Abstract
Background and Purpose:
Metformin is used for the management of type 2 diabetes mellitus (T2DM). Metformin is being tested clinically as an anti-cancer agent. Metformin concentrations safely achievable in human solid tissues including tumors are unknown. Metformin concentration in tissue compartments as a function of dose will inform rational dosing in preclinical models and interpretation of clinical results.
Experimental Approach:
Subjects with solid tumors to be treated by resection and either (A) willingness to take metformin for 7–10 days before surgery, or (B) taking metformin for T2DM were eligible. Whole blood, plasma, tumor, tumor-adjacent uninvolved tissue, and subcutaneous adipose tissue were obtained for LC-MS/MS to measure metformin concentrations.
Key Results:
All subjects had primary lung tumors. Metformin dose was significantly correlated with drug concentrations in all tissues analyzed. Inter-subject metformin concentrations varied by over two orders of magnitude. Metformin concentrations were significantly higher in tumor tissues and lower in adipose tissues compared to other tissues. Concentrations in blood and plasma were significantly correlated with concentrations in solid tissues.
Conclusion and Implications:
Metformin accumulates in cellular compartments. Concentrations observed in plasma, blood, lung, and tumor tissues in subjects treated with doses FDA-approved for T2DM are lower than those typically used in tissue culture studies. However, such tissue concentrations are in line with those found within cultured cells treated with supra-pharmacological doses of metformin. Given the large inter-subject variability in metformin concentrations, it is imperative to determine whether there is an association between tissue metformin concentration and anti-cancer activity in humans.
Keywords: metformin, cancer, tissue distribution
Introduction
Metformin is a member of the biguanide class of drugs commonly used to treat type 2 diabetes mellitus (T2DM). Metformin lowers fasting blood glucose concentrations by suppressing hepatic gluconeogenesis and increasing glucose uptake and utilization in muscle and visceral tissues. However, the molecular mechanism of action of metformin is unclear. Metformin activates 5’ AMP-activated protein kinase (AMPK) to modulate metabolic and lipid signaling, which may occur via (i) direct binding to AMPK, (ii) inhibition of mitochondrial complex I (NADH oxidoreductase) to suppress ATP production and cause an energy deficit in cells, (iii) increasing cellular redox state, and/or (iv) an AMPK-independent mechanism (2–5). Metformin has also been shown to have anti-cancer effects in preclinical models and epidemiological studies of humans with T2DM, wherein metformin increased survival compared to treatment with insulin or other anti-diabetes agents (6–9). Furthermore, metformin-treated T2DM patients with cancer have longer survival than non-diabetics with cancer, and metformin treatment lowers the risk of developing cancer (7,10–15). Despite metformin having an unclear mechanism of anti-cancer activity, the epidemiological data led to the initiation of many clinical trials studying whether metformin is an effective anti-cancer agent in non-diabetic patients (6,16).
Adding to the incomplete understanding of the mechanism of anti-cancer action of metformin is the literature relating to its tissue distribution. With a twice-daily (BID) oral dose of 750 mg of metformin (extended release formulation), the terminal plasma elimination half-life is approximately 6.2 h (17). Absorption of metformin occurs predominantly in small intestine and is mediated through plasma membrane monoamine transporters. However, this absorption is often incomplete, resulting in substantial inter-patient variability in metformin bioavailability (mean ± SD of 55% ± 16%), and causing considerable inter-individual variability in its pharmacokinetics (18). Steady-state metformin plasma concentrations have been reported in the nanomolar and micromolar ranges (19,20). While no metformin accumulation in plasma was observed in patients treated with multiple doses, metformin plasma concentration decay is bi-phasic, with a rapid initial decay (alpha) phase indicative of distribution perhaps into red blood cells, and a second more prolonged elimination (beta) phase possibly due to slow uptake and release from a peripheral tissue compartment (18,21). Metformin exists as a hydrophilic cationic species at physiological pH, and as such its passive diffusion into tissues is potentially limited. Nonetheless, metformin is widely distributed throughout bodily tissues and is imported into cells in liver and other peripheral tissues mainly through organic cation transporters 1 and 3 (SLC22A1 and SLC22A3) (22). Metformin is not metabolized and poorly bound to plasma proteins; approximately 90% of a dose is eliminated from the body by renal excretion within 24 h (23).
Despite the contradictory literature on the molecular mechanism of action of metformin, and the widely variable pharmacokinetics in humans, a number of clinical trials attempting to provide proof of concept for direct anti-tumor activity of metformin in non-diabetic patients have been launched and early results are conflicting (6,16). For example, Yee et al. reported that the addition of metformin modestly increased the pathologic complete response rate to neoadjuvant ganitumab in breast cancer patients (24). El-Khayat et al. found that the addition of metformin significantly increased the rate of pathologic complete response to neoadjuvant chemotherapy in breast cancer (25). In contrast, Lopez-Bonet et al. observed that the addition of metformin did not alter the pathologic complete response rate of HER2-positive breast tumors treated with neoadjuvant trastuzumab, anthracycline, and taxane (26). A study in ovarian cancer patients revealed that metformin increased the proportion of cancer cells that co-expressed the stem cell-like markers aldehyde dehydrogenase (ALDH) and CD133 (27). The OCOG-ALMERA study showed that metformin decreased the efficacy of chemoradiotherapy in patients with locally advanced non-small cell lung cancer compared to chemoradiotherapy alone (28). In considering potential causes of variability between clinical trial results, one major problem is that the majority of these trials did not measure the concentrations of metformin achieved in the vascular compartment or peripheral tissues. A significant proportion of preclinical evidence supporting metformin as an effective anti-cancer agent was generated using supra-pharmacological drug concentrations (5–50 mM) that are not known to be achievable in humans. To better understand the potential anti-cancer effects of metformin in humans, and to identify clinically relevant drug concentrations to inform future preclinical studies and the interpretation of findings reported thus far, it is crucial to determine the concentrations of metformin achieved in human tissue compartments. If the insulin-independent effects of metformin are predominantly responsible for its anti-cancer activity [as some preclinical studies have suggested (29)], then measuring the concentrations of metformin achieved in tumor tissues, and determining whether those concentrations are sufficient to activate AMPK, are steps to aid the understanding of conflicting clinical trial results. Therefore, to determine achievable metformin concentrations and their relationships between different tissue compartments in humans, we undertook a window-of-opportunity clinical study with the objectives of determining the intra-tumor concentrations of metformin in patients with solid tumors of thoracic origin, and the concentrations of metformin in adipose tissue, tumor-adjacent normal tissue, plasma, and whole blood.
