Abstract
Marcia hiantina (Mollusca, Bivalvia) (Lamarck, 1818), is an edible clam mainly distributed along the tropical coastal regions. Recent researches have demonstrated that clams can possess compounds, including polysaccharides, with a wide range of biological actions including antioxidant, immunomodulatory and antitumor activities. Here an α-glucan was isolated from M. hiantina by hot water, purified by anion exchange chromatography, and its structure was characterized by a combination of multiple nuclear magnetic resonance (NMR) methods (1D 1H, 1H-1H COSY, 1H-1H TOCSY, 1H-1H NOESY, 1H-13C HSQC and 1H-13C HSQC-NOESY spectra), gas chromatography-mass spectrometry, and high performance size exclusion chromatography (HPSEC). The analysis from NMR, monosaccharide composition, methylation analyses and HPSEC combined with multi-angle light scattering (MALS) of M. hiantina-derived α-glycan confirmed a branched polysaccharide exclusively composed of glucose (Glc), mostly 4-linked in its backbone, branched occasionally at 6-positions, and having a molecular weight of ~ 570 kDa. The mollusk α-glucan was subjected to four cell-based assays: (i) viability of three cell lines (RAW264.7, HaCaT, and HT-29), (ii) activity on lipopolysaccharide (LPS)-induced prostaglandin production in RAW264.7 cells, (iii) inhibitory activities of in H2O2- and LPS-induced reactive oxygen species (ROS) production in HMC3 cells, and (iv) HaCaT cell proliferation. Results have indicated no cytotoxicity, potent inhibition of both H2O2- and LPS-induced ROS, and potent cell proliferative activity.
Keywords: Bivalve, Glucan, Mollusk, Marcia hiantina, Methylation analysis, NMR
Introduction
Carbohydrates together with proteins, lipids, and nucleic acids, comprise the main biomacromolecules of living beings. These macromolecules share a common structural feature: they are composed of small subunits. Research of these compounds is large because of their potential functions as nutraceutical and/or bioactive agents that assist human health care [1, 2]. Among these biomacromolecules, carbohydrates are by far the most abundant and most structurally complex class. Sugars are not only structural elements or involved in energy storage, but also involved in signaling events of cell-cell and cell-matrix interactions [3-5]. The most abundant carbohydrates in nature are the polymers of glucose (Glc) namely glucans. Examples of these polysaccharides are cellulose, starch and glycogen. The first two are of plant origin, while the last one is of animal origin.
In the most recent decades, polysaccharides from different terrestrial or marine sources have been extensively investigated as potential therapeutic agents, and questions about biodegradability, non-toxic nature, and biocompatibility have arisen [6]. Polysaccharides from marine sources have particularly been of more interest due to their potential biomedical uses in many systems such as immunomodulation, antivirus, anticoagulation, antitumor, antioxidation, anti-inflammation, and antithrombosis [7, 8]. Interestingly, bivalve mollusks have been widely investigated as potential sources for new nutraceuticals and bioactive compounds [9]. Consumption of mollusks as daily dietary elements have indicated enhancement of immunity, and reduced risk of ailments [10]. In recent years, research has appeared on the polysaccharides from the bivalve Cyclina sinensis, which has been reported to have a significant immunostimulatory activity [11]. In another example, a new 2-sulfated β-galactan from the clam species Meretrix petechialis has shown potential anti-HIV activity [12].
Marcia hiantina (Lamarck, 1818), or hiant venus, is a mollusk bivalve belonging to the family Veneridae (venus clams), as recorded by World Register of Marine Species [13-18]. Over 500 species of venus clams are known, most of which are edible, and many of which are used as food sources [19]. M. hiantina is widely distributed along the Indo-West Pacific from the Gulf of Aden to Papua New Guinea, north to southern Japan and south to Queensland. Up to this report, no description regarding the isolation and structural characterization of the main polysaccharides of M. hiatina has been published.
In this work, we isolated and purified the main carbohydrate from M. hiantina by hot water extraction and anion exchange chromatography (AEC), and characterized its structure by multiple nuclear magnetic resonance (NMR) methods (1D 1H, 1H-1H COSY, 1H-1H TOCSY, 1H-1H NOESY, 1H-13C HSQC and 1H-13C HSQC-NOESY spectra), together with gas chromatography (GC)-mass spectrometry (MS), and high performance size exclusion chromatography (HPSEC) and multi-angle light scattering (MALS) techniques. Results have confirmed an α-glucan composed of a backbone of 4-linked α-Glc units and occasionally branched at C-6. The mollusk α-glucan was studied in four cell-based assays: cell viability, activity on lipopolysaccharide (LPS)-induced prostaglandin expression, inhibitory effect over H2O2- and LPS-induced reactive oxygen species (ROS) production, and cell proliferation. Results have indicated no cytotoxicity, inhibition of both H2O2- and LPS-induced ROS, and cell proliferative activity.
Results and discussion
Fractionation, preliminary 1D 1H NMR and molecular weight analyses
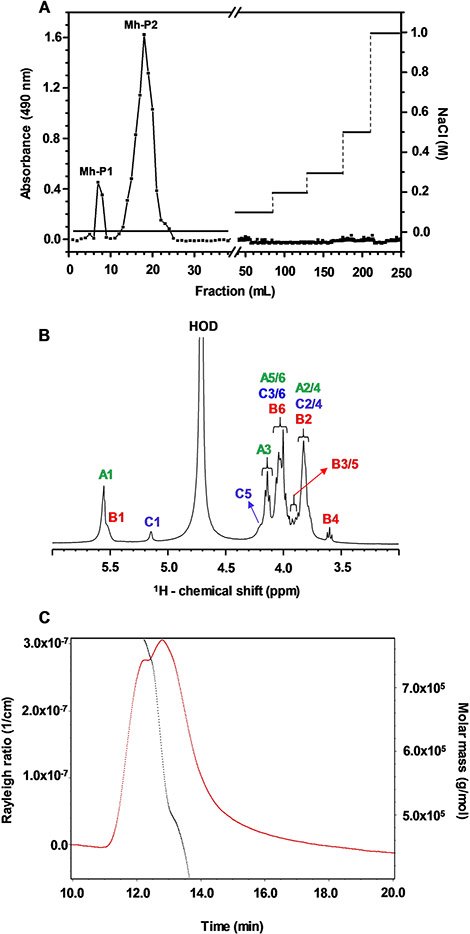
The crude polysaccharide from M. hiantina, obtained by hot water extraction, was fractionated by AEC using a DEAE column with step-wise NaCl elution system (Fig. 1 A). The fractions were detected by the phenol-sulfuric assay [20] through absolute absorbance at 490 nm. Two fractions, Mh-P1 and Mh-P2, were differently eluted from the non-salt portion of the gradient (Fig. 1 A). This indicates that Mh-P1 and Mh-P2 are both composed of neutral monosaccharides and their different retention times suggest different structures.
Fig. 1.
