Abstract
The tripartite complex AcrAB-TolC is the major RND pump in Escherichia coli and other Enterobacteriaceae. It consists of the AcrB transporter, which is embedded in the inner membrane, the AcrA adapter located in the periplasm, and the channel protein TolC responsible for the transport of substrates towards the extracellular environment. Besides conferring resistance to many classes of antibiotics, AcrAB plays a role in the pathogenesis and virulence of several bacterial pathogens. Here we report that the AcrAB pump heavily affects the infection process of the LF82 strain, the prototype of Adherent-Invasive Escherichia coli (AIEC) which are highly abundant in the ileal mucosa of Chron disease patients. We found that the deletion of genes encoding AcrA and/or AcrB leads to decreased survival of LF82 within macrophages. Ectopic AcrAB expression in a acrAB defective mutant restores the wild type condition. Furthermore, we demonstrate that inhibition of AcrB and replacement of the transporter with an unfunctional AcrB also interfere with bacterial viability inside macrophages. Overall, these data suggest a pivotal role of the AcrAB efflux pump in bacteria-host cell interactions also in AIEC.
Subject terms: Bacteriology, Microbial genetics, Pathogens
Introduction
AcrAB belongs to the resistance nodulation cell-division (RND) efflux pump (EP) family, which is primarily responsible for resistance to a broad class of antibiotics contributing to both intrinsic and horizontally acquired multidrug resistance (MDR)1–4. It represents the major EP in Escherichia coli, Salmonella, and other Enterobacteriaceae where it confers resistance to a large panel of different compounds including antibiotics, dyes, detergents, bile salts, and host-derived molecules5–8. AcrAB-TolC is a tripartite EP composed by an inner membrane transporter, AcrB, an outer membrane channel, TolC and a periplasmic adaptor protein (PAP), AcrA9. The latter protein, in addition to establishing the link between AcrB and TolC, transmits the conformational changes of AcrB to TolC, contributing to the opening of the channel10,11. This EP exerts its function by exploiting the proton motive force represented by the electrochemical potential of H+ across the inner membrane. The energy transduction and substrate transport are spatially separated inside the AcrB transporter12–14. AcrB is an asymmetric homotrimer in which any protomer can assume, not necessarily in a synchronous mode, different conformational states identified as loose (L), tight (T) or open (O)15,16. It has been demonstrated that the periplasmic loop of AcrB is responsible for the substrate specificity of the entire tripartite complex17. AcrB recognizes its specific substrate through a periplasmic access pocket (AP) in the loose state, while in the tight state the substrate binds to a deeper pocket (DP)15,18. AcrAB and its homologues are more widespread than other RND systems and their capability to extrude a large number of different substrates makes them the most clinically relevant among RND EPs3,19,20.
An increasing number of data highlights the role of several MDR EPs, including AcrAB, in different steps of the bacterial pathogencity process, spanning survival within the host cells, biofilm formation, and toxin extrusion21–25. In this context the AcrAB EP has been described to be relevant for the virulence phenotype of several bacterial pathogens. In particular, in S. enterica it has been shown that the deletion of acrB or acrA, as well as the lack of a functional AcrB transporter, impairs epithelial cell invasion and reduces bacterial virulence in in vivo infection models21,26,27. In Enterobacter cloacae Complex (ECC), among the many RND EPs present in the genome, only AcrAB is involved in bacterial pathogenicity. Indeed, E. cloacae acrB or acrA defective strains display attenuated virulence in a Galleria melonella28 and in mouse infection models29. Moreover, inactivation of acrB leads to decreased virulence in Klebsiella pneumoniae, as demonstrated by the low number of defective bacteria recovered from the spleen of infected mice as compared to the wt strain30. Furthermore, Moraxella catarrhalis mutants defective in single components of the AcrAB-OprM EP show a decreased infection ability of cell monolayers, suggesting that all components are required for the full invasion of human nasopharyngeal cells in vitro31. In Neisseria gonorrhoeae the MtrCDE RND efflux system, homologous to the AcrAB-TolC, is required for survival of the pathogen in the female genital tract32. The role of AcrAB has been also demonstrated in plant pathogens as Erwinia amylovora where AcrAB is an efflux complex required for virulence33.
Adherent-Invasive Escherichia coli (AIEC) is a group of pathogenic E. coli isolated at high frequency in patients suffering of Chron’s disease (CD), a severe intestinal inflammatory disease34. Several molecular and cellular studies support a role of AIEC in CD by promoting inflammation via stimulation of the immune system35–37. The AIEC phenotype relies on the ability of the bacteria to adhere and invade intestinal epithelial cells and to survive and replicate extensively inside macrophages without inducing cell death38–40. While many reports stress the relevance of AcrAB in the virulence of several bacterial pathogens, no data are currently available concerning the possible involvement of this EP in the AIEC pathogenicity program. Therefore, in the present study, we have investigated on the role of the AcrAB EP during the infection process of LF82, the prototype strain of AIEC34. We have analysed the intracellular viability of LF82 derivatives lacking either the entire acrAB operon or the single acrA and acrB genes. The results from infections of Caco-2 intestinal epithelial cells and of THP-1-derived macrophages suggest that the loss of AcrAB components greatly affects bacterial survival in the macrophage environment, while viability in epithelial cells is only very marginally touched. The requirement of AcrB transporter activity for intramacrophage survival was confirmed by loss of function approaches using either an EP inhibitor or a LF82 strain expressing an unfunctional AcrB form in the infection experiments. Overall, our data indicate that also in AIEC the AcrAB EP is required for the full expression of pathogenicity.
