Abstract
Somatic activating mutations of PIK3CA are associated with development of vascular malformations (VMs). Here, we describe a microfluidic model of PIK3CA-driven VMs consisting of human umbilical vein endothelial cells expressing PIK3CA activating mutations embedded in three-dimensional hydrogels. We observed enlarged, irregular vessel phenotypes and the formation of cyst-like structures consistent with clinical signatures and not previously observed in cell culture models. Pathologic morphologies occurred concomitant with up-regulation of Rac1/p21-activated kinase (PAK), mitogen-activated protein kinase cascades (MEK/ERK), and mammalian target of rapamycin (mTORC1/2) signaling networks. We observed differential effects between alpelisib, a PIK3CA inhibitor, and rapamycin, an mTORC1 inhibitor, in mitigating matrix degradation and network topology. While both were effective in preventing vessel enlargement, rapamycin failed to reduce MEK/ERK and mTORC2 activity and resulted in hyperbranching, while inhibiting PAK, MEK1/2, and mTORC1/2 mitigates abnormal growth and vascular dilation. Collectively, these findings demonstrate an in vitro platform for VMs and establish a role of dysregulated Rac1/PAK and mTORC1/2 signaling in PIK3CA-driven VMs.
A microfluidic model of vascular malformations demonstrates clinical phenotypes and a mechanism of disease progression.
INTRODUCTION
Vascular malformations (VMs) are a class of rare genetic disorders associated with localized developmental abnormalities of venous, arterial, capillary, or lymphatic vessels (1, 2). Histologically, VMs are characterized by a complex of structural lesions with enlarged, irregular lumens that are lined with vascular endothelial cells, surrounded by uneven and disorganized extracellular matrix (ECM) (1, 3). These lesions are thought to be congenital, often progress in severity with time, are associated with vascular obstruction and impaired drainage, and can be life-threatening (1–4). Treatment for VMs, especially for complex lymphatic malformations that occur in multiple body sites such as generalized lymphatic anomaly (GLA) and Gorham-Stout disease (GSD), is difficult and rarely curative. Current interventional approaches rely on surgery, sclerotherapy (5–7), and a limited repertoire of drugs (2, 7, 8).
VMs are caused by somatic mutations in genes involved in vasculature development, and the same genes are commonly implicated in malignant tumor angiogenesis (2, 4). Specific surface receptors including vascular endothelial growth factor receptor 3, Notch, ephrin type-B receptor 4, activin receptor–like kinase 1, and endoglin receptor ligands, including bone morphogenetic protein 9, angiopoietin-2, and vascular endothelial growth factor C, and signaling proteins, including phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), Ras–guanosine triphosphatase (GTPase), and Raf proto-oncogenes, among others, have been associated with one or more types of malformations (2, 4). In particular, somatic activating “hotspot” mutations in the PIK3CA gene have been identified to be the cause of the majority (~80%) of cystic lymphatic malformations (9, 10), and several studies have further associated these mutations with GLA, capillary malformations, and venous malformations (11–15). PIK3CA encodes the catalytic p110α subunit of phosphatidylinositol-3-kinase (PI3K), a component of PI3K lipid kinases that is involved in cell proliferation, migration, contractility, survival, and metabolism regulation (16). In response to receptor tyrosine kinase signaling, PI3K catalyzes the production of phosphatidylinositol 3,4,5-bisphosphate [PtdIns(3,4,5)P3 or PIP3] in the plasma membrane (16). PIP3 subsequently recruits pleckstrin homology domain–harboring proteins including pyruvate dehydrogenase kinase 1 (PDK1), mammalian target of rapamycin complex 2 (mTORC2), and protein kinase B (PKB/AKT) to the plasma membrane (16–18), triggering phosphorylation of AKT by PDK1 and mTORC2 and activation of AKT1 and mTORC1 signaling networks (16, 18). In addition to controlling cell growth and proliferation through mTOR signaling, PI3K also regulates cell polarization and migration through interactions with transforming protein RhoA-GTPase/Rac1-GTPase signaling pathways (19, 20).
Several in vitro and in vivo animal models have been developed to study the pathobiology and to assess treatment efficacy in PIK3CA-related VMs. It has recently been shown that clonal expression of PIK3CA-activating mutation in the lymphatic endothelium promotes vessel overgrowth and the formation of VMs in mice (13–15, 21–23), demonstrating a causative role of PIK3CA-activating mutations in the pathogenesis of VMs. Characterization of these mouse- and patient-derived lymphatic endothelial cells revealed consistent molecular alterations associated with dysregulated PI3K/AKT/mTOR signaling, including inhibition of tumor suppressor tuberin, and phosphorylation and hyperactivation of AKT1 and mTORC1/2 signaling. These signaling changes are associated with increased cell size, cell proliferation, and angiogenesis (10, 15, 22). Despite these established cellular phenotypes, the respective contributions of cellular, molecular, and mechanical factors to disease progression are difficult to isolate and control in vivo, and how each of these associated signaling axes might contribute to VMs remains unclear.
Identification of altered PI3K/mTOR signaling has led to the use of mTOR and PI3K inhibitors in treating VMs. Rapamycin (sirolimus), an mTOR inhibitor, is the most commonly used drug for treating VMs (15, 24). Recent studies from animal models of PIK3CA-mutated breast cancer and PIK3CA-driven lymphatic malformations led to successful repurposing of a PIK3CA-specific inhibitor (alpelisib) in treating lymphatic malformations (23, 25). Treatment with alpelisib was shown to restore healthy cellular phenotypes and prevent development of and promote regression of vascular lesions in mouse models and in many patients (23, 25). In contrast, treatment with rapamycin may be effective at restricting the growth of lesions but appears to have limited efficacy in complete lesion remission (23, 25). Although both drugs can be effective in treating VMs, the cellular processes that govern growth inhibition and reversal of enlarged vessels remain largely unknown.
Here, we generated human umbilical vein endothelial cells (HUVECs) expressing a PIK3CA-activating mutation (PIK3CAE542K) and incorporated PIK3CAE542K HUVECs in three-dimensional (3D) vascular networks embedded in physiologic ECM within microfluidic devices. We demonstrate that mutant endothelial cells are hyperproliferative and develop vascular networks with complex, irregular, and enlarged structures, closely resembling clinical phenotypes. These phenotypes were driven by endothelial cell–autonomous hyperactivation of PI3K signaling and were associated with increased cell proliferation and matrix proteolysis in mutant vascular networks. We observed that broad-spectrum inhibition of endothelial cell proliferation and matrix proteolysis mitigated lesion formation but did not rescue alterations in vascular morphology. In agreement with clinical data, we demonstrated that alpelisib is more effective than rapamycin in preventing the development of malformed vasculature as rapamycin prevented vascular enlargement but resulted in vascular hyperbranching. We investigated the signaling effects of alpelisib inhibition and demonstrated that in contrast to rapamycin, alpelisib led to suppression of both mTORC2-dependent AKT1 phosphorylation and mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) signaling in PIK3CAE542K cells. We further identified Rac1/PAK (p21-activated kinase) and MEK/ERK activation as key effectors of PIK3CA hyperactivation and use our model to demonstrate that targeting PAK, MEK/ERK, and mTORC1/2 signaling are effective in improving malformed vessel structure.
RESULTS
Endothelial cells expressing PIK3CA-activating mutations demonstrate unique cytoskeletal phenotypes
To investigate the effects of PIK3CA-activating mutations on vascular endothelial cells, we generated HUVECs expressing PIK3CAE542K-IRES-GFP or PIK3CAE545K-IRES-GFP, PI3K-activating mutations detected in patients with VM syndromes (9–15, 26), by lentiviral transduction. As controls, we infected HUVECs with the same lentiviral vector encoding green fluorescent protein (GFP) or wild-type PIK3CA (PIK3CAWT; figs. S1 and S2). GFP expression was used to determine transduction efficiency for each cell line (fig. S2A), and we observed similar levels of p110⍺ expression in PIK3CAWT, PIK3CAE542K, and PIK3CA545K cells (fig. S1C). Western blot analysis of phospho-AKT1 (p-AKT1) and phospho-ribosomal S6 kinase (S6K) confirmed that PI3K/AKT/mTOR signaling was up-regulated in HUVECs expressing PIK3CA-activating mutations (fig. S1D). Immunostaining with vascular endothelial–cadherin (VE-cadherin) and phalloidin demonstrated distinct cytoskeletal and adhesive phenotypes, with cells expressing PIK3CAE542K or PIK3CAE545K characterized by diminished and discontinuous VE-cadherin adhesions, increased formation of actin stress fibers, and decreased cortical actin localization (fig. S1, A and B). Consistent with previous reports (21), we observed that HUVECs expressing PIK3CA mutations were larger in area than controls (fig. S1, A and B) and proliferated at an increased rate as measured by 5-ethynyl-2′-deoxyuridine (EdU) incorporation (fig. S3). While there are no substantial changes in the percent population of senescence cells in PIK3CAE542K or PIK3CAE545K when compared to controls, we observed elevated β-galactosidase activity in HUVECs that were larger in cell size (fig. S2B).
Microfluidic model recapitulates clinical phenotypes of VMs
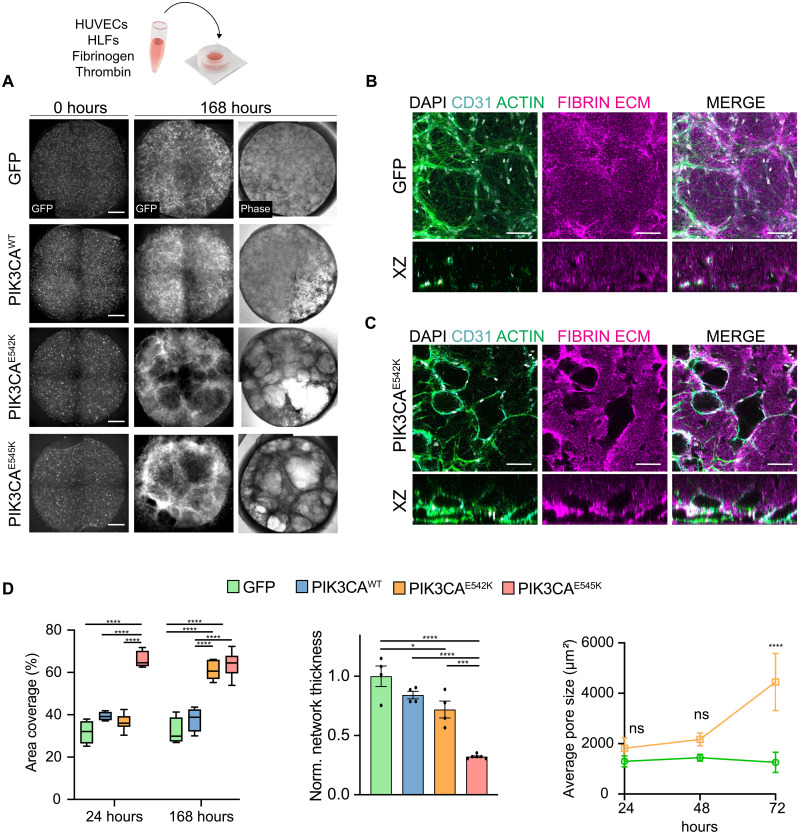
While overall these phenotypes are consistent with previous reports on the effects of expressing PIK3CA-activating mutations on the cell shape and cytoskeletal organization (21), it is unclear how these changes relate to the pathophysiology and clinical presentation of VMs, including disorganized vessel organization and cyst formation. To relate these cellular phenotypic changes to microvascular topology, we generated 3D vascular networks by coculturing HUVECs and stromal human lung fibroblasts (HLFs) within fibrin hydrogels (Fig. 1A). To visualize vascular network formation for each cell type over time, we imaged the GFP channel using laser scanning confocal microscopy every 24 hours. Notably, while GFP and PIK3CAWT cells formed interconnected networks that are well distributed in 3D, PIK3CAE542K and PIK3CAE545K cells formed irregular vascular networks with expanded vessel diameter that are akin to histological hallmarks of vascular lesions in patients (Fig. 1, A to C). The lack of organization in vascular networks was accompanied by an increase in the projected area covered by PIK3CAE542K and PIK3CAE545K cells within tissue constructs when compared to controls (Fig. 1D). Notably, when imaging the fibrin matrix, we observed the presence of cavities that enlarge over time in mutant vascular networks (Fig. 1D and fig. S4, A and B). Close inspection of spaces void of ECM revealed that these cavities were composed of dilated vessel enveloped by a continuous layer of endothelial cells (Fig. 1C). The formation of voids coincides with increased levels of secreted proteases in the conditioned media of PIK3CAE542K cells (fig. S5), implicating a role of dysregulated matrix proteolysis in mutant cells.
