Highlights
-
•
Genome-scale CRISPR-Cas9 knockout screening may provide new insights into the mechanisms underlying clinical radioresistance. However, there are few published reports of screening for radiosensitivity and resistance genes in nasopharyngeal carcinoma(NPC) using CRISPR/Cas9 libraries.
-
•
Our study used genome-wide CRISPR/Cas9-sgRNA library virus to screen for functional genes associated with radiosensitivity and radioresistance in NPC. It may contribute to the discovery of new therapeutic targets for NPC.
-
•
Nine genes involved in the radiosensitivity or radioresistance of NPC: four genes for radiosensitivity (FBLN5, FAM3C, MUS81, and DNAJC17), two genes for radioresistance (CDKN2AIP, SP1), two potential radioresistant genes (TOMM20, SNX22), and a potential radiosensitive gene (CALD1). Meanwhile, the Fanconi anemia pathway and the TGF-beta signaling pathway possibly contributed to radiosensitivity or radioresistance in NPC.
Keywords: Nasopharyngeal carcinoma, Radiosensitivity, Radiaoresistance, CRISPR-cas9, Functional gene
Abstract
Background
Genome-scale CRISPR-Cas9 knockout screening may provide new insights into the mechanism underlying clinical radioresistance in nasopharyngeal carcinoma (NPC), which is remain largely unknown. Our objective was to screen the functional genes associated with radiosensitivity and radioresistance in NPC, laying a foundation for further research on its functional mechanismand.
Methods
CRISPR-Cas9 library lentivirus screening in radiation-treated NPC cells was combined with second-generation sequence technology to identify functional genes, which were further validated in radioresistant NPC cells and patient tissues.
Results
Eleven radiosensitive and radioresistant genes were screened. Among these genes, the expression of FBLN5, FAM3C, MUS81, and DNAJC17 were significantly lower and TOMM20, CDKN2AIP, SNX22, and SP1 were higher in the radioresistant NPC cells (C666-1R, 5-8FR) (p < 0.05). CALD1 was highly expressed in C666-1R. Furthermore, we found knockout of FBLN5, FAM3C, MUS81 and DNAJC17 promoted the proliferation of NPC cells, while CDKN2AIP and SP1 had the opposed results (p < 0.05). This result was verified in NPC patient tissues. Meanwhile, KEGG analysis showed that the Fanconi anemia pathway and the TGF-β signaling pathway possibly contributed to radiosensitivity or radioresistance in NPC.
Conclusions
Nine genes involved in the radiosensitivity or radioresistance of NPC: four genes for radiosensitivity (FBLN5, FAM3C, MUS81, and DNAJC17), two genes for radioresistance (CDKN2AIP, SP1), two potential radioresistant genes (TOMM20, SNX22), and a potential radiosensitive gene (CALD1). Genome-scale CRISPR-Cas9 knockout screening for radiosensitive and radioresistant genes in NPC may provide new insights into the mechanisms underlying clinical radioresistance to improve the efficacy of radiotherapy for NPC.
Introduction
Nasopharyngeal carcinoma (NPC) is a malignant tumor that occurs on the top and lateral wall of nasopharyngeal cavity [1]. The tumor originates from the epithelial cells covering the nasopharyngeal surface [2]. NPC is a geographically relevant tumor with a particular global distribution and complex etiology, with higher incidence in Southern China, Southeast Asian countries, Northern and Northeastern Africa, Alaska of the United States, and Western Canada [3]. Due to the anatomical limitations and high radiosensitivity, radiotherapy is the primary treatment for NPC [4]. Despite the improvement of technology of radiotherapy and the updating of equipment, the prognosis of locally advanced NPC is still poor, with a 5-year overall survival rate of 60% [5], [6], [7], [8]. The resistance of cancer cells to radiation is one of the main reasons for treatment failure. Therefore, increasing the sensitivity of tumor cells to radiotherapy is the key to improve the efficacy of radiotherapy for NPC. How to screen and identify functional genes for radiosensitivity and radioresistance in NPC in an efficient, accurate and high-throughput manner, to deeply understand the radiosensitivity and radioresistance of NPC and their mechanisms, and to improve the sensitivity of tumor cells to radiotherapy are the current problems that need to be addressed.