Results
The lack of AcrAB affects AIEC LF82 survival inside macrophages
In a previous work24 we have analyzed the expression profile of all MDR EPs present in AIEC both under laboratory conditions and during the invasion of epithelial cells and macrophages. The results indicate that the expression of the LF82 acrA gene, selected as reporter of the behavior of the acrAB operon, is very high both under laboratory conditions as well as in intracellular bacteria. On account of the pivotal role of the AcrAB pump in pathogenic bacteria we asked whether this EP is specifically involved in the infection process of the AIEC LF82 strain. To this end, using the one-step method of gene inactivation41, we generated a LF82 derivative carrying a deletion of the entire acrAB operon. The extent of the deletion in LF82 ΔacrAB was verified by sequencing. In LB medium or in tissue DMEM and RPMI culture media the ΔacrAB derivative had a generation time similar to the parental strain (Supplementary Fig. S1). As expected, it showed higher susceptibility to the antibiotics tested (erythromycin, ciprofloxacin, tetracycline) and bile salts (Table 1).
Table 1.
MICs of AcrAB substrates for LF82 and acrA, acrB or both and complementary strains.
| Strain | MIC of substratea (µg/ml) | |||
|---|---|---|---|---|
| ERY | CIP | TET | BS | |
| LF82 (wt) | 200 | 0.25 | 1 | > 1500 |
| LF82 ΔacrAB | 25 | < 0.001 | 0.25 | 750 |
| LF82 ΔacrB | 25 | < 0.001 | 0.25 | 750 |
| LF82 ∆acrA | 12.5 | < 0.001 | 0.25 | 750 |
| LF82 ∆acrAB pacrAB | 200 | 0.2 | 1 | > 1500 |
| LF82 ΔacrB pacrB | 200 | 0.2 | 1 | > 1500 |
| LF82 ΔacrB pacrBD408A | 25 | 0.0015 | 0.25 | 750 |
aERY erythromycin, CIP ciprofloxacin, TET tetracycline, BS bile salts.
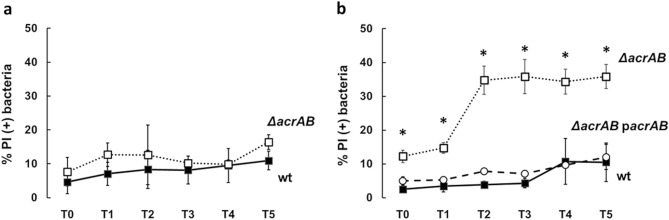
Infection of Caco-2 epithelial cells and THP-1-derived macrophages were carried out with LF82 or its acrAB deletion mutant. To monitor the survival in the intracellular environment, bacteria were recovered at several time points (T0, T1, T2, T3, T4 and T5) post infection (p.i.), and stained with propidium iodide (PI) and DAPI. Subsequently the amount of PI positive bacteria, corresponding to dead cells, was evaluated by fluorescence microscopy. As shown in Fig. 1a most LF82 bacteria, either wt or ∆acrAB, infecting Caco-2 cells remain viable throughout the infection period, indicating that lack of AcrAB does not significantly affect the survival of LF82 in the epithelial cell environment. On the contrary, as compared to the LF82 wt strain the absence of AcrAB induces a significant increase of bacteria that die within the THP-1-derived macrophages. The percentage of PI positive LF82 ΔacrAB is already very high at early stages of infection (T0 and T1) and reaches dramatically higher values two hours p.i. (T2). Indeed, at this time point the PI positive LF82 ΔacrAB cells are more than 30% of the total population. This value remains constant during successive infection time points (Fig. 1b). To confirm the direct contribution of AcrAB to the viability of LF82 in the macrophage environment, we performed infection experiments using the LF82 ΔacrAB complemented with pacrAB, a recombinant plasmid containing the acrAB operon (Supplementary Table S1). The ectopic expression of the acrAB operon fully restores the wt phenotype in the ΔacrAB derivative. Indeed, we observe a percentage of dead bacteria comparable to that of the wt at each time points of infection (Fig. 1b). These data suggest that in AIEC the efflux pump AcrAB plays a significant role during the pathogenic process. It is reasonable to speculate that AcrAB allows LF82 to resist the acidic environment of macrophages, probably by exporting toxic metabolites and/or signaling molecules necessary to maintain the homeostasis of gene expression involved in different metabolic pathways.
Figure 1.
The AcrAB pump affects the intracellular viability of LF82. The viability of intracellular LF82 (black square) and LF82 ΔacrAB (open square) bacteria recovered from Caco-2 epithelial cells (a) and LF82 (black square), LF82 ΔacrAB (open square), LF82 ΔacrAB pacrAB (open circle) recovered from THP-1-derived macrophages (b) at 0, 1, 2, 3, 4, and 5 h p.i. (Referred to as T0, T1, T2, T3, T4 and T5, respectively) was assessed by DAPI/PI double staining. The values are expressed as percentage of PI (+) dead bacteria relative to DAPI (+) bacteria. The results are the average of at least three independent experiments. Error bars represent the SD. The statistical significance was determined by a two tailed student’s t test. *p ≤ 0.01.
