Abstract
Clostridium perfringens is a ubiquitous spore-forming anaerobic pathogen that is frequently associated with enteric disease in chickens. Moreover, enterotoxin-producing C. perfringens has high zoonotic potential as well as serious public health concerns due to the emanation of food-borne intoxication. The present study was designed to isolate, identify, and toxinotype C. perfringens from both healthy and cases of necrotic or ulcerative enteritis chickens. A total of 110 samples were collected from July 2019 to February 2021. Among the samples, 38 (34.5%, 95% CI: 26.39–43.83) were positive for C. perfringens and were obtained from broiler 21 (33.3%, 95% CI: 22.91–45.67), Sonali 9 (34.6%, 95% CI: 19.31–53.88), and layer 8 (38%, 95% CI: 20.68–59.20). C. perfringens was highly prevalent (35.7%, 95% CI: 25.48–47.44) in enteritis chickens compared with healthy ones. In multiplex PCR toxinotyping, 34 (89.4%) isolates were identified as C. perfringens type A by the presence of the alpha toxin gene (cpa). Moreover, in addition to the cpa gene, 3 (14.3%, 95% CI: 4.14–35.48) broiler and 1 (11.1%, 95% CI: 0.01–45.67) Sonali isolates harbored the enterotoxin gene (cpe) and were classified as type F. However, none of the isolates carried genes encoding beta (cpb), epsilon (etx), iota (iap), or beta-2 (cpb2) toxins. Multivariable logistic regression analysis identified the following variables such as; “previously used litter materials” (OR 21.77, 95% CI 2.22–212.66, p ≤ 0.008); intestinal lesions, “presence of ulceration” (OR 30.01, 95% CI 3.02–297.91, p ≤ 0.004); “ballooned with gas” (OR 24.74, 95% CI 4.34–140.86, p ≤ 0.001) and “use of probiotics” (OR 5.24, 95% CI 0.74–36.75, p ≤ 0.095) act as risk factors for C. perfringens colonization in chicken gut. This is the first study of molecular toxinotyping of C. perfringens from healthy and enteric-diseased chickens in Bangladesh, which might have a potential food-borne zoonotic impact on human health.
1. Introduction
C. perfringens is an anaerobic, spore forming enteric pathogen that causes both clinical and subclinical enteric disease in chickens. The most severe clinical form of enteric disease is necrotic enteritis, which is characterized by a ballooned, friable intestine with necrosis of the intestinal mucosa that is often covered by a tan-to-yellow pseudomembrane [1]. In addition, due to secondary bacterial infection and a roughened intestinal mucosal surface, it appears like a Turkish towel [1]. The clinical form of the disease is associated with a huge economic burden [2], and the subclinical form of the disease significantly reduces the growth performance of chickens by causing extensive damage to the gut epithelial layer [3]. The principal mechanism of disease manifestation by C. perfringens is associated with the release of six major extracellular toxins, which are described as alpha (α), beta (β), epsilon (ε), iota (ι), enterotoxin (cpe), and NetB. Based on the production of the abovementioned toxins, C. perfringens is classified into seven toxinogenic types, A to G [4, 5]. In the toxinogenic typing scheme, type A and all other types of C. perfringens produce alpha (a) toxin. In addition to alpha (α) toxin, type B produces beta (β) and epsilon (ε) toxins, type C produces beta (β) toxin, type D produces epsilon (ε) toxin, type E produces iota (ι) toxin, type F produces enterotoxin (cpe), and type G produces NetB toxin [4, 5]. Among all the C. perfringens types, type F is most frequently associated with food poisoning in humans and causes food-borne illness by producing an enterotoxin (cpe) [4–7]. Still, to date, it is positioned as the third most common food poisoning agent in the industrialized world [2].
Previously, the typing methodology for C. perfringens was based on a traditional toxin neutralization bioassay using mice or guineapigs [8]. Later, these typing schemes were replaced by enzyme-linked immunosorbent assays (ELISA) [9]. But, in the last few decades, C. perfringens typing schemes were accurately replaced by more convenient multiplex-polymerase chain reaction (M-PCR) assays targeting toxin-encoding genes [5, 10–13].
Although clinical necrotic enteritis cases are frequently found in poultry farms in Bangladesh, they are all diagnosed based only on clinical signs and postmortem findings. To date, this is the first study on the molecular identification of C. perfringens, which is circulating in chicken flocks in Bangladesh. As a result, the purpose of this study was to determine the phenotypic and toxinogenic typing of C. perfringens isolates from healthy and necrotic enteritis-infected chickens in Bangladesh.
2. Materials and Methods
2.1. Samples
We collected intestinal swabs and intestinal contents from broiler, Sonali (a crossbreed of Fayoumi and Rhode Island Red), and layer chickens from July 2019 to February 2021. Samples were collected from chickens that were brought to the poultry practitioners for postmortem examinations at the Department of Pathology and Parasitology and the Department of Physiology, Biochemistry, and Pharmacology at Chattogram Veterinary and Animal Sciences University (CVASU). The samples only included chickens that showed enteric disease, including necrotic or ulcerative lesions during postmortem examination (Figures 1(a) and 1(b)). Additionally, specimens were collected from apparently healthy birds from live-broiler markets that were kept for selling. Intestinal swabs and intestinal contents (Figures 1(c) and 1(d)) were collected in 10 mL of cooked meat medium (Oxoid Limited, Hampshire, England) and immediately transported on ice to the Department of Microbiology and Veterinary Public Health laboratory for isolation and identification of C. perfringens.