CRISPR is an acquired immune mechanism used by bacteria to resist the invasion of foreign genetic material. It has the advantages of simple design, strong specificity, high efficiency, and the ability to produce multiple types of editing results at target sites [9]. The CRISPR-Cas9 gene-editing system consists of the Cas9 protein (or other homologous proteins) with an endonuclide function and a single guide RNA (sgRNA). Inactivated caspase-dead mutants of Cas9 (dCas9) fuse or recruit transcription factors and act on transcription start sites (TSS) to regulate gene transcription. When sgRNA targets individual genes, CRISPR can be considered as an efficient gene-editing tool [10]. When sgRNA targets whole genome sequences, CRISPR is upgraded as a genome-wide screening tool [11,12]. Gene editing in mammalian cells using CRISPR-Cas9 was first reported in 2012 [13]. CRISPR-Cas9, as an efficient and convenient new generation gene-editing technology, has been successfully applied in transgenic yeast [14], fruit fly [15], mouse [16], and other model organisms [17]. In recent years, CRISPR-Cas9 technology has made great progress. Through the design of an sgRNA hybridization libraries, CRISPR cleavage targets can cover the whole genome with higher specificity. Thus, CRISPR-Cas9 can be used for functional deletion screening by inducing gene mutation and functional acquisition screening by activating transcription [18]. By repressing gene expression at the DNA level, we can study phenotypes that require gene knockout to appear.
Radiation resistance of NPC is a major issue that seriously affects the prognosis of patients. Traditional studies based on specific gene function analysis have reported many pathways or mechanisms related to the radiosensitivity of NPC, these include ATM/ATR pathway, mTORC1 pathway, Erk pathway, PI3K-Akt pathway and some miRNA molecules, etc [19], [20], [21], [22]. There is not systematic mechanism of radiosensitivity and radioresistance in NPC at present. The omics-based screening analysis technology can systematically and extensively explore the functional genes and pathways related to radiosensitivity and radioresistance, and provide a more comprehensive understanding of the mechanisms of radiosensitization and radiation resistance. The Cas9 library is an important tool for high-throughput screening based on gene function. However, there are few published reports regarding the use of CRISPR-Cas9 library to screen radiosensitivity and sexual resistance genes in NPC. In this study, genome-wide CRISPR-Cas9-sgRNA library virus was used to screen for radiosensitive and radioresistant functional genes in NPC. It provides a basis for systematically exploring the mechanism of radiosensitivity and radioresistance in NPC.
Materials and methods
CRISPR-Cas9 library Lentivirus screening of key functional genes in NPC combined with second-generation sequencing technology to identify specific sgRNA
genome-wide CRISPR-Cas9 gene knockout screening was performed to identify genes associated with radiosensitivity and radioresistance in the NPC cell line. We transfected C666-1 cells with a genome-scale CRISPR knocked out (GeCKO V2.0 Pooled Library of Shanghai Gikco Company, LTD.) v2.0 Pooled Library. The library contained 19,050 encoding genes and 1,864 microRNA genes. The GeCKO plasmid library constructed by Zhang feng laboratory of Broad institute [23,24] was used in our experiment, and the sgDNA library contained 6 different sgRNAs for each coding gene and 4 different sgRNAs for each miRNA. The coverage and uniformity of the library sgRNA were checked, and QC testing for lentivirus was carried out comprehensively in accordance with the requirements of the FDA and the Chinese Pharmacopeia for viral vaccines and other preparations. The GeCKO system single vector virus sgRNA, Cas9, and puromycin(puro) were used to selectively markers into the cells, and C666-1 cell were infected by Cas9 library virus and cultured. The cells were divided into three groups for radioresistance screening after successfully infection. One group of samples was used as the control, and early library infection was the starting control for genetic change trend analysis. The other two groups were exposed to doses of 0 Gy/1f and 2 Gy/1f respectively, and cell samples were collected 7 and 14 days after irradiation. PCR amplification was performed using genomic DNA of living cells in the sgRNA coding region, followed by high-throughput sequencing analysis [23].
SgRNAs corresponding to genes that regulate radiosensitivity can be selectively enriched or lost. To eliminate as much as possible random changes or false positive or false negative changes caused by corresponding genes affecting cell proliferation, two-time point samples were set up in the experiment, and the samples in the early stage of library infection were retained for trend correction. Genes with the same trend at both time points were more likely to be associated with radiosensitivity. Our CRISPR screening experiments was performed in three biological replicates.