The transporter AcrB and the adaptor protein AcrA both contribute to the fitness of AIEC LF82 inside macrophages
It is known that both gene products of the acrAB operon contribute to the export of substrates3. Indeed, the inner membrane transport protein AcrB is critical for the substrate specificity and the periplasmic adapter protein AcrA is important for connecting AcrB to the TolC channel and transmitting AcrB conformational changes to TolC3,9. Based on the results obtained with the LF82 ΔacrAB we asked to what extent AcrB and AcrA contribute to the LF82 survival inside macrophages. The entire acrA or acrB genes were deleted and the growth properties and susceptibility to selected antibiotics of LF82 ΔacrB and LF82 ΔacrA were assessed (Supplementary Fig. S1 and Table 1). After infecting THP-1-derived macrophages with LF82 ΔacrB or LF82 ΔacrA the viability of intracellular bacteria compared to the wt was analyzed at different time points p.i. As shown in Fig. 2 throughout the infection period the percentage of LF82 ΔacrB PI positive bacteria is higher than in the wt. Indeed, as previously observed in the case of LF82 ΔacrAB, the fraction of dead LF82 ΔacrB bacteria is much higher than that of the parental strain starting from one hours p.i. (T1) and reaches 40% at five hours p.i. (T5). In addition, Fig. 2 shows that also the lack of AcrA leads to reduced bacterial viability as compared to the wt: the percentage of dead intracellular LF82 ΔacrA bacteria is around 20% at T1 and T2 p.i and reaches 27% at five hours p.i. (T5). All together these data reveal an important role of the AcrB transporter in the survival of LF82 within the macrophages, quite possibly as a consequence of a unique transport function for specific host-derived compounds. Moreover, the results suggest that AcrA also contributes, though at a lesser extent, to the fitness of LF82 inside macrophages.
Figure 2.
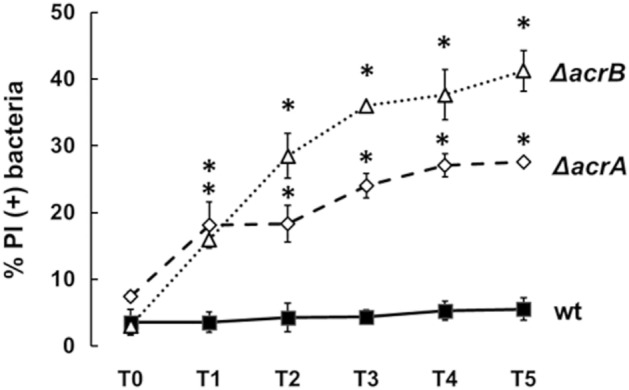
Both the AcrB and AcrA components contribute to survival of LF82 within macrophages. Intracellular LF82 (black square), LF82 ΔacrB (open triangle), and LF82 ΔacrA (open diamond) bacteria were recovered from THP-1 differentiated into macrophages at 0, 1, 2, 3, 4 and 5 h p.i. (referred to as T0, T1, T2, T3, T4 and T5, respectively) and stained with DAPI/PI. The values are expressed as percentage of PI (+) dead bacteria relative to DAPI (+) bacteria. The results are the average of at least three independent experiments. Error bars represent the SD. The statistical significance was determined by a two tailed student’s t test. *p ≤ 0.01.
The AcrB loss of efflux function is responsible for the reduced survival of AIEC LF82 inside macrophages
AcrB is a large abundant transmembrane protein that spans the inner membrane of Gram-negative bacteria. Thus, to ascertain whether the reduced viability of LF82 ΔacrB and ΔacrAB mutants inside macrophages was due to the absence of this sizeable membrane component or to the loss of its specific efflux function, we inhibited the AcrB activity or used an LF82 strain expressing an unfunctional form of AcrB.
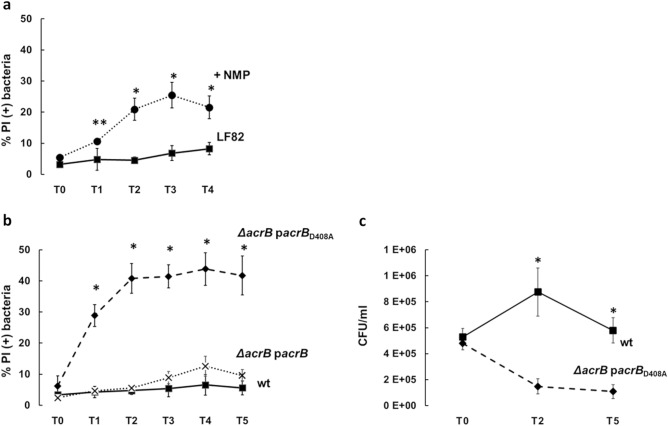
AcrB function was inhibited by treatment with 1-(1-naphthyl-methyl)-piperazine (NMP), a well characterized efflux pump inhibitor (EPI) in Gram-negative bacteria42, used at 100 µg/ml, a concentration known to induce an inhibitory effect on drug efflux in commensal E. coli43. We verified whether NMP has any influence on the behavior of bacterial and THP-1 single cultures. The inhibitor did not affect LF82 viability nor had toxic effects on THP-1 cells as assessed by measuring LDH (Lactate dehydrogenase) release in the medium (data not shown). To investigate on the phenotype induced by AcrB inhibition we carried out infection experiments of THP-1 cells differentiated into macrophages with the LF82 wt strain in the presence or absence of NMP. The inhibitor (100 µg/ml) was added to the bacterial cells in RPMI medium, just before exposing them to the macrophages and the effect of the treatment was monitored every hour for four hours. As shown in Fig. 3a, AcrB inhibition significantly affects the viability of intracellular bacteria. A mild increase of intracellular dead bacteria (10%) in the presence of NMP was observed already at one hour p.i. (T1). This proportion reaches 21% at two hours p.i. (T2) and further rises (25%) at time point T3. At four hours p.i. the percentage of intracellular dead bacteria slightly decreases although it remains significantly higher than in the absence of the NMP inhibitor. Interestingly, the AcrB transporter function clearly influences the behavior of LF82 bacteria facing the host cell environment, as bacterial viability in the extracellular environment is comparable with or without the NMP compound (data not shown). By and large, the data we obtained from AcrB inhibition experiments follow those from experiments performed with the LF82 ΔacrB mutant, indicating that the efflux function of AcrB is required for LF82 survival inside the macrophage phagolysosome.