(A) DEAE-Sephacel column chromatogram eluted with stepwise system of increasing NaCl concentrations and the polysaccharide was measured using the phenol-sulfuric method. (B) 1D 1H NMR (δH expansion from 6.0 to 3.0 ppm) spectrum of Mh-P2 acquired on 500 MHz spectrometer at 50 °C. (C) size-exclusion chromatography (SEC)-MALS elution of Mh-P2 showing MW distribution (black) and Rayleigh ratio (1/cm) (red) versus elution time
The one-dimensional (1D) 1H NMR spectra of Mh-P1 and Mh-P2, recorded at 25 °C at 500 MHz, are shown respectively in Figures S1 and S2. As discussed further, the decision was made to perform detailed structural and functional analyses solely on Mh-P2 due to its higher yield (Mh-P1:Mh-P2 5:95 ratio based on NMR of the mixture, and DEAE chromatogram). The anomeric 1H chemical shifts (δH) of units A, B and C in Mh-P2 were respectively 5.33, 5.32 and 4.89 ppm at room temperature (25 °C) (Figure S2). We noticed that the anomeric 1H signal of residue C (δH at 4.89 ppm) was very close to the residual water signal (HOD). Another 1D 1H NMR spectrum was collected at 50 °C (Fig. 1B) to move the C1 peak further from the HOD signal and to uncover any potential 1H signal that could be overlapping with HOD and thus not previously noted at the spectrum recorded at 25 °C. No additional signals were seen with the increased temperature indicating that all 1H NMR signals of Mh-P2 are detectable for further NMR assignment. Since the anomeric 1H of residue C (δH at 5.14 ppm) resonates far away from the HOD peak at 50 °C, and all peaks are present for assignment at this temperature (Fig. 1B), 50 °C was chosen for all further NMR experiments of Mh-P2 in this work.
As described in detail below, a series of two-dimensional (2D) NMR spectra was collected and properly assigned on Mh-P2 aiding recognition of the 1D 1H NMR peaks of Fig. 1B. The anomeric 1H region of this spectrum (4.4–5.6 ppm) showed three signals, two of which were partially overlapping (A1 and B1 with δH respectively at 5.55 and 5.54 Table 1), while the third one was well resolved (C1 with δH at 5.14 ppm, Table 1), indicating the presence of three glycosyl units. The ring proton signals of these three residues were all condensed between δH at 3.76 and 4.20 ppm, except for the well resolved up-field triplet (δH at 3.59 ppm) situated outside the ring 1H bulk region (Fig. 1B).
Table 1.
1H and 13C chemical shifts (ppm) of composing Glc units in the polysaccharide from M. hiantina at 50 °C
| Unit Structure |
1H and 13C chemical shiftsa, δ (ppm) |
|||||
|---|---|---|---|---|---|---|
| H1 C1 |
H2 C2 |
H3 C3 |
H4 C4 |
H5 C5 |
H6/H6’ C6 |
|
| Residue A | 5.55 | 3.81 | 4.13 | 3.81 | 4.00 | 4.00/3.88 |
| →4)-α-Glc-(1→ | 100.1 | 71.9 | 73.7 | 77.6 | 71.6 | 60.8 |
| Residue B | 5.54 | 3.78 | 3.88 | 3.59 | 3.91 | 4.03/3.88 |
| α-Glc-(1→ | 100.1 | 71.9 | 73.1 | 69.9 | 71.6 | 60.8 |
| Residue C | 5.14 | 3.76 | 4.09 | 3.76 | 4.20 | 4.05/3.86 |
| →4,6)-α-Glc-(1→ | 98.9 | 71.9 | 73.7 | 78.9 | 70.6 | 68.1 |
Chemical shifts are relative to external trimethylsilylpropionoic acid at 0 ppm for 1H and methanol for 13C. The relative down-field 13C resonances which are used as diagnostic peaks of the glycosidic linkages are highlighted in bold
The molecular weight (MW) analysis was performed on Mh-P2 as the major polysaccharide in M. hiantina using HPSEC-MALS (Fig. 1 C). The Mh-P2 peak was eluted between dextran standards of MW ~ 100 kDa and 500 kDa (actual MW by HPSEC-MALS of 649 kDa (±2.9%)) (Figure S3). HPSEC-MALS detected a single, monodisperse peak for Mh-P2 with a MW of 573 kDa (± 4.67%).
Monosaccharide composition and glycosidic linkage analyses
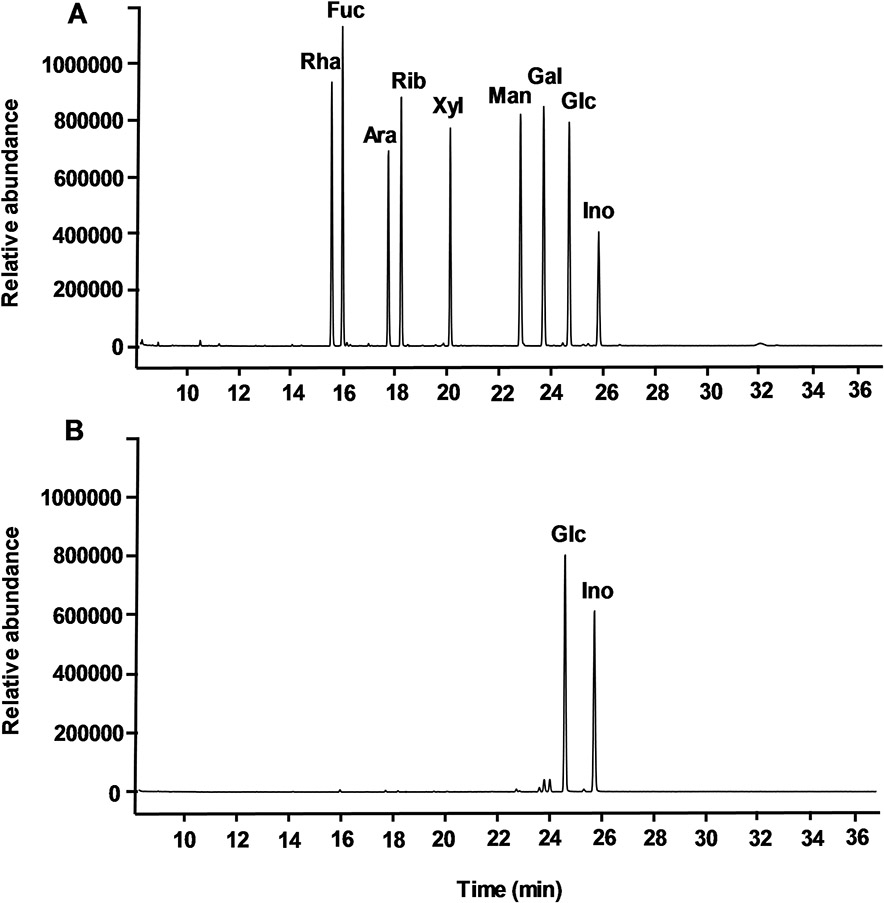
The monosaccharide composition of Mh-P1 and Mh-P2 were analyzed by GC after full hydrolysis and derivatization as alditol acetates. The GC chromatogram of nine standard monosaccharides (rhamnose, fucose, arabinose, ribose, xylose, mannose, galactose, glucose, and inositol), derivatized in the same manner, was used to compare their retention times with the products from Mh-P1 (Figure S4) and Mh-P2 (Fig. 2). Inositol was used as internal standard. The GC chromatogram of Mh-P1 and Mh-P2 showed only one major peak, other than the reference inositol peak, which was identified as Glc, indicating a glucan polysaccharide for both fractions.
Fig. 2.