Methods
Study IRB approval and ethical conduct
This study was approved by the Dartmouth College Committee for the Protection of Human Subjects. The study was registered on clinicaltrials.gov (NCT03477162) and conducted in accordance with ICH Good Clinical Practice. The study was monitored by the Norris Cotton Cancer Center Data, Safety Monitoring, and Accrual Committee (DSMAC). Informed consent was obtained from participants included in the study before performing any study-related procedures. All study procedures involving human participants were in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Study participant recruitment and eligibility
Study participants were recruited from the clinics of the Norris Cotton Cancer Center at Dartmouth between January 2019 and April 2021. Major study inclusion criteria were: confirmed or suspected malignant solid tumor of thoracic origin with the intent to treat or biopsy by surgery as the standard of care; absolute neutrophil count ≥1,500/mm3; platelet count ≥75,000/mm3; bilirubin ≤1.5x the upper limit of the normal range (ULN); alanine aminotransferase and aspartate aminotransferase ≤3x ULN; either estimated glomerular filtration rate (eGFR) >60 mL/min/1.73 m2 (by CKD-EPI equation) or estimated creatinine clearance >60 mL/min. Study participant exclusion criteria included: a history of diabetes mellitus currently being treated without metformin; a history of alcoholism or binge (ethanol) drinking disorder; the need for a radiographic analysis with an iodinated contrast agent during the presurgical treatment period; a history of reactive hypoglycemia; known hypersensitivity to metformin; active or a history of lactic acidosis, metabolic acidosis, or diabetic ketoacidosis; current pregnancy or breastfeeding.
Study design
Subjects with T2DM being treated with metformin for a clinical indication at the time of study enrollment were eligible and continued metformin treatment as clinically indicated at their prescribed dose during the presurgical study period. Subjects without T2DM were treated with metformin extended release (Glucophage XR) 750 mg once daily (QD) for 4 days, then 750 mg twice daily (BID) for 3 to 6 days for a total of 7 to 10 days of metformin treatment prior to surgical tumor resection. The last dose of metformin was taken within one day of surgery. Subjects were asked to record metformin administration times including on the day before surgery in a diary. On the morning of their thoracic surgery, ~20 mL of venous blood was drawn into EDTA-treated tubes within 5 h (mean ± SD of 2.45 ± 1.40 h) before surgical resection of the lung tumor, adjacent uninvolved normal lung tissue, and subcutaneous adipose tissue. Blood was either frozen within 2 h of collection at −80°C, or centrifuged at 4°C at 1,000 x g for 30 min. to isolate plasma and plasma aliquots were subsequently frozen at −80°C. Solid tissues were flash-frozen in liquid nitrogen as ~5-mm fragments within 2 h of collection and stored at −80°C. A subject undergoing surgery who did not receive presurgical metformin treatment was enrolled to provide control tissues.
Metformin measurement in whole blood, plasma, and solid tissues
Solid tissue specimens were weighed, thawed, and homogenized in deionized water using a NextAdvance bullet blender and stainless steel beads. Metformin concentrations were measured by liquid chromatography with tandem mass spectrometry using a modification of the assay method described previously (30). Protein was precipitated from samples (50 μL of whole blood or plasma; 5 mg of homogenized tissue) with 200 μL acetonitrile containing 200 ng/mL phenformin as the internal standard. Supernatants were collected and dried to completion under nitrogen at 50°C. Dried samples were suspended in 50 μL water for injection onto an Accucore HILIC 50 mm x 2.1 mm, 2.6-μm column with 10 mm x 2.1 mm HILIC guard cartridge. Isocratic conditions (25% 100 mM ammonium formate pH 3.2 and 75% acetonitrile) over 3 min were utilized at 0.35 mL/min on a Dionex Ultimate 3000 HPLC system. After sample injection, the column was washed with deionized water + 0.1% formic acid, and then equilibrated back to isocratic conditions. A TSQ Vantage tandem quadrupole mass spectrometer with a HESI-II probe, operated in multiple reaction monitoring mode, was used for positive ion detection. Xcalibur software was used for data acquisition and processing. MS/MS detection was conducted monitoring 130.09 → 71.100 m/z (collision energy 20 V) for metformin and 206.07 → 60.11 m/z (collision energy 16 V) for phenformin. S-Lens RF amplitudes for metformin and phenformin were 40 and 60 V respectively. Source parameters were spray voltage 2000 V, vaporizer temperature 275°C, capillary temperature 350°C, sheath gas 43 rel. units, and aux gas 4 rel. units. The lower limit of quantification (<LLOQ) of the metformin assay was 3 ng/mL. Metformin calibration standards were linear over the range 3–3,000 ng/mL in whole blood and plasma (n=8 non-zero standards), 3–1,000 ng/mL in lung tissue (n=7 non-zero standards), and 3–300 ng/mL in adipose tissue (n=6 non-zero standards) using control tissues from the subject not treated with metformin. Only calibration standards within ±15% of the nominal value, or ±20% for the LLOQ, were used for quantitation. At least 75% of the calibrations standards were retained for the regression analysis. This method was validated using quality controls of 10, 100, and 1,000 ng/mL metformin with n≥4 for each concentration as appropriate for the calibration range of each matrix. The coefficients of variation (CV) were 2–9% for plasma, 1–8% for whole blood, 1–6% for adipose tissue, and 1–5% for lung tissue. Accuracy ranged from 99–106% in plasma, 100–110% in whole blood, 103–111% in adipose tissue, and 92–103% in lung tissue. Lung tissue served as a surrogate matrix for tumor tissue for assay validation. Metformin concentrations were measured in triplicates of unknown whole blood, plasma, and homogenized tissues using 3 injections per sample.
