Abstract
Microorganisms are exceptional sources of a wide array of unique natural products and play a significant role in drug discovery. During the golden era, several life-saving antibiotics and anticancer agents were isolated from microbes; moreover, they are still widely used. However, difficulties in the isolation methods and repeated discoveries of the same molecules have caused a setback in the past. Artificial intelligence (AI) has had a profound impact on various research fields, and its application allows the effective performance of data analyses and predictions. With the advances in omics, it is possible to obtain a wealth of information for the identification, isolation, and prediction of the targets of secondary metabolites. In this review, we discuss drug discovery based on natural products from microorganisms with the help of AI and machine learning.
Introduction
Microorganisms are well known to produce structurally diverse secondary metabolites that are widely used in clinical settings for treating various clinical conditions, such as cancer, infectious disease, and inflammation.1 Conversely, they are also used in various other sectors, such as agriculture (as herbicides and insecticides), the food sector (as nutraceuticals), enzyme inhibitors, and for bioremediation, which uses natural products (NPs) directly or develops molecules derived from their scaffolds.2,3 Compared with synthetic molecules, NPs offer specific features in terms of structural complexity and scaffold diversity.4 The discovery of NPs has also revealed previously unknown targets in cells. For instance, rapamycin, which was isolated from a strain of Streptomyces hygroscopicus, has resulted in the identification of the mechanistic target of the rapamycin (mTOR) cell signaling pathway.5
Artificial intelligence (AI) uses computers to perform complex functions, analyse large datasets, and interpret them based on algorithms.6 AI has been used widely in various research fields and industries for decision-making and processing tasks because it provides efficient analysis and faster results with reduced human error and at times uncovers data structures difficult to obtain from other sources.7 Recently, AI has received increased attention and is being used by chemists to perform various tasks in drug discovery, as well as to identify molecular properties, process automation, plan synthetic routes, and predict the bioactivity of molecules.8–10 Based on the recent prolific growth in machine learning (ML) and the wealth of information in cloud computing in the form of databases and repositories, researchers can now gain access to big data and integrate AI/ML approaches into their tasks.
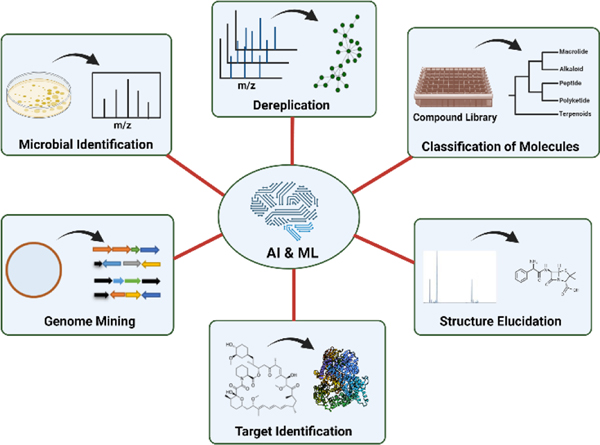
Despite the unparalleled role of NPs in drug discovery, this approach has various challenges, such as the isolation, screening, purification, and structural characterization of the NPs derived from microbial sources.11 However, in the past two decades, the repetitive identification of existing and already known NPs, the demand for resources, and the time-consuming nature of the tasks have curbed interest in NPs among researchers and industries.12 With the advancement of genomics, proteomics, metabolomics, and other omics technologies recently, it is now possible to obtain a wealth of information to identify the biosynthetic dark matter.13,14 AI/ML in the field of NPs has been growing, to analyse the extensive amount of data stemming from the omics techniques (Figure 1) and open the microbial Pandora’s box for the discovery of bioactive molecules.
Figure 1:
Application of AI/ML to various areas of microbial natural product drug discovery.
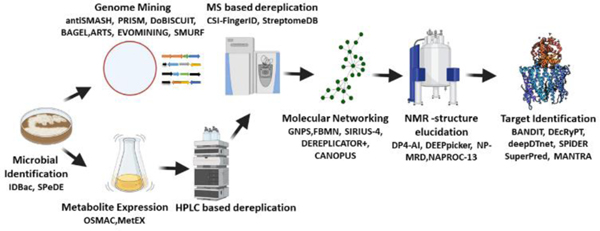
This review features the existing and emerging AI- and ML-based tools in various stages of the investigation of NPs from microorganisms. (Figure 2) We will highlight the techniques available to identify the microbes and prioritize them based on their genome and metabolite potentials. Subsequently, we will discuss fast dereplication, which is one of the major challenges in NP discovery, together with the tools available for this type of analysis. Furthermore, we will address the expedited elucidation of the structure of compounds and the identification of their targets with the aid of AI/ML. Finally, we will discuss the development of new powerful tools and the integration of multiple techniques that will speed up NP discovery, thus leading to a boom in the identification of potent drug candidates in the future.
Figure 2:
Various stages of natural product drug discovery with the corresponding available AI/ML tools.
2. Application of AI/ML in natural product discovery
2.1. Selection of organism and Taxonomic Identification.
The selection of organisms is the preliminary step in NP discovery. Certain species, such as actinomycetes, have been among the most prolific sources of pharmaceutical candidates in the past.12 However, the overmining of this limited resource has led to the repeated rediscovery of known compounds and has exhausted the identification of novel molecules in this setting.15 Although the isolation of NPs is very laborious and challenging, careful selection of underexplored microorganisms16 from untapped environments, such as marine sources17 and symbiotic sponges,18 increases the chance of identifying molecules with different scaffolds. In addition to cultured microorganisms, nearly 99% of microbial species are uncultured in the lab and hold promise in the search for new NPs. This has led to the identification of potent antibiotics, such as teixobactin19 and lassomycin,20 using specialized culture techniques.
The classical approaches in bacterial identification according to taxonomy are time-consuming and misleading; however, with the advent of the omics and ML techniques, it is possible to predict microbes efficiently.21 Although Gram staining is the gold-standard technique for the initial classification of bacteria, it is a highly time-intensive and manual-dependent activity. In contrast, using convolutional neural networks (CNNs), researchers were able to classify different shapes of Gram-positive and Gram-negative bacteria via imaging with high confidence.22 This technique can be further extended to various microorganisms, for their identification and classification using ML tools. DNA-based identification is the most accurate method of classification of various microorganisms, as in the identification of DNA from bacteria, which can also be distinguished based on the specialized metabolites they produce. In the past, the ability to correlate microbial identity with signature metabolites was limited, even with access to the vast amount of data generated by mass spectrometry. However, recently, researchers developed a technique termed IDBac with the help of ML to classify microbes based on their proteins and specialized metabolites using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS).23 Using this approach, those authors could discriminate Bacillus subtilis at the strain level based on its ability to produce cyclic peptide antibiotics and a group of Micromonospora with 99% sequence similarity with high confidence. Another algorithm called SPeDE also facilitates the identification of microbes at taxonomic resolution from a mass spectral dataset of both culture-dependent and -independent samples.24 MALDI-TOF is a powerful tool that is known for its versatility and is used in various fields with the advantage of being relatively easy to operate, fast, and accurate. The high-throughput capacity of MALDI-TOF combined with ML tools allows the rapid identification of microbial communities compared with traditional biochemical or molecular biology techniques.25 Hence, in the future, rare and underexplored microbes can be identified directly from samples with the help of ML-assisted MALDI-TOF, which will accelerate the process of candidate selection for NP screening and isolation. Another interesting application of MALDI is imaging MS (IMS), which has been used to map the spatial distribution of various secondary metabolites.26–29
2.2. Genome mining with the aid of AI/ML
The use of genome mining for secondary metabolite identification has been rapidly increasing in recent years with the advent of next-generation sequencing techniques, followed by bioinformatics pipelines.30 Although NPs are highly diverse in structure, their biosynthetic machinery, which is known as biosynthetic gene clusters (BGCs), is highly conserved in the microbes that fall under the class of polyketide synthases (PKSs),31 nonribosomally synthesized peptides (NRPs),32 ribosomally synthesized and post-translationally modified peptides, alkaloids,33 and terpenes.34 The technique begins with the identification of existing and novel BGCs from the genome sequences and further characterization of novel gene clusters, to complete the analysis. To perform this type of complex analysis using big data, ML algorithms are widely used to predict the BGC assembly lines and predict the putative encoded structure from the sequence.35 With the help of BGC databases36–41 and computational tools,42–49 NPs can be predicted based on previously characterized pathways (Table 1). Using one such tool, antiSMASH, which employs profile hidden Markov models (pHMMs) to identify the BGC, a novel polyketide named formicamycin (Figure 3) has been isolated.50 In another study, a potent antituberculous compound, gladiolin (Figure 3), was isolated with the help of genome mining from Burkholderia gladioli, which is a previously unknown source of NPs, in a patient with cystic fibrosis.52 More recently, a new class of previously unknown cryptic BGCs, i.e., lanthipeptides,51 was identified with the help of ML and deep learning (DL) strategies.
Table 1.
