Abstract
BACKGROUND:
Substance use disorders (SUDs) are associated with a variety of co-occurring psychiatric disorders and other SUDs, which partly reflects genetic pleiotropy. Polygenic risk scores (PRSs) and phenome-wide association studies are useful in evaluating pleiotropic effects. However, the comparatively low prevalence of SUDs in population samples and the lack of detailed information available in electronic health records limit these data sets’ informativeness for such analyses.
METHODS:
We used the deeply phenotyped Yale-Penn sample (n = 10,610 with genetic data; 46.3% African ancestry, 53.7% European ancestry) to examine pleiotropy for 4 major substance-related traits: alcohol use disorder, opioid use disorder, smoking initiation, and lifetime cannabis use. The sample includes both affected and control subjects interviewed using the Semi-Structured Assessment for Drug Dependence and Alcoholism, a comprehensive psychiatric interview.
RESULTS:
In African ancestry individuals, PRS for alcohol use disorder, and in European individuals, PRS for alcohol use disorder, opioid use disorder, and smoking initiation were associated with their respective primary DSM diagnoses. These PRSs were also associated with additional phenotypes involving the same substance. Phenome-wide association study analyses of PRS in European individuals identified associations across multiple phenotypic domains, including phenotypes not commonly assessed in phenome-wide association study analyses, such as family environment and early childhood experiences.
CONCLUSIONS:
Smaller, deeply phenotyped samples can complement large biobank genetic studies with limited phenotyping by providing greater phenotypic granularity. These efforts allow associations to be identified between specific features of disorders and genetic liability for SUDs, which help to inform our understanding of the pleiotropic pathways underlying them.
Individuals with substance use disorders (SUDs) are at an increased risk of comorbid psychiatric and medical disorders (1). However, the etiologic factors underlying comorbidity are not well understood. Large-scale genome-wide association studies (GWASs) have identified common risk markers for many SUDs (2–6) and established a pattern of genetic correlations among SUDs and between SUDs and other traits. This growing body of evidence suggests that there are common loci or biological pathways that contribute to the risk for multiple SUDs and psychiatric disorders. Identifying pleiotropic loci and pathways could provide insight into the etiologies of co-occurring disorders, advancing efforts to categorize, prevent, and treat SUDs and co-occurring medical and psychiatric conditions.
The large samples required to identify variants of generally small effect are often characterized by phenotypic information that is neither purpose collected nor detailed. This trade-off between sample size and depth of phenotyping limits clinically meaningful insights into disease biology (7). Furthermore, the selection of a phenotype for GWAS—if not limited by the phenotype data available—requires assumptions regarding the most representative or informative traits. Thus, phenome scans (8) or phenome-wide association studies (PheWASs) (9) complement GWASs by testing the phenome in a hypothesis-free manner.
PheWASs have been most commonly implemented using data from electronic health records (EHRs), where ICD codes are converted to a simplified dataset that contains case-control status for more than 1800 diseases (9). A recent PheWAS of genetic liability for SUDs, represented by polygenic risk scores (PRSs), identified cross-trait associations across multiple phenotypic domains in EHR data (10). However, as with GWASs, PheWASs of EHR data are limited by their reliance on ICD diagnoses and minimal phenotyping. PheWASs have also been performed on data extracted from epidemiological studies or clinical trials (11,12), an approach that allows testing of subthreshold (with respect to diagnosis) and non–diagnosis-based phenotypes.
The Yale-Penn sample, recruited for genetic studies of SUDs, was deeply phenotyped using the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA). This comprehensive psychiatric interview schedule assesses physical, psychosocial, and psychiatric manifestations of SUDs and co-occurring psychiatric disorders (13,14). It includes more than 3500 items representing demographic information, lifetime diagnostic criteria for DSM-IV (15) and DSM-5 (16) SUDs and DSM-IV (15) psychiatric disorders, psychosocial history, medical history, and a detailed substance use history. The SSADDA yields reliable diagnoses and criterion counts for SUDs and psychiatric disorders (13,14).
The detailed information available on the Yale-Penn sample enables insights into the shared genetic etiology of a variety of substance use and psychiatric traits. The dataset has been used to conduct GWASs (17–21), gene-by-environment studies (22,23), and phenotypic investigations (24,25). Because the Yale-Penn dataset includes nearly equal numbers of African (AFR) and European (EUR) ancestry individuals, analyses can be conducted in both population groups.
Here, we describe the selection of a subset of the data points collected with the SSADDA to create a PheWAS dataset based on the Yale-Penn sample. Using PRSs for alcohol use disorder (AUD), opioid use disorder (OUD), smoking initiation (SMK), and lifetime cannabis use (CAN), we demonstrated the utility of this dataset for evaluating SUD pleiotropy. Furthermore, we identified associations that contributed to our understanding of the shared genetic etiology and phenotypic comorbidity of these traits.
METHODS AND MATERIALS
Yale-Penn Dataset
Participants (N = 16,715) were recruited at 5 sites in the United States for genetic studies of cocaine, opioid, and alcohol dependence following institutional review board approval at each site. Participants gave written informed consent prior to data collection. Cases were identified through addiction treatment facilities, inpatient and outpatient psychiatric services, and advertisements in local media and screened for the presence of ≥1 of the 3 SUD diagnoses. Some cocaine- or opioid-dependent individuals were recruited as probands of small nuclear families and, when available, their affected and unaffected parents and siblings were also recruited. Unaffected control subjects were recruited from nonpsychiatric outpatient medical settings and through advertisements.
Semi-Structured Assessment for Drug Dependence and Alcoholism
The SSADDA, which comprises 24 modules, yields up to 3727 data points (depending on skip patterns) that assess the physical, psychological, social, and psychiatric manifestations of SUDs, psychiatric disorders, and environmental covariates likely to affect SUDs. Computer-assisted administration of the SSADDA allows the interviewer to enter participants’ responses directly.
Demographic information elicited with the SSADDA includes gender, age, height, weight, education, employment, and relationship status. Environmental variables, such as adverse childhood experiences, are also assessed. Medical history for common diseases is assessed by a yes/no response to “Have you been diagnosed with …” followed by a series of medical disorders that the participant is asked to endorse.
The SSADDA’s semi-structured format, accompanied by rigorous training and quality control procedures (13), allows a carefully trained nonclinician interviewer to assess diagnostic criteria and disorders. It queries age of symptom onset, severity, duration, and craving for the major drugs of abuse that yield DSM-IV diagnoses of nicotine dependence, dependence or abuse of other substances, mood disorders (major depressive disorder, bipolar disorder), schizophrenia, mania, conduct disorder, antisocial personality disorder (ASPD), attention-deficit/hyperactivity disorder, suicidality, anxiety disorders (panic disorder, agoraphobia, social anxiety disorder, obsessive-compulsive disorder, generalized anxiety disorder, posttraumatic stress disorder [PTSD]), and gambling disorder. Recoding of criteria to accord with DSM-5 also makes it possible to generate DSM-5 SUD, but not psychiatric, diagnoses.