Statistics
Statistical analysis of metformin concentration was carried out in three directions: (a) identification of the dose-concentration relationship between concentration in five tissue compartments (plasma, whole blood, adipose, lung, and tumor) as a function of dose via linear regression analysis; (b) pairwise correlations between the five compartments; (c) building increasing-complexity predictive models for estimation of tumor metformin concentration based on dose and measurement in plasma and whole blood. The linear regression model with all measurements of metformin concentration and zero intercept was used since zero metformin in all compartments is expected with zero dose (this hypothesis has been tested by statistical means – the intercept in all five models was statistically insignificant, p>0.05). A saturated nonlinear dose-response relationship model was considered, but we used a linear model because (i) the scatter of metformin concentrations is high, and (ii) there is no indication of saturation (Fig. 1). The uncertainty of tumor metformin concentration was quantified by the standard error of prediction and the coefficient of determination (squared correlation coefficient, R2) that determines the proportion of variance of metformin concentration in the tumor by dose and predictors based on circulating concentration (31). We used Pearson correlation coefficient (r) to quantify the correlation, and the coefficient of determination (R2) to measure the quality of prediction of metformin concentration in tissue given the administered dose. To account for variation across subjects, a linear mixed model was employed estimated in the statistical package R by the function lme from package nlme (32). Using random effect, the mixed model accounts for the inter-subject heterogeneity (33) of metformin concentrations when treated with the same daily dose. We also tested the hypothesis that metformin formulation and/or frequency of administration produces an effect on (a) the slope or (b) heterogeneity variance for each tissue type using the R function anova.
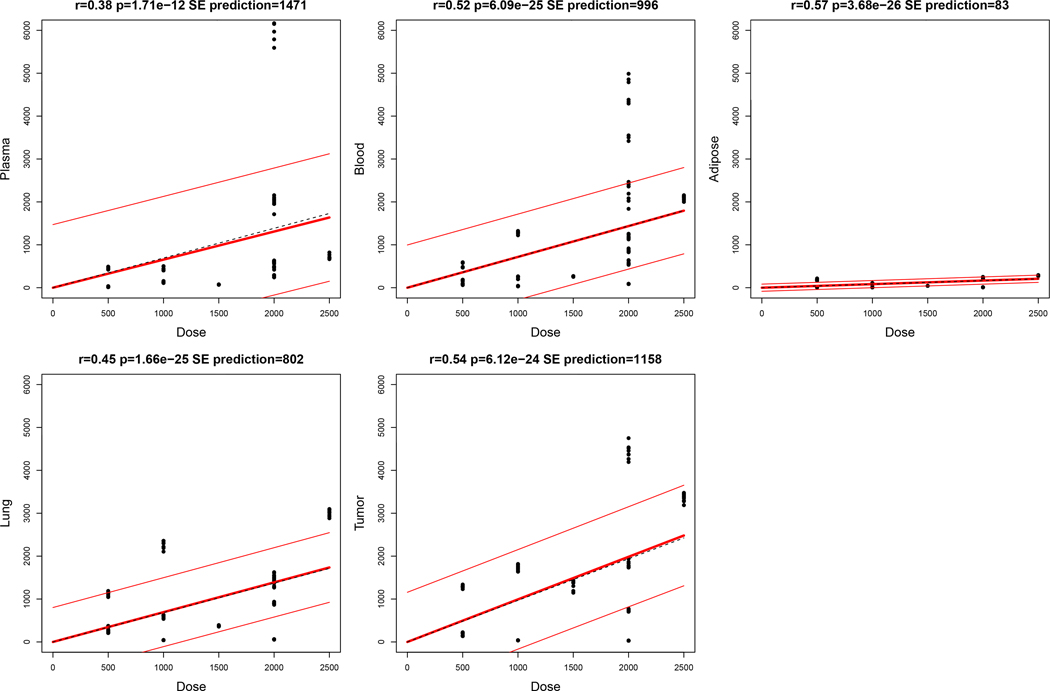
Fig. 1-. Daily dose of metformin correlates with tissue concentrations.
Concentrations of metformin in plasma, blood, adipose, lung tissue, and tumor tissue available from 12, 14, 7, 12, and 11 subjects were compared to daily dose. Correlation testing included all replicates with metformin concentrations ≥LLOQ. Regression lines (thick) are plotted ± standard error of prediction by regression (thin). p-value of slopes were determined by t-test compared to the null hypothesis.