GC chromatograms of (A) derivatives of nine standard monosaccharides, and (B) composing monosaccharides from Mh-P2. Peaks are labeled as Rha for rhamnose, Fuc for fucose, Ara for arabinose, Rib for ribose, Xyl for xylose, Man for mannose, Gal for galactose, Glc for glucose and Ino for inositol
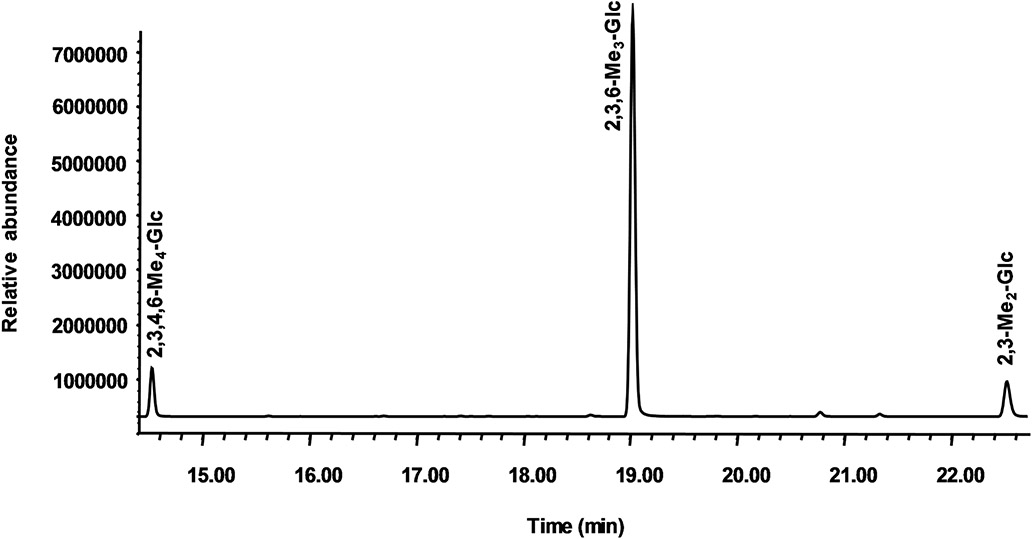
The partially methylated alditol acetates (PMAAs), obtained from Mh-P1 and Mh-P2 by methylation, depolymerization, reduction, and acetylation, were analyzed by GC-MS to investigate the profile of glycosidic bonds. The relative area ratio of file PMAA peaks along with their retention times and file major fragments used to identify the linkages are shown in Table S1 for Mh-P1 and Table 2 for Mh-P2. Six PMAAs were found in Mh-P1 (Figure S5), which were (1,5-Ac2-1-d-2,3,4,6-Me4-Glcol), (1, 3, 5-Ac3-1-d-2,4,6-Me3-Glcol), (1, 5, 6-Ac3-1-d-2,3 4-Me3-Glcol), (1,4,5-Ac3-1-d-2,3,6-Me3-Glcol), (1,3,5,6-Ac4-1-d-2,4-Me2-Glcol), and (1,4,5,6-Ac4-1-d-2,3-Me2-Glcol) corresponding respectively to [Glc-(1→],[→3)-Glc-(1→],[→6)-Glc-(1→],[→4)-Glc-(1→],[→3,6)-Glc-(1→], and [→4,6)-Glc-(1→] linkages, with a ratio of 1.0:0.1:4.4:5.2:0.2:0.6 (Table S1). Three PMAAs were found in Mh-P2 (Fig. 3) which were (1,5-Ac2-1-d-2,3,4,6-Me4-Glcol), (1,4,5-Ac3-1-d-2,3,6-Me3-Glcol), and (1,4,5,6-Ac4-1-d-2,3-Me2-Glcol) corresponding respectively to [Glc-(1→],[→4)-Glc-(1→], and [→4,6)-Glc-(1→] linkages, with a percentage ratio of 9:81:10 (Table 2). The near equal intensity of terminal Glc and 4,6-Glc indicates that terminal Glc represents the termini of the side chains since the intensity of the non-reducing end of the backbone is negligible as expected for a very large polysaccharide.
Table 2.
GC-MS results from methylation analysis on Mh-P2.
| Reten tion time (min) |
Methylated sugara |
Mass frag ments (m/z) |
Peak area ratio |
Percentageb (%) |
Linkage |
|---|---|---|---|---|---|
| 14.6 | 2,3,4,6-Me4-Glc | 118, 161, 162, 205 | 1.0 | 9.1 | Glc-(1→ |
| 19.0 | 2,3,6-Me3-Glc | 118, 233 | 8.8 | 80.8 | →4)-Glc-(1→ |
| 22.5 | 2.3-Me2-Glc | 118, 261 | 1.1 | 10.1 | →4,6)-Glc-(1→ |
After the methylation reaction, the polysaccharide was hydrolyzed, and the monosaccharide products were analyzed as their alditol acetate derivatives
The proportion of the methylated acetates are based on the integral of each peak compared relatively to the sum of integrals of the three derivatives
Fig. 3.
GC chromatogram of the linkage analysis on Mh-P2. Peaks identified and confirmed by NMR-based analysis are 2,3,4,6-Me4-Glc (reducing end Glc), 2,3,6-Me3-Glc (4-linked Glc), and 2,3-Me2-Glc (4,6-linked branching Glc)
Structural characterization by 2D through-bond NMR spectroscopy
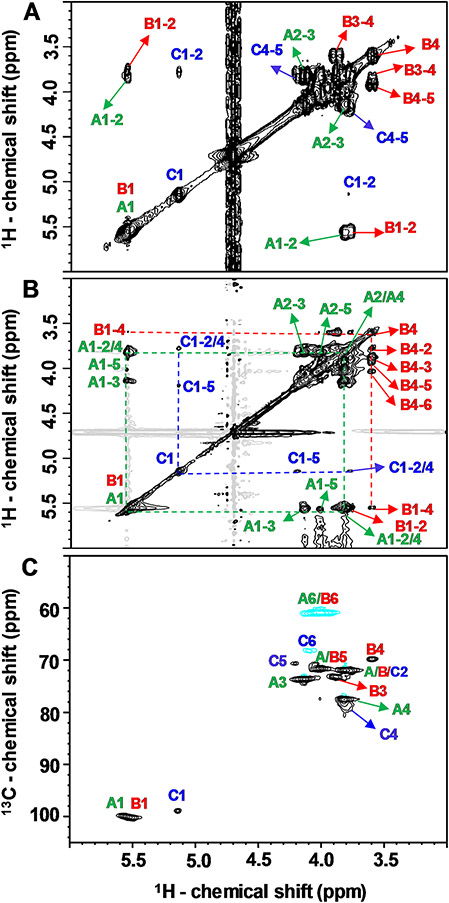
1H and 13C chemical shifts of units A-C of Mh-P2 (Table 1) were obtained by complete signal assignments of a series of 2D NMR spectra, including 1H-1H correlation spectroscopy (COSY) (Fig. 4 A), 1H-1H total correlation spectroscopy (TOCSY) (Fig. 4B), and 1H-13C heteronuclear single quantum coherence (HSQC) (Fig. 4 C). Two and three-bond short-range 1H-1H correlations of geminal and vicinal carbon-bound hydrogens were determined by a 1H-1H COSY spectrum (Fig. 4 A). Different spin-systems were identified by long-range correlations between all 1H within the same Glc ring using1H-1H TOCSY spectrum (Fig. 4B). All 1H-13C pairs of the three Glc units were recognized and assigned in the 1H-13C HSQC spectrum (Fig. 4 C). The detailed NMR signal assignments for assessment of both 1H and 13C resonances of the three Glc units of Mh-P2 is described below per residue type.
Fig. 4.