Figure 3.
The lack of AcrB function causes loss of LF82 viability inside macrophages. (a) Effect of NMP on LF82 survival inside macrophages. THP-1-derived macrophages were infected with LF82 untreated (black square) or treated (black circle) with 100 µg/ml NMP. (b) Impact of the presence of the unfunctional AcrB D408A trasporter on LF82 survival within macrophages THP-1 derived macrophages were infected with LF82 (black square) and LF82 ∆acrB complemented with either the wild type (x) or the unfunctional (black diamond) AcrB D408A transporter. In (a) and (b) intracellular bacteria were recovered at the indicated time points and double stained with DAPI/PI. The values are expressed as percentage of PI (+) dead cells relative to DAPI (+) cells. (c) Viable counts (CFU/ml) of intracellular LF82 (black square) and LF82 ∆acrB complemented with the unfunctional AcrB D408A transporter (black diamond). At the indicated time points, infected THP-1 cells were lysed and dilutions of intracellular bacteria plated on LB agar. The results shown are the average of at least three independent experiments. Error bars represent the SD. The statistical significance was determined by a two tailed student’s t test. *p ≤ 0.01; **p ≤ 0.05.
To further determine whether the observed decrease in viability of intracellular bacteria was due to loss of AcrB function, we constructed a LF82 derivative incapable of efflux activity via AcrB. To this end we first generated the pacrB plasmid carrying the acrB gene (Supplementary Table S1). Then, the A at position 1223 of acrB was replaced with C in the plasmid pacrB giving rise to the D408A amino acid substitution in the AcrB protein (pacrBD408A). It has been demonstrated that in Salmonella typhimurium the D408A substitution disrupts proton translocation in the pump and abolishes efflux activity27. Both pacrB and pacrBD408A plasmids were used to transform the LF82 ΔacrB strain, obtaining the LF82 ΔacrB pacrB and LF82 ΔacrB pacrBD408A derivatives, respectively. The latter strain displays a membrane protein composition similar to that of the wt, with the AcrB protein lacking the transporter function. To evaluate if the amino acid substitution could affect bacterial fitness, we analyzed the growth properties of LF82 and LF82 ΔacrB pacrBD408A in LB medium. The results show that LF82 ΔacrB pacrBD408A has a similar generation time as the wt, indicating that the mutation does not confer a detectable growth defect in LB medium (Supplementary Fig. S1). We also determined the susceptibility of the LF82 ΔacrB pacrBD408A strain to bile salts and antibiotics. As for the LF82 ΔacrB mutant, also LF82 ΔacrB pacrBD408A is hypersusceptible to bile salts, with a twofold dilution decrease in the MIC as compared to the parental strain (Table 1). As expected, the susceptibility of the LF82 ΔacrB pacrB derivative to bile salts and antibiotics matches that of wt strain.
To assess whether the LF82 ΔacrB strain expressing an unfunctional AcrB still showed impaired survival ability, we compared the viability of LF82 wt with that of LF82 ΔacrB pacrB and LF82 ΔacrB pacrBD408A derivatives during macrophage infection by determining the percentage of PI positive intracellular bacteria at different time points p.i. As shown in Fig. 3b already at one hour p.i. (T1) cell death caused by the expression of the unfunctional AcrB D408A is almost seven-fold higher (28%) than in the wild type (4%). It further increases (40%) at two hours p.i. (T2) and remains at approximately this value throughout the infection period. Importantly, the viability curve of intracellular LF82 ΔacrB pacrB overlaps that of the parental strain. The phenotype conferred by the unfunctional AcrB D408A was further corroborated by determining the viable bacteria by the CFU assay. As shown in Fig. 3c, the CFU/ml at representative time points (T0, T2 and T5) nicely mirror the survival curves defined by the “Live and Dead” assay. These results further confirm that the efflux activity associated with AcrB is critical for LF82 persistence and multiplication inside macrophages, ruling out the possibility that the effect observed in the absence of AcrB might be due to the simple lack of this large and abundant inner membrane protein.
Discussion
We present evidence supporting the crucial role of AcrAB EP in the infection process of a prototype AIEC strain, LF82. In particular, our data, based on infection experiments with mutant strains lacking efflux pump components AcrB and/or AcrA, clearly indicate that both proteins are required for AIEC persistence and replication inside THP-1 derived macrophages. Furthermore, chemical inhibition of AcrB transporter activity in the wt LF82 and loss of function experiments point to a pivotal role of the AcrB efflux function in the survival of AIEC inside the macrophage phagolysosome.