Figure 1.
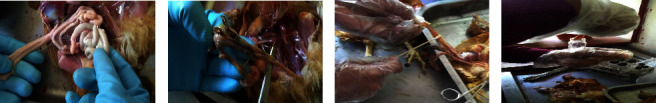
Postmortem examinations of chickens revealed the presence of intestinal lesions like (a) intestine balloons with gas, (b) ulceration, (c) collection of swab samples, and (d) intestinal contents for the isolation and identification of C. perfringens.
2.2. Isolation and Identification of C. perfringens
For primary enrichment of C. perfringens, cooked meat medium containing samples was incubated anaerobically at 37°C for 48 hrs in an anaerobic jar (Oxoid Limited, Thermo Fisher Scientific Inc., UK) with an anaerobic GasPak (Oxoid Limited, Hampshire, England). The pre-enriched samples were then inoculated onto 5% bovine blood agar with added colistin sulfate (2 mg/litter) and incubated anaerobically for 24 hours. The characteristic colonies with double zones of beta hemolysis (inner zone: complete hemolysis; outer zone: partial hemolysis) were presumptively identified as C. perfringens. For further confirmation, the suspected bacterial colonies were subjected to Gram staining (Gram positive, large bacilli) and the catalase test (negative). Two to three subcultures were performed to obtain the pure culture, and finally, the pure colonies were subcultured on brain heart infusion broth (BHI) (Oxoid Limited, Thermo Fisher Scientific Inc., UK). All the isolates were preserved at −80°C for further analysis.
2.3. Extraction of DNA and Identification of C. perfringens Isolates by PCR
The preparation of template DNA was performed according to previously published literature [14]. Briefly, three to five pure bacterial colonies were suspended in 150 μl of ultrapure water in a 1.5 ml microcentrifuge tube, boiled at 100°C for 10 min, and immediately cooled at −20°C for five minutes. Finally, the cell lysates were centrifuged at 12,000 g for 5 minutes, and 100 μl of the supernatant containing template DNA was transferred into a microcentrifuge tube and stored at −20°C for further analysis. A species-specific primer (16S rRNA gene) was used for confirmation of C. perfringens [15]. A total of 25 µl of the PCR reaction was prepared with 12.5 µl of Taq PCR MasterMix (Qiagen, Merelbeke, Belgium), 1 µl (20 picomole/µl) of both forward and reverse primer, 4 µl (5 ng/µl) of template DNA, and 6.5 µl of nuclease-free water. Amplification was performed in the thermal cycler (DLAB Scientific Inc., USA) with the PCR program consisting of initial denaturation at 95°C for 15 min, followed by 94°C for 30 sec, 53°C for 1.5 min, and 72°C for 1.5 min for 35 cycles, and final extension at 72°C for 10 min.
2.4. Toxinotyping by PCR
After confirmation of the C. perfringens species, all positive isolates were selected for toxinotyping using five different toxin genes α (cpa), β (cpb), ε(etx), ί(iap), and enterotoxin (cpe); their primer sequences are listed in Table 1. Multiplex PCR (m-PCR) was performed for specific amplification of the toxinotyping gene, which was described in a previous study [11]. For m-PCR, a total volume of 50 µl reaction was prepared, which comprises 25 µl of Taq PCR Master Mix, 1 µl (20 pmol/µl) of each primer for six toxin genes, 4 µl of C. perfringens confirmed genomic DNA, and 9 µl of nuclease-free water for every sample. A uniplex PCR was performed for the detection of β2 (cpb-2) and enterotoxin (cpe) gene, comprising 25 µl of the reaction volume. Also, the similar PCR condition was followed to perform the uniplex PCR. Finally, all of the PCR products were separated on an ethidium bromide-stained 1.5% agarose gel (Oxoid Limited, Thermo Fisher Scientific Inc., UK) and visualized using UV lights in a gel documentation system (Alphalmager, Alpha Innotech, San Leandro, CA, USA). Previously confirmed C. perfringens isolates were used as positive controls, and nuclease-free water was used as a negative control.
Table 1.