(A) 1H-1H COSY, (B) 1H-1H TOCSY (δH-δH expansions from 6.0–3.0 to 3.0–6.0 ppm), and (C) 1H-13C HSQC (δH-δC expansions from 6.0–50.0 to 3.0–102.5 ppm) spectra of Mh-P2 acquired on 500 MHz spectrometer at 50 °C
Assignments of residue A
All assignments of residue A were represented in green in all figures. The anomeric 1H1 (δH1 at 5.55 ppm, Table 1) was the starting point to identify the other ring 1H signals by tracking the connections 1H1-1H2, 1H2-1H3, 1H3-1H4, 1H4-1H5 and 1H5-1H6 in the 1H-1H COSY spectrum (Fig. 4A), and the whole spin-system in 1H-1H TOCSY spectrum (Fig. 4B). The non-anomeric ring 1H resonances were assigned as δH2 3.81, δH33 4.13, δH4 3.81, δH5 4.00, δH6 4.00 and δH6, 3.88 ppm (Table 1). The corresponding 13C resonances of all assigned 1H signals were determined from the 1H-13C correlations in the 1H-13C HSQC spectrum (Fig. 4 C), and were assigned respectively as δC1 100.09, δC2 71.92, δC3 73.69, δC4 77.61, δC5 71.61 and δC6 60.88 ppm. The HSQC spectrum showed down-field 13C resonances of C1 and C4 (Table 1), suggesting that residue A is likely 4-linked Glc unit. According to many reported researches, the anomeric regions of α-configuration appear in 5.1–5.8 ppm for anomeric protons and 98–101 ppm for anomeric carbons, while β-configuration relatively corresponds to 4.3–4.8 ppm for anomeric protons and 103.0-106.0 ppm for anomeric carbons [21-24]. Accordingly, residue A was identified as →4)-α-Glc-(1→ sugar residue.
Assignments of residue B
Red color was chosen to represent residue B. The anomeric 1H of residue B (δH1 at 5.54 ppm) was overlapped with residue A anomeric 1H1 (δH1 at 5.55 ppm, Table 1). Although this resonance overlaps markedly, we were able to assign residue B due to the uniqueness of its up-field 1H4 peak (δH1 at 3.59 ppm) with a triplet splitting pattern, which was a part of the residue B spin-system in the 1H-1H TOCSY spectrum (Fig. 4B). By summing up the cross-peak information of 1H1-1H2, 1H2-1H3, 1H3-1H4, 1H4-1H5 and 1H5-1H6 from the 1H-1H COSY spectrum (Fig. 4A), and the cross-peaks of the whole spin-system in the 1H-1H TOCSY spectrum (Fig. 4B), the ring 1H chemical shifts were assigned as δH1 5.54, δH2 3.78, δH3 3.88, δH4 3.59, δH5 3.91, δH6 4.03 and δH6, 3.88 ppm (Table 1). The corresponding 13C resonances of all assigned 1H signals were deduced from the 1H-13C HSQC spectrum (Fig. 4 C), and were assigned as δC1 100.09, δC2 71.92, δC3 73.16, δC4 69.9, δC5 71.6 and δC6 60.88 ppm (Table 1). The lack of down-field displacement of its 13C resonances except C1 suggests that residue B is likely the non-reducing end 1-linked Glc unit and was identified as α-Glc-(1→ sugar residue.
Assignments of residue C
It is the blue assignments. The well-resolved anomeric 1H1 of residue C (δH1 5.14 ppm. Table 1) was the starting point to assign the other ring 1H in the 1H-1H COSY (Fig. 4A), and the 1H-1H TOCSY spectra (Fig. 4B) giving rise to the following resonances: δH2 3.76, δH3 4.09, δH4 3.76, δH5 4.20, δH6 4.05 and δH6, 3.86 ppm (Table 1). A well-resolved anomeric 13C of residue C (δC1, 98.9 ppm) was identified in the HSQC spectrum (Fig. 4 C), and the other ring 13C were assigned as δC2 71.92, δC3 73.69, δC4 78.90, δC5 70.66 and δC6 68.15 ppm (Table 1) with three down-field 13C resonances, C1, C4 and C6, suggesting that residue C is the 4,6-linked Glc unit, which is the branching site in the backbone of M. hiantina polysaccharide and was identified as →4,6)-α-Glc-(1→ residue.
Structural characterization by 2D through-space NMR spectroscopy
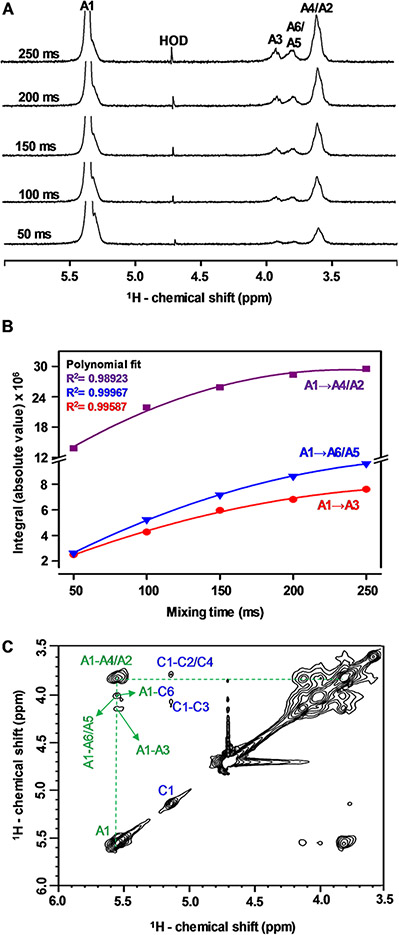
Further NMR signal assignments were made by the through-space 1H-1H connectivities generated through the 1H-1H nuclear Overhauser effect spectroscopy (NOESY) spectrum and through the cross-peaks generated through the 1H-13C HSQC-NOESY spectrum. To choose the optimal mixing time, a series of selective 1D 1H-1H NOESY experiments was performed at different mixing times (Fig. 5). The anomeric 1H1 signal of unit A was selected and successive spectra were acquired at mixing times of 50, 100, 150, 200 and 250 ms (Fig. 5 A). The integrals (absolute values) of the resultant 1H-1H NOE signals were determined and plotted as a function of mixing times. The build-up curves of 1H-1H NOE signals are shown in Fig. 5B. The integrals of the NOE signals increase as the mixing time goes up. The mixing time of 150 ms was chosen for the 1H-1H NOESY (Fig. 5 C) and 1H-13C HSQC-NOESY spectra (Figure S6), as the point inside the linear region prior to the maximum intensity, to avoid spin diffusion.
Fig. 5.
(A) Selective 1D 1H-1H NOE (δH expansion from 6.0 to 3.0 ppm) spectra at different mixing times (50, 100, 150, 200, 250 ms), (B) 1H-1H NOE build up curves at the different mixing times, and (C) 1H-1H NOESY (δH-δH expansions from 6.0–3.5 to 3.5–6.0 ppm) spectrum of Mh-P2 acquired on 500 MHz spectrometer at 50 °C and 150 ms mixing time
The 2D 1H-1H NOESY (Fig. 5 C) and 1H-13C HSQC-NOESY (Figure S6) spectra were used together to assign the inter- and intra-residue 1H-1H connectivities through space. In the 1H-1H NOESY spectrum (Fig. 5 C), the anomeric 1H1 of residue A (δH 5.55 ppm, Table 1) showed three 1H-1H NOE cross-peaks classified as strong, medium and weak, depending on their signal intensities which is primarily dictated by abundance and internuclear distances. The strong connectivity was the A4/A2 (δH 3.81 ppm) (resolved later in the 1H-13C HSQC-NOESY spectrum, Figure S6), the medium was related to A3 (δH 4.13 ppm), and the weak was related to A5/A6 (δH 4.00 ppm).
The overlapped connectivities of A1 with A4/A2 and A5/A6, were resolved in the 1H-13C HSQC-NOESY spectrum (Figure S6). Residue A showed four 1H-1H NOE signals in the 1H-13C HSQC spectrum: two strong cross-peaks (A1-A2 and A1-A4), a weak cross-peak (A1-A3), and a very weak cross-peak (A1-A6). All these through-space connectivities are indicated in green in Figure S6. It is obvious that the strong cross peak found between H1 and H4 of residue A is not only for an intra-residue but also an inter-residue connection due to the strength of connectivity, revealing the presence of inter-residual (1→4)–glycosidic linkage of the backbone structure made of residue A.