As a major efflux pump in Escherichia coli, Salmonella, and other Enterobacteriaceae AcrAB-TolC is often used as model of the RND family, the main class of efflux pumps (EPs) involved in the emergence of multidrug resistance (MDR). The physiological role of the AcrAB EP is much more complex than that of a simple antibiotic export system as it is known to confer resistance to a variety of compounds (e.g. dyes, detergents, and host-derived molecules)1,9. In particular, in E. coli AcrAB contributes to the colonization and adaptation of the bacteria to the intestinal tract by exporting bile salts8,44, and its function has been associated with the virulence of several pathogenic bacteria21,29,30,32,33,45. Among different E. coli pathogens, AIEC represent a specific group associated with Chron’s disease, a severe intestinal inflammation syndrome. AIEC colonize intestinal epithelial cells promoting the disruption of the intestinal barrier34,39. They also enter macrophages, persisting and replicating within phagolysosomes without triggering cell death40. Persistence inside this harsh environment requires prompt bacterial adaptation, which implies the ability to keep the bacterium free of dangerous compounds. In this respect, we have previously reported how the expression of MDR EPs encoded by AIEC strain LF82 is specifically modulated during the infection process. In particular, we found that the expression levels of fsr and mdtL, which encode MDR EPs of the MFS family, and mdtE, which encodes an EP of the RND family, are highly expressed in macrophages24. We were also able to prove that the MdtEF EP is involved in increasing the fitness of AIEC inside macrophages.
In the present study we demonstrate that AcrAB contributes to the survival of AIEC LF82 in the macrophage environment as evidenced by increased bacterial death after infection of THP1-derived macrophages with a AcrAB-defective mutant (LF82 ΔacrAB). The percentage of dead LF82 ΔacrAB bacteria is higher than that of the wt strain throughout the infection period considered. The capability of ectopically expressed acrAB genes to restore the intracellular viability in LF82 ΔacrAB confirms the contribution of the AcrAB EP to the fitness of AIEC in the macrophage niche and highlights the relevance of this EP in such a hostile environment.
It is known that AcrA and AcrB have specific functions and contribute to the full functionality of the EP. The evidence presented here indicates that the inactivation of AcrA or AcrB both leads to decreased bacterial viability in macrophages. These data are in agreement with the results obtained in other pathogenic bacteria showing that each component of the AcrAB complex plays a specific role in the virulence, pathogenesis and survival within infected host cells26,29,45. Interestingly, we observed that the percentage of dead LF82 ΔacrA cells during macrophage infection is lower than that of the LF82 ΔacrB strain. It has been recently reported that the AcrB transporter is able to interact with the periplasmic adapter protein AcrE46, which might functionally compensate the absence of AcrA thus explaining the milder death percentage of LF82 ΔacrA and highlighting the predominant role of AcrB in the extrusion of toxic compounds present in the phagolysosome. The relevance of AcrB is further strengthened by the inactivation of the EP achieved by adding a specific inhibitor (NMP) during infection of macrophages. The presence of NMP impairs the LF82 viability at very early time point of infection (Fig. 3a). We were also able to rule out that the inability of LF82 ΔacrAB and LF82 ΔacrB to survive inside macrophages is due to a profound alteration of the inner membrane because of the lack of an abundant constituent such as AcrB. Indeed, complementation of LF82 ΔacrB with an AcrB protein (AcrB D408A) lacking the efflux activity does not recover the wild-type viability (Fig. 3b,c).
Overall, our data support the view that in order to better survive intracellularly LF82 cells require the ability of AcrB to select and bind different substrates, thus allowing them to escape the dangerous compounds present in the inhospitable macrophage environment. It has been reported that in Salmonella the AcrAB EP is involved in the infection of both macrophages and epithelial cells, being required for adhesion to and invasiveness of host cells21,26,27. Here we mainly focused on the requirement of the AcrAB EP for LF82 survival inside host cells, demonstrating that its activity is needed for AIEC persistence and replication within the human THP-1-derived macrophages, allowing a successful infection. As for the role of AcrAB during the infection of epithelial cells we find that it does not influence the survival of LF82 within Caco-2 cells. The evidence we have presented here confirms the clear involvement of AcrAB in the interactions between host cell and bacteria and extends the range of bacterial pathogens that exploit the AcrAB function to cope with the hurdles encountered in the intracellular environment.
Methods
Bacterial strains, plasmids and growth conditions
Bacterial strains and plasmids used in this study are listed in Supplementary Table S1. Escherichia coli LF82 is an AIEC strain isolated from a chronic ileal lesion of a Chron Disease patient34. E. coli DH10b has been used as the recipient in cloning experiments. Strains LF82 ΔacrAB, LF82 ΔacrB, and LF82 ΔacrA have been constructed using the one-step method of gene inactivation41 by transforming LF82 pKD46 with an amplicon obtained using pKD13 or pKD4 as template and the oligo pairs ABF/ABR for the acrAB deletion, BF-BR for the acrB deletion and AF-AR for the acrA deletion (Supplementary Tables S1 and S2). Plasmids pacrAB and pacrB were obtained by cloning the acrAB and acrB genes respectively into pGIP7, a pACYC184-derived vector carrying the ptac promoter47 (Supplementary Table S1). The acrAB and acrB amplicons obtained using LF82 as template and oligo pairs pACacrABF/pACacrABR and pACacrBF/pACacrBR respectively (Supplementary Table S2) were digested with BamHI and cloned into pGIP7 downstream the ptac promoter. Plasmids pacrAB and pacrB and the size of the of acrAB, acrB and acrA deletions have been verified by DNA sequencing (Biofab, Rome). The D408A mutation of acrB was obtained by a Site-Directed Mutagenesis System (GENEART®) using the oligo pair pACacrB-D408A F/pACacrB-D408A R (Supplementary Table S2). In particular, we replaced the A at position 1223 (from the ATG of acrB) with C, generating a codon change from GAC to GCC (D408A). The presence of the correct mutation on pacrBD408A has been verified by DNA sequencing (Biofab, Rome).
