Abstract
ZBTB7A belongs to a small family of transcription factors having three members in humans (7A, 7B, and 7C). They share a BTB/POZ protein interaction domain at the amino end and a zinc-finger DNA-binding domain at the carboxyl end. They control the transcription of a wide range of genes, having varied functions in hematopoiesis, oncogenesis, and metabolism (in particular glycolysis). ZBTB7A-binding profiles at gene promoters contain a consensus G(a/c)CCC motif, followed by a CCCC sequence in some instances. Structural and mutational investigations suggest that DNA-specific contacts with the four-finger tandem array of ZBTB7A are formed sequentially, initiated from ZF1–ZF2 binding to G(a/c)CCC before spreading to ZF3–ZF4, which bind the DNA backbone and the 3′ CCCC sequence, respectively. Here, we studied some mutations found in t(8;21)-positive acute myeloid leukemia patients that occur within the ZBTB7A DNA-binding domain. We determined that these mutations generally impair ZBTB7A DNA binding, with the most severe disruptions resulting from mutations in ZF1 and ZF2, and the least from a frameshift mutation in ZF3 that results in partial mislocalization. Information provided here on ZBTB7A–DNA interactions is likely applicable to ZBTB7B/C, which have overlapping functions with ZBTB7A in controlling primary metabolism.
Keywords: ZBTB7A, C2H2 zinc-finger transcription factors, ZBTB7A somatic mutations, AML t(8;21), cancer metabolism
Abbreviations: AML, acute myeloid leukemia; CAST, cyclic amplification and selection of target; EGFP, enhanced GFP; FP, fluorescence polarization; GST, glutathione-S-transferase; MDACC, MD Anderson Cancer Center; NLS, nuclear location signal; PDB, Protein Data Bank; TCEP, Tris(2-carboxyethl)phosphine; ZF, zinc-finger
ZBTB7A is a member of BTB/POZ protein interaction domain–containing C2H2 zinc-finger (ZF) proteins (Fig. 1A). This protein is also known as FBI (for factor that binds to the inducer of short transcripts (1)), LRF (for leukemia/lymphoma-related factor (2, 3)), OCZF (for osteoclast-derived zinc finger (4)), and POKEMON (for POK erythroid myeloid ontogenic factor (5)), reflecting its multifaceted functions. It is part of a family of three proteins (in humans ZBTB7A, ZBTB7B, and ZBTB7C) that are very similar at the amino end (BTB/POZ) and the ZF-containing carboxyl end, though differing substantially in between (Fig. S1A). ZBTB7A represses the transcription of a wide range of genes (6, 7), including the repression of HIV-1 short transcripts (8); silencing of the tumor suppressor ARF (5), which is expressed from an alternative reading frame of the CDKN2A gene; and repression of glycolytic genes (SLC2A3 [solute carrier family 2, member 3], PFKP, and PKM) (9) and fetal hemoglobin genes (10, 11, 12). Very recently, increased levels of ZBTB7A were shown to modulate host genes related to oxidative responses, leading to coronavirus persistence rather than cell death (13).
Figure 1.
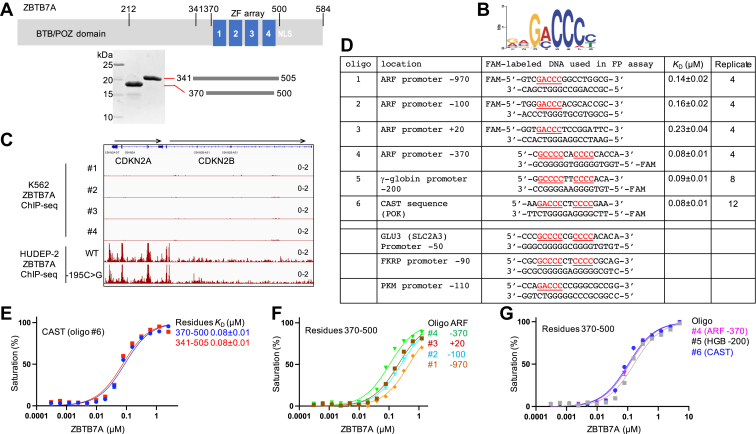
ZBTB7A binds at promoter of p14ARF.A, schematic diagram of ZBTB7A protein and two recombinant fragments of DNA-binding domain used in the study. B, ZBTB7A-binding motif (taken from the study of Ref. (10)). C, a snapshot of ZBTB7A binding at the promoter of the CDKN2A gene (which gives rise to p14ARF protein); taken from Gene Expression Omnibus under accession GSE103445. Chromatin immunoprecipitation sequencing (ChIP-Seq) data in K562 cells (four replicates) and HUDEP-2 (WT and -195C>G mutant), normalized to track scale (0–2) shown to the right. D, a list of oligonucleotides (#1–#6) used in the study. The KD value represents the mean ± SD of N number of independent determinations as listed. E–G, the DNA-binding assay of ZBTB7A fragments against varied sequences, using fluorescence polarization.
Somatic mutations of ZBTB7A have been identified routinely in acute myeloid leukemia (AML) patients, associated with the t(8;21) translocation (14, 15, 16, 17, 18, 19). This t(8;21) (q22;q22) translocation results in a RUNX1–RUNX1T1 fusion gene (20, 21), which is one of the most frequent chromosomal aberrations in AML. Despite the causative role of the RUNX1–RUNX1T1 fusion in leukemia initiation, this fused gene alone is insufficient to induce leukemia (22, 23, 24, 25, 26). In fact, ectopic expression of wildtype ZBTB7A prevents RUNX1–RUNX1T1-mediated clonal expansion of human CD34+ cells (27), and retroviral expression of wildtype ZBTB7A inhibits cell growth in the t(8;21) translocation–positive AML cell line Kasumi-1 (15) (possibly by inhibiting glycolysis, see Discussion section). Recent studies of a population mutational spectrum have suggested that additional genetic lesions, including ZBTB7A mutations, are required for development of RUNX1–RUNX1T1-associated diseases (16, 19) as well as in a variety of solid tumors (28).
Given the importance of understanding the role of naturally occurring ZBTB7A mutations in disease development, we studied selected ZBTB7A mutations within the C-terminal ZF DNA-binding domain and their effects on binding to sequence elements from the ZBTB7A-responsive promoters. We used biophysical methods to investigate the detailed interactions of the ZBTB7A DNA-binding domain with relevant promoter elements and determined precisely how known human ZBTB7A mutations interfere with DNA binding and cellular localization. In sum, our study provides significant insights into DNA sequence recognition by both native and disease-associated mutants of the human transcription factor ZBTB7A.
Results
ZBTB7A DNA-binding domain binds multiple p14ARF promoter sequences
Previous ZBTB7A chromatin immunoprecipitation sequencing experiments were carried out in human K562 (myelogenous leukemia), HUDEP-2 (umbilical erythroid progenitor), and hematopoietic stem and progenitor–derived cells (10, 11). These studies identified ZBTB7A-binding profiles containing a consensus G(a/c)CCC motif (Fig. 1B) and detected a strong peak of ZBTB7A occupancy in the promoter of the CDKN2A gene (p14ARF) in HUDEP-2 cells but not in K562 cells (Fig. 1C; this difference is discussed later). The human p14ARF promoter region contains multiple putative ZBTB7A-binding sites (5), so we chose the three at −970, −100, and +20 that include the GaCCC motif (oligos #1–3 in Fig. 1D; position numbers are relative to the first transcribed base). We chose three additional oligonucleotides for comparison. One is the −370 p14ARF site, which includes a GcCCC motif (oligo #4), which resembles the −200 element of the fetal globin promoter (oligo #5) in that both contain an 11 bp segment with two stretches of C:G base pairs separated by two random base pairs. Finally, we included a ZBTB7A consensus binding sequence (oligo #6), which had been identified by cyclic amplification and selection of targets (CAST) (5), which combines the two features together: a GaCCC motif as well as a segment of four C:G base pairs.