Effect in cell-based assays
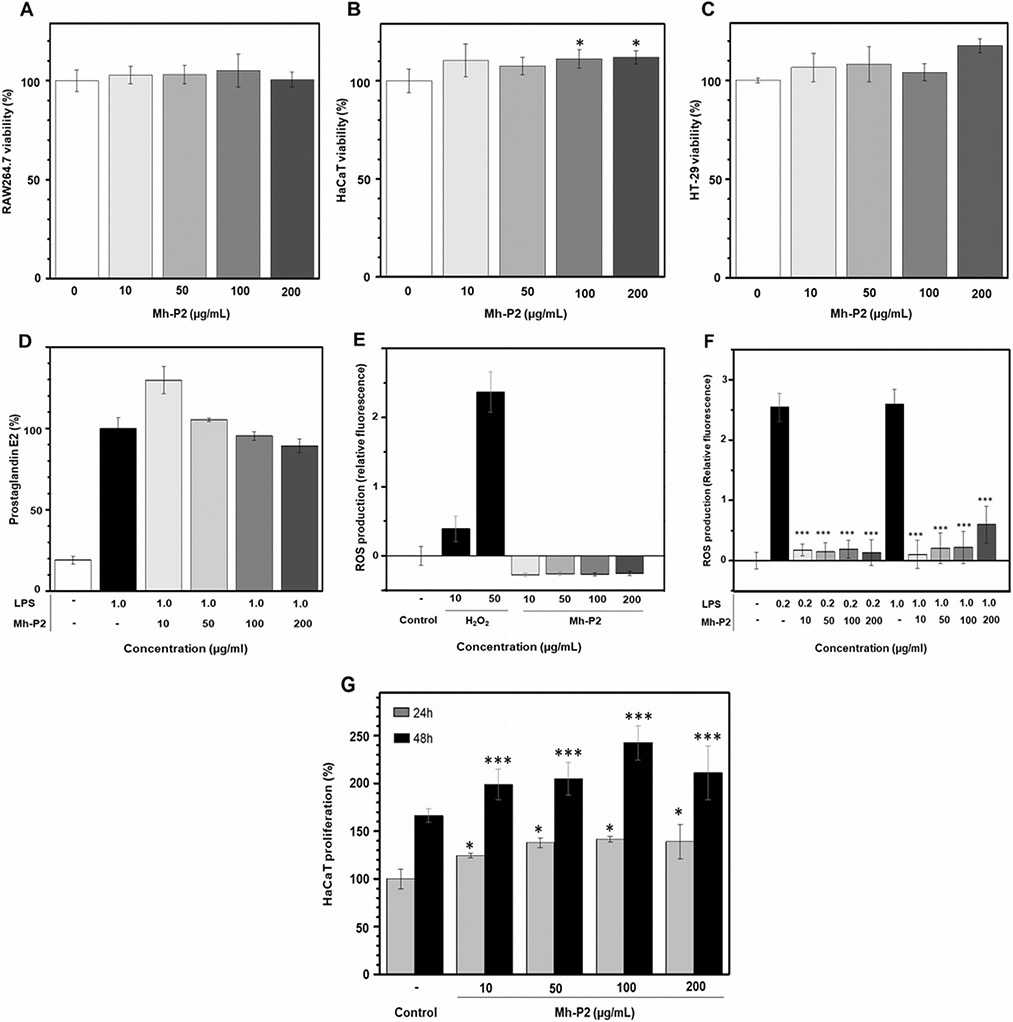
Four different concentrations of Mh-P2 (10, 50, 100 and 200 μg/mL) were used in cell viability assays using three cell lines: mouse monocyte/macrophage cells (RAW264.7) (Fig. 6 A), immortalized human keratinocyte cells (HaCAT) (Fig. 6B), and human colorectal adenocarcinoma cells epithelial morphology (HT-29) (Fig. 6 C). The results showed that Mh-P2 at the concentrations 10–200 μg/mL had no effect on the viability of RAW264.7 and no toxicity (Fig. 6 A). Mh-P2 at 100–200 μg/mL had significant increase (*p<0.05) on the viability and was not toxic to HaCaT cells (Fig. 6B). Mh-P2 showed no anticancer effect against the HT-29 cells (Fig. 6 C).
Fig. 6.
Mh-P2 was subjected to four cell-based assays: (A-C) cell viability in three cell lines. (A) RAW264.7. (B) HaCaT, and (C) HT-29, (D) activity on LPS-induced prostaglandin expression in RAW264.7 cells, inhibitory actions against (E) H2O2- and (F) LPS-induced ROS production in HMC3 cells, and (G) HaCaT cell proliferation (significance level: *p < 0.05, **p < 0.01 and ***p < 0.001)
Prostaglandin E2 inhibitory activity of Mh-P2 at the concentrations 10–200 μL on LPS-induced prostaglandin E2 upregulation in RAW264.7 (Fig. 6D) showed that Mh-P2 does not interfere in this activity. Mh-P2 was tested as inhibitor of ROS production in HMC3 cells using H2O2 (Fig. 6E) and LPS (Fig. 6 F) as positive control. Results have indicated that Mh-P2 is a potent anti-oxidant agent since the ROS production was substantially reduced in the presence of Mh-P2 at all concentrations tested (***p < 0.001). Mh-P2 was tested as cell proliferative agent on HaCaT cells exposed 24 and 48 h to the mollusk glucan (Fig. 6G). Data have indicated that Mh-P2 is a potent HaCaT proliferative compound at all concentrations tested. A commercially available α-glucan (glycogen) extracted from oyster was also assayed as positive control and results have indicated significantly equal activity of Mh-P2 and this control. 1D 1H NMR spectrum of this standard showed similar profile of the one from Mh-P2 validating therefore the positive control.
Conclusion
In this work, we have isolated, purified and studied the structure and potential biological actions of an α-glucan (Mh-P2) from the mollusk M. hiantina. Extraction was made by hot water while purification was performed by AEC technique. Mh-P2 was extracted from the whole body of M. hiatina, and information about specific tissue-distribution or tissue-abundancy was not reached in this work. Structural characterization was performed by multiple NMR methods, together with GC-MS and HPSEC-MALS. Results have confirmed two α-glucan fractions in M. hiantina: Mh-P1 of low abundance (5%) is mainly composed of 6- or 4-linked α-Glc units, and Mh-P2 as the major water-soluble polysaccharide (95%) composed of a backbone of 4-linked α-Glc units occasionally branched at C-6. Based on structural similarities between the oyster glycogen and Mh-P2, Mh-P2 is likely to be a storage polysaccharide in the mollusk, but future investigations remains to be performed to confirm this hypothesis. Mh-P2 was biologically investigated in four cell-based assays: cell viability, activity on LPS-induced prostaglandin expression, inhibition of H2O2- and LPS-induced ROS production, and cell proliferation. Positive potent effects in cell proliferation and anti-oxidant assays were observed at all concentrations tested. Medical effects of Mh-P2 are unlikely to occur if the mollusk M. hiantina is consumed as food since the α-glucan here reported is likely to be degraded by the mammalian digestive system into Glc monosaccharides, as commonly happens to α-glucans, including glycogens, ingested in the diet.