List of the AI/ML tools available for various phases of natural product identification and drug leads
| Task | Tool | Features | Ref |
|---|---|---|---|
| Microbial Identification with AI/ML tools | |||
| MALDI-TOF analysis | IDBac | Bioinformatics pipeline that integrates both intact protein and metabolite for detection | 23 |
| SpeDE | Identification based on unique features instead of global similarity | 24 | |
| Genome Mining AI/ML tools | |||
| BGC databases | antiSMASH database | Popular and comprehensive resource on secondary metabolite BGC | 36 |
| DoBISCUIT | Curated and literature-based collection of PKS and NRPS biosynthetic gene clusters | 37 | |
| IMG-ABC | Largest database of curated BGC from microbial genomes and metagenomes | 38 | |
| MIBiG | Collection of large curated BGC | 39 | |
| ClusterMine360 | Curated database of BGCs including produced compound(s), taxonomic information | 40 | |
| Bactibase | Integrated open-access database of bacterial antimicrobial peptides/bacteriocins | 41 | |
|
BGC Identification from Genomes
BGC databases |
antiSMASH | Most widely used tool for BGC detection based on profile Hidden Markov Models (pHMMs) | 42 |
| PRISM | BGC identification along with cheminformatic dereplication and biological activity | 43 | |
| BAGEL | Mining tool for ribosomally synthesized and post-translationally modified peptides (RIPPs) | 44 | |
| ARTS | Prioritization of the most promising BGCs encoding antibiotics with novel modes of action | 45 | |
| EvoMining | Identify secondary metabolite biosynthetic gene clusters (BGCs) based on phylogenomics | 46 | |
| SMURF | HMM-based BGC identification tool from fungi | 47 | |
| MIPS-CG | Identify completely novel BGCs using genome data in fungus alone | 48 | |
| DeepBGC | Deep learning genome-mining strategy for BGC cluster prediction | 49 | |
| BGC identification from Metagenome | MetaBGC | A read-based algorithm for the detection of BGCs directly in metagenomic sequencing data | 54 |
| eSNaPDA | Surveying and Mining BGCs from Metagenomes also take into account metadata | 57 | |
| Metabolite production and expression | |||
| Elicitor screening | MetEx | UPLC–MS-guided high-throughput elicitor screening | 70 |
| Natural product dereplication and structure elucidation with help of AI/ML | |||
| Databases | DNP | Structure database containing over 226,000 NPs with physical and chemical properties | 76 |
| MarinLit | Database of the marine natural products (Not open access) | 77 | |
| StreptomeDB | Database of NP isolated from streptomyces with chemical and biological information | 78 | |
| NPEdia | Database for Natural Products | 79 | |
| NPAtlas | Online database of microbial-derived natural products with structures and features | 80 | |
| MS based dereplication/Identification | GNPS | Online repository for untargeted MS/MS data with sample information | 96 |
| FBMN | Incorporates isotope patterns and retention time along with MN | 102 | |
| DEREPLICATOR+ | Molecular Network combined with dereplication workflow | 105 | |
| Bioactive-MN | MN guided bioassay-guided fractionation of bioactive compound(s) | 106 | |
| SIRIUS-4 | Molecular structure identification from MS/MS | 110 | |
| CANOPUS | Predict structure exclusively for which neither spectral nor reference data are available | 112 | |
| MetGem | Molecular Networks Based on the t-SNE Algorithm | 113 | |
| MESSAR | Automated prediction of metabolite substructures from tandem mass spectra | 114 | |
| Moldiscovery | Molecule identification by probabilistic model with their mass spectra | 115 | |
| FALCON | Density-based clustering of MS/MS spectra for unsupervised structure prediction | 116 | |
| SIMILE | Significant Interrelation of MS/MS Ions via Laplacian Embedding to predict the structural relationships of compounds | 117 | |
| MolNetEnhancer | Enhanced Molecular Networks by Integrating Metabolome Mining and Annotation Tools | 118 | |
| COSMIC | High-confidence structural annotation of metabolites absent from spectral libraries | 119 | |
| NMR based structure elucidation/dereplication | NAPROC-13 | A database for dereplication of NPs in mixtures based on C13 NMR | 120 |
| NP-MRD | A huge structural and NMR database of nearly 41,000 NP | 121 | |
| DP4-AI | Automated NMR data analysis for structure prediction | 122 | |
| DEEP picker | Deconvolution of complex two-dimensional NMR spectra based on DNN | 123 | |
| MixONat | Software for the Dereplication of Mixtures Based on 13C NMR Spectroscopy | 124 | |
| ELINA | 1H NMR based identification of bioactive compounds in a mixture prior to purification | 125 | |
| SMART-Miner | A convolutional neural network-based metabolite identification from NMR spectra | 126 | |
| SMART 2.0 | NMR-based machine learning tool for annotation of molecularly diverse Natural Products | 127 | |
| Integrated approach | NPClassifier | A Deep Neural Network-Based Structural Classification Tool for Natural Products | 128 |
| GNP | Identify polyketides and NRP using genome and LC-MS/MS | 129 | |
| NRPminer | NRP identification by Integrating genomics and metabolomics dataset | 130 | |
| NRPquest | Integrates genomics and metabolomics for NRP discovery | 131 | |
| DEREP-NP | Database for dereplication from MS and NMR Experiments | 132 | |
| ML-based target identification | deepDTnet | Target identification by deep learning from heterogeneous networks | 133 |
| BANDIT | Bayesian ML approach that integrates multiple data types to predict drug binding targets | 134 | |
| SPiDER | ML tool using self-organizing maps built from various features for target prediction | 135 | |
| DEcRyPT | Machine Intelligence workflow-based target prediction | 136 | |
| SuperPred | Drug classification and target prediction using 2D, Fragment, and 3D similarity | 137 | |
| MANTRA2.0 | Mechanism of action prediction using transcriptional profiles. | 138 | |
| Openchem | A Deep Learning Toolkit for Computational Chemistry and Drug Design | 139 | |
| DeepTox | Toxicity Prediction using Deep Learning | 140 | |
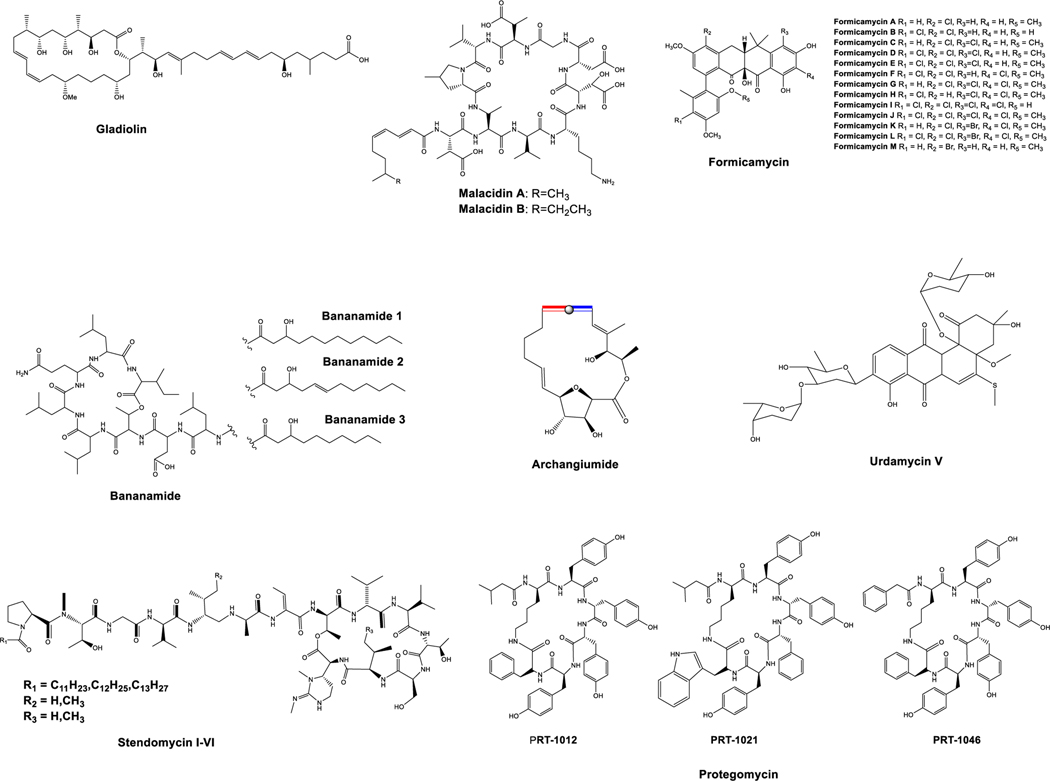
Figure 3:
Novel natural products predicted with the support of AI/ML tools.
Conventionally, the process of NP isolation uses a “grind and find” approach, which involves culturing the microorganism followed by purification and structure elucidation; however, with the advent of genome mining and ML/DL-based approaches, novel metabolites have been isolated from uncultured organisms.52 For instance, the combination of the two strategies has led to the discovery of the antibiotic malacidin from the global microbiome using heterologous expression without culturing the organism.53 A computational algorithm based on hidden Markov models (HMMs) is available for BGC identification from metagenomic samples, which allows the identification of interesting molecules from the human microbiome.54,55 In many cases, most of the BGCs remain silent, without expression, which hinders the production of secondary metabolites; nevertheless, using elicitors (e.g., small molecules and coculture), it is possible to predict the biosynthetic genes and express them with the help of ML tools.56 One of the major obstacles to NP discovery is the identification of secondary metabolites from unconventional sources because of the lack of cultivation of the microbes. However, with the emergence of metagenomics and ML, it is now possible to predict NPs in environmental or biological niches using specialized ML tools.54,57
2.3. AI/ML tools for Metabolite production and expression
Many microorganisms, such as those in the genera Streptomyces and Myxococcus, have been predicted to have large secondary metabolite BGCs with the advent of genome sequencing and bioinformatics. However, they usually do not code for NPs and remain as silent gene clusters.58 Therefore, various genome engineering techniques have been applied to activate those silent gene clusters, such as cocultivation,59 one strain many compounds,60 elicitors,61 ribosome engineering,62 chemical epigenetics,63 epigenetic modification,64 overexpression of transcription factors,65 and heterologous expression,66 which have had huge success in identifying new compounds. Despite the success in the control of parameters such as growth and strain engineering, media optimization remains challenging.67 To overcome this hurdle, various AI/ML techniques have been developed to control and monitor the production of metabolites. A study reported by Neythen et al. has used deep reinforcement learning, an approach from AI, for the control of cocultures in a continuous bioreactor.68 Using this approach, those authors were able to optimize the output of the coculture bioprocess by controlling various parameters. This type of study can be considered for controlling various factors in the production of NPs. Another study reported by Fei et al. used a high-throughput method to activate the silent BGCs in various organisms.69 The authors screened elicitors to induce secondary metabolite production with the help of IMS in nearly 500 conditions. Using this approach, they identified a new glycopeptide from Amycolatopsis keratiniphila, NRRL B24117, with the help of laser-ablation-coupled electrospray ionization MS. Although this approach can perform HTS to overcome the drawback of IMS and to analyze complex datasets, Brett et al. have developed a work tool for Metabolomics Explorer (MetEx)that enables users quickly and intuitively to analyze complex liquid chromatography (LC)-MS and metabolomics datasets.70
2.4. Dereplication of NPs with AI/ML techniques
During the golden era of NP development, several drug candidates were identified, most of which are still widely used for treating various diseases and infections.71 However, in the late 20th century, NP discovery started slowing down because of the repeated isolation of known compounds.72 To overcome this issue, fast identification of the known secondary metabolites is necessary, to reduce the analytical time and resources.73 Dereplication is a key process in the quick identification of previously known compounds in microbial extracts.74 Microbial extracts contain various compounds; therefore, the use of dereplication techniques helps eliminate redundancy and provides knowledge regarding novel compounds. To perform this highly efficient and robust task, ML tools with high accuracy are required. Previously, the dereplication techniques were carried out using high-performance liquid chromatography connected with a UV or photodiode array (PDA) detector with an automated bioassay and inbuilt library databases.75 However, structural information is lacking when using UV/PDA-based detection, and a more powerful instrument is required to capture additional spectral properties of the compounds.