Oligonucleotide primer sequences and amplified PCR product sizes were used for the detection of C. perfringens and its toxin genes.
| Name of bacteria and toxins | Target gene | Primers name | Primer sequences (5′-3′) | PCR product size (bp) | Ref |
|---|---|---|---|---|---|
| C. perfringens | 16S RNA | ClPer-1 | TAACCTGCCTCATAGAGT | 481 | [15] |
| ClPer-2 | TTTCACATCCCACTTAATC | ||||
|
| |||||
| Alpha (α) | cpa | CPAlphaF | GCTAATGTTACTGCCGTTGA | 324 | [11] |
| CPAlphaR | CCTCTGATACATCGTGTAAG | ||||
| Beta (β) | cpb | CPBetaF3 | GCGAATATGCTGAATCATCTA | 195 | |
| CPBetaR3 | GCAGGAACATTAGTATATCTTC | ||||
| Epsilon (ε) | etx | CPEpsilonF | TGGGAACTTCGATACAAGCA | 376 | |
| CPEpsilonR2 | AACTGCACTATAATTTCCTTTTCC | ||||
| Iota (ι) | iap | CPIotaF2 | AATGGTCCTTTAAATAATCC | 272 | |
| CpIotaR | TTAGCAAATGCACTCATATT | ||||
| Beta-2 (β2) | cpb2 | CPBeta2totalF2 | AAATATGATCCTAACCAACAA | 548 | |
| CPBeta2totalR | CCAAATACTCTAATYGATGC | ||||
| Enterotoxin | cpe | CPEnteroF | TTCAGTTGGATTTACTTCTG | 485 | |
| CPEnteroR | TGTCCAGTAGCTGTAATTT | ||||
2.5. Data Analysis
All targeted demographic as well as postmortem data was recorded into a Microsoft Excel 2010 spread sheet. The prevalence was calculated by considering the number of positive C. perfringens isolates as the numerator, divided by the number of chickens sampled as the denominator. Chi-square test was performed to find out the association between the binary result of C. perfringens and the farm and chicken factors. Firstly, univariable logistic regression analysis was performed to identify possible risk factors, and subsequently, any factor having a p-value of ≤0.20 was selected to build the further multivariable logistic regression model. Any variables with a p-value of 0.05 were considered significant and kept in the final model. All descriptive and analytical analyses were performed using STATA®13.0 software [16]. Finally, the representative heat map was constructed using Graphpad Prism (version 7.05).
3. Results
3.1. Samples
A total of 110 intestinal samples were collected from broiler (63), Sonali (26), and layer (21) chickens; 40 samples from healthy broiler and 70 from enteric-diseased chicken with various intestinal lesions.
3.2. Prevalence of C. perfringens
A total of 38 (34.5%, 95% CI: 26.39 to 43.83) PCR-confirmed C. perfringens isolates were recovered from 110 chickens (Table 2). Of them, 21 (33.3%, 95% CI: 22.91 to 45.67) isolates were obtained from broiler, 9 (34.6%, 95% CI: 19.31 to 53.88) from Sonali, and 8 (38%, 95% CI: 20.68 to 59.20) from layer chickens (Figure 2). Among the C. perfringens isolates, 13 (32.5%, 95% CI: 20.01 to 48.06) were recovered from healthy broiler chickens, and 25 (35.7%, 95% CI: 25.48 to 47.44) were from diseased chickens (Table 2).
Table 2.
Distribution of C. perfringens isolates and its toxins from different chickens.
| Chickens | No. of samples | PCR positive isolates | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Live bird market (healthy) | Clinical case (diseased) | Cpa(%) | α | β | ε | ι | β2 | Etb(%) | |
| Broiler | 40 | 23 | 21 (33.3) | 21 | 0 | 0 | 0 | 0 | 3 (14.3) |
| Sonali | 0 | 26 | 9 (34.6) | 9 | 0 | 0 | 0 | 0 | 1 (11.1) |
| Layer | 0 | 21 | 8 (38.0) | 8 | 0 | 0 | 0 | 0 | 0 |
| Total | 110 | 38 (34.5) | 38 | 0 | 0 | 0 | 0 | 4 (10.5) | |
a Clostridium perfringens, benterotoxin.
Figure 2.
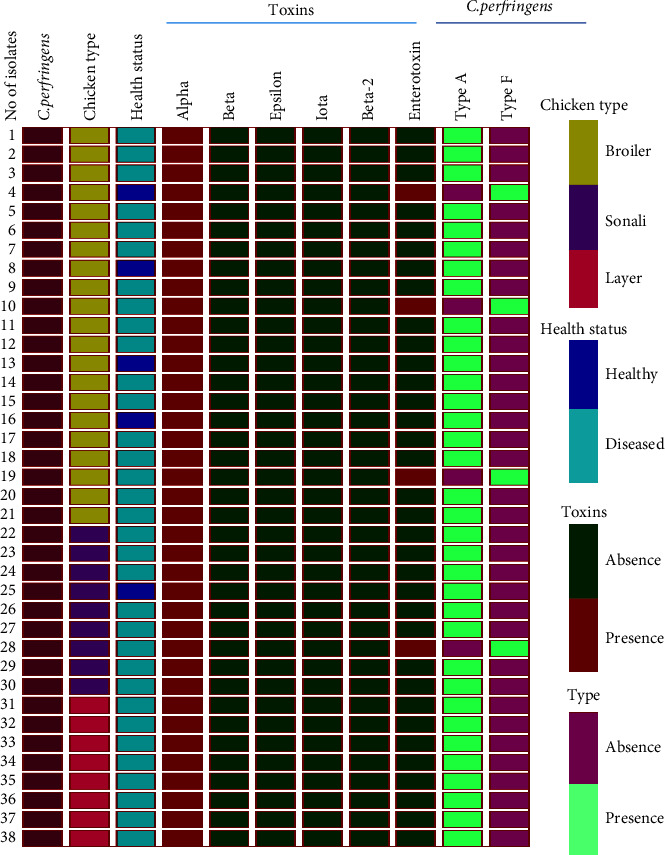
Heat map illustrates the chicken types and their health status as well as the distribution of different C. perfringens types and their toxins.