Results
Study population
Twenty participants with primary lung cancers were enrolled into this study. Five subjects were not evaluable. Fourteen subjects completed presurgical treatment with metformin (Table 1). One participant provided negative control tissue specimens and did not receive metformin. While the study was designed to enroll 18 evaluable subjects, the study was terminated prior to completion of enrollment due to a shortage of funding. Among the 14 evaluable participants treated with metformin, 13 participants (93%) had T2DM and were receiving clinically indicated metformin doses at the time of study entry. Among the 14 metformin-treated subjects, whole blood, plasma, tumor, lung, and adipose tissue specimens were available from 14, 14, 11, 12, and 11 subjects respectively.
Table 1.
Subject demographics.
| Characteristic | Metformin-treated subjects (n=14) |
Untreated control subject (n=1) |
|---|---|---|
|
| ||
| Age (yr) | ||
| median | 70.5 | 75 |
| range (min, max) | 62, 80 | -- |
|
| ||
| Sex | ||
| male, n (%) | 5 (36) | 1 (100) |
| female, n (%) | 9 (64) | -- |
|
| ||
| Body weight | ||
| median | 87.5 | 97.2 |
| range (min, max) | 56.2, 117.5 | -- |
|
| ||
| Body-mass index | ||
| median | 29.7 | 29.9 |
| range (min, max) | 22.7, 46.6 | -- |
|
| ||
| History of T2DM, n (%) | 13 (93) | 0 (0) |
|
| ||
| Subject taking metformin at time of study entry, n (%) | 13 (93) | 0 (0) |
|
| ||
| Subjects enrolled | 19 | 1 |
| Subjects evaluable | 14 | 1 |
| Subjects non-evaluable | 5 | -- |
| Reason for non-evaluability: | ||
| withdrew due to adverse events* | 1 | -- |
| did not complete study treatment | 2 | -- |
| did not complete drug diary | 2 | -- |
One non-T2DM subject withdrew due to self-reported nausea and diarrhea that the subject attributed to study-prescribed metformin.
Metformin concentrations varied widely between subjects and were correlated with dose
Metformin was detected in plasma from 12/14 study participants treated with metformin (Table 2). Among 11/12 of those subjects, the CV across plasma triplicates was 4% to 11%, and samples from the twelfth subject (#14) with relatively low plasma metformin concentrations had a CV of 30%. We detected a wide inter-subject range of metformin concentrations. For plasma metformin concentrations, the detected range was <3–6,179 ng/mL (<0.02–47.84 μM) across 12 subjects, and 2 subjects had concentrations below the lower limit of quantification (<LLOQ) of the assay (Table 2). Most subjects were taking metformin at the time of study entry to treat TD2M, and their doses, formulations, and schedules were tailored to their individual clinical needs. We therefore explored whether the inter-subject variability in metformin dose (500 to 2,500 mg/day), schedule (QD or BID), formulation [standard release (SR) or extended release (ER)] (Table 2), and time since last dose could underlie the differences in plasma concentrations. While we found a significant correlation between daily metformin dose and metformin concentrations across all tissues by linear regression (Fig. 1), time since last dose was not significantly correlated with plasma concentrations.
Table 2.
Metformin dose, time post-dose for sample procurement, and blood and tissue concentrations.
| Subject ID | Metformin dose and schedule; formulation | Metformin daily dose | Subject on metformin at time of enrollment | Duration of metformin treatment for T2DM (yr) | Time from last metformin dose to surgery (h) | Plasma metformin conc. (ng/mL) | Whole blood metformin conc. (ng/mL) | Derived intracellular metformin conc. (ng/mL) | Lung tumor tissue metformin conc. (ng/g) | Normal lung tissue metformin conc. (ng/g) | Adipose tissue metformin conc. (ng/g) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 500 mg QD; ER | 500 mg | Y | 1.97 | 26.6 | 18 ± 1.4 | 175 ± 13.2 | 157 ± 3.8 | 212 ± 9.9 | 230 ± 19.4 | <LLOQ |
| 12 | 500 mg QD, ER | 500 mg | Y | 7.03 | 24.4 | 459 ± 27.8 | 514 ± 55.4 | 55 ± 9.1 | 1290 ± 29.5 | 1126 ± 46.1 | 186 ± 15.4 |
| 14 | 500 mg QD, SR | 500 mg | Y | 0.24 | 29.0 | 25 ± 7.7 | 84 ± 14.1 | 59 ± 4.7 | 145 ± 7.2 | 330 ± 38.3 | <LLOQ |
| 6 | 500 mg BID; SR | 1000 mg | Y | 2.84 | 15.3 | <LLOQ | 41 ± 4.9 | n/a | 40 ± 1.4 | 44 ± 2.2 | <LLOQ |
