of 104 cells using the CELLQuest program on a FACS Calibur flow cytometer (Becton–Dickinson, San Jose, CA). Clumped cells were excluded from the cell cycle distribution analysis by gating.
Gene expression analysis from Oncomine database
Data regarding NRF2 mRNA expression in normal and tumor specimen, and ABCC1 (MRP1) and G6PD mRNA expression in drug-resistant and drug-sensitive cancer cell lines was retrieved from ONCOMINE database (www.oncomine.org).
Supplementary figures
NRF2 is unduly expressed and associated with cell growth in HNSCC.
(A) The levels of total NRF2 (t-NRF2) and phospo-NRF2 (Ser40) proteins were detected by western blotting in HNSCC cell lines. β-Actin was used as the internal control. (B) The effect of NRF2-knockdown on the viability of HSC-3 cells. Data are presented as the mean ± S.D. from three independent experiments (***p < 0.001 vs. siControl).
Gossypol does not impair NRF2 mRNA level in HNSCC cells.
The expression levels of NRF2 mRNA did not change after gossypol treatment. The levels of NRF2 mRNA were detected by RT-qPCR after incubation with 2.5, 5, 10 μM gossypol for 24 h in HSC-3 cells.
Effects of gossypol on cell viability of non-small cell lung cancer cell line NCI–H460.
(A) Dose–response curve for viability using methylene blue assay in NCI–H460 cells exposed to gossypol for 72 h and the IC50 value was calculated. The percentage of viable cells was calculated compared to the control cells. (B) Dose-dependent effects of gossypol on protein levels of NRF2 and NRF2-downstream proteins, such as AKR1C2 and PRDX1. NCI–H460 cells were incubated with the indicated concentrations of gossypol for 24 h. The changes of NRF2, AKR1C2, and PRDX1 protein levels were evaluated by western blot assay. β-Actin was used as the internal control.
Gossypol induces the autophagy in HSC-3 cells.
HSC-3 cells were treated with the indicated concentration of gossypol. After 24 h, cells were lysed and the levels of LC3-I and LC3-II were determined were detected by western blotting. The β-actin was used as the loading control. The cells treated with 50 μM chloroquine (CQ) for 24 h was used as a positive control.
Gossypol induces cell cycle arrest at the G2/M phase in HNSCC cells.
HSC-3 cells were incubated with various concentrations of gossypol (0, 2.5, and 5 μM) for 24 and 48 h, and the cell cycle distribution was determined by flow cytometry followed by PI staining. Bar graphs show the percentages of HSC-3 cells in G0/G1, S, and G2/M phases after treatment with gossypol for 24 h (A) and 48 h (B), respectively.
Gossypol reduces G6PD mRNA levels in HNSCC cells.
The expression levels of G6PD mRNA were decreased after gossypol treatment. The levels of G6PD mRNA were detected by RT-qPCR after incubation with 0, 2.5, 5, 10 μM gossypol for 24 h in HSC-3 cells.
Supplementary tables
Table S1.
Antibody list
| Antibody | Assay | Host | Information |
|---|---|---|---|
| α-Tubulin | Immunoblot | mouse | GTX628802 (GeneTex) |
| β-Actin | Immunoblot | mouse | GTX109639 (GeneTex) |
| AKR1C1 | Immunoblot | mouse | H00001645-A01 (Abnova) |
| G6PD | Immunoblot | rabbit | 8866S (Cell Signaling Technology) |
| Lamin B1 | Immunoblot | mouse | ab20396 (Abcam) |
| MRP1/ABCC1 | Immunoblot | rabbit | 72202S (Cell Signaling Technology) |
| NRF2 | Immunoblot | rabbit | ab62352 (Abcam) |
| p-NRF2(S40) | Immunoblot | rabbit | ab76026 (Abcam) |
| NQO1 | Immunoblot | mouse | 3187S (Cell Signaling Technology) |
| PARP | Immunoblot | rabbit | GTX100573 (GeneTex) |
| PRDX1 | Immunoblot | rabbit | ARP48454_P050 (Aviva Systems Biology) |
Table S2.