Growth kinetics of LF82 and its derivatives in different media (LB, DMEM or RPMI) were measured using a CLARIOstar plate reader (BMG LABTECH, Offenburg, Germany). When required, Congo Red was added (0.01%) to Trypticase soy agar to monitor the Congo Red phenotype (CR+). Antibiotics and inhibitors were used at the following concentrations: ampicillin 30 µg/ml, chloramphenicol 25 μg/ml, ciprofloxacin 0.0015 to 0.25 μg/ml; erythromycin 25 to 200 μg/ml; kanamycin 30 μg/ml; gentamicin 10 μg/ml or 100 μg/ml for infection procedures; tetracycline 0.25 to 1 μg/ml; 1-(1-naphthyl-methyl)-piperazine (NMP) 100 μg/ml. Bile salts were used at concentrations ranging from 750 to 1500 μg/ml when required.
General procedures
DNA purification, restriction, cloning, plasmid transformation and gel electrophoresis were perfomed as described23,47. Oligonucleotides were based on the LF82 genome48 and are listed in Supplementary Table S2. PCR reactions were set up with DreamTaq DNA polymerase (Thermo Fisher Scientific) or Ex taq DNA polymerase (Takara). DNA and protein sequence comparisons were performed using the BLAST Server (National Center for Biotechnology Information).
Determination of MICs of different compounds
MICs were determined following previously established protocols as recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (EUCAST 2000). In particular, strains LF82, LF82 ΔacrAB, LF82 ΔacrA, LF82 ΔacrB and LF82 ΔacrAB pacrAB, LF82 ΔacrB pacrB and LF82 ΔacrB pacrBD408A were inoculated into 3 ml of LB and grown at 37 °C with shaking for 16 h. Cultures were then diluted to OD600 0.02 in LB and 100 µl aliquots were transferred to microtiter wells containing 100 µl of the compounds to be tested (at appropriate concentrations). After 16 h incubation at 37 °C growth was estimated by measuring the OD600 of each well. At least two biological replicates were performed.
Cell cultures and infections
The THP-1 cell line was grown in RPMI 1640 (Gibco) medium containing 10% heat-inactivated fetal bovine serum (FBS) (Euroclone), 0.05 IU/ml penicillin and 0.05 IU/ml streptomycin (PS), referred to as RF10, at 37 °C in a humidified 5% CO2 atmosphere. Before bacterial infection, THP-1 monocytes were differentiated into macrophages. Cells were seeded in 6-well tissue culture plates (Falcon) at 1.0 × 106 cells/well in RF10 supplemented with 50 nM PMA. After 48 h, PMA containing medium was removed and cells left for further 24 h in RF10. Two hours before bacterial addition, RF10 was replaced with fresh RPMI without serum and antibiotics. The human epithelial colorectal adenocarcinoma Caco-2 cell line was grown in Dulbecco minimal essential medium (DMEM) (Gibco) containing 10% FBS and PS, referred to as DF10, at 37 °C in a humidified 5% CO2 atmosphere. For bacterial infection cells were seeded in 6-well tissue culture plates (Falcon) at 4.0 × 105 cells/well in growth medium. After 48 h cells were serum-starved over-night in DMEM supplemented with 0.5% FBS and PS (DF0.5). Two hours before bacterial infection, the medium was replaced with fresh DMEM without serum and antibiotics. Bacteria were added to the cell cultures at MOI 100. The plates were then centrifuged (15 min, 750×g) and incubated 30 min (THP-1) or 45 min (Caco-2) at 37 °C under 5% CO2 atmosphere to allow bacteria to enter the cells. Finally, extracellular bacteria were removed by washing three times with PBS. This point was taken as time zero (T0). Fresh medium (RPMI or DMEM) containing gentamicin (100 μg/ml) was added to kill extracellular bacteria, and infected cells were incubated at 37 °C up to 4 or 5 h.
Live and dead assay and viable bacterial count
Intracellular dead bacteria were evaluated by staining the entire population with DAPI (Sigma) and labelling dead cells with Propidium Iodide (PI) (Sigma). At the indicated time points, infected cells were lysed by adding 1% Triton X-100 in 1 × PBS. The cell lysate was pelleted at 13,000 rpm for 5 min. The pellet containing intracellular bacteria was washed once in 1 × PBS and suspended in 1 × PBS containing 10 μg/ml DAPI and 15 μM PI. Samples were incubated 20 min at room temperature in the dark. Bacteria were centrifuged at 13,000 rpm for 5 min and washed once with 1 × PBS. Pellet was resuspended in 20 μl 1× PBS containing 50% glycerol, 5 μl of the stained bacteria were added to the glass slide and overlaid with the coverslip for immediate observation. Stained cells were examined using the NIKON ECLIPSE 50i fluorescence microscope. Single images were recorded on QICAM FAST 1394 camera (Qimaging) and imported to ImageJ software for processing (Supplementary Fig. S2).
To define the number of viable intracellular bacteria, macrophage infection was carried out as describe above. At the indicated time points, after lysing the host cells with 1% Triton X-100, intracellular bacteria were collected, washed and resuspended in PBS. Serial dilutions of the bacterial suspensions were plated on LB agar plates to calculate the CFU/ml.