Variable Selection and Data Cleaning
We used an interactive consensus process among study clinicians (EEH, JR, HRK), a data scientist (HX), and a geneticist (RLK) to reduce the number of variables to 689 for use in PheWASs. Variables that were considered informative for genetic studies and nonduplicative were retained, and the data were cleaned to ensure consistency across categories. Full details of variable selection and data cleaning are available in Supplemental Methods in Supplement 1.
Case and Control Definitions
Participants who met diagnostic criteria for a disorder were coded as cases and those who met no diagnostic criteria for the disorder were coded as controls. Subthreshold cases, e.g., those meeting at least one, but less than the required number of criteria for a diagnosis, were excluded from further analyses for that disorder. For symptom variables, participants who endorsed an individual symptom were considered cases and those who did not were controls. Unanswered items were coded as “NA,” and individuals were considered as neither a case nor a control for that phenotype.
Genotyping, Imputation, and PRSs
Yale-Penn samples were genotyped in 3 batches using the Illumina HumanOmni1-Quad microarray, the Illumina Human-CoreExome array, or the Illumina Multi-Ethnic Global array (Illumina, Inc.). Genotyping quality control has been described in detail previously (19,20,26). Genotype data were imputed using the Michigan Imputation Server (27) with the 1000 Genomes phase 3 reference panel (28). Further details are available in Supplemental Methods in Supplement 1.
PRSs were calculated for AUD (5), OUD (3), SMK (2), and CAN (4) using PRS–Continuous Shrinkage software (29) (Table S1 in Supplement 2). We used the PRS–Continuous Shrinkage software “auto” option to estimate the parameters of shrinkage priors and fixed the random seed to 1 for replicable results. We matched available ancestry summary statistics in all analyses (e.g., an AFR GWAS for AUD was used to calculate AUD PRS in AFR Yale-Penn individuals).
Statistical Analysis
For the PheWAS, we fitted logistic regression models for binary traits and linear regression models for continuous traits, adjusting for sex, age, and the top 10 principal components within each genetic ancestry. Binary phenotypes with fewer than 100 cases or 100 controls and continuous phenotypes with fewer than 100 individuals within each ancestral group were excluded for that group. A Bonferroni correction was applied within each ancestral group to account for multiple testing (AFR p < 8.7 × 10−5, EUR p < 7.9 × 10−5). To further examine the pleiotropic effects identified in PheWAS analyses, we conducted supplementary PheWAS for each PRS: 1) in cases for the corresponding SUD, 2) in controls for the corresponding SUD, and 3) covarying for the other SUD PRSs.
RESULTS
Sample
The PheWAS dataset comprises 689 variables in 25 phenotypic categories: 8 for substance use, 14 for psychiatric disorder, and 3 (demographics, environment, and medical) for other features. Table 1 shows the demographic and clinical features of the analytic sample and Table S2 in Supplement 2 shows the case counts for all diagnoses. The sample with genetic data available (n = 10,610) was 55.6% male (AFR: 54.9%, EUR: 56.2%) and included 4918 AFR participants (998 or 20.3% with no SUD diagnosis) and 5692 EUR participants (1370 or 24.1% with no SUD diagnosis). The mean number of SUD diagnoses in the sample was 2.44 (SD = 1.97) for DSM-IV and 2.28 (SD = 1.82) for DSM-5. We focused on individuals with ≥1 SUD diagnosis for DSM-IV alcohol dependence (AD); opioid dependence (OD); tobacco dependence (TD); or cannabis dependence; or DSM-5 AUD, OUD, or cannabis use disorder (CUD), comprising 3813 AFR (38.6% female) and 4294 EUR (37.1% female) individuals (average age = 40.1 years, SD = 11.0). There is a high degree of co-occurrence of SUD diagnoses in both population groups (Figure 1; Tables S3–S5 in Supplement 2).
Table 1.
Demographic and Clinical Features
| Ancestry | DSM-IV Diagnosis | DSM-5 Diagnosis | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tobacco Dependence | Alcohol Dependence | Opioid Dependence | Cannabis Dependence | Alcohol Use Disorder | Opioid Use Disorder | Cannabis Use Disorder | ||||||||
| n (% Female) | Age, Mean ± SD | n (% Female) | Age, Mean ± SD | n (% Female) | Age, Mean ± SD | n (% Female) | Age, Mean ± SD | n (% Female) | Age, Mean ± SD | n (% Female) | Age, Mean ± SD | n (% Female) | Age, Mean ± SD | |
| African | ||||||||||||||
| Cases | 2679 (40.2%) | 42.2 ± 9.2 | 2631 (35.7%) | 42.0 ± 9.6 | 938 (35.4%) | 44.5 ± 8.7 | 1402 (28.5%) | 40.3 ± 9.8 | 3215 (36.3%) | 41.7 ± 9.6 | 1014 (34.9%) | 44.5 ± 8.7 | 2082 (30.7%) | 40.7 ± 9.7 |
| Controls | 1534 (57.3%) | 39.1 ± 11.6 | 1470 (64.2%) | 40.7 ± 11.4 | 3848 (47.9%) | 40.5 ± 10.4 | 2532 (56.0%) | 41.7 ± 10.6 | 1395 (65.4%) | 40.6 ± 11.5 | 3839 (48.0%) | 40.5 ± 10.4 | 2376 (58.2%) | 41.7 ± 10.7 |
| European | ||||||||||||||
| Cases | 3156 (37.7%) | 38.4 ± 11.6 | 2945 (34.1%) | 39.1 ± 11.6 | 2276 (34.7%) | 36.5 ± 10.4 | 1628 (28.7%) | 35.1 ± 10.7 | 3669 (34.5%) | 38.6 ± 11.7 | 2394 (34.2%) | 36.5 ± 10.5 | 2416 (28.7%) | 36.2 ± 11.1 |
| Controls | 1983 (54.5%) | 42.9 ± 14.7 | 1813 (61.2%) | 42.0 ± 15.0 | 3246 (51.0%) | 42.1 ± 14.2 | 3082 (55.1 %) | 42.5 ± 13.8 | 1617 (62.6%) | 42.8 ± 15.1 | 3213 (51.3%) | 42.1 ± 14.2 | 2846 (57.1%) | 42.7 ± 14.0 |
Figure 1.