2.4.1. Mass spectrometry-based dereplication using AI/ML
MS is a technique that has been widely used recently for dereplication in NPs because of its sensitivity, accuracy, and rapidity. Another major advantage of MS is its ability to gain a large amount of structural information from a trace amount of sample using an untargeted approach.14 The combination of mass information with UV/PDA can readily identify compounds with the help of databases such as Dictionary of Natural Products76 (http://dnp.chemnetbase.com/intro/), MarinLit77 (https://marinlit.rsc.org/), StreptomeDB78 (http://www.pharmbioinf.uni-freiburg.de/streptomedb), NPEdia79 (http://www.cbrg.riken.jp/npedia/), and The Natural Products Atlas80 (https://www.npatlas.org/). Using this approach, secondary metabolites from various actinomycetes have been dereplicated.81 LC coupled with MS can achieve high-throughput screening of metabolites; however, the analysis of the data in an efficient way remains challenging. Moreover, this requires researchers manually to search various datasets, such as UV signatures, mass spectra, and microorganisms in different databases, which are far from comprehensive.14 ML-based approaches could be a good solution for the in-line identification of NPs using spectral information without manual support against the available databases.
Although MS plays an important role in the identification and dereplication of NPs, it has several drawbacks and major problems arise regarding the overlapping parent molecular masses of various metabolites based on MS spectra alone.82,83 Therefore, a more efficient MS-based dereplication technique, such as tandem MS, is required and can increase the sensitivity of the detection of compounds based on MS/MS fragmentation.84 However, the analysis of MS/MS data is a cumbersome and intensive manual task, and an automated untargeted metabolomics pipeline is thus warranted to identify the metabolites efficiently. Recently, using various ML tools and algorithms, it was possible to interpret high-resolution mass spectra with reduced noise.85 Several AI/ML-based tool has been developed for mass spectral data processing and analysis such as MZmine86, Metaboanlayst87, MS-Dial88, Decon2LS89, XCMS90,THRASH91 and some are available as part of commercial vendor packages such as XCalibur (Thermo Fisher), MassHunter (Agilent), and using those metabolites has been predicted with high confidence manually.92 Metabolomics databases that are available based on MS/MS patterns are Massbank93, Metlin94, LMSD95, MoNA (https://mona.fiehnlab.ucdavis.edu/), Massbank (https://massbank.eu/MassBank/) and GNPS96, But in terms of microbial NPs identification, these are not widely used due to the scarcity of spectral data of natural products with the exception of GNPS96.
Recently, molecular networking (MN) has received widespread attention in the NP community for the dereplication and delineation of novel secondary metabolites from various sources with minimal manual interference. This approach was first introduced in 2012 for metabolite analysis from a set of living microbial colonies,97 yielding results that were comparable to the DNA sequencing of environmental samples to study microbial communities.98 MN is a computational technique that interprets the complex dataset that arises from MS analysis and visualizes it in the form of a network.99 To enable the analysis of MN, a crowdsourced library of reference spectra from a large number of compounds has been deposited from various communities and is available for analysis in GNPS96 (Global Natural Products Social Molecular Networking (http://gnps.ucsd.edu)). MN can identify compounds based on MS/MS spectral similarities and can also link the unknown molecules with related ones by exploiting similar fragmentation patterns. MN has been recognized for its high success rate and is becoming a routine tool for dereplication. For example, using MN and indexing 260 strains with ecologically diverse origins, the Pseudomonas-specialized metabolome led to the discovery of poaeamide B and bananamides (Figure 3).100 In another study, two novel chlorinated metabolites, isoconulothiazole B and conulothiazole C, were isolated from cyanobacteria using the MN strategy.101
Moreover, based on MN, further developments have been made to render the road toward the identification of NPs more straightforward. Using classical MN, various features have been incorporated with MS/MS, and feature-based MN (FBMN) has been introduced.102 It can efficiently distinguish isomers based on chromatography and ion mobility, while also facilitating spectral annotations and quantifications, thereby enabling robust analyses. Further, during ionization molecules form different adduct which limits the library annotation in MN to overcome this bottleneck Ion Identity Molecular Networking (IIMN) was developed.103 This feature improved the network connectivity for structurally related molecule and can be used to reveal unknown ion-ligand complexes. Very recently to identify bioactive compounds a scalable native metabolomics approach integrating non-targeted liquid chromatography tandem mass spectrometry, and simultaneous detection of protein binding via native mass spectrometry was developed.104 Using this integrated technique, rivulariapeptolides a family of serine protease inhibitors with nanomolar potency was identified and such approach could be central importance for drug discovery in future.
Hosein et al. have developed DEREPLICATOR+, an algorithm that can aid in the identification of NP classes such as NRPs, polyketides, terpenes, benzenoids, alkaloids, and flavonoids.105 A common problem in NP identification is the isolation of active compounds during bioassay-guided purification from the extract. To overcome this hurdle, bioactivity-based MN, which integrates bioinformatics workflow to map the bioactive score using MN, was developed.106 Using this approach, antiviral compounds were isolated from extracts of Euphorbia dendroides, for which a classical bioassay-guided fractionation procedure had previously failed.106 Further, a versatile, open-access platform NP Analyst was developed as a user friendly web-based infrastructure enabling NP community to analyze without the need for intense data processing.107 Although in the past MN could only be done via the web with GNPS, now many off-line tools such as MZmine3.086, MS-DIAL88, Metaboseek108, NetID109 and commercial software like Compound Discoverer (Thermo Scientific) have the ability to perform MN without the online platform making it easier.
Although mass spectral analytical tools are available for the identification of known compounds from databases, predicting the structure of unknown metabolites is a very challenging task. However, with the advent of ML, it is improving fast. Bocker et al. developed a tool (SIRIUS 4) that can identify the structure based on MS/MS datasets using a support vector machine.110 Further, advancing SIRIUS 4, ZODIAC, a network-based algorithm for the de novo annotation of database-independent molecular formulas was developed by the same group.111 Employing Bayesian statistics and Gibbs sampling it ensures fast processing in practice and is found to be better than SIRIUS by 16.5 fold. Using such ML tools novel molecular formula can be annotated. In another study that used a Deep Neural Network (DNN), a computational tool (class assignment and ontology prediction using MS, CANOPUS) was developed that could predict unknown metabolites for which spectral and structural reference data were not available.112 Similar to CANOPUS, a high-confidence structural annotation tool COSMIC based on SVM was developed.119 MS2DeepScore, which is an ML- supported mass spectral similarity-predicting algorithm was developed that allowed clustering, to identify metabolites similar to GNPS.96,141 Further, FALCON116 a density-based clustering of MS/MS spectra116, MS2LDA combined with Mass2Motif142 an unsupervised substructure discovery platform143 and Significant Interrelation of MS/MS Ions via Laplacian Embedding (SIMILE)117 are also available to predict the structural relationships of compounds. MN-based approaches for dereplication can be carried out with high success and can be further employed for the structural elucidation of novel compounds in the future with the support of the ML approaches developed recently.112–115,119
2.4.2. AI for the NMR-based structure elucidation/dereplication of NPs
The structural elucidation of molecules is a challenging problem in NP research. Although X-ray crystallography provides unambiguous structural information, it is often impeded by the requirement of a single crystal, and the limited amount of the isolated molecule restricts its wide application.144 Nuclear magnetic resonance (NMR) is a universally employed spectroscopy method that allows NP chemists to deduce molecular structures from spectra.145 Computer-aided structural elucidation (CASE) still plays a marginal role in this setting, although it was one of the earlier applications of AI.146 Although databases for NMR are available (NAPROC-13,120 CH-NMR-NP (https://www.j-resonance.com/en/nmrdb/), BMRB,147 and Spektraris NMR),148 they have several drawbacks and, thus, do not truly satisfy the requirements of NP communities.149. To overcome this issue, NP-MRD,121 which is an NMR database including over 41,000 NP compounds from >7400 different living species with various features, was introduced very recently.121 This database is still under development; however in the future, it will allow automated dereplication and CASE to be performed much more efficiently.
To assist the structure elucidation and perform dereplication, ML tools and software, such as logic for structure elucidation,150 ACD/Structure elucidator,151 Mestrelab Mnova,152 and Computer-aided Spectral Assignment,153 were developed and have aided NP identification.154,155 Recently, a robust AI-powered structure-prediction tool (DP4-AI)122 was developed and allowed the successful assignment of the structure of complex NPs.156–158 Using the CNN-based based approach NMR-based machine learning tool “Small Molecule Accurate Recognition Technology” (SMART 2.0) for mixture analysis and characterization of new natural products were developed.127 This led to the identification of a new chimeric swinholide-like macrolide, symplocolide A, as well as the annotation of swinholide A, samholides A–I, and several new derivatives. In another study, SMART-Miner a metabolite identification tool from the 1H-13C HSQC NMR spectra with the support of CNN was developed. The model was trained on 657 chemical entities collected from HMDB and BMRB to subsequently identify those molecules in complex mixtures with an accuracy of 88%.