| 11 | 500 mg BID; SR | 1000 mg | Y | 3.57 | 24.3 | 127 ± 13.8 | 241 ± 28.6 | 114 ± 6.5 | n/a | 596 ± 33.3 | 70 ± 1.9 |
| 8 | 1000 mg QD; SR | 1000 mg | Y | 0.04 | 20.9 | 442 ± 39.5 | 1285 ± 29.1 | 843 ± 8.3 | 1725 ± 65.8 | 2269 ± 89.1 | 111 ± 2.4 |
| 5 | 750 mg BID; ER | 1500 mg | N | --- | 16.5 | 76 ± 2.3 | 267 ± 5.9 | 191 ± 2.9 | 1344 ± 106.2 | 376 ± 10.5 | 45 ± 1.9 |
| 7 | 1000 mg BID; ER | 2000 mg | Y | 5.35 | 23.6 | <LLOQ | 89 ± 2.3 | n/a | 33 ± 1.0 | 63 ± 2.9 | <LLOQ |
| 3 | 1000 mg BID; SR | 2000 mg | Y | 7.63 | 14.8 | 474 ± 21.3 | 1192 ± 47.8 | 718 ± 8.3 | 1799 ± 69.3 | 1427 ± 110.8 | 210 ± 4.1 |
| 4 | 1000 mg BID; SR | 2000 mg | Y | 14.06 | n/a | 272 ± 23.6 | 869 ± 31.7 | 597 ± 7.4 | 737 ± 26.3 | 902 ± 19.9 | n/a |
| 9 | 1000 mg BID; SR | 2000 mg | Y | 1.30 | 14.0 | 2005 ± 127.0 | 2243 ± 215.2 | 238 ± 18.5 | n/a | n/a | n/a |
| 10 | 1000 mg BID; SR | 2000 mg | Y | n/a | n/a | 604 ± 23.2 | 588 ± 41.8 | n/a | 4445 ± 164.5 | 1492 ± 95.8 | 247 ± 4.5 |
| 13 | 1000 mg BID; SR | 2000 mg | Y | 9.09 | n/a | 6179 ± 352.0 | 4238 ± 608.8 | n/a | n/a | n/a | n/a |
| 1 | 1500 mg in AM, 1000 mg in PM; SR | 2500 mg | Y | 3.09 | 21.5 | 719 ± 48.2 | 2093 ± 50.1 | 1374 ± 9.9 | 3365 ± 93.5 | 3013 ± 75.0 | 285 ± 9.9 |
| 15 | 0 mg | 0 mg | N | --- | --- | <LLOQ | <LLOQ | n/a | n/a | <LLOQ | <LLOQ |
Subjects are grouped are rows are shaded by daily dose. Metformin concentrations (conc.) are presented as mean ± SD. Derived intracellular metformin conc. was calculated as the difference between blood and plasma conc. QD- once daily; BID- twice daily; SR- standard release; ER- extended release; AM- morning; PM- evening; n/a- specimen or data not available/calculable; Y- yes; N- no; LLOQ- lower limit of quantification.
It is critical to determine metformin concentrations in tumor tissue if this drug is to be used as an anti-cancer agent. Sampling blood to measure metformin concentrations is less invasive than sampling tumor tissue. Thus, we evaluated regression models to predict tumor tissue metformin concentration informed by circulating whole blood or plasma metformin concentrations. We determined that a linear equation incorporating the 3 variables of dose, plasma concentration, and whole blood concentration resulted in the most accurate model with the least standard error (Fig. S1).
Metformin accumulation varies between tissue compartments
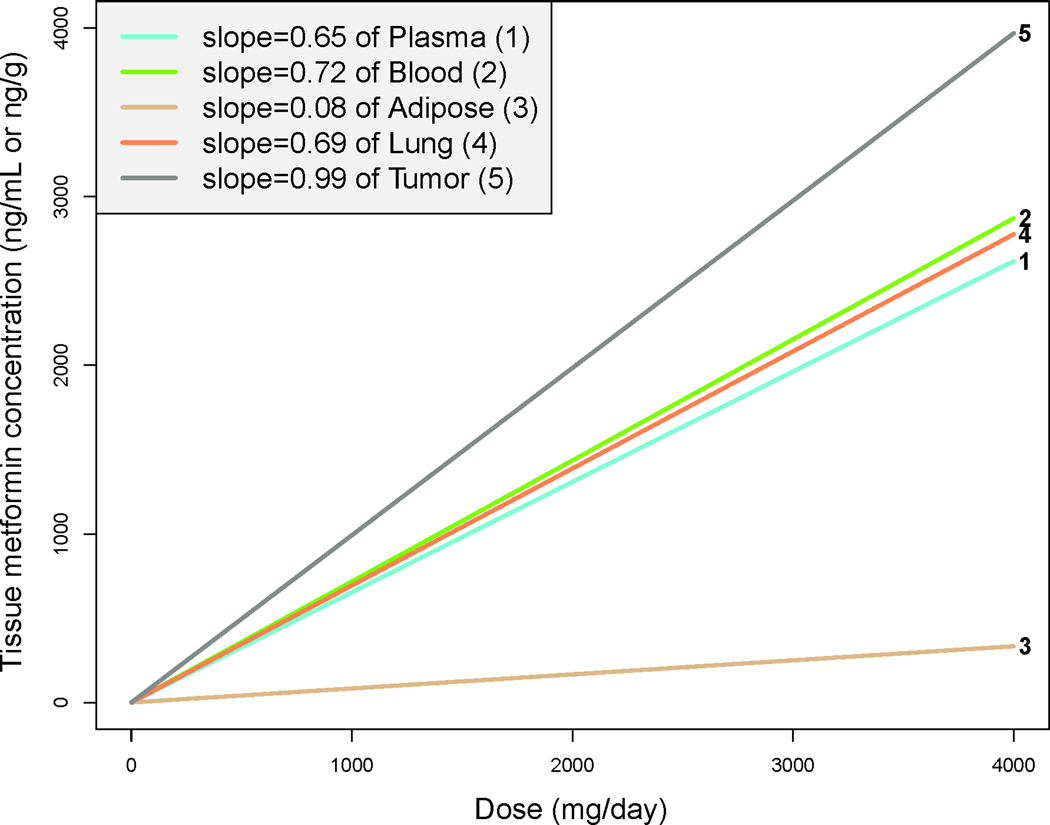
Lung tumor tissue metformin concentrations ranged from 33–4,445 ng/g [0.25–34.41 μM; median 1290 ng/g (9.99 μM)], and normal lung tissue concentrations ranged from 44–3,013 ng/g [0.34–23.33 μM; median 749 ng/g (5.80 μM)] (Table 2). We compared metformin concentrations between the five tissue compartments. In almost all cases, we found significant correlations between plasma, blood, and solid tissues (all r ≥0.63, all p≤0.05; Fig. S2). In adipose tissue from 7/11 subjects with detectable metformin concentrations, these ranged from 45 to 285 ng/g [0.35 to 2.21 μM; median across 11 subjects was 70 ng/g (0.54 μM)], with lower metformin accumulation compared to the other cellular tissues (i.e., blood, tumor, lung). To compare metformin accumulation between tissue compartments, we applied a linear mixed-effects model to estimate metformin accumulation. Modeling confirmed that metformin concentrations increased with dose, and accumulation is most and least prominent in tumor and adipose tissues, respectively (Fig. 2). Adjustment for metformin formulation and dosing frequency did not significantly affect models (Fig. S3). Accordingly, metformin concentrations were significantly lower in adipose tissue compared to the other four tissue compartments (all p<0.0001), and significantly higher in tumor tissue compared to all other compartments analyzed (all p<0.003) (Fig. S4). Furthermore, metformin levels were higher in whole blood than plasma in 12/14 subjects, enabling calculation of intracellular drug levels in blood (Table 2). These findings collectively indicate that metformin accumulates in cellular tissues (whole blood, lung, and tumor, but not adipose), and that plasma concentrations correlate with metformin accumulation in such tissues.