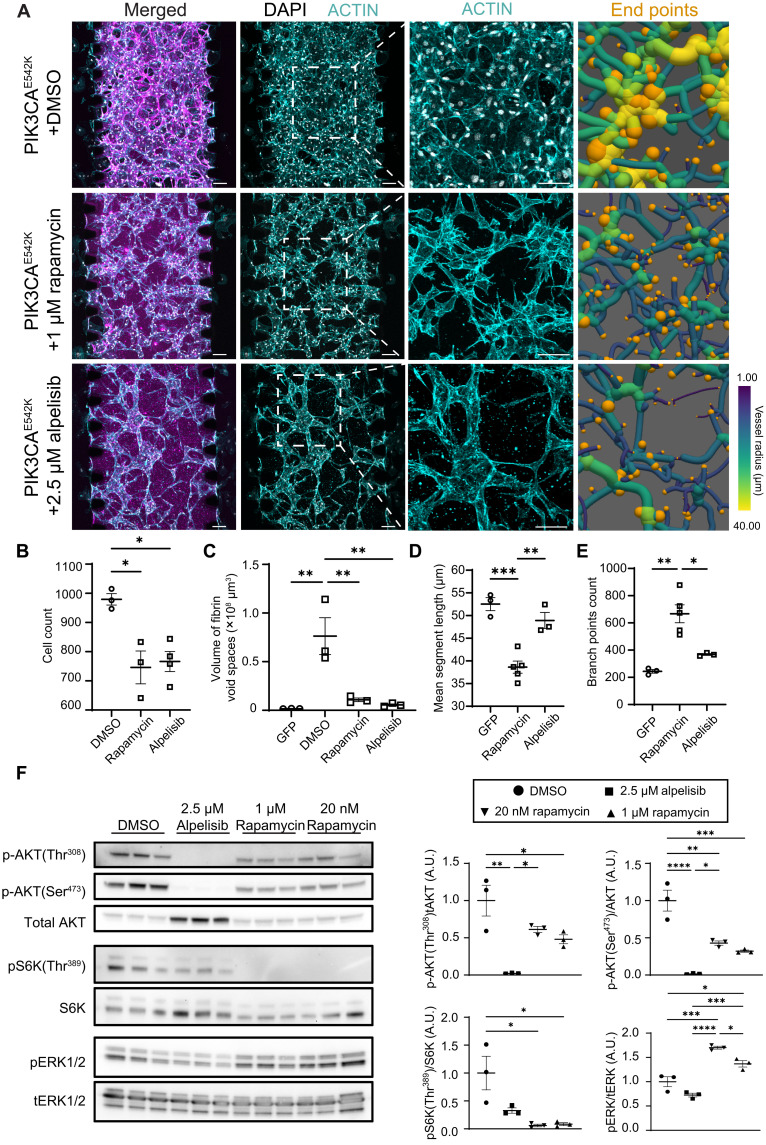
Fig. 1. 3D culture of endothelial cells expressing constitutively active PIK3CA isoforms recapitulates irregular and enlarged vessel phenotypes associated with VMs.
(A) Control cells and endothelial cells expressing PIK3CA-activating mutations cultured in 3D fibrin matrices and imaged at 0 and 168 hours after seeding. Scale bars, 1000 μm. (B and C) Representative confocal images of control and PIK3CAE542K vascular networks. Vascular networks were labeled with fibrin (magenta), 4′,6-diamidino-2-phenylindole (DAPI) (white), actin (green), and endothelial cell–specific CD31 staining (cyan). Vascular networks were fixed 3 days after seeding. Scale bars, 100 μm. (D) Quantification of images in (A). (n = 3 to 5 independent experiments; mean ± SD; one-way analysis of variance (ANOVA) with Tukey post test, *P ≤ 0.05; ***P ≤ 0.001; ****P ≤ 0.0001). ns, not significant.
Endothelial cell–autonomous processes contribute to pathologic network formation
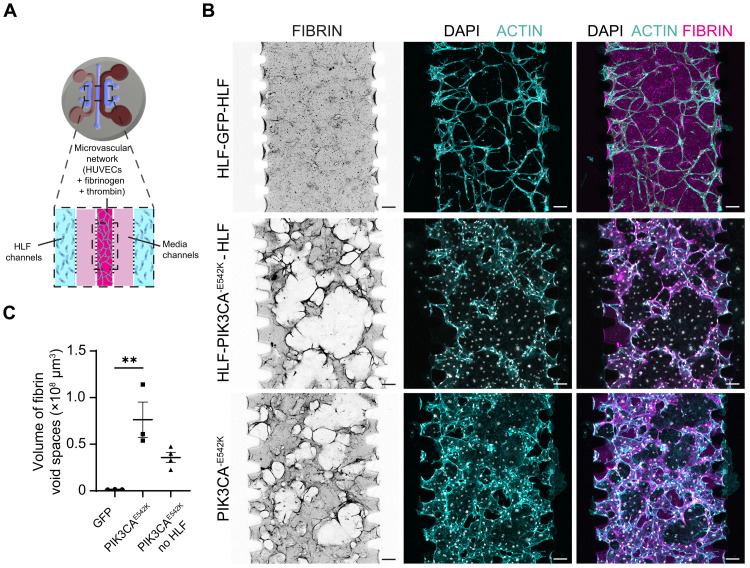
We next sought to evaluate the role of stromal-endothelial cell interactions in the development of malformed vessel structures observed in 3D culture. Using a multichannel microfluidic device (27), we seeded HUVECs and stromal fibroblasts in separate spatial compartments and quantified the effects of compartmentalized stromal-vascular network coculture on vessel morphology (Fig. 2A). Consistent with the results when stromal and endothelial cells were embedded within the same hydrogel, we observed abnormal growth and increased vessel dilation in PIK3CA-mutant vascular networks when compared to controls (Fig. 2, B and C), suggesting that direct interaction with stromal cells is not essential for initiating VMs. In addition, complete removal of stromal HLFs from the culture system modestly reduced PIK3CAE542K vascular volume but did not prevent the development of vascular lesions as quantified by the total volume of fibrin void cavities (Fig. 2, B and C, and fig. S6). Although reduction of growth factor concentration in the culture medium decreased void formation, the volume of fibrin voids remained significantly increased compared to GFP control (fig. S6). Further, we generated mosaic vascular networks by coculturing HUVECs expressing LifeAct-mRuby with GFP control or PIK3CAE542K, and we observed localized ECM remodeling and focal development of lesion only in the immediate vicinities of PIK3CA-mutant cells within mosaic vascular networks (fig. S7). Together, these data suggest that while malformation growth may be regulated by nonautonomous signaling between endothelial cells and surrounding cell types, the development of VMs are driven mostly by cell-autonomous PI3K activation in endothelial cells.
Fig. 2. PIK3CAE542K functions largely cell-autonomously in driving vascular overgrowth and vessel enlargement.
(A) Schematic of microfluidic device used for culturing endothelial cells and HLFs in separate spatial compartments. (B) Confocal images (maximum intensity projections of the bottom half of vascular networks) of GFP and PIK3CAE542K vascular networks labeled with fibrin (magenta), DAPI (white), and actin (cyan). Vascular networks were generated by coculturing endothelial cells and HLFs in different spatial compartments (HLF–endothelial cells–HLF) or by monoculture of PIK3CAE542K cells in all three compartments. Vascular networks were fixed 3 days after seeding. Scale bars, 100 μm. (C) Volumetric quantification of void spaces in fibrin hydrogel (n = 3; means ± SEM; one-way ANOVA with Tukey post test, **P ≤ 0.01).
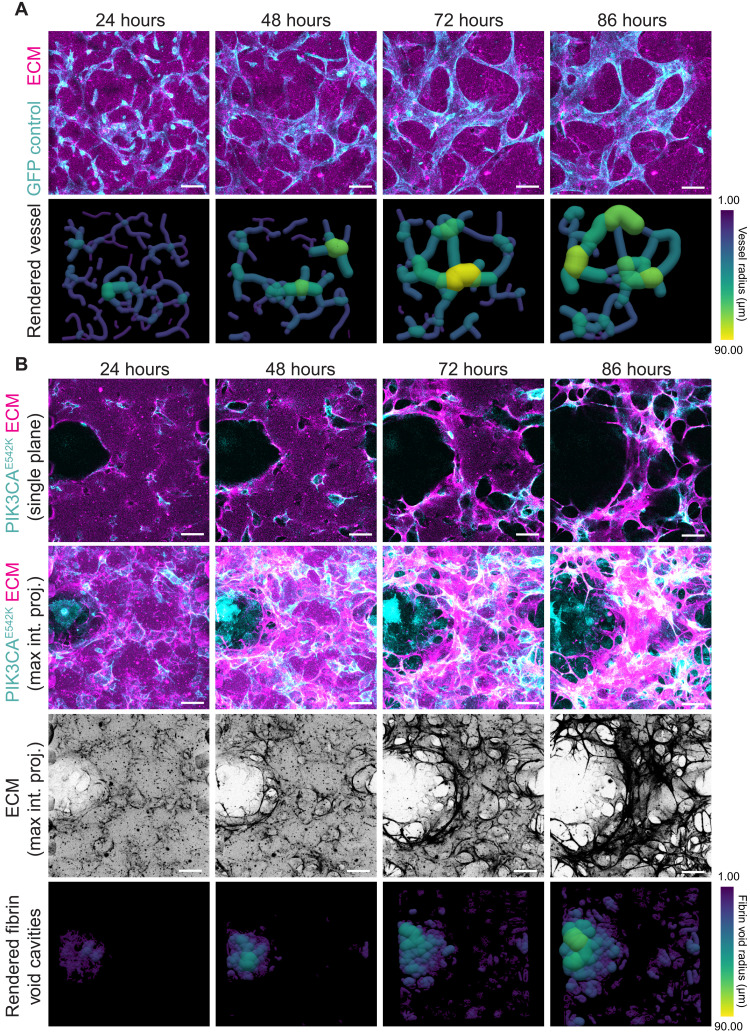
To gain further insights into the mechanisms driving vessel enlargement, we performed live cell imaging of vascular networks cultured in fluorescently labeled fibrin hydrogel. This allowed us to capture dynamics of cell-cell and cell-ECM interactions in control and PIK3CAE542K cells over an 86-hour period of vascular network formation (Fig. 3 and movie S1). Consistent with results from fixed samples, GFP cells formed an interconnected vascular network with a relatively uniform distribution of vessel diameter (Fig. 3A and movie S1). We observed local ECM remodeling as reflected by increased fibrin intensity in the vicinity of control vascular network (movie S1). In comparison to control, we found excessive ECM remodeling and degradation, which are associated with lumen expansion in PIK3CAE542K vascular networks (Fig. 3B and movie S1). We conclude from these data that PIK3CA-driven VMs result from abnormal proliferation and dysregulated matrix degradation of mutant endothelial cells.
Fig. 3. Temporal progression of VM-like phenotypes in PIK3CAE542K vascular networks.
