Abstract
T cell engineering has changed the landscape of cancer immunotherapy. Chimeric antigen receptor T cells have demonstrated a remarkable efficacy in the treatment of B cell malignancies in hematology. However, their clinical impact on solid tumors has been modest so far. T cells expressing an engineered T cell receptor (TCR-T cells) represent a promising therapeutic alternative. The target repertoire is not limited to membrane proteins, and intrinsic features of TCRs such as high antigen sensitivity and near-to-physiological signaling may improve tumor cell detection and killing while improving T cell persistence. In this review, we present the clinical results obtained with TCR-T cells targeting different tumor antigen families. We detail the different methods that have been developed to identify and optimize a TCR candidate. We also discuss the challenges of TCR-T cell therapies, including toxicity assessment and resistance mechanisms. Last, we share some perspectives and highlight future directions in the field.
Engineering T cells with T cell receptors is a promising strategy to treat solid tumors.
INTRODUCTION
Immunotherapy has revolutionized the therapeutic management of cancers in the past decade. Seven immune checkpoint inhibitors (ICIs) were Food and Drug Administration (FDA)–approved in more than 85 oncology indications in only 7 years (1). However, a large fraction of patients do not benefit from ICI due, in part, to the scarcity of tumor-specific effector T cells. This limitation could be overcome with adoptive cell transfer (ACT), which consists in the infusion of antigen-specific T cells in an amount much greater than what can be observed with endogenous response (2). Different ACT techniques are being developed, including tumor-infiltrating lymphocyte (TIL) therapy, T cell receptor–engineered T (TCR-T) cell therapy, and chimeric antigen receptor T (CAR-T) cell therapy. The ACT initially developed was based on the isolation of tumor-specific TILs for ex vivo expansion and reinfusion into the patient (3). While effective in certain cancer types such as melanoma, this approach was only feasible for resectable tumors from which enough T cells could be isolated and amplified (4). TCR-T and CAR-T cell therapies consist in genetically engineered T cells, modified to express a receptor directed against a tumor antigen. CAR-T cell therapies were a considerable breakthrough in hematological cancers, with six therapies now FDA-approved, targeting CD19 or B cell maturation antigen (5, 6). However, the clinical efficacy of CAR-T cells in solid tumors has been much less rewarding, with multiple obstacles including the scarcity of available antigens, tumor heterogeneity, or tumor immunosuppression (6). Advanced solid tumors are also characterized by a desmoplastic stroma and an aberrant vascularization, resulting in hypoxia and altered nutrient availability (7, 8). TCR-T cell therapy represents an alternative that offers several advantages. First, the repertoire of targetable antigens for TCR-T cell therapy is larger than for CAR-T cells. Indeed, because of the nature of the TCR, TCR-T cells can recognize epitopes derived from both membrane and intracellular proteins and presented by the major histocompatibility complex (MHC), while CAR-T cells are limited to targeting cell surface antigens. However, the antigen recognition for TCR-T cells is restricted to the human leucocyte antigen (HLA) allele presenting the epitope, thus restricting the number of patients who can benefit from a given TCR-T cell therapy. Second, the epitope density required to induce activation is lower for TCR-T cells than for classical CAR-T cells (1 to 50 versus 103 epitopes per cell, respectively) (9). This increased sensitivity may improve tumor cell detection and killing. Last, the high avidity of TCR-T cells may also improve their efficacy, and the lower affinity of TCRs for their target compared to CARs may allow each TCR-T cell to “scan” and eliminate several antigen-presenting tumor cells.
To date, compelling clinical data were published for TCR-T cell therapies in solid cancers. In the following review, we perform a synthesis of the different types of tumor antigens now being targeted with TCR-T cell therapy in the clinic and new promising antigens developed at the preclinical stage. We review the different strategies to identify tumor-specific TCRs and optimize their expression and the efficacy of TCR-T cell therapy. We also discuss the challenges that need to be addressed to improve the safety and efficacy of this approach. Last, we give some perspectives for future research, presenting promising strategies to improve persistence of engineered T cells in vivo or to develop allogeneic approaches.
THE REPERTOIRE OF TARGETABLE ANTIGENS IN CLINICAL TRIALS
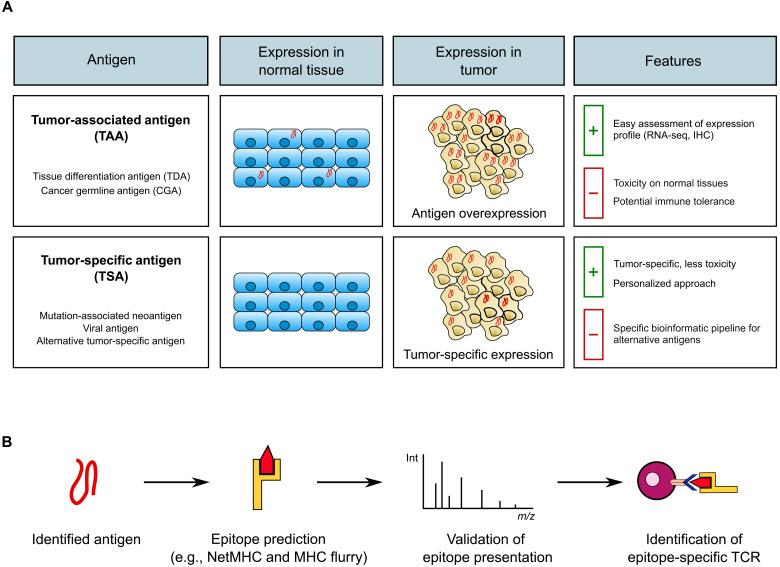
Antigen selection is a key point in the development of safe and efficient TCR-T cell therapies. The ideal antigen would be expressed selectively and homogeneously in tumor cells and generate epitopes presented on MHC class I molecules on their surface. Two major classes of tumor antigens are now considered in clinical trials: tumor-associated antigens (TAAs) and tumor-specific antigens (TSAs) (Fig. 1) (4). Clinical trials for which results have been published are presented in Table 1.
Fig. 1. Choice of target antigen in TCR-T cell therapy.
(A) Tumor-associated antigens (TAAs) are overexpressed in tumor versus normal tissues, whereas tumor-specific antigens (TSAs) are expressed exclusively in the tumor. Pros and cons of each antigen type are displayed in the last column. IHC, immunohistochemistry. (B) After identifying the antigen, epitopes need to be predicted on the basis of data-driven bioinformatic tools (examples: NetMHC and MHC Flurry) and their presence in tumor cells confirmed by peptidomics/immunopeptidomics. These steps are followed by the identification of epitope-specific TCRs. Int, intensity; m/z, mass/charge ratio.
Table 1. List of published TCR-T cell clinical trials.
Symbols: (), response duration in months after treatment; +, ongoing. Abbreviations: TDAs, tissue differentiation antigens; CGAs, cancer germline antigens; CR, complete response; PR, partial response by RECIST criteria; CEA, carcinoembryonic antigen; gp100, glycoprotein 100; HPV, human papillomavirus; MAGE-A, melanoma-associated antigen; MART-1, melanoma antigen recognized by T cells 1; NY-ESO-1, New York esophageal squamous cell carcinoma-1; MPNST, malign peripheral nerve sheath tumors; NSCLC, non–small cell lung cancer; CRS, cytokine release syndrome; n.s., not specified; KO-KI, knockout–knock-in; N.A., not available.
| Antigen type | Target antigen | Epitope | HLA | Cancer type | Vector | Number of cells | Number of patients | Clinical trial | Phase | Objective response rate (ORR) (%) | Clinical response (months) | Toxicities related to TCR-T cells (%) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TDAs | MART-1 | AAGIGILTV | HLA-A*02:01 | Melanoma | Retrovirus | 0.5 to 34 × 109 | 17 | n.s. | n.s. | 2 (12%) | 2 PR (20, 21) | None | Morgan et al. (10) |
| MART-1 | AAGIGILTV | HLA-A*02:01 | Melanoma | Retrovirus | 0.15 to 10 × 1010 | 20 | NCT00509288 | II | 6 (30%) | 6 PR (3, 4, 9, 17+, 17+) | Skin rash: 14 (70%) | Johnson et al. (11) | |
| Uveitis: 11 (55%) | |||||||||||||
| Hearing loss: 10 (50%) | |||||||||||||
| MART-1 | EAAGIGILTV | HLA-A*02:01 | Melanoma | Retrovirus | 0.6 to 4.8 × 109 | 13 | NCT00910650 | II | 0 | Skin rash: 3 (23%) | Chodon et al. (12) | ||
| CRS: 2 (15%) | |||||||||||||
| MART-1 | EAAGIGILTV | HLA-A*02:01 | Melanoma | Retrovirus | 0.05 to 4.56 × 109 | 12 | NCT02654821 | I/IIa | 2 (16.7%) | 2 PR (4.1, 7.1) | CRS/sepsis: 1 (8%) | Rohaan et al. (13) | |
| Dermatitis: 10 (83%) | |||||||||||||
| Uveitis: 2 (17%) | |||||||||||||
| Hearing loss: 4 (33%) | |||||||||||||
| gp100 | KTWGQYWQV | HLA-A*02:01 | Melanoma | Retrovirus | 0.18 to 11 × 1010 | 16 | NCT00509496 | II | 3 (16%) | 1 CR (14+) | Skin rash: 15 (94%) | Johnson et al. (11) | |
| Uveitis: 4 (25%) | |||||||||||||
| 2 PR (4, 3) | Hearing loss: 5 (31%) | ||||||||||||
| CEA | IMIGVLVGV | HLA-A*02:01 | Colorectal cancer | Retrovirus | 2 to 4 × 108 | 3 | NCT00923806 | I | 1 (33%) | 1 PR (6) | Severe transient colitis: 3 (100%) | Parkhurst et al. (14) | |
| CGAs | MAGE-A3 | KVAELVHFL | HLA-A*02:01 | Melanoma | Retrovirus | 2.8 to 7.9 × 1010 | 9 (7 + 1 + 1) | NCT01273181 | I/II | 5 (56%) | 1 CR (15+) | Severe neurologic toxicity: 3 (33%) including 2 deaths | Morgan et al. (16) |
| Synovial sarcoma | 4 PR (4, 4, 5, 12+) | ||||||||||||
| Esophageal cancer | |||||||||||||
| MAGE-A3 | EVDPIGHLY | HLA-A*01 | Melanoma | Lentivirus | 2.4 to 5.3 × 109 | 2 (1 + 1) | NCT01350401 | I | 0 | Severe cardiac toxicity and death: 2 (100%) | Linette et al. (17) | ||
| Myeloma | NCT01352286 | ||||||||||||
| MAGE-A3 | HLA-DPB1*0401 | Metastatic solid tumors | Retrovirus | 0.01 to 123 × 109 | 17 | NCT02111850 | I | 4 (23.5%) | 1 CR (29+) 3 PR (4, 4, 18+) | Liver toxicity: 2 (12%) | Lu et al. (18) | ||
| MAGE-A4 | NYKRCFPVI | HLA-A*24:02 | Esophageal cancer | Retrovirus | 0.2 to 5 × 109 | 10 | UMIN000002395 | I | 0 | none | Kageyama et al. (19) | ||
| MAGE-A4 | GVYDGREHTV | HLA-A*02 | Advanced solid tumors | Lentivirus | 0.12 to 10 × 109 | 38 | NCT03132922 | I | 9 (23.7%) | 9 PR | CRS: 9 (50%) | Hong et al. (20) | |
| MAGE-A10 | GLYDGMEHL | HLA-A*02:01 or HLA-A*02:06 | NSCLC | Lentivirus | 0.1 to 6.77.109 | 11 | NCT02592577 | I | 1 (9%) | 1 PR (1) | CRS: 3 (27%) | Blumenschein et al. (21) | |
| Neurotoxicity: 1 (9%) | |||||||||||||
| NY-ESO-1 | SLLMWITQC | HLA-A*02:01 | Melanoma | Retrovirus | 0.16 to 13 × 1010 | 17 (11 + 6) | NCT00670748 | I | 5 (45%) 4 (67%) | 2 CR (20+, 22+) | None | Robbins et al. (22) | |
| Synovial sarcoma | 3 PR (3, 8, 9+) | ||||||||||||
| 4 PR (5, 8, 10, 18) | |||||||||||||
| NY-ESO-1 | SLLMWITQC | HLA-A*02:01 | Melanoma | Retrovirus | 0.9 to 13 × 1010 | 38 (20 + 16) | NCT00670748 | II | 11 (55%) 11 (61%) | 4 CR (24, 40+, 54+, 58+) | None | Robbins et al. (23) | |
| 7 PR (3, 3, 5, 6+, 8, 10, 28) | |||||||||||||
| Synovial sarcoma | 1 CR (6) | ||||||||||||
| 10 PR (3, 3, 4, 5, 7, 8, 10, 11, 18, 47+) | |||||||||||||
| NY-ESO-1 | SLLMWITQC | HLA-A*02:01 | Melanoma | Retrovirus | 1 × 109 | 10 | NCT02070406 | I | 2 (20%) | 2 PR | CRS: 1 (10%) | Nowicki et al. (24) | |
| Synovial sarcoma | |||||||||||||
| Liposarcoma | NCT01697527 | ||||||||||||
| Osteosarcoma | |||||||||||||
| MPNST | |||||||||||||
| NY-ESO-1 | SLLMWITQC | HLA-A*02:01 HLA-A*02:06 | Synovial sarcoma | Lentivirus | 0.4 to 14.4 × 109 | 12 | NCT01343043 | I/II | 6 (50%) | 1 CR (8) | CRS: 5 (42%) | D’Angelo et al. (25) | |
| 5 PR (4, 7, 8, 18) | |||||||||||||
| NY-ESO-1SLLMWITQC | HLA-A*02:01 HLA-A*02:06 | Synovial sarcoma | Lentivirus | 2.67 × 109 | 30 | NCT01343043 | I/II | 9 (30%) | 9 PR (2-13) | n.s. | Ramachandran et al. (26) | ||
| NY-ESO-1(CRISPR -Cas9)SLLMWITQC | HLA-A*02:01 | Liposarcoma Myeloma | Lentivirus | 0.6 to 7.1 × 108 | 3 | NCT03399448 | I | 0 | None | Stadtmauer et al. (27) | |||
| Viral antigens | HPV16-E6 | TIHDIILECV | HLA-A*02:01 | HPV16-positive epithelial cancer | Retrovirus | 0.1 to 13.4 × 1010 | 12 | NCT02280811 | I/II | 2 (17%) | 2 PR (3, 6) | None | Doran et al. (38) |
| HPV16-E7 | YMLDLQPET | HLA-A*02:01 | HPV16-positive epithelial cancer | Retrovirus | 0.1 to 12.1010 | 12 | NCT02858310 | I | 6 (50%) | 6 PR (3, 4, 4, 8, 8, 9) | None | Nagarsheth et al. (39) | |
| HBV | N.A. | HLA-A*02 or HLA-Cw0801 | HBV-HCC | Electroporation | 1 × 104/kg to 5 × 106/kg | 8 | NCT03899415 | I | 1 | 1 PR (27.7) | Liver toxicity: 1 (12%) | Meng et al. (40) | |
| MCPyV | KLLEIAPNC | HLA-A*02:01 | Merkel cell carcinoma | Lentivirus | 1 to 9 × 108 | 5 | NCT03747484 | I | 1 (25%) | n.s. | None | Veatch et al. (41) | |
| Neo-antigens | TP53 | HMTEVVRHC | HLA-A*02:01 | Metastatic breast cancer | Retrovirus | 5.3 × 1010 | 1 | NCT03412877 | I | 1 (100%) | 1 PR (6) | CRS | Kim et al. (34) |
| KRAS G12D | GADGVGKSA | HLA-C*08:02 | Metastatic pancreatic cancer | Retrovirus | 1.6.1010 | 1 | IND 27501 | I | 1 (100%) | 1 PR (6+) | None | Leidner et al. (35) | |
| GADGVGKSAL | |||||||||||||
| Mutation-associated neoantigens (CRISPR-Cas9 KO-KI) | Multiple HLA class I | Metastatic solid tumors | Electroporation | 0.13 to 4.109 | 16 | NCT03970382 | I | 0 | CRS: 1 (6%) | Foy et al. (37) | |||
| Neurotoxicity: 1 (6%) | |||||||||||||
Tumor-associated antigens
TAAs are antigens overexpressed in cancers but with a limited expression in normal tissues. Their expression can be restricted to tissues of tumor origin [tissue differentiation antigens (TDAs)] or to germline tissues [cancer germline antigens (CGAs)] (4). TAAs are attractive therapeutic targets because they are often shared between patients. However, because of their expression in normal tissues, although at a low level, they may be associated with some on-target off-tumor toxicity. In addition, high-affinity–specific T cells may be eliminated during thymic negative selection, making the identification of potent TCRs more difficult.