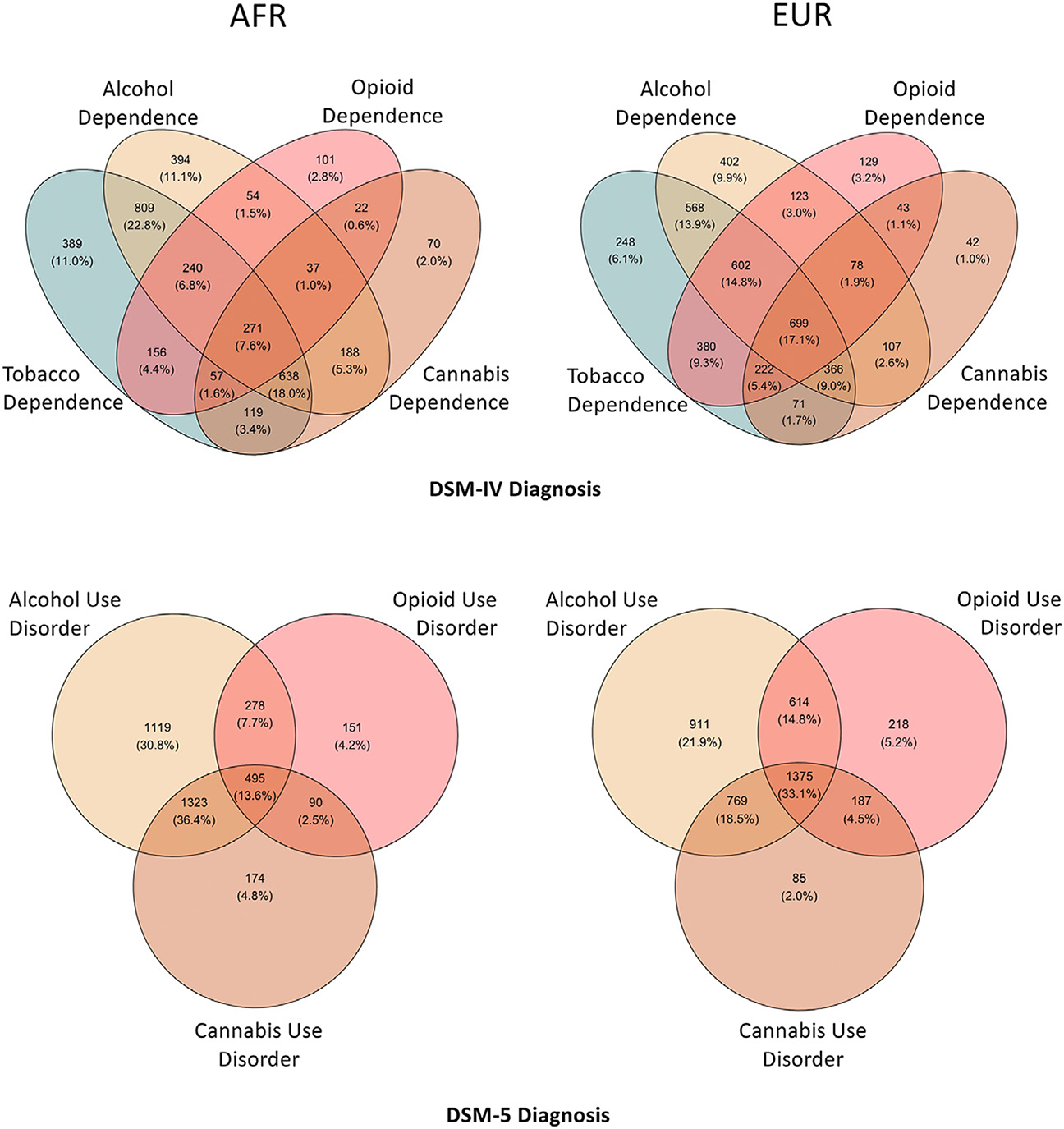
Comorbidity among substance use disorders in the Yale-Penn dataset. Venn diagrams show the comorbidity among DSM-IV substance dependence diagnoses (top) and DSM-5 use disorder diagnoses (bottom) in African (AFR) individuals (left) and European (EUR) individuals (right). Number and percentage of individuals with overlapping disorders is labeled in each section.
Primary Associations of SUD PRSs
In AFR individuals, PRS for AUD (PRSAUD) was significantly associated with DSM-IV AD (odds ratio [OR] = 1.20, p = 7.0 × 10−6), DSM-5 AUD diagnosis (OR = 1.21, p = 1.8 × 10−6), and DSM-5 AUD criterion count (β = 0.30, p = 5.4 × 10−7) (Figure 2A; Table S6 in Supplement 2). PRS for OUD (PRSOUD) and smoking initiation (PRSSMK) were not significantly associated with either respective diagnosis or any other phenotypes (Figure 2A; Tables S7 and S8 in Supplement 2). We could not generate PRS for cannabis lifetime use (PRSCAN) in AFR individuals because the discovery data were limited to EUR ancestry.
Figure 2.
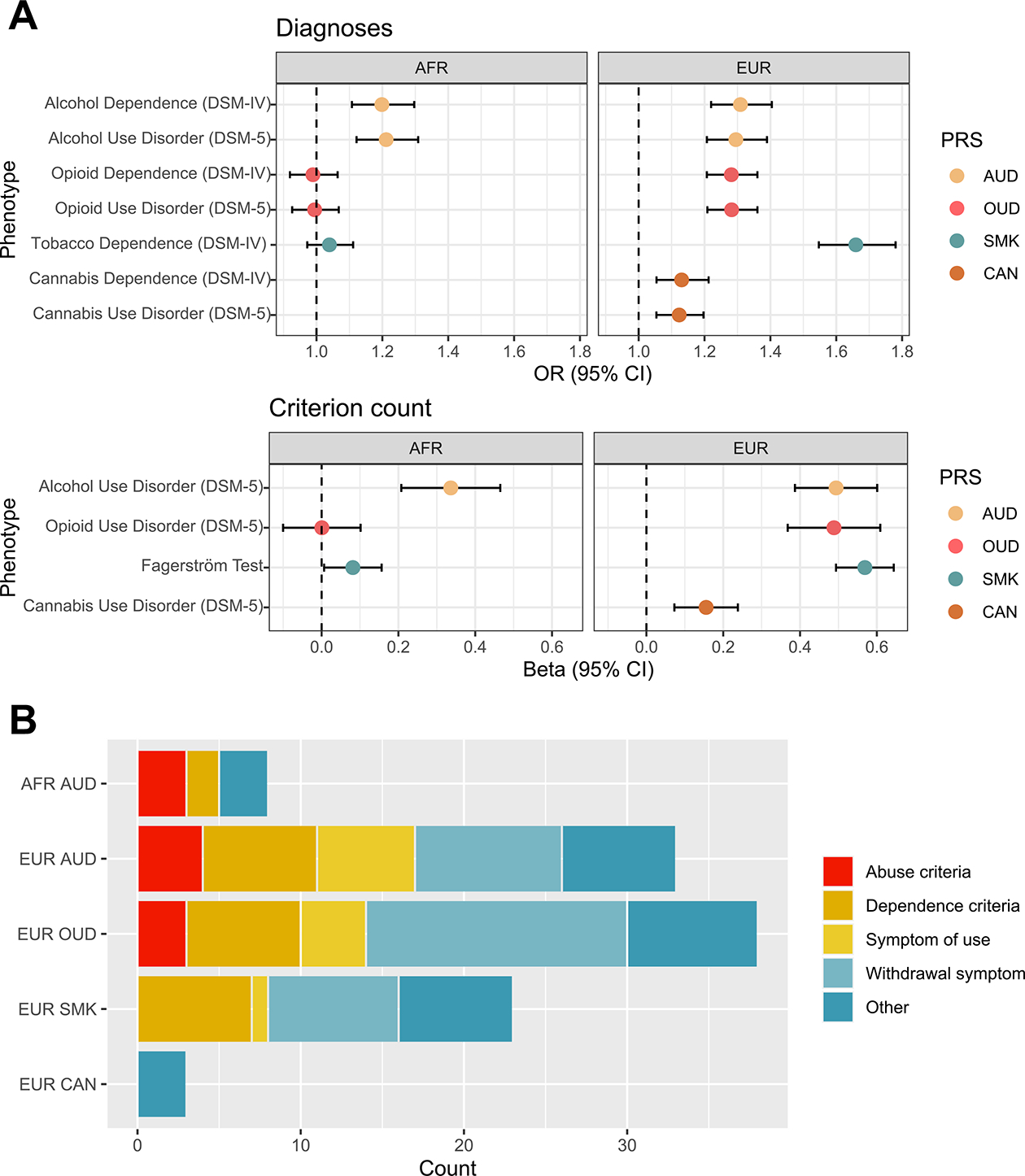
Substance use disorder polygenic risk score (PRS) primary associations. (A) Forest plot showing odds ratios and 95% confidence intervals for associations between each PRS and the primary DSM-IV and DSM-5 diagnosis in African (AFR) individuals (left) and European (EUR) individuals (right). (B) Stacked bar plot showing the number and type of phenotypes involving the same substance associated with each PRS. AUD, alcohol use disorder; CAN, lifetime cannabis use; OR, odds ratio; OUD, opioid use disorder; SMK, smoking initiation.