(A) Time-lapse confocal images (maximum intensity projections) of GFP control endothelial cells (cyan) cultured in fibrin ECM (magenta) and corresponding volume rendering of segmented vascular networks. Scale bars, 100 μm. (B) Time-lapse images (single-plane and maximum intensity projections) of PIK3CAE542K cells (cyan) cultured in fibrin ECM (magenta) (top), maximum intensity projections of fibrin ECM (middle), and corresponding 3D rendering of segmented ECM void spaces. Scale bars, 100 μm.
Inhibition of cell proliferation and matrix degradation attenuates VM phenotypes
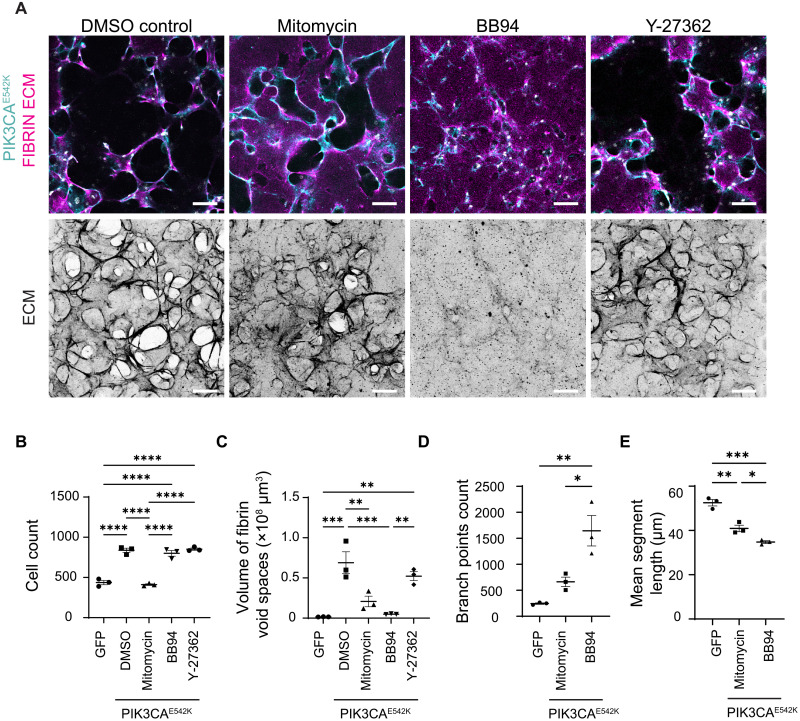
To study the role of endothelial cell proliferation in vascular enlargement, we inhibited cell proliferation by treating PIK3CAE542K cells with mitomycin C. After 3 days of culture, the number of cells within mutant vascular networks was similar to that observed in the control networks (Fig. 4, A and B). Visual examination of ECM and quantification of vascular structure showed a reduction in ECM remodeling and volume of vascular lesions with mitotic inhibition (Fig. 4, A and C). Despite the attenuated severity in ECM degradation, treatment with the mitotic inhibitor did not fully rescue the vascular topology driven by PIK3CAE542K expression (Fig. 4, A, D, and E). As compared to GFP control, mitotically inhibited PIK3CAE542K vascular networks exhibited enlarged vessels and increased vascular branching (Fig. 4, A and D). Treatment with batimastat (BB-94), a broad-spectrum metalloproteinase inhibitor, reduced fibrin void formation (Fig. 4, A to C) and did not affect cell viability (fig. S8), and similar results were observed with aprotinin treatment (fig. S4C), suggesting that endothelial cell-mediated matrix degradation drives void formation. A hyperbranching morphology similar to mitotically inhibited networks was observed upon treatment with BB-94 (Fig. 4, A to E), implying that inhibition of cell proliferation or matrix degradation individually is not sufficient to rescue healthy microvascular topology. As elevated Rho kinase signaling is implicated in the development of cerebral cavernous malformations (CCMs) (28–30), a type of VMs in the brain and spinal cord, we tested whether the development of VMs in our model required Rho kinase activity by inhibiting PIK3CAE542K cells with Y27362, a Rho kinase inhibitor. While we observed modest attenuation in ECM remodeling with Y27362 treatment, the morphologies of mutant vascular networks treated with Y27362 were indistinguishable from those of the untreated control (Fig. 4, A to C), suggesting that targeting RhoA activity and cellular contractility is not effective in preventing PIK3CA-driven VMs. Collectively, these results highlight that proliferation, matrix degradation, cytoskeletal dynamics, and tissue contractility are dysregulated in PIK3CAE542K vascular networks, but targeting them individually does not rescue the phenotype, suggesting that intervention is necessary upstream of these cellular phenotypes.
Fig. 4. Inhibition of endothelial cell proliferation and matrix proteolysis mitigates but is not sufficient to rescue malformed vascular architecture.
(A) Representative images of PIK3CAE542K vascular networks cultured for 3 days with dimethyl sulfoxide (DMSO), BB94 (1 μM), or Y-27362 (10 μM). For mitotic inhibition, PIK3CAE542K cells were pretreated with mitomycin-C (0.01 mg/ml) before culturing in fibrin hydrogel. Scale bars, 100 μm. (B to E) Cell count (B), volumetric quantification of ECM (C), vessel branching (D), and vessel segment length (E) of GFP control and PIK3CAE542K vascular networks (n = 3; means ± SEM; one-way ANOVA with Tukey post test, *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001).
Inhibition of PIK3CA rescues network topology, while inhibition of mTOR signaling results in network hyperbranching
Rapamycin and alpelisib, a PI3Kα isoform-specific inhibitor, have both been demonstrated to be effective in treating patients with PIK3CA-driven VM (15, 24, 25). Because a comparative study in a PIK3CA mouse model demonstrated that alpelisib is more effective than rapamycin in improving organ abnormalities (23), we sought to test and compare their efficacies in treating VMs in our tissue engineered human vascular network model (Fig. 5 and fig. S5, A and C). We observed reduced cell proliferation (Fig. 5B and fig. S8B) and drastic reduction in ECM proteolysis as measured by the volume of fibrin void cavities with either rapamycin or alpelisib treatments (Fig. 5C). We further observed a dose-dependent rescue of vascular network topology with treatment with alpelisib (fig. S9A). While both drugs prevented the development of vascular lesions as measured by the volume of void spaces in fibrin hydrogel (Fig. 5C), we observed significant differences in treatment responses as manifested by vascular branching patterns (Fig. 5, A, D, and E). In particular, PIK3CAE542K networks treated with rapamycin exhibited a vascular hyperbranching phenotype, as demonstrated by the formation of network with shorter vessel segments (Fig. 5, A and D) and the increased number of vessel branches and end points (Fig. 5, A and E). Consistent with a previous report (23), treatment with alpelisib effectively reduced mTORC2 activity as evidenced by reduced AKT1 phosphorylation at Ser473 by Western blot analysis, but rapamycin was not effective in inhibiting mTORC2 activity (Fig. 5F). Further, we observed reduced ERK1/2 phosphorylation in PIK3CAE542K cells treated with alpelisib, but not with rapamycin (Fig. 5F), consistent with previous results in fibroblasts and cancer cells (31) and suggesting that in addition to mTORC2 signaling, MEK/ERK activity is differentially regulated by these inhibitors. Overall, these results suggest that cross-talk between PI3K and MEK/ERK signaling may contribute to VMs.
Fig. 5. Rapamycin treatment improves VM but leads to vascular hyperbranching.
(A) Representative images (maximum intensity projections) of PIK3CAE542K vascular networks treated with DMSO, rapamycin (1 μM) or alpelisib (2.5 μM) for 3 days. Vascular networks were labeled with fibrin (magenta), DAPI (white), and actin (cyan). End points (orange) were highlighted in 3D renderings of segmented vessels. Scale bars, 100 μm. (B to E) Quantification of cell number (B), ECM morphology (C), mean vessel length (D), and vessel branching (E) in GFP control, rapamycin-treated PIK3CAE542K vascular networks, or alpelisib-treated PIK3CAE542K vascular networks (n ≥ 3; means ± SEM; one-way ANOVA with Tukey post test). (F) Western blot and quantification of relative p-AKT1(Thr308), p-AKT1(Ser473), p-S6K(Thr389), and p-ERK1/2(Thr202/Tyr204) levels in PIK3CAE542K cells treated with DMSO, rapamycin, or alpelisib for 24 hours (n ≥ 3; mean ± SEM; one-way ANOVA with Tukey post test, *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001).
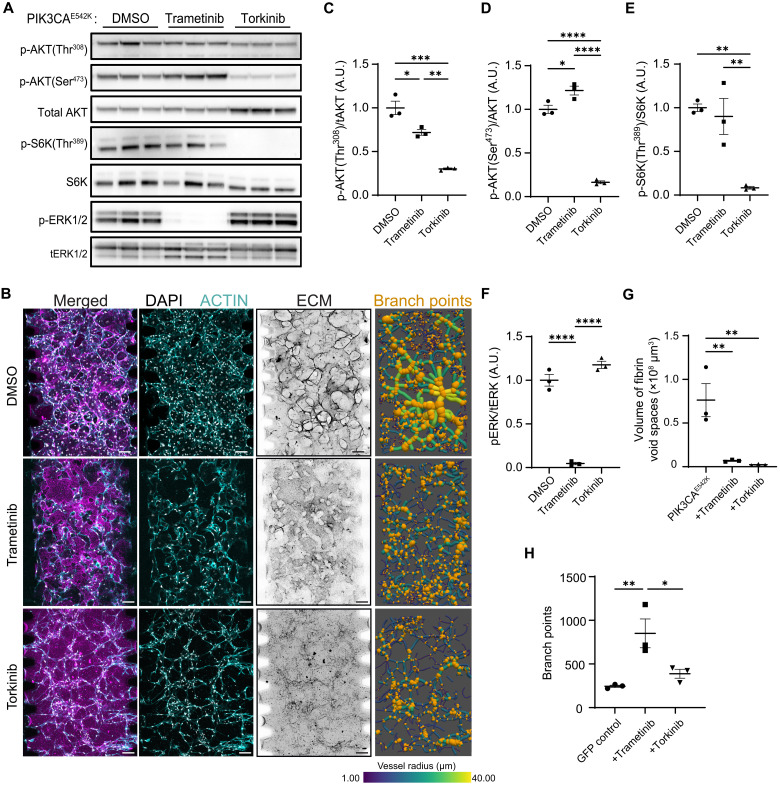
Targeting elevated MEK1/2 signaling with trametinib reduces vascular lesions but results in vascular hyperbranching
To test the relevance of MEK/ERK activation in PIK3CA-driven VMs, we treated PIK3CAE542K vascular network with trametinib (Fig. 6 and fig. S9C), a MEK1/2 kinase inhibitor that has proven to be effective in improving VMs in patients with activating MAP2K1 or KRAS mutations and in KRAS-driven mouse models of GSD (32–34). While reduced ERK1/2 phosphorylation is distinctive of trametinib treatment (Fig. 6, A and F), trametinib-treated PIK3CAE542K vascular networks displayed a reduced vascular lesions and hyperbranched vascular network morphology that is comparable to the inhibition of mTORC1 with rapamycin (Fig. 6, B and G to H, and fig. S10). These results indicate that MEK/ERK and mTORC1 signaling pathways share overlapping roles in the regulation of vascular growth as inhibition of either MEK/ERK or mTORC1 signaling alleviates vascular overgrowth but results in abnormal hyperbranched vascular networks.
Fig. 6. Treatment with trametinib (MEK1/2 inhibitor) and torkinib (mTORC1/2 inhibitor) improves and rescues vascular phenotypes in PIK3CAE542K vascular networks, respectively.