Tissue differentiation antigens
Clinical trials targeting TDAs, such as melanoma antigen recognized by T cells 1 (MART-1) (10–13), glycoprotein 100 (gp100) (11), or carcinoembryonic antigen (CEA) (14), showed some clinical responses, but several toxicities were described because of their low expression in normal tissues. In the first clinical trial evaluating MART-1–specific TCR-T cells in patients with melanoma, the objective response rate (ORR) did not exceed 12% (2 of 17) (10). To improve the clinical response, an affinity-enhanced TCR-recognizing MART-1 was tested in two other clinical trials. Although clinical response was slightly improved in one of the trials, with ORR of 30% (6 of 20) and 0% (0 of 13), respectively, several serious cutaneous, ocular, and auditive toxicities were described because of low expression of MART-1 in normal melanocytes (11, 12). More recently, a clinical trial using a different TCR for MART-1 had to be prematurely terminated because of severe toxicities as described before and the death of one patient (13). Similar results were observed with TCR-T cells targeting gp100. The ORR was 16% in patients with melanoma (3 of 16), but, as previously observed, many cutaneous, ocular, or auditive adverse events were reported (11). It should be mentioned in this context that tebentafusp, a soluble affinity-enhanced TCR specific for gp100 fused to an anti-CD3 single-chain variable fragment, has been approved for the treatment of HLA-A*02:01–positive adult patients with unresectable metastatic uveal melanoma, on the basis of the results of a phase 3 randomized trial showing a substantial overall survival benefit. Toxicity was mostly mild to moderate, with most treatment-related adverse events classified as either skin-related (because of gp100-positive melanocytes) or cytokine-mediated (because of T cell activation) (15). In clinical trials evaluating TCR-T cells targeting CEA, one of the three patients with colorectal cancer experienced a partial response, but side effects such as severe inflammatory colitis were described in all patients (14). Other targets, such as mesothelin in pancreatic cancer, are now being tested in the clinic (NCT04809766).
Cancer germline antigens
Most of the TCR-T cell clinical trials targeting CGAs, also called “cancer testis antigens,” focus on members of the melanoma-associated antigen (MAGE-A) protein family (16–21) and New York esophageal squamous cell carcinoma-1 (NY-ESO-1) (22–27). The first two clinical trials targeting two different MHC class I–restricted epitopes derived from MAGE-A3 led to severe and lethal toxic effects caused by cross-reactivity (recognition of an unrelated epitope). In the first one, an objective response was observed in 56% of patients (five of nine), but MAGE-A3 TCR-T cells cross-reacted with MAGE-A12, a protein expressed in the brain, causing severe neurotoxicities and two deaths (16). The second trial was prematurely stopped because of cardiac toxicities and the death of the two treated patients. It was retrospectively determined that MAGE-A3–specific TCR-T cells also recognized an epitope derived from TITIN, a protein expressed in cardiomyocytes (17, 28). More recently, better clinical results were obtained with MHC class II–restricted TCR targeting MAGE-A3, with 25.3% ORR (4 of 17) and no major toxicities (18). MAGE-A4 was also targeted using TCR-T cells. No clinical response was observed in a first trial performed in patients with esophageal cancer (19). However, very encouraging results were recently reported with affinity-enhanced TCR-T cells targeting a different MAGE-A4 epitope (afamitestine autoleucel), especially in sarcoma. At the time of presentation of the phase 2 trial SPEARHEAD-1, the evaluable population included 33 patients with synovial sarcoma and 4 patients with myxoid/round cell liposarcoma. The ORR was 39.4%, with a disease control rate of 84.8%. Two complete responses were observed in patients with synovial sarcoma. Toxicity was manageable with cytokine release syndrome (CRS) in 22 patients (59%) (20, 29). Preliminary clinical results of a strategy targeting MAGE-A10 in patients with non–small cell lung cancer were less encouraging because only 1 of the 11 patients experienced a transient partial response (ORR, 9%), and adverse events including CRS and neurotoxicities were described (21).
TCR-T cells targeting NY-ESO-1 showed promising results in clinical trials, especially in melanoma and synovial sarcoma. Among 107 patients treated in five clinical trials, the average response rate was 47% (ORR between 20 and 67%) with 8 complete responses and 40 partial responses without major toxicities (23–26). In a recent clinical trial using NY-ESO-1 TCR-T cells, endogenous TCR and programmed cell death 1 (PD-1) were knocked out using the CRISPR-Cas9 genome editing tool. Although no clinical response was observed among the three treated patients, in vivo persistence of the CRISPR-engineered T cells was increased (36 weeks versus 1 week) compared to other trials studying NY-ESO-1 (27). While showing promising results, the prevalence of NY-ESO-1 expression is still limited in metastatic cancers, and its tumor expression is often heterogeneous (30). Other CGAs are being tested in the clinic, such as KK-LC-1 (NCT03778814 and NCT05035407) or PRAME (NCT03686124 and NCT02743611), with no clinical results yet published.
Tumor-specific antigens
TSA, also called neoantigens, are proteins exclusively expressed by tumor cells because they are linked to the tumorigenesis process (mutations and viral induction). Targeting these neoantigens with immunotherapy presents a very limited risk of toxicity because they are not expressed by normal tissues. Moreover, high-avidity T cells specific for these neoantigens are not eliminated during the negative thymic selection and can be isolated from patient tumor or healthy donor peripheral blood (31).
Mutation-associated neoantigens
These neoantigens result from nonsynonymous mutations linked to cancer-initiating genetic events or to global genetic instability (32). “Public” neoepitopes refer to epitopes derived from frequently mutated driver genes, such as TP53, KRAS, or PIK3CA (31), that will be shared between different patients with one specific HLA allele. TCR-T cells targeting these public neoantigens are now being tested in clinical trials. In a recent study, a screening was performed to identify neoepitopes derived from shared TP53 mutations and corresponding specific TCRs. One patient with breast cancer was treated with TCR-T cells targeting a p53R175H neoepitope and experienced a partial response with limited toxicity. However, the patient progressed after 6 months because of loss of class I MHC expression (33, 34). For KRAS, a single-patient investigational new drug application was performed to evaluate the safety and tolerability of KRASG12D-specific TCR-T cells in a patient with pancreatic cancer. After 6 months, tumor regression was still ongoing with no toxicity described, and functional TCR-engineered T cells were persisting in the circulation (35). TCR-T cells targeting the KRASG12V mutant (Mut) are also being tested in the clinic (NCT03190941). Last, screening methods were used to identify TCRs that recognize a Mut PIK3CA public neoantigen shared among HLA-A*03:01 patients. Engineered Mut PIK3CA–specific TCR-T cells showed an antitumor response against established tumors in vivo in mice bearing PIK3CA-Mut tumor but not wild-type PIK3CA tumors (36). Together, these results demonstrate the clinical interest of targeting neoantigens from mutated cancer drivers with TCR-T cells therapy. However, identifying epitope containing the public mutation with the HLA restriction limits the list of potential target epitopes. This issue may be partially addressed by targeting passenger mutations that occur in genes that do not intervene in the carcinogenic process due to cancer genetic instability. The development of a personalized TCR-T cell therapy is challenging and even more complex than for cancer vaccines. A pioneering clinical study has recently been published, demonstrating the feasibility of using CRISPR gene editing to create personalized TCR-T cells (37). For each of 16 trial participants, neoantigen-specific TCR were isolated, cloned, and validated from each individual’s blood. The two endogenous TCR genes from the patients’ own T cells were deleted, and the sequences encoding the selected neoantigen-specific TCR were simultaneously inserted. Patients received up to three different TCR-T cells in a cell dose escalation. No major safety concerns were observed. The authors showed that the TCR-T cells migrated to the tumors. Five patients experienced stable disease, and the other 11 had disease progression as best response on therapy. Despite moderate clinical activity, this study paves the way for the development of optimized personalized TCR-T cell therapies. Combination with anti–PD-1 antibody is now being tested in the clinic (NCT03970382 and NCT04520711).
Viral antigens
Some cancers can be induced by viral infections, such as human papilloma virus (HPV)–, hepatitis B virus (HBV)–, Merkel cell polyomavirus (MCPyV)–, or Epstein-Barr virus (EBV)–associated cancers. E6 and E7 viral antigens expressed in HPV-associated cancers have been targeted in clinical trials. In the first trial using HPV16-E6–specific TCR-T cells, clinical responses were reported in 17% of patients (2 of 12) without apparent toxicities (38). In the second trial using HPV16-E7 TCR-T cells, partial responses were observed in 50% of patients (6 of 12) up to 9 months after treatment with no notable toxicities. Nevertheless, tumor escape due to decrease of antigen presentation was described in several patients (39). Recently, HBV-specific TCR-T cells targeting HBV-associated hepatocellular carcinoma were tested in the clinic. One of the eight patients (ORR, 12.5%) experienced a partial response lasting 27.7 months, with minor toxicities described (40). Preliminary results of MCPyV-specific TCR-T cells for the treatment of PD-1 inhibitor-refractory metastatic Merkel cell carcinoma showed a 25% ORR, with one of the five patients presenting a mixed response. However, other effective therapies have been administered at the same time, making it difficult to interpret the results (41). TCR-T cells targeting other viral proteins are now tested in the clinic, such as latent membrane proteins (LMP1 and LMP2) in EBV-related nasopharyngeal carcinoma (NCT03925896, NCT04509726, and NCT03648697). Even if the applications are limited to some cancers, viral antigens benefit from the inherent tumor specificity with no toxicity toward normal tissues described in current clinical trials. In addition, they can be shared between patients with the same viral-induced cancer and HLA type (9).