Previously, we used a ZBTB7A recombinant fragment including residues 370 to 500, which encompassed the four-finger DNA-binding domain, in a study of binding to the fetal globin promoter (12). During the course of that study, AlphaFold (DeepMind) (29) released protein structure predictions for the human proteome including ZBTB7A, which predicted two additional folded helices preceding the ZF domain (Fig. S1, B and C). We thus generated a longer fragment, including residues 341 to 505, that covers the N-terminal putative helices plus five additional amino acids (including two basic residues) at the C terminus (Fig. 1A). However, using fluorescence polarization (FP), we found that both fragments bind the 16-bp CAST sequence (oligo #6) equally well (Fig. 1E). The more N- and C-proximal additions to the ZF region contributed little to the binding affinities. Specifically, the dissociation constant KD values (∼80 nM) were approximately the same for the two constructs.
Next, we compared the binding affinities for the four p14ARF promoter sequences (oligos #1–4) (Fig. 1F). Oligos #1 to 3, each containing a single GaCCC element, exhibited 2 to 3 times reduced binding affinity relative to that of oligo #4. Oligo #4, taken from the ARF promoter −370 site, has the same affinity (KD ∼ 80 nM) as that of the CAST oligo and the −200 site from the fetal globin promoter (Fig. 1G). All three higher-affinity oligos (#4–6) share the same features of a 5′ G(a/c)CCC motif and a 3′ C:G-rich segment. This enhanced affinity is understandable given that the predicted recognition sequence for a four-ZF array is approximately 12 bp, using the rule of one-finger recognizing three adjacent DNA base pairs (30, 31). A 5-bp GaCCC motif should be bound by just two fingers, which would presumably result in reduced binding affinity relative to sequences bound by all four fingers.
In addition to p14ARF promoter, others showed that ZBTB7A binds to the promoters of glycolytic genes (SLC2A3, PFKP, and PKM) (9), which contain the G(a/c)CCC motif (Fig. 1D). SLC2A3 encodes the glucose transmembrane transporter GLUT3 (32). Once inside the cell, glucose is converted to pyruvate via a cascade of ten chemical reactions (33) (glycolysis; Fig. S2). In addition to the three characterized glycolytic genes, we found that ZBTB7A occupies the promoters of all ten glycolytic genes in K562 or HUDEP-2 cells or both (Fig. S2). This control of glycolytic genes is independent from that of globin genes, as mutant HUDEP-2 cells harboring the -195C-to-G substitution at the γ-globin gene promoter showed no effect on the binding activity of ZBTB7A at the promoters of the glycolytic genes (Fig. S2).
Furthermore, an RNA-Seq analysis of ZBTB7A knockout K562 cells revealed increased expression of the genes for glucose transporters (SLC2A1 and SLC2A3), phosphoglycerate mutases (PGM2 and PGM3), and enolase (ENO2). However, there was no change in expression of the kinase genes involved in glycolysis—hexokinase (HK1), pyruvate kinase (PKM), and phosphofrutokinase (PFKP and PFKM) (27).
Structure of ZBTB7A DNA-binding domain in complex with GACCC element
To gain insight into the structural basis for ZBTB7A recognition of the GaCCC motif, we cocrystallized the four-finger DNA-binding domain (residues 370–500) with three 15-bp DNA oligos, each containing a single GACCC with 5′ overhangs or blunt ends (Fig. 2). These complexes crystallized in three different space groups, and the structures were determined to resolutions of 2.25, 2.85, and 3.09 Å, respectively (Table S1). In space groups C2221 (Protein Data Bank [PDB] ID: 7N5W) and P21212 (PDB ID: 7N5U), we observed the three fingers ZF1–ZF3, whereas ZF4 was disordered (Fig. 2, A and B). In both space groups, ZF1 and ZF2 lie in the DNA major groove, whereas ZF3 points away from DNA in varied directions.
Figure 2.
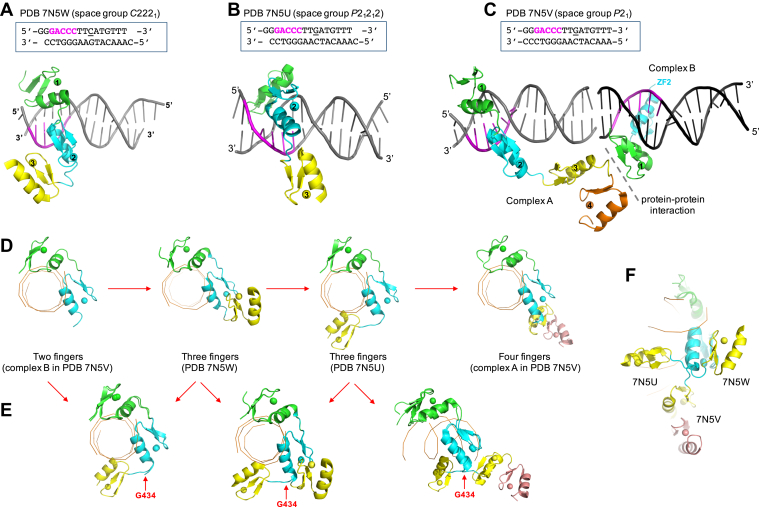
Structures of ZBTB7A DNA-binding domain in complex with GaCCC. In all cases, ZF1 is shown in green, ZF2 in cyan, ZF3 in yellow, and ZF4 in salmon. A–C, ZBTB7A–GaCCC complexes in space group C2221 (Protein Data Bank [PDB] ID: 7N5W) (A), space group P21212 (PDB ID: 7N5U) (B), and space group P21 (PDB ID: 7N5V) (C). In the P21 structure, there are two complexes (A and B). D, a view along the DNA axis of ZBTB7A–GaCCC complexes for structures consisting of two-finger (ZF1–2), three-finger (ZF1–3), and four-finger (ZF1–4). E, pairwise comparisons of different conformations of ZF3 (colored in yellow) with a switch point at Gly434 of the linker between ZF2 and ZF3. F, the observed three different conformations of ZF3. ZF, zinc finger.
In space group P21, we observed two protein–DNA complexes (Fig. 2C). Complex A contains all four fingers, with ZF1–ZF2 binding in the DNA major groove and ZF3 and ZF4 pointing away from DNA (Fig. 2C). The ZF3–ZF4 pair is stabilized via protein–protein interactions with complex B, which harbors only the first two fingers (Fig. 2C). In sum, we observed four ZBTB7A–DNA complexes that included a GaCCC element (Fig. 2D). In all four complexes, ZF1–ZF2 were very similar, having an rmsd of <1 Å. The main differences between these four complexes involved crystal packing interactions with neighboring molecules, resulting in different conformations of ZF3–ZF4, some of which were disordered and lacked electron density.
In the absence of specific contacts, ZF3–ZF4 adopted various conformations (Fig. 2E), and this was achieved via a series of rotations of main-chain torsion angles involving the glycine residue in the linker between the two fingers (i.e., Gly434 between ZF2 and ZF3). This glycine—the most flexible of amino acids—is conserved in the corresponding linkers between pairs of fingers (bright red in Fig. 3A). In fact, these glycines are very highly conserved among vertebrate orthologs of ZBTB7A as phylogenetically distant from humans as whale sharks (Fig. S1D). An equivalent conformational switch point within the linker was previously identified in the 11-finger protein CTCF (34), suggesting that the flexibility of the linker between pair of fingers plays a critical role, perhaps allowing the multifinger array greater versatility in its sequence recognition, as well as in binding varied lengths of DNA duplex.
Figure 3.