To analyze the two-dimensional NMR spectra, a DNN-based approach for peak picking and spectral deconvolution (DEEP Picker) was developed very recently.123 In another study, various classes of NPs were predicted using ML from 13C-NMR spectroscopic data.159 NMR is relatively less explored for dereplication compared with HRMS because of its sensitivity; nevertheless, it can offer high accuracy in terms of the prediction of stereoisomers and the detection of all organic compounds in a mixture.160 Recently, using 13C-NMR, a dereplication software (MixONat)124 was developed that allowed the distinction of structurally close NPs, including stereoisomers, and aided the identification of xanthones in Calophyllum brasiliense.124,161 In another study that used 1H-NMR, Grienke et al. developed a workflow ELINA (Eliciting Nature’s Activities) based on a heterocovariance analysis, which can detect chemical features that correlate with bioactivity before isolation; using this approach, the authors discovered lanostane triterpenes from the extract of the fungus Fomitopsis pinicola.125
2.5. Integrated approach for NP discovery using AI/ML
Multiple strategies have been developed over the years for NP prioritization, and a combination of various approaches (e.g., genomic, metabolomic, taxonomic, spectral information, and bioactivity) can be used as a factor for ranking before the downstream process of purification and structure elucidation of NPs.162 More recently, Kim et al. developed NPClassifier,128 which is a tool that can classify NPs using a DL approach. They have been categorized into three hierarchical levels based on the pathway and chemical properties; moreover, structural details can be classified using this NP, which indicates its applicability for drug discovery and the elucidation of biological interactions. In another study, an automated genome-guided NP discovery tool, with the support of an LC-MS/MS dataset, was developed that could automatically predict, combinatorialize, and identify polyketides and NRPs from crude extracts.129 Hosein et al. developed NRPquest, which is an ML tool that integrates MS and Genome Mining for Nonribosomal Peptide (NRP) discovery.130 Similarly, another tool (NRPminer) was developed very recently that combined both genomics and metabolomics to identify novel NRPs; using this approach, four unknown NRP families were identified from microbes and human microbiota and shown to exhibit antiparasitic activity.131 By integrating genomics and metabolomics focusing on NRPs, several novel protegomycin derivatives from a previously unknown NP source (X. doucetiae and X. poinarii) were identified (Figure 3).131 A study reported by Kleigrewe et al. integrated metabolomics and genome analysis to discover NPs from cyanobacteria; using this innovative approach, the authors discovered a new class of di- and trichlorinated acyl amide columbamides with cannabinomimetic activity.163 Previously, we combined genome mining with MN and identified urdamycin E and a novel derivative, urdamycin V (Figure 3), from Streptomyces spp., which induce cell death by inhibiting mTOR in cancerous cells.164,165 Carlos et al. developed a database (DEREP-NP) to dereplicate metabolites efficiently by integrating MS and NMR spectra.132 Another study that combined NMR-based profiling with genome mining led to the discovery of the allenic macrolide Archangiumide (Figure 3) from Myxobacterium.166 Using MS-guided genome mining, which connects the chemotypes of peptide NPs with their BGCs by iteratively matching de novo tandem MS, a new NP peptidogenomics approach was developed.167 Using this combined approach, five new stendomycin analogues were identified that differed in the acyl chain and in valine or isoleucine substitutions at positions 5 and 13 from S. hygroscopicus ATCC 53653 (Figure 3).
3. Bioactivity and Target Identification of NPs with AI/ML techniques
One of the challenges in the development of NP-based drug candidates is the identification of their mechanism of action and side effects, which is a costly and lengthy process.168,169 Because of the enormous structural diversity and broad chemical spaces, the bioactivity of NPs is discovered based on phenotypic effects or via high-throughput phenotypic screening.170,171 To identify the targets experimentally, chemical genomics172,173 and chemical proteomics174 approaches are generally used; however, although they can validate the targets they are often laborious and time-consuming processes.133 To overcome this, computational approaches can narrow down the large search space of the targets.175 There are three computational approaches and, in addition to the traditional structure-based176 and ligand-based target identification methods,177 ML-based approaches have numerous advantages and can be promising strategies for NP target identification.178 To identify drug targets, Madhukar et al. developed BANDIT,134 a Bayesian machine-learning approach that integrates multiple data types to predict drug binding targets.134 Using this approach, the authors predicted the targets of nearly 4,000 compounds with 90% accuracy and further validated 14 novel microtubule inhibitors. In another study aimed at identifying drug–target interactions (DTIs), a CNN-based tool, NeoDTI, was developed.179 NeoDTI mines large-scale graph data and automatically learns the topology-preserving representations of drugs and targets, to facilitate DTI prediction with compound–protein binding affinity. Using such approaches, the drug targets of NPs can be identified, which can accelerate the drug-discovery platform. In another study, a DL toolkit, “Openchem,” which is based on the PyTorch framework, was developed for drug design and computational chemistry.139 It can enable drug discovery and molecular modeling applications using DL algorithms. This DL-based approach can help in various tasks in NP discovery, such as their physical properties and structure–activity relation. A recent study reported by Walker and Clardy described an ML-based approach to predict the biological activity of NPs using genome mining without isolation.180 The authors used ML classifiers to predict antibacterial or antifungal activity based on known NP BGCs with an accuracy of 80%.
The SPiDER ML tool merges the concept of self-organizing maps, consensus scoring, and statistical analysis to successfully identify targets for both known drugs and computer-generated molecular scaffolds; moreover, using this method, off-target fenofibrate-related compounds were identified.135 Furthermore, to increase the confidence, the Drug–Target Relationship Predictor (DEcRyPT) machine intelligence workflow, which uses regression random forest technology as an orthogonal learning approach to self-organizing maps, was developed.136 Using this ML tool, the targets of β-lapachone were identified and validated as potent modulators of 5-lipoxygenase.136 SuperPred137 provides drug classification and target prediction considering features such as 2-D, Fragment, and 3-D similarity and adapting concepts of the basic local alignment search tool (BLAST) algorithm.137,181 These ML approaches can innovate the drug target identification process and serve as an alternative powerful strategy to chemoproteomics. Another study reported by Carrella et al. developed MANTRA 2.0, which is a transcriptional profile-based drug target identification that uses a microarray dataset.138 By uploading the gene expression profile of the compound in cell lines, an ML-based automated pipeline revealed its mechanism of action based on the transcriptional signature of existing drugs.138 Despite the advantages of the ML tools, they can sometimes be inaccurate and only the previously studied targets can be predicted with further target validation.182,183 In the drug-discovery process, one of the key criteria for candidate molecules is that they have fewer adverse effects; however, numerous time- and cost-intensive in vitro and in vivo studies are required to assess toxicity.184 Computational toxicology can be effectively used to screen a large number of compounds without the use of time-consuming animal studies; nevertheless, this approach has severe drawbacks in terms of accuracy.185 To overcome this issue, a recent study reported a DL pipeline, “DeepTox,” which exhibited a high accuracy of toxicity prediction.140 Such a DL-based approach can be utilized in the future effectively to predict the toxicity of NPs and to tweak molecules with less adverse effects.
4. Conclusions and future perspectives
NPs from microorganisms and their molecular frameworks have a long tradition for many drug leads and are still widely used for treating various diseases and infections.182,186,187 The bioprospecting of the NP leads is challenging because of the amount of data generated and technical barriers, such as screening, isolation, characterization, and target identification. AI approaches can be used to address these problems and uncover hidden patterns by employing algorithms and decreasing the analytical time, resources, and costs required to identify NPs.188 As proof of concept, recently, a highly effective antibiotic (halicin) with an entirely new mechanism of action was identified from the ZINC15 database using a DL approach.10 AI can help prioritize the microbes for screening based on their taxonomic novelty and genomes regarding the ability to produce novel NPs. Furthermore, it can help rapid dereplication and assist in the identification of active compounds using LC-HRMS and NMR.
Several NPs were isolated during the golden age of NPs, but most of them have been neglected or are limited by specific bioactivity with the discovery of various lead compounds at similar times.1,189 However, the surge of antimicrobial resistance and technological advancements have rekindled the interest in NPs as drug leads and repurposing is being assessed.190 The cyclic peptide griselimycin was identified in 1960 from Streptomyces191 and exhibited potent antituberculous activity, but was neglected; however, very recently, it was modified and introduced into the drug-development pipeline.192 Similarly, another NP, chrysomycin A, which is a rare C-aryl glycoside, was first discovered over 60 years ago and has anticancer activity193,194 with no further studies; however, recently, it was reported as inhibiting multidrug-resistant tuberculosis effectively (MDR-TB).195,196 Drug repurposing and alternate bioactivity prediction are cost-effective processes compared with drug discovery; nevertheless, they are quite challenging. To overcome this drawback, AI/ML can be used for candidate selection.197 Furthermore, AI can also assist in macromolecular target identification in a fast and effective manner.
A big obstacle in the full-fledged implementation of AI in NP research is the lack of integrated and curated databases.198 Most of the data, such as taxonomic, structural, genomic, and metabolomic data, for the specific compounds are not available compiled in the form of databases and presented in the form of scientific literature, which is very difficult to access and analyze manually.198,199 Hence, an integrated approach is required for the effective analysis of NPs, as is a single algorithm for the management of the entire process of NP discovery alone. By addressing these issues, the common problems associated with AI, such as errors and repeatability, can be controlled in the learning process from reliable datasets.200–202 With the worsening drug-resistance scenario and the increase in the number of new infections, the search for novel NPs is essential. Nature is extremely generous to mankind by providing diverse compounds over the centuries to cure diseases. With the advent of technological advancements and AI, can we expect a new golden era of NP drug discovery?