Alternative TSAs
Alternative processes can generate TSAs derived from the noncoding genome; from alternate open reading frames; or from aberrant transcription, translation, or posttranslational modifications (31, 42–44), and are referred to as “alternative tumor-specific antigens” (ATSAs) (42). One example of ATSA is represented by mutational frameshift neoantigens, which derive from peptides generated by frameshift insertions/deletions (INDELs). Roudko et al. (45) identified that patients with microsatellite instability–high tumors shared tumor-specific frameshift mutations resulting from INDELs within microsatellite sequences. Neoepitopes derived from these frameshift mutations are shared between patients and present a strong immunogenicity in vitro. Tumor antigens can also arise from abnormal mRNA splicing, such as intron retention or exon-exon junctions (44). Aberrant translation can also generate ATSA. As an example, Charpentier et al. (46) identified specific melanoma neoantigens, MELOE-1 and MELOE-2, derived from internal ribosome entry site–dependent translation of meloe long noncoding RNA. These neoantigens, for which specific TILs from patients with melanoma have been identified, are highly immunogenic in vitro. Development of identification techniques for alternative open reading frame or translation of “noncoding” sequences enabled the discovery of so-called “cryptic antigens” (43). Using mass spectrometry (MS) analysis of MHC class I immunopeptidome, Laumont et al. (47) estimated that cryptic antigens represented ~10% of MHC class I epitopes in leukemia patients. More recently, Ouspenskaia et al. (48) performed the ribosome profiling of 29 primary healthy and cancer samples and cell lines to create a database for MS identification of cryptic antigens. Results showed that new or unannotated open reading frames (nuORFs) contribute to 1.5 to 2.2% of MHC class I immunopeptidome in 10 cancer samples, with 50% of nuORFs detected in more than 1 sample. Retroelements also constitute a source of ATSA (42). Human endogenous retroviruses (HERVs) result from ancient retroviral infections and represent 8% of the human genome. Usually epigenetically silenced in normal tissues, HERVs can be reactivated in tumors due to DNA demethylation. Some HERVs are exclusively expressed in tumors, such as HERV-E in renal cell carcinoma (RCC). After showing preclinical antitumoral activity, HERV-E TCR-T cells are now being tested in the clinic in patients with RCC (49). Although no clinical data are available for these unconventional antigens, these findings increase the repertoire of potential targetable tumor antigens for TCR-T cell therapy.
TCR IDENTIFICATION AND OPTIMIZATION
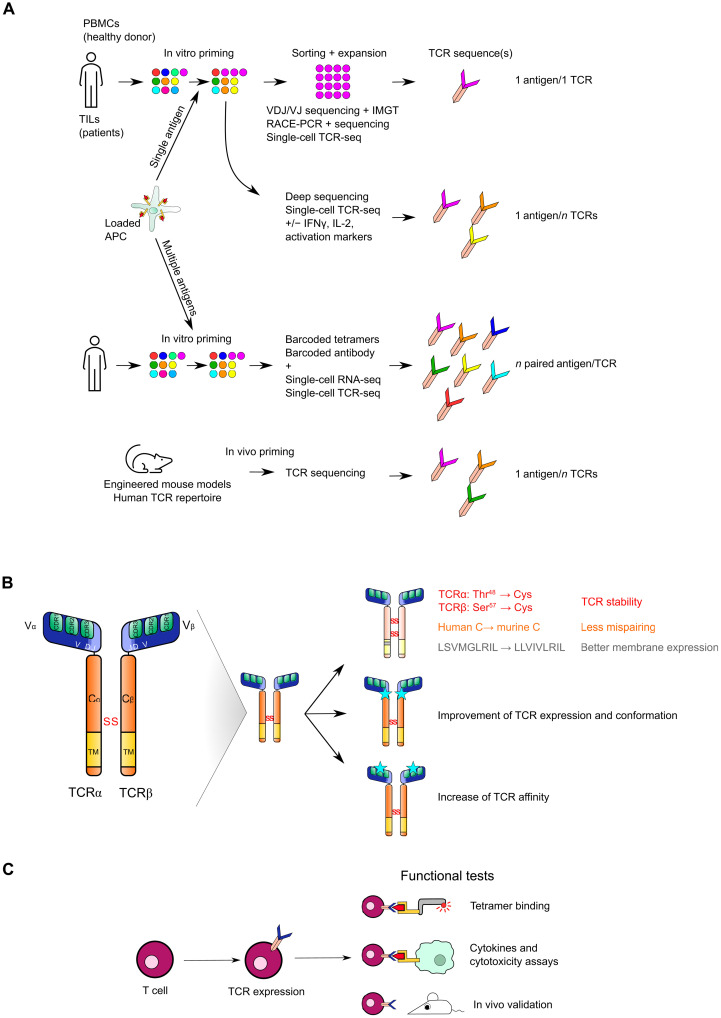
Identification of an epitope-specific TCR is complex given the extent of the TCR repertoire and the characteristics of the interaction between the TCR and the peptide-MHC (pMHC) complex. Indeed, it is estimated that one TCR can recognize up to 106 different epitopes (50), and one epitope can be recognized by several TCRs (51). Identification techniques of pMHC-specific TCRs are based on in vitro induction of a T cell response in a specific HLA context. T cells can be isolated directly from a tumor or from patient’s or healthy donors’ blood. The first step typically consists in performing antigen stimulation of T cells (T cell priming) to enrich the population with antigen-specific T cells after clonal expansion (Fig. 2A). Then, epitope-specific T cells can be sorted and amplified in vitro. TCR sequences can be determined by TCRα and TCRβ sequencing of isolated T cells after rapid amplification of cDNA ends–polymerase chain reaction. (52–54). More recently, some groups combined VDJ sequencing and reference sequences from the ImMunoGeneTics database to identify functional TCRs (55, 56). The development of single-cell RNA sequencing (scRNA-seq) focused on TCR sequences (scTCR-seq) has also facilitated the identification of TCRs from primed T cells. ScTCR-seq was combined with deep sequencing to identify TCR sequences of T cells expressing high levels of activation markers after antigenic stimulation, therefore selecting highly functional TCRs early in the process (57, 58). Combination of barcoded tetramers, barcoded antibodies, and scRNA-seq allowed the identification of functional TCRs with peptide-specific activation signatures (59). The evolution of these techniques opens the path to simultaneously identify TCRs specific for different epitopes through the transduction of tandem minigenes in antigen-presenting cells (APCs) or by pulsing APCs with peptide libraries (58, 60). Moreover, the recent advances in in vivo mouse models with a humanized T cell repertoire created new opportunities to identify human epitope-specific TCRs after in vivo priming/vaccination (61, 62). One advantage of these models is the increased likelihood of identifying high-affinity TCRs in animals where the absence of antigen expression prevented negative thymic selection (63). However, TCR identification from human T cells may limit the risk of selecting self-reactive T cells.
Fig. 2. TCR identification, optimization, and validation.
(A) T cells from healthy donor’s peripheral blood mononuclear cells (PBMCs) or patient’s TILs can be primed by antigen-presenting cells (APCs) loaded with one or several epitopes or antigens to stimulate the activation and expansion of specific T cells. TCRs from these specific T cells can be identified by sequencing T cell bulk or single cells. RACE-PCR, rapid amplification of cDNA ends–polymerase chain reaction; IFN-γ, interferon-γ. (B) TCR optimization can be performed by modifying the constant regions of both TCRα and TCRβ chains to prevent mispairing and increase TCR expression and stability. More recently, mutations in the variable regions but outside CDR regions have been shown to improve TCR expression and stability. TCR affinity maturation aims at increasing TCR affinity. (C) Once the TCR has been identified, in vitro and in vivo functional assays validate the specificity and the efficacy of the engineered T cells.
Once the TCR sequence has been identified, optimization is often performed to improve TCR expression and/or affinity (Fig. 2B). For instance, some TAA-specific TCRs may have a low affinity due to elimination of high-affinity T cells specific for these self-antigens during thymic selection (64). Because T cell cytotoxic activity depends on TCR affinity (65), different methods were developed to improve TCR affinity, such as random mutation of the pMHC recognition site, phage display, or identification of high-affinity TCRs from immunization of humanized mice with the target epitope. Nonetheless, increasing TCR affinity is a double-edged sword (64). The capacity of T cells to eliminate a tumor cell presenting the epitope of interest depends on the cytotoxic response at the immune synapse and on TCR-pMHC interaction lability that allows T cells to eliminate several targets successively. A TCR with too strong affinity may lead to the early exhaustion of T cells following target antigen recognition, limiting successive tumor cell recognition. In addition, modifying TCR sequence may also bypass the negative selection that occurs in the thymus and thus create unanticipated cross-reactivity with self-antigens, as detailed below (11, 12, 14, 16, 17).
High TCR expression and proper assembly is also key in the generation of TCR-T cells. Recent work highlighted the importance of the global TCR conformation and especially the interactions between the variable and constant regions in proper TCR expression. Thomas et al. (66) demonstrated that the high variability of TCR expression and assembly, despite equivalent initial characterization, relies on specific residues at the structural interface between the variable and the constant regions. Replacing suboptimal residues at specific positions by optimal amino acids resulted in homogenization of TCR expression levels. The impact on TCR specificity was not addressed. Other TCR sequence engineering can improve expression and stability of the exogenous/transgenic TCR when expressed in T cells. Highlighted by Rosenberg’s team, these modifications increase the functionality of the engineered T cells (Fig. 2B). Substitution of human constant regions of both TCRα and TCRβ chains by the corresponding murine constant regions is essential to avoid mispairing and does not lead to toxicities in the clinic (67). Indeed, without these substitutions, exogenous TCRα and TCRβ chains can respectively pair with endogenous TCRβ and TCRα chains, leading to the creation of new and potentially cross-reactive TCRs. The addition of a second disulfide bond in the murine constant region increases exogenous TCR stability at the membrane (68), while addition of a hydrophobic sequence within the α chain transmembrane region facilitates membrane expression (69).
Recently, new approaches with CRISPR-Cas9 technology were used to replace the endogenous TCR with the transgenic TCR. Genetic knockout (KO) of both endogenous α and β chains circumvents the mispairing issues and prevents competitive binding to the CD3 complex with the endogenous TCR (70, 71). However, although no serious adverse events have been described in clinical trials using genome editing so far, recent findings highlight the need to carefully assess genome integrity when using the CRISPR-Cas9 technology (72). Endogenous TCR KO can be combined with different gene delivery methods. Retroviral or lentiviral vectors are commonly used. These viral methods lead to the random integration of n copies of the TCR transgene in the host genome. Using homology DNA repair instead of lentiviral transduction for the knock-in (KI) of the transgenic TCR at the T cell receptor α constant (TRAC) locus limits the number of transgenic copies to one, resulting in a more homogenous product with decreased risk of oncogenic genetic events (73) and a physiological expression of transgenic TCR (74). However, the insertion rate is very low with this approach (5 to 10% of T cells). Recently, endogenous TCR KO by CRISPR-Cas9 technology was combined with adeno-associated virus (AAV) transduction to perform a KI of the TCR transgene at the TRAC locus. This enabled to substantially increase the insertion rate (60 to 70% of CD3+ T cells) while maintaining a physiological expression of the exogenous TCR. These KI TCR-T cells showed increased functional avidity and reduced cross-reactivity in vivo (75). These results are similar to those obtained with CAR-T cells, where the insertion of the CAR transgene at the TRAC locus by AAV vectors enhanced CAR-T cell potency. It also induced an effective recycling of the receptor, contributing to delayed exhaustion of the engineered T cells (76, 77). Nonviral gene delivery is developing rapidly with the promises of faster and less expensive development, the possibility to modify several endogenous genes at one time and an improved safety. In the recent publication of Foy et al. (37) reporting the engineering of personalized TCR-T cells, the two TCRα and TCRβ chains were removed using CRISPR-Cas9 technology, and the sequences of the neoantigen-specific TCR were simultaneously inserted using a nonviral approach, with a KI efficacy reaching 23% (range, 11.4 to 46.8%) with the optimized process developed during the course of the study.
The selected T cell engineering strategy needs to be validated in functional assays to evaluate TCR-T cell efficiency and safety in vitro and in vivo (Fig. 2C) (64, 78). Multimer binding assays validate proper TCR assembly and conformation. Assessment of cytotoxicity and cytokine release in coculture experiments with target cells and tumor cells allows assessing TCR-T cell efficacy, specificity, and avidity. Administration of TCR-T cells in engineered mouse models is a prerequisite to validate TCR-T cell viability, functionality, and persistence in vivo (tumor cell infiltration and formation of a memory compartment). A detailed review of all the models and options available to evaluate efficacy and safety has been made in two publications of the T2EVOLVE consortium (79, 80).