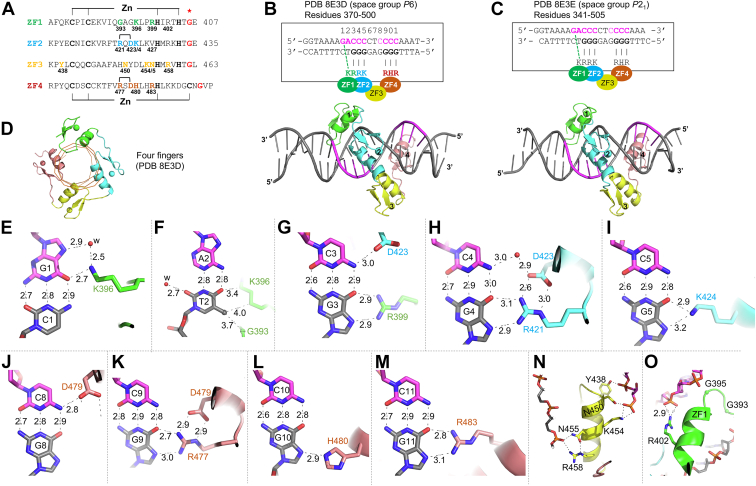
Structures ofZBTB7A in complex with CAST sequence containing two stretches of C:G-rich sequence: details of ZBTB7A–base interactions.A, sequence alignment of the four ZFs of ZBTB7A, highlighting the zinc ligands (C2H2 in ZF1–3 and C3H1 in ZF4), DNA base–interacting residues (colored to match ZF1, ZF2, and ZF4), DNA–phosphate interactions by ZF3, the Arg–Glu (RD) electrostatic pair in ZF2 and ZF4, and the glycine switch point of the linkers between the two ZF units (asterisk; bright red). B and C, ZBTB7A–CAST complexes using blunt-end DNA (Protein Data Bank ID: 8E3D) or 5′ overhang DNA (PDB ID: 8E3E). The 11-bp recognition sequence is numbered by position #s of 1 to 11. Basic residues of ZF1, ZF2, and ZF4 making base-specific interactions are colored as in A. D, a view of the ZBTB7A–CAST complex with four fingers wrapped along the DNA. E–M, examples of base-specific contacts, taken from PDB 8E3D. Interatomic distances are measured in Å. E, Lys396 of ZF1 interacts with guanine G1 (as defined in B). F, the methyl group of thymine T2 is involved in van der Waals contacts. G, Arg399 of ZF1 and Asp423 of ZF2 interact with the C3:G3 base pair. H, Arg421 of ZF2 interacts with guanine G4. Asp423 forms an ion pair with Arg421. I, Lys424 of ZF2 interacts with guanine G5. J, Asp479 of ZF4 is in contact with cytosine C8. K, Arg477 of ZF4 interacts with guanine G9. Asp479 forms an ion pair with Arg479. L, His480 of ZF4 interacts with guanine G10. M, Arg483 of ZF4 interacts with guanine G11. N, ZF3 makes a number of DNA–phosphate interactions. O, Gly395 and Arg402 of ZF1 are in close contact with DNA–phosphate groups. CAST, cyclic amplification and selection of target; ZF, zinc finger.
Structure of ZBTB7A DNA-binding domain in complex with CAST sequence
Next, we cocrystallized the CAST sequence (oligo #6), which contains both a 5′ GaCCC motif and 3′ four C:G bp, in complex with the short (residues 370–500) and the long fragments (residues 341–505) of ZBTB7A. As noted previously, both fragments bind the DNA equally (Fig. 1E), and both fragments crystallized, in the space groups of P6 and P21, respectively (Fig. 3, B and C). The complex structures were determined to resolutions of 2.62 and 2.99 Å, respectively (Table S1). Although crystallized in two different space groups, the two structures are highly similar, with an rmsd of <0.9 Å over 109 pairs of Cα atoms. The additional N-terminal residues included in the longer fragment were disordered, and we did not observe the AlphaFold-predicted helices (Fig. S1C).
All four ZF units occupy and wrap along the DNA major groove (Fig. 3D). The first two fingers, ZF1 and ZF2, interact with the 5′ sequence GaCCC, ZF3 provides a spacer (via sequence-independent DNA phosphate binding), and ZF4 interacts with the 3′ sequence (CCCC). As in conventional protein–DNA interactions (35, 36), seven C:G base pairs (at bp positions 1, 3, 4, 5, 9, 10, and 11; labeled in Fig. 3B) were recognized by lysine, arginine, and histidine residues via direct hydrogen bonds with the O6 and N7 atoms of guanines: guanine G1 by Lys396 of ZF1 (Fig. 3E), G3 by Arg399 of ZF1 (Fig. 3G), G4 by Arg421 of ZF2 (Fig. 3H), G5 by Lys424 of ZF2 (Fig. 3I), G9 by Arg477 of ZF4 (Fig. 3K), G10 by His480 of ZF4 (Fig. 3L), and G11 by Arg483 of ZF4 (Fig. 3M). Two negatively charged residues, Asp423 of ZF2 and Asp479 of ZF4, provide a hydrogen bond with cytosines C3 and C8, respectively (Fig. 3, G and J). The A:T base pair at position 2 forms van der Waals contacts with the Cα atom of Gly393 and the side chain aliphatic carbon of Lys396 (via the methyl group of T2) (Fig. 3F). All these DNA-contacting residues of the human ortholog are fully conserved among other vertebrate orthologs of ZBTB7A (Fig. S1D), and 9 of 10 are conserved among the human ZBTB7A family members ZBTB7B and 7C (Fig. S1A). Among the 11 bp covered by the four ZF units, two base pairs at positions 6 and 7 are not involved in direct interactions with the DNA bases, and these correspond to the position of ZF3 (which makes base-independent backbone contacts).
Among the base-specific interactions, seven of them are in the bottom strand (T2, G3, G4, G5, G9, G10, and G11), whereas the G1, C3, and C8 are in the top strand. In general, more-specific DNA-binding proteins recognize both strands (37). The cross-strand interaction of Lys396–G1 has been previously observed in Lys413 of KLF4 and Lys328 of ZNF410 (38, 39). The ability to switch, between Lys396–G1 interaction in GaCCC and Lys396–G2 interaction of the opposite strand of GcCCC (12) (akin to the Lys424–G5 interaction [Fig. 3I]), allows ZBTB7A to bind sequences that vary at the second position of G(a/c)CCC. In addition, the cross-strand interaction between Asp423–C3 and Asp47–C8 are common among the ZF units containing an Arg–Asp electrostatic stabilization pair, for example, Arg421–Asp423 of ZF2 (Fig. 3H) and Arg477–Asp479 of ZF4 (Fig. 3K).
Most importantly, our structural characterization of ZBTB7A in complex with DNA elements illustrated that ZF1–ZF2 binding of G(a/c)CCC is nearly identical in structure with either the full consensus sequence or just the 5′ part. In addition, ZBTB7A displayed the greatest reduction in binding affinity in response to single-base pair substitutions in the DNA sequence recognized by ZF1 and ZF2, followed by a smaller impact on binding in DNA recognized by ZF4, and the least in DNA protected by ZF3 (12). These observations suggest that the specific contacts with the ZF array (in this case, four fingers) are formed sequentially; putatively starting with ZF1–ZF2 binding to GaCCC before spreading to ZF3–ZF4, which respectively bind to the backbone and 3′ end of CCCC sequence. However, it is also possible that ZF3 binding contributes to nonspecific DNA association during the period of short-range linear diffusion (sliding) while searching for specific binding sites (40, 41, 42).
The involvement of two fingers versus four fingers might reflect the observed differences in DNA-binding affinities (Fig. 2D). Although ZF3 is not involved in DNA base–specific interactions, it did contribute direct contacts with the DNA phosphate groups via five residues (Tyr438, Asn450, Lys454, Asn455, and Arg458; Fig. 3N). All five of these phosphate-binding residues are fully conserved among vertebrate orthologs (Fig. S1D), and in human ZBTB7C, though there are two substitutions in ZBTB7B (N450S and R458H; Fig. S1A). ZF3 also forms interfinger interactions with ZF2 and ZF4. These ZF3-mediated interactions are important for positioning the succeeding ZF4 appropriately into the DNA major groove.