In EUR individuals, PRSAUD and PRSOUD were significantly associated with their respective DSM-IV and DSM-5 diagnoses and DSM-5 criterion counts (PRSAUD: DSM-IV AD OR = 1.30, p = 1.3 × 10−13, DSM-5 AUD OR = 1.29, p = 6.7 × 10−13, DSM-5 criterion count β = 0.47, p = 2.3 × 10−20; PRSOUD: DSM-IV OD OR = 1.28, p = 2.9 × 10−16, DSM-5 OUD OR = 1.28, p = 3.1 × 10−16, DSM-5 criterion count β = 0.49, p = 3.3 × 10−15) (Figure 2A; Tables S9 and S10 in Supplement 2). Similarly, PRSSMK was significantly associated with the DSM-IV diagnosis of TD (OR = 1.67, p = 1.7 × 10−46) and the Fagerström Test for Nicotine Dependence score (β = 0.57, p = 7.3 × 10−49) (Figure 2A; Table S11 in Supplement 2). PRSCAN was only nominally associated with the respective criterion count (β = 0.16, p = 2.2 × 10−4) and DSM diagnoses (DSM-IV cannabis dependence: OR = 1.13, p = 5.0 × 10−4; DSM-5 CUD: OR = 1.12, p = 3.0 × 10−4) (Figure 2A; Table S12 in Supplement 2).
Associations With Phenotypes Involving the Same Substance
PRSs associated with their respective substance use diagnosis were also associated with other phenotypes for the same substance. PRSAUD was associated with 11 alcohol phenotypes in AFR individuals (Figure 2B; Table S6 in Supplement 2) and 36 alcohol phenotypes in EUR individuals (Figure 2B; Table S9 in Supplement 2). In AFR, PRSAUD was associated with 3 of the 4 alcohol abuse criteria, including “continued use despite social/interpersonal problems” (OR = 1.20, p = 1.1 × 10−7), which was more significantly associated with PRSAUD compared with the diagnosis itself. PRSAUD was also significantly associated with frequent alcohol use, alcohol abuse, “ever had blackout,” and 2 of the 7 DSM-IV AD criteria (“unsuccessful efforts to decrease use” and “used more than intended”). In EUR, PRSAUD was associated with each AUD diagnostic criterion and with “sought treatment,” frequent use, age of first use, “ever had blackout,” 9 withdrawal symptoms (e.g., “depressed mood”), and 6 symptoms of heavy use (e.g., “depression”). Of these, 10 remained significant in a case-only analysis, including criterion count and “sought treatment.”
Among EUR individuals, PRSOUD was associated with 41 opioid phenotypes (Figure 2B; Table S10 in Supplement 2), including “time spent obtaining/using” (OR = 1.30, p = 5.34 × 10−18) and “ever used opioids” (OR = 1.28, p = 1.9 × 10−16)—both associations more significant than with the diagnosis. PRSOUD was significantly associated with 10 OD and abuse criteria (“legal problems” being the exception [OR = 1.04, p = .44]). PRSOUD was also significantly associated with “sought treatment”; frequent use; 4 symptoms of heavy use; and 16 withdrawal symptoms, the most significant being “depressed mood” (OR = 1.26, p = 3.7 × 10−14). None of these remained significant in a case-only analysis.
Among EUR individuals, PRSSMK was associated with 25 tobacco phenotypes (Figure 2B; Table S11 in Supplement 2), including all 7 TD criteria. Following the top associations with the Fagerström Test for Nicotine Dependence score and the DSM-IV diagnosis of TD, the most significant association was “smoked over 100 cigarettes lifetime” (OR = 1.62, p = 8.6 × 10−45). Associations were also found with frequent tobacco use; “sought use”; “ever used tobacco”; age at first use; “health problems”; and 8 withdrawal symptoms, the most significant being “irritability” (OR = 1.44, p = 5.7 × 10−33). In case-only analysis, no phenotypes survived correction.
Although only nominally associated with the diagnosis of CUD in EUR individuals, PRSCAN (based on a lifetime measure of cannabis use) was significantly associated with 3 other cannabis phenotypes (Figure 2B; Table S12 in Supplement 2): “ever used” (OR = 1.20, p = 1.3 × 10−6), “regularly use” (OR = 1.15, p = 4.9 × 10−6), and cannabis abuse (OR = 1.14, p = 2.0 × 10−5). None of these survived correction in the case-only analysis.
Phenome-wide Analyses
The PheWAS of PRS in AFR individuals identified no significant associations that passed Bonferroni correction in other phenotypic domains (Figure 3; Tables S6–S8 in Supplement 2). However, we identified multiple significant associations across phenotypic domains in EUR individuals (Figure 3; Tables S9–S12 in Supplement 2), and for all 4 PRSs, the largest number were with other substance use phenotypes.
Figure 3.