(A) Western blot analysis of relative p-AKT1(Thr308), p-AKT1(Ser473), p-S6K(Thr389), and p-ERK1/2(Thr202/Tyr204) levels in PIK3CAE542K cells treated with the indicated inhibitors. (B) Representative confocal images (maximum intensity projections) of PIK3CAE542K treated with DMSO, trametinib (MEK1/2 inhibitor, 1 μM) or torkinib (mTORC1/2 inhibitor, 5 μM) for 3 days. Scale bars, 100 μm. (C to F) Quantification of relative p-AKT1(Thr308), p-AKT1(Ser473), p-S6K(Thr389), and p-ERK1/2 levels in (A) (n ≥ 3; means ± SEM; one-way ANOVA with Tukey post test). (G and H) Quantification of matrix (G) and vessel branching morphology (H) in (B) (n ≥ 3; mean ± SEM; one-way ANOVA with Tukey post test, *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001).
Dual inhibition of mTORC1 and mTORC2 rescues network topology
Given the known role of mTORC2 signaling in regulating actin cytoskeletal dynamics (35) and our observation that rapamycin treatment led to incomplete suppression of mTORC2 signaling, we postulated that vascular hyperbranching is a consequence of dysregulated mTORC2 signaling. To test this, we treated PIK3CAE542K vascular networks with torkinib, an adenosine 5′-triphosphate–competitive mTORC1/2 inhibitor (36, 37), and compared the resulting vascular morphology to that of the control GFP, alpelisib, rapamycin, or trametinib-treated PIK3CAE542K vascular network (Fig. 6H and fig. S10). Treatment with torkinib reduced phosphorylation of AKT1-Ser473 (Fig. 6, A and D) and prevented the development of vascular lesions and hyperbranching phenotypes (Fig. 6, B, G, and H, and fig. S10). Further, when compared to trametinib, treatment with torkinib had a greater effect on suppressing protease secretion (fig. S5), implicating a role of mTORC1/2 signaling in matrix degradation. Collectively, these results are consistent with previous observations demonstrating that mTORC2 activates AKT1 via phosphorylation at Ser473 to regulate endothelial cell sprouting and migration through regulation of actin cytoskeletal and focal adhesion dynamics (38) and indicates a combined role of mTORC1-dependent overgrowth and mTORC2-dependent cytoskeletal defects in the development of malformed vascular networks.
Elevated Rac1 activity contributes to VM
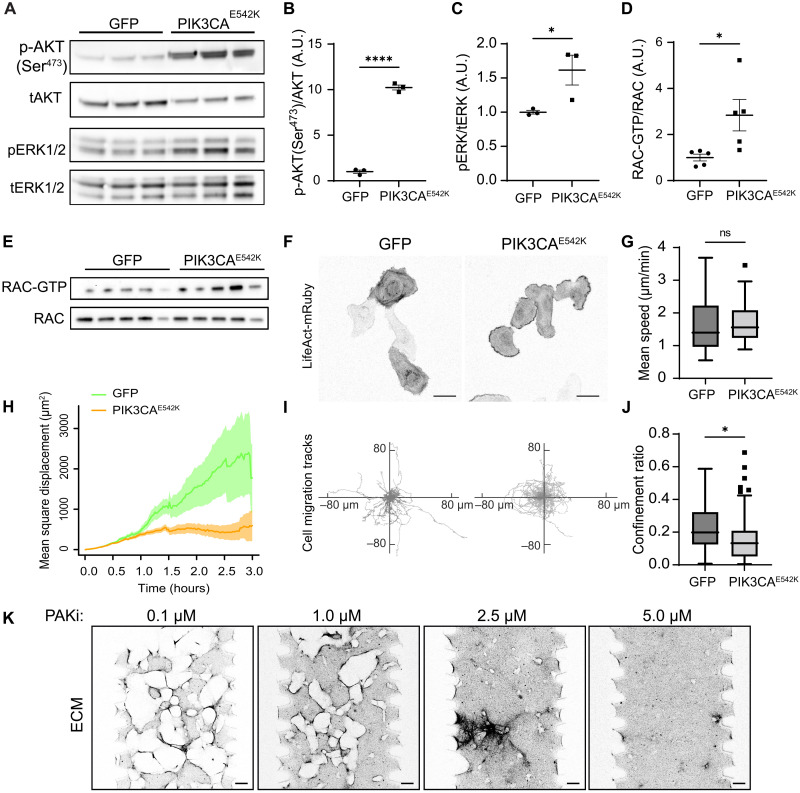
Previous studies have demonstrated the role of dysregulated small GTPase signaling and MEK/ERK activation in the development of complex lymphatic malformations (34, 39–43). One potential explanation for increased MEK/ERK signaling could be elevated small GTPase activity in PIK3CAE542K cells. To explore this hypothesis, we examined the effect of alpelisib treatment on Ras, Rac, and Cdc42-GTPase activities in PIK3CAE542K cells and found that treatment with alpelisib resulted in reduced Rac-GTP level (fig. S9B). Consistent with this observation, we found higher levels of ERK1/2 phosphorylation and Rac-GTP in PIK3CAE542K cells when compared to control (Fig. 7, A to E). As Rac1-GTPase plays a critical role in mediating actin polymerization and membrane protrusion at the leading edge of migrating cells, we performed time-lapse imaging and investigated the effect of PIK3CA mutation on endothelial cell migration (Fig. 7, F to J, and movie S2). Consistent with cellular phenotypes associated with Rac1 activation (44, 45), we found that PIK3CAE542K cells produced persistent actin-rich membrane ruffles around cell edges (Fig. 7F and movie S2). Further, we observed clear differences in cell migration behavior, where control cells migrate more persistently than cells expressing PIK3CAE542K mutation (Fig. 7I). To quantify this effect, we calculated the mean migration speed (Fig. 7G), mean square displacement (Fig. 7H), and the confinement ratio of control and mutant cells (Fig. 7J). While we observed no significant difference in migration speed, PIK3CAE542K cells showed significantly reduced mean square displacement and confinement ratios as defined by net displacement over total traveled distance (Fig. 7, H to J), demonstrating that persistent cell migration was inhibited with constitutive PI3K activation in endothelial cells expressing PIK3CA mutation. We further tested the effect of Rac-GTPase and PAK1 inhibition on phospho-ERK1/2 level. Treatment with Rac-GTPase inhibitor EHT1864 or PAK inhibitor PF-3758039 reduced ERK1/2 phosphorylation (fig. S11A). Further, treatment with PAK inhibitor PF-3758039 resulted in a dose-dependent reduction of vascular lesions as measured by volume of fibrin void spaces in PIK3CAE542K vascular networks (Fig. 7K and fig. S11C). Notably, we observed changes in 3D vascular network morphology where vascular networks appeared sheet-like and flattened with higher doses of PAK inhibitions (fig. S11B). All doses of PAK inhibition led to a similar reduction in endothelial cell number, indicating that these dose-dependent morphology changes are not solely mediated by alteration in endothelial cell proliferation (figs. S8B and S11D). Together, these results confirm that both MEK/ERK and Rac-GTPase activities are up-regulated with PI3K activation and suggest that excessive Rac/PAK and MEK/ERK signaling may be targetable in PIK3CA-driven VMs.
Fig. 7. Endothelial cells expressing PIK3CA mutation display molecular signatures of excessive Rac1 and MEK/ERK signaling.
(A to C) Western blot analysis (A) and quantification of relative p-AKT1(Ser473) (B) and p-ERK1/2(Thr202/Tyr204) (C) levels in serum-starved GFP control and PIK3CAE542K endothelial cells (n = 3; means ± SEM; two-tailed unpaired Student’s t test, *P ≤ 0.05; ****P ≤ 0.0001). (D and E) Quantification (D) and Western blot (E) of active Rac1/2/3 level in GFP control and PIK3CAE542K endothelial cells. Active Rac1/2/3 was isolated using a glutathione S-transferase–tagged p21-binding domain of PAK1 (n = 5; means ± SEM; two-tailed unpaired Student’s t test, *P ≤ 0.05). (F) Representative images of GFP and PIK3CAE542K cells expressing LifeAct-mRuby, cultured in 2D. Note the presence of actin-rich membrane ruffles at the periphery of PIK3CAE542K cells. Scale bars, 50 μm. (G to J) Mean migration speed (G), mean square displacement (H), migration trajectories (I), and confinement ratios as defined by net displacement over total traveled distance (J) of GFP and PIK3CAE542K cells (n = 57 and 60 tracks, Tukey box and whisker, two-tailed unpaired Student’s t test, *P ≤ 0.05). (K) Representative confocal images of ECM surrounding PIK3CAE542K vascular networks treated with 0.1 to 5.0 μM of PAK inhibitor, PF-3758309. Scale bars, 100 μm.
DISCUSSION
Here, we generated vascular networks with HUVECs expressing a PIK3CA-activating mutation (figs. S1 and S2) and demonstrated that the mutant endothelial cells develop into dilated vascular networks with complex and irregular lumens that resemble clinical hallmarks observed in patients with VMs (Figs. 1 to 3). These marked phenotypes observed in 3D culture are notable in comparison to the relatively mild differences observed for the same cells in 2D cell culture (figs. S1 to S3). We found that vessel enlargement and cyst development are driven largely by endothelial cell–autonomous defects that are associated with increased endothelial cell proliferation (figs. S3, S6, and S7) and dysregulated cell–mediated matrix proteolysis and tissue mechanics (Figs. 2 and 4 and fig. S5). Using our model, we tested the efficacies of rapamycin, an mTORC1 inhibitor, and alpelisib, a PIK3CA-specific inhibitor, in treating VMs (Fig. 5 and fig. S9C). We analyzed the resulting vascular network geometry in 3D and found that while both inhibitors prevented vessel enlargement and improved vascular network morphology, treatment with rapamycin resulted in vessel hyperbranching (Fig. 5 and fig. S10C). The E542K and E545K variants both result in lysine substitution in the helical domain of PIK3CA that weaken the interaction between p110α and p85 regulatory subunits, resulting in constitutive activation of PI3K signaling. Given the similarities in molecular pathophysiology of these variants and the similar downstream signaling observed here, we focused our experimental manipulations on the E542K variant with the expectation that results will be broadly applicable to E545K.
We observed that rapamycin was ineffective in suppressing phosphorylation of the mTORC2 target, AKT1 Ser473 (Fig. 5F), when compared to alpelisib. mTOR serine/threonine kinase is a downstream effector of PI3K signaling and functions through two multiprotein complexes, mTORC1 and mTORC2 (46). While mTOR is known to regulate endothelial cell proliferation and angiogenesis (47–49) and inhibition of AKT has been shown to inhibit progression of malformations in the retina of mice harboring PIK3CAH1047 mutations (50), the specific manner in which mTORC1 and mTORC2 contribute to the cellular processes involved in angiogenesis remains to be defined. Using our model, we showed that inhibition of mTORC1 with rapamycin resulted in vascular hyperbranching (Fig. 5, A and E, and fig. S10C). Notably, dual inhibition of mTORC1/2 rescued vessel hyperbranching (Fig 6, B and H, and fig. S10C), indicating a specific role of mTORC2 in regulating vessel length and branchpoint selection during vessel formation. We found that proteases secretion was up-regulated in PIK3CAE542K cells when compared to control (fig. S5). Notably, treatments with alpelisib or torkinib broadly reduced and normalized the levels of secreted urokinase-type plasminogen, kallikreins 3, 5, 6, 11, and 13, proteases that are known to promote plasminogen activation and fibrinolysis (51). Together, our data point to a role of mTORC2-dependent regulation of protease secretion during vascular remodeling.