The radiation dose for the NPC cell line
The NPC cell line C666-1was irradiated with x-rays, and given 0, 1, 2, or 4Gy/1f, respectively. The cell growth curve of NPC cells after irradiation was determined by the colony formation and proliferation assay CCK8, and the irradiation dose at which the cell growth inhibition rate reached 40% was selected as the optimal irradiation dose.
The screening dose and time of puromycin
The NPC cells c666-1 were treated with puromycin at different concentrations of 1ug/mL, 2ug/ml, 3ug/ml, 4ug/ml and 5ug/ml for 0-48h. The concentration and duration of puromycin were selected under the condition that the cell mortality rate >90%.
Bioinformatics analysis
The original sequencing data were extracted, filtered, and treated with low quality to obtain clean data. Then the expression abundance of the corresponding sequence was obtained after quality control and compared with the sgRNA sequence of the GeCKO library gene. All sgRNAs were then enriched for analysis. Candidate genes were obtained by sequencing of sgRNA enrichment in cell samples collected 7 and 14 days after irradiation. The effective genes showed a certain degree of enrichment in the cell samples collected 7 days after irradiation and a higher degree of enrichment in the cell samples collected 14 days after irradiation, thus obtaining a list of genes related to radiation sensitivity and radiation resistance of NPC cells after irradiation. Through gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes(KEGG) pathway analyses, genes with high enrichment that were closely related to radiosensitivity and radioresistance were selected.
Establishment of radioresistant NPC cells
NPC cell lines c666-1 and 5-8F were purchased from the Cell Center of Central South University, Changsha, China. In order to establish cells with significant resistance to radiation(abbreviated C666-1R and 5-8FR), C666-1 and 5-8F cells were irradiated at progressively higher doses. All subsequent experiments used exponentially growing cells. The radiation dose was increased to 4, 6, 8, and 10Gy. Each dose was irradiated twice, with a total dose of 60Gy.The remaining cells were cultured and subcultured more than five times. The C666-1 and 5-8F cells that were identified as radioresistant cell were named C666-1R (resistant cells C666-1) and 5-8FR (resistant cells 5-8F). The resistance of ability of resistant cells was verified by colony formation. Subsequent experiments were conducted using C666-1R and 5-8FR within 5-10 generations after the termination of the final round of radiation exposure.
A single gene knockout stable cell line with low expression was constructed
Cas9 technology was used to knockout key genes to construct stable cell lines of C666-1 and CNE1 with low expression of candidate genes. The same technology was used to establish the control cell lines. The sgRNA sequences and diversity of candidate genes are listed in Table S1. Firstly, cell suspensions with a density of 3-5*10^4 /ml were inoculated in a six-well plate (Corning, USA) for 16-24h. After the cell confluence was 20-30%, the corresponding virus venom and infection enhancement solution were added according to the cell multiplicities of infection (MOI) and virus titer, and cultured for 12-16h. The conventional culture solution was then replaced and cultured. The efficiency of the infection was observed by fluorescence microscopy after approximately 72h. After the infection, puromycin was used for screening. The corresponding total RNA and proteins were extracted to verify the differential expression in proteins of candidate genes.
Cell proliferation assay
In order to verify the function of the candidate genes identified from GeCKO screening, we use the CCK8 assay to detect the proliferative ability of the stable cells with single gene knockout after radiotherapy. Cells at the logarithmic growth stage were digested with 0.125% trypsin (containing 0.02% EDTA) and then inoculated into 96-well plates with 1000 cells per well in triplicate. After the cells were attached to the wall, the optimal irradiation dose was applied. After 0,12, 24, 36, 48, 60, and 72 h of irradiation, 10ul of the CCK8 reagent was added to each well and incubated at 37°C for 2 h, and the absorbance (OD value) of the plate was measured at a wavelength of 450nm. Each data was repeated at least three times.
Clone formation experiment
The cells were inoculated in a 60 mm culture dish (Corning). After attached to the wells, the cells were irradiated at different doses (0, 2, 4, 6, 8, and l0Gy) and then cultured for 14 days. Thereafter, the culture medium was discarded, and the cells were washed twice with PBS and fixed with methanol for 15 min. The fixative solution was discarded and stained with crystal violet reagent (concentration 0.1%). The number of colonies greater than 50 cells was counted under the microscope.
