Abstract
Liquid-liquid phase separation (LLPS) is involved in various dynamic biological phenomena. In epithelial cells, dynamic regulation of junctional actin filaments tethered to the apical junctional complex (AJC) is critical for maintaining internal homeostasis against external perturbations; however, the role of LLPS in this process remains unknown. Here, after identifying a multifunctional actin nucleator, cordon bleu (Cobl), as an AJC-enriched microtubule-associated protein, we conducted comprehensive in vitro and in vivo analyses. We found that apical microtubules promoted LLPS of Cobl at the AJC, and Cobl actin assembly activity increased upon LLPS. Thus, microtubules spatiotemporally regulated junctional actin assembly for epithelial morphogenesis and paracellular barriers. Collectively, these findings established that LLPS of the actin nucleator Cobl mediated dynamic microtubule-actin cross-talk in junctions, which fine-tuned the epithelial barrier.
Phase separation of the actin nucleator Cobl coordinates microtubules and actin filaments to fine-tune epithelial barriers.
INTRODUCTION
Liquid-liquid phase separation (LLPS) is essential for the dynamic regulation of many biological phenomena owing to its ability to quickly concentrate specific sets of proteins with diverse constituent stoichiometries to create membraneless subcompartments (1, 2). LLPS of zonula occludens-1 (ZO-1) has recently been shown to play a role in the “formation” of the epithelial barrier (3–5); however, the role of LLPS in the “fine-tuning” of the epithelial barrier, which is critical for maintaining internal homeostasis against ever-changing external environments, remains unclear.
In polarized and differentiated epithelial cells, the apical junctional complex (AJC), which mainly consist of tight junctions (TJs) and adherens junctions (AJs) in vertebrates, is primarily responsible for cell-to-cell adhesion, and its dynamic nature plays central roles in fine-tuning the properties of epithelial cell sheets (5–10). Dynamic regulation of cytoskeleton-related components associated with the AJC is essential for dynamic AJC regulation, and junctional actin filaments (AFs), which align around the AJC in a belt-like arrangement to form the circumferential ring (CR), are particularly critical (11, 12). It should also be noted that, as epithelial cells differentiate, a subset of noncentrosomal microtubules (MTs) become closely associated with the AJC, including the apical MT network that associates with TJs in a side-by-side manner (13–15), although their precise role in the AJC regulation remains elusive.
Here, we found that MTs facilitate LLPS of cordon bleu (Cobl), a vertebrate-specific tandem Wiskott-Aldrich syndrome protein-homology 2 (WH2)–based actin nucleator with context-dependent AF severing activity (16–18), in epithelial cell-cell junctions. Notably, this LLPS-mediated phenomenon enabled MTs to dynamically regulate actin assembly within the CR and thus spatiotemporally regulate epithelial properties, which contributes to the understanding of a mechanism underlying the fine-tuning of epithelial function.
RESULTS
Cobl was identified as an AJC-enriched, MT-associated protein
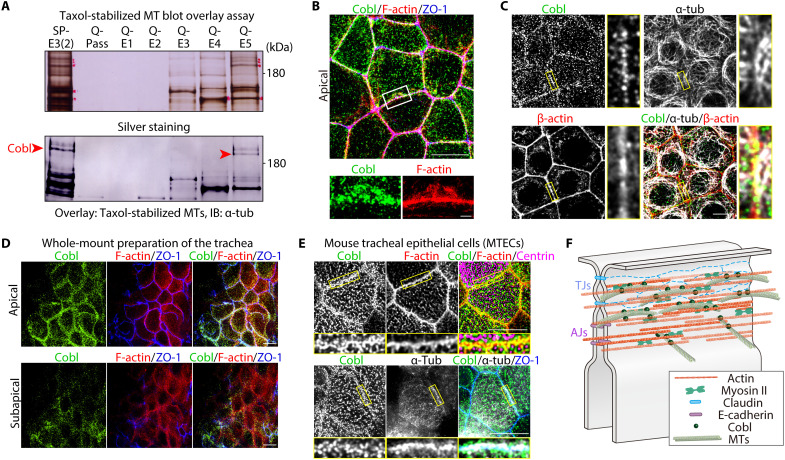
From taxol-stabilized MT blot overlay assays using our original AJC-enriched bile canaliculi (BC) fraction (fig. S1, A and B) (15), we identified Cobl based on its amino acid sequence (Fig. 1A). In cultured Eph4 mammary gland epithelial cells, Cobl localized as small clusters at the apical surface and along cell-cell junctions and was closely associated with both the CR (Fig. 1B and movie S1) and apical MTs (Fig. 1C), which were dynamically linked with the AJC via their sides and tips (fig. S1C and movie S2). Exogenous full-length Cobl (CoblFL) tagged with Venus (Venus-CoblFL) localized similarly in Cobl-revertant (REV) cells, in which Venus-Cobl was stably expressed in Cobl-knockout (Cobl-KO) cells (fig. S1D), and immunoprecipitation analysis confirmed the associations between Cobl, α-tubulin, and β-actin (fig. S1E).
Fig. 1. Cobl was identified as an AJC-enriched, MT-associated protein.
(A) Membrane overlay assays of taxol-stabilized MTs using the AJC-enriched BC fraction (see also fig. S1). Immunoblotted bands marked by red arrowheads correspond to Cobl. (B) Representative superresolution micrographs of immunostained Eph4 epithelial cells in the apical plane (see also movie S1). Scale bar, 10 μm (low magnification) and 1 μm (high magnification). (C) In Eph4 cells, Cobl was closely associated with both actin and MTs at cell-cell junctions. Scale bar, 10 μm. (D) In the in vivo trachea, similarly to cultured Eph4 epithelial cells, Cobl localized to cell-cell junctions at the apical and subapical planes. Scale bar, 10 μm. (E) Cobl was also closely associated with both F-actin and MTs at the AJC in mouse tracheal epithelial cells (MTECs). Scale bar, 5 μm. (F) Schematic drawing of Cobl localization at the AJC in association with actomyosin-based CR and MTs.
To further characterize Cobl localization within cell-cell junctions, we quantified Cobl colocalization via immunofluorescence using CR (i.e., F-actin, myosin IIA, and myosin IIB), TJ (i.e., ZO-1 and occludin), and AJ markers (i.e., E-cadherin, α-catenin, and vinculin) (fig. S2, A and B). Results showed that Cobl colocalized best with CR markers, particularly F-actin, followed by AJ and TJ markers (fig. S2C). Consistently, along the apicobasal axis, Cobl colocalized well with F-actin–based CRs around TJs and AJs (fig. S2, D and E). Furthermore, immunoelectron microscopic analyses of REV cells revealed that Venus-Cobl immunogold particles accumulated in CR regions along the AJC (fig. S2F), confirming that Cobl localized at AJC-associated CR in cell-cell junctions. Serial coimmunoprecipitation assays revealed that Cobl coimmunoprecipitated with various AJC components, such as ZO-1 (fig. S3, A and B), α-catenin, and E-cadherin (fig. S3C). KO of ZO-1 and double KO of ZO-1 and ZO-2 reduced the enrichment of Cobl in the AJC but did not completely release Cobl from the AJC (fig. S3, D and E). Given that Cobl localization to the AJC in ZO-1 KO cells was restored by the expression of Cobl-binding ZO-1 domains (fig. S3F), but not by non–Cobl-binding ZO-1 domain expression (fig. S3G), the binding affinity between Cobl and ZO-1 played an important role in Cobl localization to the AJC but was not completely necessary. Further analysis revealed that total perturbation of the AJC integrity caused Cobl to be completely released from the AJC (fig. S3H). Therefore, we concluded that the overall affinity between Cobl and AJC components, rather than a single protein within AJC components, was crucial in recruiting Cobl condensates to the AJC.
Next, we examined the localization of Cobl in other cultured and in vivo epithelial cells. In both cultured Madin-Darby canine kidney II cells (fig. S4A) and CSG120/7 cells (epithelial cells derived from mouse submandibular gland malignant neoplasms) (fig. S4B), Cobl also localized as small clusters at the apical surface and along the AJC in association with both MTs and AFs. Furthermore, Cobl showed similar localization in the in vivo tracheas (Fig. 1D) and was associated with both AFs and MTs at the AJC in mouse tracheal epithelial cells (Fig. 1E), collectively indicating that Cobl is an AJC-enriched MT- and AF-associated protein in both various cultured and in vivo epithelial cells (Fig. 1F).
Last, we analyzed in vivo mouse non-epithelial tissues [embryonic day 9.5 (E9.5) mouse embryos and hippocampal neuronal cells], in which Cobl was previously shown to play an important role (16). In E9.5 mouse embryos, Cobl preferentially localized to the apical side of the floor plate, where α-tubulin and β-actin also accumulated (fig. S4C). In hippocampal neuronal cell primary cultures, Cobl accumulated along the axon, and magnified microscopic images of the growth cone showed that Cobl, α-tubulin, and β-actin were closely associated (fig. S4D). Together, these results demonstrated that Cobl associated with both AFs and MTs in various epithelial and non-epithelial tissues, suggesting the importance of these associations in the biological function of Cobl.
Cobl formed LLPS-mediated biomolecular condensates in cells
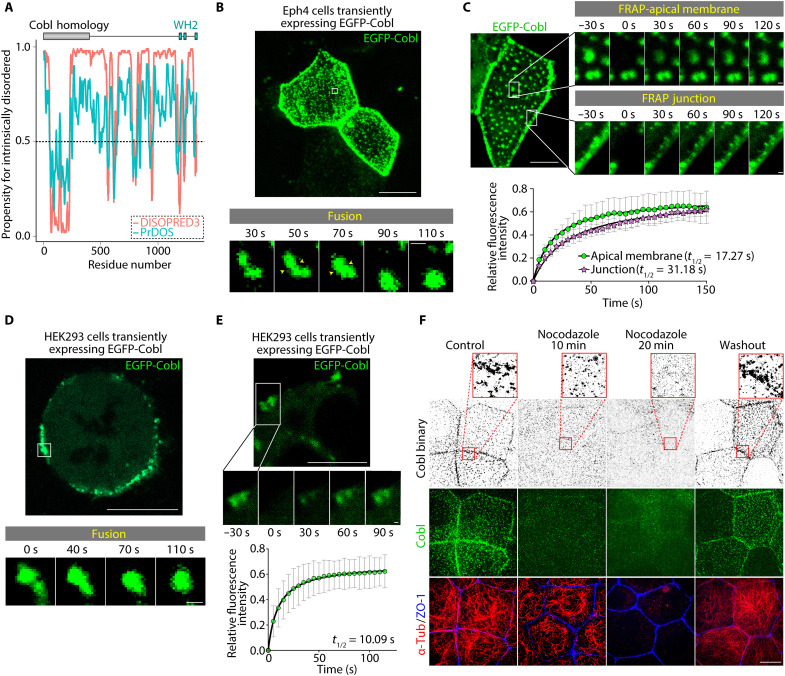
The observed Cobl clusters were round in shape and varied in size, and computational prediction indicated that Cobl was rich in intrinsically disordered regions (Fig. 2A). Therefore, we next examined the role of LLPS in Cobl cluster formation. Live imaging analyses revealed that in Eph4 cells, Cobl clusters were dynamic and underwent fusion within seconds (Fig. 2B and movie S3). In fluorescence recovery after photobleaching (FRAP), the fluorescent intensities of Cobl clusters recovered rapidly after photobleaching (Fig. 2C), further indicating that they were formed via LLPS. Notably, Cobl turnover within AJC-associated condensates was slightly slower than that within apical membrane-associated condensates (Fig. 2C and movie S4), suggesting that additional interactions with AJC components unique to AJC-associated condensates may underlie this difference. In human embryonic kidney 293 (HEK293) cells, enhanced green fluorescent protein (EGFP)–tagged CoblFL (EGFP-CoblFL) overexpression induced membrane-bound Cobl condensates in some cells (Fig. 2, D and E, and movie S5). By contrast, EGFP-tag overexpression alone resulted in pancellular localization in both Eph4 and HEK293 cells (fig. S5A), collectively indicating that formation of LLPS-mediated condensates was a common feature of Cobl, which was conserved across various cells.
Fig. 2. Cobl underwent LLPS in an MT-dependent manner in cells.
(A) Prediction of internally disordered regions within Cobl. The results of DISOPRED3 (http://bioinf.cs.ucl.ac.uk/psipred) and PrDOS (https://prdos.hgc.jp), which are popular applications that accurately predict internally disordered regions, are shown. Higher propensity scores indicated an increased likelihood that the residue is intrinsically disordered. Considering that a cutoff of 0.5 is frequently used to determine intrinsically disordered residues, Cobl had an abundance of regions that were likely to be intrinsically disordered. (B) In Eph4 cells, EGFP-Cobl condensates underwent fusion within seconds (see also movie S3). Scale bar, 10 μm (low magnification) and 0.5 μm (high magnification). (C) The fluorescence intensities of EGFP-Cobl condensates recovered rapidly, with t1/2 values of 17.27 s (apical membrane) and 31.18 s (junction) (see also movie S4). Scale bar, 10 μm (low magnification) and 0.5 μm (high magnification). N = 4, each. (D) In HEK293 cells, exogenous EGFP-Cobl formed membrane-bound condensates (see also movie S5). Scale bar, 10 μm (low magnification) and 0.5 μm (high magnification). (E) The fluorescence intensities of EGFP-Cobl condensates recovered rapidly, with a t1/2 of 10.09 s. Scale bar, 10 μm (low magnification) and 0.5 μm (high magnification). N = 4. (F) Treatment with 2 μM nocodazole dissolved Cobl condensates in WT cells. Scale bar, 10 μm. Points represent means, and error bars represent SDs.