In vitro studies have demonstrated a role of mTORC2 in controlling actin cytoskeleton rearrangement and cell spreading in fibroblasts (35). More recently, studies of mTOR signaling in neurons further showed that mTORC2 plays a vital role in controlling neuron size, morphology and synaptic function (52, 53). This is thought to be mediated through regulation of actin cytoskeleton rearrangements and protrusion dynamics via mTORC2-Tiam1-Rac1 signaling (52). Notably, our data show that both mTORC2 and Rac1 activities are elevated in endothelial cells expressing PIK3CAE542K mutation (Fig. 7, A to E). At the cellular level, we observed increased cell spreading, excess membrane protrusion, and cell migration defects that are consistent with Rac1 activation (Fig. 7, F to J). Previous work in an inducible model demonstrates that inactivation of PI3K⍺ increases actomyosin contractility through myosin phosphatase target subunit 1–mediated dephosphorylation of myosin light chain 2, resulting in increased actin stress fiber formation and impaired junctional remodeling (54). Furthermore, it has been shown that a balance of Rac1 and myosin activity is necessary for the formation and stability of endothelial adherens junctions (55), and together, these results suggest that PIK3CA signaling could regulate the relative balance of Rac1 versus myosin activity. Collectively, our results point to excess mTORC2 and Rac1/Pak signaling leading to dysregulated actin cytoskeletal dynamics and endothelial cell motility in cells expressing PIK3CA-activating mutations.
Most of the mutations identified in VMs occur within key components of two major growth factor–induced signaling pathways, PI3K-AKT-mTOR and RAS-MAPK-ERK (2, 4). While both pathways regulate vascular development and tumor angiogenesis, a mechanistic understanding of how activating mutations in PI3K and MAPK signaling lead to the formation of cysts associated with VMs remains elusive. Our study found increased levels of activated Rac and phosphorylated-ERK1/2 in PIK3CAE542K cells when compared to controls (Fig. 7A). A recent study using endothelial cells derived from patients demonstrated that the level of pERK 1/2 varied among patients, with a subset demonstrating elevated levels (56), and previous work has demonstrated a dose-dependent up-regulation of MAPK/ERK in HUVECs expressing PIK3CA-activating mutations (12) and in human induced pluripotent stem cells expressing endogenous levels of PIK3CA-H1047R (57). Further, treatment of mutant cells with alpelisib, a PIK3CA-specific inhibitor, decreased Rac-GTP and phospho-ERK1/2 levels (figs. S5F and S9B), suggesting a role of dysregulated Rac/PAK/MAPK signaling in PIK3CA-driven VMs. Although MAPK pathway is canonically known to be activated by Ras/Raf signaling, there is evidence in the oncology and immunology fields to demonstrate that Raf/MEK/ERK signaling could be activated by PI3K, mediated by Rac1/Cdc42/PAK and independent of Ras-Raf interaction (58, 59). Reducing MAPK activities with trametinib, a MEK1/2 inhibitor, prevented vessel enlargement but resulted in hyperbranched vessel morphology that is similar to vascular networks treated with rapamycin (Fig. 6, B and H, and fig. S10C). This is expected, as MAPK scaffolds are known to regulate and cross-inhibit several nodes of mTORC1 signaling (60), and the level of mTORC2-dependent AKT1-Ser473 phosphorylation remains unchanged with MEK1/2 inhibition (Fig. 6, A and D). Collectively, our findings demonstrate a functional redundancy in the ERK1/2 and mTORC1 signaling pathways in regulating endothelial cell proliferation and vessel enlargement and that inhibition of cell growth through ERK1/2 or mTORC1 alone is not sufficient to rescue cytoskeletal perturbations that are regulated by mTORC2/Rac1/PAK signaling. These findings are in line with the two-hit model of CCMs, where loss of function mutations of the CCM complex that result in increased MEKK3–ERK5–Krüeppel-like factor signaling and a subsequent gain of function mutation in PIK3CA signaling synergize to promote aggressive CCM lesions (61, 62).
The tissue engineered VM model described here enables in vitro vascular disease modeling, comparative drug studies, and drug-specific pathway discovery, but an important limitation of our studies is that overexpression of PIK3CA mutant isoforms may accelerate pathogenesis and resulted in nonphysiological phenotypes. With growing interest in combination therapies for the treatment of various tumors and interest in repurposing oncology drugs for the treatment of VMs, this platform could provide an important tool for dose optimization studies or for preclinical screening of synergistic or antagonistic effects of combination treatments. While we demonstrate the model’s utility in elucidating cross-talk between PI3K/AKT/mTOR, MEK/ERK, and Rac1/PAK signaling and illustrate changes in underlying vessel morphology in response to targeted inhibition of these signaling axes, a more systematic screening and chemogenomic study against other PIK3CA-activating variants and other drivers of VMs would shed light on the targetable mechanisms of VMs. Another important limitation to our platform is that the rate of degradation far exceeds ECM synthesis and deposition, and the lack of immune and stromal cells precludes the rigorous study of healing processes or whether any of the compounds can reverse VM phenotypes. The development of VMs is a complex and multifactorial processes, involving cytoskeletal reorganization, ECM remodeling, proliferation, and potentially changes in endothelial cell contractility. Understanding how each of the PI3K-associated signaling axes contribute to these distinct cell behaviors identifies key signaling nodes relevant to not only PIK3CA-driven VMs but also other types of VMs. We anticipate that the approach described here will motivate the adaptation of vasculature-on-chip models for use in future studies of vascular disease and patient-specific drug screening.
MATERIALS AND METHODS
Cell culture
HUVECs (Lonza) were cultured in growth medium (EGM-2, Lonza). HLFs (Lonza) were cultured in growth medium (FGM-2, Lonza), and human embryonic kidney (HEK)–293T cells (Clonetech) were grown in high-glucose Dulbecco’s modified Eagle’s medium (Hyclone) supplemented with 10% fetal bovine serum (Hyclone) and 1% penicillin/streptomycin (Life Technologies). All cells were cultured at 37°C and 5% CO2 in a humidified incubator.
Antibodies and cell biology reagents
Anti–VE-cadherin (F-8) was from Santa Cruz Biotechnology. Anti-CD31 (89C2) was from Cell Signaling Technologies. 4′,6-Diamidino-2-phenylindole (DAPI), rhodamine phalloidin (1 mg/ml), Alexa Fluor 647 phalloidin (1 mg/ml), and Alexa Fluor–conjugated secondary antibodies were from Life Technologies. For antibody concentrations, see the “Immunofluorescence” and “Western blot and small GTPase activity assays” sections below. Rapamycin, alpelisib (BYL719), Y-27362 2HCl, trametinib (GSK1120212), batimastat (BB-94), EHT1864 2HCl, and PF-3758309 were from Selleckchem.
Cloning and lentiviral infection
Plasmids encoding pHAGE-EF1aL-eGFP-W (Addgene, plasmid #126686, a gift from D. Kotton), pHAGE-PIK3CA-E542K-ires-GFP, pHAGE-PIK3CA-E545K-ires-GFP (Addgene, plasmid nos. 116479 and 116485, gifts from G. Mills and K. Scott), pHAGE-PIK3CA-WT-ires-GFP, and pLenti.PGK.LifeAct-Ruby.W (Addgene, plasmid no. 51009, a gift from R. Lansford) were used for lentivirus generation. Q5 site–directed mutagenesis (New England Biolabs) was performed on pHAGE-PIK3CA-E542K-ires-GFP to generate pHAGE-PIK3CA-WT-ires-GFP. For lentivirus production, HEK-293T cells were transfected with pHAGE-plasmid, pSPAX2 plasmid, and pMD2.G plasmid (Addgene, plasmid nos. 12260 and 12259, gifts from D. Trono) using calcium phosphate transfection. Lentiviral supernatants were collected 2 days after transfection and concentrated using PEG–IT (System Biosciences, Palo Alto, CA) viral precipitator. Concentrated lentivirus was resuspended in phosphate-buffered saline (PBS). Serial dilutions of lentiviral particles were performed to determine the optimal concentration for transduction. Concentrated lentiviruses were subsequently added to 100,000 HUVECs cultured in a six-well plate. Transduction efficiency was determined by GFP immunofluorescence. Transduced HUVECs were used between passages 5 and 11.
Polydimethylsiloxane wells for network formation
To generate wells for vascular networks, polydimethylsiloxane (PDMS; Sylgard 184, Dow-Corning) was mixed at 10:1 base:curing agent, poured into the lid of a 150-mm culture dish, and degassed in a dessicator under house vacuum for 1 hour. PDMS was cured overnight at 60°C. Cured PDMS was then removed from the lid, and a 6-mm biopsy punch and razor were used to create 1 cm by 1 cm squares of PDMS with a 6-mm-diameter hole at the center. Clear tape and isopropanol were used to clean the PDMS surfaces and glass coverslip. PDMS and coverslips were treated with oxygen plasma (Harrick Plasma) for 30 s, bonded, and incubated at 100°C for 20 min. Poly-l-lysine in water [0.01% (w/v); Sigma-Aldrich] was added to the interior of the wells and incubated at room temperature (RT) for 5 min before 3× washes with sterile, distilled (DI)–H2O. One percent (w/v) glutaraldehyde in water was then added to wells and incubated at RT for 5 min before 3× washes with sterile DI-H2O and incubated overnight in DI-H2O on a laboratory orbital shaker overnight at RT. Clean, surface-treated wells were then wrapped in aluminum foil, autoclaved, dried at 100°C for 20 min, and placed in sterile wells of a 24-well plate.
Hydrogel formation and seeding in PDMS wells
Fresh solutions of fibrinogen from bovine plasma (Millipore Sigma) were prepared before each seeding by dissolving fibrinogen powder in Dulbecco’s phosphate-buffered saline (DPBS; Millipore Sigma) to a concentration of 5 mg/ml and incubating at 37°C for 30 min before filter sterilization with a 0.2-μm syringe filter (Millipore Sigma). Thrombin from bovine plasma was solubilized in DPBS to a concentration of 100 U/ml, aliquoted, frozen at −80°C, and thawed immediately before each seeding. HUVECs and HLFs were lifted from culture flasks with 0.25% (w/v) trypsin-EDTA (Thermo Fisher Scientific), centrifuged at 200g for 5 min. HUVECs and HLFs were each resuspended at 13 ×106 cells/ml of EGM-2. For each well, 50 μl of cells in fibrin prepolymer were prepared by mixing 25 μl of fibrinogen (5 mg/ml), 11.5 μl of 13 × 106 HUVECs/ml EGM-2, 11.5 μl of 13 × 106 HLFs/ml EGM-2, 0.5 μl of thrombin (100 U/ml), and 1.5 μl of EGM-2 for final cell concentrations of 3 × 106 cells/ml in the total slurry. After mixing with a pipette to avoid bubbles, a 50-μl slurry was added to each PDMS well, incubated for 15 min at 37°C in a cell culture incubator, with inversion every few minutes to allow cells to be distributed in 3D. A total of 2.5 ml of fresh EGM-2 was added into each well of the 24-well plate containing a PDMS well, and media were changed daily.
Microfluidic device fabrication
Microfluidic devices were fabricated using a protocol adapted from previous approaches (63–65). Briefly, film masks (Fineline) were printed from technical drawings made with AutoCAD (Autodesk). Silicon wafers (100-mm single side polish, test grade, University Wafers) were stripped with 5% (v/v) hydrofluoric acid (Sigma-Aldrich) and laminated with 250-μm-thick photoresist epoxy laminates (SUEX-250, DJ Microlaminates) using a laminator heated to 65°C with a rolling speed of 1 foot/min. After postlamination bakes at 70°C for 15 min and 95°C for 5 min, laminated wafers were then illuminated through the transparency mask with columnated 350-nm light for a total exposure of 1000 mJ. After postexposure bakes at 70° and 95°C, uncross-linked photoresist stripped using propylene glycol monomethyl ether acetate (Millipore Sigma) developer and dried with isopropanol. After plasma treatment for 30 s, trichloro(1H,1H,2H,2H-perfluorooctyl)silane (Sigma-Aldrich) was vapor-deposited onto the patterned silicon wafers (referred to subsequently as silicon masters). Individual devices were then fabricated using standard soft lithography. PDMS (10:1 base:curing agent) was poured onto silicon masters, degassed using house vacuum, and cured at 60°C overnight. Cured PDMS was then removed from the master, cut and trimmed with a razor, and media and hydrogel ports were punched out with biopsy punches (6 and 1.5 mm, respectively). The surface of the PDMS were cleaned with tape and isopropanol. Cleaned PDMS devices and glass coverslips were then treated with oxygen plasma for 30 s, bonded, and incubated at 100°C for 20 min. Before use, devices were sterilized with ultraviolet treatment for 20 min.