Real-time RT-PCR
Total RNA was extracted using TRIzol. mRNAs were retroactively transcribed according to the protocol recommended by the Mona detection kit (GeneCopoeia lNC. S MD, USA). The mRNA was quantitatively detected using Nano Drop (Thermo Fisher Scientific, Madision, USA). The synthetic mRNA primers were designed by Takara Bio (Takara Bio, San Francisco, CA), and the primers of elven known mRNAs were purchased from GeneCopoeia (Guangzhou, China). The primer details are listed in Table S2. Real-time qRT-PCR was performed on a CFX96 TouchTM System (Bio-Rad, Hercules, CA, USA). The internal control was ACTIN. The 2 − Δ Δ CT method was used to calculate the relative expression. Each data was repeated at least three times.
Western Blot (WB)
Cells were collected and total protein was extracted with Radio Immunoprecipitation Assay (RIPA) and Phenyl methane sulfonyl fluoride (PMSF). A bicinchoninic acid (BCA) protein quantification kit (Beyotime, China, Shanghai) was used to determine the protein concentration. The protein was mixed with the loading buffer and denatured by heating at 100°C, and then subjected to SDS-PAGE. The protein was then transferred to a PVDF membrane (Millipore, Billerica, MA, USA). The membrane was cut according to molecular weight, sealed with 5% skim milk, and incubated with primary and secondary antibodies. Relative protein levels were quantified using actin as a reference. Grayscale analysis was performed using ImageJ software (Amersham Biosciences), and independent samples t-test was used for statistical evaluation(p < 0.05 was considered significant). Each experiment was repeated at least three times.
Patients and samples
Tissue samples were obtained from 15 radiotherapy resistant NPC patients and 15 radiotherapy sensitive NPC patients at the First Affiliated Hospital of Guangxi Medical University. Radioresistant patients were defined as those who were evaluated as stable disease (SD) or progressive disease (PD) at three months after radiotherapy and recurrence within 6 months, while radiosensitive patients as those who were complete remission (CR) or partial remission (PR) at three months after radiotherapy. Samples were collected before the patients were treated with radiotherapy. All the tissue specimens were fixed in 4% formalin and embedded in paraffin.
Immunohistochemistry (IHC) for radiosensitive and radioresistant genes expression quantitation
Briefly, tissue sections were incubated with a polyclonal antibody overnight at 4°C after antigen retrieval, followed by incubation with a biotinylated secondary antibody and an avidin-biotin peroxidase complex (ZSGB-Bio, Beijing, China). Then DAB chromogenic substrate solution (ZSGB-Bio, Beijing, China) was added to the slides to form an immunoreaction. Harris hematoxylin was used for counterstaining. IHC results were interpreted by two independent pathologists who were blinded to the clinicopathological information.
Statistical analysis
Data are expressed as the mean ± standard deviation. All experiments were repeated at least three times. Independent sample t-test and repeated measure ANOVA were the statistical analyses specified for each experiment. P < 0.05 was considered statistically significant.
Results
Key functional genes of NPC screened by the Genome-scale CRISPR-Cas9 and second-generation sequencing technology