Materials and methods
Materials
Specimen samples of the mollusk M. hiantina were collected from Northern Iloilo, Philippines and were identified by taxonomic classification. The shells were removed, and their meat was washed with deionized water and frozen at −20 °C prior to the polysaccharide extraction. DEAE Sephacel (#17-0500-01) was obtained from GE Healthcare Bio-Sciences (Uppsala, Sweden). NMR tubes (Norell, 3 mm high throughput 7 inches) were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). Deuterium oxide (D2O, 99.9%) was purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA, USA). Dialysis molecular porous membrane tubing (MWCO: 12–14 kDa, flat width: 32 mm, vol/length: 3.3 ml/cm) was purchased from Spectrum Laboratories (CA, USA). Chemicals and reagents, ethanol, sodium dihydrogen phosphate (NaH2PO4), trifluoroacetic acid (TFA), sodium hydroxide, sodium borohydride (NaBH4), ammonium hydroxide, glacial acetic acid, anhydrous acetic anhydride, 1-methylimidazole and methyl iodide hydrogen peroxide (H2O2) all were of analytical grade. Separation of alditol acetates and partially methylated alditol acetates was performed on a 30 m Supelco SP-2331 bonded phase fused silica capillary column.
Mouse monocyte/macrophage cell line (RAW264.7), and human colorectal adenocarcinoma (HT-29) cells were obtained from the Korean Cell Line Bank (KCLB). Human keratinocyte (HaCaT) cells were obtained from Professor Kim Hyeung-Rak (Department of Food Science and Nutrition, Pukyong National University, Busan, Korea). Human microglial cells (HMC3) (#CRL-3304) were purchased from the American Type Culture Collection (ATCC), (Manassas, VA, USA)
Dulbecco modified Eagle medium high glucose and low glucose (DMEM), Eagle minimum essential medium (EMEM) and fetal bovine serum (FBS) were purchased from HyClone (MA, USA). Penicillin/streptomycin (#15,140,122) and RPMI 1640 medium (#11,875,093) were purchased from Gibco (CA, USA). CellTiter-96® aqueous cell proliferation assay kit was purchased from Promega (Madison, WI, USA). Counting Kit-8 (CCK-8, CK04-11) was purchased from Dojindo (Kumamoto, Japan).
Prostaglandin E2 kit (#514,010) was purchased from Cayman Chemicals (Ann Arbor, MI, USA). The reactive oxygen species (ROS) indicator, chloromethyl derivative of 2′,7′-dichlorodihydrofluorescein diacetate (CM-H2DCFDA) (#C6827), was purchased from Thermo Fisher Scientific (Waltham, MA, USA). CM-H2DCFDA was detected on a CLARIOstar plate reader (BMG Labtech, Ortenberg, Germany). Lipopolysaccharide (LPS) from Escherichia coli O55:B5 (γ-irradiated, BioXtra), and oyster glycogen (#G8751) were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA).
Methods
Isolation of the M. hiantina-derived polysaccharide
Frozen samples were separately thawed at room temperature and thoroughly washed with deionized water. Each sample was placed in an autoclave with deionized water (1.7 kg: 5 L deionized water) and heated at 80 °C for 1 h. Ethyl alcohol was then added to the cooled extract at a volume ratio of 1:0.5 (water: ethyl alcohol). Sodium chloride was added to the solution to give a final concentration of 0.5 M and the mixture was left overnight at 4.0 °C for precipitation. The pellet was not further examined since its 1D 1H NMR spectrum showed a very complex mixture of (likely more hydrophobic) components. The supernatant was recovered, and ethyl alcohol was added in a volume ratio of 1:0.075 (water: ethyl alcohol). Sodium hydroxide was added to the supernatant solution up to a concentration of 0.5 M and the resulting suspension was centrifuged at 3000 rpm for 20 min. The precipitate was then re-dissolved in ethyl alcohol: deionized water (1:0.75), neutralized with 0.5 M NaH2PO4 in 1:0.75 (deionized water: ethyl alcohol), and centrifuged at 3000 rpm for 20 min. The supernatant was dialyzed against deionized water using dialysis membrane (MWCO: 12–14 kDa), and then lyophilized to obtain a crude M. hiantina-derived polysaccharide.
Purification
The isolated crude polysaccharide was fractionated using DEAE Sephacel column. The polysaccharide (100 mg) was dissolved in the lowest amount of deionized water and then loaded onto the DEAE Sephacel column (2.5 cm × 30 cm) followed by stepwise elution with deionized water, 0.1, 0.2, 0.3, 0.5 and 1 M sodium chloride (NaCl) of 3 times column volume, respectively. The eluate was at a flow rate of 20 mL/hr. Each fraction (5 mL/tube) was collected and the carbohydrate content was measured by the phenol-sulfuric acid method [20]. And the two fractions eluted with deionized water were collected and lyophilized to obtain two purified polysaccharides Mh-P1 and Mh-P2.
NMR spectroscopy analysis
The purified polysaccharide Mh-P2 (20 mg) was dissolved in 300 μl D2O (99.9%) for the NMR analysis. One-dimensional 1H and two-dimensional 1H-1H homonuclear or 1H-13C heteronuclear experiments were recorded including 1H-1H COSY, 1H-1H TOCSY, 1H-13C HSQC, 1H-1H NOESY, and 1H-13C HSQC-NOESY. Acquisition parameters of 1H-1H COSY experiment were as follows: number of scans (NS): 8, size of free induction decays (TD): 1024 and 256 for F1 and F2 respectively, acquisition time: 0.20 and 0.05 s for T1 and T2 respectively. Acquisition parameters of 1H-1H TOCSY experiment: NS: 8, TD: 1024 and 128 for F1 and F2 respectively, acquisition time: 0.15 and 0.0007 s for T1 and T2 respectively. Acquisition parameters of 1H-13C HSQC experiment: NS: 4, TD: 1024 and 256 for F1 and F2 respectively, acquisition time: 0.34 and 0.04 s for T1 and T2 respectively. Series of selective 1D 1H-1H NOESY experiments were performed at different mixing times (50, 100, 150, 200, 250) to select the best one to perform 1H-1H NOESY and 1H-13C HSQC-NOESY experiments. Acquisition parameters of 1H-1H NOESY experiment were NS: 8, TD: 2048 and 256 for F1 and F2 respectively, mixing time: 150 milliseconds, acquisition time: 0.29 and 0.04 s for T1 and T2 respectively. Acquisition parameters of 1H-13C HSQC-NOESY experiment: [NS: 16, TD: 2048 and 1024 for F1 and F2 respectively, mixing time: 150 milliseconds, acquisition time: 0.29 and 0.04 s for T1 and T2 respectively. Spectra were recorded at 25 or 50 °C on a Bruker 500 MHz spectrometer equipped with a cryogenic probe, except for the 1D selective 1H-1H NOESY experiments which were recorded at 25 °C on a Bruker 400 MHz spectrometer. The acquired NMR data were processed using TopSpin 4.0 software.
MW analysis
HPSEC by using an Ultimate 3000 high performance liquid chromatography system (Thermo Scientific, Sunnyvale, CA) with an Acquity BEH SEC column (200 Å, 1.7 μm, 4.6 mm X 300 mm. Waters, MA) was performed to measure the MW of M. hiantina polysaccharide. The eluent was monitored by MALS using a DAWN HELEOS II MALS detector (Wyatt Technologies Co., Santa Barbara, CA). The instrument was calibrated using polystyrene in toluene for the MALS detector and by bovine serum albumin for MALS/RI detectors. Isocratic elution of the sample was employed using 50 mM ammonium acetate on the SEC column at the flow rate of 0.2 mL/min. Different MW dextran sulfates (~ 100 kDa and > 500 kDa, Sigma Aldrich Co.) were used to validate the accuracy of the MW measurement.