Hydrogel formation and seeding in microfluidic devices
Hydrogels were prepared following the same protocol as the PDMS wells with the exception of different cell concentrations for each channel. For the HUVEC channel, HUVECs were resuspended at a concentration of 5.5 × 106 HUVECs/ml in EGM-2 media and added to an equal volume of fibrinogen (5 mg/ml) diluted in PBS. To visualize ECM degradation and vascular lesion formation, 1.5 mg/ml of Alexa Fluor 64–conjugated fibrinogen (Thermo Fisher Scientific) was added to the hydrogel. After mixing, 1 U of thrombin was added to the slurry and mixed before injecting into the device. Devices were then flipped upside down and incubated for 15 min at 37°C in a cell culture incubator, inverting every few minutes to ensure even distribution of cells in 3D. The same protocol was used for preparing HLF-fibrin hydrogels for the HLF channels with the exception that the HLF suspension was prepared at a density of 2.75 × 106 HLFs/ml in EGM-2 media. Media were then changed daily and devices were fixed 3 days after seeding. For mitotic inhibition, HUVECs plated onto 10-cm dishes were incubated with mitomycin-C (0.01 mg/ml; source) for 2.5 hours at 37°C. Mitomycin-C–treated HUVECs were trypisinized and seeded into microfluidic devices as described above.
Immunofluorescence
To observe network assembly over time, PDMS wells loaded with HUVECs expressing GFP (see the “Cloning and lentiviral infection” section, above) were imaged daily with a laser scanning confocal microscope (FV3000, Olympus) with a 488-nm laser diode. Z-stacks of the gel volume were taken at ×10 magnification [U Plan S-Apo, 0.4–numerical aperture (NA) air objective, Olympus]. For end point immunostaining, PDMS wells or devices were fixed with 4% paraformaldehyde (Millipore Sigma-Aldrich) in PBS containing calcium and magnesium (PBS++) at 37°C for 15 min. After rinsing twice with PBS++, devices were left on a laboratory rocker for 24 hours in PBS++ to wash. Cells were then permeabilized with 0.3% Triton X-100 (Millipore Sigma) for 10 min at RT, and nonspecific antibody binding was blocked with 2% (w/v) bovine serum albumin (BSA) in PBS++ for 24 hours at RT. Wells and devices were then sealed in tissue culture plates and kept at 4°C until antibodies were added. Samples cultured in microfluidic devices were imaged daily at ×4 magnification (U Plan Fluor, 0.13-NA air objective, Olympus) on a wide-field microscope and fixed 3 days after seeding. For immunostaining, cells were permeabilized with 0.3% Triton X-100. Primary antibodies against VE-cadherin (1:200, v/v; mouse anti-human F-8, Santa Cruz Biotechnology) or CD31 (1:1000, v/v; mouse, anti-human CD31, Cell Signaling Technologies) were diluted in 2% BSA in PBS++ and applied overnight on a laboratory rocker at 4°C. Cells were then rinsed three times over 1 hour with PBS++, and secondary antibodies (goat anti-mouse immunoglobulin G conjugated to Alexa Fluor 647 or Cy3, Thermo Fisher Scientific) were diluted in 2% BSA in PBS++ (1:200, v/v) and applied to devices at RT for 1 hour on a laboratory rocker before rinsing three times over 30 min with PBS++. F-actin was labeled with Alexa Fluor 488 or rhodamine-phalloidin (Thermo Fisher Scientific), and the nucleus was labeled with DAPI (Thermo Fisher Scientific) diluted in PBS++ (1:200 and 1:1000, v/v, respectively) for 1 hour at RT before rinsing three times over 30 min with PBS++. Images were acquired with an Olympus FV3000 laser scanning confocal microscope with a 30× U Plan S-Apo N 1.05-NA silicone oil immersion objective, and images were adjusted for brightness and contrast using ImageJ. For computational image processing used to measure cell and network topology, see the “Quantification of vascular network topology and fibrin void regions” section below.
Western blot and small GTPase activity assays
Cells were cultured in complete media unless otherwise noted. For measuring relative changes in phospho-AKT, phospho-S6K, and phospho-ERK level in response to inhibitors, HUVECs were cultured in media with inhibitor for 24 hours. Cells were lysed on ice with radioimmunoprecipitation assay buffer (Thermo Fisher Scientific) containing Halt protease and phosphatase inhibitor (Thermo Fisher Scientific). Clarified lysates were resolved on Novex 4 to 12% bis-tris gel (Thermo Fisher Scientific) and transferred to a polyvinylidene difluoride membrane. Standard immunoblotting protocols were performed for Western blotting with primary antibodies used at the following concentrations: anti-p110α (1:1000; Cell Signaling Technology, #4249), anti–p-AKT-Thr308 (1:1000, Cell Signaling Technology, #9275), anti–p-AKT-Ser473 (1:1000; Cell Signaling Technology, #9271), anti-AKT (1:1000; Cell Signaling Technology, #9272), anti–pS6K-Thr389 (1:1000; Cell Signaling Technology, #9205), anti-S6K (1:1000; Cell Signaling Technology, #9202), anti–pERK1/2-Thr202/Tyr204 (1:1000; Cell Signaling Technology, #9101), anti-ERK1/2 (1:1000; Cell Signaling Technology), anti-Rac1/2/3 (1:1000; Cell Signaling Technology, #2465), and anti–glyceraldehyde-3-phosphate dehydrogenase (1:2000; Cell Signaling Technology, #2118). Horseradish peroxidase–conjugated secondary antibodies and SuperSignal West Femto or Clarity Western ECL chemiluminescent substrate were used for detection. Western blot images were quantified with Fiji/ImageJ. Proteome profiler human protease array (R&D Systems, #ARY021B) was performed according to the manufacturer’s instructions. Ras, Rac, and Cdc42 pulldown assays (Cytoskeleton Inc.) were performed according to the provided instruction manual. Average pixel intensity of each analyte and background signal were quantified using Fiji/ImageJ. Average pixel intensity was subtracted with average background signal. Expression level of each analyte was normalized by setting the lowest average pixel intensity and the highest average pixel intensity of each dataset to 0 and 100, respectively. Lysates from confluent cells cultured in complete media were used to compare Rac–guanosine 5′-triphosphate (GTP) level in GFP control and PIK3CA mutant cells. For alpelisib treatment, confluent cells cultured overnight in reduced serum conditions were stimulated with complete media with or without alpelisib for 10 min. Cells were rinsed once with cold PBS++ and lysed with cold lysis buffer [50 mM tris (pH 7.5), 10 mM MgCl2, 0.3 M NaCl, 2% IGEPAL, and protease and phosphatase inhibitor cocktails, Cytoskeleton Inc.]. Lysates were sonicated at 3 W on ice for 10 s and clarified at 14,000g for 5 min. Protein concentration was quantified with bicinchoninic acid assay (Thermo Fisher Scientific), and the protein concentration and volume were equalized with cell lysis buffer. Five hundred micrograms of lysate was incubated with 10 μg of PAK-p21 bidning domain (PAK-PBD) or RAF-Ras binding domain (RAF-RBD) beads on a rotator for an hour at 4°C. Bead pellets were washed three times with wash buffer [25 mM tris (pH 7.5), 30 mM MgCl2, and 40 mM NaCl, Cytoskeleton Inc.] and extracted with 2× NuPAGE lithium dodecyl sulfate (LDS) containing 100 mM dithiothreitol. For quantification, active RAC and RAS1 were normalized to total RAC and RAS1, respectively.
Live cell imaging, migration tracking, and quantification
HUVECs transduced with LifeAct-mRuby were cultured overnight on 35-mm dishes (Ibidi). Live imaging of microvascular networks was performed with an Olympus FV3000 resonant scanning confocal system equipped with a stage-top incubator. Images were taken at ×10 magnification (U Plan S-Apo, 0.4-NA air objective, Olympus), every 5 min for a period of 72 hours. Live imaging of cell migration was carried out on the same system but with a Plan S-Apo 30×/1.05-NA silicone oil (Olympus) objective. Images were captured every minute for 2 hours. Tracking of endothelial cells was performed using the cellpose TrackMate (66–68) module on Fiji. A pretrained cytoplasm model with an estimated 30-μm cell diameter was used for segmenting cells. To confirm tracking accuracy, cell migration trajectories were visually inspected. Migration tracks were plotted with MATLAB. Mean migration speed and confinement ratios were plotted, and statistical analyses were performed using GraphPad Prism. Mean square displacement plot was generated using MotilityLab (69).
β-Galactosidase staining
HUVECs were plated onto 12-well plate at a density of 25,000 cells per well. Cells were fixed 24 hours after seeding and stained for β-galactosidase activity (Cell Signaling Technology, #9860S) according to the manufacturer’s protocol. Images were captured by wide-field microscopy. Cells with positive β-galactosidase staining were quantified using manual thresholding in Fiji/ImageJ. The same threshold value was used for all images.
Junctional and actin quantification
Adherens junction and actin content were quantified using methods previously described, with modifications (70). To quantify adherens junction structure and assembly of HUVECs cultured in 2D, grayscale images of HUVECs immunostained with VE-cadherin were binarized on a threshold based on the mean cytosolic intensity of VE-cadherin for each image. A region of interest (ROI) was drawn within the cytosol of a cell for each image, the mean intensity within the ROI was determined, and the threshold for binarization was set to 2× this mean intensity value. Hot pixel noise was removed using the “Despeckle” command in ImageJ. To remove perinuclear signal, the DAPI channel for each image was binarized, dilated 3×, and inverted to create a mask that was then applied to the despeckled, binarized VE-cadherin images. Junctional area was defined as the total number of nonzero pixels. To quantify cortical actin, the intensity of Alexa Fluor 488 phalloidin–labeled cells was plotted along lines drawn between the centroids in the nuclei of neighboring cells. Local intensity peaks were measured to determine the location of stress fibers and cortical actin. Cortical actin was defined as the area under the peak at cell-cell junctions normalized to total area under the curve.
Cell count and area quantification
Cell area was quantified using a custom Cell Profiler pipeline. Briefly, the DAPI channel was used for segmenting individual cell nuclei for both cell count and area quantification. The binary mask containing the segmented nuclei was used as a reference for segmenting cells. Thresholding for cells was based on actin staining and was determined using a minimum cross entropy thresholding method.
Quantification of vascular network topology and fibrin void regions
Confocal z-stacks (4-μm step size, captured from top to bottom of the HUVEC channel within microfluidic devices) were used for measurement of vascular network topology. Rolling z projections (average over five z sections, Fiji) were performed to increase the signal-to-noise ratio in the images. Maximal intensity rolling z projections were performed for DAPI and actin channels, and minimum intensity rolling z projection was performed for the fibrin ECM channel. Contrast enhancement (0.35% of saturated pixels) was used to normalize image brightness across the z-stack. The processed z-stacks were exported as image sequences for 2D segmentation in CellProfiler. DAPI and actin channels were used for segmenting vascular networks. An adaptive minimum cross-entropy thresholding method (size of adaptive window = 250 pixels) was used for segmenting vascular networks. Briefly, a binary mask containing segmented nuclei was used as a reference for segmenting vascular networks labeled with actin. Binarized images were reconstructed into a z-stack and used as input images for 3D skeletonization, pruning, and vasculature analysis in VesselVio (71). To quantify fibrin void regions, processed fibrin images were binarized using adaptive thresholding (size of adaptive window = 100 pixels, Sauvola thresholding method). Binarized images were inverted and reassembled into z-stacks. Volume filling and 3D void volume analysis of binarized fibrin void regions was performed in VesselVio (71). Because of inaccuracies in skeletonization algorithm in extracting topologies of cystic and dilated mutant vascular networks, the branch and end points computations of mutant networks treated with dimethyl sulfoxide condition were not included in statistical comparisons between treatments.