Effects of ZBTB7A mutations on DNA binding and localization
We next considered naturally occurring disease-associated mutations within the ZBTB7A DNA-binding domain (Fig. 4A). In a t(8;21)-AML study, recurring ZBTB7A mutations were identified in 23% of 56 patients, including missense and truncating mutations resulting in alteration or loss of one or more ZF units (15). Furthermore, ∼43% of ZBTB7A missense mutations recorded in human cancer genomic databases are located in the ZF DNA-binding domain (28). We considered G395C and R402H/C of ZF1, K424T of ZF2, and R458fs of ZF3 (fs indicates a frameshift mutation) in the context of the DNA-binding domain (residues 370–500). These residues are indicated in Figure 4A. To avoid potential crosscontamination from the chromatography system, we expressed and purified manually the five mutants and wildtype proteins as glutathione-S-transferase (GST) fusions using fresh glutathione agarose beads (Fig. 4A). The mutants G395C, R402C, and K424T have similar solubility, whereas R402H has reduced solubility and R458fs has increased solubility (comparing lanes 3 and 4 in Fig. 4B).
Figure 4.
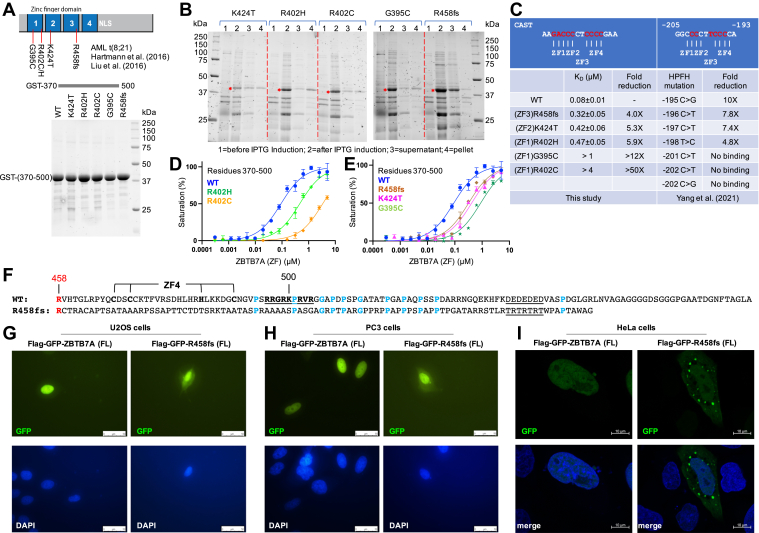
The effects of AML t(8;21)-associated ZBTB7A mutations on DNA binding and localization.A, five somatic mutations of ZBTB7A and GST-tagged DNA-binding domain proteins used in the DNA-binding assays. B, five mutants were simultaneously expressed and purified together with wildtype protein using six sets of GST affinity chromatography. The ∗ indicates the induced protein band. C, summary of FP-based binding assays. Each KD value is presented as fitted value ± SD against 15 data points for every protein variant. The effects on ZBTB7A binding of hereditary persistence of fetal hemoglobin (HPFH)–associated mutations at the −200 element of the fetal globin promoter are included for comparison (two rightmost columns) (12). D and E, FP binding in two separate plots for clarity. F, sequence alignment of wildtype and R458fs mutant. G and H, images of fluorescence microscopy (Nikon Eclipse E800) showing the localization patterns of FLAG-GFP-ZBTB7A full-length proteins or R458fs mutant in U2OS and PC3 cells, with nuclei being stained with 4′,6-diamidino-2-phenylindole (DAPI). The scale bar represents 50 μm. I, images of confocal microscopy (Zeiss LSM880) showing the localization patterns in HeLa cells. The scale bar represents 10 μm. AML, acute myeloid leukemia; FP, fluorescence polarization; GST, glutathione-S-transferase.
In comparison with wildtype protein, under constant conditions, ZBTB7A mutants demonstrated approximately four times reduced binding for R458fs, followed by mutations at K424T and R402H (∼5–6×), G395C (>12×), and R402C (>50×) (Fig. 4C). The reductions in binding were the greatest with respect to ZF1 mutants (G395C and R402C) and the least with the ZF3 mutant (R458fs) (Fig. 4D and E). In comparison, ZBTB7A showed no binding to DNA containing single-base pair substitutions in the base pairs recognized by R399 of ZF1 and R421 of ZF2, with lesser effects of substitutions in the DNA recognized by ZF4, and the least for substitutions in the segment protected by ZF3 (12) (Fig. 4C).
We note that Arg402 is located in the helix of ZF1 and electrostatically interacts with a DNA backbone phosphate (Fig. 3O). Substituting this arginine with Cys or His would ablate the direct contact. Because histidine can carry a positive charge, the reduction in R402H binding is milder than that of the cysteine substitution (R402C). Like Arg402, substitution of Lys424 with the smaller and uncharged threonine (K424T) would ablate the base-specific interaction with guanine G5 (Fig. 3I). Similarly, a cancer-associated mutation of Lys365-to-Thr (K365T) in ZF4 of CTCF, found in endometrial cancer cells (43), results in reduced DNA binding (34). Gly395 is located in the top of the loop prior to the ZF1 helix and is positioned deeply into the major groove of DNA (Fig. 3O). Its substitution with cysteine (G395C) could yield repulsion between DNA and the mutant protein. A previous study revealed that the R402H/C missense mutants, an N-terminal frameshift mutation (A175fs), and a C-terminal truncation (R377X) just prior to the ZF DNA-binding domain, each failed to repress a luciferase reporter containing ZBTB7A-binding elements derived from the ARF promoter (in contrast to wildtype ZBTB7A) (15).
Interestingly, the mutation that had the least impact (∼4×) was the frameshift mutation of R458fs of ZF3 (a single nt deletion of the last nt of the CGC R458 codon, at c.1374del, which eliminates translation of the entire ZF4 element). The reduced DNA binding is similar in strength to that with the substrates containing only the GaCCC motif (which requires the binding of only ZF1–2), in comparison to the CAST sequence (Fig. 1D). This observation agrees with previous data showing that removing ZF4 or both ZF3 and ZF4 did not eliminate DNA binding (8, 12). We note that the region immediately after ZF4 contains an arginine/lysine-rich sequence (497-RRGRKPRVR-504) that potentially could serve as a nuclear location signal (NLS) (44), and the frameshift mutation would remove this signal (Fig. 4F).
We examined the localization of ZBTB7A and its corresponding R458fs variant protein, by transiently expressing GFP-ZBTB7A or GFP-R458fs in three human cell lines: osteosarcoma-derived U2OS, prostate cancer–derived PC3, and cervical carcinoma–derived HeLa (Fig. 4, G–I). As seen previously, WT ZBTB7A localized exclusively to the nuclei (15, 45), whereas R458fs showed a diffused pattern with localization in both the nucleus and cytoplasm. Thus, we conclude that the R/K-rich sequence after ZF4 is a functional NLS that is required for the complete nuclear localization of ZBTB7A, and that the frameshift mutation R458fs results in partial mislocalization (suggesting that ZBTB7A might possess more than one NLS). An early study suggested that deletion of ZF1 or ZF2 completely abolished nuclear localization, suggesting that the same ZFs primarily responsible for DNA binding are also required for subcellular targeting (45). Interestingly, using confocal microscopy, we observed that R458fs variant proteins form cytoplasmic speckles in some HeLa cells (Fig. 4I), at least superficially reminiscent of stress granules (46).
This R/K-rich sequence is partially conserved in ZBTB7B and ZBTB7C and may serve the same function in those proteins (Fig. S1A). Interestingly, while R458fs eliminated ZF4 and the NLS, the frameshift mutation preserves the ten structural residues (proline and glycine) flanking the NLS (Fig. 4F). This sequence conservation after a frameshift is quite striking, but nothing about the sequence surrounding the deletion suggests it would be a hot spot for indels, such that conservation in the alternative reading frame would have been selected for. The altered cytoplasmic localization of R458fs is the same as observed for A175fs and R377X mutants in transiently transfected U2OS cells (15).