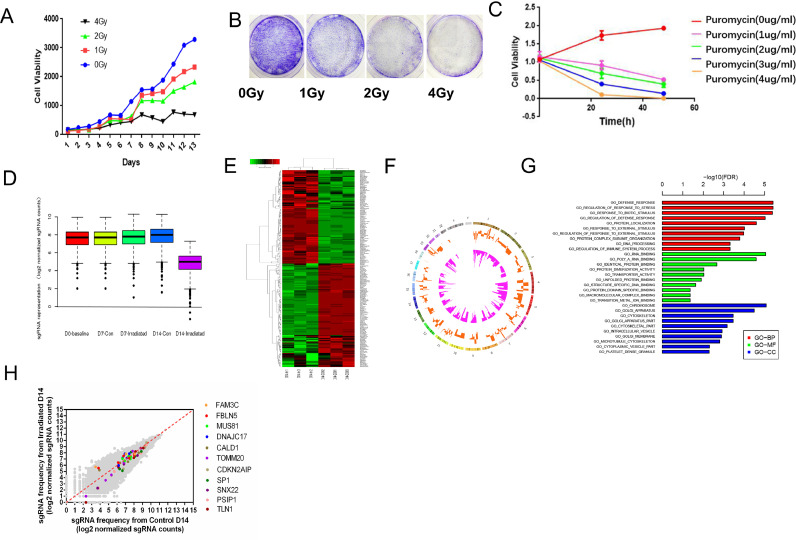
The quality inspection results showed that the coverage and uniformity of the library sgRNA were good. The quality inspection test indicated that all indicators met the GMP standard and could be used for subsequent experiments (Fig. S1.A-B, Table S3-6). In this experiment, when MOI=3, Function MOI=0.42, the virus titer was the best. Therefore, MOI=3 was selected as the virus infection dose in this study (Function MOI=OD(Puromycin+)/OD(Puromycin-)) (Fig. S2.A-B). The colony formation and CCK8 experiment in C666-1 cell line showed that the cells were exposed to doses of 2Gy/1f then the cell growth inhibition rate reached 40%, so 2Gy/1f of irradiation was the optimal irradiation dose for C666-1 cells (Fig. 1A-B, Fig. S3). After treatment with puromycin at a concentration of 3ug/ml for 48h, the cell-killing efficiency was close to 100%. Therefore, 3ug/ml was selected as the working concentration of puromycin in this study (Fig. 1C). We infected the NPC cell line C666-1 using the condition CRISPR/Cas9-sgRNA library virus and provided 2Gy/1f of irradiation. The nuclear genomes of the irradiated and unirradiated groups were extracted on the 7th and 14th day after irradiation respectively, and the second-generation sequencing was performed [23].
Fig. 1.
CRISPR-Cas9 library lentivirus screening in radiation-treated nasopharyngeal carcinoma cells combined with second-generation sequencing technology to identify specific sgRNA. (A) CCK8 assays in C666-1 cells for 13 consecutive days after single irradiation with different doses (0, 1, 2, and 4Gy) were used for determing radiation dose. (B) Colony formation of different doses in C666-1. (B) CCK8 assays were used to determine the time and concentration of puromycin. (D) SgRNA enrichment box diagram. (E) Cluster analysis diagram of differential genes and (F) Circor analysis diagram, including the top 5 upregulated genes: FBLN5, FAM3C, MUS81, DNAJC17, and CALD1, and top 6 downregulated genes: TOMM20, CDKN2AIP, SNX22, PSIP1, SP1, and TLN1. (G) GO function analysis cluster diagram after 14 days of irradiation. (H) SgRNA frequency from irradiated Day14 of 11 key functional genes (FBLN5, FAM3C, MUS81, DNAJC17, CALD1; TOMM20, CDKN2AIP, SNX22, PSIP1, SP1, TLN1).
CRISPR-Cas9-sgRNA library virus filtering the results of the analysis indicate that the effective gene will be showed a certain degree of enrichment in collected cells after 7 days after irradiation, and showed higher abundance after 14 days (Fig. 1D). Combining bioinformatics with cluster analysis of differential genes, 210 genes closely related to radiosensitivity and radioresistance in NPC were identified, among which 79 genes were upregulated and 131 genes were downregulated (Fig. 1E, F). The 210 genes selected were analyzed by GO analysis to understand their roles in biological processes, molecular functions, and cellular components (Fig. 1G). For further study, we selected genes with high enrichment that were closely related to radiosensitivity and radioresistance, including top 5 upregulated genes: FBLN5, FAM3C, MUS81, DNAJC17 and CALD1, and top 6 downregulated genes: TOMM20, CDKN2AIP, SNX22, PSIP1, SP1, and TLN1(Fig. 1H). The key genes information from the results of high-throughput sequencing analysis are shown in Table S7.