Cobl-MT interactions promoted Cobl LLPS
Unexpectedly, in Eph4 cells, nocodazole-induced MT depolymerization gradually reduced the number of Cobl condensates (Fig. 2F and fig. S5B), suggesting that Cobl-MT interactions promoted Cobl LLPS. In contrast to nocodazole treatment, Cobl still existed as condensates when AFs were forced to depolymerize under latrunculin (fig. S5C). Consistently, taxol-induced MT stabilization marginally increased the number of Cobl condensates (fig. S5D). Therefore, we next examined the effects of MTs on Cobl LLPS in vitro. Using HEK293 transfection (fig. S6A), we first purified a FLAG peptide (FLAG)– and EGFP-tagged CoblFL protein (FLAG-EGFP-CoblFL), and a FLAG- and EGFP-tagged tandem-WH2 domain–containing truncated Cobl protein [designated as Cobl D5 domain (CoblD5; amino acids 1001 to 1337); FLAG-EGFP-CoblD5].
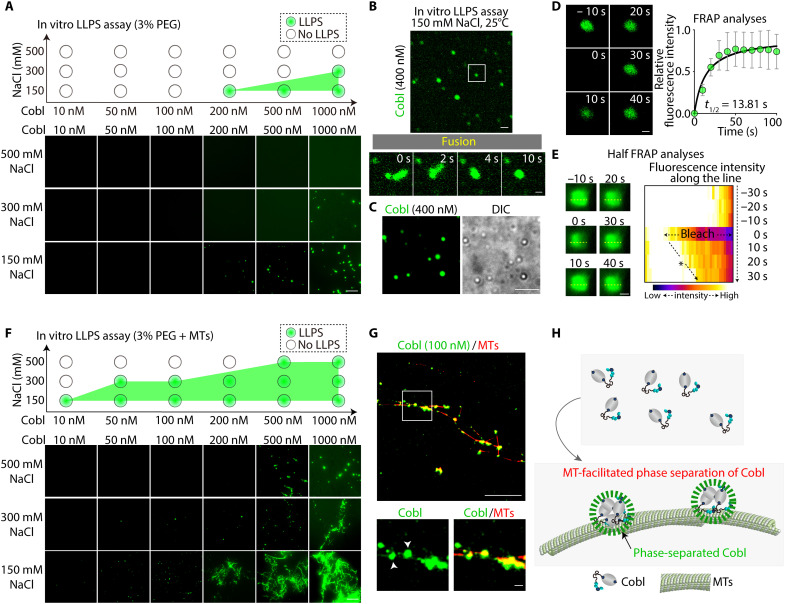
Counting of blanching steps by single-molecule total internal reflection fluorescence (TIRF) imaging revealed that both FLAG-EGFP-CoblFL and FLAG-EGFP-CoblD5 existed as monomers, at least at low concentrations (5 nM; movie S6 and fig. S6B). Subsequent in vitro LLPS assay revealed that in an LLPS buffer containing 3% polyethylene glycol (PEG) and 150 mM NaCl (pH 7.4; 25°C), FLAG-EGFP-CoblFL condensates spontaneously appeared at a concentration above 200 nM (Fig. 3A). Condensate formation was critically dependent on ionic strength and temperature, with FLAG-EGFP-CoblFL condensate formation being suppressed at higher NaCl concentrations (Fig. 3A) and lower temperatures (fig. S6C). Live imaging analyses revealed that these FLAG-EGFP-CoblFL condensates fused within seconds (Fig. 3, B and C, and movie S7). FRAP analyses validated the dynamic diffusion between the inside and outside of the condensates (t1/2 = 13.81 s; Fig. 3D), and half-FRAP analyses demonstrated even faster diffusion within the condensates (t1/2 = 10.00 s; Fig. 3E and fig. S6D).
Fig. 3. Cobl underwent LLPS in an MT-dependent manner in vitro.
(A) In vitro LLPS assay of Cobl. The tendency for Cobl to undergo LLPS was shown to be critically dependent on NaCl concentration, with higher NaCl concentrations resulting in a lower tendency. Protein concentration: FLAG-EGFP-CoblFL, 10, 50, 100, 200, 500, or 1000 nM. Scale bar, 10 μm. (B) Cobl condensates underwent fusion within seconds in vitro (see also movie S7). Scale bar, 10 μm (low magnification) and 0.5 μm (high magnification). Protein concentration: FLAG-EGFP-CoblFL, 400 nM. (C) Representative confocal and differential interference contrast (DIC) images of phase-separated Cobl condensates in vitro. Protein concentration: FLAG-EGFP-CoblFL, 400 nM. Scale bar, 5 μm. (D) Fluorescence intensities of in vitro Cobl condensates recovered rapidly, with a t1/2 of 13.81 s. Protein concentration: FLAG-EGFP-CoblFL, 400 nM. Scale bar, 0.5 μm. N = 4. (E) Half FRAP assay of larger Cobl condensates in vitro with a kymograph along the yellow dotted line. Fluorescence intensities of the bleached region recovered from the unbleached side with a t1/2 of 10.00 s (see also fig. S6D). Protein concentration: FLAG-EGFP-CoblFL, 1 μM. Scale bar, 1 μm. (F) In vitro LLPS assay of Cobl in the presence of MTs. The tendency for Cobl to undergo LLPS was apparently increased. Protein concentration: FLAG-EGFP-CoblFL, 10, 50, 100, 200, 500, or 1000 nM and MTs, 8 μM. Scale bar, 10 μm. (G) Cobl accumulated along HiLyte 647–labeled MTs and formed condensates on MTs (white arrowheads). Protein concentration: FLAG-EGFP-CoblFL, 100 nM and HiLyte 647–labeled MT, 8 μM. Scale bar, 10 μm (low magnification) and 0.5 μm (high magnification). (H) Schematic drawing of MT-facilitated phase separation of Cobl. Points represent means, and error bars represent SDs.
Notably, MTs lowered the concentration of FLAG-EGFP-CoblFL required for LLPS (Fig. 3F). In addition, using 10% HiLyte 647–labeled MTs, we confirmed that in the presence of MTs, FLAG-EGFP-CoblFL formed condensates primarily on MTs (Fig. 3G). Consistent with this, low-angle shadowing electron microscopy (EM) revealed that FLAG-tagged CoblFL (FLAG-CoblFL) monomers adopted a “dumbbell” shape in solution (fig. S6E), whereas several FLAG-CoblFL molecules constituted condensates on MTs (fig. S6F). Furthermore, it was revealed that in the presence of MTs, FLAG-EGFP-CoblFL were able to form condensates even in a buffer without a crowding agent (i.e., PEG), whereas FLAG-EGFP-CoblFL alone could not undergo LLPS without a crowding agent (fig. S6G), collectively demonstrating that Cobl-MT interactions promoted CoblFL LLPS (Fig. 3H). In addition, FLAG-EGFP-CoblD5 did not form condensates at concentrations up to 3 μM (fig. S6H), confirming that CoblFL, not CoblD5, inherently underwent LLPS.
Next, we conducted Cobl domain analyses using EGFP-tagged Cobl domains, designated as D1 to D5 domains (i.e., EGFP-CoblD1 to EGFP-CoblD5) (fig. S7A). A coimmunoprecipitation assay revealed that Cobl bound α-tubulin via its D1, D3, and D4 domains (fig. S7B), and a high-speed cosedimentation assay revealed that Cobl bound AFs only via the tandem WH2 domain–containing D5 domain (fig. S7C). Notably, in Cobl-KO cells transiently expressing Cobl domains, none of Cobl domains (i.e., EGFP-CoblD1 to EGFP-CoblD5) underwent LLPS (fig. S7D). However, EGFP-tagged Cobl lacking a D5 domain (EGFP-CoblΔD5), which cannot bind AFs but can multivalently bind α-tubulin, formed small round condensates similarly to EGFP-CoblFL (fig. S7D). This result suggested that the Cobl-MT interaction, when multivalent, promoted Cobl LLPS, whereas the Cobl-actin interaction was dispensable for Cobl LLPS in cells.
Cobl-KO impaired actin assembly within the CR in cultured epithelial cells, leading to defective apical constriction and impaired paracellular barrier function
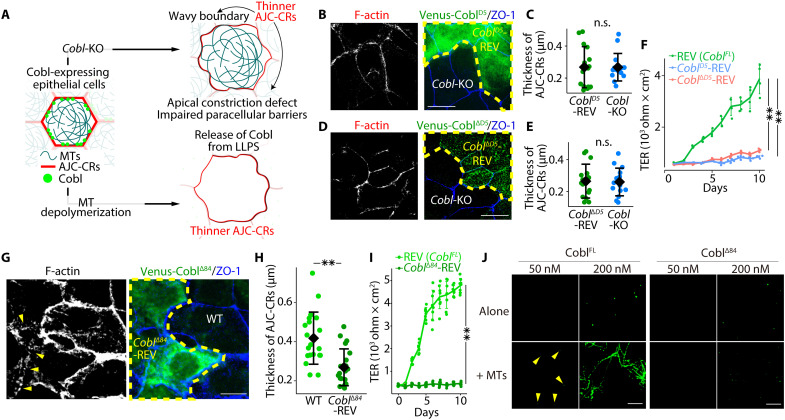
To examine the function of Cobl in epithelial cells, we first analyzed Cobl-KO Eph4 cells (fig. S8A). We found that ZO-1–delineated boundaries between cells were more tortuous in Cobl-KO cells than in Cobl-expressing [wild-type (WT) and REV] cells (Fig. 4A), as demonstrated by the persistence length of the ZO-1–delineated boundaries (fig. S8B). In addition, CR thickness measurements (fig. S8C) in cocultures of Cobl-KO and Cobl-expressing (WT or REV) cells revealed that the CR was significantly thinner in Cobl-KO cells than in WT (Fig. 4, B and C) and REV cells (Fig. 4, D and E), which was further confirmed via EM analysis (Fig. 4F). It is known that ras homolog family member A (RhoA), particularly active RhoA, is a key regulator for actin assembly in the CR (19), but we found no differences in junctional RhoA and active RhoA levels between Cobl-KO and WT cells (fig. S8D). In addition, junctional accumulation of myosin IIA and myosin IIB, the main constituents of myosin motors in the CR (20), was decreased (fig. S8, E and F). Myosin light chain (MLC) phosphorylation was also down-regulated (fig. S8G), but the Rho-associated coiled-coil containing kinase (ROCK) pathway, which controls MLC phosphorylation levels (19), was unchanged (fig. S8, H and I). Collectively, these results confirmed that CR integrity was critically disturbed in Cobl-KO cells compared with Cobl-expressing (WT or REV) cells.
Fig. 4. Cobl-KO and MT depolymerization impaired junctional actin assembly, causing defective apical constriction and reduced paracellular barrier function.
(A) Representative confocal micrographs of WT, Cobl-KO, and REV cells. Scale bar, 10 μm. (B) Cocultures of WT and Cobl-KO cells. Scale bar, 10 μm. (C) CR thickness, as determined by the method described in fig. S8C. **P < 0.01 (unpaired t test). N = 20 cells each. (D) Cocultures of REV and Cobl-KO cells. Scale bar, 10 μm. (E) CR thickness. **P < 0.01 (unpaired t test). N = 20 cells each. (F) EM analysis showed that the CR, indicated by yellow arrows in the EM images, was thinner in Cobl-KO cells than in Cobl-expressing (WT and REV) cells, as confirmed by subsequent quantification. Scale bar, 100 nm. **P < 0.01 (unpaired t test). N = 6 cells, each. (G) Heatmap of the WT and Cobl-KO cell surface area in cocultures. Scale bar, 10 μm. (H) Surface areas of WT and Cobl-KO cells. **P < 0.01 (Mann-Whitney U test). N = 3 micrographs each. (I) Daily changes in TER in WT, Cobl-KO, and REV cells. **P < 0.01 (Mann-Whitney U test). N = 8 trials each. (J) Fluxes of fluorescein or FITC-labeled dextran ranging in size from 0.4 to 40 kDa. *P < 0.05 (unpaired t test). N = 4 trials each. (K) WT cells treated with 2 μM nocodazole. Conditions are the same as in Fig. 2F. Scale bar, 10 μm. (L) Temporal changes in CR thickness and irregularity of the ZO-1–delineated boundary under 2 μM nocodazole treatment. N = 20 cells (actin thickness) and N = 50 cells (persistence length). Diamonds in dot plots, solid lines in line graphs, and bars in bar graphs represent means, and error bars or shaded areas represent SDs.