CHALLENGES OF TCR-T CELL THERAPIES
Toxicity prediction
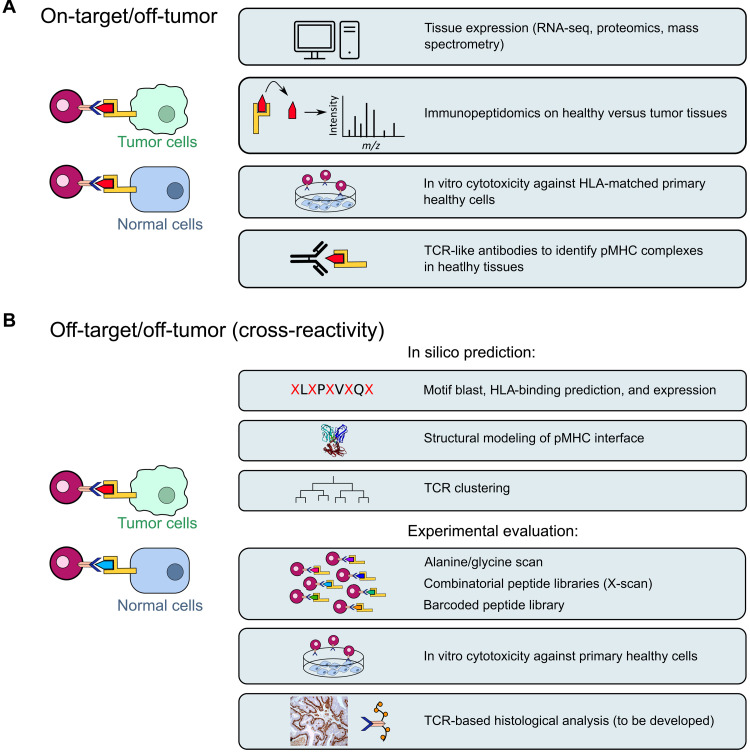
Several toxicities associated with TCR-T cells have been described in the clinic. On-target off-tumor toxicities are linked to target antigen expression in normal tissues and are mostly associated with TAAs. In clinical trials targeting MART-1 and gp100 with TCR-T cell therapy, ocular, cutaneous, and auditive toxicities were due to TAA expression in melanocytes (11, 12). Another study described severe acute colitis in patients treated with TCR-T cells targeting CEA due to its expression on normal intestinal cells (14). Evaluation of TCR-T cell–related on-target off-tumor toxicity includes bioinformatic analysis of transcriptomic and proteomic databases, immunopeptidomics, and in vitro or ex vivo assays to assess the capacity of TCR-T cells to recognize normal cells or tissues (Fig. 3A). The recent development of CAR-T cells targeting pMHC complexes (81) may also allow to develop pMHC-specific antibodies for histological assessment of epitope presentation in tumor versus normal tissues.
Fig. 3. Methods to assess the potential toxicities of TCR-T cell therapies.
(A) Methods to anticipate on-target off-tumor toxicities (presence of the targeted epitope in normal tissues). (B) Methods to anticipate off-target off-tumor toxicities (presence of a cross-reactive epitope in normal tissues).
Off-target off-tumor toxicities or cross-reactivity are linked to the TCR recognition of a different antigen than the targeted antigen on normal cells. Cross-reactivities have been reported in clinical trials using affinity-enhanced TCR-T cells, as previously mentioned. Indeed, TCR-T cells targeting MAGE-A3 also recognized an epitope derived from MAGE-A12 or TITIN proteins, leading to the death of four patients (16, 17, 28). Therefore, it is essential to include a strategy evaluating TCR cross-reactivity at the preclinical level, especially when TCR sequences have been modified to enhance affinity (Fig. 3B). Initial assessment of cross-reactive epitopes can consist in the consecutive replacement of each peptide residue by an alanine residue (alanine scan) to identify residues essential for TCR-pMHC interactions (82). These essential residues can also be defined by bioinformatics as any combination of five to six amino acids of the 9- or 10-mer epitope (83). Peptide motifs composed of essential amino acids are then blasted against the human proteome to identify other proteins containing similar motifs that can serve as epitopes binding HLA molecules. This method succeeded in identifying retrospectively the cross-reactivity between MAGE-A3 and TITIN epitopes responsible for fatal cardiac toxicities. However, it does not take into account cross-reactivities that result from the replacement of residues at the same position by amino acids with similar physicochemical properties (84). In this context, other teams developed a mutational positioning scan (X-scan) with a peptide library where each epitope residue is consecutively replaced by every other amino acid (85, 86). For each positive hit, peptide profiles are then searched in the human proteome to identify potential cross-reactive peptides. Unbiased methods have also been developed to assess TCR cross-reactivity. Positional scanning synthetic combinatorial libraries (PS-SCLs) consist of trillions of peptides where each natural amino acid is consecutively fixed at each position and the rest of the peptide is composed of a random combination of all other existing amino acids (87–89). Results from PS-SCL screening are then compared to protein databases to predict potential cross-reactive epitopes.
Recent studies performed high-throughput screening using peptide libraries and barcoded tetramers to test the recognition of thousands of peptides from the human proteome by selected TCRs (51, 90). Although powerful, these screenings can only assess cross-reactivity for a subset of epitopes from the whole proteome. Other strategies based on three-dimensional (3D) modeling of pMHC-TCR complexes and structure-guided analysis were developed, alone or in combination with previous approaches to predict potential cross-reactivity considering the whole proteome as accurately as possible (91). Structure-guided design has been used to optimize TCR affinity with limited impact on specificity using x-ray crystal structure datasets to train prediction algorithms (92–94). Conversely, Hellman et al. (95) used structure-guided design to improve TCR specificity (limiting cross-reactivity) without altering TCR affinity. Other groups collected a dataset of known cross-reactivities and crystallographic structures to refine a 3D structural model, allowing prediction of cross-reactivity based on the similarity of electrostatic surfaces at the TCR-pMHC interface (96, 97). Another dataset of TCR-pMHC interfaces was harnessed to develop and train an algorithm that demonstrated that TCR cross-reactivity with peptides without similar physicochemical properties was linked to structural flexibility of the pMHC complex (98, 99). More recently, a deep learning–based prediction algorithm succeeded in predicting cross-reactivity with high sensitivity (100). Combining 3D modeling tools and peptide libraries associated with barcoded dextramers, Bentzen et al. (51) identified TCR fingerprints and epitope motifs underlying TCR-pMHC interaction. They observed that different TCRs targeting the same epitope mostly have different fingerprints, suggesting that each TCR-pMHC interaction should be considered unique. This concept was challenged by studies seeking to identify similarities within TCR targeting the same epitope (TCR clustering), where complementarity-determining region (CDR) similarities were used to regroup TCRs (101–104). As these studies aim at predicting antigens for a given TCR, they may provide useful information to assess cross-reactivity. Nonetheless, no clinical validation of these predictive approaches has been published. Implementation of the existing TCR-pMHC databases such as ATLAS (105) will be necessary to improve the prediction accuracy of these models. Additional and complementary approaches such as assessment of TCR-T cell cytotoxicity on primary cell lines (86) or assessment of the presence of pMHC complexes in situ in different tissues (immunopeptidomics and TCR-based histological analysis) may also help anticipate toxicities. A rationalized conception of TCR optimization must be implemented considering TCR affinity and avidity as well as efficiency and safety of TCR-T cells (64, 106).
Identification of resistance mechanisms
Primary and secondary resistance mechanisms to TCR-based immunotherapies have been described (9). Primary resistance mechanisms may be mainly represented by a low or a heterogenous expression of the target antigen in tumor cells or by intrinsic resistance of the tumor cells to T cell–mediated cytotoxicity (107). Another possible issue is the administration of most T cells with a late memory phenotype after the in vitro expansion step, leading to more exhaustion and less persistence than the infusion of T cell with a stem cell–like or an early memory phenotype (108). Regarding TCR-T cells, secondary or acquired resistance mechanisms represent the main concern. Up-regulation of immune checkpoint ligands at the surface of tumor cells can impair the expansion and functionality of the transferred T cells by activating immune checkpoint receptors, leading to exhaustion. The main escape mechanism to TCR-T cell therapies is the loss or decrease of MHC class I molecules on tumor cells, preventing the recognition of the target epitope by TCR-T cells. Loss of MHC class I expression can result from different mechanisms including deletion or mutations of HLA genes themselves, mutations of β-2-microglobuline or genes involved in antigen presentation, loss of heterozygosity, or epigenetic silencing of HLA genes in tumor cells. HLA-negative tumor cells would be positively selected during TCR-T treatment. Loss of HLA heterozygosity (HLA LOH) was recently described in two clinical trials using TILs specific for KRAS (109) or P53 (110) mutational neoepitopes. Another clinical trial targeting P53 mutations in a patient with breast cancer showed relapse 6 months after TCR-T cell therapy, with tumor cells expressing intact P53 and presenting LOH of chromosome 6 containing the HLA-A*02:01 locus (34). Epigenetic silencing of HLA genes was also described in a clinical trial where, after relapse, no mutation or LOH was detected by tumor sequencing, but treatment with the hypomethylating agent azacytidine restored the expression of HLA molecules (111). A recent clinical study using TCR-T cells targeting HPV-16 E7 antigen in HPV-16+ epithelial cancers showed that resistance to T cell therapies involved several actors of the antigen presentation process and of the interferon pathway, with a patient demonstrating loss of TAP1, TAP2, and IFNGR. Nonresponding or relapsed patients also showed impaired HLA-A*02:01 expression (39).
PERSPECTIVES: NEXT STEPS IN T CELL ENGINEERING
As TCR-T cell therapies develop as a promising tool to target a wide panel of tumor antigens in solid tumors, many hurdles remain and need to be overcome, such as T cell–mediated toxicities, resistance mechanisms, and accessibility. To limit the risk of T cell activation against normal cells, it is theoretically possible to apply the strategies of logic gates proposed for CAR-T cells, where T cell activation or inhibition is conditioned by the integration of two signals instead of one. Logic gates “A AND B” are based on combinatorial antigen recognition and trigger the activation of the T cells only if both antigens A and B are concomitantly expressed by tumors cells. In “A NOT B” logic gates, normal cells that express an antigen B absent on tumor cells, in addition to the targeted antigen A, are protected (112). This approach is more challenging for TCRs compared to CARs, especially for the A AND B gates, but inhibitory signaling platforms have recently been developed (A NOT B gates) (113). Suicide gene systems aiming at creating a kill switch in engineered T cells as a safeguard mechanism are also an efficient strategy to stop unanticipated adverse events (114).
Besides toxicities, TCR-T cell therapies face many resistance mechanisms impairing their efficiency, as described above. Combination treatment of TCR-engineered T cells with ICIs can be achieved to avoid T cell exhaustion. In a clinical trial targeting P53 neoantigens (34), some patients were given pembrolizumab after the observation of a high fraction of PD-1+ antigen–specific T cells after infusion (34). Another clinical trial is testing a combination between MCPyV TCR-T cells and anti–PD-1/programmed cell death-ligand 1 treatments (41). Chapuis and colleagues (115) combined ACT of autologous antigen-specific T cells with cytotoxic T lymphocyte–associated antigen 4 blockade and demonstrated a durable clinical response, although the patient was refractory to both treatments individually. Last, gene modifications developed for CAR-T cell therapies to overcome tumor-related immunosuppression could also be used for TCR-T cells, such as PD-1 disruption, PD-1–CD28 chimeric constructs, or dominant negative transforming growth factor–β receptor type 2 (116). In addition to synthetic biology, combination with approaches targeting the tumor microenvironment (7, 8) or modified cytokines resistant to hypoxic and acidic conditions prevalent in tumors may improve the efficiency of adoptive T cell therapies (117).
One of the main hurdles faced in T cell therapy clinical trials is the short persistence of adoptively transferred T cells related to the rapid exhaustion of engineered T cells (115). Generating engineered T cells with a stem cell memory phenotype (Tscm) at the time of infusion improves T cell persistence in vivo and long-term antitumor efficacy (118). It is possible to differentiate and expand Tscm in vitro starting from naive precursors (119). The supplementation of T cell culture medium with interleukin-7 (IL-7) and IL-15 enables the expansion of Tscm defined as CD45RA+ CD45RO+ CCR7+ CD62L+ CD95+ IL7RA+ (108). IL-21 can also support the Tscm phenotype of T cells during expansion (120). Although IL-2 is less efficient at maintaining a Tscm phenotype during the expansion phase, patient undergoing adoptive T cell therapy can be injected with IL-2 to support T cell proliferation and survival in vivo (115).
TCR-T cell therapies developed so far rely on engineering of autologous T cells. Similar to CAR-T cells, there would be many advantages in developing allogeneic approaches, such as immediate availability, possible standardization of the product, time for multiple cell modifications, easier redosing, and reduced cost (116). By deleting both endogenous TCRα and TCRβ chains, insertion of the transgenic TCR at the TRAC locus would avoid the risk of graft-versus-host disease. This technology should be combined with strategies to limit the rejection of the allogeneic T cells by the host immune system, such as partial HLA matching or gene editing (HLA class I deletion combined with natural killer cell inhibition) to generate universal T cells (116, 121).
In conclusion, TCR-T cell therapy has already shown very encouraging results in solid tumors, including cancers responding poorly to current immunotherapies, such as sarcomas. The complexity of this therapeutic strategy is associated with many challenges. However, a better selection of TSAs and optimization of T cell engineering should reduce the risk of toxicity while increasing antitumor efficacy. Combination with therapeutics able to improve T cell homing (oncolytic viruses and radiation therapy) or increase the activity and persistence of the infused T cells (checkpoint inhibitors, cytokines, and cancer vaccines) may further increase the therapeutic potential of TCR-T cells. One can expect that this approach, like next-generation CAR-T cells, will change the natural history of cold tumors and provide a solution to a great therapeutic need.