Discussion
ZBTB7A as a proto-oncogene/tumor suppressor
Earlier studies suggested a proto-oncogene function for ZBTB7A, owing to its ability to repress tumor suppressor genes p14ARF and Rb in various tissues and transgenic mice (5, 47). While the importance of p14ARF inactivation in cancer is less clear (at least in some cases, see Ref. (48)), in patients with t(8;21)-positive AML, the ZBTB7A mutations are predicted to derepress p14ARF. Nevertheless, a tumor-suppressive role of p14ARF in t(8;21)-positive AML has not been established. We note a differing occupancy by ZBTB7A in the CDKN2A promoter in HUDEP-2 and K562 cells; where CDKN2A specifies the p14ARF protein (Fig. 1C). We do not know whether this is just an inconsistency of ZBTB7A occupancy between the two cell lines, or if instead, the repressor binding is no longer detectable once p14ARF is fully expressed in K562 cells. An opposite observation was made regarding ZBTB7A occupancy at the γ-globin gene promoter: ZBTB7A binds in K562 cells but not in HUDEP-2 cells (11).
In more recent studies, including work in t(8;21)-positive AML (15, 27), a tumor suppressor function of ZBTB7A has been suggested: wildtype ZBTB7A prevents cancer progression, whereas ZBTB7A deletion and somatic mutations promote cancer progression (9, 28, 49). The suggestion is that ZBTB7A has this effect via its transcriptional repression of genes for glycolytic enzymes. Metabolic reprogramming, in which glycolysis is elevated, is associated with a variety of cancers (50, 51, 52). ZBTB7A represses the transcription of several genes implicated in glycolysis (SLC2A3, PFKP, and PKM) in several human cancer cells (9); whereas ZBTB7A knockdown in colon carcinoma cells HCT116 yielded increased glycolysis and proliferation (9); and loss of ZBTB7A sensitizes K562 cells to glycolysis inhibition (27). On top of this, our exploration of data for the other glycolytic genes (Fig. S2) suggest that the effects of ZBTB7A involve all ten genes. Thus, mutations of ZBTB7A in t(8;21)-positive AML will likely increase glycolysis and help tumor cells to produce more energy for growth.
ZBTB7A and AML
While ZBTB7A is thus relevant to AML, fewer than a quarter of t(8;21)-positive AML patients (among 56 patients) carry ZBTB7A mutations (15). In ZBTB7A-WT patients, the expression level of ZBTB7A is unknown. Because high expression of wildtype ZBTB7A was associated with a favorable outcome in patients with cytogenetically normal AML (15), increased expression of nonmutant ZBTB7A, via alterations of ZBTB7A upstream regulators (53) and/or epigenetic changes (54), might prevent or slow down disease progression in t(8;21)-positive AML patients. This possibility deserves exploration.
Comparison to other ZF proteins
Here, we focused our work on DNA-binding domain of ZBTB7A and provided significant insights into DNA sequence recognition by both the native protein and disease-associated mutants. ZBTB7A contains four ZFs, and the first two fingers are essential for the binding of the G(a/c)CCC motif (8, 12), whereas the last two fingers contribute more modestly to overall DNA-binding affinity. The two-finger array might be the minimal unit for efficient DNA binding by ZF proteins. For example, the two fingers of Zfp57 recognize the TGCCGC element at the imprinting control regions (55), and the two fingers of HIC2 recognize TGCCAA in erythroid BCL11A enhancers (56). However, unlike the conventional two-finger protection of six base pairs, ZF1–ZF2 of ZBTB7A only recognizes five base pairs, owing to small side chains (G393 of ZF1 and V427 of ZF2; Fig. 3A) that, in more conventional ZFs, have bulkier and polar/charged residues engaging in base-specific contacts. On the other hand, the variability in ZF3–ZF4 is common for the ends of ZF arrays when they are less involved in direct DNA binding, for example, HIC2 (56), ZFP568 (57), and CTCF (34).
As the DNA-recognition residues are conserved among the three family members (Fig. S1A), the information provided here on ZBTB7A–DNA interactions is most likely to be directly applicable to ZBTB7B and ZBTB7C, which might have overlapping functions with ZBTB7A in controlling primary metabolism. Mutations of ZBTB7B (also known as ThPOK for T-helper-inducing POZ-Krüppel-containing protein) have been associated with pathways that control energy intake and expenditure in obesity (58). ZBTB7C (also known as Kr-POK for kidney cancer–related POZ and Krüppel-containing protein) is involved in the regulation of fatty acid biosynthesis (59) and blood glucose (60).
Future directions and summary
Our current structural work is limited to the DNA-binding domain. In the longer term, it is essential to understand the protein conformation of full-length ZBTB7A, and whether domains outside the ZF domain affect the structure and DNA binding of the ZF domain. Our study showed that residues 341 to 380, preceding ZF domain, do not fold into helical structures (under our conditions, and in contrast to AlphaFold predictions) and contribute little to DNA binding. They are also not well conserved. Another current limitation to our knowledge is that we do not know whether ZBTB7A mutations have dominant negative effects. Continued work may also reveal whether substitutions of the base-interacting residues (e.g., K424T) gain alternative binding sites.
In summary, our work illuminated the complex nature of ZBTB7A as a double-edged sword. As aforenoted, the mutations or deletion of ZBTB7A in t(8;21)-positive AML probably increases glycolysis and helps tumor cells to produce more energy for growth, though the same mutations or deletion of ZBTB7A could disrupt protein–DNA interactions at the fetal globin promoter to alleviate fetal globin silencing and treat β-hemoglobinopathies (61). This combination of advantages or disadvantages is probably typical for a master transcription factor that controls the expression of many genes.
Experimental procedures
Structure quality and refinement statistics are listed in Table S1. The plasmids used for protein expression in Escherichia coli are listed in Table S2. All DNA oligonucleotides used are listed in Table S3.
Protein expression and purification
The fragments of human ZBTB7A (residues 370–500, pXC2222; residues 341–505, pXC2311) were cloned into pGEX-6P-1 vector with a GST fusion tag. Both plasmids were separately transformed into E. coli strain BL21-Codon-plus (DE3)-RIL. Bacteria were grown in lysogeny broth (62) at 37 °C until reaching midlog phase (absorbance at 600 nm between 0.4 and 0.5), at which point the incubation temperature was set to 16 °C and 25 μM ZnCl2 was added to the cell culture. When the culture temperature reached 16 °C and an absorbance reached ∼0.8 at 600 nm, addition of 0.2 mM IPTG induced gene expression for 20 h. Cell harvesting and protein purification were carried out at 4 °C through a three-column chromatography protocol, conducted in a BIO-RAD NGC system. Cells were collected by centrifugation, and the pellet was suspended in lysis buffer (20 mM Tris–HCl [pH 7.5], 5% glycerol, 0.5 mM Tris(2-carboxyethl)phosphine [TCEP], supplemented with 0.5 M NaCl and 25 μM ZnCl2). Cells were lysed by sonication, and 0.3% (w/v) polyethylenimine was slowly titrated into the cell lysate before centrifugation (63). The debris was removed by centrifugation for 30 min at 47,000g, and the supernatant was loaded onto a 5 ml GSTrap column (GE Healthcare). The resin was washed with lysis buffer, and bound protein was eluted in 100 mM Tris–HCl [pH 8.0], 0.5 M NaCl, 5% glycerol, 0.5 mM TCEP, and 20 mM glutathione (reduced form). The proteins were digested with PreScission protease (produced in-house) to remove the GST fusion tag. The cleaved proteins were loaded onto a 5 ml Heparin column (GE Healthcare). The proteins were eluted by NaCl gradient from 0.25 to 1 M in the lysis buffer. The peak fractions were pooled, concentrated, loaded onto a HiLoad 16/60 Superdex S200 column (GE Healthcare), and equilibrated with 0.25 M NaCl-supplemented lysis buffer. The preparations were frozen and stored at −80 °C for crystallography and DNA-binding assays.
The mutants, in the context of GST fusion (residues 370 to 500), containing substitutions of G395C (pXC2282), R402H (pXC2280), R402C (pXC2281), R458fs (pXC2283), or K424T (pXC2279), were generated using PCR-based mutagenesis and confirmed by sequencing. To avoid potential crosscontamination from the chromatography system, we expressed and purified manually the five mutants and wildtype proteins as GST fusions using fresh glutathione agarose beads. Samples were then flash frozen and stored at −80 °C for DNA-binding assays.