Next, we focused on epithelial cellular processes, for which actin assembly within the CR is critical. Coculture of Cobl-KO and Cobl-expressing cells revealed that the apical surface area became significantly smaller in Cobl-expressing (WT and REV) cells than in Cobl-KO cells (Fig. 4, G and H, and fig. S9, A and B), and the ratio of the apical area to the basal area was also significantly smaller in Cobl-expressing REV cells than in Cobl-KO cells (fig. S9, C and D). Furthermore, transepithelial electric resistance (TER), an indicator of paracellular barrier function against ions, was significantly reduced (Fig. 4I), and fluxes of 0.4-kDa fluorescein and 4- to 40-kDa fluorescein isothiocyanate (FITC)–labeled dextran, indicators of paracellular barrier function to small molecules, were significantly increased in Cobl-KO cells compared with Cobl-expressing (WT and REV) cells (Fig. 4J). These results indicated that Cobl-KO–induced actin assembly impairment within the CR critically affected epithelial function.
Notably, in WT cells, as nocodazole-induced MT depolymerization released Cobl from LLPS, the CR gradually became thinner, and cell-cell boundaries became more irregular (Fig. 4, K and L). In contrast, nocodazole treatment did not alter the thickness of the CR in Cobl-KO cells (fig. S9E). Therefore, we concluded that Cobl-KO led to impaired actin assembly within the CR in cultured epithelial cells, subsequently leading to apical constriction and paracellular barrier impairment, and MT depolymerization in WT cells phenocopied Cobl-KO cells via its effect on Cobl (Fig. 5A). In contrast to the pronounced effect of MTs on Cobl, there were no differences in MT organization between Cobl-KO and WT cells (fig. S9F).
Fig. 5. Neither CoblD5, CoblΔD5, nor CoblΔ84 rescued the phenotype of Cobl-KO cells.
(A) Schematic drawing of the effects of Cobl-KO and MT depolymerization on cultured epithelial cells. (B) Cocultures of Cobl-KO and CoblD5-REV cells. (C) CoblD5 alone did not rescue the phenotype of Cobl-KO cells (i.e., thinner CRs). n.s., not significant (Mann-Whitney U test). N = 16 cells each. Scale bar, 10 μm. (D) Cocultures of Cobl-KO and CoblΔD5-REV cells. (E) CoblΔD5 alone did not rescue the phenotype of Cobl-KO cells (i.e., thinner CRs). n.s., not significant (unpaired t test). N = 15 cells each. Scale bar, 10 μm. (F) Daily changes in TER in REV, CoblD5-REV, and CoblΔD5-REV cells. **P < 0.01 (unpaired t test). N = 4 trials each. (G) Cocultures of WT and CoblΔ84-REV cells revealed that the CR was still thinner in CoblΔ84-REV cells (yellow arrowheads) than in WT cells. Scale bar, 10 μm. (H) CR thickness. **P < 0.01 (Mann-Whitney U test). N = 20 cells, each. (I) Daily changes in TER in REV and CoblΔ84-REV cells. **P < 0.01 (unpaired t test). N = 8 trials, each. (J) In vitro LLPS assay of CoblFL and CoblΔ84 with and without MTs. Similar to the case of CoblFL, CoblΔ84 alone was able to undergo LLPS at a concentration of 200 nM. In the presence of MTs, CoblΔ84 did not undergo LLPS at a concentration of 50 nM in contrast to the case of CoblFL (yellow arrowheads). Scale bar, 10 μm. Protein concentration: FLAG-EGFP-CoblFL, 50 or 200 nM; FLAG-EGFP-CoblΔ84, 50 or 200 nM; and MTs, 8 μM. Diamonds in dot plots and solid lines in line graphs represent means, and error bars represent SDs.
Last, to separately examine the effects of the Cobl-MT and Cobl-actin interactions on Cobl function in cultured epithelial cells, we established and analyzed CoblD5-REV cells, in which Venus-tagged CoblD5 (Venus-CoblD5), capable of binding AFs, was stably expressed in Cobl-KO cells, and CoblΔD5-REV cells, in which Venus-tagged CoblΔD5 (Venus-CoblΔD5), capable of binding α-tubulin multivalently and undergoing LLPS in cells, was stably expressed in Cobl-KO cells (fig. S9G). Similar to Cobl-KO cells, both CoblD5-REV and CoblΔD5-REV cells have shown thinner AFs within CRs (Fig. 5, B to E) and the paracellular barrier impairment (Fig. 5F), indicating that neither CoblD5 nor CoblΔD5 could rescue the phenotype of Cobl-KO cells. Therefore, these results also confirmed that both multivalent Cobl-MT interactions with the resulting LLPS and Cobl-actin interactions were both indispensable for Cobl function in cultured epithelial cells.
Alternation of Cobl function impaired junctional actin accumulation and epithelial paracellular barrier function in the stomach and induced gastritis in mice
To investigate the in vivo role of Cobl, we conducted CRISPR-Cas9–mediated genome editing in fertilized mouse eggs using guide RNA designed to target exon1 of the Cobl gene. Genetic analyses revealed that the resulting progeny exhibited an 84–base pair (bp) deletion (CoblΔ84/Δ84 mice) in the region that included the junction of exon 1 and intron 1 in the Cobl gene (fig. S10, A to C).
Before analyzing CoblΔ84/Δ84 mice, we characterized CoblΔ84 in cells and in vitro. First, we examined the effect of 84-bp deletion in cultured epithelial cells. We established CoblΔ84-REV cells, in which Venus-CoblΔ84 was stably expressed in Cobl-KO cells (fig. S10D). Unexpectedly, CoblΔ84 neither formed condensates nor localized at the AJC (fig. S10E). Accordingly, CoblΔ84-REV cells had thinner CRs and impaired paracellular barrier function (Fig. 5, G to I). Next, we conducted in vitro LLPS assays using FLAG- and EGFP-tagged CoblΔ84 (FLAG-EGFP-CoblΔ84). FLAG-EGFP-CoblΔ84 was able to make condensates at 200 nM concentration in LLPS buffer (Fig. 5J), indicating that the ability to undergo LLPS did not apparently differ between CoblFL and CoblΔ84. However, unexpectedly, although FLAG-EGFP-CoblΔ84 still had an intact D3 domain with binding affinity to α-tubulin, the effects of MTs on LLPS markedly differed between FLAG-EGFP-CoblFL and FLAG-EGFP-CoblΔ84 because FLAG-EGFP-CoblΔ84 did not undergo LLPS at 50 nM concentration even in the presence of MTs, and the number of condensates did not differ on the basis of the presence or absence of MTs at 200 nM concentration (Fig. 5J). These results collectively indicated that MTs could not promote CoblΔ84 LLPS, leading to the altered phenotype observed in the CoblΔ84-REV cells.
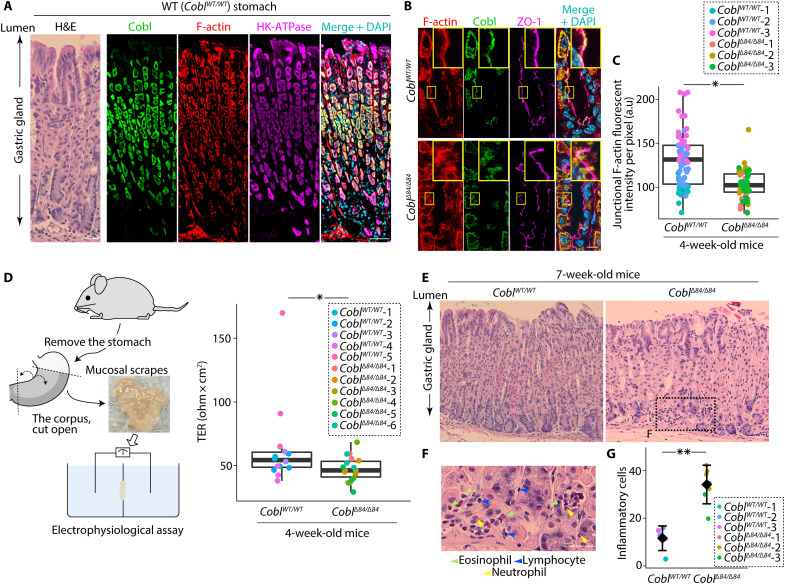
Next, to assess the effects of CoblΔ84 on epithelial cells in vivo, we examined the stomach because we can electrophysiologically measure the paracellular barrier function of the stomach epithelia, which is assumed to cause gastritis when impaired (10). Cobl was sufficiently expressed in the stomach (fig. S11A). Note that Western blot analyses of the stomach epithelia revealed that the CoblΔ84/Δ84 mice were Cobl-knockdown mice (Supplementary Text and fig. S11B). In the WT stomach, Cobl was expressed primarily in hydrogen potassiumadenosine triphosphatase (HK-ATPase)–positive parietal cells (Fig. 6A) and accumulated along the ZO-1–delineated AJC in Cobl-positive cells (Fig. 6B). In 4-week-old CoblΔ84/Δ84 mice, the F-actin fluorescent intensity of Cobl-positive cells was significantly reduced (Fig. 6, B and C). Although there were no differences in AJC-related protein expression compared with that in WT mice (fig. S11C) and we could not detect any apparent infiltration of inflammatory cells (fig. S11D) or changes in the proportion of chief to parietal cells (fig. S11E), serial electrophysiological examinations revealed that the TER of the stomach epithelia of 4-week-old CoblΔ84/Δ84 mice was significantly lower than those of 4-week-old WT mice (Fig. 6D, Supplementary Text, and fig. S11, F and G). Further histochemical analyses conducted in 7-week-old mice revealed that more inflammatory cells infiltrated the stomachs of 7-week-old CoblΔ84/Δ84 mice than those of 7-week-old WT mice (Fig. 6, E to G), and inflammatory gene expression was up-regulated (fig. S11, H and I). Therefore, we concluded that CoblΔ84/Δ84 mice showed reduced junctional F-actin accumulation within the CR in Cobl-positive epithelial cells and impaired epithelial paracellular barrier function in the stomach, which resulted in gastritis.
Fig. 6. Alternation of Cobl caused impaired epithelial permeability in the stomach and induced gastritis.
(A) In mouse stomachs, Cobl colocalized with F-actin in stomach epithelial cells that were positive for HK-ATPase (a marker for parietal cells). Scale bar, 50 μm. (B) Junctional F-actin accumulation was disturbed in CoblΔ84/Δ84 mice with a representative portion being enlarged in the top right. (C) Quantification of junctional F-actin accumulation. *P < 0.05 (Mann-Whitney U test). N = 3 mice each. (D) TER of the stomach. *P < 0.05 (Mann-Whitney U test). N = 5 (WT) and 6 (CoblΔ84/Δ84) mice. (E) Representative H&E staining of the gastric glands from WT and CoblΔ84/Δ84 mice. Scale bar, 50 μm. (F) Higher magnification image of inflammatory cell infiltration (green, blue, and yellow arrowheads) in the lamina propria mucosae of CoblΔ84/Δ84 mice. Scale bar, 50 μm. (G) Quantification of infiltrating inflammatory cells. **P < 0.01 (unpaired t test). N = 3 mice, each. In box plots, solid lines represent medians, boxes represent interquartile ranges, and error bars extending from the box represent the range of data within 1.5 times the interquartile range. In a dot plot, diamonds represent means, and error bars represent SDs.
In vitro, MTs directed Cobl activity toward actin assembly by inhibiting its AF-severing function and facilitating its AF-nucleating activity
We then examined Cobl function in vitro. Although no in vitro studies have evaluated CoblFL, CoblD5 has been shown to exert AF nucleation and AF-severing activities at low and high concentrations, respectively (17).
Regarding Cobl AF-severing activity, TIRF AF-severing assays (fig. S12A) revealed that, while no apparent filament-severing activity was observed at concentrations less than 100 nM, FLAG-CoblFL severed AFs efficiently at concentrations of above 200 nM (Fig. 7, A and B, and fig. S12, B and C). Notably, in the presence of 8 μM MTs, 400 nM CoblFL did not sever AFs (Fig. 7, A and B, and movie S8).
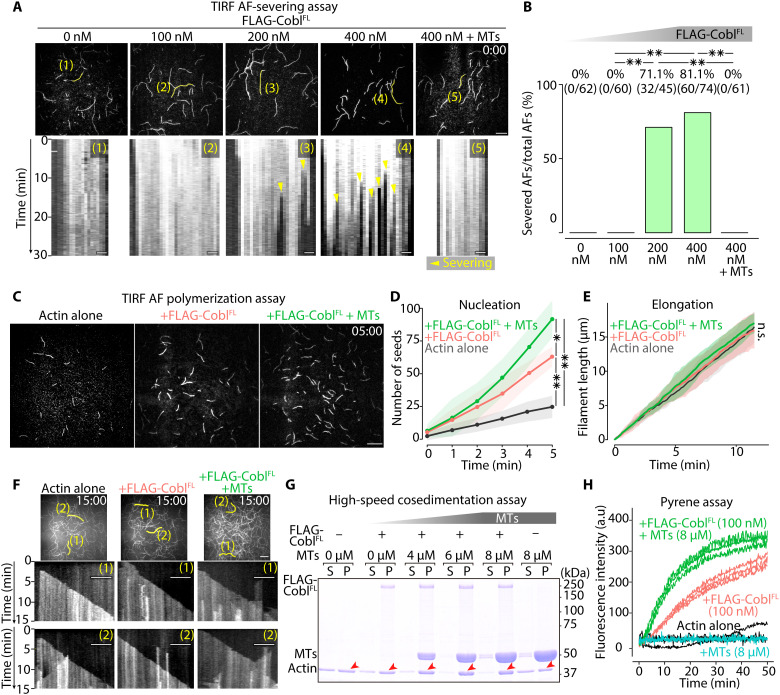
Fig. 7. MTs biased Cobl activity toward actin assembly.