LDH cytotoxicity assay
To verify that the NMP inhibitor does not affect cell viability, cytotoxicity was measured by using CyQUANT™ LDH Cytotoxicity Assay Kit (Invitrogen). THP-1-derived macrophages were grown in RPMI with or without NMP (100 µg/ml) in tissue culture dish (35 X 10 mm, Falcon) at 1.0 × 106 cells/well in growth medium and LDH activity in the medium was determined after 2- or 4-h treatment following manufacturer instructions. Samples containing maximum LDH activity and spontaneous LDH activity were also prepared as well as the LDH positive control. The absorbance has been measured at 490 nm and 680 nm with a CLARIOstar plate reader (BMG LABTECH, Offenburg, Germany) and the percentage of cytotoxicity was calculated according to the manufacturer’s instructions.
Statistical analyses
The statistical difference between the percentage of dead intracellular bacteria of the LF82 wt strain and its derivative strains at each time point was determined by a two-tailed student’s t-test. The experiments were run at least in triplicate.
Supplementary Information
Acknowledgements
We gratefully acknowledged the late Arlette Darfeuille Michaud and her lab for providing strain LF82. This research was supported by a grant from the Italian Ministry of Foreign Affairs and International Cooperation (JP21GR04). The funders have no role in study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conceived and designed the experiments: G.F., M.G., B.C. Performed the experiments: G.F., M.P. Analyzed the data: G.F., M.G., G.P. Contributed reagents/materials/analysis tools: B.C., G.P. Wrote the paper: G.F., M.G., B.C. All the authors review the manuscript.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Milena Grossi, Email: milena.grossi@uniroma1.it.
Bianca Colonna, Email: bianca.colonna@uniroma1.it.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-29817-0.
References
- 1.Blair JM, Piddock LJ. Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: An update. Curr. Opin. Microbiol. 2009;12:512–519. doi: 10.1016/j.mib.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Nikaido H. Multidrug resistance in bacteria. Annu. Rev. Biochem. 2009;78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du D, et al. Multidrug efflux pumps: Structure, function and regulation. Nat. Rev. Microbiol. 2018;16:523–539. doi: 10.1038/s41579-018-0048-6. [DOI] [PubMed] [Google Scholar]
- 4.Henderson PJF, et al. Physiological functions of bacterial ‘multidrug’ efflux pumps. Chem. Rev. 2021;121:5417–5478. doi: 10.1021/acs.chemrev.0c01226. [DOI] [PubMed] [Google Scholar]
- 5.Ma D, et al. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 6.Zgurskaya HI, Nikaido H. Bypassing the periplasm: Reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1999;96:7190–7195. doi: 10.1073/pnas.96.13.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikaido E, Yamaguchi A, Nishino K. AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J. Biol. Chem. 2008;283:24245–24253. doi: 10.1074/jbc.M804544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urdaneta V, Casadesús J. Adaptation of Salmonella enterica to bile: Essential role of AcrAB-mediated efflux. Environ. Microbiol. 2018 doi: 10.1111/1462-2920.14047. [DOI] [PubMed] [Google Scholar]
- 9.Alav I, et al. Structure, assembly, and function of tripartite efflux and type 1 secretion systems in gram-negative bacteria. Chem. Rev. 2021;121:5479–5596. doi: 10.1021/acs.chemrev.1c00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure. 2006;14:577–587. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lobedanz S, et al. A periplasmic coiled-coil interface underlying TolC recruitment and the assembly of bacterial drug efflux pumps. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4612–4617. doi: 10.1073/pnas.0610160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pos KM. Drug transport mechanism of the AcrB efflux pump. Biochim. Biophys. Acta Proteins Proteom. 2009;1794:782–793. doi: 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 13.Zhang XC, Liu M, Han L. Energy coupling mechanisms of AcrB-like RND transporters. Biophys. Rep. 2017;3:73–84. doi: 10.1007/s41048-017-0042-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, et al. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. Elife. 2017;6:1–19. doi: 10.7554/eLife.24905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramaswamy VK, Vargiu AV, Malloci G, Dreier J, Ruggerone P. Molecular rationale behind the differential substrate specificity of bacterial RND multi-drug transporters. Sci. Rep. 2017;7:1–18. doi: 10.1038/s41598-017-08747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tam HK, et al. Allosteric drug transport mechanism of multidrug transporter AcrB. Nat. Commun. 2021;12:1–10. doi: 10.1038/s41467-021-24151-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elkins CA, Nikaido H. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominately by two large periplasmic loops. J. Bacteriol. 2002;184:6490–6498. doi: 10.1128/JB.184.23.6490-6499.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Webber A, Ratnaweera M, Harris A, Luisi BF, Ntsogo Y. A model for allosteric communication in drug transport by the AcrAB-TolC tripartite efflux pump. Antibiotics. 2022;11:52. doi: 10.3390/antibiotics11010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piddock LJV. Multidrug-resistance efflux pumps—Not just for resistance. Nat. Rev. Microbiol. 2006;4:629. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 20.Alvarez-Ortega C, Olivares J, Martínez JL. RND multidrug efflux pumps: What are they good for? Front. Microbiol. 2013;4:1–11. doi: 10.3389/fmicb.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buckley AM, et al. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol. 2006;8:847–856. doi: 10.1111/j.1462-5822.2005.00671.x. [DOI] [PubMed] [Google Scholar]