Fig. 2-. A linear mixed-effects model predicts metformin concentration by dose.
A linear mixed-effects model was applied accounting for inter-subject variation to predict tissue metformin concentration compared to daily dose. Lines are numbered according to tissue site in key.
Discussion and Conclusions
Our study confirmed prior observations that plasma and whole blood metformin concentrations vary widely among patients treated with metformin at doses approved for the management of T2DM. We provide the first report of metformin concentrations measured in solid tumor and normal tissues in humans, and show that solid tissue concentrations vary between patients and correlate with whole blood and plasma concentrations.
The finding of similar metformin concentrations in cellular compartments (i.e., whole blood, tumor, or normal lung vs. plasma) supports data from a prior study that showed that metformin area under the curve is similar between plasma and erythrocytes even though the drug is more rapidly cleared from plasma (34). Importantly, our findings indicate that achievable metformin concentrations in plasma, tumor, and normal lung are about 100- to 1,000-fold lower than those used in published tissue culture studies (8,35,36). However, the metformin concentrations we found in the tissues of our study participants are also found within in vitro cultured cells treated with pharmacological or supra-pharmacological doses of metformin. Furthermore, treatment of cultured cells with those supra-pharmacological concentrations induced AMPK activation (9,20), suggesting that (a) pharmacologically achievable concentrations in human tumor and normal lung tissues are relevant to published data from in vitro cellular studies, (b) drug transport may be a limiting factor in getting drug into cultured cells, and (c) higher concentrations of metformin may elicit effects independently of AMPK and ATP production. It is noteworthy that relatively high concentrations (~5 mM) of metformin are required to inhibit Complex I in isolated mitochondria (4).
We observed a wide inter-subject range (<3–6,179 ng/mL; <0.02–47.84 μM) of plasma concentrations of metformin [median 450 ng/mL (3.49 μM)]. This variability is consistent with a report describing plasma metformin concentrations of 0.08–6.5 μM (mean = 2.8 μM) in women treated for ≥14 days with metformin 1500 mg/day (administered as 500 mg three times daily) (20). In addition, a report of subjects treated with the relatively high (but clinically utilized) oral dose of 2,000 mg/day metformin (administered as 1,000 mg twice a day) for ≥3 months revealed a mean plasma metformin concentration of 576 ng/mL (4.46 μM) (range of 54–4,133 ng/mL, 0.42–32.00 μM) (19). While the mean plasma concentration detected in our study was ~2.7 times that reported previously (20), the median plasma concentration observed in our data is more in line with the previous finding possibly due to greater variance in our cohort, longer treatment durations, and/or higher dosing of some participants in our study. In our study, the variability in time from (a) the last metformin dose to (b) the procurement of blood, plasma, and tissues was driven by the practical limitations of undertaking such sampling in the environment of surgical operating room schedules, and the performance and duration of operations meant to be curative for lung cancer.
In parallel to the wide variability in plasma metformin concentrations, tumor and normal lung tissue concentrations showed broad ranges. However, lung and tumor tissue metformin concentrations were strongly correlated (r=0.76, p=0.007, Fig. S2), indicating that these healthy and diseased tissues have similar capacities for metformin disposition. Adipose tissue showed lower metformin accumulation compared to the other cellular tissues (i.e., blood, tumor, lung) (Fig. 2), suggesting that hydrophilic metformin with a pKa of 12.4 is thus ionized at physiological pH. With a log P variably estimated at −0.92 to −1.8, metformin does not accumulate in fat due to low fat solubility and its ionization state at physiological pH. Collectively, these observations support the hypothesis that metformin accumulates within cellular tissues with the exception of adipose tissue.