Primers used for quantitative real-time RT-PCR
| Gene | Sequence | Accession no. | Product size (bp) | |
|---|---|---|---|---|
| hAKR1C1 | FP | CATGCCTGTCCTGGGATTT | NM_001353.6 | 109 |
| RP | AGAATCAATATGGCGGAAG | |||
| hAKR1C2 | FP | GCAGCGCATCAGACAGAA | NM_001354.6 | 104 |
| RP | CAAGGGTCAAATATCGCACA | |||
| hAKR1C3 | FP | CATTGGGGTGTCAAACTTCA | NM_003739.6 | 112 |
| RP | CCGGTTGAAATACGGATGAC | |||
| hG6PD | FP | TGACCTGGCCAAGAAGAAGA | NM_000402.4 | 185 |
| RP | CAAAGAAGTCCTCCAGCTTG | |||
| hHO-1 | FP | TTCAGAAGGGCCAGGTGA | NM_002133.3 | 78 |
| RP | CCTCAAGGGCCACATAGAT | |||
| hNRF2 | FP | CAGCGACGGAAAGAGTATGA | NM_006164.5 | 200 |
| RP | TGGGCAACCTGGGAGTAG | |||
| hNQO1 | FP | ATGTATGACAAAGGACCCTTCC | NM_000903.3 | 88 |
| RP | TCCCTTGCAGAGAGTACATGG | |||
| hPRDX1 | FP | CACTGACAAACATGGGGAAGT | NM_002574.4 | 82 |
| RP | TTTGCTCTTTTGGACATCAGG | |||
| hRPLP0 | FP | TGGTCATCCAGCAGGTGTTCGA | NM_001002 | 119 |
| RP | ACAGACACTGGCAACATTGCGG |
Funding Statement
This study was supported by Research Grants MOST 109-2320-B-400-012, 109-2314-B-400-046, 108-2314-B-400-007, and MOST 107-2314-B-400-020 from Ministry of Science and Technology, Taiwan; and by NHRI-BP-109-PP-02 from National Health Research Institutes, Taiwan.
Footnotes
Conflict of interest
The authors declare no competing financial interests.
Authors’ contributions
Ya-Chu Tang and Hsin-Huei Chang contributed to design and conduct of the main experiments, process and analyze the experimental data, and prepare manuscript drafts. Huang-Hui Chen provided suggestions for the experiment design and assisted the experiments. Jau-Ying Yao assisted part of the in vitro experiment. Yu-Tsen Chen carried out the initial screening. Yung-Jen Chuang and Jang-Yang Chang provided suggestions for the experiment design. Ching-Chuan Kuo initiated the research idea, guided research directions, supervised research project, and edited the manuscript.
Funding
This study was supported by Research Grants MOST 109-2320-B-400-012, 109-2314-B-400-046, 108-2314-B-400-007, and MOST 107-2314-B-400-020 from Ministry of Science and Technology, Taiwan; and by NHRI-BP-109-PP-02 from National Health Research Institutes, Taiwan.
References
- 1. McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, et al. The Cap’n’Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–307. [PubMed] [Google Scholar]
- 2. Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rojo de la Vega M, Chapman E, Zhang DD. NRF2 and the hallmarks of cancer. Cancer Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bai X, Chen Y, Hou X, Huang M, Jin J. Emerging role of NRF2 in chemoresistance by regulating drug-metabolizing enzymes and efflux transporters. Drug Metab Rev. 2016;48:541–67. doi: 10.1080/03602532.2016.1197239. [DOI] [PubMed] [Google Scholar]
- 5. Xue D, Zhou X, Qiu J. Emerging role of NRF2 in ROS-mediated tumor chemoresistance. Biomed Pharmacother. 2020;131:110676. doi: 10.1016/j.biopha.2020.110676. [DOI] [PubMed] [Google Scholar]
- 6. Zhu J, Wang H, Chen F, Fu J, Xu Y, Hou Y, et al. An overview of chemical inhibitors of the Nrf2-ARE signaling pathway and their potential applications in cancer therapy. Free Radic Biol Med. 2016;99:544–56. doi: 10.1016/j.freeradbiomed.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 7. Chen CC, Chu CB, Liu KJ, Huang CY, Chang JY, Pan WY, et al. Gene expression profiling for analysis acquired oxaliplatin resistant factors in human gastric carcinoma TSGH-S3 cells: the role of IL-6 signaling and Nrf2/AKR1C axis identification. Biochem Pharmacol. 2013;86:872–87. doi: 10.1016/j.bcp.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 8. Chen HH, Chang HH, Chang JY, Tang YC, Cheng YC, Lin LM, et al. Enhanced B-Raf-mediated NRF2 gene transcription and HATs-mediated NRF2 protein acetylation contributes to ABCC1-mediated chemoresistance and glutathione-mediated survival in acquired topoisomerase II poison-resistant cancer cells. Free Radic Biol Med. 2017;113:505–18. doi: 10.1016/j.freeradbiomed.2017.10.375. [DOI] [PubMed] [Google Scholar]