FP
The FP experiments (63, 64) were carried out to measure the DNA-binding affinity, using a Synergy 4 Microplate Reader (BioTek). Aliquots of 6-carboxy-fluorescein-labeled DNA duplexes (5 nM final concentration; sequences indicated in Figs. 1 and 4) were incubated in 20 μl with varied amount of proteins (0–2.5 μM) in 20 mM Tris–HCl (pH 7.5), 225 mM NaCl, 5% glycerol, and 0.5 mM TCEP for 10 min at room temperature. The mixture was transferred to a black opaque 384-well plate before measurement. The data were processed using GraphPad Prism (version 8.0; GraphPad Software, Inc) with equation [mP] = [maximum mP] × [C]/(KD +[C]) + [baseline mP], in which mP is millipolarization and [C] is protein concentration. The KD value for each protein–DNA interaction was derived from N replicate experiments (as indicated in Fig. 1D). For those binding curves that did not reach saturation, the lower limit of the binding affinity was estimated. Raw FP data are shown in Fig. S3.
Protein localization
For the plasmid expressing enhanced GFP (EGFP)-ZBTB7A (pXC2315), the complementary DNA fragment encoding EGFP was amplified by PCR from pLJM1-EGFP vector (purchased from Addgene) with forward and reverse primers (Table S3) and subcloned into the pcDNA3-ZBTB7A vector (12) using HindIII and EcoRI restriction sites. The plasmid expressing mutant EGFP-ZBTB7A-R458fs (pXC2327) was generated using PCR-based mutagenesis in the context of pXC2315 plasmid. The resulting constructs were verified by sequencing.
For plasmid transfection, cells (PC3, U2OS, and HeLa) were grown on coverslips and then transfected with EGFP-ZBTB7A or EGFP-R458fs plasmid using Lipofectamine 3000 transfection reagent (Invitrogen; catalog no.: L3000) following the manufacturer’s instructions. At 24 h post-transfection, cells were washed twice with PBS and fixed in 4.0% paraformaldehyde solution in PBS for 10 min at room temperature. After washing with PBS (3×, 5 min each), cells were incubated with 4′,6-diamidino-2-phenylindole solution (Thermo Fisher Scientific) in 1:800 dilution in PBS for 10 min at room temperature, rinsed with dH2O twice, and then mounted with mounting solution (Vector Laboratories; H-1200). Transfected PC3 and U2OS cells were imaged with a Nikon Eclipse E800 Fluorescence Microscope, whereas transfected HeLa cells were mounted with ProLong Gold Antifade Mountant (Life Technologies; catalog no.: P36930) and imaged via confocal microscopy (Zeiss LSM880) at 63× (1.4 numerical aperture) objective.
X-ray crystallography
The purification of ZBTB7A DNA-binding domain (residues 370–500) used for cocrystallization was described recently (12). The DNA oligonucleotides (Table S1) were synthesized by Integrated DNA Technologies. The concentrated ZBTB7A fragment (0.9 mM) was mixed with oligonucleotides (annealed in 10 mM Tris–HCl [pH 7.5] and 50 mM NaCl) at a molar ratio 1:1.2 of protein:DNA and incubated on ice for 30 min. An Art Robbins Gryphon Crystallization Robot was used to set up screens of the sitting drop at ∼19 °C via vapor diffusion. The complex crystals (PDB ID: 7N5V) were obtained under the condition of 0.1 M Bis–Tris (pH 6.5), 20% (w/v) PEG monomethyl ether 5000. The complex crystals (PDB ID: 7N5U) were grown under the condition of 4% (v/v) tacsimate (pH 5.0) and 12% (w/v) PEG3350. The complex crystals (PDB ID: 7N5W) were grown under the condition of 0.1 M sodium malonate (pH 6.0) and 10% (w/v) PEG3350. The complex crystals (PDB ID: 8E3D) were grown under the condition of 0.2 M potassium chloride and 20% (w/v) PEG3350. Finally, the complex crystals (PDB ID: 8E3E) of ZBTB7A residues 341 to 505 were grown under the condition of 0.1 M sodium malonate (pH 7.0) and 12% (w/v) PEG3350.
Crystals were flash frozen using 20% (v/v) ethylene glycol as the cryoprotector. The X-ray diffraction data were collected at SER-CAT 22-ID beamline of the Advanced Photon Source at Argonne National Laboratory utilizing an X-ray beam at 1.0 Å wavelength. The diffraction data were measured quantitatively and processed with HKL2000 (65). The structure (PDB ID: 7N5U) was determined by ZnSAD (single-wavelength anomalous dispersion) to give an interpretable map with initial figure-of-merit of 0.45 (Table S1). Anomalous signal was examined using the PHENIX Xtriage module, which reported an anomalous signal to 5 Å for the dataset utilized for our initial phasing. The PHENIX AutoSol module (66) found three zinc atom positions and gave a density-modified map with an R-factor of 0.40. The initial electron density showed recognizable molecular features of the β-sheets and α-helices of the ZFs and helical DNA. Reinserting the zinc positions into AutoSol and utilizing the full resolution of the dataset gave a better map allowing for our initial model build.
This structure of PDB 7N5U allowed molecular replacement for the other structures with careful manual (re)building of ZF3 and ZF4 as needed. Molecular replacement for beginning determination of other structures was performed with the PHENIX PHASER module (67) and further model building commenced with COOT (68). Some single ZFs had weak electron density, and their manual placement in the map was necessary. Zinc positions were verified by anomalous difference maps using the diffraction data where Bijvoet mates were separated. Manual building proceeded in COOT to (re)build the protein and the DNA duplex. COOT was also utilized for corrections between PHENIX refinement rounds.
All structure refinements were performed by PHENIX Refine, with 5% randomly chosen reflections for validation by R-free values. Structure quality was analyzed during PHENIX refinements and later validated by the PDB validation server (Table S1). Molecular graphics were generated using PyMol (Schrödinger, LLC).
Data availability
All plasmids generated in this study are available with a completed materials transfer agreement. The X-ray structures (coordinates and structure factor files) of the ZBTB7A ZF domain with bound DNA have been deposited to PDB and are publicly available as of the date of publications. Accession numbers are PDB 7N5U, PDB 7N5V, PDB 7N5W, PDB 8E3D, and PDB 8E3E.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interests with the contents of this article.
Acknowledgments
We thank Prof Merlin Crossley of University of New South Wales for providing the initial ZBTB7A construct; Ms Yu Cao of MD Anderson Cancer Center (MDACC) for technical assistance throughout the study; Dr Collene R. Jeter of MDACC for help with confocal microscopy and Dr Margarida A. Santos of MDACC for comments on the article. We thank the beamline scientists of the Southeast Regional Collaborative Access Team at the Advanced Photon Source, Argonne National Laboratory, USA. The use of the Southeast Regional Collaborative Access Team is supported by its member institutions and equipment grants from the US National Institutes of Health (grant nos.: S10_RR25528, S10_RR028976, and S10_OD027000). Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract W-31-109-Eng-38. The work was supported by the US National Institutes of Health (grant no.: R35GM134744) and the Cancer Prevention and Research Institute of Texas (grant no.: RR160029). The use of the Flow Cytometry and Cell Imaging Core facility is supported by CPRIT grant RP170628.
Author contributions
X. Z. and X. C. conceptualization; X. C. methodology; B. L., Y. H., and R. M. B. formal analysis; R. R., J. R. H., and Q. C. investigation; X. C. writing–original draft; R. M. B. writing–review & editing; J. Y. validation; X. Z. supervision; X. Z. project administration; X. C. funding acquisition.
Funding and additional information
X. C. is a CPRIT Scholar in Cancer Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Eric Fearon
Contributor Information
Xing Zhang, Email: xzhang21@mdanderson.org.
Xiaodong Cheng, Email: xcheng5@mdanderson.org.