(A) Representative micrographs at the time when FLAG-CoblFL was added (t = 0 min) to AFs (see also movie S8), with respective kymographs showing the time course of the marked filaments (yellow lines). Protein concentration: Alexa Fluor 488–labeled G-actin, 0.5 μM; FLAG-CoblFL, 0, 100, 200, or 400 nM; and MTs, 0 or 4 μM. Scale bar, 10 μm (micrographs) and 1 μm (kymographs). (B) Severing probability quantified at 30 min after FLAG-CoblFL addition. **P < 0.01 (Fisher’s exact test). (C) Representative micrographs at 5 min after initiating polymerization (see also movie S9). Protein concentration: Alexa Fluor 488–labeled G-actin, 1 μM; FLAG-CoblFL, 0 or 100 nM; and MTs = 0 or 8 μM. Scale bar, 10 μm. (D) Time course of the number of seeds [per one TIRF movie (81.92 μm2)] for 5 min after initiating the polymerization. **P < 0.01 and *P < 0.05 (unpaired t test). N = 3 trials each. (E) Change in AF length over time (unpaired t test). N = 15 filaments each. (F) Kymographs of the marked filaments (yellow lines in representative micrographs) demonstrated similar elongation rates. Scale bar, 10 μm (micrographs) and 5 μm (kymographs). (G) High-speed cosedimentation assay showing a change in the actin pellet (P) fraction (red arrowheads; see also fig. S13D) in the presence of Cobl and MTs. Protein concentration: G-actin, 1 μM; FLAG-CoblFL, 0 or 100 nM; and MTs, 0, 4, 6, or 8 μM. (H) MTs increased the slope of the Cobl actin polymerization curves. Protein concentration: pyrene-labeled G-actin, 1 μM; FLAG-CoblFL, 0 or 100 nM; and MTs, 0 or 8 μM. N = 3 trials each. Solid lines represent means, and shaded areas represent SDs.
Regarding Cobl AF nucleation activity, TIRF AF polymerization assays (fig. S12D) revealed that 100 nM FLAG-CoblFL increased the number of actin seeds, with MTs further promoting this effect, whereas the elongation rate was similar despite the addition of FLAG-CoblFL and MTs (Fig. 7, C to F, fig. S12E, and movie S9). It was also confirmed that MTs alone did not affect AF polymerization (fig. S13, A to C, and movie S10). High-speed cosedimentation assays revealed that 100 nM FLAG-CoblFL increased the filament fraction, with MTs further increasing the filament fraction in a concentration-dependent manner in the presence of FLAG-CoblFL (Fig. 7G and fig. S13D). Pyrene assays showed that MTs shortened the lag phase and increased the slope of the CoblFL actin polymerization curves (Fig. 7H). Collectively, MTs inhibited the AF-severing function of Cobl and facilitated its AF-nucleating activity.
Last, to further clarify the relationship between MTs, Cobl LLPS, and AF nucleation activity, after confirming that CoblFL, but not CoblD5, formed condensates at 100 nM in the presence of MTs (fig. S13E), we performed a comparative TIRF AF polymerization assay using CoblFL and CoblD5. Results showed that MTs up-regulated the AF-nucleating activity of CoblFL, but not that of CoblD5 (fig. S13, F and G, and movie S11). Therefore, we concluded that MTs facilitated Cobl AF-nucleating activity by promoting Cobl LLPS.
Cobl condensate formation directly enhanced its AF nucleating activity
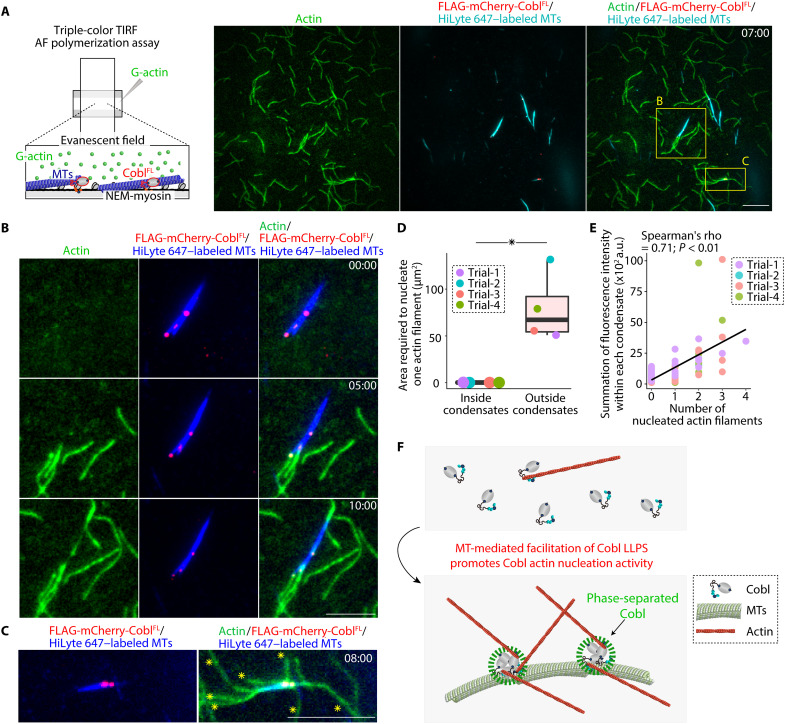
Last, we conducted triple-color TIRF AF polymerization assays using 1 μM 20% Alexa Fluor 488–labeled G-actin, 100 nM mCherry-tagged FLAG-CoblFL (mCherry-FLAG-CoblFL), and 4 μM 10% HiLyte 647–labeled MTs (Fig. 8A and movie S12). AFs were nucleated via mCherry-FLAG-CoblFL condensates from both the MT lateral sides and tips, with mCherry-FLAG-CoblFL remaining at the end of the elongating AFs (Fig. 8B and movie S13), and large mCherry-FLAG-CoblFL condensates on MTs nucleated several AFs (Fig. 8C and movie S14). Negative staining electron microscopic analyses also supported these observations (fig. S13H). AF nucleation efficacy quantification revealed that condensate formation significantly enhanced Cobl actin nucleation activity (Fig. 8D). The fluorescent intensity within each condensate was significantly correlated with the number of AFs nucleated from each condensate (Fig. 8E), collectively suggesting that Cobl condensate formation directly enhanced its AF-nucleating activity, and the efficacy of actin nucleation remained constant once Cobl formed condensates (Fig. 8F).
Fig. 8. MT-mediated Cobl LLPS resulted in MT-dependent promotion of Cobl actin nucleation.
(A) Representative TIRF micrographs at 7 min, obtained using triple-color TIRF microscopic analyses (see also movie S12). Protein concentration: Alexa Fluor 488–labeled G-actin, 1 μM; FLAG-mCherry-CoblFL, 100 nM; and HiLyte 647–labeled MTs, 4 μM. Scale bar, 10 μm. (B) MTs elongated AFs directly via CoblFL condensates (see also movie S13). Scale bar, 5 μm. (C) Large Cobl condensates could elongate several AFs, as indicated by yellow asterisks (see also movie S14). Scale bar, 5 μm. (D) The area required to nucleate one AF. *P < 0.05 (Mann-Whitney U test). N = 4 trials, each. (E) Correlation between the number of AFs from each condensate and the fluorescence intensity within each condensate. (F) Schematic drawing of the mechanism underlying MT-mediated facilitation of Cobl actin nucleation activity. In a box plot, solid lines represent medians, boxes represent interquartile ranges, and error bars extending from the box represent the range of data within 1.5 times the interquartile range.
DISCUSSION
In this study, by comprehensive in vitro and in vivo analyses, we showed that in epithelial junctions, MTs facilitated Cobl LLPS, which directed Cobl activity toward actin assembly and spatiotemporally promoted junctional actin assembly to fine-tune epithelial morphogenesis and paracellular barriers. LLPS can sequester and concentrate a specific set of proteins to facilitate reactions within condensates while slowing reactions outside condensates (1). Considering the binding affinity between Cobl and G-actin, phase-separated Cobl may increase the concentration of a localized G-actin reservoir within condensates, leading to efficient AF nucleation, while lowering the probability that free Cobl molecules will encounter AFs, thereby reducing Cobl’s severing activity. In addition, LLPS-induced increases in dwell time, which enhances the activity of signaling cascades consisting of slow, multistep processes (2, 21), may also play important roles, given that Cobl-mediated actin nucleation has been proposed to consist of multistep processes, including slow growth at the pointed end followed by conformational changes within the actin-Cobl nucleus and rapid growth at the barbed end (22). A previous live imaging analyses of non-epithelial cells showed that AFs could elongate from Cobl puncta, with increases in fluorescence intensity before AF elongation (23, 24). Therefore, these results collectively suggest that Cobl uses LLPS to control its own activity in various cells.
Our findings also expanded our knowledge regarding the biological roles of MTs in promoting LLPS and actin assembly. As drivers of LLPS, weak, multivalent protein-protein (or protein-RNA) interactions and/or intrinsically disordered regions are important. Moreover, as facilitators of LLPS, limiting the diffusion of molecules to a two-dimensional plane also plays an important role (25, 26). Therefore, we believe that restricting Cobl diffusion to the MT surface underlies the MT-mediated facilitation of Cobl LLPS. In addition, the conformational change in Cobl upon binding to MTs may also predispose it to undergo LLPS. Notably, several other MT-associated proteins, including disease-associated tau, which can form disease-causing aggregates under prolonged LLPS, form condensates on MTs (27). Therefore, further studies on the role of MTs in promoting LLPS will also be important for understanding the mechanism of various diseases. Regarding MT-facilitated AF nucleation, previous studies have suggested that MT plus-ends can facilitate actin network assembly via the interactions between mDia1 and MT plus-end tracking proteins, including adenomatous polyposis coli and end-binding protein 1 (28, 29). Therefore, our results advance our current understanding by showing that MT lateral sides, in addition to MT plus-ends, can promote actin assembly via phase-separated Cobl.
In conclusion, we showed that apical MTs facilitated the LLPS of the actin nucleator Cobl to accelerate Cobl-mediated actin assembly at the AJC for epithelial morphogenesis and paracellular barriers. We believe that our results, which revealed the central role of LLPS-mediated MT-actin cross-talk, contribute to elucidating the mechanisms through which epithelial cells dynamically fine-tune their properties and provide clues for the development of therapies aimed at the spatiotemporal control of epithelial properties.
MATERIALS AND METHODS
Cell lines
Eph4 mouse mammary gland epithelial cells (RRID: CVCL_0073) were provided by E. Reichmann (Research Institute of Molecular Pathology, Vienna, Austria). Madin-Darby canine kidney II cells were provided by M. Murata (National Institute for Physiological Sciences, Okazaki, Japan), and CSG120/7 cells were provided by W. Birchmeier (Max-Delbruck-Center for Molecular Medicine, Berlin, Germany). HEK293 cells (RRID: CVCL_0045) were purchased from the American Type Culture Collection (Manassas, VA, USA, catalog no. CRL-1573). Eph4 and HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Nissui Pharmaceutical, Tokyo, Japan; catalog no. 05919) supplemented with 10% fetal bovine serum (FBS; Nichirei Biosciences, Tokyo, Japan; catalog no. 171012) and 2 mM l-glutamine (Wako, Osaka, Japan; catalog no. 073-05391) at 37°C under a 5% CO2 atmosphere. For chemical treatment, the cells were incubated in DMEM containing 2 μM nocodazole (Sigma-Aldrich, St. Louis, MO, USA; catalog no. M1404) for the times indicated in the text.
Primary cell cultures
Primary hippocampal cultures were obtained from E15 embryos of WT female C57BL/6J mice (RRID: IMSR_JAX:000664). The hippocampus was isolated, digested with Hepes-buffered saline [HBS; 20 mM Hepes-NaOH (pH 7.3), 140 mM NaCl, and 1 mM CaCl2] containing 0.125% trypsin (Nacalai Tesque, Kyoto, Japan; catalog no. 35555-54) and deoxyribonuclease I (0.1 mg/ml; Sigma-Aldrich, St. Louis, MO, USA; catalog no. 10104159001) for 10 min at 37°C, and dissociated gently by passage through a sterilized Pasteur pipette. Digestion was stopped by adding FBS to a final concentration of 10%. Cells were pooled via centrifugation at 800 rpm for 5 min and resuspended in minimum essential medium (Thermo Fisher Scientific, Waltham, MA, USA; catalog no. 11095080) containing 5% FBS, B27 (Thermo Fisher Scientific, Waltham, MA, USA; catalog no. 17504044), and GlutaMAX (Thermo Fisher Scientific, Waltham, MA, USA; catalog no. 35050061). The cells were plated on poly-l-lysine–coated glass coverslips (1 mg/ml) and cultured at 37°C under a 5% CO2 atmosphere. After 48 hours, the medium was replaced with Neurobasal Medium (Thermo Fisher Scientific, Waltham, MA, USA; catalog no. 21103049) supplemented with B27 and GlutaMAX.