Acknowledgments
VJS acknowledges MEXT for fellowship support. We express sincere thanks for a by JSPS KAKENHI (grant numbers 20H05936 and 21H04705) to H.S. This work was also supported by NIH award R01CA236350 to H. S.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Atanasov AG, Zotchev SB, Dirsch VM, International Natural Product Sciences Taskforce and C. T. Supuran, Nat. Rev. Drug Discov, 2021, 20, 200–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katz L. and Baltz RH, J. Ind. Microbiol. Biotechnol, 2016, 43, 155–176. [DOI] [PubMed] [Google Scholar]
- 3.Bachmann BO, Van Lanen SG and Baltz RH, J. Ind. Microbiol. Biotechnol, 2014, 41, 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grabowski K, Baringhaus K-H and Schneider G, Nat. Prod. Rep, 2008, 25, 892–904. [DOI] [PubMed] [Google Scholar]
- 5.Sabatini DM, Proc. Natl. Acad. Sci. U. S. A, 2017, 114, 11818–11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skinnider M, Wang F, Pasin D, Greiner R, Foster L, Dalsgaard P. and Wishart DS, ChemRxiv,, DOI: 10.26434/chemrxiv.14644854.v1. [DOI] [Google Scholar]
- 7.Jiménez-Luna J, Grisoni F. and Schneider G, Nature Machine Intelligence, 2020, 2, 573–584. [Google Scholar]
- 8.Baum ZJ, Yu X, Ayala PY, Zhao Y, Watkins SP and Zhou Q, J. Chem. Inf. Model, 2021, 61, 3197–3212. [DOI] [PubMed] [Google Scholar]
- 9.Choudhary N, Bharti R. and Sharma R, Materials Today: Proceedings,, DOI: 10.1016/j.matpr.2021.09.428. [DOI] [Google Scholar]
- 10.Stokes JM, Yang K, Swanson K, Jin W, Cubillos-Ruiz A, Donghia NM, MacNair CR, French S, Carfrae LA, Bloom-Ackermann Z, Tran VM, Chiappino-Pepe A, Badran AH, Andrews IW, Chory EJ, Church GM, Brown ED, Jaakkola TS, Barzilay R. and Collins JJ, Cell, 2020, 181, 475–483. [DOI] [PubMed] [Google Scholar]
- 11.Silver Lynn L, Clin. Microbiol. Rev, 2011, 24, 71–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyddiard D, Jones GL and Greatrex BW, FEMS Microbiol. Lett,, DOI: 10.1093/femsle/fnw084. [DOI] [PubMed] [Google Scholar]
- 13.Hautbergue T, Jamin EL, Debrauwer L, Puel O. and Oswald IP, Nat. Prod. Rep, 2018, 35, 147–173. [DOI] [PubMed] [Google Scholar]
- 14.Bouslimani A, Sanchez LM, Garg N. and Dorrestein PC, Nat. Prod. Rep, 2014, 31, 718–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genilloud O, Nat. Prod. Rep, 2017, 34, 1203–1232. [DOI] [PubMed] [Google Scholar]
- 16.Gerth K, Bedorf N, Höfle G, Irschik H. and Reichenbach H, J. Antibiot, 1996, 49, 560–563. [DOI] [PubMed] [Google Scholar]
- 17.Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR and Fenical W, Angew. Chem. Int. Ed Engl, 2003, 42, 355–357. [DOI] [PubMed] [Google Scholar]
- 18.Rust M, Helfrich EJN, Freeman MF, Nanudorn P, Field CM, Rückert C, Kündig T, Page MJ, Webb VL, Kalinowski J, Sunagawa S. and Piel J, Proc. Natl. Acad. Sci. U. S. A, 2020, 117, 9508–9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C. and Lewis K, Nature, 2015, 517, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gavrish E, Sit CS, Cao S, Kandror O, Spoering A, Peoples A, Ling L, Fetterman A, Hughes D, Bissell A, Torrey H, Akopian T, Mueller A, Epstein S, Goldberg A, Clardy J. and Lewis K, Chem. Biol, 2014, 21, 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hugenholtz P, Chuvochina M, Oren A, Parks DH and Soo RM, ISME J, 2021, 15, 1879–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith KP, Kang AD and Kirby JE, J. Clin. Microbiol,, DOI: 10.1128/JCM.01521-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark CM, Costa MS, Sanchez LM and Murphy BT, Proc. Natl. Acad. Sci. U. S. A, 2018, 115, 4981–4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dumolin C, Aerts M, Verheyde B, Schellaert S, Vandamme T, Van der Jeugt F, De Canck E, Cnockaert M, Wieme AD, Cleenwerck I, Peiren J, Dawyndt P, Vandamme P. and Carlier A, mSystems, 2019, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauget M, Valot B, Bertrand X. and Hocquet D, Trends Microbiol, 2017, 25, 447–455. [DOI] [PubMed] [Google Scholar]
- 26.Esquenazi E, Yang Y-L, Watrous J, Gerwick WH and Dorrestein PC, Nat. Prod. Rep, 2009, 26, 1521–1534. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y-L, Xu Y, Straight P. and Dorrestein PC, Nat. Chem. Biol, 2009, 5, 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez DJ, Haste NM, Hollands A, Fleming TC, Hamby M, Pogliano K, Nizet V. and Dorrestein PC, Microbiology, 2011, 157, 2485–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esquenazi E, Coates C, Simmons L, Gonzalez D, Gerwick WH and Dorrestein PC, Mol. Biosyst, 2008, 4, 562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baltz RH, J. Ind. Microbiol. Biotechnol,, DOI: 10.1093/jimb/kuab044. [DOI] [Google Scholar]
- 31.Nivina A, Yuet KP, Hsu J. and Khosla C, Chem. Rev, 2019, 119, 12524–12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hai Y, Huang A. and Tang Y, J. Nat. Prod, 2020, 83, 593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mullowney MW, McClure RA, Robey MT, Kelleher NL and Thomson RJ, Nat. Prod. Rep, 2018, 35, 847–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baunach M, Franke J. and Hertweck C, Angew. Chem. Int. Ed Engl, 2015, 54, 2604–2626. [DOI] [PubMed] [Google Scholar]
- 35.Scherlach K. and Hertweck C, Nat. Commun, 2021, 12, 3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blin K, Pascal Andreu V, de Los Santos ELC, Del Carratore F, Lee SY, Medema MH and Weber T, Nucleic Acids Res, 2019, 47, D625–D630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ichikawa N, Sasagawa M, Yamamoto M, Komaki H, Yoshida Y, Yamazaki S. and Fujita N, Nucleic Acids Res, 2013, 41, D408–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palaniappan K, Chen I-MA, Chu K, Ratner A, Seshadri R, Kyrpides NC, Ivanova NN and Mouncey NJ, Nucleic Acids Research, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kautsar SA, Blin K, Shaw S, Navarro-Muñoz JC, Terlouw BR, van der Hooft JJJ, van Santen JA, Tracanna V, Suarez Duran HG, Pascal Andreu V, Selem-Mojica N, Alanjary M, Robinson SL, Lund G, Epstein SC, Sisto AC, Charkoudian LK, Collemare J, Linington RG, Weber T. and Medema MH, Nucleic Acids Res, 2020, 48, D454–D458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conway KR and Boddy CN, Nucleic Acids Res, 2013, 41, D402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammami R, Zouhir A, Le Lay C, Ben Hamida J. and Fliss I, BMC Microbiol, 2010, 10, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH and Weber T, Nucleic Acids Res, 2019, 47, W81–W87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skinnider MA, Dejong CA, Rees PN, Johnston CW, Li H, Webster ALH, Wyatt MA and Magarvey NA, Nucleic Acids Res, 2015, 43, 9645–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Jong A, van Hijum SAFT, Bijlsma JJE, Kok J. and Kuipers OP, Nucleic Acids Res, 2006, 34, W273–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mungan MD, Alanjary M, Blin K, Weber T, Medema MH and Ziemert N, Nucleic Acids Res, 2020, 48, W546–W552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sélem-Mojica N, Aguilar C, Gutiérrez-García K, Martínez-Guerrero CE and Barona-Gómez F, Microb Genom,, DOI: 10.1099/mgen.0.000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH and Fedorova ND, Fungal Genet. Biol, 2010, 47, 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeda I, Umemura M, Koike H, Asai K. and Machida M, DNA Res, 2014, 21, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hannigan GD, Prihoda D, Palicka A, Soukup J, Klempir O, Rampula L, Durcak J, Wurst M, Kotowski J, Chang D, Wang R, Piizzi G, Temesi G, Hazuda DJ, Woelk CH and Bitton DA, Nucleic Acids Research, 2019, 47, e110–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin Z, Munnoch JT, Devine R, Holmes NA, Seipke RF, Wilkinson KA, Wilkinson B. and Hutchings MI, Chemical Science, 2017, 8, 3218–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kloosterman AM, Cimermancic P, Elsayed SS, Du C, Hadjithomas M, Donia MS, Fischbach MA, van Wezel GP and Medema MH, PLoS Biol, 2020, 18, e3001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller SJ and Clardy J, Nat. Chem, 2009, 1, 261–263. [DOI] [PubMed] [Google Scholar]
- 53.Hover BM, Kim S-H, Katz M, Charlop-Powers Z, Owen JG, Ternei MA, Maniko J, Estrela AB, Molina H, Park S, Perlin DS and Brady SF, Nat Microbiol, 2018, 3, 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugimoto Y, Camacho FR, Wang S, Chankhamjon P, Odabas A, Biswas A, Jeffrey PD and Donia MS, Science, 2019, 366. [DOI] [PubMed] [Google Scholar]
- 55.Donia MS and Fischbach MA, Science, 2015, 349, 1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Banf M, Zhao K. and Rhee SY, Bioinformatics, 2019, 35, 3178–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reddy BVB, Milshteyn A, Charlop-Powers Z. and Brady SF, Chem. Biol, 2014, 21, 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bader CD, Panter F. and Müller R, Biotechnol. Adv, 2020, 39, 107480. [DOI] [PubMed] [Google Scholar]