3.3. Toxin Typing and Distribution of C. perfringens
All C. perfringens isolates (38) encode the alpha toxin gene (cpa) (Table 2), and 34 (89.4%) are classified as type A (Figure 2). The type A C. perfringens was distributed among the broiler, Sonali, and layer chickens, which were 18 (47.3%), 8 (21.0%), and 8 (21.0%), respectively. In addition to the alpha toxin gene (cpa), 4 (10.5%) isolates also harbored the enterotoxin gene (cpe), which is classified as type F (Figure 2). Moreover, these enterotoxin-producing isolates were obtained from Sonali (1) and broiler (3) chickens (Figure 2). Of the three enterotoxin-positive isolates from broiler chickens, one isolate was recovered from healthy chickens, and the remaining two isolates were found in diseased chickens. None of the isolates encoded beta, iota, epsilon, or beta 2-toxin genes (Figure 2).
3.4. Risk Factors Associated with the Harboring of C. perfringens in Enteric Diseased Chicken
The univariable analysis identified five potential risk factors (p ≤ 0.20) associated with the harboring of C. perfringens in enteric diseased chickens (Table 3). In the subsequent multivariable analysis, three variables were identified as significant risk factors associated with the presence of C. perfringens. The significantly associated variables were: “previously used litter materials” (OR 21.77, 95% CI 2.22–212.66, p ≤ 0.008), intestinal lesions “ulceration” (OR 30.01, 95% CI 3.02–297.91, p ≤ 0.004) and “ballooned with gas” (OR 24.74, 95% CI 4.34–140.86, p ≤ 0.000), and “use of probiotics” (OR 5.24, 95% CI 0.74–36.75, p ≤ 0.095) (Table 4).
Table 3.
Univariable logistic regression analysis of risk factors for the presence of C. perfringens in diseased chicken.
| Variables | Covariable | No. of chickens | No. of chickens positive for C. perfringens(%) | 95% CIa | OR (95% CI) | p-valueb |
|---|---|---|---|---|---|---|
| Farm location | Hilly area | 6 | 1 (16.67) | 0.42–64.12 | Reference | 0.309 |
| Plain land | 64 | 24 (37.50) | 25.70–50.49 | 3 (0.33–27.23) | ||
|
| ||||||
| Birds type | Layer | 21 | 8 (38.10) | 18.10–61.56 | 1.16 (0.35–3.84) | 0.964 |
| Broiler | 23 | 8 (34.78) | 16.37–57.26 | 1 (0.30–3.27) | ||
| Sonali | 26 | 9 (34.62) | 17.21–55.66 | Reference | ||
|
| ||||||
| Flock sizec | Small | 9 | 3 (33.33) | 7.48–70.07 | 1.3 (0.23–7.31) | 0.674 |
| Medium | 18 | 5 (27.78) | 9.69–53.48 | Reference | ||
| Large | 43 | 17 (39.53) | 24.97–55.59 | 1.7 (0.13–1.07) | ||
|
| ||||||
| Chicken aged | Chick | 3 | 0 (0.00) | — | 1.71 (0.02–2.23) | 0.619 |
| Pullet | 4 | 2 (50.00) | 6.75–93.24 | 4 (0.21–75.65) | ||
| Finisher | 5 | 1 (20.00) | 0.50–71.64 | Reference | ||
| Hen | 14 | 6 (42.86) | 17.66–71.13 | 3 (0.26–34.19) | ||
| Starter | 20 | 6 (30.00) | 11.89–54.27 | 1 (−) | ||
| Grower | 24 | 10 (41.67) | 22.10–63.35 | 2.85 (0.27–29.56) | ||
|
| ||||||
| Feed types | Homemade | 7 | 3 (42.86) | 9.89–81.59 | 0.73 (0.14–3.65) | 0.907 |
| Mash | 15 | 5 (33.33) | 11.82–61.61 | Reference | ||
| Pellet | 48 | 17 (35.42) | 22.16–50.54 | 0.66 (0.10–4.20) | ||
|
| ||||||
| Litter materials | No use | 11 | 3 (27.27) | 6.02–60.97 | 3.37 (0.46–24.28) | 0.006b |
| New | 20 | 2 (10.00) | 1.23–61.69 | Reference | ||
| Previously used | 39 | 20 (35.71) | 34.78–67.58 | 9.47 (1.93–46.46) | ||
|
| ||||||
| Materials type | Straw | 5 | 3 (60.00) | 14.66–94.72 | 4 (0.43–37.10) | 0.606 |