Monosaccharide composition analysis
Glycosyl composition analysis was performed by GC-MS of the alditol acetates (AAs) as described by Peña et al. [25]. Briefly, the sample (300 μg) was hydrolyzed in 2 M TFA for 2 h in a Teflon-lined screw-cap tube at 120 °C, 20 mM myo-inositol was added as internal standard to both sample and standards. Then the dried hydrolysate was reduced with NaBH4 in ammonium hydroxide for 1.5 h at 40–45 °C. The unreacted NaBH4 was decomposed by adding glacial acetic acid. Then, the reaction product was mixed with 1-methylimidazole and anhydrous acetic anhydride for 30 min at 40–45 °C. The reaction terminated by adding 2 ml of deionized water, 1 ml dichloromethane was added, and the aqueous layer was discarded. Deionized water was added again to the organic layer and again the aqueous layer was discarded. This step was repeated four more times to ensure complete removal of water-soluble salts which could interfere with the downstream analysis. The resulting alditol acetates were analyzed on an Agilent 7890 A GC interfaced to a 5975 C MSD (mass selective detector, electron impact ionization mode). Separation of neutral monosaccharides was performed on a 30 m Supelco SP-2331 bonded phase fused silica capillary column. The monosaccharide composition of M. hiantina-derived polysaccharides Mh-P1 and Mh-P2 were identified by comparing the retention times with standard monosaccharides including rhamnose, fucose, arabinose, ribose, xylose, mannose, glucose and galactose.
Glycosyl linkage analysis
Glycosyl linkage analysis was performed by GC-MS of the PMAA derivatives produced form the samples. The procedure is a slight modification of the one described by Willis et al. [26]. Briefly, permethylation of the sample was affected by two rounds of treatment with sodium hydroxide (15 min) and methyl iodide (30 min). The samples were then hydrolyzed using 2 M TFA (2 h in sealed tube at 120 °C), reduced with NaBD4, and acetylated using acetic anhydride/TFA. The resulting PMAAs were analyzed on an Agilent 7890 A GC interfaced to a 5975 C MSD (mass selective detector, electron impact ionization mode); separation was performed on a 30 m Supelco SP-2331 bonded phase fused silica capillary column for the neutral residues and an EC-1 column for the amino containing residue.
Cell culture
The cell lines RAW 264.7, HaCaT and HT-29 were cultured in DMEM, high and low glucose media (HyClone, SH30243.01) respectively for RAW 264.7 and HaCaT, and HT-29 in RPMI-1640 medium, all supplemented with 10% FBS (HyClone, SH30084.03) and 1% penicillin/streptomycin (Gibco, 15,140,122, CA, USA). The cells were subsequently incubated at 37 °C, 95% humidity in an atmosphere of 5% CO2.
Cell viability assay on RAW264.7, HaCaT and HT-29 cells
The RAW264.7, HaCaT and HT-29 cells viability assays were conducted using a CellTiter-96® aqueous cell proliferation assay kit (Promega, Madison WI). Cells were cultured in 96-well plates at a density of 1 × 104 cells/well for 24 h at 37 °C in 5% CO2. After media removal, 100 μl of Mh-P2 were diluted in media and incubated for 24 h at 37 °C in 5% CO2. The media was removed from the 96-well plates using suction. MTS solution at a dilution of 1/10 with FBS-free DMEM (high for RAW264.7 and low for the other two cell lines) was added to 100 μL of each well and the cells were incubated for 1 h at 37 °C. Absorbance was measured at 490 nm with a microplate reader (Molecular Devices, VersaMax ELISA Microplate Reader, USA). The mean optical density (OD) of 4 wells in the indicated groups was used to calculate the percentage of cell viability as the following: percentage of cell viability = (ODtreatment group−ODblank group)/(ODcontrol group−ODblank group)×100%. The experiment was performed in quadruplicate.
Prostaglandin E2 inhibitory assay on RAW264.7 cells
The RAW 264.7 cells were seeded on 96-well plates at a concentration of 2 × 105 cells/well and incubated for 24 h at 37 °C in 5% CO2. The cells were treated with serum-free DMEM (high) containing Mh-P2 in the present of 100 ng/mL of LPS for 24 h. Then, the medium was collected and analyzed for prostaglandin E2 using a commercial enzyme immunoassay kit (Prostaglandin E2 EIA Kit, Item No. 514,010, Ann Arbor, Michigan, USA). The collected medium was loaded at 50 μl into a 96-well plate coated with goat anti-mouse. After that, 50 μl of primary antibody solution and 50 μl of prostaglandin E2 conjugate were added, incubated at 4 °C for 18 h, rinsed 5 times with wash buffer and 200 μl of Ellman’s reagent were added. Incubation at room temperature, shaking for 1 h, was followed. The plate was read at a wavelength of 420 nm.
Proliferation activity on human keratocyte cells (HaCaT)
The cell proliferation assay was conducted using Cell Counting Kit-8 (CCK-8, Dojindo Mol. Tech., Rockville., MD, USA). HaCaT cells were cultured in 96-well plates at a density of 5 × 103 cells/well for 24 h at 37 °C in 5% CO2. After removing medium 100 μl of Mh-P2 diluted in the medium were added and incubated for 24 or 48 h at 37 °C in 5% CO2. The medium was then removed from the 96-well plate. CCK-8 solution at a dilution of 1/10 with FBS-free DMEM (low) was added to 100 μL in each well and the cells were incubated for 1 h at 37 °C in 5% CO2. Absorbance was measured at 450 nm in a microplate reader (Molecular Devices, VersaMax ELISA Microplate Reader, USA). The mean optical density (OD) of 4 wells in the indicated groups was used to calculate the percentage of cell proliferation as the following: percentage of cell proliferation=(ODtreatment group−ODblank group)/(ODcontrol group−ODblank group)×100%. The experiment was performed in quadruplicate.
ROS assay on HMC3 cells
For the H2O2- or LPS-induced ROS assays, HMC3 cells were seeded on a 96-well plate at a density of 20k/well in EMEM complete media. The cells were treated for 24 h at 37 °C in 5% CO2 with the following concentrations of Mh-P2 (10, 50, 100 and 200 μg/mL) in absence or presence of H2O2 (10 or 50 μM), and LPS (0.2 or 1.0 μg/mL). Media alone was used as a negative control. All samples were run with six technical replicates per plate over three plates (n = 3 independent observations). The ROS indicator, CM-H2DCFDA (#C6827; Thermo Fisher Scientific, Waltham, MA, USA) was applied at 10 μM (final conc.) for 1 h at 37°C, 5% CO2. Following the incubation, the cells were washed with DPBS, followed by addition of 100 μL of HBSS. The ROS production was measured on a CLARIOstar plate reader (BMG Labtech, Ortenberg, Germany) at an excitation/emission spectra of 485/528 nm.
Statistical analysis
All results obtained using RAW 264.7, HaCaT, HT-29 and HMC3 cells were expressed as mean ± SD (standard deviation) and the experiments were performed in quadruplicate. Significant differences were analyzed using t-test (Excel 2020, Microsoft, Redmond, WA, USA) and P-values < 0.05 were considered significant with the significance level of *p < 0.05, **p < 0.01 and ***p < 0.001.