Acknowledgments
Author contributions: Writing: E.B., C.G., N.C., and S.D.; conceptualization, validation, review, and editing: N.C. and S.D.
Competing interests: E.B. received a PhD CIFRE grant from ErVaccine Technologies, N.C. is an employee of ErVaccine Technologies, and S.D. is the founder and chairman of ErVaccine Technologies. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper.
REFERENCES AND NOTES
- 1.J. A. Beaver, R. Pazdur, The wild west of checkpoint inhibitor development. N. Engl. J. Med. 386, 1297–1301 (2022). [DOI] [PubMed] [Google Scholar]
- 2.M. Mallet, R. E. Boulos, V. Alcazer, P. Bonaventura, Y. Estornes, N. Chuvin, S. Depil, Tumour burden and antigen-specific T cell magnitude represent major parameters for clinical response to cancer vaccine and TCR-engineered T cell therapy. Eur. J. Cancer Oxf. Engl. 171, 96–105 (2022). [DOI] [PubMed] [Google Scholar]
- 3.S. A. Rosenberg, P. Spiess, R. Lafreniere, A new approach to the adoptive immunotherapy of cancer with tumor-infiltrating lymphocytes. Science 233, 1318–1321 (1986). [DOI] [PubMed] [Google Scholar]
- 4.F. Manfredi, B. C. Cianciotti, A. Potenza, E. Tassi, M. Noviello, A. Biondi, F. Ciceri, C. Bonini, E. Ruggiero, TCR redirected T cells for cancer treatment: Achievements, hurdles, and goals. Front. Immunol. 11, 1689 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.S. A. Holstein, M. A. Lunning, CAR T-cell therapy in hematologic malignancies: A voyage in progress. Clin. Pharmacol. Ther. 107, 112–122 (2020). [DOI] [PubMed] [Google Scholar]
- 6.L. Zhao, Y. J. Cao, Engineered T cell therapy for cancer in the clinic. Front. Immunol. 10, 2250 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.A. Antoñana-Vildosola, S. R. Zanetti, A. Palazon, Enabling CAR-T cells for solid tumors: Rage against the suppressive tumor microenvironment. Int. Rev. Cell Mol. Biol. 370, 123–147 (2022). [DOI] [PubMed] [Google Scholar]
- 8.A. Rodriguez-Garcia, A. Palazon, E. Noguera-Ortega, D. J. Powell, S. Guedan, CAR-T cells hit the tumor microenvironment: Strategies to overcome tumor escape. Front. Immunol. 11, 1109 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.S. S. Chandran, C. A. Klebanoff, T cell receptor-based cancer immunotherapy: Emerging efficacy and pathways of resistance. Immunol. Rev. 290, 127–147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.R. A. Morgan, M. E. Dudley, J. R. Wunderlich, M. S. Hughes, J. C. Yang, R. M. Sherry, R. E. Royal, S. L. Topalian, U. S. Kammula, N. P. Restifo, Z. Zheng, A. Nahvi, C. R. de Vries, L. J. Rogers-Freezer, S. A. Mavroukakis, S. A. Rosenberg, Cancer regression in patients after transfer of genetically engineered lymphocytes. Science 314, 126–129 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.L. A. Johnson, R. A. Morgan, M. E. Dudley, L. Cassard, J. C. Yang, M. S. Hughes, U. S. Kammula, R. E. Royal, R. M. Sherry, J. R. Wunderlich, C.-C. R. Lee, N. P. Restifo, S. L. Schwarz, A. P. Cogdill, R. J. Bishop, H. Kim, C. C. Brewer, S. F. Rudy, C. VanWaes, J. L. Davis, A. Mathur, R. T. Ripley, D. A. Nathan, C. M. Laurencot, S. A. Rosenberg, Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood 114, 535–546 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.T. Chodon, B. Comin-Anduix, B. Chmielowski, R. C. Koya, Z. Wu, M. Auerbach, C. Ng, E. Avramis, E. Seja, A. Villanueva, T. A. McCannel, A. Ishiyama, J. Czernin, C. G. Radu, X. Wang, D. W. Gjertson, A. J. Cochran, K. Cornetta, D. J. L. Wong, P. Kaplan-Lefko, O. Hamid, W. Samlowski, P. A. Cohen, G. A. Daniels, B. Mukherji, L. Yang, J. A. Zack, D. B. Kohn, J. R. Heath, J. A. Glaspy, O. N. Witte, D. Baltimore, J. S. Economou, A. Ribas, Adoptive transfer of MART-1 T-cell receptor transgenic lymphocytes and dendritic cell vaccination in patients with metastatic melanoma. Clin. Cancer Res. 20, 2457–2465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M. W. Rohaan, R. Gomez-Eerland, J. H. van den Berg, M. H. Geukes Foppen, M. van Zon, B. Raud, I. Jedema, S. Scheij, R. de Boer, N. A. M. Bakker, D. van den Broek, L. M. Pronk, L. G. Grijpink-Ongering, A. Sari, R. Kessels, M. van den Haak, H. A. Mallo, M. Karger, B. A. van de Wiel, C. L. Zuur, C. W. Duinkerken, F. Lalezari, J. V. van Thienen, S. Wilgenhof, C. U. Blank, J. H. Beijnen, B. Nuijen, T. N. Schumacher, J. B. A. G. Haanen, MART-1 TCR gene-modified peripheral blood T cells for the treatment of metastatic melanoma: A phase I/IIa clinical trial. Immuno-Oncol. Technol. 15, 100089 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.M. R. Parkhurst, J. C. Yang, R. C. Langan, M. E. Dudley, D.-A. N. Nathan, S. A. Feldman, J. L. Davis, R. A. Morgan, M. J. Merino, R. M. Sherry, M. S. Hughes, U. S. Kammula, G. Q. Phan, R. M. Lim, S. A. Wank, N. P. Restifo, P. F. Robbins, C. M. Laurencot, S. A. Rosenberg, T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol. Ther. J. Am. Soc. Gene Ther. 19, 620–626 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.P. Nathan, J. C. Hassel, P. Rutkowski, J.-F. Baurain, M. O. Butler, M. Schlaak, R. J. Sullivan, S. Ochsenreither, R. Dummer, J. M. Kirkwood, A. M. Joshua, J. J. Sacco, A. N. Shoushtari, M. Orloff, J. M. Piulats, M. Milhem, A. K. S. Salama, B. Curti, L. Demidov, L. Gastaud, C. Mauch, M. Yushak, R. D. Carvajal, O. Hamid, S. E. Abdullah, C. Holland, H. Goodall, S. Piperno-Neumann; IMCgp100-202 Investigators , IMCgp100-202 investigators, overall survival benefit with tebentafusp in metastatic uveal melanoma. N. Engl. J. Med. 385, 1196–1206 (2021). [DOI] [PubMed] [Google Scholar]
- 16.R. A. Morgan, N. Chinnasamy, D. Abate-Daga, A. Gros, P. F. Robbins, Z. Zheng, M. E. Dudley, S. A. Feldman, J. C. Yang, R. M. Sherry, G. Q. Phan, M. S. Hughes, U. S. Kammula, A. D. Miller, C. J. Hessman, A. A. Stewart, N. P. Restifo, M. M. Quezado, M. Alimchandani, A. Z. Rosenberg, A. Nath, T. Wang, B. Bielekova, S. C. Wuest, N. Akula, F. J. McMahon, S. Wilde, B. Mosetter, D. J. Schendel, C. M. Laurencot, S. A. Rosenberg, Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J. Immunother. 36, 133–151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.G. P. Linette, E. A. Stadtmauer, M. V. Maus, A. P. Rapoport, B. L. Levine, L. Emery, L. Litzky, A. Bagg, B. M. Carreno, P. J. Cimino, G. K. Binder-Scholl, D. P. Smethurst, A. B. Gerry, N. J. Pumphrey, A. D. Bennett, J. E. Brewer, J. Dukes, J. Harper, H. K. Tayton-Martin, B. K. Jakobsen, N. J. Hassan, M. Kalos, C. H. June, Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood 122, 863–871 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Y.-C. Lu, L. L. Parker, T. Lu, Z. Zheng, M. A. Toomey, D. E. White, X. Yao, Y. F. Li, P. F. Robbins, S. A. Feldman, P. van der Bruggen, C. A. Klebanoff, S. L. Goff, R. M. Sherry, U. S. Kammula, J. C. Yang, S. A. Rosenberg, Treatment of patients with metastatic cancer using a major histocompatibility complex class II-restricted T-cell receptor targeting the cancer germline antigen MAGE-A3. J. Clin. Oncol. 35, 3322–3329 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S. Kageyama, H. Ikeda, Y. Miyahara, N. Imai, M. Ishihara, K. Saito, S. Sugino, S. Ueda, T. Ishikawa, S. Kokura, H. Naota, K. Ohishi, T. Shiraishi, N. Inoue, M. Tanabe, T. Kidokoro, H. Yoshioka, D. Tomura, I. Nukaya, J. Mineno, K. Takesako, N. Katayama, H. Shiku, Adoptive transfer of MAGE-A4 T-cell receptor gene-transduced lymphocytes in patients with recurrent esophageal cancer. Clin. Cancer Res. 21, 2268–2277 (2015). [DOI] [PubMed] [Google Scholar]
- 20.D. S. Hong, B. A. Van Tine, A. J. Olszanski, M. L. Johnson, D. A. Liebner, T. Trivedi, Q. Lin, E. Elefant, R. Dryer-Minnerly, J.-M. Navenot, D. Williams, I. R. Ramachandran, P. M. Fracasso, E. Norry, M. O. Butler, Phase I dose escalation and expansion trial to assess the safety and efficacy of ADP-A2M4 SPEAR T cells in advanced solid tumors. J. Clin. Oncol. 38, 102 (2020). [Google Scholar]
- 21.G. R. Blumenschein, S. Devarakonda, M. Johnson, V. Moreno, J. Gainor, M. J. Edelman, J. V. Heymach, R. Govindan, C. Bachier, B. Doger de Spéville, M. J. Frigault, A. J. Olszanski, V. K. Lam, N. Hyland, J.-M. Navenot, S. Fayngerts, Z. Wolchinsky, R. Broad, D. Batrakou, M. M. Pentony, J. P. Sanderson, A. Gerry, D. Marks, J. Bai, T. Holdich, E. Norry, P. M. Fracasso, Phase I clinical trial evaluating the safety and efficacy of ADP-A2M10 SPEAR T cells in patients with MAGE-A10+ advanced non-small cell lung cancer. J. Immunother. Cancer 10, e003581 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.P. F. Robbins, R. A. Morgan, S. A. Feldman, J. C. Yang, R. M. Sherry, M. E. Dudley, J. R. Wunderlich, A. V. Nahvi, L. J. Helman, C. L. Mackall, U. S. Kammula, M. S. Hughes, N. P. Restifo, M. Raffeld, C.-C. R. Lee, C. L. Levy, Y. F. Li, M. El-Gamil, S. L. Schwarz, C. Laurencot, S. A. Rosenberg, Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J. Clin. Oncol. 29, 917–924 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.P. F. Robbins, S. H. Kassim, T. L. N. Tran, J. S. Crystal, R. A. Morgan, S. A. Feldman, J. C. Yang, M. E. Dudley, J. R. Wunderlich, R. M. Sherry, U. S. Kammula, M. S. Hughes, N. P. Restifo, M. Raffeld, C.-C. R. Lee, Y. F. Li, M. El-Gamil, S. A. Rosenberg, A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: Long-term follow-up and correlates with response. Clin. Cancer Res. 21, 1019–1027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.T. S. Nowicki, B. Berent-Maoz, G. Cheung-Lau, R. R. Huang, X. Wang, J. Tsoi, P. Kaplan-Lefko, P. Cabrera, J. Tran, J. Pang, M. Macabali, I. P. Garcilazo, I. B. Carretero, A. Kalbasi, A. J. Cochran, C. S. Grasso, S. Hu-Lieskovan, B. Chmielowski, B. Comin-Anduix, A. Singh, A. Ribas, A pilot trial of the combination of transgenic NY-ESO-1-reactive adoptive cellular therapy with dendritic cell vaccination with or without Ipilimumab. Clin. Cancer Res. 25, 2096–2108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.S. P. D’Angelo, L. Melchiori, M. S. Merchant, D. Bernstein, J. Glod, R. Kaplan, S. Grupp, W. D. Tap, K. Chagin, G. K. Binder, S. Basu, D. E. Lowther, R. Wang, N. Bath, A. Tipping, G. Betts, I. Ramachandran, J.-M. Navenot, H. Zhang, D. K. Wells, E. Van Winkle, G. Kari, T. Trivedi, T. Holdich, L. Pandite, R. Amado, C. L. Mackall, Antitumor activity associated with prolonged persistence of adoptively transferred NY-ESO-1 c259T cells in synovial sarcoma. Cancer Discov. 8, 944–957 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.I. Ramachandran, D. E. Lowther, R. Dryer-Minnerly, R. Wang, S. Fayngerts, D. Nunez, G. Betts, N. Bath, A. J. Tipping, L. Melchiori, J.-M. Navenot, J. Glod, C. L. Mackall, S. P. D’Angelo, D. M. Araujo, W. A. Chow, G. D. Demetri, M. Druta, B. A. Van Tine, S. A. Grupp, A. R. Abdul Razak, B. Wilky, M. Iyengar, T. Trivedi, E. V. Winkle, K. Chagin, R. Amado, G. K. Binder, S. Basu, Systemic and local immunity following adoptive transfer of NY-ESO-1 SPEAR T cells in synovial sarcoma. J. Immunother. Cancer 7, 276 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.E. A. Stadtmauer, J. A. Fraietta, M. M. Davis, A. D. Cohen, K. L. Weber, E. Lancaster, P. A. Mangan, I. Kulikovskaya, M. Gupta, F. Chen, L. Tian, V. E. Gonzalez, J. Xu, I.