| Cage | 11 | 3 (27.27) | 6.02–60.97 | Reference | ||
| Rice husk | 15 | 6 (40.00) | 16.33–67.02 | 1.33 (0.30–5.88) | ||
| Saw dust | 39 | 13 (33.33) | 19.08–50.21 | 1.77 (0.33–9.55) | ||
|
| ||||||
| Floor type | Muddy | 5 | 2 (40.00) | 5.27–85.33 | 1.70 (0.24–11.95) | 0.475 |
| Semi pucca | 32 | 9 (28.13) | 13.74–46.74 | Reference | ||
| Concrete | 33 | 14 (42.42) | 25.47–60.78 | 1.88 (0.66–5.29) | ||
|
| ||||||
| Water source | Pond | 1 | 0 (0.00) | — | Omitted | 0.730 |
| Tube well | 29 | 10 (34.48) | 17.93–54.33 | Reference | ||
| Tank water | 40 | 15 (37.50) | 22.72–54.19 | 1.14 (0.42–3.09) | ||
|
| ||||||
| Vaccination | Yes | 61 | 22 (36.07) | 24.16–49.37 | 1.12 (0.25–4.96) | 0.873 |
| No | 9 | 3 (33.33) | 7.48–70.07 | Reference | ||
|
| ||||||
| Deworming | Yes | 25 | 9 (36.00) | 17.97–57.47 | 1.01 (0.36–2.82) | 0.970 |
| No | 45 | 16 (35.56) | 21.86–51.21 | Reference | ||
|
| ||||||
| Use of antibiotics | Yes | 54 | 23 (42.59) | 29.23–56.79 | 5.19 (1.07–25.13) | 0.027 |
| No | 16 | 2 (12.50) | 1.55–38.34 | Reference | ||
|
| ||||||
| Use of probiotics | Yes | 13 | 7 (53.85) | 25.13–80.77 | 2.52 (0.74–8.60) | 0.131 |
| No | 57 | 18 (31.58) | 19.90–45.24 | Reference | ||
|
| ||||||
| Use of feed enzymes | Yes | 31 | 12 (38.71) | 21.84–57.81 | 1.26 (0.47–3.37) | 0.641 |
| No | 39 | 13 (33.33) | 19.08–50.21 | Reference | ||
|
| ||||||
| Use of feed supplements | Yes | 63 | 24 (38.10) | 26.14–51.20 | 3.69 (0.41–32.57) | 0.212 |
| No | 7 | 1 (14.29) | 0.36–57.87 | Reference | ||
|
| ||||||
| Intestinal lesions | Ballooned with gas | 5 | 2 (40.00) | 5.27–85.33 | 6.22 (0.72–53.37) | 0.001b |
| Ulceration | 13 | 5 (38.46) | 13.85–68.42 | 5.83 (1.13–29.85) | ||
| Necrosis | 21 | 15 (71.43) | 47.82–88.71 | 23.33 (5.09–106.81) | ||
| Hemorrhage | 31 | 3 (9.68) | 2.04–25.75 | Reference | ||
a-Confidence interval, b-significance, which is ≤0.05, c-flock size which are small (≤500), medium (501–1000) and large (≥1000), d-chicken age: in case of broiler and Sonali, [starter (1–21 days), grower (22–42) and finisher (≥42 days)]; in case of layer, [chick (1–28 days), pullet (29–105 days), and hen (≥105 days)].
Table 4.
Multivariable logistic regression analysis of risk factors for the presence of C. perfringens in diseased chicken.
| Outcome variable | Explanatory variable | Description | OR∗(95%CI) | p-value |
|---|---|---|---|---|
| Clostridium perfringens | Litter materials | No use | 5 | 0.218 |
| New | 1 | Reference | ||
| Previously used | 21.77 (2.22–212.66) | 0.008 | ||
| Intestinal lesions | Hemorrhage | 1 | Reference | |
| Ulceration | 30.01 (3.02–297.91) | 0.004 | ||
| Necrosis | 3.17 (0.22–44.04) | 0.388 | ||
| Ballooned with gas | 24.74 (4.34–140.86) | 0.001 | ||
| Use of probiotics | No | 1 | Reference | |
| Yes | 5.24 (0.74–36.75) | 0.095 |
∗ Odds ratio.
4. Discussion
C. perfringens are spore-forming bacilli that commonly inhabit soil, poultry litter, and are also harbored as gut pathogens in chickens and other animals [17, 18]. The different toxinotypes cause a wide variety of significant systemic and enteric diseases in different species of animals, including chickens. Another significant aspect is that it is involved in food-borne intoxication (food-borne zoonosis), which evolved from consumption of different raw and canned foods, particularly chicken meat and meat products [19].