Mouse tracheal epithelial cell (MTEC) cultures were prepared from the tracheas of 8-week-old WT male C57BL/6J mice. After enzymatic digestion, cells were pooled via centrifugation at 400g for 10 min at 4°C and then resuspended in MTEC basic medium consisting of DMEM/F-12 (Lonza, Basel, Switzerland; catalog no. 12-719), 3.6 mM sodium bicarbonate, penicillin (100 U/ml), streptomycin (100 μg/ml), amphotericin B (0.25 μg/ml; Wako, Osaka, Japan; catalog no. 161-23181), and 10% FBS. After culturing for 3.5 hours at 37°C under a 5% CO2 atmosphere, nonadherent cells were collected by centrifugation, resuspended using the bronchial epithelial cell growth BulletKit (Lonza, Basel, Switzerland; catalog no. CC-3170), counted, and seeded at 1 × 105 cells/cm2 onto 12-mm-diameter transwell filters (Corning, Corning, NY, USA; catalog no. 3460) coated with collagen type I (50 μg/ml; Corning, Corning, NY, USA; catalog no. 354236). After 4 days of culture at 37°C under a 5% CO2 atmosphere, the air-liquid interface (ALI) stage was created using PneumaCult (STEMCELL Technologies, Vancouver, BC, Canada; catalog no. 05001) in accordance with the manufacturer’s instructions. For immunofluorescence, transwell filters with an MTEC sheet (ALI, 50 days) were cut out and used.
Animals
All procedures involving animals strictly followed the institutional guideline and were approved by the Institutional Animal Care and Use Committee of Osaka University (reference number: FBS-17-003 and FBS-17-005). WT chicks, purchased from local farms, were used for the preparation of AJC-enriched BC fractions, as described previously (30). WT C57BL/6J mice were purchased from Japan SLC (Shizuoka, Japan). Three-week-old WT male C57BL/6J mice were used for immunostaining of whole-mount tracheas. Eight-week-old WT male C57BL/6J mice were used for MTEC cultures. Pregnant WT C57BL/6J mice were used for neural tube immunostaining and primary hippocampal cultures. Seven-week-old male WT and Cobl gene-edited male C57BL/6J mice generated in the laboratory of Integrative Biological Science, Institute for Frontier Life and Medical Sciences, Kyoto University, as described below, were used for hematoxylin and eosin (H&E) staining of stomach tissues. Four-week-old WT and Cobl gene-edited male C57BL/6J mice were used for electrophysiological assays and immunostaining of stomach tissues.
Taxol-stabilized MT blot overlay assays
The AJC-enriched BC fraction was prepared from 2-day-old chick livers as detailed previously (30). It was then suspended in a hypotonic solution [10 mM Hepes-NaOH (pH 7.5), 1 mM NaHCO3, and leupeptin (4 μg/ml); Sigma-Aldrich, St. Louis, MO, USA; catalog no. L2884] and ultracentrifuged at 100,000g at 4°C for 30 min. The pellet was suspended in buffer A [10 mM Hepes-NaOH (pH 7.5), 1 mM EGTA, 6 M urea, leupeptin (4 μg/ml), and 10 mM p-amidinophenylmethanesulfonyl fluoride; Sigma-Aldrich, St. Louis, MO, USA; catalog no. A6664] and ultracentrifuged at 100,000g at 4°C for 60 min. The supernatant was applied to an SP-Sepharose column (GE Healthcare, Chicago, IL, USA; catalog no. 17072901; flow-through fraction, “SP-Pass”), and fractions eluted between 100 and 150 mM NaCl were collected in buffer A to obtain “SP-E3(1),” “SP-E3(2),” and “SP-E3(3).” The “SP-E3(2)” fraction was applied to a Q-Sepharose column (GE Healthcare, Chicago, IL, USA; catalog no. 17051001; flow-through fraction, “Q-Pass”), and fractions were eluted with 50 mM NaCl in buffer A to obtain “Q-E1,” “Q-E2,” “Q-E3,” “Q-E4,” and “Q-E5.” The fractions were subjected to SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (Merck Millipore, Burlington, MA, USA; catalog no. IPVH00010). The membranes were blocked with 1% bovine serum albumin (BSA; Nacalai Tesque, Kyoto, Japan; catalog no. 01281-26) in Britton-Robinson buffer 80 (BRB80) buffer [80 mM Pipes-NaOH (pH 6.9), 1 mM MgCl2, and 1 mM EGTA] at room temperature for 60 min and then incubated with MT solution in the presence of 5% skim milk. For MT solution, tubulin (1 mg/ml) purified from porcine brain was polymerized at 37°C in a polymerization buffer [80 mM Pipes-NaOH (pH 6.8, 3 mM MgCl2, 1 mM EGTA, 1 mM guanosine 5′-triphosphate (GTP), and 10% dimethyl sulfoxide; Nacalai Tesque, Kyoto, Japan; catalog no. 09659-14] for 60 min and then diluted 22-fold with 20 μM taxol (Sigma-Aldrich, St. Louis, MO, USA; catalog no. T1912) containing BRB80 buffer. After washing three times with BRB80 buffer at 37°C for 5 min, the membranes were fixed with 10% trichloroacetic acid in Milli-Q deionized water (Merck Millipore, Burlington, MA, USA) at 4°C for 10 min and then washed three times with tris-buffered saline [10 mM tris-HCl (pH 7.5) and 150 mM NaCl] at room temperature for 5 min each. Proteins binding to polymerized MTs were detected using mouse anti–α-tubulin antibodies (Sigma-Aldrich, St. Louis, MO, USA; catalog no. T9026; RRID: AB_477593), followed by biotin-conjugated anti-mouse immunoglobulin G (IgG) antibodies (Vector Laboratories, Burlingame, CA, USA; catalog no. BA-9200; RRID: AB_2336171) and alkaline phosphatase–conjugated streptavidin (Promega, Madison, WI, USA; catalog no. V5591) using nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate visualization (Thermo Fisher Scientific, Waltham, MA, USA; catalog no. 34042). The corresponding single band at above 180 kDa in the silver-stained polyacrylamide gel was cut, and its amino acid sequence was determined through Edman degradation.
SDS-PAGE immunoblotting
Samples were mixed with 1×, 2×, or 6× SDS-sample buffer [final concentration: 50 mM tris-HCl (pH 6.8), 2% SDS, 10% glycerol, 2% β-mercaptoethanol, and 0.02% bromophenol blue]; boiled at 98°C for 10 min; separated on 7.5, 12.5, 4 to 7.5, or 10 to 20% polyacrylamide gels; and transferred to PVDF membranes. The PVDF membranes were blocked with 5% skim milk or 5% BSA for 30 min, probed with primary antibodies at room temperature for 60 to 120 min, and then incubated with horseradish peroxidase–conjugated secondary antibodies at room temperature for 30 to 60 min. Immunoblotted bands were visualized with enhanced chemiluminescence (Merck Millipore, Burlington, MA, USA; catalog no. WBKLS).
Immunofluorescence
Cultured cells plated on glass coverslips or transwell filters were fixed with cold methanol at −20°C for 10 min or fixed with 1% formaldehyde (Nacalai Tesque, Kyoto, Japan; catalog no. 16223-55)–containing HBS at room temperature for 8 min. For whole-mount trachea preparations, the trachea was longitudinally opened along the ventral midline and fixed with 4% paraformaldehyde (PFA; Nacalai Tesque, Kyoto, Japan; catalog no. 02890-45)–containing HBS at room temperature for 2 hours. For neural tube immunostaining, E9.5 mouse embryos were collected, fixed with 4% PFA-containing HBS at 4°C for 24 hours, cryoprotected with 10 to 30% sucrose, embedded in optimal cutting temperature (O.C.T.) compound (Sakura Finetek Japan, Tokyo, Japan; catalog no. 4583), frozen in liquid N2, and cut into approximately 7-μm-thick sections. For immunostaining of the stomach, the stomach was removed from 4-week-old mice, embedded in O.C.T. compound, frozen in liquid N2, and cut into approximately 7-μm-thick sections. The samples were treated with 0.25% Triton X-100 (Nacalai Tesque, Kyoto, Japan; catalog no. 12967-45) in HBS at room temperature for 5 min and washed three times with HBS. After soaking in HBS containing 1% BSA at room temperature for 10 min, the samples were treated with primary antibodies, such as mouse monoclonal anti–ZO-1, guinea pig polyclonal anti-Cobl, rat monoclonal anti-occludin, and rat polyclonal anti-ROCK1 antibodies (31–34), at room temperature for 60 to 180 min, washed three times with HBS, and incubated with secondary antibodies at room temperature for 60 min [in some experiments, rhodamine-phalloidin (Thermo Fisher Scientific, Waltham, MA, USA; catalog no. R415, RRID: AB_2572408) was added to detect F-actin]. The samples were washed three times with HBS, briefly soaked with Milli-Q deionized water, and mounted with Faramount Mounting Medium (Agilent, Santa Clara, CA, USA; catalog no. S3025). Immunofluorescent micrographs were taken using a spinning disk confocal superresolution microscope (SpinSR10; Olympus, Tokyo, Japan) equipped with silicon oil-immersion objective lenses (UPLSAPO40XS, UPLSAPO60XS2, or UPLSAPO100XS; Olympus, Tokyo, Japan), a CSU-W1 SoRa-Unit (Yokogawa, Tokyo, Japan), an ORCA-Flash4.0 V3 digital CMOS camera (Hamamatsu Photonics, Shizuoka, Japan), and an incubation chamber (37°C, 5% CO2, 85% humidity; Tokai Hit, Shizuoka, Japan). Information for the antibodies used in this study is supplied in table S1.
Recombinant DNA plasmid construction
Mouse Cobl cDNA was amplified using polymerase chain reaction (PCR) from a cDNA library derived from mouse brain (Takara Bio, Shiga, Japan; catalog no. 9528). Mouse Cobl cDNA was subcloned into the pCAGGS-Venus vector (provided by A. Miyawaki, RIKEN Brain Science Institute, Saitama, Japan), pCAGGS-FLAG vector (provided by A. Kubo, Keio University, Tokyo, Japan), pCAGGS-EGFP vector, and pmCherry-C1 vector (Takara Bio, Shiga, Japan; catalog no. 632524) using In-Fusion cloning (Takara Bio, Shiga, Japan; catalog no. 638909) to generate pCAGGS-Venus-CoblFL, pCAGGS-FLAG-CoblFL, pCAGGS-EGFP-CoblFL, and pmCherry-CoblFL, respectively. EGFP-CoblFL and mCherry-CoblFL, which were cloned from pCAGGS-EGFP-CoblFL and mCherry-CoblFL, respectively, were subcloned into the pCAGGS-FLAG vector using In-Fusion cloning to generate pCAGGS-FLAG-EGFP-CoblFL and pCAGGS-FLAG-mCherry-CoblFL, respectively. pEGFP-CoblD1, pEGFP-CoblD2, pEGFP-CoblD3, pEGFP-CoblD4, and pEGFP-CoblD5 were provided by M. M. Kessels and B. Qualmann (Jena University Hospital, Jena, Germany) (32). CoblD5, CoblΔD5, and CoblΔ84, which were cloned from pCAGGS-Venus-CoblFL, were subcloned into the pCAGGS-Venus vector to obtain pCAGGS-Venus-CoblD5, pCAGGS-Venus-CoblΔD5, and pCAGGS-Venus-CoblΔ84, respectively. CoblΔ84 was also subcloned into the pCAGGS-EGFP vector followed by the pCAGGS-FLAG vector to obtain pCAGGS-FLAG-EGFP-CoblΔ84. The EB1 gene, which was cloned from RFP-EB1 (provided by Y. Mimori-Kiyosue, RIKEN Center for Biosystems Dynamics Research, Hyogo, Japan), was subcloned into the pCAGGS-Venus vector using In-Fusion cloning to generate pCAGGS-Venus-EB1. pX330-U6-chimeric_BB-CBh-hSpCas9 was provided by F. Zhang [Broad Institute of Massachusetts Institute of Technology (MIT) and Harvard, Cambridge, MA, USA] through Addgene (Addgene, plasmid #42230) (35). The construction of pX330-U6-chimeric-CoblKO1 and pX330-U6-chimeric-CoblKO2 is described below. Information for recombinant DNAs used in this study is listed in table S2. Sequence information for oligonucleotides generated in this study is provided in table S3.
Transient and stable transfection
Transient transfection of each DNA construct was performed using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA; catalog no. L3000001) for Eph4 cells and PEI Max (Polysciences, Warrington, PA, USA; catalog no. 24765-1) for HEK293 cells following the manufacturer’s instructions. To establish stable transfectants, the transfected Eph4 cells were selected by incubation in medium containing G418 (500 μg/ml; Nacalai Tesque, Kyoto, Japan; catalog no. 108321-42-2), and cell clones derived from single cells were picked up.