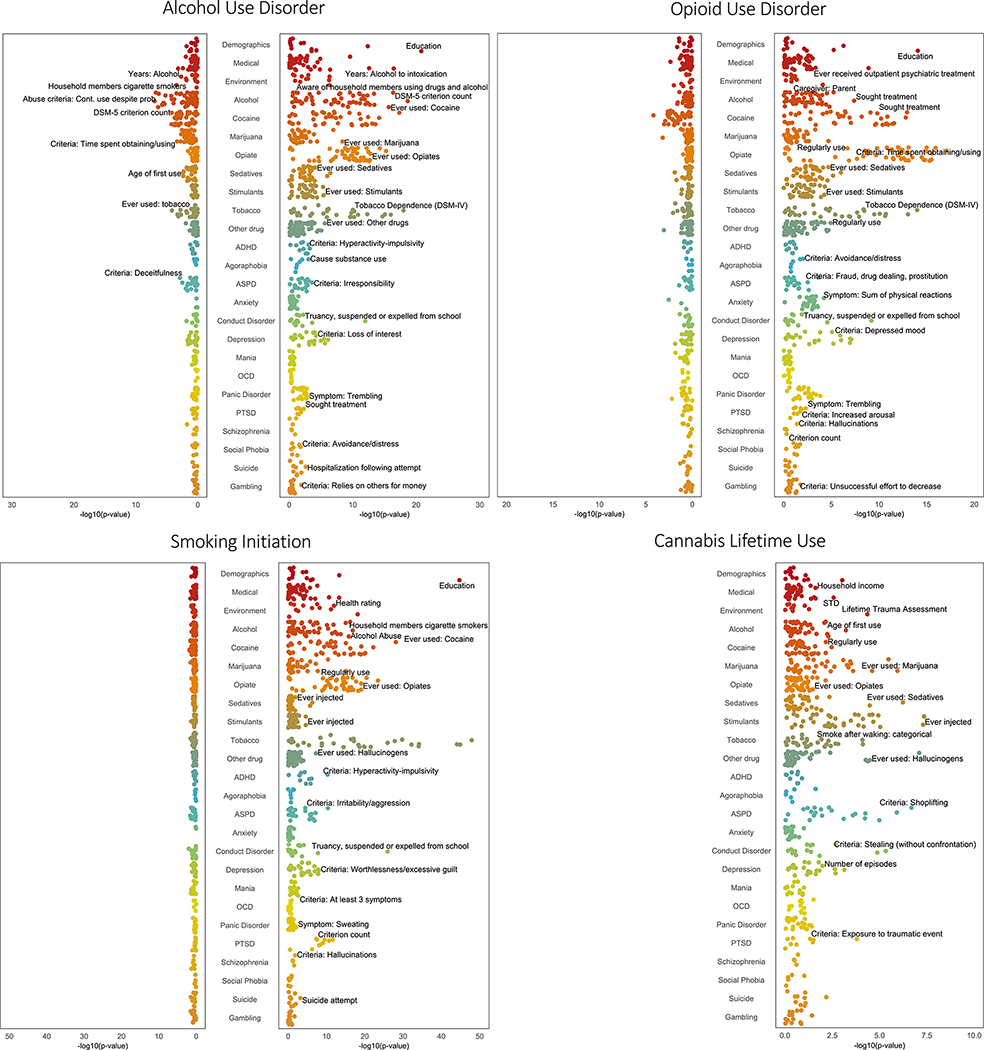
Phenome-wide association studies of substance use disorder polygenic risk scores. Phenome-wide association study results for each polygenic risk score in African individuals (left panel of each graph) and European individuals (right panel of each graph). Within each category, the top associated phenotype was labeled if it passed false discovery rate correction. ADHD, attention-deficit/hyperactivity disorder; ASPD, antisocial personality disorder; OCD, obsessive-compulsive disorder; PTSD, posttraumatic stress disorder; STD, sexually transmitted disease.
PRSAUD was associated with 126 phenotypes in 12 categories, including all 7 substance use categories, the most significant of which was DSM-IV TD (OR = 1.35, p = 6.0 × 10−19) and “ever used cocaine” (OR = 1.34, p = 3.1 × 10−18). Of these, 21 remained significant in a case-only analysis and 36 remained significant in the control-only analysis. The “ever used cocaine” phenotype was significant in the case-only analysis, but not in the control-only analysis. Both case-only and control-only analyses showed significant associations with DSM-IV TD and DSM-IV OD.
PRSOUD was associated with 76 phenotypes in 12 categories, including all 7 substance use categories, with the most significant being DSM-IV TD (OR = 1.28, p = 6.0 × 10−15), “sought treatment for cocaine use” (OR = 1.25, p = 7.7 × 10−14), and Fagerström Test for Nicotine Dependence score (β = 0.27, p = 1.4 × 10−13). Although none of these remained significant in the case-only analysis, in the control-only analysis, there were 5 significant substance use phenotypes, including DSM-IV TD.
PRSSMK was associated with 168 phenotypes in 15 categories, including all 7 substance use categories. The most significant substance use phenotype was “ever used cocaine” (OR = 1.47, p = 5.1 × 10−29), which remained significant in the control-only analysis. PRSCAN was associated with 23 phenotypes in 7 categories, though unlike the other PRSs, it was associated with phenotypes in only 4 of the 7 substance use categories. The most significant substance use phenotypes were “ever injected stimulants” (OR = 1.19, p = 6.1 × 10−8) and “ever used” stimulants (OR = 1.18, p = 6.6 × 10−8), hallucinogens (OR = 1.18, p = 8.8 × 10−8), or sedatives (OR = 1.16, p = 6.5 × 10−7). Both “ever injected” and “ever used” stimulants remained significant in the control-only, but not the case-only, analysis.
The psychiatric phenotype most significantly associated with PRSAUD, PRSOUD, and PRSSMK was “truancy, suspended or expelled from school” in the conduct disorder domain (PRSAUD: OR = 1.27, p = 7.7 × 10−13; PRSOUD: OR = 1.22, p = 4.5 × 10−10; PRSSMK: OR = 1.44, p = 7.9 × 10−27). This finding is driven by the association in controls for each substance, with a stronger association in control-only analyses than case-only analyses. Both PRSAUD and PRSOUD were associated with multiple depression-related phenotypes, including the major depressive disorder criterion count (PRSAUD: β = 0.24, p = 4.5 × 10−6; PRSOUD: β = 0.25, p = 5.3 × 10−7). The second most significant phenotype for PRSSMK was the criterion count for PTSD (β = 0.21, p = 3.1 × 10−12), which was more significant in the control-only analysis (β = 0.15, p = 8.9 × 10−5) than the case-only analysis (β = 0.12, p = 5.9 × 10−3). PRSSMK was also associated with phenotypes for depression, ASPD, and attention-deficit/hyperactivity disorder. PRSCAN was associated with 4 phenotypes in the ASPD domain, including the ASPD diagnosis (OR = 1.21, p = 3.1 × 10−5) and 2 in the conduct disorder domain: “stealing (without confrontation)” (OR = 1.18, p = 3.4 × 10−6) and “persistent pattern of behavior” (OR = 1.18, p = 2.3 × 10−6).