- 22.Blanco P, et al. Bacterial multidrug efflux pumps: Much more than antibiotic resistance determinants. Microorganisms. 2016;4:1–19. doi: 10.3390/microorganisms4010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasqua, M. et al. The MFS efflux pump EmrKY contributes to the survival of Shigella within macrophages. Sci. Rep. (2019). [DOI] [PMC free article] [PubMed]
- 24.Fanelli G, Pasqua M, Colonna B, Prosseda G, Grossi M. Expression profile of multidrug resistance efflux pumps during intracellular life of Adherent-Invasive Escherichia coli strain LF82. Front. Microbiol. 2020;11:1935. doi: 10.3389/fmicb.2020.01935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasqua M, et al. Host–bacterial pathogen communication: The wily role of the multidrug efflux pumps of the MFS family. Front. Mol. Biosci. 2021;8:1–13. doi: 10.3389/fmolb.2021.723274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blair JMA, La Ragione RM, Woodward MJ, Piddock LJV. Periplasmic adaptor protein AcrA has a distinct role in the antibiotic resistance and virulence of Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 2009;64:965–972. doi: 10.1093/jac/dkp311. [DOI] [PubMed] [Google Scholar]
- 27.Wang-Kan X, et al. Lack of AcrB efflux function confers loss of virulence on Salmonella enterica serovar Typhimurium. MBio. 2017;8:1–14. doi: 10.1128/mBio.00968-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guérin F, et al. Pumps in Enterobacter cloacae Complex. Antimicrob. Agents Chemother. 2016;60:2373–2382. doi: 10.1128/AAC.02840-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pérez A, et al. Involvement of the AcrAB-TolC efflux pump in the resistance, fitness, and virulence of Enterobacter cloacae. Antimicrob. Agents Chemother. 2012;56:2084–2090. doi: 10.1128/AAC.05509-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padilla E, et al. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 2010;54:177–183. doi: 10.1128/AAC.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spaniol V, Bernhard S, Aebi C. Moraxella catarrhalis AcrAB-OprM efflux pump contributes to antimicrobial resistance and is enhanced during cold shock response. Antimicrob. Agents Chemother. 2015;59:1886–1894. doi: 10.1128/AAC.03727-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jerse AE, et al. A gonococcal efflux pump system enhances bacterial survival in a female mouse model of genital tract infection. Infect. Immun. 2003;71:5576–5582. doi: 10.1128/IAI.71.10.5576-5582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burse A, Weingart H, Ullrich MS. The phytoalexin-inducible multidrug efflux pump AcrAB contributes to virulence in the fire blight pathogen, Erwinia amylovora. Mol. Plant Microbe Interact. 2004;17:43–54. doi: 10.1094/MPMI.2004.17.1.43. [DOI] [PubMed] [Google Scholar]
- 34.Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn’s disease. Infect. Immun. 1999;67:4499–4509. doi: 10.1128/IAI.67.9.4499-4509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carvalho FA, et al. Crohn’s disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J. Exp. Med. 2009;206:2179–2189. doi: 10.1084/jem.20090741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bretin A, et al. Activation of the EIF2AK4-EIF2A/eIF2α-ATF4 pathway triggers autophagy response to Crohn disease-associated adherent-invasive Escherichia coli infection. Autophagy. 2016;12:770–783. doi: 10.1080/15548627.2016.1156823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Douadi C, et al. Anti-TNF agents restrict Adherent-invasive Escherichia coli replication within macrophages through modulation of Chitinase 3-like 1 in patients with Crohn’s disease. J. Crohns Colitis. 2022;16:1140–1150. doi: 10.1093/ecco-jcc/jjab236. [DOI] [PubMed] [Google Scholar]
- 38.Glasser AL, et al. Adherent invasive Escherichia coli strains from patients with Crohn’s disease survive and replicate within macrophages without inducing host cell death. Infect. Immun. 2001;69:5529–5537. doi: 10.1128/IAI.69.9.5529-5537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaler CR, Elhenawy W, Coombes BK. The unique lifestyle of Crohn’s disease-associated adherent-invasive Escherichia coli. J. Mol. Biol. 2019;431:2970–2981. doi: 10.1016/j.jmb.2019.04.023. [DOI] [PubMed] [Google Scholar]
- 40.Demarre GL, et al. The Crohn’s disease-associated Escherichia coli strain LF82 relies on SOS and stringent responses to survive, multiply and tolerate antibiotics within macrophages. PLoS Pathog. 2019;15:2090. doi: 10.1371/journal.ppat.1008123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lomovskaya O, et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrob. Agents Chemother. 2001;45:105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bohnert JA, Kern WV. Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob. Agents Chemother. 2005;49:849–852. doi: 10.1128/AAC.49.2.849-852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thanassi DG, Cheng LW, Nikaido H. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 1997;179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webber MA, et al. The global consequence of disruption of the AcrAB-TolC efflux pump in Salmonella enterica includes reduced expression of SPI-1 and other attributes required to infect the host. J. Bacteriol. 2009;191:4276–4285. doi: 10.1128/JB.00363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McNeil HE, et al. Identification of binding residues between periplasmic adapter protein (PAP) and RND efflux pumps explains PAP-pump promiscuity and roles in antimicrobial resistance. PLoS Pathog. 2019;15:1–28. doi: 10.1371/journal.ppat.1008101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leuzzi A, et al. Role of the SRRz/Rz1 lambdoid lysis cassette in the pathoadaptive evolution of Shigella. Int. J. Med. Microbiol. 2017;307:268–275. doi: 10.1016/j.ijmm.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 48.Miquel S, et al. Complete genome sequence of Crohn’s disease-associated adherent-invasive E coli strain LF82. PLoS ONE. 2010;5:e12714. doi: 10.1371/journal.pone.0012714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.