We and others undertook preclinical pharmacokinetic studies in animals to attempt to define achievable tissue concentrations of metformin. Dowling et al. showed that mouse models in which metformin elicits anti-cancer activity (at a dose of ~0.5 mg/kg/day in drinking water) provided metformin plasma concentrations of 297–16,274 ng/mL (0.45–24.3 μM) that are known to be generally achievable in human plasma. We found that a lower dosing regimen (~0.1 mg/kg/day in drinking water) in mice also produced a robust anti-cancer effect in breast cancer xenografts and activated AMPK in these tumors (9). Therein, mouse tumor tissue metformin concentrations ranged from 80–114 ng/g (0.62–0.89 μM; mean ± SD of 98 ± 12 ng/g, 0.76 ± 0.09 μM). In parallel, our studies in cultured breast cancer cells revealed that treatment with 1 μM metformin – approximately the lowest concentration required to robustly elicit AMPK activation in these cells – for 24 h yielded intracellular drug concentrations of 1.5 ± 0.5 μM (mean ± SD). Our collective data strongly suggest that metformin concentrations achievable in mice and cultured cells are sufficient to affect cellular energetics and activate AMPK, and are also achievable in human tissue using metformin dosing regimens that are FDA-approved for the treatment of T2DM.
Supplementary Material
WHAT IS ALREADY KNOWN ABOUT THIS SUBJECT
Metformin use is associated with improved cancer outcome in patients with concomitant type 2 diabetes mellitus (T2DM) and cancer, and this drug is now being widely tested in non-diabetic cancer patients. Metformin concentrations that provide ant-cancer effects have been determined in preclinical studies, but concentrations achievable in solid tumors in humans have not been reported.
WHAT THIS STUDY ADDS
In subjects treated with metformin at doses approved for T2DM, metformin concentrations found in plasma, blood, lung, and tumor tissues are lower than those typically used in tissue culture studies. However, metformin accumulated in cellular compartments, and metformin concentrations known to affect cellular energetics and activate AMPK in preclinical studies were achieved in human tissues. Blood and solid tissue concentrations of metformin varied widely between subjects.
Acknowledgments:
This work was supported by NIH [Dartmouth Clinical and Translational Science Institute UL1TR001086 (JDP) from the National Center for Advancing Translational Sciences (NCATS); R01CA211869 to TWM; Dartmouth College Norris Cotton Cancer Center Support Grant P30CA023108] and The Dartmouth-Hitchcock Cancer Research Fellows Program (JDP). We thank the following Norris Cotton Cancer Center Shared Resources for their support: Pathology; Clinical Pharmacology; Office of Clinical Research. Author contributions: JDP and TWM designed the study; RA, JDP, and TWM wrote the protocol; KF and JDP recruited subjects and procured tissue specimens; KF, ST, and TWM collected subject data; DBP, DBN, and LDL performed pharmacological analyses; ED performed statistical analyses; all authors contributed to manuscript composition and approval of the final manuscript.
Footnotes
Nomenclature of Targets and Ligands: Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20 (1).
PRINCIPAL INVESTIGATOR STATEMENT: Joseph D. Phillips, Department of Medicine, Dartmouth-Hitchcock Medical Center is the PI responsible for this clinical study.
Conflicts of interest: LDL is a consultant to G1 Therapeutics and 7 Hills Pharma LLC and has received clinical trial support from Bristol Myers Squibb, AbbVie, Bayer Pharmaceuticals, and AstraZeneca.
Data availability statement:
The data that support the findings of this study are available from the corresponding authors upon reasonable request. Some data may not be made available because of privacy or ethical restrictions.
References
- 1.Alexander SPH, Fabbro D, Kelly E, Mathie A, Peters JA, Veale EL, et al. THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Enzymes. British journal of pharmacology 2019;176 Suppl 1:S297–S396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalender A, Selvaraj A, Kim SY, Gulati P, Brule S, Viollet B, et al. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab 2010;11(5):390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wheaton WW, Weinberg SE, Hamanaka RB, Soberanes S, Sullivan LB, Anso E, et al. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. eLife 2014;3:e02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madiraju AK, Erion DM, Rahimi Y, Zhang XM, Braddock DT, Albright RA, et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature 2014;510(7506):542–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madiraju AK, Qiu Y, Perry RJ, Rahimi Y, Zhang XM, Zhang D, et al. Metformin inhibits gluconeogenesis via a redox-dependent mechanism in vivo. Nature medicine 2018;24(9):1384–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chae YK, Arya A, Malecek MK, Shin DS, Carneiro B, Chandra S, et al. Repurposing metformin for cancer treatment: current clinical studies. Oncotarget 2016;7(26):40767–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans JM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD. Metformin and reduced risk of cancer in diabetic patients. Bmj 2005;330(7503):1304–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viollet B, Guigas B, Sanz Garcia N, Leclerc J, Foretz M, Andreelli F. Cellular and molecular mechanisms of metformin: an overview. Clin Sci (Lond) 2012;122(6):253–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hampsch RA, Wells JD, Traphagen NA, McCleery CF, Fields JL, Shee K, et al. AMPK Activation by Metformin Promotes Survival of Dormant ER(+) Breast Cancer Cells. Clin Cancer Res 2020;26(14):3707–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bodmer M, Meier C, Krahenbuhl S, Jick SS, Meier CR. Long-term metformin use is associated with decreased risk of breast cancer. Diabetes Care 2010;33(6):1304–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decensi A, Puntoni M, Goodwin P, Cazzaniga M, Gennari A, Bonanni B, et al. Metformin and cancer risk in diabetic patients: a systematic review and meta-analysis. Cancer prevention research 2010;3(11):1451–61. [DOI] [PubMed] [Google Scholar]
- 12.Jiralerspong S, Palla SL, Giordano SH, Meric-Bernstam F, Liedtke C, Barnett CM, et al. Metformin and pathologic complete responses to neoadjuvant chemotherapy in diabetic patients with breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2009;27(20):3297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li D, Yeung SC, Hassan MM, Konopleva M, Abbruzzese JL. Antidiabetic therapies affect risk of pancreatic cancer. Gastroenterology 2009;137(2):482–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Libby G, Donnelly LA, Donnan PT, Alessi DR, Morris AD, Evans JM. New users of metformin are at low risk of incident cancer: a cohort study among people with type 2 diabetes. Diabetes Care 2009;32(9):1620–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright JL, Stanford JL. Metformin use and prostate cancer in Caucasian men: results from a population-based case-control study. Cancer causes & control : CCC 2009;20(9):1617–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camacho L, Dasgupta A, Jiralerspong S. Metformin in breast cancer - an evolving mystery. Breast cancer research : BCR 2015;17:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bristol-Myers_Squibb_Company. Glucophage (metformin hydrochloride) [package insert]. US Food and Drug Administration website https://wwwaccessdatafdagov/drugsatfda_docs/label/2017/020357s037s039,021202s021s023lblpdf Revised April 2017 Accessed Nov 22, 2021.