- 9. Panieri E, Buha A, Telkoparan-Akillilar P, Cevik D, Kouretas D, Veskoukis A, et al. Potential applications of NRF2 modulators in cancer therapy. Antioxidants (Basel) 2020:9. doi: 10.3390/antiox9030193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin H, Qiao Y, Yang H, Nan Q, Qu W, Feng F, et al. Small molecular Nrf2 inhibitors as chemosensitizers for cancer therapy. Future Med Chem. 2020;12:243–67. doi: 10.4155/fmc-2019-0285. [DOI] [PubMed] [Google Scholar]
- 11. Bollong MJ, Yun H, Sherwood L, Woods AK, Lairson LL, Schultz PG. A small molecule inhibits deregulated NRF2 transcriptional activity in cancer. ACS Chem Biol. 2015;10:2193–8. doi: 10.1021/acschembio.5b00448. [DOI] [PubMed] [Google Scholar]
- 12. Singh A, Venkannagari S, Oh KH, Zhang YQ, Rohde JM, Liu L, et al. Small molecule inhibitor of NRF2 selectively intervenes therapeutic resistance in KEAP1-deficient NSCLC tumors. ACS Chem Biol. 2016;11:3214–25. doi: 10.1021/acschembio.6b00651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang YC, Hsiao JR, Jiang SS, Chang JY, Chu PY, Liu KJ, et al. c-MYC-directed NRF2 drives malignant progression of head and neck cancer via glucose-6-phosphate dehydrogenase and transketolase activation. Theranostics. 2021;11:5232–47. doi: 10.7150/thno.53417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen HH, Chen YT, Huang YW, Tsai HJ, Kuo CC. 4-Keto-pinoresinol, a novel naturally occurring ARE activator, induces the Nrf2/HO-1 axis and protects against oxidative stress-induced cell injury via activation of PI3K/AKT signaling. Free Radic Biol Med. 2012;52:1054–66. doi: 10.1016/j.freeradbiomed.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 15. Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem. 2002;277:42769–74. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 16. Wang X, Howell CP, Chen F, Yin J, Jiang Y. Gossypol–a polyphenolic compound from cotton plant. Adv Food Nutr Res. 2009;58:215–63. doi: 10.1016/S1043-4526(09)58006-0. [DOI] [PubMed] [Google Scholar]
- 17. Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, Moran CA, et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res. 2010;16:3743–53. doi: 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Noman ASM, Parag RR, Rashid MI, Islam S, Rahman MZ, Chowdhury AA, et al. Chemotherapeutic resistance of head and neck squamous cell carcinoma is mediated by EpCAM induction driven by IL-6/p62 associated Nrf2-antioxidant pathway activation. Cell Death Dis. 2020;11:663. doi: 10.1038/s41419-020-02907-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matthews JH, Liang X, Paul VJ, Luesch H. A complementary chemical and genomic screening approach for druggable targets in the Nrf2 pathway and small molecule inhibitors to overcome cancer cell drug resistance. ACS Chem Biol. 2018;13:1189–99. doi: 10.1021/acschembio.7b01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hayes JD, McMahon M, Chowdhry S, Dinkova-Kostova AT. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid Redox Signal. 2010;13:1713–48. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- 21. Qin S, Hou DX. Multiple regulations of Keap1/Nrf2 system by dietary phytochemicals. Mol Nutr Food Res. 2016;60:1731–55. doi: 10.1002/mnfr.201501017. [DOI] [PubMed] [Google Scholar]