For PRSAUD, PRSOUD, and PRSSMK, the most significant association with a demographic phenotype was a negative association with educational attainment (PRSAUD: β = −0.21, p = 3.2 × 10−21; PRSOUD: β = −0.16, p = 1.7 × 10−14; PRSSMK: β = −0.31, p = 3.8 × 10−45), evident in both the case-only and control-only analyses for all PRSs except the PRSOUD case-only analysis. These 3 PRSs were also negatively associated with household income and positively associated with the number of outpatient psychiatric treatments. Both PRSAUD and PRSSMK were positively associated with childhood environmental variables, including “aware of household members using drugs or alcohol” and “frequent use of drugs/alcohol in household.” PRSAUD was also associated with phenotypes in the medical section, such as “alcohol used to intoxication” (β = 0.95, p = 2.5 × 10−17) and “health rating” (higher score = poorer health; β = 0.08, p = 2.4 × 10−7). PRSSMK (OR = 1.18, p = 3.0 × 10−8) and PRSCAN (OR = 1.13, p = 3.0 × 10−5) were positively associated with lifetime trauma, although these were not significant in case-only or control-only analyses. PRSOUD was associated with not having had a parent as the main caregiver (PRSOUD: OR = 0.84, p = 5.5 × 10−5).
In a supplementary analysis of PheWAS associations for each PRS, with other SUD PRSs as additional covariates (Tables S13–S19 in Supplement 2), the specificity of most PRSs for the corresponding substances increased. For instance, PRSAUD in EUR was associated with 162 phenotypes at a Bonferroni-corrected p value, with 36 (22%) alcohol related. Covarying for the other PRSs yielded 36 significant associated phenotypes, of which 31 (86%) were alcohol related. Similar proportional increases were seen for PRSOUD (35% of associated phenotypes were opioid related compared with 81% when covarying for other SUD PRSs), with smaller proportional increases for PRSSMK (13%–17%) and PRSCAN (12%–17%).
DISCUSSION
PheWAS is valuable for exploring cross-trait associations of phenotypes with genetic liability for specific disorders, though to date, most PheWASs have used high-level EHR phenotypes typically limited to clinical diagnosis. Here, we describe a dataset for PheWAS derived from the Yale-Penn sample, ascertained using a detailed psychiatric interview whose administration included multiple quality control procedures (13). We selected features to reduce 3727 variables to 689 variables that are informative for genetic analysis. We refined cases and controls for each binary variable by applying methods commonly used in EHR PheWASs (9), removing subthreshold individuals who met ≥1 criteria, but not the full diagnosis. We identified novel phenotypic associations with PRSs for 4 substance use traits, particularly for subthreshold criteria or pertinent symptoms available only in a deeply phenotyped sample such as that derived from the Yale-Penn study.
The SSADDA is a useful assessment tool. Diagnoses made using a semi-structured interview following careful training procedures, with prespecified criteria and strict quality control methods, yield valid diagnoses (30) that likely are more accurate than those derived from EHR billing codes. As expected, many of the PRSs were associated with their respective primary diagnoses, supporting the validity of the approach. Likely due to the comparatively small GWAS discovery sample, PRSCAN was not associated with the primary diagnoses. Despite similar AFR and EUR target sample sizes, there were few associations for PRSs in the AFR sample, reflecting the lack of power for AFR in the parent GWASs.
The detailed information obtained with the SSADDA makes it possible to evaluate the impact of genetic risk for substance use traits on a variety of other substance-related traits not typically available in EHRs. In AFR, the criterion most strongly associated with PRSAUD was “continued use despite social/interpersonal problems,” which was the second strongest association in EUR. Although in DSM-IV, this is an alcohol abuse criterion, factor analysis has shown that it loads on the same factor as the 7 AD criteria, and in item response theory analysis, it is among those with the greatest information value (31). The DSM-IV substance abuse criterion “legal problems” had the fewest significant associations with any of the respective PRSs. This is consistent with the results of twin and epidemiological studies in which the legal criterion has the lowest loading of the DSM-IV criteria and low discriminatory power (32,33), supporting its omission from DSM-5 (34). Our results also showed several associations with craving, a criterion that was added to DSM-5, which although not reported by all individuals with SUDs, in some studies is predictive of relapse (35,36) and thus a target of pharmacological and psychosocial treatments (37–39). Furthermore, in our case-only analyses of PRSAUD, many of the associations with phenotypes related to the primary substance remained, suggesting that greater PRS is associated with greater severity of the phenotype.
In addition to the association of PRSs with primary substance-related phenotypes, each PRS also showed multiple associations with other substance use traits. The high genetic correlation among substance use traits (2–6) suggests that this likely reflects true shared genetic effects. However, given the high levels of substance-related comorbidity in the Yale-Penn sample, the results likely also reflect phenotypic correlation and ascertainment bias. To address this, we ran a control-only analysis, which for many SUD diagnoses showed associations of the PRS for one substance (e.g., AUD) with diagnoses for other SUDs (e.g., DSM-IV TD) even among controls (e.g., without AD or AUD). This suggests that the association is not driven by phenotypic correlation. Whereas covarying the other SUD PRSs yielded greater specificity for the primary substance, at least some of the cross-trait findings are likely due to shared genetic etiology, with each PRS contributing additional substance-specific risk.
We replicated associations between genetic risk for SUDs and other traits, including psychiatric diagnoses. The association between AUD and major depressive disorder has been identified in a PheWAS of AUD PRS (10) and problematic alcohol use PRS (6) and in the analysis of genetic correlations between AUD and depression (6). Here, we dissect this by identifying associations of both PRSAUD and PRSOUD and specific features of major depression, including low mood and difficulty concentrating. The psychiatric phenotype most significantly associated with PRSAUD, PRSOUD, and PRSSMK was “truancy, suspended or expelled from school” in the conduct disorder domain. Interestingly, in our control-only analysis, this association remains, again suggesting a potential direct association between PRS for substance use and this conduct disorder phenotype even when SUDs are absent. Consistent with phenotypic studies showing positive correlations between substance use and antisocial behaviors (40), we also observed significant associations for all 4 EUR PRSs with other criteria of ASPD and conduct disorder, such as shoplifting, fraud, and cheating. Previous studies have identified genetic correlations between PTSD and SUDs (41). Among EUR subjects, we observed a significant association of PRSSMK with PTSD criteria, lifetime trauma assessment, and seeking treatment for PTSD. Similarly, PRSCAN was significantly associated with lifetime trauma assessment. These relationships help to elucidate the features that underlie these common co-occurring symptoms and disorders.