- 59.Stroe MC, Netzker T, Scherlach K, Krüger T, Hertweck C, Valiante V. and Brakhage AA, Elife,, DOI: 10.7554/eLife.52541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bode HB, Bethe B, Höfs R. and Zeeck A, Chembiochem, 2002, 3, 619–627. [DOI] [PubMed] [Google Scholar]
- 61.Seyedsayamdost MR, Proc. Natl. Acad. Sci. U. S. A, 2014, 111, 7266–7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shima J, Hesketh A, Okamoto S, Kawamoto S. and Ochi K, J. Bacteriol, 1996, 178, 7276–7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asai T, Yamamoto T, Shirata N, Taniguchi T, Monde K, Fujii I, Gomi K. and Oshima Y, Org. Lett, 2013, 15, 3346–3349. [DOI] [PubMed] [Google Scholar]
- 64.Mao X-M, Xu W, Li D, Yin W-B, Chooi Y-H, Li Y-Q, Tang Y. and Hu Y, Angew. Chem. Int. Ed Engl, 2015, 54, 7592–7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang T, Wan J, Zhan Z, Bai J, Liu B. and Hu Y, Acta Pharm Sin B, 2018, 8, 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Biggins JB, Liu X, Feng Z. and Brady SF, J. Am. Chem. Soc, 2011, 133, 1638–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramzi AB, Baharum SN, Bunawan H. and Scrutton NS, Front. Bioeng. Biotechnol, 2020, 8, 608918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Treloar NJ, Fedorec AJH, Ingalls B. and Barnes CP, PLoS Comput. Biol, 2020, 16, e1007783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu F, Wu Y, Zhang C, Davis KM, Moon K, Bushin LB and Seyedsayamdost MR, Nat. Chem. Biol, 2019, 15, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Covington BC and Seyedsayamdost MR, ACS Chem. Biol,, DOI: 10.1021/acschembio.1c00737. [DOI] [Google Scholar]
- 71.Shen B, Cell, 2015, 163, 1297–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jensen PR, Chavarria KL, Fenical W, Moore BS and Ziemert N, Journal of Industrial Microbiology and Biotechnology, 2014, 41, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ito T. and Masubuchi M, The Journal of Antibiotics, 2014, 67, 353–360. [DOI] [PubMed] [Google Scholar]
- 74.Gaudêncio SP and Pereira F, Natural Product Reports, 2015, 32, 779–810. [DOI] [PubMed] [Google Scholar]
- 75.Hook DJ, More CF, Yacobucci JJ, Dubay G. and O’Connor S, J. Chromatogr, 1987, 385, 99–108. [DOI] [PubMed] [Google Scholar]
- 76.Buckingham J, Dictionary of natural products, supplement 3: Third supplement, CRC Press, London, England, 1996. [Google Scholar]
- 77.Blunt JW, Carroll AR, Copp BR, Davis RA, Keyzers RA and Prinsep MR, Nat. Prod. Rep, 2018, 35, 8–53. [DOI] [PubMed] [Google Scholar]
- 78.Moumbock AFA, Gao M, Qaseem A, Li J, Kirchner PA, Ndingkokhar B, Bekono BD, Simoben CV, Babiaka SB, Malange YI, Sauter F, Zierep P, Ntie-Kang F. and Günther S, Nucleic Acids Res, 2021, 49, D600–D604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tomiki T, Saito T, Ueki M, Konno H, Asaoka T, Suzuki R, Uramoto M, Kakeya H. and Osada H, J Comput Aid Chem, 2006, 7, 157–162. [Google Scholar]
- 80.van Santen JA, Jacob G, Singh AL, Aniebok V, Balunas MJ, Bunsko D, Neto FC, Castaño-Espriu L, Chang C, Clark TN, Cleary Little JL, Delgadillo DA, Dorrestein PC, Duncan KR, Egan JM, Galey MM, Haeckl FPJ, Hua A, Hughes AH, Iskakova D, Khadilkar A, Lee J-H, Lee S, LeGrow N, Liu DY, Macho JM, McCaughey CS, Medema MH, Neupane RP, O’Donnell TJ, Paula JS, Sanchez LM, Shaikh AF, Soldatou S, Terlouw BR, Tran TA, Valentine M, van der Hooft JJJ, Vo DA, Wang M, Wilson D, Zink KE and Linington RG, ACS Cent Sci, 2019, 5, 1824–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mehetre GT, Vinodh JS, Burkul BB, Desai D, Santhakumari B, Dharne MS and Dastager SG, RSC Advances, 2019, 9, 9850–9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Caesar LK, Kellogg JJ, Kvalheim OM and Cech NB, J. Nat. Prod, 2019, 82, 469–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hubert J, Nuzillard J-M and Renault J-H, Phytochemistry Reviews, 2017, 16, 55–95. [Google Scholar]
- 84.Hoffmann T, Krug D, Hüttel S. and Müller R, Anal. Chem, 2014, 86, 10780–10788. [DOI] [PubMed] [Google Scholar]
- 85.Kaur P. and O’Connor PB, Journal of the American Society for Mass Spectrometry, 2006, 17, 459–468. [DOI] [PubMed] [Google Scholar]
- 86.Pluskal T, Castillo S, Villar-Briones A. and Oresic M, BMC Bioinformatics, 2010, 11, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pang Z, Zhou G, Ewald J, Chang L, Hacariz O, Basu N. and Xia J, Nat. Protoc,, DOI: 10.1038/s41596-022-00710-w. [DOI] [PubMed] [Google Scholar]
- 88.Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, VanderGheynst J, Fiehn O. and Arita M, Nat. Methods, 2015, 12, 523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jaitly N, Mayampurath A, Littlefield K, Adkins JN, Anderson GA and Smith RD, BMC Bioinformatics, 2009, 10, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smith CA, Want EJ, O’Maille G, Abagyan R. and Siuzdak G, Anal. Chem, 2006, 78, 779–787. [DOI] [PubMed] [Google Scholar]
- 91.Horn DM, Zubarev RA and McLafferty FW, J. Am. Soc. Mass Spectrom, 2000, 11, 320–332. [DOI] [PubMed] [Google Scholar]
- 92.Kumar V, Kumar AA, Joseph V, Dan VM, Jaleel A, Kumar TRS and Kartha CC, Mol. Cell. Biochem, 2020, 463, 147–160. [DOI] [PubMed] [Google Scholar]
- 93.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, Oda Y, Kakazu Y, Kusano M, Tohge T, Matsuda F, Sawada Y, Hirai MY, Nakanishi H, Ikeda K, Akimoto N, Maoka T, Takahashi H, Ara T, Sakurai N, Suzuki H, Shibata D, Neumann S, Iida T, Tanaka K, Funatsu K, Matsuura F, Soga T, Taguchi R, Saito K. and Nishioka T, J. Mass Spectrom, 2010, 45, 703–714. [DOI] [PubMed] [Google Scholar]
- 94.Smith CA, O’Maille G, Wa EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R. and Siuzdak G, Ther. Drug Monit, 2005, 27, 747–751. [DOI] [PubMed] [Google Scholar]
- 95.Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH Jr, Murphy RC, Raetz CRH, Russell DW and Subramaniam S, Nucleic Acids Res, 2007, 35, D527–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, Porto C, Bouslimani A, Melnik AV, Meehan MJ, Liu W-T, Crüsemann M, Boudreau PD, Esquenazi E, Sandoval-Calderón M, Kersten RD, Pace LA, Quinn RA, Duncan KR, Hsu C-C, Floros DJ, Gavilan RG, Kleigrewe K, Northen T, Dutton RJ, Parrot D, Carlson EE, Aigle B, Michelsen CF, Jelsbak L, Sohlenkamp C, Pevzner P, Edlund A, McLean J, Piel J, Murphy BT, Gerwick L, Liaw C-C, Yang Y-L, Humpf H-U, Maansson M, Keyzers RA, Sims AC, Johnson AR, Sidebottom AM, Sedio BE, Klitgaard A, Larson CB, Torres-Mendoza CABP,D, Gonzalez DJ, Silva DB, Marques LM, Demarque DP, Pociute E, O’Neill EC, Briand E, Helfrich EJN, Granatosky EA, Glukhov E, Ryffel F, Houson H, Mohimani H, Kharbush JJ, Zeng Y, Vorholt JA, Kurita KL, Charusanti P, McPhail KL, Nielsen KF, Vuong L, Elfeki M, Traxler MF, Engene N, Koyama N, Vining OB, Baric R, Silva RR, Mascuch SJ, Tomasi S, Jenkins S, Macherla V, Hoffman T, Agarwal V, Williams PG, Dai J, Neupane R, Gurr J, Rodríguez AMC, Lamsa A, Zhang C, Dorrestein K, Duggan BM, Almaliti J, Allard P-M, Phapale P, Nothias L-F, Alexandrov T, Litaudon M, Wolfender J-L, Kyle JE, Metz TO, Peryea T, Nguyen D-T, VanLeer D, Shinn P, Jadhav A, Müller R, Waters KM, Shi W, Liu X, Zhang L, Knight R, Jensen PR, Palsson BO, Pogliano K, Linington RG, Gutiérrez M, Lopes NP, Gerwick WH, Moore BS, Dorrestein PC and Bandeira N, Nat. Biotechnol, 2016, 34, 828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, Moore BS, Laskin J, Bandeira N. and Dorrestein PC, Proc. Natl. Acad. Sci. U. S. A, 2012, 109, E1743–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aron AT, Gentry EC, McPhail KL, Nothias L-F, Nothias-Esposito M, Bouslimani A, Petras D, Gauglitz JM, Sikora N, Vargas F, van der Hooft JJJ, Ernst M, Kang KB, Aceves CM, Caraballo-Rodríguez AM, Koester I, Weldon KC, Bertrand S, Roullier C, Sun K, Tehan RM, Boya P CA, Christian MH, Gutiérrez M, Ulloa AM, Tejeda Mora JA, Mojica-Flores R, Lakey-Beitia J, Vásquez-Chaves V, Zhang Y, Calderón AI, Tayler N, Keyzers RA, Tugizimana F, Ndlovu N, Aksenov AA, Jarmusch AK, Schmid R, Truman AW, Bandeira N, Wang M. and Dorrestein PC, Nat. Protoc, 2020, 15, 1954–1991. [DOI] [PubMed] [Google Scholar]