- 22. Qu Z, Sun J, Zhang W, Yu J, Zhuang C. Transcription factor NRF2 as a promising therapeutic target for Alzheimer’s disease. Free Radic Biol Med. 2020;159:87–102. doi: 10.1016/j.freeradbiomed.2020.06.028. [DOI] [PubMed] [Google Scholar]
- 23. Boo YC. Natural Nrf2 modulators for skin protection. Antioxidants (Basel) 2020:9. doi: 10.3390/antiox9090812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robledinos-Antón N, Fernández-Ginés R, Manda G, Cuadrado A. Activators and inhibitors of NRF2: a review of their potential for clinical development. Oxid Med Cell Longev. 2019;2019:9372182. doi: 10.1155/2019/9372182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ouyang WC, Liao YW, Chen PN, Lu KH, Yu CC, Hsieh PL. Hinokitiol suppresses cancer stemness and oncogenicity in glioma stem cells by Nrf2 regulation. Cancer Chemother Pharmacol. 2017;80:411–9. doi: 10.1007/s00280-017-3381-y. [DOI] [PubMed] [Google Scholar]
- 26. Zeng Y, Ma J, Xu L, Wu D. Natural product gossypol and its derivatives in precision cancer medicine. Curr Med Chem. 2019;26:1849–73. doi: 10.2174/0929867324666170523123655. [DOI] [PubMed] [Google Scholar]
- 27. Bauer JA, Trask DK, Kumar B, Los G, Castro J, Lee JS, et al. Reversal of cisplatin resistance with a BH3 mimetic, (-)-gossypol, in head and neck cancer cells: role of wild-type p53 and Bcl-xL. Mol Cancer Therapeut. 2005;4:1096–104. doi: 10.1158/1535-7163.MCT-05-0081. [DOI] [PubMed] [Google Scholar]
- 28. Heist RS, Fain J, Chinnasami B, Khan W, Molina JR, Sequist LV, et al. Phase I/II study of AT-101 with topotecan in relapsed and refractory small cell lung cancer. J Thorac Oncol. 2010;5:1637–43. doi: 10.1097/JTO.0b013e3181e8f4dc. [DOI] [PubMed] [Google Scholar]
- 29. Stein MN, Hussain M, Stadler WM, Liu G, Tereshchenko IV, Goodin S, et al. A phase II study of AT-101 to overcome Bcl-2–mediated resistance to androgen deprivation therapy in patients with newly diagnosed castration-sensitive metastatic prostate cancer. Clin Genitourin Cancer. 2016;14:22–7. doi: 10.1016/j.clgc.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sonpavde G, Matveev V, Burke JM, Caton JR, Fleming MT, Hutson TE, et al. Randomized phase II trial of docetaxel plus prednisone in combination with placebo or AT-101, an oral small molecule Bcl-2 family antagonist, as first-line therapy for metastatic castration-resistant prostate cancer. Ann Oncol. 2012;23:1803–8. doi: 10.1093/annonc/mdr555. [DOI] [PubMed] [Google Scholar]
- 31. Bulut G, Atmaca H, Karaca B. Trastuzumab in combination with AT-101 induces cytotoxicity and apoptosis in Her2 positive breast cancer cells. Future Oncol. 2020;16:4485–95. doi: 10.2217/fon-2019-0521. [DOI] [PubMed] [Google Scholar]
- 32. Hsieh YS, Chu SC, Huang SC, Kao SH, Lin MS, Chen PN. Gossypol reduces metastasis and epithelial-mesenchymal transition by targeting protease in human cervical cancer. Am J Chin Med. 2021;49:181–98. doi: 10.1142/S0192415X21500105. [DOI] [PubMed] [Google Scholar]
- 33. Huang SF, Chu SC, Hsu LS, Tu YC, Chen PN, Hsieh YS. Antimetastatic effects of gossypol on colon cancer cells by targeting the u-PA and FAK pathways. Food Func. 2019;10:8172–81. doi: 10.1039/c9fo01306g. [DOI] [PubMed] [Google Scholar]
- 34. Lu MD, Li LY, Li PH, You T, Wang FH, Sun WJ, et al. Gossypol induces cell death by activating apoptosis and autophagy in HT-29 cells. Mol Med Rep. 2017;16:2128–32. doi: 10.3892/mmr.2017.6804. [DOI] [PubMed] [Google Scholar]