Supplementary Material
Funding
This work was supported by funds from the National Institutes of Health [1P20GM130460-01A1-7936 (VHP, and JSS), 1R03NS110996-01A1 (VHP and JSS), and R01 DA052851 (JJP)], the University of Mississippi (VHP). The linkage and monosaccharide analyses were supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences, grant number DE-SC0015662 to Parastoo Azadi at the Complex Carbohydrate Research Center. Bernadeth F. Ticar acknowledges the CHED DARE TO (Commission on Higher Education Discovery-Applied Research and Extension for Trans-interdisciplinary Opportunities) research grant for the K to 12 Transition Program of the Philippines for providing the research funds for Iloilo Science and Technology University, Iloilo City, Iloilo, Philippines. Sunmarine Biotech Co. Ltd., commissioned by Bernadeth F. Ticar, is properly acknowledged for some of the biological analyses. The content of the information does not necessarily reflect the position, or the policy of the sponsors and no official endorsement should be inferred.
Abbreviations
- δC
carbon chemical shift
- δH
proton chemical shift
- AEC
anion exchange chromatography
- COSY
correlation spectroscopy
- DMEM
Dulbecco’s modified Eagle’s medium
- EMEM
Eagle minimum essential medium
- FBS
fetal bovine serum
- GC
gas chromatography
- HPSEC
high performance size exclusion chromatography
- HSQC
heteronuclear single-quantum coherence
- LPS
lipopolysaccharide
- MW
molecular weight
- NMR
nuclear magnetic resonance
- NOESY
nuclear Overhauser effect spectroscopy
- MALS
multi-angle light scattering
- Mh-P1
fraction 1 derived from M. hiantina crude polysaccharide
- Mh-P2
fraction 2 derived from M. hiantina crude polysaccharide
- MS
mass spectrometry
- PMAA
partially methylated alditol acetate
- ROS
reactive oxygen species
- SEC
size exclusion chromatography
- TOCSY
total correlation spectroscopy
Footnotes
Conflict interest statement All authors declare that there is no conflict of interest.
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s10719-022-10092-6.
Data Availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1.Carbajal-Valenzuela IA, Apolonio–Hernandez NM, Gutierrez-Chavez DV, et al. : Biological macromolecules as nutraceuticals. Biol. Macromol 97–138 (2022). 10.1016/B978-0-323-85759-8.00001-4 [DOI] [Google Scholar]
- 2.Olatunde A, Bahattab O, Rauf A, et al. : The importance of biological macromolecules in biomedicine. Biol. Macromol 53–68 (2022). 10.1016/B978-0-323-85759-8.00003-8 [DOI] [Google Scholar]
- 3.Brandley BK, Schnaar RL: Cell-surface carbohydrates in cell recognition and response. J. Leukoc. Biol 40, 97–111 (1986). 10.1002/jlb.40.1.97 [DOI] [PubMed] [Google Scholar]
- 4.Ohtsubo K, Marth JD: Glycosylation in Cellular Mechanisms of Health and Disease. Cell. 126, 855–867 (2006) [DOI] [PubMed] [Google Scholar]
- 5.Hart GW, Copeland RJ: Glycomics hits the big time. Cell. 143, 672–676 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song R, Murphy M, Li C, et al. : Current development of biodegradable polymeric materials for biomedical applications. Drug Design, Development and Therapy 12:3117–3145 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim SK, Li YX: Medicinal benefits of Sulfated Polysaccharides from Sea vegetables. Adv. Food Nutr. Res 64, 391–402 (2011). 10.1016/B978-0-12-387669-0.00030-2 [DOI] [PubMed] [Google Scholar]
- 8.Olafsdottir ES, Ingolfsdottir K: Polysaccharides from Lichens-Structural characteristics. Planta Med. 67, 199–208 (2001) [DOI] [PubMed] [Google Scholar]
- 9.Benkendorff K: Molluscan biological and chemical diversity: secondary metabolites and medicinal resources produced by marine molluscs. Biol. Rev 85, 757–775 (2010) [DOI] [PubMed] [Google Scholar]
- 10.Chakraborty K, Joy M: High-value compounds from the molluscs of marine and estuarine ecosystems as prospective functional food ingredients: An overview. Food Research International 137(2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong Y, Cao C, Ai C, et al. : Structural characterization and immunostimulatory activity of a glucan from Cyclina sinensis. Int. J. Biol. Macromol 161, 779–786 (2020). 10.1016/j.ijbiomac.2020.06.020 [DOI] [PubMed] [Google Scholar]
- 12.Amornrut C, Toida T, Imanari T, et al. : A new sulfated β-galactan from clams with anti-HIV activity. Carbohydr. Res 321, 121–127 (1999). 10.1016/S0008-6215(99)00188-3 [DOI] [PubMed] [Google Scholar]
- 13.World Register of Marine Species Marcia hiantina: (Lamark, 1818). In: http://www.marinespecies.org/aphiaphp?p=taxdetails&id=216583
- 14.Vine P: Red Sea Invertebrates. Immel Publishing, London: (1986) [Google Scholar]
- 15.Huber M: Compendium of Bivalves. A full-color Guide to 3,300 of the World’s Marine Bivalves. A Status on Bivalvia After 250 Years of Research. ConchBooks, Hackenheim: (2010) [Google Scholar]
- 16.Leung K, Lui K, Wai T, et al. : Study on the Soft Shore in Hoi Ha Wan Marine Park. Final Report. the Agriculture & Fisheries and Conservation Department, The Hong Kong SAR Government; (2006) [Google Scholar]
- 17.Gould AA Description of new shells collected by the United States North Pacific Exploring Expedition. Proceedings of the Boston Society of Natural History (1861) [Google Scholar]
- 18.Liu JY, [Ruiyu]: Checklist of Marine Biota of China seas, vol. 1267. China Science Press; (2008) [Google Scholar]
- 19.Cox LR Superfamily Veneracea, in:Treatise_on_invertebrate_paleontology: part N, Mollusca 6, Bivalvia. 2:670–690 (1969) [Google Scholar]
- 20.DuBois Michel, Gilles KA, Hamilton JK, et al. : Colorimetric method for determination of sugars and related substances. Anal. Chem 28, 350–356 (1956). 10.1021/ac60111a017 [DOI] [Google Scholar]
- 21.Coxon B Carbon-13 nuclear magnetic resonance spectroscopy of food-related disaccharides and trisaccharides. Developments in food carbohydrate v. 2 (1980) [Google Scholar]
- 22.Bradbury JH, Jenkins GA: Determination of the structures of trisaccharides by 13 C-n.m.r. spectroscopy. Carbohydr. Res 126, 125–156 (1984). 10.1016/0008-6215(84)85131-9 [DOI] [PubMed] [Google Scholar]
- 23.Bock K, Thøgersen H Nuclear Magnetic Resonance Spectroscopy in the Study of Mono- and Oligosaccharides. Annual Reports on NMR Spectroscopy 13:1–57. (1983). 10.1016/S0066-4103(08)60307-5 [DOI] [Google Scholar]
- 24.King-Morris MJ, Serianni AS: Carbon-13 NMR studies of [1-13 C]aldoses: empirical rules correlating pyranose ring configuration and conformation with carbon-13 chemical shifts and carbon-13/carbon-13 spin couplings. J. Am. Chem. Soc 109, 3501–3508 (1987). 10.1021/ja00246a001 [DOI] [Google Scholar]
- 25.Peña MJ, Tuomivaara ST, Urbanowicz BR, et al. : Methods for structural characterization of the products of cellulose- and xyloglucan-hydrolyzing enzymes. In: Methods in Enzymology, pp. 121–139. Academic Press Inc. (2012) [DOI] [PubMed] [Google Scholar]
- 26.Willis LM, Stupak J, Richards MR, et al. : Conserved glycolipid termini in capsular polysaccharides synthesized by ATP-binding cassette transporterdependent pathways in Gram-negative pathogens. Proc. Natl. Acad. Sci. U S A 110, 7868–7873 (2013). 10.1073/pnas.1222317110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.