Live imaging of cells
Eph4 and HEK293 cells transiently expressing Venus-EB1, mCherry-ZO-1 (36), or EGFP-CoblFL were grown on 35-mm glass-bottom dishes (AGC Techno Glass, Shizuoka, Japan; catalog no. 3961-035), preincubated in FluoroBrite DMEM (Thermo Fisher Scientific, Waltham, MA, USA; catalog no. A1896701) supplemented with 10% FBS and 2 mM l-glutamine at 37°C under a 5% CO2 atmosphere for 1 hour, and processed for live imaging. Cells were observed for the indicated times using a spinning disk confocal superresolution microscope (SpinSR10; Olympus, Tokyo, Japan) with a silicon oil-immersion objective lens (UPLSAPO100XS; Olympus, Tokyo, Japan).
Immunoelectron microscopy
Cells were fixed in 4% formalin for 8 min at room temperature, followed by treatment with 0.05% saponin in HBS for 10 min. After blocking for 30 min in blocking buffer containing 0.05% saponin in Block Ace (DS Pharma Biomedical Co. Ltd., Osaka, Japan), the cells were washed in HBS and incubated with the mixture of rabbit anti-GFP antibody (Thermo Fisher Scientific, Waltham, MA, USA; catalog no. A-6455; RRID: AB_221570) and Can Get Solution A (TOYOBO, Osaka, Japan; catalog no. NKB-501) at 37°C for 60 min. After three washes in HBS, the cells were incubated with the secondary antibody, FluoroNanogold anti-rabbit IgG (Nanoprobes, Yaphank, NY, USA; catalog no. 7404-0.5ML), and mixed with an equal volume of Can Get Solution A at room temperature for 2 hours. The cells were fixed in a solution containing 1% glutaraldehyde in 0.1 M Hepes buffer (pH 7.4) at room temperature for 1 hour. After two washes in 0.1 M Hepes buffer (pH 7.4) for 5 min, two washes in 50 mM Hepes buffer (pH 5.8) for 5 min, and two washes in Milli-Q water, the cells were mounted in reagent from the HQ Silver Enhancement Kit (Nanoprobes, Yaphank, NY, USA; catalog no. 2012-45ML), fixed with 1% OsO4 in 0.1 M Hepes buffer (pH 7.4) on ice for 1 hour, washed in Milli-Q water three times, incubated in 0.5% uranyl acetate in Milli-Q water, dehydrated, and embedded in Poly/Bed 812 (Polysciences, Warrington, PA, USA; catalog no. 08791-500).
Immunoprecipitation of Cobl-KO cells transfected with EGFP-tag or EGFP-Cobl
Cobl-KO Eph4 epithelial cells transfected with pCAGGS-EGFP or pCAGGS-EGFP-CoblFL on three 10-cm dishes were washed three times with HBS-taxol [20 mM Hepes-NaOH (pH 7.4), 140 mM NaCl, 1 mM CaCl2, and 1 mM taxol]. Thereafter, the cells were scraped with 300 μl of HBS-taxol, homogenized (4000 rpm × 30 strokes with a tight-fitting Dounce homogenizer), and centrifuged (20,400g at 28°C) for 10 min. The supernatant was incubated with 30 μl of protein A-Sepharose bead slurry (GE Healthcare, Chicago, IL, USA; catalog no. 17-0780-01) conjugated in advance with mouse anti-GFP antibodies (Thermo Fisher Scientific, Waltham, MA, USA; catalog no. A-11120; RRID: AB_221568) or total mouse IgG (Southern Biotech, Birmingham, AL, USA; cat. no. 0107–01; RRID: AB_2732898) as a control at room temperature for 120 min. After five washes with HBS-taxol, the immunoprecipitation beads were subjected to SDS-PAGE and immunoblotting.
Quantification of colocalization using immunostained micrographs
We obtained individual cell-outlined binary images of ZO-1–immunostained micrographs using a plugin in ImageJ (Automated Multicellular Tissue Analysis) developed by the Advanced Digital Microscopy Core Facility at the Institute for Research in Biomedicine (Barcelona, Spain; freely available at http://adm.irbbarcelona.org). Then, we dilated the lines outlining each cell using “dilate” function in ImageJ to establish regions of interest (ROIs) representing CR regions. Last, we calculated Pearson’s correlation coefficients within each ROI of the CR region using the “JACoP” plugin (freely available at https://imagej.net/plugins/jacop) in ImageJ.
FRAP and half FRAP analyses
For FRAP analyses of Cobl condensates in cells, Eph4 and HEK293 cells transiently expressing EGFP-CoblFL were grown on 35-mm glass-bottom dishes, preincubated in FluoroBrite DMEM supplemented with 10% FBS and 2 mM l-glutamine at 37°C under a 5% CO2 atmosphere for 1 hour, and processed for FRAP analyses. For FRAP and half FRAP analyses of Cobl condensates in vitro, FLAG-EGFP-CoblFL proteins were purified as described below. FRAP and half FRAP analyses were performed using a confocal laser scanning microscope (LSM 880; Carl Zeiss, Oberkochen, Germany) with a Plan-Apochromat 63×/1.4 Oil DIC M27 objective lens (Carl Zeiss, Oberkochen, Germany).
Purification of Cobl proteins
HEK293 cells (70% confluent, grown on 10-cm dishes) were transfected with pCAGGS-FLAG-CoblFL, pCAGGS-FLAG-CoblD1, pCAGGS-FLAG-CoblD2, pCAGGS-FLAG-CoblD3, pCAGGS-FLAG-CoblD4, pCAGGS-FLAG-CoblD5, pCAGGS-FLAG-EGFP-CoblFL, pCAGGS-FLAG-EGFP-CoblD5, pCAGGS-FLAG-EGFP-CoblΔ84, or pCAGGS-FLAG-mCherry-Cobl (10 μg per dish) using PEI MAX in accordance with the manufacturer’s instructions. At 72 hours after transfection, the cells in each dish were scraped with 200 μl of ice-cold HBS, homogenized (4000 rpm × 30 strokes with a tight-fitting Dounce homogenizer), and centrifuged (20,400g at 4°C) for 30 min. The supernatant was incubated with 100 μl of anti-Flag M2 affinity gel beads (Sigma-Aldrich, St. Louis, MO, USA; catalog no. A2220) at 4°C for 4 hours. After incubation, the beads were washed 10 times with ice-cold HBS and eluted with 3× FLAG peptide (Sigma-Aldrich, St. Louis, MO, USA; catalog no. F4799) in ice-cold Milli-Q deionized water or G buffer [2 mM tris-HCl (pH 8.0), 0.1 mM CaCl2, 0.2 mM adenosine 5′-triphosphate (ATP), 0.5 mM dithiothreitol (DTT; Nacalai Tesque, Kyoto, Japan; catalog no. 14128-46), if protein was used in subsequent cosedimentation assays, TIRF assays, and pyrene actin polymerization assays], following the manufacturer’s instructions. Protein solutions were dialyzed in a cellulose dialysis membrane soaked in ice-cold Milli-Q deionized water or G buffer (if protein was used in subsequent cosedimentation assays, TIRF assays, and pyrene actin polymerization assays) to remove the 3× FLAG peptide at 4°C overnight and ultracentrifuged at 300,000g at 4°C for 20 min. The purity of the supernatant was assessed using SDS-PAGE and Coomassie brilliant blue (CBB) staining.
Counting of bleaching steps
Cover glasses (24 mm by 32 mm; Matsunami Glass, Osaka, Japan; catalog no. C024321) were cleaned by four rounds of 30-min sonication in 0.5 M KOH at room temperature, followed by sonication for 30 min in 0.5 M HCl, two rounds of 30-min sonication in Milli-Q deionized water, and two rounds of 30-min sonication in absolute ethanol. Ten microliters of purified CoblFL and CoblD5 proteins in Milli-Q deionized water (final concentration in 20 μl; 5 nM) was mixed with 5 μl of BRB80 with taxol (BRB80T) buffer [BRB80T; 80 mM Pipes-NaOH (pH 6.9), 1 mM MgCl2, 1 mM EGTA, 1 mM GTP, and 1 mM taxol], and 5 μl of 4× LLPS buffer that did not contain PEG [final concentration in 20 μl: 20 mM Hepes-NaOH (pH 7.4) and 150 mM NaCl] was placed onto the prewashed cover glass (24 mm by 32 mm) and sandwiched with a flint glass slide (26 mm by 76 mm; Matsunami Glass, Osaka, Japan; catalog no. S024410). TIRF time-lapse observation was conducted using an Eclipse Ti inverted microscope (Nikon, Tokyo, Japan) equipped with an oil-immersion objective lens (CFI Apochromat TIRF 100XC Oil; Nikon, Tokyo, Japan), an Ixon3 electron multiplying charge coupled device (EMCCD) camera (Andor, Belfast, UK), and a Nikon perfect focus system. The point where fluorescence intensity suddenly decreased was defined as a bleaching step, and the number of bleaching steps was counted manually.
In vitro LLPS assay
In vitro LLPS assays were conducted in a total volume of 60 μl. Thirty microliters of FLAG-EGFP-CoblFL protein in Milli-Q deionized water (final concentration in 60 μl: 10, 50, 100, 200, 400, 500, or 1000 nM) was mixed with 15 μl of MTs in BRB80T buffer (final concentration in 60 μl: 0 or 8 μM) and 15 μl of 4× LLPS buffer [final concentration in 60 μl: 20 mM Hepes-NaOH (pH 7.4), NaCl (150, 300, or 500 mM), and 3% PEG] and incubated for 30 min at temperatures described in the figure legends. These solutions were placed onto cover glasses (24 mm by 32 mm), sandwiched with a flint glass slide (26 mm by 76 mm), and sealed with a transparent manicure to prevent evaporation. Thereafter, micrographs were taken using a fluorescence microscope (BX51 or BX53; Olympus, Tokyo, Japan) or a spinning disk confocal superresolution microscope (SpinSR10; Olympus, Tokyo, Japan).
Coimmunoprecipitation
HEK293 cells were transiently transfected with 3 μg of pCAGGS-EGFP-CoblFL, pEGFP-CoblD1, pEGFP-CoblD2, pEGFP-CoblD3, pEGFP-CoblD4, and pEGFP-CoblD5 on 6-cm dishes. Cells were then washed three times with HBS-taxol and scraped into 300 μl of radioimmunoprecipitation assay (RIPA) buffer containing 1 mM taxol [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% Nonidet P-40 (Nacalai Tesque, Kyoto, Japan; catalog no. 9016-45-9), 2 mM Na3VO4, 50 mM NaF, and 1 mM taxol]. The cell lysate was clarified using centrifugation (20,400g, 28°C, and 10 min), incubated with 20 μl of protein A-Sepharose bead slurry, and conjugated in advance with mouse anti-GFP antibodies or total mouse IgG as a control at room temperature for 120 min. After five washes with RIPA buffer, the immunoprecipitation beads were subjected to SDS-PAGE and immunoblotting.
Low-angle shadowing EM
A 1:1 mixture of MTs in BRB80T buffer (final concentration: 4 μM) and FLAG-CoblFL in Milli-Q deionized water (final concentration: 100 nM) was mixed with an equal amount of glycerol, sprayed on mica, and dried under a high vacuum. Then, the samples were shadowed with platinum, and the replica of the molecules was imaged using a JEM1400 Plus electron microscope (JEOL, Tokyo, Japan).
AF cosedimentation assay
Cosedimentation assays were conducted in a total volume of 10 μl. Specifically, 7.5 μl of FLAG-CoblD1, FLAG-CoblD2, FLAG-CoblD3, FLAG-CoblD4, FLAG-CoblD5, and FLAG-CoblFL (final concentration in 10 μl: 5 μM) were mixed with 2.5 μl of phalloidin-stabilized AFs (final concentration in 10 μl: 5 μM) in a reaction buffer [15 mM Hepes-NaOH (pH 7.4), 200 mM NaCl, 25 mM KCl, 0.75 mM EGTA, 500 μM MgCl2, 50 μM ATP, and 0.75 mM DTT]. After 30-min incubation at 25°C, samples were ultracentrifuged at 50,000g at 25°C for 10 min in a TLA-100 rotor (Beckman, Brea, CA, USA; catalog no. #343840). Then, the supernatants and pellets were subjected to SDS-PAGE and CBB staining.
Generation of Cobl-KO, REV, CoblD5-REV, CoblΔD5-REV, and CoblΔ84-REV cells
The target sequence, as determined using CRISPRdirect (freely available at https://crispr.dbcls.jp/), was 5′-GGACGCGCCGCGTGCACTGGCGG-3′ and 5′-GCCCGTGCTCCGCCACCTCCTGG-3′. After the oligo 5′-caccGGACGCGCCGCGTGCACTGG-3′ was annealed with 5′-aaacCCAGTGCACGCGGCGCGTCC-3′ and 5′-caccGCCCGTGCTCCGCCACCTCC-3′ was annealed with 5′-aaacGGAGGTGGCGGAGCACGGGC-3′, the annealed oligos were subcloned into the Bbs I site of the pX330-U6-Chimeric_BB-CBh-hSpCas9 plasmid using Bbs I (New England Biolabs, Ipswich, MA, USA; catalog no. R0539) and T4 DNA ligase (Promega, Madison, WI, USA; catalog no. M1801) to generate pX330-U6-chimeric-CoblKO1 and pX330-U6-chimeric-CoblKO2 plasmids. Eph4 cells were transfected with pX330-U6-chimeric-CoblKO1 or pX330-U6-chimeric-CoblKO2 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA; catalog no. 11668019). Stable transfectants (Cobl-KO cells) were established as described above. Stable transfectants of pCAGGS-Venus-CoblFL, pCAGGS-Venus-CoblΔD5, pCAGGS-Venus-CoblD5, and pCAGGS-Venus-CoblΔ84 in Cobl-KO cells were subsequently established and designated as REV cells.