- 35. Benvenuto M, Mattera R, Masuelli L, Taffera G, Andracchio O, Tresoldi I, et al. (±)-Gossypol induces apoptosis and autophagy in head and neck carcinoma cell lines and inhibits the growth of transplanted salivary gland cancer cells in BALB/c mice. Int J Food Sci Nutr. 2017;68:298–312. doi: 10.1080/09637486.2016.1236077. [DOI] [PubMed] [Google Scholar]
- 36. Voss V, Senft C, Lang V, Ronellenfitsch MW, Steinbach JP, Seifert V, et al. The pan-Bcl-2 inhibitor (−)-gossypol triggers autophagic cell death in malignant glioma. Mol Cancer Res : MCR. 2010;8:1002–16. doi: 10.1158/1541-7786.MCR-09-0562. [DOI] [PubMed] [Google Scholar]
- 37. Hu ZY, Sun J, Zhu XF, Yang D, Zeng YX. ApoG2 induces cell cycle arrest of nasopharyngeal carcinoma cells by suppressing the c-Myc signaling pathway. J Transl Med. 2009;7:74. doi: 10.1186/1479-5876-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim SK, Yang JW, Kim MR, Roh SH, Kim HG, Lee KY, et al. Increased expression of Nrf2/ARE-dependent anti-oxidant proteins in tamoxifen-resistant breast cancer cells. Free Radic Biol Med. 2008;45:537–46. doi: 10.1016/j.freeradbiomed.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 39. Lee W-J, Cheng T-C, Yen Y, Fang C-L, Liao Y-C, Kuo C-C, et al. Tea polyphenol epigallocatechin-3-gallate inhibits cell proliferation in a patient-derived triple-negative breast cancer xenograft mouse model via inhibition of proline-dehydrogenase-induced effects. J Food Drug Anal. 2021:29. doi: 10.38212/2224-6614.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Esmaeili MA. Combination of siRNA-directed gene silencing with epigallocatechin-3-gallate (EGCG) reverses drug resistance in human breast cancer cells. J Chem Biol. 2016;9:41–52. doi: 10.1007/s12154-015-0144-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rottenberg S, Disler C, Perego P. The rediscovery of platinum-based cancer therapy. Nat Rev Cancer. 2021;21:37–50. doi: 10.1038/s41568-020-00308-y. [DOI] [PubMed] [Google Scholar]
- 42. Lu Y, Li J, Dong CE, Huang J, Zhou HB, Wang W. Recent advances in gossypol derivatives and analogs: a chemistry and biology view. Future Med Chem. 2017;9:1243–75. doi: 10.4155/fmc-2017-0046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NRF2 is unduly expressed and associated with cell growth in HNSCC.
(A) The levels of total NRF2 (t-NRF2) and phospo-NRF2 (Ser40) proteins were detected by western blotting in HNSCC cell lines. β-Actin was used as the internal control. (B) The effect of NRF2-knockdown on the viability of HSC-3 cells. Data are presented as the mean ± S.D. from three independent experiments (***p < 0.001 vs. siControl).
Gossypol does not impair NRF2 mRNA level in HNSCC cells.
The expression levels of NRF2 mRNA did not change after gossypol treatment. The levels of NRF2 mRNA were detected by RT-qPCR after incubation with 2.5, 5, 10 μM gossypol for 24 h in HSC-3 cells.
Effects of gossypol on cell viability of non-small cell lung cancer cell line NCI–H460.
(A) Dose–response curve for viability using methylene blue assay in NCI–H460 cells exposed to gossypol for 72 h and the IC50 value was calculated. The percentage of viable cells was calculated compared to the control cells. (B) Dose-dependent effects of gossypol on protein levels of NRF2 and NRF2-downstream proteins, such as AKR1C2 and PRDX1. NCI–H460 cells were incubated with the indicated concentrations of gossypol for 24 h. The changes of NRF2, AKR1C2, and PRDX1 protein levels were evaluated by western blot assay. β-Actin was used as the internal control.
Gossypol induces the autophagy in HSC-3 cells.