-Y. Jung, J. J. Melenhorst, G. Plesa, J. Shea, T. Matlawski, A. Cervini, A. L. Gaymon, S. Desjardins, A. Lamontagne, J. Salas-Mckee, A. Fesnak, D. L. Siegel, B. L. Levine, J. K. Jadlowsky, R. M. Young, A. Chew, W.-T. Hwang, E. O. Hexner, B. M. Carreno, C. L. Nobles, F. D. Bushman, K. R. Parker, Y. Qi, A. T. Satpathy, H. Y. Chang, Y. Zhao, S. F. Lacey, C. H. June, CRISPR-engineered T cells in patients with refractory cancer. Science 367, eaba7365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.B. J. Cameron, A. B. Gerry, J. Dukes, J. V. Harper, V. Kannan, F. C. Bianchi, F. Grand, J. E. Brewer, M. Gupta, G. Plesa, G. Bossi, A. Vuidepot, A. S. Powlesland, A. Legg, K. J. Adams, A. D. Bennett, N. J. Pumphrey, D. D. Williams, G. Binder-Scholl, I. Kulikovskaya, B. L. Levine, J. L. Riley, A. Varela-Rohena, E. A. Stadtmauer, A. P. Rapoport, G. P. Linette, C. H. June, N. J. Hassan, M. Kalos, B. K. Jakobsen, Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci. Transl. Med. 5, 197ra103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.S. P. D’Angelo, B. A. Van Tine, S. Attia, J.-Y. Blay, S. J. Strauss, C. M. Valverde Morales, A. R. Abdul Razak, E. Van Winkle, T. Trivedi, S. Biswas, D. Williams, E. Norry, D. M. Araujo, SPEARHEAD-1: A phase 2 trial of afamitresgene autoleucel (formerly ADP-A2M4) in patients with advanced synovial sarcoma or myxoid/round cell liposarcoma. J. Clin. Oncol. 39, 11504 (2021). [Google Scholar]
- 30.S. P. Kerkar, Z.-F. Wang, J. Lasota, T. Park, K. Patel, E. Groh, S. A. Rosenberg, M. M. Miettinen, MAGE-A is more highly expressed than NY-ESO-1 in a systematic immunohistochemical analysis of 3668 cases. J. Immunother. 39, 181–187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.V. Leko, S. A. Rosenberg, Identifying and targeting human tumor antigens for T cell-based immunotherapy of solid tumors. Cancer Cell 38, 454–472 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.F. Kast, C. Klein, P. Umaña, A. Gros, S. Gasser, Advances in identification and selection of personalized neoantigen/T-cell pairs for autologous adoptive T cell therapies. Onco. Targets. Ther. 10, 1869389 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.P. Malekzadeh, A. Pasetto, P. F. Robbins, M. R. Parkhurst, B. C. Paria, L. Jia, J. J. Gartner, V. Hill, Z. Yu, N. P. Restifo, A. Sachs, E. Tran, W. Lo, R. P. T. Somerville, S. A. Rosenberg, D. C. Deniger, Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J. Clin. Invest. 129, 1109–1114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.S. P. Kim, N. R. Vale, N. Zacharakis, S. Krishna, Z. Yu, B. Gasmi, J. J. Gartner, S. Sindiri, P. Malekzadeh, D. C. Deniger, F. J. Lowery, M. R. Parkhurst, L. T. Ngo, S. Ray, Y. F. Li, V. Hill, M. Florentin, R. V. Masi, B. C. Paria, N. Levin, A. Bera, E. A. Hedges, A. Choi, P. D. Chatani, A. Y. Parikh, S. Levi, S. Seitter, Y.-C. Lu, Z. Zheng, T. D. Prickett, L. Jia, J. M. Hernandez, C. D. Hoang, P. F. Robbins, S. L. Goff, R. M. Sherry, J. C. Yang, S. A. Rosenberg, Adoptive cellular therapy with autologous tumor-infiltrating lymphocytes and T-cell receptor–Engineered T cells targeting common p53 neoantigens in human solid tumors. Cancer Immunol. Res. 10, 932–946 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R. Leidner, N. Sanjuan Silva, H. Huang, D. Sprott, C. Zheng, Y.-P. Shih, A. Leung, R. Payne, K. Sutcliffe, J. Cramer, S. A. Rosenberg, B. A. Fox, W. J. Urba, E. Tran, Neoantigen T-cell receptor gene therapy in pancreatic cancer. N. Engl. J. Med. 386, 2112–2119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.S. S. Chandran, J. Ma, M. G. Klatt, F. Dündar, C. Bandlamudi, P. Razavi, H. Y. Wen, B. Weigelt, P. Zumbo, S. N. Fu, L. B. Banks, F. Yi, E. Vercher, I. Etxeberria, W. D. Bestman, A. Da Cruz Paula, I. S. Aricescu, A. Drilon, D. Betel, D. A. Scheinberg, B. M. Baker, C. A. Klebanoff, Immunogenicity and therapeutic targeting of a public neoantigen derived from mutated PIK3CA. Nat. Med. 28, 946–957 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.S. P. Foy, K. Jacoby, D. A. Bota, T. Hunter, Z. Pan, E. Stawiski, Y. Ma, W. Lu, S. Peng, C. L. Wang, B. Yuen, O. Dalmas, K. Heeringa, B. Sennino, A. Conroy, M. T. Bethune, I. Mende, W. White, M. Kukreja, S. Gunturu, E. Humphrey, A. Hussaini, D. An, A. J. Litterman, B. B. Quach, A. H. C. Ng, Y. Lu, C. Smith, K. M. Campbell, D. Anaya, L. Skrdlant, E. Y.-H. Huang, V. Mendoza, J. Mathur, L. Dengler, B. Purandare, R. Moot, M. C. Yi, R. Funke, A. Sibley, T. Stallings-Schmitt, D. Y. Oh, B. Chmielowski, M. Abedi, Y. Yuan, J. A. Sosman, S. M. Lee, A. J. Schoenfeld, D. Baltimore, J. R. Heath, A. Franzusoff, A. Ribas, A. V. Rao, S. J. Mandl, Non-viral precision T cell receptor replacement for personalized cell therapy. Nature, 10.1038/s41586-022-05531-1 (2022). [DOI] [PMC free article] [PubMed]
- 38.S. L. Doran, S. Stevanović, S. Adhikary, J. J. Gartner, L. Jia, M. L. M. Kwong, W. C. Faquin, S. M. Hewitt, R. M. Sherry, J. C. Yang, S. A. Rosenberg, C. S. Hinrichs, T-cell receptor gene therapy for human papillomavirus-associated epithelial cancers: A first-in-human, phase I/II study. J. Clin. Oncol. 37, 2759–2768 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.N. B. Nagarsheth, S. M. Norberg, A. L. Sinkoe, S. Adhikary, T. J. Meyer, J. B. Lack, A. C. Warner, C. Schweitzer, S. L. Doran, S. Korrapati, S. Stevanović, C. L. Trimble, J. A. Kanakry, M. H. Bagheri, E. Ferraro, S. H. Astrow, A. Bot, W. C. Faquin, D. Stroncek, N. Gkitsas, S. Highfill, C. S. Hinrichs, TCR-engineered T cells targeting E7 for patients with metastatic HPV-associated epithelial cancers. Nat. Med. 27, 419–425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.F. Meng, J. Zhao, A. T. Tan, W. Hu, S.-Y. Wang, J. Jin, J. Wu, Y. Li, L. Shi, J.-L. Fu, S. Yu, Y. Shen, L. Liu, J. Luan, M. Shi, Y. Xie, C.-B. Zhou, R. W. Wong, W. Lu-En, S. Koh, A. Bertoletti, T. Wang, J.-Y. Zhang, F.-S. Wang, Immunotherapy of HBV-related advanced hepatocellular carcinoma with short-term HBV-specific TCR expressed T cells: Results of dose escalation, phase I trial. Hepatol. Int. 15, 1402–1412 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.J. Veatch, K. Paulson, Y. Asano, L. Martin, B. Lee, E. T. Hall, S. Bhatia, P. Nghiem, A. Chapuis, Merkel polyoma virus specific T-cell receptor transgenic T-cell therapy in PD-1 inhibitor refractory Merkel cell carcinoma. J. Clin. Oncol. 40, 9549–9549 (2022). [Google Scholar]
- 42.C. C. Smith, S. R. Selitsky, S. Chai, P. M. Armistead, B. G. Vincent, J. S. Serody, Alternative tumour-specific antigens. Nat. Rev. Cancer 19, 465–478 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.C. M. Laumont, C. Perreault, Exploiting non-canonical translation to identify new targets for T cell-based cancer immunotherapy. Cell. Mol. Life Sci. 75, 607–621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.C. Chong, G. Coukos, M. Bassani-Sternberg, Identification of tumor antigens with immunopeptidomics. Nat. Biotechnol. 40, 175–188 (2021). [DOI] [PubMed] [Google Scholar]
- 45.V. Roudko, C. C. Bozkus, T. Orfanelli, C. B. McClain, C. Carr, T. O’Donnell, L. Chakraborty, R. Samstein, K. Huang, S. V. Blank, B. Greenbaum, N. Bhardwaj, Shared immunogenic poly-epitope frameshift mutations in microsatellite unstable tumors. Cell 183, 1634–1649.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.M. Charpentier, M. Croyal, D. Carbonnelle, A. Fortun, L. Florenceau, C. Rabu, M. Krempf, N. Labarrière, F. Lang, IRES-dependent translation of the long non coding RNA meloe in melanoma cells produces the most immunogenic MELOE antigens. Oncotarget 7, 59704–59713 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.C. M. Laumont, T. Daouda, J.-P. Laverdure, É. Bonneil, O. Caron-Lizotte, M.-P. Hardy, D. P. Granados, C. Durette, S. Lemieux, P. Thibault, C. Perreault, Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames. Nat. Commun. 7, 10238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.T. Ouspenskaia, T. Law, K. R. Clauser, S. Klaeger, S. Sarkizova, F. Aguet, B. Li, E. Christian, B. A. Knisbacher, P. M. Le, C. R. Hartigan, H. Keshishian, A. Apffel, G. Oliveira, W. Zhang, S. Chen, Y. T. Chow, Z. Ji, I. Jungreis, S. A. Shukla, S. Justesen, P. Bachireddy, M. Kellis, G. Getz, N. Hacohen, D. B. Keskin, S. A. Carr, C. J. Wu, A. Regev, Unannotated proteins expand the MHC-I-restricted immunopeptidome in cancer. Nat. Biotechnol. 40, 209–217 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R. Nadal, E. Cherkasova, S. Barisic, D. Granadier, G. Aue, B. Wells, G. Hawks, T. Hughes, R. Shalabi, D. Stroncek, S. Highfill, G. Scurti, L. Chen, R. Reger, M. Nishimura, R. Childs, A phase I study of HERV-E TCR transduced autologous T Cells (HERV-E TCR T Cells) in patients (pts) with metastatic clear cell renal cell carcinoma (mccRCC). Ann. Oncol. 29, viii329 (2018). [Google Scholar]
- 50.L. Wooldridge, J. Ekeruche-Makinde, H. A. van den Berg, A. Skowera, J. J. Miles, M. P. Tan, G. Dolton, M. Clement, S. Llewellyn-Lacey, D. A. Price, M. Peakman, A. K. Sewell, A single autoimmune T cell receptor recognizes more than a million different peptides. J. Biol. Chem. 287, 1168–1177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.A. K. Bentzen, L. Such, K. K. Jensen, A. M. Marquard, L. E. Jessen, N. J. Miller, C. D. Church, R. Lyngaa, D. M. Koelle, J. C. Becker, C. Linnemann, T. N. M. Schumacher, P. Marcatili, P. Nghiem, M. Nielsen, S. R. Hadrup, T cell receptor fingerprinting enables in-depth characterization of the interactions governing recognition of peptide-MHC complexes. Nat. Biotechnol. 36, 1191–1196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ö. Çınar, B. Brzezicha, C. Grunert, P. M. Kloetzel, C. Beier, C. A. Peuker, U. Keller, A. Pezzutto, A. Busse, High-affinity T-cell receptor specific for MyD88 L265P mutation for adoptive T-cell therapy of B-cell malignancies. J. Immunother. Cancer 9, e002410 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.B. Y. Jin, T. E. Campbell, L. M. Draper, S. Stevanović, B. Weissbrich, Z. Yu, N. P. Restifo, S. A. Rosenberg, C. L. Trimble, C. S. Hinrichs, Engineered T cells targeting E7 mediate regression of human papillomavirus cancers in a murine model. JCI Insight. 3, e99488 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. Arnaud, J. Chiffelle, R. Genolet, B. Navarro Rodrigo, M. A. S. Perez, F. Huber, M. Magnin, T. Nguyen-Ngoc, P. Guillaume, P. Baumgaertner, C. Chong, B. J. Stevenson, D. Gfeller, M. Irving, D. E. Speiser, J. Schmidt, V. Zoete, L. E. Kandalaft, M. Bassani-Sternberg, S. Bobisse, G. Coukos, A. Harari, Sensitive identification of neoantigens and cognate TCRs in human solid tumors. Nat. Biotechnol. 40, 656–660 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.P. Mercier-Letondal, C. Marton, M. Deschamps, C. Ferrand, C. Vauchy, C. Chenut, A. Baguet, O. Adotévi, C. Borg, J. Galaine, Y. Godet, Isolation and characterization of an HLA-DRB1*04-restricted HPV16-E7 T cell receptor for cancer immunotherapy. Hum. Gene Ther. 29, 1202–1212 (2018). [DOI] [PubMed] [Google Scholar]