- 99.Vincenti F, Montesano C, Di Ottavio F, Gregori A, Compagnone D, Sergi M. and Dorrestein P, Front. Chem, 2020, 8, 572952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nguyen DD, Melnik AV, Koyama N, Lu X, Schorn M, Fang J, Aguinaldo K, Lincecum TL Jr, Ghequire MGK, Carrion VJ, Cheng TL, Duggan BM, Malone JG, Mauchline TH, Sanchez LM, Kilpatrick AM, Raaijmakers JM, De Mot R, Moore BS, Medema MH and Dorrestein PC, Nat Microbiol, 2016, 2, 16197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Teta R, Sala GD, Esposito G, Via CW, Mazzoccoli C, Piccoli C, Bertin MJ, Costantino V. and Mangoni A, Org Chem Front, 2019, 6, 1762–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nothias L-F, Petras D, Schmid R, Dührkop K, Rainer J, Sarvepalli A, Protsyuk I, Ernst M, Tsugawa H, Fleischauer M, Aicheler F, Aksenov AA, Alka O, Allard P-M, Barsch A, Cachet X, Caraballo-Rodriguez AM, Da Silva RR, Dang T, Garg N, Gauglitz JM, Gurevich A, Isaac G, Jarmusch AK, Kameník Z, Kang KB, Kessler N, Koester I, Korf A, Le Gouellec A, Ludwig M, Martin H C, McCall L-I, McSayles J, Meyer SW, Mohimani H, Morsy M, Moyne O, Neumann S, Neuweger H, Nguyen NH, Nothias-Esposito M, Paolini J, Phelan VV, Pluskal T, Quinn RA, Rogers S, Shrestha B, Tripathi A, van der Hooft JJJ, Vargas F, Weldon KC, Witting M, Yang H, Zhang Z, Zubeil F, Kohlbacher O, Böcker S, Alexandrov T, Bandeira N, Wang M. and Dorrestein PC, Nat. Methods, 2020, 17, 905–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schmid R, Petras D, Nothias L-F, Wang M, Aron AT, Jagels A, Tsugawa H, Rainer J, Garcia-Aloy M, Dührkop K, Korf A, Pluskal T, Kameník Z, Jarmusch AK, Caraballo-Rodríguez AM, Weldon KC, Nothias-Esposito M, Aksenov AA, Bauermeister A, Albarracin Orio A, Grundmann CO, Vargas F, Koester I, Gauglitz JM, Gentry EC, Hövelmann Y, Kalinina SA, Pendergraft MA, Panitchpakdi M, Tehan R, Le Gouellec A, Aleti G, Mannochio Russo H, Arndt B, Hübner F, Hayen H, Zhi H, Raffatellu M, Prather KA, Aluwihare LI, Böcker S, McPhail KL, Humpf H-U, Karst U. and Dorrestein PC, Nat. Commun, 2021, 12, 3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reher R, Aron AT, Fajtová P, Stincone P, Liu C, Ben Shalom IY, Bittremieux W, Wang M, Matos-Hernandez ML, Alexander KL, Caro-Diaz EJ, Naman CB, Hughes CC, Dorrestein PC, O’Donoghue AJ, Gerwick WH and Petras D, bioRxiv, 2021. [Google Scholar]
- 105.Mohimani H, Gurevich A, Shlemov A, Mikheenko A, Korobeynikov A, Cao L, Shcherbin E, Nothias L-F, Dorrestein PC and Pevzner PA, Nat. Commun, 2018, 9, 4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nothias L-F, Nothias-Esposito M, da Silva R, Wang M, Protsyuk I, Zhang Z, Sarvepalli A, Leyssen P, Touboul D, Costa J, Paolini J, Alexandrov T, Litaudon M. and Dorrestein PC, J. Nat. Prod, 2018, 81, 758–767. [DOI] [PubMed] [Google Scholar]
- 107.Lee S, van Santen JA, Farzaneh N, Liu DY, Pye CR, Baumeister TUH, Wong WR and Linington RG, ACS Cent. Sci, 2022, 8, 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Helf MJ, Fox BW, Artyukhin AB, Zhang YK and Schroeder FC, Nat. Commun, 2022, 13, 782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen L, Lu W, Wang L, Xing X, Chen Z, Teng X, Zeng X, Muscarella AD, Shen Y, Cowan A, McReynolds MR, Kennedy BJ, Lato AM, Campagna SR, Singh M. and Rabinowitz JD, Nat. Methods, 2021, 18, 1377–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dührkop K, Fleischauer M, Ludwig M, Aksenov AA, Melnik AV, Meusel M, Dorrestein PC, Rousu J. and Böcker S, Nat. Methods, 2019, 16, 299–302. [DOI] [PubMed] [Google Scholar]
- 111.Ludwig M, Nothias L-F, Dührkop K, Koester I, Fleischauer M, Hoffmann MA, Petras D, Vargas F, Morsy M, Aluwihare L, Dorrestein PC and Böcker S, Nat Mach Intell, 2020, 2, 629–641. [Google Scholar]
- 112.Dührkop K, Nothias L-F, Fleischauer M, Reher R, Ludwig M, Hoffmann MA, Petras D, Gerwick WH, Rousu J, Dorrestein PC and Böcker S, Nat. Biotechnol, 2021, 39, 462–471. [DOI] [PubMed] [Google Scholar]
- 113.Olivon F, Elie N, Grelier G, Roussi F, Litaudon M. and Touboul D, Anal. Chem, 2018, 90, 13900–13908. [DOI] [PubMed] [Google Scholar]
- 114.Liu Y, Mrzic A, Meysman P, De Vijlder T, Romijn EP, Valkenborg D, Bittremieux W. and Laukens K, PLoS One, 2020, 15, e0226770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cao L, Guler M, Tagirdzhanov A, Lee Y-Y, Gurevich A. and Mohimani H, Nat. Commun, 2021, 12, 3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bittremieux W, Laukens K, Noble WS and Dorrestein PC, Rapid Commun. Mass Spectrom, 2021, e9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Treen DGC, Wang M, Xing S, Louie KB, Huan T, Dorrestein PC, Northen TR and Bowen BP, Nat. Commun, 2022, 13, 2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ernst M, Kang KB, Caraballo-Rodríguez AM, Nothias L-F, Wandy J, Chen C, Wang M, Rogers S, Medema MH, Dorrestein PC and van der Hooft JJJ, Metabolites, 2019, 9, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hoffmann MA, Nothias L-F, Ludwig M, Fleischauer M, Gentry EC, Witting M, Dorrestein PC, Dührkop K. and Böcker S, Nat. Biotechnol,, DOI: 10.1038/s41587-021-01045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.López-Pérez JL, Therón R, del Olmo E. and Díaz D, Bioinformatics, 2007, 23, 3256–3257. [DOI] [PubMed] [Google Scholar]
- 121.Wishart DS, Sayeeda Z, Budinski Z, Guo A, Lee BL, Berjanskii M, Rout M, Peters H, Dizon R, Mah R, Torres-Calzada C, Hiebert-Giesbrecht M, Varshavi D, Varshavi D, Oler E, Allen D, Cao X, Gautam V, Maras A, Poynton EF, Tavangar P, Yang V, van Santen JA, Ghosh R, Sarma S, Knutson E, Sullivan V, Jystad AM, Renslow R, Sumner LW, Linington RG and Cort JR, Nucleic Acids Res,, DOI: 10.1093/nar/gkab1052. [DOI] [Google Scholar]
- 122.Howarth A, Ermanis K. and Goodman JM, Chem. Sci, 2020, 11, 4351–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li D-W, Hansen AL, Yuan C, Bruschweiler-Li L. and Brüschweiler R, Nat. Commun, 2021, 12, 5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bruguière A, Derbré S, Dietsch J, Leguy J, Rahier V, Pottier Q, Bréard D, Suor-Cherer S, Viault G, Le Ray A-M, Saubion F. and Richomme P, Anal. Chem, 2020, 92, 8793–8801. [DOI] [PubMed] [Google Scholar]
- 125.Grienke U, Foster PA, Zwirchmayr J, Tahir A, Rollinger JM and Mikros E, Sci. Rep, 2019, 9, 11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kim HW, Zhang C, Cottrell GW and Gerwick WH, Magn. Reson. Chem,, DOI: 10.1002/mrc.5240. [DOI] [PubMed] [Google Scholar]
- 127.Reher R, Kim HW, Zhang C, Mao HH, Wang M, Nothias L-F, Caraballo-Rodriguez AM, Glukhov E, Teke B, Leao T, Alexander KL, Duggan BM, Van Everbroeck EL, Dorrestein PC, Cottrell GW and Gerwick WH, J. Am. Chem. Soc, 2020, 142, 4114–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim HW, Wang M, Leber CA, Nothias L-F, Reher R, Kang KB, van der Hooft JJJ, Dorrestein PC, Gerwick WH and Cottrell GW, J. Nat. Prod, 2021, 84, 2795–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Johnston CW, Skinnider MA, Wyatt MA, Li X, Ranieri MRM, Yang L, Zechel DL, Ma B. and Magarvey NA, Nat. Commun, 2015, 6, 8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mohimani H, Liu W-T, Kersten RD, Moore BS, Dorrestein PC and Pevzner PA, J. Nat. Prod, 2014, 77, 1902–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Behsaz B, Bode E, Gurevich A, Shi Y-N, Grundmann F, Acharya D, Caraballo-Rodríguez AM, Bouslimani A, Panitchpakdi M, Linck A, Guan C, Oh J, Dorrestein PC, Bode HB, Pevzner PA and Mohimani H, Nat. Commun, 2021, 12, 3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zani CL and Carroll AR, J. Nat. Prod, 2017, 80, 1758–1766. [DOI] [PubMed] [Google Scholar]
- 133.Zeng X, Zhu S, Lu W, Liu Z, Huang J, Zhou Y, Fang J, Huang Y, Guo H, Li L, Trapp BD, Nussinov R, Eng C, Loscalzo J. and Cheng F, Chem. Sci, 2020, 11, 1775–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Madhukar NS, Khade PK, Huang L, Gayvert K, Galletti G, Stogniew M, Allen JE, Giannakakou P. and Elemento O, Nat. Commun, 2019, 10, 5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Reker D, Rodrigues T, Schneider P. and Schneider G, Proc. Natl. Acad. Sci. U. S. A, 2014, 111, 4067–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rodrigues T, Werner M, Roth J, da Cruz EHG, Marques MC, Akkapeddi P, Lobo SA, Koeberle A, Corzana F, da Silva Júnior EN, Werz O. and Bernardes GJL, Chem. Sci, 2018, 9, 6899–6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nickel J, Gohlke B-O, Erehman J, Banerjee P, Rong WW, Goede A, Dunkel M. and Preissner R, Nucleic Acids Res, 2014, 42, W26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Carrella D, Napolitano F, Rispoli R, Miglietta M, Carissimo A, Cutillo L, Sirci F, Gregoretti F. and Di Bernardo D, Bioinformatics, 2014, 30, 1787–1788. [DOI] [PubMed] [Google Scholar]
- 139.Korshunova M, Ginsburg B, Tropsha A. and Isayev O, J. Chem. Inf. Model, 2021, 61, 7–13. [DOI] [PubMed] [Google Scholar]
- 140.Klambauer G, Unterthiner T, Mayr A. and Hochreiter S, Toxicol. Lett, 2017, 280, S69. [Google Scholar]
- 141.Huber F, van der Burg S, van der Hooft JJJ and Ridder L, J. Cheminform, 2021, 13, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rogers S, Ong CW, Wandy J, Ernst M, Ridder L. and van der Hooft JJJ, Faraday Discuss, 2019, 218, 284–302. [DOI] [PubMed] [Google Scholar]
- 143.van der Hooft JJJ, Wandy J, Young F, Padmanabhan S, Gerasimidis K, Burgess KEV, Barrett MP and Rogers S, Anal. Chem, 2017, 89, 7569–7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Buevich AV and Elyashberg ME, J. Nat. Prod, 2016, 79, 3105–3116. [DOI] [PubMed] [Google Scholar]
- 145.Reynolds WF, Pharmacognosy, 2017, 567–596. [Google Scholar]
- 146.Lindsay RK, Applications of Artificial Intelligence for Organic Chemistry: The DENDRAL Project, McGraw-Hill Book Company, 1980. [Google Scholar]