In the present study, the overall prevalence of C. perfringens in chicken is 34.5%, but in some countries, particularly Jordan and Egypt, it is higher than 40% [20, 21]. Besides, the prevalence of C. perfringens in broiler chicken was 33.3% in our study, which is slightly higher than the recent findings of Praveen Kumar et al. [22] and Zhang et al. [23]. They described the prevalence as 21.97% and 23.1% in broiler chicken in India and China, respectively. This variation may be due to geographical conditions as well as the earlier contaminated soil and unchanged bedding materials of the poultry farm. As C. perfringens is a spore-forming pathogen that can survive in soil for an extended period of time, chickens are frequently infected. On the other hand, chickens that harbor C. perfringens continuously shed it in the poultry litter and environment, and it acts as a potential source of infection for chickens. Cross-infection through droppings, litter materials, poultry waste products, feed, and water is a major route of infection for chickens and other animals [23, 24]. In addition, the prevalence of C. perfringens may increase significantly when chickens have a concurrent clinical or subclinical coccidial infection in the gut [25].
Toxinotyping of C. perfringens by multiplex PCR assay displayed the presence of the alpha toxin gene in all isolates and the absence of other major lethal toxin genes except the enterotoxin (cpe) gene, which was expressed by type F isolates. The current findings demonstrate that 34 (89.4%) of the PCR-confirmed C. perfringens isolates are type A. This study revealed that C. perfringens type A is the most dominant toxin type circulating both in healthy and diseased chickens. These findings are strongly supported by several previous studies [2, 20, 23, 26]. However, none of the C. perfringens isolates carried the cpb, etx, or iap toxin genes, which indicate the absence of C. perfringens toxinotypes B, C, D, and E in this study. Similar findings were also previously reported [26–28], showing that C. perfringens obtained from chickens encoded only the alpha toxin gene. In addition to the four major toxinotyping toxins, β2 toxin (cpb2) was also absent in all chicken isolates, which were considered minor or nonessential virulence factors for disease manifestation in chicken [29].
Although C. perfringens is a significant food-borne zoonotic pathogen, very limited microbiological and epidemiological studies have been reported in Bangladesh. In our study, 4 (10.5%) of the type F isolates harbored the enterotoxigenic gene (cpe), which is a potential virulence determinant of C. perfringens for food poisoning in humans [4, 5, 19, 30]. The low incidence of enterotoxin in chicken may be due to sample source variation as well as the genetic absence of an encoded gene. However, in addition to food-borne intoxication, the enterotoxin producing C. perfringens strains also frequently cause neonatal diarrhea [31]. Furthermore, the enterotoxin gene (cpe) encoded by C. perfringens type F isolates clearly poses the risk of human food-borne intoxication as well as food-borne zoonosis [4, 5, 32]. Finally, it is recognized that any C. perfringens isolates with alpha toxin and enterotoxin have a significant impact on the production of necrotic enteritis in chickens and also cause fatal food-borne enteric disease in humans. However, in the current study, we classified C. perfringens from A to F, with the exception of type G, which harbored a net B toxin.
We identified previously used litter materials as a potential risk factor for the colonization of C. perfringens in chickens. Besides, the presence of associated lesions, such as ulceration and ballooned gas in the chicken gut, is also asserted as a colonization factor. Long-term and repeated use of the same litter materials in poultry houses may harbor the spores of C. perfringens [23, 33] and act as an initial trigger of exposure. The presence of intestinal lesions may involve the initial intestinal damage caused by coccidial pathogens [34] and subsequent colonization of C. perfringens. Finally, C. perfringens releases various toxins and enzymes responsible for gut lesions in chicken. Moreover, further details on the sequencing of C. perfringens isolates helped to find out the mechanisms of colonization and toxin production, as well as the control of necrotic enteritis in chickens and food-borne zoonosis in humans.
5. Conclusions
This study describes the prevalence and toxinotyping of C. perfringens isolated from chicken in Bangladesh. We found that 33.3% of broiler, 34.6% Sonali, and 38.0% of layer chickens tested positive for C. perfringens. It is interesting that, among the enteric-diseased and healthy chickens, the prevalence of C. perfringens was higher in the enteric-diseased birds (35.7%). In the present investigation, alpha toxin-producing C. perfringens type A was the predominant genotype, and relatively considerable numbers of isolates were type F (10.5%) that carried the enterotoxin gene (cpe). Previously used litter materials were identified as risk factors that were associated with a higher prevalence of C. perfringens in chickens.
Acknowledgments
The authors would like to thank the Department of Pathology and Parasitology, and the Department of Physiology, Biochemistry and Pharmacology, at Chattogram Veterinary and Animal Sciences University (CVASU) for supporting the collection of the clinical specimens from chickens after completing the postmortem. This study was financially supported by the Director, Research and Extension, CVASU and Ministry of Science and Technology, Bangladesh.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Ethical Approval
Ethical approval was obtained from the institutional ethical approval committee of Chattogram Veterinary and Animal Sciences University (Approval no. CVASU/Dir (R&E) EC/2019/12 (5/9)) to implement the study.