HSC-3 cells were treated with the indicated concentration of gossypol. After 24 h, cells were lysed and the levels of LC3-I and LC3-II were determined were detected by western blotting. The β-actin was used as the loading control. The cells treated with 50 μM chloroquine (CQ) for 24 h was used as a positive control.
Gossypol induces cell cycle arrest at the G2/M phase in HNSCC cells.
HSC-3 cells were incubated with various concentrations of gossypol (0, 2.5, and 5 μM) for 24 and 48 h, and the cell cycle distribution was determined by flow cytometry followed by PI staining. Bar graphs show the percentages of HSC-3 cells in G0/G1, S, and G2/M phases after treatment with gossypol for 24 h (A) and 48 h (B), respectively.
Gossypol reduces G6PD mRNA levels in HNSCC cells.
The expression levels of G6PD mRNA were decreased after gossypol treatment. The levels of G6PD mRNA were detected by RT-qPCR after incubation with 0, 2.5, 5, 10 μM gossypol for 24 h in HSC-3 cells.
Table S1.
Antibody list
| Antibody | Assay | Host | Information |
|---|---|---|---|
| α-Tubulin | Immunoblot | mouse | GTX628802 (GeneTex) |
| β-Actin | Immunoblot | mouse | GTX109639 (GeneTex) |
| AKR1C1 | Immunoblot | mouse | H00001645-A01 (Abnova) |
| G6PD | Immunoblot | rabbit | 8866S (Cell Signaling Technology) |
| Lamin B1 | Immunoblot | mouse | ab20396 (Abcam) |
| MRP1/ABCC1 | Immunoblot | rabbit | 72202S (Cell Signaling Technology) |
| NRF2 | Immunoblot | rabbit | ab62352 (Abcam) |
| p-NRF2(S40) | Immunoblot | rabbit | ab76026 (Abcam) |
| NQO1 | Immunoblot | mouse | 3187S (Cell Signaling Technology) |
| PARP | Immunoblot | rabbit | GTX100573 (GeneTex) |
| PRDX1 | Immunoblot | rabbit | ARP48454_P050 (Aviva Systems Biology) |
Table S2.
Primers used for quantitative real-time RT-PCR
| Gene | Sequence | Accession no. | Product size (bp) | |
|---|---|---|---|---|
| hAKR1C1 | FP | CATGCCTGTCCTGGGATTT | NM_001353.6 | 109 |
| RP | AGAATCAATATGGCGGAAG | |||
| hAKR1C2 | FP | GCAGCGCATCAGACAGAA | NM_001354.6 | 104 |
| RP | CAAGGGTCAAATATCGCACA | |||
| hAKR1C3 | FP | CATTGGGGTGTCAAACTTCA | NM_003739.6 | 112 |
| RP | CCGGTTGAAATACGGATGAC | |||
| hG6PD | FP | TGACCTGGCCAAGAAGAAGA | NM_000402.4 | 185 |
| RP | CAAAGAAGTCCTCCAGCTTG | |||
| hHO-1 | FP | TTCAGAAGGGCCAGGTGA | NM_002133.3 | 78 |
| RP | CCTCAAGGGCCACATAGAT | |||
| hNRF2 | FP | CAGCGACGGAAAGAGTATGA | NM_006164.5 | 200 |
| RP | TGGGCAACCTGGGAGTAG | |||
| hNQO1 | FP | ATGTATGACAAAGGACCCTTCC | NM_000903.3 | 88 |
| RP | TCCCTTGCAGAGAGTACATGG | |||
| hPRDX1 | FP | CACTGACAAACATGGGGAAGT | NM_002574.4 | 82 |
| RP | TTTGCTCTTTTGGACATCAGG | |||
| hRPLP0 | FP | TGGTCATCCAGCAGGTGTTCGA | NM_001002 | 119 |
| RP | ACAGACACTGGCAACATTGCGG |
Data Availability Statement
The analysis of NFE2L2 (NRF2), ABCC1 (MRP1) and G6PD mRNA expression levels (Figs. 1A, 5A and 6A and S1) were obtained from the Oncomine (https://www.oncomine.org/). All data generated or analyzed during this study were included in this published article and its supplementary information file. All data in this study is available from the corresponding author upon reasonable request.