- 147.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H. and Markley JL, Nucleic Acids Res, 2008, 36, D402–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fischedick JT, Johnson SR, Ketchum REB, Croteau RB and Lange BM, Phytochemistry, 2015, 113, 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.McAlpine JB, Chen S-N, Kutateladze A, MacMillan JB, Appendino G, Barison A, Beniddir MA, Biavatti MW, Bluml S, Boufridi A, Butler MS, Capon RJ, Choi YH, Coppage D, Crews P, Crimmins MT, Csete M, Dewapriya P, Egan JM, Garson MJ, Genta-Jouve G, Gerwick WH, Gross H, Harper MK, Hermanto P, Hook JM, Hunter L, Jeannerat D, Ji N-Y, Johnson TA, Kingston DGI, Koshino H, Lee H-W, Lewin G, Li J, Linington RG, Liu M, McPhail KL, Molinski TF, Moore BS, Nam J-W, Neupane RP, Niemitz M, Nuzillard J-M, Oberlies NH, Ocampos FMM, Pan G, Quinn RJ, Reddy DS, Renault J-H, Rivera-Chávez J, Robien W, Saunders CM, Schmidt TJ, Seger C, Shen B, Steinbeck C, Stuppner H, Sturm S, Taglialatela-Scafati O, Tantillo DJ, Verpoorte R, Wang B-G, Williams CM, Williams PG, Wist J, Yue J-M, Zhang C, Xu Z, Simmler C, Lankin DC, Bisson J. and Pauli GF, Nat. Prod. Rep, 2019, 36, 35–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nuzillard J-M and Plainchont B, Magn. Reson. Chem, 2018, 56, 458–468. [DOI] [PubMed] [Google Scholar]
- 151.Elyashberg ME, Blinov KA, Williams AJ, Molodtsov SG, Martin GE and Martirosian ER, J. Chem. Inf. Comput. Sci, 2004, 44, 771–792. [DOI] [PubMed] [Google Scholar]
- 152.Cobas C, Seoane F, Vaz E, Bernstein MA, Dominguez S, Pérez M. and Sýkora S, Magn. Reson. Chem, 2013, 51, 649–654. [DOI] [PubMed] [Google Scholar]
- 153.Nuzillard J-M and Massiot G, Anal. Chim. Acta, 1991, 242, 37–41. [Google Scholar]
- 154.Williams RB, O’Neil-Johnson M, Williams AJ, Wheeler P, Pol R. and Moser A, Org. Biomol. Chem, 2015, 13, 9957–9962. [DOI] [PubMed] [Google Scholar]
- 155.Bakiri A, Plainchont B, de Paulo Emerenciano V, Reynaud R, Hubert J, Renault J-H and Nuzillard J-M, Mol. Inform,, DOI: 10.1002/minf.201700027. [DOI] [PubMed] [Google Scholar]
- 156.Cooper JK, Li K, Aubé J, Coppage DA and Konopelski JP, Org. Lett, 2018, 20, 4314–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.MacGregor CI, Han BY, Goodman JM and Paterson I, Chemical Communications, 2016, 52, 4632–4635. [DOI] [PubMed] [Google Scholar]
- 158.Snyder KM, Sikorska J, Ye T, Fang L, Su W, Carter RG, McPhail KL and Cheong PH-Y, Org. Biomol. Chem, 2016, 14, 5826–5831. [DOI] [PubMed] [Google Scholar]
- 159.Martínez-Treviño SH, Uc-Cetina V, Fernández-Herrera MA and Merino G, J. Chem. Inf. Model, 2020, 60, 3376–3386. [DOI] [PubMed] [Google Scholar]
- 160.Vignoli A, Ghini V, Meoni G, Licari C, Takis PG, Tenori L, Turano P. and Luchinat C, Angew. Chem. Int. Ed Engl, 2019, 58, 968–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Silva-Castro LF, Derbré S, Le Ray AM, Richomme P, García-Sosa K. and Peña-Rodriguez LM, Phytochem. Anal, 2021, 32, 1102–1109. [DOI] [PubMed] [Google Scholar]
- 162.Wolfender J-L, Litaudon M, Touboul D. and Queiroz EF, Nat. Prod. Rep, 2019, 36, 855–868. [DOI] [PubMed] [Google Scholar]
- 163.Kleigrewe K, Almaliti J, Tian IY, Kinnel RB, Korobeynikov A, Monroe EA, Duggan BM, Di Marzo V, Sherman DH, Dorrestein PC, Gerwick L. and Gerwick WH, J. Nat. Prod, 2015, 78, 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dan VM, J S V, C J S, Sanawar R, Lekshmi A, Kumar RA, Santhosh Kumar TR, Marelli UK, Dastager SG and Pillai MR, ACS Chem. Biol, 2020, 15, 780–788. [DOI] [PubMed] [Google Scholar]
- 165.Dan VM, Muralikrishnan B, Sanawar R, J S V, Burkul BB, Srinivas KP, Lekshmi A, Pradeep NS, Dastager SG, Santhakumari B, Santhoshkumar TR, Kumar RA and Pillai MR, Sci. Rep, 2018, 8, 2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hu J-Q, Wang J-J, Li Y-L, Zhuo L, Zhang A, Sui H-Y, Li X-J, Shen T, Yin Y, Wu Z-H, Hu W, Li Y-Z and Wu C, Org. Lett, 2021, 23, 2114–2119. [DOI] [PubMed] [Google Scholar]
- 167.Kersten RD, Yang Y-L, Xu Y, Cimermancic P, Nam S-J, Fenical W, Fischbach MA, Moore BS and Dorrestein PC, Nat. Chem. Biol, 2011, 7, 794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen X, Wang Y, Ma N, Tian J, Shao Y, Zhu B, Wong YK, Liang Z, Zou C. and Wang J, Signal Transduct Target Ther, 2020, 5, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Dai L, Li Z, Chen D, Jia L, Guo J, Zhao T. and Nordlund P, Pharmacol. Ther, 2020, 216, 107690. [DOI] [PubMed] [Google Scholar]
- 170.Feher M. and Schmidt JM, ChemInform, 2003, 34. [Google Scholar]
- 171.Moffat JG, Vincent F, Lee JA, Eder J. and Prunotto M, Nat. Rev. Drug Discov, 2017, 16, 531–543. [DOI] [PubMed] [Google Scholar]
- 172.Zon LI and Peterson RT, Nature Reviews Drug Discovery, 2005, 4, 35–44. [DOI] [PubMed] [Google Scholar]
- 173.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet J-P, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES and Golub TR, Science, 2006, 313, 1929–1935. [DOI] [PubMed] [Google Scholar]
- 174.Lum KM, Sato Y, Beyer BA, Plaisted WC, Anglin JL, Lairson LL and Cravatt BF, ACS Chem. Biol, 2017, 12, 2671–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Langley GR, Adcock IM, Busquet F, Crofton KM, Csernok E, Giese C, Heinonen T, Herrmann K, Hofmann-Apitius M, Landesmann B, Marshall LJ, McIvor E, Muotri AR, Noor F, Schutte K, Seidle T, van de Stolpe A, Van Esch H, Willett C. and Woszczek G, Drug Discov. Today, 2017, 22, 327–339. [DOI] [PubMed] [Google Scholar]
- 176.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS and Olson AJ, J. Comput. Chem, 2009, 30, 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ and Shoichet BK, Nature Biotechnology, 2007, 25, 197–206. [DOI] [PubMed] [Google Scholar]
- 178.Luo Y, Zhao X, Zhou J, Yang J, Zhang Y, Kuang W, Peng J, Chen L. and Zeng J, Nat. Commun, 2017, 8, 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wan F, Hong L, Xiao A, Jiang T. and Zeng J, Bioinformatics, 2019, 35, 104–111. [DOI] [PubMed] [Google Scholar]
- 180.Walker AS and Clardy J, J. Chem. Inf. Model, 2021, 61, 2560–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Altschul SF, Gish W, Miller W, Myers EW and Lipman DJ, J. Mol. Biol, 1990, 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 182.Rodrigues T, Reker D, Schneider P. and Schneider G, Nat. Chem, 2016, 8, 531–541. [DOI] [PubMed] [Google Scholar]
- 183.Jeon J, Nim S, Teyra J, Datti A, Wrana JL, Sidhu SS, Moffat J. and Kim PM, Genome Med, 2014, 6, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hefti FF, BMC Neurosci, 2008, 9 Suppl 3, S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rusyn I. and Daston GP, Environ. Health Perspect, 2010, 118, 1047–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Demain AL, J. Ind. Microbiol. Biotechnol, 2014, 41, 185–201. [DOI] [PubMed] [Google Scholar]
- 187.Sahayasheela VJ, Yu Z, Hirose Y, Pandian GN, Bando T. and Sugiyama H, Bull. Chem. Soc. Jpn, 2022, 95, 693–699. [Google Scholar]
- 188.Li G, Lin P, Wang K, Gu C-C and Kusari S, Trends in Cancer, 2021. [DOI] [PubMed] [Google Scholar]
- 189.Brahmachari G, Discovery and Development of Therapeutics from Natural Products Against Neglected Tropical Diseases, Elsevier, 2019. [Google Scholar]
- 190.Theuretzbacher U, Outterson K, Engel A. and Karlén A, Nat. Rev. Microbiol, 2020, 18, 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Terlain B. and Jean-Pierre T, Bull. Soc. Chim. Fr, 1971, 1971, 2349–2356. [PubMed] [Google Scholar]
- 192.Kling A, Lukat P, Almeida DV, Bauer A, Fontaine E, Sordello S, Zaburannyi N, Herrmann J, Wenzel SC, König C, Ammerman NC, Barrio MB, Borchers K, Bordon-Pallier F, Brönstrup M, Courtemanche G, Gerlitz M, Geslin M, Hammann P, Heinz DW, Hoffmann H, Klieber S, Kohlmann M, Kurz M, Lair C, Matter H, Nuermberger E, Tyagi S, Fraisse L, Grosset JH, Lagrange S. and Müller R, Science, 2015, 348, 1106–1112. [DOI] [PubMed] [Google Scholar]
- 193.Strelitz F, Flon H. and Asheshov IN, J. Bacteriol, 1955, 69, 280–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Jain SK, Pathania AS, Parshad R, Raina C, Ali A, Gupta AP, Kushwaha M, Aravinda S, Bhushan S, Bharate SB and Vishwakarma RA, RSC Adv, 2013, 3, 21046–21053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Muralikrishnan B, Dan VM, Vinodh JS, Jamsheena V, Ramachandran R, Thomas S, Dastager SG, Santhosh Kumar K, Lankalapalli RS and Kumar RA, RSC Adv, 2017, 7, 36335–36339. [Google Scholar]
- 196.Wu F, Zhang J, Song F, Wang S, Guo H, Wei Q, Dai H, Chen X, Xia X, Liu X, Zhang L, Yu J-Q and Lei X, ACS Cent Sci, 2020, 6, 928–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tanoli Z, Vähä-Koskela M. and Aittokallio T, Expert Opin. Drug Discov, 2021, 16, 977–989. [DOI] [PubMed] [Google Scholar]
- 198.Cech NB, Medema MH and Clardy J, Nat. Prod. Rep, 2021, 38, 1947–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.van Santen JA, Kautsar SA, Medema MH and Linington RG, Nat. Prod. Rep, 2021, 38, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bender A. and Cortés-Ciriano I, Drug Discovery Today, 2021, 26, 511–524. [DOI] [PubMed] [Google Scholar]
- 201.Anom BY, Ethics, Medicine and Public Health, 2020, 15, 100568. [Google Scholar]
- 202.Vatansever S, Schlessinger A, Wacker D, Kaniskan HÜ, Jin J, Zhou M-M and Zhang B, Med. Res. Rev, 2021, 41, 1427–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]