In EUR individuals, both PRSAUD and PRSSMK were positively associated with childhood environmental variables reflecting substance use at home, whereas PRSOUD was associated with not having a parent as the main caregiver, which capture aspects of a family history of substance use. However, although a family history of an SUD is associated with many substance use outcomes, it is not wholly overlapping with genetic risk, and the use of both sources of information can yield a fuller measure of risk (42). These findings raise an important theoretical question that cannot be answered with the data available here: namely, do associations of PRSs with features such as trauma, truancy, education, and parental substance use reflect intergenerational effects (i.e., “genetic nurture”) or do they refute typical assumptions that the genetic and environmental components in gene-by-environment interactions are uncorrelated (i.e., adverse environments are evenly distributed across the range of PRS)?
A limitation of the Yale-Penn sample is its comparatively small size given the resource-intensive recruitment and ascertainment activities. Thus, we believe that the use of deeply phenotyped samples is complementary to that of biobank data, which are more amenable to gene discovery. Unlike EHR-based genetic studies, our study is cross-sectional and therefore lacks a longitudinal perspective. We were able to calculate PRSs and conduct a PheWAS in both AFR and EUR ancestry individuals by selecting the majority of the GWASs from the Million Veteran Program, a large and diverse biobank. However, the Million Veteran Program comprises veterans who are predominantly male and older and who have high rates of medical comorbidity and thus differ from the target sample. Our prioritization of large GWASs also led to differences in the phenotypes selected—2 for SUDs (AUD and OUD) and 2 for substance use (SMK and CAN). Because SUDs and substance use have related but distinct genetic etiologies (43), this may have led to differences in associations between the PRSs.
GWASs of SUDs and related traits are limited by a lack of deep phenotyping, a trade-off with the large samples needed to provide adequate statistical power to identify common variants of small effect. This study relied on a carefully constructed diagnostic interview that enabled us to conduct analyses of both primary substance use traits, results of which validated the effort, and a wealth of phenotypic data not captured in EHR-based biobanks (e.g., individual diagnostic criteria and symptoms, age of onset, and environmental variables). The continued growth of ancestrally diverse biobanks will provide opportunities to compare the performance of PRSs with those reported here. Additional enriched, deeply phenotyped samples are needed to support such efforts.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grants (Grant Nos. AA028292 [to RLK], DA046345 [to HRK], and AA026364 [to JG]) and the Veterans Integrated Service Network 4 Mental Illness Research, Education and Clinical Center.
Footnotes
DISCLOSURES
HRK is a member of advisory boards for Dicerna Pharmaceuticals, Sophrosyne Pharmaceuticals, and Enthion Pharmaceuticals; a consultant to Sobrera Pharmaceuticals; the recipient of research funding and medication supplies for an investigator-initiated study from Alkermes; and a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was supported in the past 3 years by Alkermes, Dicerna, Ethypharm, Lundbeck, Mitsubishi, and Otsuka. JG and HRK hold U.S. patent 10,900,082 titled: “Genotype-guided dosing of opioid agonists,” issued January 26, 2021. All other authors report no biomedical financial interests or potential conflicts of interest.
A previous version of this article was published as a preprint on medRxiv: https://www.medrxiv.org/content/10.1101/2022.02.09.22270737v1.
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.biopsych.2022.08.010.
Contributor Information
Rachel L. Kember, Department of Psychiatry, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania Mental Illness Research, Education and Clinical Center, Corporal Michael J. Crescenz VA Medical Center, Philadelphia, Pennsylvania.
Emily E. Hartwell, Department of Psychiatry, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania Mental Illness Research, Education and Clinical Center, Corporal Michael J. Crescenz VA Medical Center, Philadelphia, Pennsylvania.
Heng Xu, Department of Psychiatry, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania.
James Rotenberg, Mental Illness Research, Education and Clinical Center, Corporal Michael J. Crescenz VA Medical Center, Philadelphia, Pennsylvania.
Laura Almasy, Department of Biomedical and Health Informatics, Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania; Department of Genetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania.
Hang Zhou, Department of Psychiatry, Yale University School of Medicine, New Haven, Connecticut; VA Connecticut Healthcare System, West Haven, Connecticut.
Joel Gelernter, Department of Psychiatry, Yale University School of Medicine, New Haven, Connecticut; VA Connecticut Healthcare System, West Haven, Connecticut; Departments of Genetics and Neuroscience, Yale University School of Medicine, New Haven, Connecticut.
Henry R. Kranzler, Department of Psychiatry, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania Mental Illness Research, Education and Clinical Center, Corporal Michael J. Crescenz VA Medical Center, Philadelphia, Pennsylvania.
REFERENCES
- 1.Ross S, Peselow E (2012): Co-occurring psychotic and addictive disorders: Neurobiology and diagnosis. Clin Neuropharmacol 35:235–243. [DOI] [PubMed] [Google Scholar]
- 2.Xu K, Li B, McGinnis KA, Vickers-Smith R, Dao C, Sun N, et al. (2020): Genome-wide association study of smoking trajectory and meta-analysis of smoking status in 842,000 individuals. Nat Commun 11:5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou H, Rentsch CT, Cheng Z, Kember RL, Nunez YZ, Sherva RM, et al. (2020): Association of OPRM1 functional coding variant with opioid use disorder: A genome-wide association study [published correction appears in JAMA Psychiatry 2021;78:224]. JAMA Psychiatry 77:1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasman JA, Verweij KJH, Gerring Z, Stringer S, Sanchez-Roige S, Treur JL, et al. (2018): GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia [published correction appears in Nat Neurosci 2019;22:1196]. Nat Neurosci 21:1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kember RL, Vickers-Smith R, Zhou H, Xu H, Dao C, Justice AC, et al. (2021): Genetic underpinnings of the transition from alcohol consumption to alcohol use disorder: Shared and unique genetic architectures in a cross-ancestry sample. medRxiv. 10.1101/2021.09.08.21263302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H, Sealock JM, Sanchez-Roige S, Clarke TK, Levey DF, Cheng Z, et al. (2020): Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat Neurosci 23:809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yehia L, Eng C (2019): Largescale population genomics versus deep phenotyping: Brute force or elegant pragmatism towards precision medicine. NPJ Genom Med 4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones R, Pembrey M, Golding J, Herrick D (2005): The search for genotype/phenotype associations and the phenome scan. Paediatr Perinat Epidemiol 19:264–275. [DOI] [PubMed] [Google Scholar]