- 56.A. Gros, E. Tran, M. R. Parkhurst, S. Ilyas, A. Pasetto, E. M. Groh, P. F. Robbins, R. Yossef, A. Garcia-Garijo, C. A. Fajardo, T. D. Prickett, L. Jia, J. J. Gartner, S. Ray, L. Ngo, J. R. Wunderllich, J. C. Yang, S. A. Rosenberg, Recognition of human gastrointestinal cancer neoantigens by circulating PD-1+ lymphocytes. J. Clin. Invest. 129, 4992–5004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Y.-C. Lu, Z. Zheng, P. F. Robbins, E. Tran, T. D. Prickett, J. J. Gartner, Y. F. Li, S. Ray, Z. Franco, V. Bliskovsky, P. C. Fitzgerald, S. A. Rosenberg, An efficient single-cell RNA-Seq approach to identify neoantigen-specific T cell receptors. Mol. Ther. 26, 379–389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Y.-C. Lu, Z. Zheng, F. J. Lowery, J. J. Gartner, T. D. Prickett, P. F. Robbins, S. A. Rosenberg, Direct identification of neoantigen-specific TCRs from tumor specimens by high-throughput single-cell sequencing. J. Immunother. Cancer 9, e002595 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.K.-Y. Ma, A. A. Schonnesen, C. He, A. Y. Xia, E. Sun, E. Chen, K. R. Sebastian, Y.-W. Guo, R. Balderas, M. Kulkarni-Date, N. Jiang, High-throughput and high-dimensional single-cell analysis of antigen-specific CD8+ T cells. Nat. Immunol. 22, 1590–1598 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.M. Parkhurst, A. Gros, A. Pasetto, T. Prickett, J. S. Crystal, P. Robbins, S. A. Rosenberg, Isolation of T-cell receptors specifically reactive with mutated tumor-associated antigens from tumor-infiltrating lymphocytes based on CD137 expression. Clin. Cancer Res. 23, 2491–2505 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.L.-P. Li, J. C. Lampert, X. Chen, C. Leitao, J. Popović, W. Müller, T. Blankenstein, Transgenic mice with a diverse human T cell antigen receptor repertoire. Nat. Med. 16, 1029–1034 (2010). [DOI] [PubMed] [Google Scholar]
- 62.M. J. Moore, M. Zhong, J. Hansen, H. Gartner, C. Grant, M. Huang, F. M. Harris, N. Tu, N. A. Bowerman, K. H. Edelmann, T. Barry, O. Herbin, C.-S. Tay, D. J. DiLillo, C. E. Decker, N. Levenkova, J. Shevchuk, A. Dhanik, K. A. Meagher, A. Karr, J. Roos, W. Lee, D. Suh, M. Eckersdorff, T. C. Meagher, M. Koss, L. Esau, M. A. Sleeman, R. Babb, G. Chen, C. A. Kyratsous, W. T. Poueymirou, J. R. McWhirter, V. A. Voronina, C. Guo, C. Gurer, G. D. Yancopoulos, A. J. Murphy, L. E. Macdonald, Humanization of T cell–mediated immunity in mice. Sci. Immunol. 6, eabj4026 (2021). [DOI] [PubMed] [Google Scholar]
- 63.M. Obenaus, C. Leitão, M. Leisegang, X. Chen, I. Gavvovidis, P. van der Bruggen, W. Uckert, D. J. Schendel, T. Blankenstein, Identification of human T-cell receptors with optimal affinity to cancer antigens using antigen-negative humanized mice. Nat. Biotechnol. 33, 402–407 (2015). [DOI] [PubMed] [Google Scholar]
- 64.D. Campillo-Davo, D. Flumens, E. Lion, The quest for the best: How TCR affinity, avidity, and functional avidity affect TCR-engineered T-cell antitumor responses. Cell 9, 1720 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.S. Zhong, K. Malecek, L. A. Johnson, Z. Yu, E. Vega-Saenz de Miera, F. Darvishian, K. McGary, K. Huang, J. Boyer, E. Corse, Y. Shao, S. A. Rosenberg, N. P. Restifo, I. Osman, M. Krogsgaard, T-cell receptor affinity and avidity defines antitumor response and autoimmunity in T-cell immunotherapy. Proc. Natl. Acad. Sci. U.S.A. 110, 6973–6978 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.S. Thomas, F. Mohammed, R. M. Reijmers, A. Woolston, T. Stauss, A. Kennedy, D. Stirling, A. Holler, L. Green, D. Jones, K. K. Matthews, D. A. Price, B. M. Chain, M. H. M. Heemskerk, E. C. Morris, B. E. Willcox, H. J. Stauss, Framework engineering to produce dominant T cell receptors with enhanced antigen-specific function. Nat. Commun. 10, 4451 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.C. J. Cohen, Y. Zhao, Z. Zheng, S. A. Rosenberg, R. A. Morgan, Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 Stability. Cancer Res. 66, 8878–8886 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.C. J. Cohen, Y. F. Li, M. El-Gamil, P. F. Robbins, S. A. Rosenberg, R. A. Morgan, Enhanced antitumor activity of T cells engineered to express T-cell receptors with a second disulfide bond. Cancer Res. 67, 3898–3903 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.A. Haga-Friedman, M. Horovitz-Fried, C. J. Cohen, Incorporation of transmembrane hydrophobic mutations in the TCR enhance its surface expression and T cell functional avidity. J. Immunol. 188, 5538–5546 (2012). [DOI] [PubMed] [Google Scholar]
- 70.M. Legut, G. Dolton, A. A. Mian, O. G. Ottmann, A. K. Sewell, CRISPR-mediated TCR replacement generates superior anticancer transgenic T cells. Blood 131, 311–322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.T. L. Roth, C. Puig-Saus, R. Yu, E. Shifrut, J. Carnevale, P. J. Li, J. Hiatt, J. Saco, P. Krystofinski, H. Li, V. Tobin, D. N. Nguyen, M. R. Lee, A. L. Putnam, A. L. Ferris, J. W. Chen, J.-N. Schickel, L. Pellerin, D. Carmody, G. Alkorta-Aranburu, D. del Gaudio, H. Matsumoto, M. Morell, Y. Mao, M. Cho, R. M. Quadros, C. B. Gurumurthy, B. Smith, M. Haugwitz, S. H. Hughes, J. S. Weissman, K. Schumann, J. H. Esensten, A. P. May, A. Ashworth, G. M. Kupfer, S. A. W. Greeley, R. Bacchetta, E. Meffre, M. G. Roncarolo, N. Romberg, K. C. Herold, A. Ribas, M. D. Leonetti, A. Marson, Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 559, 405–409 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.A. D. Nahmad, E. Reuveni, E. Goldschmidt, T. Tenne, M. Liberman, M. Horovitz-Fried, R. Khosravi, H. Kobo, E. Reinstein, A. Madi, U. Ben-David, A. Barzel, Frequent aneuploidy in primary human T cells after CRISPR–Cas9 cleavage. Nat. Biotechnol. 40, 1807–1813 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.T. R. Müller, S. Jarosch, M. Hammel, J. Leube, S. Grassmann, B. Bernard, M. Effenberger, I. Andrä, M. Z. Chaudhry, T. Käuferle, A. Malo, L. Cicin-Sain, P. Steinberger, T. Feuchtinger, U. Protzer, K. Schumann, M. Neuenhahn, K. Schober, D. H. Busch, Targeted T cell receptor gene editing provides predictable T cell product function for immunotherapy. Cell Rep. Med. 2, 100374 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.K. Schober, T. R. Müller, F. Gökmen, S. Grassmann, M. Effenberger, M. Poltorak, C. Stemberger, K. Schumann, T. L. Roth, A. Marson, D. H. Busch, Orthotopic replacement of T-cell receptor α- and β-chains with preservation of near-physiological T-cell function. Nat. Biomed. Eng. 3, 974–984 (2019). [DOI] [PubMed] [Google Scholar]
- 75.E. Ruggiero, E. Carnevale, A. Prodeus, Z. I. Magnani, B. Camisa, I. Merelli, C. Politano, L. Stasi, A. Potenza, B. C. Cianciotti, F. Manfredi, M. Di Bono, L. Vago, M. Tassara, S. Mastaglio, M. Ponzoni, F. Sanvito, D. Liu, I. Balwani, R. Galli, M. Genua, R. Ostuni, M. Doglio, D. O’Connell, I. Dutta, S. A. Yazinski, M. McKee, M. S. Arredouani, B. Schultes, F. Ciceri, C. Bonini, CRISPR-based gene disruption and integration of high-avidity, WT1-specific T cell receptors improve antitumor T cell function. Sci. Transl. Med. 14, eabg8027 (2022). [DOI] [PubMed] [Google Scholar]
- 76.J. Eyquem, J. Mansilla-Soto, T. Giavridis, S. J. C. van der Stegen, M. Hamieh, K. M. Cunanan, A. Odak, M. Gönen, M. Sadelain, Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.D. T. MacLeod, J. Antony, A. J. Martin, R. J. Moser, A. Hekele, K. J. Wetzel, A. E. Brown, M. A. Triggiano, J. A. Hux, C. D. Pham, V. V. Bartsevich, C. A. Turner, J. Lape, S. Kirkland, C. W. Beard, J. Smith, M. L. Hirsch, M. G. Nicholson, D. Jantz, B. McCreedy, Integration of a CD19 CAR into the TCR alpha chain locus streamlines production of allogeneic gene-edited CAR T cells. Mol. Ther. 25, 949–961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.L. Gaissmaier, M. Elshiaty, P. Christopoulos, Breaking bottlenecks for the TCR therapy of cancer. Cell 9, 2095 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.E. Donnadieu, M. Luu, M. Alb, B. Anliker, S. Arcangeli, C. Bonini, B. De Angelis, R. Choudhary, D. Espie, A. Galy, C. Holland, Z. Ivics, C. Kantari-Mimoun, M. J. Kersten, U. Köhl, C. Kuhn, B. Laugel, F. Locatelli, I. Marchiq, J. Markman, M. A. Moresco, E. Morris, H. Negre, C. Quintarelli, M. Rade, K. Reiche, M. Renner, E. Ruggiero, C. Sanges, H. Stauss, M. Themeli, J. Van den Brulle, M. Hudecek, M. Casucci, Time to evolve: Predicting engineered T cell-associated toxicity with next-generation models. J. Immunother. Cancer 10, e003486 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.S. Guedan, M. Luu, D. Ammar, P. Barbao, C. Bonini, P. Bousso, C. J. Buchholz, M. Casucci, B. De Angelis, E. Donnadieu, D. Espie, B. Greco, R. Groen, J. B. Huppa, C. Kantari-Mimoun, B. Laugel, M. Mantock, J. L. Markman, E. Morris, C. Quintarelli, M. Rade, K. Reiche, A. Rodriguez-Garcia, J. R. Rodriguez-Madoz, E. Ruggiero, M. Themeli, M. Hudecek, I. Marchiq, Time 2EVOLVE: Predicting efficacy of engineered T-cells – How far is the bench from the bedside? J. Immunother. Cancer 10, e003487 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.M. V. Maus, J. Plotkin, G. Jakka, G. Stewart-Jones, I. Rivière, T. Merghoub, J. Wolchok, C. Renner, M. Sadelain, An MHC-restricted antibody-based chimeric antigen receptor requires TCR-like affinity to maintain antigen specificity. Mol. Ther. Oncolytics 3, 1–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.C. Arber, X. Feng, H. Abhyankar, E. Romero, M.-F. Wu, H. E. Heslop, P. Barth, G. Dotti, B. Savoldo, Survivin-specific T cell receptor targets tumor but not T cells. J. Clin. Invest. 125, 157–168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.V. Jaravine, A. Mösch, S. Raffegerst, D. J. Schendel, D. Frishman, Expitope 2.0: A tool to assess immunotherapeutic antigens for their potential cross-reactivity against naturally expressed proteins in human tissues. BMC Cancer 17, 892 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.S. Frankild, R. J. de Boer, O. Lund, M. Nielsen, C. Kesmir, Amino acid similarity accounts for T cell cross-reactivity and for “holes” in the T cell repertoire. PLOS ONE 3, e1831 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.L. Wooldridge, B. Laugel, J. Ekeruche, M. Clement, H. A. van den Berg, D. A. Price, A. K. Sewell, CD8 controls T cell cross-reactivity. J. Immunol. 185, 4625–4632 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.E. C. Border, J. P. Sanderson, T. Weissensteiner, A. B. Gerry, N. J. Pumphrey, Affinity-enhanced T-cell receptors for adoptive T-cell therapy targeting MAGE-A10: Strategy for selection of an optimal candidate. Onco. Targets. Ther. 8, e1532759 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.J. Ninovasquez, A powerful combination: The use of positional scanning libraries and biometrical analysis to identify cross-reactive T cell epitopes. Mol. Immunol. 40, 1063–1074 (2004). [DOI] [PubMed] [Google Scholar]