- 18.Graham GG, Punt J, Arora M, Day RO, Doogue MP, Duong JK, et al. Clinical Pharmacokinetics of Metformin. Clin Pharmacokinet 2011;50(2):81–98. [DOI] [PubMed] [Google Scholar]
- 19.Christensen MM, Brasch-Andersen C, Green H, Nielsen F, Damkier P, Beck-Nielsen H, et al. The pharmacogenetics of metformin and its impact on plasma metformin steady-state levels and glycosylated hemoglobin A1c. Pharmacogenetics and genomics 2011;21(12):837–50. [DOI] [PubMed] [Google Scholar]
- 20.Dowling RJ, Lam S, Bassi C, Mouaaz S, Aman A, Kiyota T, et al. Metformin Pharmacokinetics in Mouse Tumors: Implications for Human Therapy. Cell Metab 2016;23(4):567–8. [DOI] [PubMed] [Google Scholar]
- 21.Timmins P, Donahue S, Meeker J, Marathe P. Steady-state pharmacokinetics of a novel extended-release metformin formulation. Clin Pharmacokinet 2005;44(7):721–9. [DOI] [PubMed] [Google Scholar]
- 22.Shu Y, Sheardown SA, Brown C, Owen RP, Zhang S, Castro RA, et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. The Journal of clinical investigation 2007;117(5):1422–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tucker GT, Casey C, Phillips PJ, Connor H, Ward JD, Woods HF. Metformin kinetics in healthy subjects and in patients with diabetes mellitus. British journal of clinical pharmacology 1981;12(2):235–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yee D, Isaacs C, Wolf DM, Yau C, Haluska P, Giridhar KV, et al. Ganitumab and metformin plus standard neoadjuvant therapy in stage 2/3 breast cancer. NPJ Breast Cancer 2021;7(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Khayat SM, Abouegylah M, Abdallah D, Geweil AG, Elenbaby AM, Zahra OS. The effect of metformin when combined with neoadjuvant chemotherapy in breast cancer patients. Medical oncology 2021;39(1):1. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Bonet E, Buxo M, Cuyas E, Pernas S, Dorca J, Alvarez I, et al. Neoadjuvant Metformin Added to Systemic Therapy Decreases the Proliferative Capacity of Residual Breast Cancer. J Clin Med 2019;8(12). [DOI] [PMC free article] [PubMed]
- 27.Brown JR, Chan DK, Shank JJ, Griffith KA, Fan H, Szulawski R, et al. Phase II clinical trial of metformin as a cancer stem cell-targeting agent in ovarian cancer. JCI Insight 2020;5(11). [DOI] [PMC free article] [PubMed]
- 28.Tsakiridis T, Pond GR, Wright J, Ellis PM, Ahmed N, Abdulkarim B, et al. Metformin in Combination With Chemoradiotherapy in Locally Advanced Non-Small Cell Lung Cancer: The OCOG-ALMERA Randomized Clinical Trial. JAMA Oncol 2021;7(9):1333–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinn BJ, Kitagawa H, Memmott RM, Gills JJ, Dennis PA. Repositioning metformin for cancer prevention and treatment. Trends Endocrinol Metab 2013;24(9):469–80. [DOI] [PubMed] [Google Scholar]
- 30.Liu A, Coleman SP. Determination of metformin in human plasma using hydrophilic interaction liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 2009;877(29):3695–700. [DOI] [PubMed] [Google Scholar]
- 31.Demidenko E. Advanced Statistics with Applications with R. Hoboken, NJ: Wiley 2020. [Google Scholar]
- 32.Demidenko E. Mixed Models: Theory and Applications with R. Wiley; (Hoboken, NJ: ) 2013. [Google Scholar]
- 33.Brown H, Prescott, R. Applied Mixed Models in Medicine (3rd ed.) Hoboken: Wiley. 2015. [Google Scholar]
- 34.Robert F, Fendri S, Hary L, Lacroix C, Andrejak M, Lalau JD. Kinetics of plasma and erythrocyte metformin after acute administration in healthy subjects. Diabetes Metab 2003;29(3):279–83. [DOI] [PubMed] [Google Scholar]
- 35.Dowling RJ, Zakikhani M, Fantus IG, Pollak M, Sonenberg N. Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells. Cancer Res 2007;67(22):10804–12. [DOI] [PubMed] [Google Scholar]
- 36.Ben Sahra I, Regazzetti C, Robert G, Laurent K, Le Marchand-Brustel Y, Auberger P, et al. Metformin, independent of AMPK, induces mTOR inhibition and cell-cycle arrest through REDD1. Cancer Res 2011;71(13):4366–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request. Some data may not be made available because of privacy or ethical restrictions.