EdU and cell viability assays
Cells were pulse-labeled with 10 μM EdU for 30 min before fixation and immunostaining. Click-iT EdU Cell Proliferation assay (Thermo Fisher Scientific) was performed according to the manufacturer’s instructions. For cell viability assay, cells were cultured for 24 hours with indicated concentration of inhibitors. After 24 hours, 2 μM calcein-AM and 4 μM propidium iodide were added onto endothelial cells. Cells were incubated at RT for 30 min before imaging.
Statistical analysis
Graphs and statistical analyses were generated in Prism 9 (GraphPad). Data were plotted as individual points with means ± SEM. Unless otherwise mentioned, statistical differences between two sets of normally distributed data were calculated using unpaired, two-tailed Student’s test. Statistical differences on more than two groups of data were determined using one-way analysis of variance (ANOVA), followed by Tukey post hoc test.
Acknowledgments
Funding: This work was supported by the National Institutes of Health (R35GM142944), by the American Heart Association (CDA857738), and by research grants from the University of Pennsylvania Orphan Disease Center in partnership with the Lymphangiomatosis and Gorham’s Disease Alliance and the Lymphatic Malformation Institute, the UNC Computational Medicine Institute, and the North Carolina Biotechnology Center (FLG-3814). W.Y.A. is supported by grants from the CLOVES Syndrome Community and the Eshelman Institute for Innovation. E.L.D., S.A.H., and C.P.W. acknowledge financial support through the Integrative Vascular Biology Training Program (T32HL69768), and E.L.D. acknowledges support from a Ruth L. Kirchstein predoctoral individual fellowship (F31HL162462). Device fabrication was performed in the Chapel Hill Analytical and Nanofabrication Laboratory (CHANL), a member of the North Carolina Research Triangle Nanotechnology Network (RTNN), which is supported by the National Science Foundation (ECCS-2025064), as part of the National Nanotechnology Coordinated Infrastructure (NNCI).
Author contributions: Conceptualization: W.J.P. and J.B. Methodology: W.Y.A., C.C., H.W., A.H.C., E.L.D., C.P.W., B.G., and W.J.P. Investigation: W.Y.A., C.C., H.W., E.L.D., D.R., S.A.H., C.P.W., and R.A. Supervision: A.J.H., B.G., M.L.K., J.B., and W.J.P. Visualization: H.W. and S.K. Writing—original draft: W.Y.A. and W.J.P. Writing—review and editing: W.Y.A., C.C., E.L.D., S.A.H., C.P.W., A.J.H., B.G., M.L.K., J.B., and W.J.P.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Legends for movies S1 and S2
Uncropped western blots
Tabular format of data from all figures
Other Supplementary Material for this : manuscript includes the following:
Movies S1 and S2
REFERENCES AND NOTES
- 1.Sadick M., Muller-Wille R., Wildgruber M., Wohlgemuth W. A., Vascular anomalies (part I): Classification and diagnostics of vascular anomalies. Rofo 190, 825–835 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Queisser A., Seront E., Boon L. M., Vikkula M., Genetic basis and therapies for vascular anomalies. Circ. Res. 129, 155–173 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Cox J. A., Bartlett E., Lee E. I., Vascular malformations: A review. Semin. Plast. Surg. 28, 58–63 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makinen T., Boon L. M., Vikkula M., Alitalo K., Lymphatic malformations: genetics, mechanisms and therapeutic strategies. Circ. Res. 129, 136–154 (2021). [DOI] [PubMed] [Google Scholar]
- 5.McCafferty I., Management of low-flow vascular malformations: Clinical presentation, classification, patient selection, imaging and treatment. Cardiovasc. Intervent. Radiol. 38, 1082–1104 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Horbach S. E., Lokhorst M. M., Saeed P., de Goüyon Matignon de Pontouraude C. M. F., Rothová A., van der Horst C. M. A. M., Sclerotherapy for low-flow vascular malformations of the head and neck: A systematic review of sclerosing agents. J. Plast. Reconstr. Aesthet. Surg. 69, 295–304 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Kalwani N. M., Rockson S. G., Management of lymphatic vascular malformations: A systematic review of the literature. J. Vasc. Surg. Venous Lymphat. Disord. 9, 1077–1082 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Ng A. T., Tower R. L., Drolet B. A., Targeted treatment of vascular anomalies. Int. J. Womens Dermatol. 7, 636–639 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luks V. L., Kamitaki N., Vivero M. P., Uller W., Rab R., Bovée J. V. M. G., Rialon K. L., Guevara C. J., Alomari A. I., Greene A. K., Fishman S. J., Kozakewich H. P. W., Maclellan R. A., Mulliken J. B., Rahbar R., Spencer S. A., Trenor C. C. III, Upton J., Zurakowski D., Perkins J. A., Kirsh A., Bennett J. T., Dobyns W. B., Kurek K. C., Warman M. L., McCarroll S. A., Murillo R., Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J. Pediatr. 166, 1048–1054.e5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osborn A. J., Dickie P., Neilson D. E., Glaser K., Lynch K. A., Gupta A., Dickie B. H., Activating PIK3CA alleles and lymphangiogenic phenotype of lymphatic endothelial cells isolated from lymphatic malformations. Hum. Mol. Genet. 24, 926–938 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Kurek K. C., Luks V. L., Ayturk U. M., Alomari A. I., Fishman S. J., Spencer S. A., Mulliken J. B., Bowen M. E., Yamamoto G. L., Kozakewich H. P. W., Warman M. L., Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am. J. Hum. Genet. 90, 1108–1115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Limaye N., Kangas J., Mendola A., Godfraind C., Schlögel M. J., Helaers R., Eklund L., Boon L. M., Vikkula M., Somatic activating PIK3CA mutations cause venous malformation. Am. J. Hum. Genet. 97, 914–921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castel P., Javier Carmona F., Grego-Bessa J., Berger M. F., Viale A., Anderson K. V., Bague S., Scaltriti M., Antonescu C. R., Baselga E., Baselga J., Somatic PIK3CA mutations as a driver of sporadic venous malformations. Sci. Transl. Med. 8, 332ra42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castillo S. D., Tzouanacou E., Zaw-Thin M., Berenjeno I. M., Parker V. E. R., Chivite I., Milà-Guasch M., Pearce W., Solomon I., Angulo-Urarte A., Figueiredo A. M., Dewhurst R. E., Knox R. G., Clark G. R., Scudamore C. L., Badar A., Kalber T. L., Foster J., Stuckey D. J., David A. L., Phillips W. A., Lythgoe M. F., Wilson V., Semple R. K., Sebire N. J., Kinsler V. A., Graupera M., Vanhaesebroeck B., Somatic activating mutations in Pik3ca cause sporadic venous malformations in mice and humans. Sci. Transl. Med. 8, 332ra43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez-Laguna L., Agra N., Ibañez K., Oliva-Molina G., Gordo G., Khurana N., Hominick D., Beato M., Colmenero I., Herranz G., Torres Canizalez J. M., Rodríguez Pena R., Vallespín E., Martín-Arenas R., Del Pozo Á., Villaverde C., Bustamante A., Ayuso C., Lapunzina P., Lopez-Gutierrez J. C., Dellinger M. T., Martinez-Glez V., Somatic activating mutations in PIK3CA cause generalized lymphatic anomaly. J. Exp. Med. 216, 407–418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fruman D. A., Chiu H., Hopkins B. D., Bagrodia S., Cantley L. C., Abraham R. T., The PI3K pathway in human disease. Cell 170, 605–635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franke T. F., Kaplan D. R., Cantley L. C., Toker A., Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science 275, 665–668 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Manning B. D., Toker A., AKT/PKB Signaling: Navigating the Network. Cell 169, 381–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang H. W., Shin M.-G., Lee S., Kim J.-R., Park W. S., Cho K.-H., Meyer T., Do Heo W., Cooperative activation of PI3K by Ras and Rho family small GTPases. Mol. Cell 47, 281–290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campa C. C., Ciraolo E., Ghigo A., Germena G., Hirsch E., Crossroads of PI3K and Rac pathways. Small GTPases 6, 71–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.di Blasio L., Puliafito A., Gagliardi P. A., Comunanza V., Somale D., Chiaverina G., Bussolino F., Primo L., PI3K/mTOR inhibition promotes the regression of experimental vascular malformations driven by PIK3CA-activating mutations. Cell Death Dis. 9, 45 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Corral I., Zhang Y., Petkova M., Ortsäter H., Sjöberg S., Castillo S. D., Brouillard P., Libbrecht L., Saur D., Graupera M., Alitalo K., Boon L., Vikkula M., Mäkinen T., Blockade of VEGF-C signaling inhibits lymphatic malformations driven by oncogenic PIK3CA mutation. Nat. Commun. 11, 2869 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venot Q., Blanc T., Rabia S. H., Berteloot L., Ladraa S., Duong J.-P., Blanc E., Johnson S. C., Hoguin C., Boccara O., Sarnacki S., Boddaert N., Pannier S., Martinez F., Magassa S., Yamaguchi J., Knebelmann B., Merville P., Grenier N., Joly D., Cormier-Daire V., Michot C., Bole-Feysot C., Picard A., Soupre V., Lyonnet S., Sadoine J., Slimani L., Chaussain C., Laroche-Raynaud C., Guibaud L., Broissand C., Amiel J., Legendre C., Terzi F., Canaud G., Targeted therapy in patients with PIK3CA-related overgrowth syndrome. Nature 558, 540–546 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams D. M., Trenor C. C. III, Hammill A. M., Vinks A. A., Patel M. N., Chaudry G., Wentzel M. S., Mobberley-Schuman P. S., Campbell L. M., Brookbank C., Gupta A., Chute C., Eile J., McKenna J., Merrow A. C., Fei L., Hornung L., Seid M., Dasgupta A. R., Dickie B. H., Elluru R. G., Lucky A. W., Weiss B., Azizkhan R. G., Efficacy and safety of sirolimus in the treatment of complicated vascular anomalies. Pediatrics 137, e20153257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delestre F., Venot Q., Bayard C., Fraissenon A., Ladraa S., Hoguin C., Chapelle C., Yamaguchi J., Cassaca R., Zerbib L., Magassa S., Morin G., Asnafi V., Villarese P., Kaltenbach S., Fraitag S., Duong J.-P., Broissand C., Boccara O., Soupre V., Bonnotte B., Chopinet C., Mirault T., Legendre C., Guibaud L., Canaud G., Alpelisib administration reduced lymphatic malformations in a mouse model and in patients. Sci. Transl. Med. 13, eabg0809 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Blesinger H., Kaulfuß S., Aung T., Schwoch S., Prantl L., Rößler J., Wilting J., Becker J., PIK3CA mutations are specifically localized to lymphatic endothelial cells of lymphatic malformations. PLOS ONE 13, e0200343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin Y., Han S., Jeon J. S., Yamamoto K., Zervantonakis I. K., Sudo R., Kamm R. D., Chung S., Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels. Nat. Protoc. 7, 1247–1259 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borikova A. L., Dibble C. F., Sciaky N., Welch C. M., Abell A. N., Bencharit S., Johnson G. L., Rho kinase inhibition rescues the endothelial cell cerebral cavernous malformation phenotype. J. Biol. Chem. 285, 11760–11764 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stockton R. A., Shenkar R., Awad I. A., Ginsberg M. H., Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J. Exp. Med. 207, 881–896 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald D. A., Shi C., Shenkar R., Stockton R. A., Liu F., Ginsberg M. H., Marchuk D. A., Awad I. A., Fasudil decreases lesion burden in a murine model of cerebral cavernous malformation disease. Stroke 43, 571–574 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carracedo A., Ma L., Teruya-Feldstein J., Rojo F., Salmena L., Alimonti A., Egia A., Sasaki A. T., Thomas G., Kozma S. C., Papa A., Nardella C., Cantley L. C., Baselga J., Pandolfi P. P., Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J. Clin. Invest. 