- 9.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. (2010): PheWAS: Demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 26:1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartwell EE, Merikangas AK, Verma SS, Ritchie MD, Regeneron Genetics Center, Kranzler HR, et al. (2022): Genetic liability for substance use associated with medical comorbidities in electronic health records of African- and European-ancestry individuals. Addict Biol 27:e13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polimanti R, Kranzler HR, Gelernter J (2016): Phenome-wide association study for alcohol and nicotine risk alleles in 26394 women. Neuropsychopharmacology 41:2688–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pendergrass SA, Brown-Gentry K, Dudek SM, Torstenson ES, Ambite JL, Avery CL, et al. (2011): The use of phenome-wide association studies (PheWAS) for exploration of novel genotype-phenotype relationships and pleiotropy discovery. Genet Epidemiol 35:410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, et al. (2005): Diagnostic reliability of the semi-structured assessment for drug dependence and alcoholism (SSADDA). Drug Alcohol Depend 80:303–312. [DOI] [PubMed] [Google Scholar]
- 14.Pierucci-Lagha A, Gelernter J, Chan G, Arias A, Cubells JF, Farrer L, Kranzler HR (2007): Reliability of DSM-IV diagnostic criteria using the semi-structured assessment for drug dependence and alcoholism (SSADDA). Drug Alcohol Depend 91:85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.American Psychiatric Association (1994): Diagnostic and Statistical Manual of Mental Disorders (DSM-IV), 4th ed. Arlington, VA: American Psychiatric Association, 886. [Google Scholar]
- 16.American Psychiatric Association (2013): Diagnostic and Statistical Manual of Mental Disorders (DSM-5), 5th ed. Arlington, VA: American Psychiatric Association. [Google Scholar]
- 17.Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR, et al. (2014): Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol Psychiatry 19:717–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu K, Kranzler HR, Sherva R, Sartor CE, Almasy L, Koesterer R, et al. (2015): Genomewide association study for maximum number of alcoholic drinks in European Americans and African Americans. Alcohol Clin Exp Res 39:1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gelernter J, Kranzler HR, Sherva R, Almasy L, Herman AI, Koesterer R, et al. (2015): Genome-wide association study of nicotine dependence in American populations: Identification of novel risk loci in both African-Americans and European-Americans. Biol Psychiatry 77:493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, et al. (2014): Genome-wide association study of alcohol dependence: Significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry 19:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao H, et al. (2014): Genome-wide association study of opioid dependence: Multiple associations mapped to calcium and potassium pathways. Biol Psychiatry 76:66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polimanti R, Levey DF, Pathak GA, Wendt FR, Nunez YZ, Ursano RJ, et al. (2021): Multi-environment gene interactions linked to the interplay between polysubstance dependence and suicidality. Transl Psychiatry 11:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sartor CE, Wang Z, Xu K, Kranzler HR, Gelernter J (2014): The joint effects of ADH1B variants and childhood adversity on alcohol related phenotypes in African-American and European-American women and men. Alcohol Clin Exp Res 38:2907–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan G, Gelernter J, Oslin D, Farrer L, Kranzler HR (2011): Empirically derived subtypes of opioid use and related behaviors. Addiction 106:1146–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peer K, Rennert L, Lynch KG, Farrer L, Gelernter J, Kranzler HR (2013): Prevalence of DSM-IV and DSM-5 alcohol, cocaine, opioid, and cannabis use disorders in a largely substance dependent sample. Drug Alcohol Depend 127:215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherva R, Wang Q, Kranzler H, Zhao H, Koesterer R, Herman A, et al. (2016): Genome-wide association study of cannabis dependence severity, novel risk variants, and shared genetic risks. JAMA Psychiatry 73:472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, et al. (2016): Next-generation genotype imputation service and methods. Nat Genet 48:1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.1000 Genomes Project Consortium; Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. (2015): A global reference for human genetic variation. Nature 526 7571:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge T, Chen CY, Ni Y, Feng YA, Smoller JW (2019): Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun 10:1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kranzler HR, Kadden RM, Babor TF, Tennen H, Rounsaville BJ (1996): Validity of the SCID in substance abuse patients. Addiction 91:859–868. [PubMed] [Google Scholar]
- 31.Rounsaville BJ, Bryant K, Babor T, Kranzler H, Kadden R (1993): Cross system agreement for substance use disorders: DSM-III-R, DSM-IV and ICD-10. Addiction 88:337–348. [DOI] [PubMed] [Google Scholar]
- 32.Edwards AC, Gillespie NA, Aggen SH, Kendler KS (2013): Assessment of a modified DSM-5 diagnosis of alcohol use disorder in a genetically informative population. Alcohol Clin Exp Res 37:443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saha TD, Chou SP, Grant BF (2006): Toward an alcohol use disorder continuum using item response theory: Results from the National Epidemiologic Survey on alcohol and related conditions. Psychol Med 36:931–941. [DOI] [PubMed] [Google Scholar]
- 34.Hasin DS, Fenton MC, Beseler C, Park JY, Wall MM (2012): Analyses related to the development of DSM-5 criteria for substance use related disorders: 2. Proposed DSM-5 criteria for alcohol, cannabis, cocaine and heroin disorders in 663 substance abuse patients. Drug Alcohol Depend 122:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stohs ME, Schneekloth TD, Geske JR, Biernacka JM, Karpyak VM (2019): Alcohol craving predicts relapse after residential addiction treatment. Alcohol Alcohol 54:167–172. [DOI] [PubMed] [Google Scholar]
- 36.Schneekloth TD, Biernacka JM, Hall-Flavin DK, Karpyak VM, Frye MA, Loukianova LL, et al. (2012): Alcohol craving as a predictor of relapse. Am J Addict 21(Suppl 1):S20–S26. [DOI] [PubMed] [Google Scholar]
- 37.Swift RM (1999): Medications and alcohol craving. Alcohol Res Health 23:207–213. [PMC free article] [PubMed] [Google Scholar]
- 38.Byrne SP, Haber P, Baillie A, Giannopolous V, Morley K (2019): Cue exposure therapy for alcohol use disorders: What can be learned from exposure therapy for anxiety disorders? Subst Use Misuse 54:2053–2063. [DOI] [PubMed] [Google Scholar]
- 39.Monti PM, Rohsenow DJ, Hutchison KE (2000): Toward bridging the gap between biological, psychobiological and psychosocial models of alcohol craving. Addiction 95(Suppl 2):S229–S236. [DOI] [PubMed] [Google Scholar]
- 40.Compton WM, Conway KP, Stinson FS, Colliver JD, Grant BF (2005): Prevalence, correlates, and comorbidity of DSM-IV antisocial personality syndromes and alcohol and specific drug use disorders in the United States: Results from the national epidemiologic survey on alcohol and related conditions. J Clin Psychiatry 66:677–685. [DOI] [PubMed] [Google Scholar]
- 41.Sheerin CM, Bountress KE, Meyers JL, Saenz de Viteri SS, Shen H, Maihofer AX, et al. (2020): Shared molecular genetic risk of alcohol dependence and posttraumatic stress disorder (PTSD). Psychol Addict Behav 34:613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaefer JD, Jang S-K, Clark DA, Deak JD, Hicks BM, Iacono WG, et al. (2021): Associations between polygenic risk of substance use and use disorder and alcohol, cannabis, and nicotine use in adolescence and young adulthood in a longitudinal twin study. Psychol Med 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez-Roige S, Palmer AA, Clarke TK (2020): Recent efforts to dissect the genetic basis of alcohol use and abuse. Biol Psychiatry 87:609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.