Calculation of persistence length and CR thickness using immunostained micrographs
For calculation of persistence length, contours between two cells were manually traced using ZO-1–immunostained micrographs of WT, Cobl-KO, and REV cells. Using Easyworm (freely available at www.chibi.ubc.ca/faculty/joerg-gsponer/gsponer-lab/software/easyworm/), a free software suite coded in MATLAB (MathWorks, Natick, MA, USA), each cell contour was fitted to parametric splines, and the persistence length (P) of each cell contour was calculated using an end-to-end distance (R) and a distance along the chain contour (L) with the formula
To calculate the thickness of CRs, a line perpendicular to each phalloidin-delineated cell-cell boundary was drawn. The signal intensity along the line was plotted using the “plot profile” function in Image J and fitted to Gaussian curves using the “Curve fitter” in Image J. The full width at half maximum of the fitted Gaussian curve was designated as the thickness of CRs. For calculation of the apical surface area, we first obtained the ROI of each cell using ZO-1–immunostained micrographs and the “Automated Multicellular Tissue Analysis” plugin (freely available at http://adm.irbbarcelona.org) in Image J. Thereafter, the area within each ROI was calculated using the “Measure” function in Image J. For calculation of the mean fluorescent intensity within the CRs of each cell, we obtained the ROIs of CRs by using the Automated Multicellular Tissue Analysis plugin and “Dilate” function in ImageJ. We then obtained the mean intensity of a protein of interest in each ROI to quantify the mean fluorescent intensity within CRs of individual cells.
EM analysis of the CR
CR thickness was analyzed using backscattered EM (BSE-SEM). The samples were fixed with 2.5% (w/v) glutaraldehyde for at least 48 hours. The fixed specimens were dehydrated with a dilution series of ethanol (65, 75, 85, 95, 99, and 100%) and propylene oxide and embedded in epoxy resin (Nacalai Tesque, Kyoto, Japan; catalog no. LUVEAK-812). The blocks were sectioned using an ultramicrotome (ARTOS 3D; Leica, Wetzlar, Germany) equipped with a diamond knife (SYM jumbo, 45°; SYNTEK, Kanagawa, Japan) to cut 100-nm serial sections. Serial sections were collected on a cleaned silicon wafer strip held by a micromanipulator (NARISHIGE, Tokyo, Japan; catalog no. MN-153) for BSE-SEM. The sections were then stained at room temperature using 2% (w/v) aqueous uranyl acetate (20 min) and Reynolds’ lead citrate (3 min) and imaged by SEM (JEOL, Tokyo, Japan; catalog no. MN-153). CR was manually traced, and thickness was quantified.
TER calculation of Eph4 cells
WT, Cobl-KO, and REV Eph4 cells were seeded at 1 × 105 cells/cm2 onto 12-mm-diameter transwell filters. Every day, the culture medium was replaced, and TER (in ohm) was measured using a Millicell-ERS epithelial volt-ohmmeter (Merck Millipore, Burlington, MA, USA; catalog no. MERS00002). TER (in ohm × cm2) was then calculated by subtracting the background value of blank filters from TER (in ohm), followed by multiplication of the surface area (1.13 cm2) of the filter.
Flux measurement of Eph4 cells
WT, Cobl-KO, or REV Eph4 cells were seeded at 1 × 105 cells/cm2 onto transwell filters (1.13-cm2 surface area, 0.4-μm pore size; Greiner Bio-One, Kremsmünster, Austria; catalog no. 665641) and were cultured for 9 days. Flux measurements of the cells were then performed using an Ussing chamber with a 5-mm-diameter pore size. The cell layer on the transwell filter was placed into the chamber, and the basal side of the chamber was filled with 4 ml of a solution [10 mM Hepes-NaOH (pH 7.4), 120 mM NaCl, 5 mM KCl, 10 mM NaHCO3, 2 mM CaCl2, 1 mM MgCl2, and 10 mM glucose]. The apical side of the chamber was filled with 4 ml of the solution containing fluorescent dyes either fluorescein sodium salt [molecular mass, 376.27 Da; Sigma-Aldrich, St. Louis, MO, USA; catalog no. F6377], FD4 (molecular mass, 3000 to 4000 Da; Sigma-Aldrich, St. Louis, MO, USA; catalog no. FD4-100MG), or FD40 (molecular mass, 40,000 Da; Sigma-Aldrich, St. Louis, MO, USA; catalog no. FD40S-1G), at a final concentration of 500 μM. The temperature was maintained at 37°C, and 100% O2 was bubbled through the solution. Thirty minutes later, 200 μl of the solution on the basal side of chamber was collected, and the fluorescence of the collected sample was determined using the SpectraMax i3x (Molecular Devices, San Jose, CA, USA) microplate reader at 485-nm excitation and 535-nm emission. For quantitative analyses, the fluorescence of a dilution series of fluorescent dyes of known concentrations was also measured.
Generation of Cobl gene-edited mice
CrisprRNAs (crRNAs) were designed using the CRISPRdirect website. The following sequences were used for crRNA synthesis (FASMAC, Kanagawa, Japan): crRNA #1(3 to 25) of Cobl, GGACGCGCCGCGTGCACTGGcgg; crRNA #2(55 to 77) of Cobl, GCCCGTGCTCCGCCACCTCCtgg; crRNA #3(10 to 32) of Cobl, GGCTTGGCCGCCAGTGCACGcgg; and crRNA #4(19 to 41) of Cobl, CTGGCGGCCAAGCCCCCCACagg. The crRNAs, trans-activating crRNA, and Cas9 protein were co-microinjected into C57BL/6J fertilized eggs, which were transferred into surrogate female mice. Correct targeting was confirmed by reading the sequences of the PCR products in the target region. The founder mice were further bred to establish the mouse lines.
Electrophysiological assays of the stomach
T.F.D prepared 4-week-old WT and CoblΔ84/Δ84 male C57BL/6J mice. We used all male mice available at the time of the experiment, and no data were excluded. A.T., blinded to the genetic modification status, conducted electrophysiological assays of the stomach using mucosal scrapes. For obtaining mucosal scrape samples, the mucosal layer of the stomach was scraped off from the inner surface of the stomachs of mice with a razor blade. All electrophysiological assays of the stomach were then conducted using a two-chamber apparatus as detailed previously (37). The sample was mounted as a flat sheet between two chambers, with an exposed area of 0.03 cm2. The chambers were stirred, and 100% O2 was bubbled through the solution. The transepithelial (i.e., transmucosal) potential was measured through 3 M KCl-agar bridges connected to calomel electrodes using a voltage-clamping device (CEZ9100; Nihon Kohden, Tokyo, Japan). After filling two chambers each with 5 ml of solution A (SolA) [10 mM Hepes-NaOH (pH 7.4), 150 mM NaCl, 2 mM CaCl2, 1 mM MgCl2, and 10 mM mannitol], we measured the resistance potential difference (in mV) when a current of 10 μA was applied across the stomach for TER (in ohm × cm2) measurement. For measurement of the dilution potential, we filled both sides of a chamber with SolA and then replaced the buffer at the apical side with solution B [10 mM Hepes-NaOH (pH 7.4), 75 mM NaCl, 2 mM CaCl2, 1 mM MgCl2, and 160 mM mannitol] and measured changes in the potential across the stomach. To measure the paracellular permeability for Lucifer Yellow (molecular mass, 457.25 Da; Nacalai Tesque, Kyoto, Japan; catalog no. 20758-22), we filled two chambers each with 3 ml of SolA, added Lucifer Yellow to the chamber at the basal side at a final concentration of 0.5 mg/ml, and measured the amount of Lucifer Yellow that had moved through the chamber at the apical side after 60 min.
Histological analysis of the stomach
T.F.D. prepared 7-week-old WT and CoblΔ84/Δ84 male C57BL/6J mice. We used all mice available at the time of the experiment, and no data were excluded. T.F.D. also conducted H&E staining and immunofluorescence staining of the stomach as previously described (37). A.T., blinded to the genetic modification status, counted the number of inflammatory cells using H&E-stained micrographs.
Quantification of proteins obtained from stomach tissues
For protein quantification of the stomach, we used mucosal scrapes because we focused on the epithelial cells in this study. Mucosal scrape samples were then subjected to SDS-PAGE immunoblotting as described above.
Quantitative real-time PCR
For quantitative real-time PCR, we used the entire stomach. Total RNA from the stomach was extracted using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA; catalog no. 15596-018) following the manufacturer’s instructions. cDNAs were then synthesized using SuperScript II reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, USA; catalog no. 18064022) according to the manufacturer’s instructions. cDNAs were mixed with SYBR Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA; catalog no. 4367659) and amplified with the QuantStudio7 Flex Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA). The threshold cycle (Ct) values were determined with QuantStudio Design and Analysis Software 2.4.3 (Thermo Fisher Scientific, Waltham, MA, USA). The expression levels of target genes were standardized to that of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) using the delta-CT method. The relative expression levels compared with those in WT mice were determined using the delta-delta-CT method.
Actin polymerization cosedimentation assay
Actin was prepared from the acetone powder of rabbit skeletal muscle by using a cycle of polymerization and depolymerization and then stored in G buffer at 4°C. Actin polymerization cosedimentation assays were conducted in a total volume of 50 μl. Actin monomer (final concentration: 1 μM) was mixed with 7.5 μl of FLAG-CoblFL in G buffer (final concentration in 50 μl: 0 or 100 nM), 12.5 μl of MTs in BRB80T buffer (final concentration in 50 μl: 0, 4, 6, or 8 μM), and 25 μl of 2× TIRF polymerization buffer [final concentrations in 50 μl: 80 mM Pipes-KOH (pH 6.9), 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1 mM ATP, 1 mM GTP, 25 mM DTT, 100 μM taxol, 20 μM ascorbic acid (Thermo Fisher Scientific, Waltham, MA, USA; catalog no. A5960), 0.5% (w/v) 400 cP of methylcellulose (Sigma-Aldrich, St. Louis, MO, USA; catalog no. M0262)] and polymerized at room temperature for 30 min within the chamber. Samples were then ultracentrifuged at 300,000g at 28°C for 20 min. Supernatants and pellets were subjected to SDS-PAGE and CBB staining. Densitometric quantification of CBB-stained bands was performed using the “Gel Analyzer” module in ImageJ.
TIRF AF severing and AF polymerization assays
Purified skeletal muscle actin was labeled with Alexa Fluor 488 succinimidyl ester (Thermo Fisher Scientific, Waltham, MA, USA; catalog no. A20100). The labeling ratio was estimated by measuring the absorbance at 290 nm for actin and 491 nm for Alexa Fluor 488 and was confirmed to be higher than 98%. Observation chambers were constructed from cover glasses (24 mm by 32 mm) attached to flint glass slides (26 mm by 76 mm) using a 5-mm strip of double-sided tape. Cover glasses were cleaned by four rounds of 30-min sonication in 0.5 M KOH at room temperature, followed by sonication for 30 min in 0.5 M HCl, two rounds of 30-min sonication in Milli-Q deionized water, and two rounds of 30-min sonication in absolute ethanol. Immediately before observation, the chamber was pretreated with 500 mM KCl solution containing 50 nM N-ethylmaleimide (NEM)-inactivated skeletal muscle myosin for 5 min, and unbound NEM-myosin was subsequently removed by washing with 50 mM KCl solution. TIRF AF severing and AF polymerization assays were conducted in a total volume of 50 μl. For TIRF AF severing assays, 20% Alexa Fluor 488–labeled actin (final concentration in 50 μl: 0.5 μM) was mixed with 25 μl of 2× TIRF polymerization buffer and polymerized within the observation chamber at room temperature for 15 min. After the chamber was washed with 50 μl of 1× TIRF-polymerization buffer, a mixture of 25 μl of FLAG-CoblFL in Milli-Q deionized water (final concentration in 50 μl: 0, 100, 200, or 400 nM), 12.5 μl of MTs in BRB80T buffer (final concentration in 50 μl: 0 or 400 nM), and 12.5 μl of 4× severing buffer [final concentration in 50 μl: 80 mM Pipes-KOH (pH 6.9), 50 mM KCl, 2 mM MgCl2, 1 mM ATP, 1 mM GTP, 100 μM taxol, 20 μM ascorbic acid, and 0.5% (w/v) 400 cP of methylcellulose] was applied. The chamber was sealed with transparent tape and observed at room temperature for 30 min. For TIRF AF polymerization assays, 20% Alexa Fluor 488–labeled actin in G buffer (final concentration in 50 μl: 1 μM) was mixed with 7.5 μl of FLAG-CoblFL in G buffer (final concentration in 50 μl: 0 or 100 nM), 12.5 μl of MTs in BRB80T buffer (final concentration in 50 μl: 0 or 8 μM), and 25 μl of 2× TIRF polymerization buffer and polymerized at room temperature for 15 min within the observation chamber. For the triple-color TIRF AF polymerization assay, 20% Alexa Fluor 488–labeled actin (final concentration in 50 μl: 1 μM) was mixed with 7.5 μl of mCherry-FLAG-CoblFL in G buffer (final concentration in 50 μl: 100 nM), 12.5 μl of 10% HiLyte 647–labeled MTs (Cytoskeleton Inc., Denver, CO, USA; catalog no. TL670M) in BRB80T buffer (final concentration in 50 μl: 4 μM), and 25 μl of 2× TIRF polymerization buffer and polymerized at room temperature for 15 min. TIRF time-lapse observation was conducted using an Eclipse Ti inverted microscope (Nikon, Tokyo, Japan) equipped with an oil-immersion objective lens (CFI Apochromat TIRF 100XC Oil; Nikon, Tokyo, Japan), an Ixon3 EMCCD camera (Andor, Belfast, UK), and a Nikon perfect focus system. The number of actin seeds was counted manually and plotted against time. Using a plugin for Image J that was developed by J. Kuhn (MIT, Cambridge, MA, USA), kymographs were made, and the length of each AF was measured and plotted against time.