118, 3065–3074 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lekwuttikarn R., Lim Y. H., Admani S., Choate K. A., Teng J. M. C., Genotype-guided medical treatment of an arteriovenous malformation in a child. JAMA Dermatol. 155, 256–257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards E. A., Phelps A. S., Cooke D., Frieden I. J., Zapala M. A., Fullerton H. J., Shimano K. A., Monitoring arteriovenous malformation response to genotype-targeted therapy. Pediatrics 146, e20193206 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Homayun-Sepehr N., McCarter A. L., Helaers R., Galant C., Boon L. M., Brouillard P., Vikkula M., Dellinger M. T., KRAS-driven model of Gorham-Stout disease effectively treated with trametinib. JCI Insight 6, e149831 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A., Hall M. N., Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6, 1122–1128 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Feldman M. E., Apsel B., Uotila A., Loewith R., Knight Z. A., Ruggero D., Shokat K. M., Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLOS Biol. 7, e38 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janes M. R., Limon J. J., So L., Chen J., Lim R. J., Chavez M. A., Vu C., Lilly M. B., Mallya S., Ong S. T., Konopleva M., Martin M. B., Ren P., Liu Y., Rommel C., Fruman D. A., Effective and selective targeting of leukemia cells using a TORC1/2 kinase inhibitor. Nat. Med. 16, 205–213 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farhan M. A., Carmine-Simmen K., Lewis J. D., Moore R. B., Murray A. G., Endothelial cell mTOR complex-2 regulates sprouting angiogenesis. PLOS ONE 10, e0135245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Couto J. A., Vivero M. P., Kozakewich H. P. W., Taghinia A. H., Mulliken J. B., Warman M. L., Greene A. K., A somatic MAP3K3 mutation is associated with verrucous venous malformation. Am. J. Hum. Genet. 96, 480–486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Couto J. A., Huang A. Y., Konczyk D. J., Goss J. A., Fishman S. J., Mulliken J. B., Warman M. L., Greene A. K., Somatic MAP2K1 mutations are associated with extracranial arteriovenous malformation. Am. J. Hum. Genet. 100, 546–554 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al-Olabi L., Polubothu S., Dowsett K., Andrews K. A., Stadnik P., Joseph A. P., Knox R., Pittman A., Clark G., Baird W., Bulstrode N., Glover M., Gordon K., Hargrave D., Huson S. M., Jacques T. S., James G., Kondolf H., Kangesu L., Keppler-Noreuil K. M., Khan A., Lindhurst M. J., Lipson M., Mansour S., O’Hara J., Mahon C., Mosica A., Moss C., Murthy A., Ong J., Parker V. E., Rivière J.-B., Sapp J. C., Sebire N. J., Shah R., Sivakumar B., Thomas A., Virasami A., Waelchli R., Zeng Z., Biesecker L. G., Barnacle A., Topf M., Semple R. K., Patton E. E., Kinsler V. A., Mosaic RAS/MAPK variants cause sporadic vascular malformations which respond to targeted therapy. J. Clin. Invest. 128, 1496–1508 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nikolaev S. I., Vetiska S., Bonilla X., Boudreau E., Jauhiainen S., Rezai Jahromi B., Khyzha N., DiStefano P. V., Suutarinen S., Kiehl T.-R., Mendes Pereira V., Herman A. M., Krings T., Andrade-Barazarte H., Tung T., Valiante T., Zadeh G., Tymianski M., Rauramaa T., Ylä-Herttuala S., Wythe J. D., Antonarakis S. E., Frösen J., Fish J. E., Radovanovic I., Somatic activating KRAS mutations in arteriovenous malformations of the brain. N. Engl. J. Med. 378, 250–261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barclay S. F., Inman K. W., Luks V. L., McIntyre J. B., al-Ibraheemi A., Church A. J., Perez-Atayde A. R., Mangray S., Jeng M., Kreimer S. R., Walker L., Fishman S. J., Alomari A. I., Chaudry G., Trenor C. C. III, Adams D., Kozakewich H. P. W., Kurek K. C., A somatic activating NRAS variant associated with kaposiform lymphangiomatosis. Genet. Med. 21, 1517–1524 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Machesky L. M., Hall A., Role of actin polymerization and adhesion to extracellular matrix in Rac- and Rho-induced cytoskeletal reorganization. J. Cell Biol. 138, 913–926 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hall A., Rho GTPases and the actin cytoskeleton. Science 279, 509–514 (1998). [DOI] [PubMed] [Google Scholar]
- 46.Liu G. Y., Sabatini D. M., mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183–203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li W., Petrimpol M., Molle K. D., Hall M. N., Battegay E. J., Humar R., Hypoxia-induced endothelial proliferation requires both mTORC1 and mTORC2. Circ. Res. 100, 79–87 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Wang S., Amato K. R., Song W., Youngblood V., Lee K., Boothby M., Brantley-Sieders D. M., Chen J., Regulation of endothelial cell proliferation and vascular assembly through distinct mTORC2 signaling pathways. Mol. Cell. Biol. 35, 1299–1313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsuji-Tamura K., Ogawa M., Inhibition of the PI3K-Akt and mTORC1 signaling pathways promotes the elongation of vascular endothelial cells. J. Cell Sci. 129, 1165–1178 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Kobialka P., Sabata H., Vilalta O., Gouveia L., Angulo-Urarte A., Muixí L., Zanoncello J., Muñoz-Aznar O., Olaciregui N. G., Fanlo L., Esteve-Codina A., Lavarino C., Javierre B. M., Celis V., Rovira C., López-Fernández S., Baselga E., Mora J., Castillo S. D., Graupera M., The onset of PI3K-related vascular malformations occurs during angiogenesis and is prevented by the AKT inhibitor miransertib. EMBO Mol. Med. 14, e15619 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emami N., Diamandis E. P., New insights into the functional mechanisms and clinical applications of the kallikrein-related peptidase family. Mol. Oncol. 1, 269–287 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang W., Zhu P. J., Zhang S., Zhou H., Stoica L., Galiano M., Krnjević K., Roman G., Costa-Mattioli M., mTORC2 controls actin polymerization required for consolidation of long-term memory. Nat. Neurosci. 16, 441–448 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomanetz V., Angliker N., Cloëtta D., Lustenberger R. M., Schweighauser M., Oliveri F., Suzuki N., Rüegg M. A., Ablation of the mTORC2 component rictor in brain or Purkinje cells affects size and neuron morphology. J. Cell Biol. 201, 293–308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Angulo-Urarte A., Casado P., Castillo S. D., Kobialka P., Kotini M. P., Figueiredo A. M., Castel P., Rajeeve V., Milà-Guasch M., Millan J., Wiesner C., Serra H., Muixi L., Casanovas O., Viñals F., Affolter M., Gerhardt H., Huveneers S., Belting H.-G., Cutillas P. R., Graupera M., Endothelial cell rearrangements during vascular patterning require PI3-kinase-mediated inhibition of actomyosin contractility. Nat. Commun. 9, 4826 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Z., Tan J. L., Cohen D. M., Yang M. T., Sniadecki N. J., Ruiz S. A., Nelson C. M., Chen C. S., Mechanical tugging force regulates the size of cell-cell junctions. Proc. Natl. Acad. Sci. U.S.A. 107, 9944–9949 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Cras T. D., Goines J., Lakes N., Pastura P., Hammill A. M., Adams D. M., Boscolo E., Constitutively active PIK3CA mutations are expressed by lymphatic and vascular endothelial cells in capillary lymphatic venous malformation. Angiogenesis 23, 425–442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Madsen R. R., Knox R. G., Pearce W., Lopez S., Mahler-Araujo B., McGranahan N., Vanhaesebroeck B., Semple R. K., Oncogenic PIK3CA promotes cellular stemness in an allele dose-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 116, 8380–8389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun H., King A. J., Diaz H. B., Marshall M. S., Regulation of the protein kinase Raf-1 by oncogenic Ras through phosphatidylinositol 3-kinase, Cdc42/Rac and Pak. Curr. Biol. 10, 281–284 (2000). [DOI] [PubMed] [Google Scholar]
- 59.Ebi H., Costa C., Faber A. C., Nishtala M., Kotani H., Juric D., Della Pelle P., Song Y., Yano S., Mino-Kenudson M., Benes C. H., Engelman J. A., PI3K regulates MEK/ERK signaling in breast cancer via the Rac-GEF, P-Rex1. Proc. Natl. Acad. Sci. U.S.A. 110, 21124–21129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mendoza M. C., Er E. E., Blenis J., The Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends Biochem. Sci. 36, 320–328 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ren A. A., Snellings D. A., Su Y. S., Hong C. C., Castro M., Tang A. T., Detter M. R., Hobson N., Girard R., Romanos S., Lightle R., Moore T., Shenkar R., Benavides C., Beaman M. M., Müller-Fielitz H., Chen M., Mericko P., Yang J., Sung D. C., Lawton M. T., Ruppert J. M., Schwaninger M., Körbelin J., Potente M., Awad I. A., Marchuk D. A., Kahn M. L., PIK3CA and CCM mutations fuel cavernomas through a cancer-like mechanism. Nature 594, 271–276 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Snellings D. A., Girard R., Lightle R., Srinath A., Romanos S., Li Y., Chen C., Ren A. A., Kahn M. L., Awad I. A., Marchuk D. A., Developmental venous anomalies are a genetic primer for cerebral cavernous malformations. Nat. Cardiovasc. Res. 1, 246–252 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim S., Lee H., Chung M., Jeon N. L., Engineering of functional, perfusable 3D microvascular networks on a chip. Lab Chip 13, 1489–1500 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Polacheck W. J., Charest J. L., Kamm R. D., Interstitial flow influences direction of tumor cell migration through competing mechanisms. Proc. Natl. Acad. Sci. U.S.A. 108, 11115–11120 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Polacheck W. J., Kutys M. L., Tefft J. B., Chen C. S., Microfabricated blood vessels for modeling the vascular transport barrier. Nat. Protoc. 14, 1425–1454 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tinevez J. Y., Perry N., Schindelin J., Hoopes G. M., Reynolds G. D., Laplantine E., Bednarek S. Y., Shorte S. L., Eliceiri K. W., TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Ershov D., Phan M.-S., Pylvänäinen J. W., Rigaud S. U., le Blanc L., Charles-Orszag A., Conway J. R. W., Laine R. F., Roy N. H., Bonazzi D., Duménil G., Jacquemet G., Tinevez J.-Y., TrackMate 7: Integrating state-of-the-art segmentation algorithms into tracking pipelines. Nat. Methods 19, 829–832 (2022). [DOI] [PubMed] [Google Scholar]
- 68.Stringer C., Wang T., Michaelos M., Pachitariu M., Cellpose: A generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021). [DOI] [PubMed] [Google Scholar]
- 69.Wortel I. M. N., Liu A. Y., Dannenberg K., Berry J. C., Miller M. J., Textor J., CelltrackR: An R package for fast and flexible analysis of immune cell migration data. ImmunoInformatics 1, 100003 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Polacheck W. J., Kutys M. L., Yang J., Eyckmans J., Wu Y., Vasavada H., Hirschi K. K., Chen C. S., A non-canonical Notch complex regulates adherens junctions and vascular barrier function. Nature 552, 258–262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bumgarner J. R., Nelson R. J., Open-source analysis and visualization of segmented vasculature datasets with VesselVio. Cell Rep. Methods 2, 100189 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Legends for movies S1 and S2
Uncropped western blots
Tabular format of data from all figures
Movies S1 and S2