Consent
Informed consent was obtained from all individual farm owners included in the study.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Conceptualization, funding acquisition, data curation, investigation, methodology, project administration, resources, validation, formal analysis, visualization, writing the original draft, and writing, reviewing and editing the manuscript were performed by EAR. TAN contributed to the investigation, methodology, reviewing, and editing the manuscript. MSI was involved in visualization, validation, and reviewing, and editing of the manuscript. HB was involved in supervision, visualization, validation, and reviewing, and editing the manuscript. MZI was involved in conceptualization, supervision, visualization, validation, and reviewing and editing of the manuscript. All authors read and approved the final manuscript.
References
- 1.Keyburn A. L., Boyce J. D., Vaz P., et al. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathogens . 2008;4(2):p. e26. doi: 10.1371/journal.ppat.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dar P. S., Wani S. A., Wani A. H., et al. Isolation, identification and molecular characterization of Clostridium perfringens from poultry in Kashmir valley, India. Journal of Entomology and Zoology Studies . 2017;5(5):409–414. [Google Scholar]
- 3.Hussein E. O. S., Ahmed S. H., Abudabos A. M., et al. Effect of antibiotic, phytobiotic and probiotic supplementation on growth, blood indices and intestine health in broiler chicks challenged with Clostridium perfringens. Animals . 2020;10(3):p. 507. doi: 10.3390/ani10030507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnston M. D., Whiteside T. E., Allen M. E., Kurtz D. M. Toxigenic profile of Clostridium perfringens strains isolated from natural ingredient laboratory animal diets. Comparative Medicine . 2022;72(1):50–58. doi: 10.30802/AALAS-CM-22-000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rood J. I., Adams V., Lacey J., et al. Expansion of the Clostridium perfringens toxin-based typing scheme. Anaerobe . 2018;53(53):5–10. doi: 10.1016/j.anaerobe.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher D. J., Miyamoto K., Harrison B., Akimoto S., Sarker M. R., McClane B. A. Association of beta2 toxin production with Clostridium perfringens type A human gastrointestinal disease isolates carrying a plasmid enterotoxin gene. Molecular Microbiology . 2005;56(3):747–762. doi: 10.1111/j.1365-2958.2005.04573.x. [DOI] [PubMed] [Google Scholar]
- 7.Hailegebreal G. A review on Clostridium perfringens food poisoning. Global Research Journal of Public Health and Epidemiology . 2017;4(3):104–109. [Google Scholar]
- 8.Smith T. J., Hill K. K., Raphael B. H. Historical and current perspectives on Clostridium botulinum diversity. Research in Microbiology . 2015;166(4):290–302. doi: 10.1016/j.resmic.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadi̇mli̇ H. H., Erganiş O., Sayin Z., Aras Z. Toxinotyping of Clostridium perfringens isolates by ELISA and PCR from lambs suspected of enterotoxemia. Turkish Journal of Veterinary and Animal Sciences . 2012;36(4):409–415. doi: 10.3906/vet-1008-437. [DOI] [Google Scholar]
- 10.Mohamed M. A., Ahmed S. Y. O., Abdel Motelib T. Y. Associated profiles of virulence gene markers in Clostridium Perfringens strains isolated from healthy and diseased broiler chickens with nectrotic enteritis. Assiut Veterinary Medical Journal . 2009;55(123):1–8. doi: 10.21608/AVMJ.2009.175005. [DOI] [Google Scholar]
- 11.Van Asten A. J., van der Wiel C. W., Nikolaou G., Houwers D. J., Gröne A. A multiplex PCR for toxin typing of Clostridium perfringens isolates. Veterinary Microbiology . 2009;136(3-4):411–412. doi: 10.1016/j.vetmic.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 12.Kupradit C., Rodtong S., Ketudat-Cairns M. Novel multiplex PCR to specifically detect bacterial foodborne pathogens. Suranaree Journal of Science and Technology . 2013;20(1):73–82. [Google Scholar]
- 13.Nguyen T. T., Vu-Khac H., Nguyen T. D. Isolation and characterization of Clostridium perfringens strains isolated from ostriches (Struthio camelus) in Vietnam. August-2020 . 2020;13(8):1679–1684. doi: 10.14202/vetworld.2020.1679-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar N. V., Sreenivasulu D., Reddy Y. N. Identification of Clostridium perfringens type C and D from enterotoxemia suspected lambs in Andhra Pradesh. Indian Veterinary Journal . 2016;93(1):29–31. [Google Scholar]
- 15.Tonooka T., Sakata S., Kitahara M., et al. Detection and quantification of four species of the genus Clostridium in infant feces. Microbiology and Immunology . 2005;49(11):987–992. doi: 10.1111/j.1348-0421.2005.tb03694.x. [DOI] [PubMed] [Google Scholar]
- 16.Diaz-Quijano F. A. A simple method for estimating relative risk using logistic regression. BMC Medical Research Methodology . 2012;12(1):14–16. doi: 10.1186/1471-2288-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saleem G. Necrotic Enteritis, Disease Induction, Predisposing Factors and Novel Biochemical Markers in Broilers Chickens . Glasgow G12 8QQ, UK: University of Glasgow; 2013. Doctoral dissertation. [Google Scholar]