Pyrene actin polymerization assay
Pyrene actin polymerization assays were conducted in 40 μl using pyrene-labeled rabbit muscle actin (Cytoskeleton Inc., Denver, CO, USA; catalog no. AP05). Briefly, 10% pyrene-labeled rabbit muscle actin in G buffer (final concentration in 40 μl: 1 μM) was mixed with 5 μl of FLAG-CoblFL in G buffer (final concentration in 40 μl: 0 or 100 nM), 10 μl of MTs in BRB80T buffer (final concentration in 40 μl: 0 or 8 μM), and 10 μl of 4× F buffer [final concentration in 40 μl: 20 mM tris-HCl (pH 7.5), 50 mM KCl, 2 mM MgCl2, and 1 mM ATP] and polymerized in a microplate for 50 min. The fluorescent signal (360-nm excitation and 420-nm emission) was scanned every 30 s for 50 min using a SpectraMax Gemini fluorescence spectrophotometer (Molecular Devices, San Jose, CA, USA).
Statistical analysis
Statistical analysis was conducted on the basis of at least three independent experiments from separate samples (i.e., biological replicates). The number of experiment replicates (N) for each statistical analysis is specified in each figure legend. The first author (K.T.), who is certified by the Japan Statistical Society (grade 2), performed statistical analyses using the statistical software R, version 3.6.1 (freely available at www.R-project.org). On the basis of the results obtained from Shapiro-Wilk tests, we used unpaired t tests, paired t tests, Mann-Whitney U tests, Fisher’s exact tests, and Spearman’s correlation coefficient, as appropriate. Data are presented as means ± SDs or medians (interquartile ranges). Results with a P value less than 0.05 were considered statistically significant.
Acknowledgments
We would like to thank T. Matsui for help in performing taxol-stabilized MT blot overlay assays; H. Kobayashi and T. Yagi for technical assistance with primary neuronal cultures; M. Uji and Y. Sugiyama for comprehensive technical assistance throughout this work; K. Sentani and W. Yasui for expert advice on the pathological findings in the stomach of CoblΔ84/Δ84 mice; A. Miyawaki, A. Kubo, M. M. Kessels, B. Qualmann, and Y. Mimori-Kiyosue for providing DNA plasmids and/or antibodies; and the Cooperative Research Program of Institute for Frontier Life and Medical Sciences, Kyoto University, for the generation of CoblΔ84/Δ84 mice. We would also like to thank Editage (www.editage.com) for editing and reviewing this manuscript for English language.
Funding: This work was supported by grants from Japan Society for the Promotion of Science of Japan to S.T. (Grant-in-Aid for Specially Promoted Research JP19H05468) and A.T. [Grant-in-Aid for Scientific Research (B) JP16H05121] and a grant from Japan Science and Technology Agency to S.T. [Core Research for Evolutionary Science and Technology (CREST) JPMJCR13W4].
Author contributions: Conceptualization: K.T., T.Y., A.T., and S.T. Methodology: K.T. and S.T. Investigation: K.T., M.K., H.K., T.Y., I.F., T.K., T.F.D., F.T., H.T., M.M., H.W., G.K., A.T., and S.T. Visualization: K.T., M.K., J.K., and S.P. Funding acquisition: A.T. and S.T. Project administration: A.T., and S.T. Supervision: A.T. and S.T. Writing (original draft): K.T. Writing (review and editing): M.K., R.T., A.T., and S.T.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Statistical source data and all data used to create figures have been deposited in Zenodo (https://doi.org/10.5281/zenodo.7481980) and are freely available under the Creative Commons Attribution 4.0 International license.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S13
Tables S1 to S3
Legends for movies S1 to S14
Other Supplementary Material for this : manuscript includes the following:
Movies S1 to S14
REFERENCES AND NOTES
- 1.Y. Shin, C. P. Brangwynne, Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 2.L. B. Case, X. Zhang, J. A. Ditlev, M. K. Rosen, Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 363, 1093–1097 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O. Beutel, R. Maraspini, K. Pombo-García, C. Martin-Lemaitre, A. Honigmann, Phase separation of zonula occludens proteins drives formation of tight junctions. Cell 179, 923–936.e11 (2019). [DOI] [PubMed] [Google Scholar]
- 4.F. G. Quiroz, V. F. Fiore, J. Levorse, L. Polak, E. Wong, H. A. Pasolli, E. Fuchs, Liquid-liquid phase separation drives skin barrier formation. Science 367, eaax9554 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.S. Citi, Cell biology: Tight junctions as biomolecular condensates. Curr. Biol. 30, R83–R86 (2020). [DOI] [PubMed] [Google Scholar]
- 6.S. Tsukita, M. Furuse, M. Itoh, Multifunctional strands in tight junctions. Nat. Rev. Mol. Cell Biol. 2, 285–293 (2001). [DOI] [PubMed] [Google Scholar]
- 7.D. Günzel, M. Fromm, Claudins and other tight junction proteins. Compr. Physiol. 2, 1819–1852 (2012). [DOI] [PubMed] [Google Scholar]
- 8.M. A. Odenwald, J. R. Turner, The intestinal epithelial barrier: A therapeutic target? Nat. Rev. Gastroenterol. Hepatol. 14, 9–21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.S. Citi, Intestinal barriers protect against disease. Science 359, 1097–1098 (2018). [DOI] [PubMed] [Google Scholar]
- 10.S. Tsukita, H. Tanaka, A. Tamura, The claudins: From tight junctions to biological systems. Trends Biochem. Sci. 44, 141–152 (2019). [DOI] [PubMed] [Google Scholar]
- 11.L. S. Rodgers, A. S. Fanning, Regulation of epithelial permeability by the actin cytoskeleton. Cytoskeleton (Hoboken) 68, 653–660 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.W. V. Graham, W. He, A. M. Marchiando, J. Zha, G. Singh, H.-S. Li, A. Biswas, M. L. D. M. Ong, Z.-H. Jiang, W. Choi, H. Zuccola, Y. Wang, J. Griffith, J. Wu, H. J. Rosenberg, Y. Wang, S. B. Snapper, D. Ostrov, S. C. Meredith, L. W. Miller, J. R. Turner, Intracellular MLCK1 diversion reverses barrier loss to restore mucosal homeostasis. Nat. Med. 25, 690–700 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.K. Kunimoto, Y. Yamazaki, T. Nishida, K. Shinohara, H. Ishikawa, T. Hasegawa, T. Okanoue, H. Hamada, T. Noda, A. Tamura, S. Tsukita, S. Tsukita, Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell 148, 189–200 (2012). [DOI] [PubMed] [Google Scholar]
- 14.E. Vasileva, S. Citi, The role of microtubules in the regulation of epithelial junctions. Tissue Barriers 6, 1539596 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.T. Yano, K. Tsukita, H. Kanoh, S. Nakayama, H. Kashihara, T. Mizuno, H. Tanaka, T. Matsui, Y. Goto, A. Komatsubara, K. Aoki, R. Takahashi, A. Tamura, S. Tsukita, A microtubule-LUZP1 association around tight junction promotes epithelial cell apical constriction. EMBO J. 40, e104712 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R. Ahuja, R. Pinyol, N. Reichenbach, L. Custer, J. Klingensmith, M. M. Kessels, B. Qualmann, Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell 131, 337–350 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.C. Husson, L. Renault, D. Didry, D. Pantaloni, M.-F. Carlier, Cordon-Bleu uses WH2 domains as multifunctional dynamizers of actin filament assembly. Mol. Cell 43, 464–477 (2011). [DOI] [PubMed] [Google Scholar]
- 18.R. Dominguez, The WH2 domain and actin nucleation: Necessary but insufficient. Trends Biochem. Sci. 41, 478–490 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A. C. Martin, B. Goldstein, Apical constriction: Themes and variations on a cellular mechanism driving morphogenesis. Development 141, 1987–1998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.M. L. Heuzé, G. H. N. Sankara Narayana, J. D’Alessandro, V. Cellerin, T. Dang, D. S. Williams, J. C. Van Hest, P. Marcq, R.-M. Mège, B. Ladoux, Myosin II isoforms play distinct roles in adherens junction biogenesis. eLife 8, e46599 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.W. Y. C. Huang, S. Alvarez, Y. Kondo, Y. K. Lee, J. K. Chung, H. Y. M. Lam, K. H. Biswas, J. Kuriyan, J. T. Groves, A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 363, 1098–1103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.X. Chen, F. Ni, X. Tian, E. Kondrashkina, Q. Wang, J. Ma, Structural basis of actin filament nucleation by tandem W domains. Cell Rep. 3, 1910–1920 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.N. E. Grega-Larson, S. W. Crawley, M. J. Tyska, Impact of cordon-bleu expression on actin cytoskeleton architecture and dynamics. Cytoskeleton (Hoboken) 73, 670–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.W. Hou, M. Izadi, S. Nemitz, N. Haag, M. M. Kessels, B. Qualmann, The actin nucleator Cobl is controlled by calcium and calmodulin. PLoS Biol. 13, e1002233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Y. G. Zhao, H. Zhang, Phase separation in membrane biology: The interplay between membrane-bound organelles and membraneless condensates. Dev. Cell 55, 30–44 (2020). [DOI] [PubMed] [Google Scholar]
- 26.S. U. Setru, B. Gouveia, R. Alfaro-Aco, J. W. Shaevitz, H. A. Stone, S. Petry, A hydrodynamic instability drives protein droplet formation on microtubules to nucleate branches. Nat. Phys. 17, 493–498 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.A. Hernández-Vega, M. Braun, L. Scharrel, M. Jahnel, S. Wegmann, B. T. Hyman, S. Alberti, S. Diez, A. A. Hyman, Local nucleation of microtubule bundles through tubulin concentration into a condensed tau phase. Cell Rep. 20, 2304–2312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D. Breitsprecher, R. Jaiswal, J. P. Bombardier, C. J. Gould, J. Gelles, B. L. Goode, Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science 336, 1164–1168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. L. Henty-Ridilla, A. Rankova, J. A. Eskin, K. Kenny, B. L. Goode, Accelerated actin filament polymerization from microtubule plus ends. Science 352, 1004–1009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.S. Tsukita, S. Tsukita, Isolation of cell-to-cell adherens junctions from rat liver. J. Cell Biol. 108, 31–41 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.M. Itoh, A. Nagafuchi, S. Yonemura, T. Kitani-Yasuda, S. Tsukita, S. Tsukita, The 220-kD protein colocalizing with cadherins in non-epithelial cells is identical to ZO-1, a tight junction-associated protein in epithelial cells: cDNA cloning and immunoelectron microscopy. J. Cell Biol. 121, 491–502 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.L. Schwintzer, N. Koch, R. Ahuja, J. Grimm, M. M. Kessels, B. Qualmann, The functions of the actin nucleator Cobl in cellular morphogenesis critically depend on syndapin I. EMBO J. 30, 3147–3159 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.M. Saitou, K. Fujimoto, Y. Doi, M. Itoh, T. Fujimoto, M. Furuse, H. Takano, T. Noda, S. Tsukita, Occludin-deficient embryonic stem cells can differentiate into polarized epithelial cells bearing tight junctions. J. Cell Biol. 141, 397–408 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.T. Nishimura, M. Takeichi, Shroom3-mediated recruitment of Rho kinases to the apical cell junctions regulates epithelial and neuroepithelial planar remodeling. Development 135, 1493–1502 (2008). [DOI] [PubMed] [Google Scholar]
- 35.L. Cong, F. A. Ran, D. Cox, S. Lin, R. Barretto, N. Habib, P. D. Hsu, X. Wu, W. Jiang, L. A. Marraffini, F. Zhang, Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Y. Yamazaki, R. Tokumasu, H. Kimura, S. Tsukita, Role of claudin species-specific dynamics in reconstitution and remodeling of the zonula occludens. Mol. Biol. Cell 22, 1495–1504 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D. Hayashi, A. Tamura, H. Tanaka, Y. Yamazaki, S. Watanabe, K. Suzuki, K. Suzuki, K. Sentani, W. Yasui, H. Rakugi, Y. Isaka, S. Tsukita, Deficiency of claudin-18 causes paracellular H+ leakage, up-regulation of interleukin-1β, and atrophic gastritis in mice. Gastroenterology 142, 292–304 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S13
Tables S1 to S3
Legends for movies S1 to S14
Movies S1 to S14