Supporting information
References
- 1.Pessler F., Pendergrast P.S., Hernandez N. Purification and characterization of FBI-1, a cellular factor that binds to the human immunodeficiency virus type 1 inducer of short transcripts. Mol. Cell. Biol. 1997;17:3786–3798. doi: 10.1128/mcb.17.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies J.M., Hawe N., Kabarowski J., Huang Q.H., Zhu J., Brand N.J., et al. Novel BTB/POZ domain zinc-finger protein, LRF, is a potential target of the LAZ-3/BCL-6 oncogene. Oncogene. 1999;18:365–375. doi: 10.1038/sj.onc.1202332. [DOI] [PubMed] [Google Scholar]
- 3.Liu C.J., Prazak L., Fajardo M., Yu S., Tyagi N., Di Cesare P.E. Leukemia/lymphoma-related factor, a POZ domain-containing transcriptional repressor, interacts with histone deacetylase-1 and inhibits cartilage oligomeric matrix protein gene expression and chondrogenesis. J. Biol. Chem. 2004;279:47081–47091. doi: 10.1074/jbc.M405288200. [DOI] [PubMed] [Google Scholar]
- 4.Kukita A., Kukita T., Ouchida M., Maeda H., Yatsuki H., Kohashi O. Osteoclast-derived zinc finger (OCZF) protein with POZ domain, a possible transcriptional repressor, is involved in osteoclastogenesis. Blood. 1999;94:1987–1997. [PubMed] [Google Scholar]
- 5.Maeda T., Hobbs R.M., Merghoub T., Guernah I., Zelent A., Cordon-Cardo C., et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433:278–285. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- 6.Constantinou C., Spella M., Chondrou V., Patrinos G.P., Papachatzopoulou A., Sgourou A. The multi-faceted functioning portrait of LRF/ZBTB7A. Hum. Genomics. 2019;13:66. doi: 10.1186/s40246-019-0252-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta S., Singh A.K., Prajapati K.S., Kushwaha P.P., Shuaib M., Kumar S. Emerging role of ZBTB7A as an oncogenic driver and transcriptional repressor. Cancer Lett. 2020;483:22–34. doi: 10.1016/j.canlet.2020.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Morrison D.J., Pendergrast P.S., Stavropoulos P., Colmenares S.U., Kobayashi R., Hernandez N. FBI-1, a factor that binds to the HIV-1 inducer of short transcripts (IST), is a POZ domain protein. Nucleic Acids Res. 1999;27:1251–1262. doi: 10.1093/nar/27.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X.S., Haines J.E., Mehanna E.K., Genet M.D., Ben-Sahra I., Asara J.M., et al. ZBTB7A acts as a tumor suppressor through the transcriptional repression of glycolysis. Genes Dev. 2014;28:1917–1928. doi: 10.1101/gad.245910.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masuda T., Wang X., Maeda M., Canver M.C., Sher F., Funnell A.P., et al. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science. 2016;351:285–289. doi: 10.1126/science.aad3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martyn G.E., Wienert B., Yang L., Shah M., Norton L.J., Burdach J., et al. Natural regulatory mutations elevate the fetal globin gene via disruption of BCL11A or ZBTB7A binding. Nat. Genet. 2018;50:498–503. doi: 10.1038/s41588-018-0085-0. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y., Ren R., Ly L.C., Horton J.R., Li F., Quinlan K.G.R., et al. Structural basis for human ZBTB7A action at the fetal globin promoter. Cell Rep. 2021;36:109759. doi: 10.1016/j.celrep.2021.109759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu X., Trimarco J.D., Williams C.A., Barrera A., Reddy T.E., Heaton N.S. ZBTB7A promotes virus-host homeostasis during human coronavirus 229E infection. Cell Rep. 2022;41:111540. doi: 10.1016/j.celrep.2022.111540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavallee V.P., Lemieux S., Boucher G., Gendron P., Boivin I., Armstrong R.N., et al. RNA-sequencing analysis of core binding factor AML identifies recurrent ZBTB7A mutations and defines RUNX1-CBFA2T3 fusion signature. Blood. 2016;127:2498–2501. doi: 10.1182/blood-2016-03-703868. [DOI] [PubMed] [Google Scholar]
- 15.Hartmann L., Dutta S., Opatz S., Vosberg S., Reiter K., Leubolt G., et al. ZBTB7A mutations in acute myeloid leukaemia with t(8;21) translocation. Nat. Commun. 2016;7:11733. doi: 10.1038/ncomms11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faber Z.J., Chen X., Gedman A.L., Boggs K., Cheng J., Ma J., et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat. Genet. 2016;48:1551–1556. doi: 10.1038/ng.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawashima N., Akashi A., Nagata Y., Kihara R., Ishikawa Y., Asou N., et al. Clinical significance of ASXL2 and ZBTB7A mutations and C-terminally truncated RUNX1-RUNX1T1 expression in AML patients with t(8;21) enrolled in the JALSG AML201 study. Ann. Hematol. 2019;98:83–91. doi: 10.1007/s00277-018-3492-5. [DOI] [PubMed] [Google Scholar]
- 18.Christen F., Hoyer K., Yoshida K., Hou H.A., Waldhueter N., Heuser M., et al. Genomic landscape and clonal evolution of acute myeloid leukemia with t(8;21): an international study on 331 patients. Blood. 2019;133:1140–1151. doi: 10.1182/blood-2018-05-852822. [DOI] [PubMed] [Google Scholar]
- 19.Opatz S., Bamopoulos S.A., Metzeler K.H., Herold T., Ksienzyk B., Braundl K., et al. The clinical mutatome of core binding factor leukemia. Leukemia. 2020;34:1553–1562. doi: 10.1038/s41375-019-0697-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rowley J.D. Identificaton of a translocation with quinacrine fluorescence in a patient with acute leukemia. Ann. Genet. 1973;16:109–112. [PubMed] [Google Scholar]
- 21.Erickson P., Gao J., Chang K.S., Look T., Whisenant E., Raimondi S., et al. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 22.Rhoades K.L., Hetherington C.J., Harakawa N., Yergeau D.A., Zhou L., Liu L.Q., et al. Analysis of the role of AML1-ETO in leukemogenesis, using an inducible transgenic mouse model. Blood. 2000;96:2108–2115. [PubMed] [Google Scholar]
- 23.Yuan Y., Zhou L., Miyamoto T., Iwasaki H., Harakawa N., Hetherington C.J., et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10398–10403. doi: 10.1073/pnas.171321298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higuchi M., O'Brien D., Kumaravelu P., Lenny N., Yeoh E.J., Downing J.R. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1:63–74. doi: 10.1016/s1535-6108(02)00016-8. [DOI] [PubMed] [Google Scholar]
- 25.Schwieger M., Lohler J., Friel J., Scheller M., Horak I., Stocking C. AML1-ETO inhibits maturation of multiple lymphohematopoietic lineages and induces myeloblast transformation in synergy with ICSBP deficiency. J. Exp. Med. 2002;196:1227–1240. doi: 10.1084/jem.20020824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schessl C., Rawat V.P., Cusan M., Deshpande A., Kohl T.M., Rosten P.M., et al. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. J. Clin. Invest. 2005;115:2159–2168. doi: 10.1172/JCI24225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Redondo Monte E., Wilding A., Leubolt G., Kerbs P., Bagnoli J.W., Hartmann L., et al. ZBTB7A prevents RUNX1-RUNX1T1-dependent clonal expansion of human hematopoietic stem and progenitor cells. Oncogene. 2020;39:3195–3205. doi: 10.1038/s41388-020-1209-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X.S., Liu Z., Gerarduzzi C., Choi D.E., Ganapathy S., Pandolfi P.P., et al. Somatic human ZBTB7A zinc finger mutations promote cancer progression. Oncogene. 2016;35:3071–3078. doi: 10.1038/onc.2015.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choo Y., Klug A. Physical basis of a protein-DNA recognition code. Curr. Opin. Struct. Biol. 1997;7:117–125. doi: 10.1016/s0959-440x(97)80015-2. [DOI] [PubMed] [Google Scholar]
- 31.Wolfe S.A., Nekludova L., Pabo C.O. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 32.Kayano T., Fukumoto H., Eddy R.L., Fan Y.S., Byers M.G., Shows T.B., et al. Evidence for a family of human glucose transporter-like proteins. Sequence and gene localization of a protein expressed in fetal skeletal muscle and other tissues. J. Biol. Chem. 1988;263:15245–15248. [PubMed] [Google Scholar]