- 18.Shrestha A., Uzal F. A., McClane B. A. Enterotoxic clostridia: Clostridium perfringens enteric diseases. Microbiology Spectrum . 2018;6(5):6–5. doi: 10.1128/microbiolspec.GPP3-0003-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stringer M. F. Clostridia in Gastrointestinal Disease . Boca Raton, Florida: CRC Press; 2018. Clostridium perfringens type A food poisoning; pp. 117–138. [DOI] [Google Scholar]
- 20.Gharaibeh S., Al Rifai R., Al-Majali A. Molecular typing and antimicrobial susceptibility of Clostridium perfringens from broiler chickens. Anaerobe . 2010;16(6):586–589. doi: 10.1016/j.anaerobe.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Osman K. M., Soliman Y. A., Amin Z. M. S., Aly M. A. K. Prevalence of Clostridium perfringens type A isolates in commercial broiler chickens and parent broiler breeder hens in Egypt. Revue Scientifique et Technique de l’OIE . 2012;31(3):931–941. doi: 10.20506/rst.31.3.2169. [DOI] [PubMed] [Google Scholar]
- 22.Praveen Kumar N., Vinod Kumar N., Karthik A. Molecular detection and characterization of Clostridium perfringens toxin genes causing necrotic enteritis in broiler chickens. Tropical Animal Health and Production . 2019;51(6):1559–1569. doi: 10.1007/s11250-019-01847-9. [DOI] [PubMed] [Google Scholar]
- 23.Zhang T., Zhang W., Ai D., et al. Prevalence and characterization of Clostridium perfringens in broiler chickens and retail chicken meat in central China. Anaerobe . 2018;54:100–103. doi: 10.1016/j.anaerobe.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Luu L., Bettridge J., Christley R. M., et al. Prevalence and molecular characterisation of Eimeria species in Ethiopian indigenous village chickens. BMC Veterinary Research . 2013;9(1):p. 208. doi: 10.1186/1746-6148-9-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allaart J. G., van Asten A. J., Gröne A. Predisposing factors and prevention of Clostridium perfringens-associated enteritis. Comparative Immunology, Microbiology and Infectious Diseases . 2013;36(5):449–464. doi: 10.1016/j.cimid.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Park J. Y., Kim S., Oh J. Y., et al. Characterization of Clostridium perfringens isolates obtained from 2010 to 2012 from chickens with necrotic enteritis in Korea. Poultry Science . 2015;94(6):1158–1164. doi: 10.3382/ps/pev037. [DOI] [PubMed] [Google Scholar]
- 27.Baums C. G., Schotte U., Amtsberg G., Goethe R. Diagnostic multiplex PCR for toxin genotyping of Clostridium perfringens isolates. Veterinary Microbiology . 2004;100(1-2):11–16. doi: 10.1016/S0378-1135(03)00126-3. [DOI] [PubMed] [Google Scholar]
- 28.Malmarugan S., Boobalan A., Dorairajan N. Necrotic Enteritis in broiler and layer farms in Tamil Nadu, India. International Journal for Agro Veterinary and Medical Sciences . 2012;6(4):241–249. doi: 10.5455/ijavms.126. [DOI] [Google Scholar]
- 29.Gholamiandekhordi A. R., Ducatelle R., Heyndrickx M., Haesebrouck F., Van Immerseel F. Molecular and phenotypical characterization of Clostridium perfringens isolates from poultry flocks with different disease status. Veterinary Microbiology . 2006;113(1-2):143–152. doi: 10.1016/j.vetmic.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 30.Miki Y., Miyamoto K., Kaneko-Hirano I., Fujiuchi K., Akimoto S. Prevalence and characterization of enterotoxin gene-carrying Clostridium perfringens isolates from retail meat products in Japan. Applied and Environmental Microbiology . 2008;74(17):5366–5372. doi: 10.1128/AEM.00783-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Athira C. K., Milton A. A. P., Reddy A., et al. Diversity of toxin-genotypes among Clostridium perfringens isolated from healthy and diarrheic neonatal cattle and buffalo calves. Anaerobe . 2018;49:99–102. doi: 10.1016/j.anaerobe.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Dolan G. P., Foster K., Lawler J., et al. An epidemiological review of gastrointestinal outbreaks associated with clostridium perfringens, North East of England, 2012–2014. Epidemiology and Infection . 2016;144(7):1386–1393. doi: 10.1017/S0950268815002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bindari Y. R., Kheravii S. K., Morton C. L., Wu S. B., Walkden-Brown S. W., Gerber P. F. Molecular detection of Eimeria species and Clostridium perfringens in poultry dust and pooled excreta of commercial broiler chicken flocks differing in productive performance. Veterinary Parasitology . 2021;291 doi: 10.1016/j.vetpar.2021.109361.109361 [DOI] [PubMed] [Google Scholar]
- 34.Adhikari P., Kiess A., Adhikari R., Jha R. An approach to alternative strategies to control avian coccidiosis and necrotic enteritis. The Journal of Applied Poultry Research . 2020;29(2):515–534. doi: 10.1016/j.japr.2019.11.005. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