- 88.V. Rubio-Godoy, V. Dutoit, Y. Zhao, R. Simon, P. Guillaume, R. Houghten, P. Romero, J.-C. Cerottini, C. Pinilla, D. Valmori, Positional scanning-synthetic peptide library-based analysis of self- and pathogen-derived peptide cross-reactivity with tumor-reactive melan-a-specific CTL. J. Immunol. 169, 5696–5707 (2002). [DOI] [PubMed] [Google Scholar]
- 89.C. Pinilla, V. Rubio-Godoy, V. Dutoit, P. Guillaume, R. Simon, Y. Zhao, R. A. Houghten, J.-C. Cerottini, P. Romero, D. Valmori, Combinatorial peptide libraries as an alternative approach to the identification of ligands for tumor-reactive cytolytic T lymphocytes. Cancer Res. 61, 5153–5160 (2001). [PubMed] [Google Scholar]
- 90.S.-Q. Zhang, K.-Y. Ma, A. A. Schonnesen, M. Zhang, C. He, E. Sun, C. M. Williams, W. Jia, N. Jiang, High-throughput determination of the antigen specificities of T cell receptors in single cells. Nat. Biotechnol. 36, 1156–1159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.N. Li, J. Yuan, W. Tian, L. Meng, Y. Liu, T-cell receptor repertoire analysis for the diagnosis and treatment of solid tumor: A methodology and clinical applications. Cancer Commun. Lond. Engl. 40, 473–483 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.V. Zoete, M. Irving, M. Ferber, M. Cuendet, O. Michielin, Structure-based, rational design of T cell receptors. Front. Immunol. 4, 268 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.B. G. Pierce, L. M. Hellman, M. Hossain, N. K. Singh, C. W. Vander Kooi, Z. Weng, B. M. Baker, Computational design of the affinity and specificity of a therapeutic T cell receptor. PLoS Comput. Biol. 10, e1003478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.K. Malecek, A. Grigoryan, S. Zhong, W. J. Gu, L. A. Johnson, S. A. Rosenberg, T. Cardozo, M. Krogsgaard, Specific Increase in potency via structure-based design of a TCR. J. Immunol. 193, 2587–2599 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.L. M. Hellman, K. C. Foley, N. K. Singh, J. A. Alonso, T. P. Riley, J. R. Devlin, C. M. Ayres, G. L. J. Keller, Y. Zhang, C. W. Vander Kooi, M. I. Nishimura, B. M. Baker, Improving T cell receptor on-target specificity via structure-guided design. Mol. Ther. 27, 300–313 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.D. A. Antunes, M. M. Rigo, M. V. Freitas, M. F. A. Mendes, M. Sinigaglia, G. Lizée, L. E. Kavraki, L. K. Selin, M. Cornberg, G. F. Vieira, Interpreting T-cell cross-reactivity through structure: Implications for TCR-based cancer immunotherapy. Front. Immunol. 8, 1210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.D. A. Antunes, J. R. Abella, S. Hall-Swan, D. Devaurs, A. Conev, M. Moll, G. Lizée, L. E. Kavraki, HLA-arena: A customizable environment for the structural modeling and analysis of peptide-HLA complexes for cancer immunotherapy. JCO Clin. Cancer Inform. 4, 623–636 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.T. P. Riley, C. M. Ayres, L. M. Hellman, N. K. Singh, M. Cosiano, J. M. Cimons, M. J. Anderson, K. H. Piepenbrink, B. G. Pierce, Z. Weng, B. M. Baker, A generalized framework for computational design and mutational scanning of T-cell receptor binding interfaces. Protein Eng. Des. Sel. PEDS. 29, 595–606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.T. P. Riley, L. M. Hellman, M. H. Gee, J. L. Mendoza, J. A. Alonso, K. C. Foley, M. I. Nishimura, C. W. Vander Kooi, K. C. Garcia, B. M. Baker, T cell receptor cross-reactivity expanded by dramatic peptide–MHC adaptability. Nat. Chem. Biol. 14, 934–942 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.T. Lu, Z. Zhang, J. Zhu, Y. Wang, P. Jiang, X. Xiao, C. Bernatchez, J. V. Heymach, D. L. Gibbons, J. Wang, L. Xu, A. Reuben, T. Wang, Deep learning-based prediction of the T cell receptor-antigen binding specificity. Nat. Mach. Intell. 3, 864–875 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.N. Thakkar, C. Bailey-Kellogg, Balancing sensitivity and specificity in distinguishing TCR groups by CDR sequence similarity. BMC Bioinformatics 20, 241 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.H. Zhang, L. Liu, J. Zhang, J. Chen, J. Ye, S. Shukla, J. Qiao, X. Zhan, H. Chen, C. J. Wu, Y.-X. Fu, B. Li, Investigation of antigen-specific T-cell receptor clusters in human cancers. Clin. Cancer Res. 26, 1359–1371 (2020). [DOI] [PubMed] [Google Scholar]
- 103.J. Glanville, H. Huang, A. Nau, O. Hatton, L. E. Wagar, F. Rubelt, X. Ji, A. Han, S. M. Krams, C. Pettus, N. Haas, C. S. L. Arlehamn, A. Sette, S. D. Boyd, T. J. Scriba, O. M. Martinez, M. M. Davis, Identifying specificity groups in the T cell receptor repertoire. Nature 547, 94–98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.P. Dash, A. J. Fiore-Gartland, T. Hertz, G. C. Wang, S. Sharma, A. Souquette, J. C. Crawford, E. B. Clemens, T. H. O. Nguyen, K. Kedzierska, N. L. La Gruta, P. Bradley, P. G. Thomas, Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature 547, 89–93 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.T. Borrman, J. Cimons, M. Cosiano, M. Purcaro, B. G. Pierce, B. M. Baker, Z. Weng, ATLAS: A database linking binding affinities with structures for wild-type and mutant TCR-pMHC complexes: Linking TCR-pMHC affinities with structure. Proteins Struct. Funct. Bioinforma. 85, 908–916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.T. T. Spear, B. D. Evavold, B. M. Baker, M. I. Nishimura, Understanding TCR affinity, antigen specificity, and cross-reactivity to improve TCR gene-modified T cells for cancer immunotherapy. Cancer Immunol. Immunother. CII. 68, 1881–1889 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.R. C. Larson, M. C. Kann, S. R. Bailey, N. J. Haradhvala, P. M. Llopis, A. A. Bouffard, I. Scarfó, M. B. Leick, K. Grauwet, T. R. Berger, K. Stewart, P. V. Anekal, M. Jan, J. Joung, A. Schmidts, T. Ouspenskaia, T. Law, A. Regev, G. Getz, M. V. Maus, CAR T cell killing requires the IFNγR pathway in solid but not liquid tumours. Nature 604, 563–570 (2022). [DOI] [PubMed] [Google Scholar]
- 108.D. H. Busch, S. P. Fräßle, D. Sommermeyer, V. R. Buchholz, S. R. Riddell, Role of memory T cell subsets for adoptive immunotherapy. Semin. Immunol. 28, 28–34 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.E. Tran, P. F. Robbins, Y.-C. Lu, T. D. Prickett, J. J. Gartner, L. Jia, A. Pasetto, Z. Zheng, S. Ray, E. M. Groh, I. R. Kriley, S. A. Rosenberg, T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med. 375, 2255–2262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.W. Lo, M. Parkhurst, P. F. Robbins, E. Tran, Y.-C. Lu, L. Jia, J. J. Gartner, A. Pasetto, D. Deniger, P. Malekzadeh, T. E. Shelton, T. Prickett, S. Ray, S. Kivitz, B. C. Paria, I. Kriley, D. S. Schrump, S. A. Rosenberg, Immunologic recognition of a shared p53 mutated neoantigen in a patient with metastatic colorectal cancer. Cancer Immunol. Res. 7, 534–543 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.K. G. Paulson, V. Voillet, M. S. McAfee, D. S. Hunter, F. D. Wagener, M. Perdicchio, W. J. Valente, S. J. Koelle, C. D. Church, N. Vandeven, H. Thomas, A. G. Colunga, J. G. Iyer, C. Yee, R. Kulikauskas, D. M. Koelle, R. H. Pierce, J. H. Bielas, P. D. Greenberg, S. Bhatia, R. Gottardo, P. Nghiem, A. G. Chapuis, Acquired cancer resistance to combination immunotherapy from transcriptional loss of class I HLA. Nat. Commun. 9, 3868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.M. A. Savanur, H. Weinstein-Marom, G. Gross, Implementing logic gates for safer immunotherapy of cancer. Front. Immunol. 12, 780399 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.D. Manry, K. Bolanos, B. DiAndreth, J.-Y. Mock, A. Kamb, Robust in vitro pharmacology of Tmod, a synthetic dual-signal integrator for cancer cell therapy. Front. Immunol. 13, 826747 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Q. Zhao, Y. Jiang, S. Xiang, P. J. Kaboli, J. Shen, Y. Zhao, X. Wu, F. Du, M. Li, C. H. Cho, J. Li, Q. Wen, T. Liu, T. Yi, Z. Xiao, Engineered TCR-T cell immunotherapy in anticancer precision medicine: Pros and cons. Front. Immunol. 12, 658753 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.A. G. Chapuis, I. M. Roberts, J. A. Thompson, K. A. Margolin, S. Bhatia, S. M. Lee, H. L. Sloan, I. P. Lai, E. A. Farrar, F. Wagener, K. C. Shibuya, J. Cao, J. D. Wolchok, P. D. Greenberg, C. Yee, T-cell therapy using interleukin-21–primed cytotoxic T-cell lymphocytes combined with cytotoxic T-cell lymphocyte antigen-4 blockade results in long-term cell persistence and durable tumor regression. J. Clin. Oncol. 34, 3787–3795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.S. Depil, P. Duchateau, S. A. Grupp, G. Mufti, L. Poirot, ‘Off-the-shelf’ allogeneic CAR T cells: Development and challenges. Nat. Rev. Drug Discov. 19, 185–199 (2020). [DOI] [PubMed] [Google Scholar]
- 117.S. Gaggero, J. Martinez-Fabregas, A. Cozzani, P. K. Fyfe, M. Leprohon, J. Yang, F. E. Thomasen, H. Winkelmann, R. Magnez, A. G. Conti, S. Wilmes, E. Pohler, M. van Gijsel Bonnello, X. Thuru, B. Quesnel, F. Soncin, J. Piehler, K. Lindorff-Larsen, R. Roychoudhuri, I. Moraga, S. Mitra, IL-2 is inactivated by the acidic pH environment of tumors enabling engineering of a pH-selective mutein. Sci. Immunol. 7, eade5686 (2022). [DOI] [PubMed] [Google Scholar]
- 118.L. Gattinoni, E. Lugli, Y. Ji, Z. Pos, C. M. Paulos, M. F. Quigley, J. R. Almeida, E. Gostick, Z. Yu, C. Carpenito, E. Wang, D. C. Douek, D. A. Price, C. H. June, F. M. Marincola, M. Roederer, N. P. Restifo, A human memory T-cell subset with stem cell-like properties. Nat. Med. 17, 1290–1297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.N. Cieri, B. Camisa, F. Cocchiarella, M. Forcato, G. Oliveira, E. Provasi, A. Bondanza, C. Bordignon, J. Peccatori, F. Ciceri, M. T. Lupo-Stanghellini, F. Mavilio, A. Mondino, S. Bicciato, A. Recchia, C. Bonini, IL-7 and IL-15 instruct the generation of human memory stem T cells from naive precursors. Blood 121, 573–584 (2013). [DOI] [PubMed] [Google Scholar]
- 120.Y. Li, M. Bleakley, C. Yee, IL-21 influences the frequency, phenotype, and affinity of the antigen-specific CD8 T cell response. J. Immunol. 175, 2261–2269 (2005). [DOI] [PubMed] [Google Scholar]
- 121.B. T. Aftab, B. Sasu, J. Krishnamurthy, E. Gschweng, V. Alcazer, S. Depil, Toward “off-the-shelf” allogeneic CAR T cells. Adv. Cell Gene Ther. 3, e86 (2020). [Google Scholar]