- 33.Bian X., Jiang H., Meng Y., Li Y.P., Fang J., Lu Z. Regulation of gene expression by glycolytic and gluconeogenic enzymes. Trends Cell Biol. 2022;32:786–799. doi: 10.1016/j.tcb.2022.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Hashimoto H., Wang D., Horton J.R., Zhang X., Corces V.G., Cheng X. Structural basis for the versatile and methylation-dependent binding of CTCF to DNA. Mol. Cell. 2017;66:711–720.e3. doi: 10.1016/j.molcel.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luscombe N.M., Laskowski R.A., Thornton J.M. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel A., Horton J.R., Wilson G.G., Zhang X., Cheng X. Structural basis for human PRDM9 action at recombination hot spots. Genes Dev. 2016;30:257–265. doi: 10.1101/gad.274928.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin M., Guo J.T. New insights into protein-DNA binding specificity from hydrogen bond based comparative study. Nucleic Acids Res. 2019;47:11103–11113. doi: 10.1093/nar/gkz963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., Olanrewaju Y.O., Zheng Y., Hashimoto H., Blumenthal R.M., Zhang X., et al. Structural basis for Klf4 recognition of methylated DNA. Nucleic Acids Res. 2014;42:4859–4867. doi: 10.1093/nar/gku134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lan X., Ren R., Feng R., Ly L.C., Lan Y., Zhang Z., et al. ZNF410 uniquely activates the NuRD component CHD4 to silence fetal hemoglobin expression. Mol. Cell. 2021;81:239–254.e8. doi: 10.1016/j.molcel.2020.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halford S.E. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem. Soc. Trans. 2009;37:343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 41.Esadze A., Iwahara J. Stopped-flow fluorescence kinetic study of protein sliding and intersegment transfer in the target DNA search process. J. Mol. Biol. 2014;426:230–244. doi: 10.1016/j.jmb.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X.W., Liu J. Observing protein one-dimensional sliding: methodology and biological significance. Biomolecules. 2021;11:1618. doi: 10.3390/biom11111618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cancer Genome Atlas Research Network. Kandoth C., Schultz N., Cherniack A.D., Akbani R., Liu Y., et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu J., Wu T., Zhang B., Liu S., Song W., Qiao J., et al. Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Commun. Signal. 2021;19:60. doi: 10.1186/s12964-021-00741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pendergrast P.S., Wang C., Hernandez N., Huang S. FBI-1 can stimulate HIV-1 Tat activity and is targeted to a novel subnuclear domain that includes the Tat-P-TEFb-containing nuclear speckles. Mol. Biol. Cell. 2002;13:915–929. doi: 10.1091/mbc.01-08-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glauninger H., Wong Hickernell C.J., Bard J.A.M., Drummond D.A. Stressful steps: progress and challenges in understanding stress-induced mRNA condensation and accumulation in stress granules. Mol. Cell. 2022;82:2544–2556. doi: 10.1016/j.molcel.2022.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeon B.N., Yoo J.Y., Choi W.I., Lee C.E., Yoon H.G., Hur M.W. Proto-oncogene FBI-1 (Pokemon/ZBTB7A) represses transcription of the tumor suppressor Rb gene via binding competition with Sp1 and recruitment of co-repressors. J. Biol. Chem. 2008;283:33199–33210. doi: 10.1074/jbc.M802935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fontana R., Ranieri M., La Mantia G., Vivo M. Dual role of the alternative reading frame ARF protein in cancer. Biomolecules. 2019;9:87. doi: 10.3390/biom9030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang G., Lunardi A., Zhang J., Chen Z., Ala U., Webster K.A., et al. Zbtb7a suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat. Genet. 2013;45:739–746. doi: 10.1038/ng.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herst P.M., Carson G.M., Eccles D.A., Berridge M.V. Bioenergetic and metabolic adaptation in tumor progression and metastasis. Front. Oncol. 2022;12:857686. doi: 10.3389/fonc.2022.857686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cassim S., Vucetic M., Zdralevic M., Pouyssegur J. Warburg and beyond: the power of mitochondrial metabolism to collaborate or replace fermentative glycolysis in cancer. Cancers (Basel) 2020;12:1119. doi: 10.3390/cancers12051119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cherkaoui S., Durot S., Bradley J., Critchlow S., Dubuis S., Masiero M.M., et al. A functional analysis of 180 cancer cell lines reveals conserved intrinsic metabolic programs. Mol. Syst. Biol. 2022;18 doi: 10.15252/msb.202211033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maeda T., Ito K., Merghoub T., Poliseno L., Hobbs R.M., Wang G., et al. LRF is an essential downstream target of GATA1 in erythroid development and regulates BIM-dependent apoptosis. Dev. Cell. 2009;17:527–540. doi: 10.1016/j.devcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alam H., Li N., Dhar S.S., Wu S.J., Lv J., Chen K., et al. HP1gamma promotes lung adenocarcinoma by downregulating the transcription-repressive regulators NCOR2 and ZBTB7A. Cancer Res. 2018;78:3834–3848. doi: 10.1158/0008-5472.CAN-17-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y., Toh H., Sasaki H., Zhang X., Cheng X. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 2012;26:2374–2379. doi: 10.1101/gad.202200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang P., Peslak S.A., Ren R., Khandros E., Qin K., Keller C.A., et al. HIC2 controls developmental hemoglobin switching by repressing BCL11A transcription. Nat. Genet. 2022;54:1417–1426. doi: 10.1038/s41588-022-01152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patel A., Yang P., Tinkham M., Pradhan M., Sun M.A., Wang Y., et al. DNA conformation induces adaptable binding by tandem zinc finger proteins. Cell. 2018;173:221–233.e2. doi: 10.1016/j.cell.2018.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turcot V., Lu Y., Highland H.M., Schurmann C., Justice A.E., Fine R.S., et al. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat. Genet. 2018;50:26–41. doi: 10.1038/s41588-017-0011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jeon B.N., Kim Y.S., Choi W.I., Koh D.I., Kim M.K., Yoon J.H., et al. Kr-pok increases FASN expression by modulating the DNA binding of SREBP-1c and Sp1 at the proximal promoter. J. Lipid Res. 2012;53:755–766. doi: 10.1194/jlr.M022178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi W.I., Yoon J.H., Song J.Y., Jeon B.N., Park J.M., Koh D.I., et al. Zbtb7c is a critical gluconeogenic transcription factor that induces glucose-6-phosphatase and phosphoenylpyruvate carboxykinase 1 genes expression during mice fasting. Biochim. Biophys. Acta Gene Regul. Mech. 2019;1862:643–656. doi: 10.1016/j.bbagrm.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 61.Weber L., Frati G., Felix T., Hardouin G., Casini A., Wollenschlaeger C., et al. Editing a gamma-globin repressor binding site restores fetal hemoglobin synthesis and corrects the sickle cell disease phenotype. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aay9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patel A., Hashimoto H., Zhang X., Cheng X. Characterization of how DNA modifications affect DNA binding by C2H2 zinc finger proteins. Methods Enzymol. 2016;573:387–401. doi: 10.1016/bs.mie.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moerke N.J. Fluorescence polarization (FP) assays for monitoring peptide-protein or nucleic acid-protein binding. Curr. Protoc. Chem. Biol. 2009;1:1–15. doi: 10.1002/9780470559277.ch090102. [DOI] [PubMed] [Google Scholar]
- 65.Otwinowski Z., Borek D., Majewski W., Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr. A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- 66.Terwilliger T.C., Adams P.D., Read R.J., McCoy A.J., Moriarty N.W., Grosse-Kunstleve R.W., et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All plasmids generated in this study are available with a completed materials transfer agreement. The X-ray structures (coordinates and structure factor files) of the ZBTB7A ZF domain with bound DNA have been deposited to PDB and are publicly available as of the date of publications. Accession numbers are PDB 7N5U, PDB 7N5V, PDB 7N5W, PDB 8E3D, and PDB 8E3E.