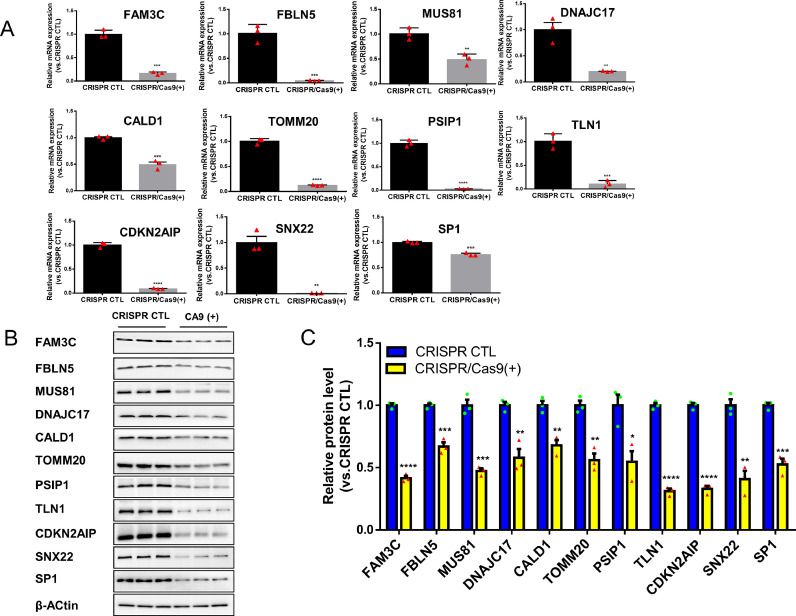
Construction of the radiotherapy resistant and the CRISPR-Cas9–mediated gene knockout cell lines
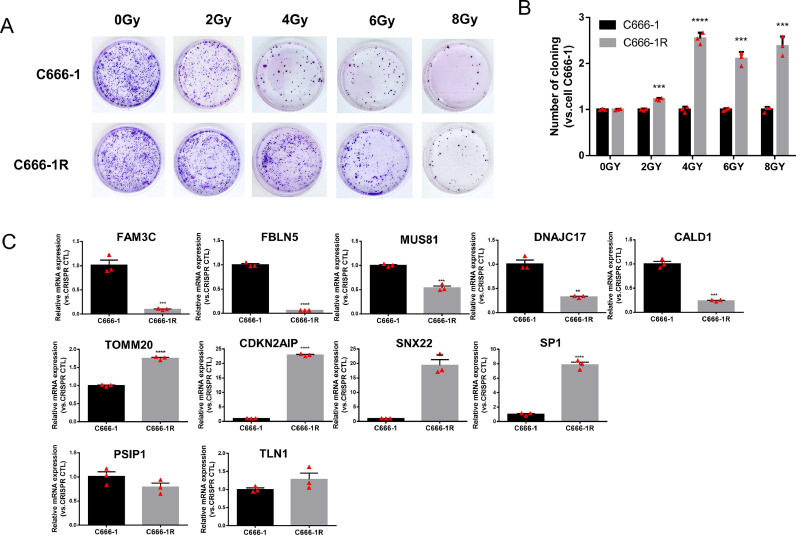
Real-time RT-PCR and WB were used to verify the successful construction of NPC cell lines with a single key gene (FBLN5, FAM3C, MUS81, DNAJC17, CALD1, TOMM20, CDKN2AIP, SNX22, PSIP1, SP1, TLN1) knockout. The mRNA expression of candidate genes in stable cell line C666-1 (Fig. 2A) and CNE1(Fig. S7) with a single key gene knockout were significantly lower than control group (p < 0.05). Western blotting results showed that the protein expression level of the C666-1 (Fig. 2B-C) and CNE1 (Fig. S6) cells with a single key gene knockout were significantly lower than control group (p < 0.05,). The fluorescence microscopy images of C666-1 and CNE1 cells with a single gene knockout, and the level of Green Fluorescent Proteins (GFP) is over 80% (Figs. S4, S5). Meanwhile C666-1R and 5-8FR cells grew slower than control groups (Figs. 4A-B, S9), with reduced colony formation ability, indicating that C666-1R and 5-8FR had stronger resistance to radiation.
Fig. 2.
CRISPR-Cas9-mediated knockdown effeciency of 11 key functional genes in C666-1 cell. (A) RT-qPCR results of individual gene knockouts in C666-1 cells infected with the CRISPR control or CRISPR /cas9 (+) virus. (B) Western blot of individual gene knockouts in C666-1 cells infected with the CRISPR control or CRISPR /cas9 (+) virus, β-actin was used as loading control. (C) Quantification of individual gene knockout band intensity in western blot. All the results were reproducible in three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001; **** p < 0.0001.
Fig. 4.
C666-1R was used to screen the radiosensitivity and radioresistance of the candidate genes. (A-B) Colony formation was used to verified the successful construction of radioresistant C666-1 cells (C666-1R). (C) qPCR was used to determine the expression levels of elven genes in C666-1 and C666-1R cells to confirm their radioresistant or radiosensitive status. Reported values were mean ± SD from three independent experiments. ** p < 0.01, *** p < 0.001; **** p < 0.0001.
Nine genes related to the radiosensitivity or radioresistance of NPC
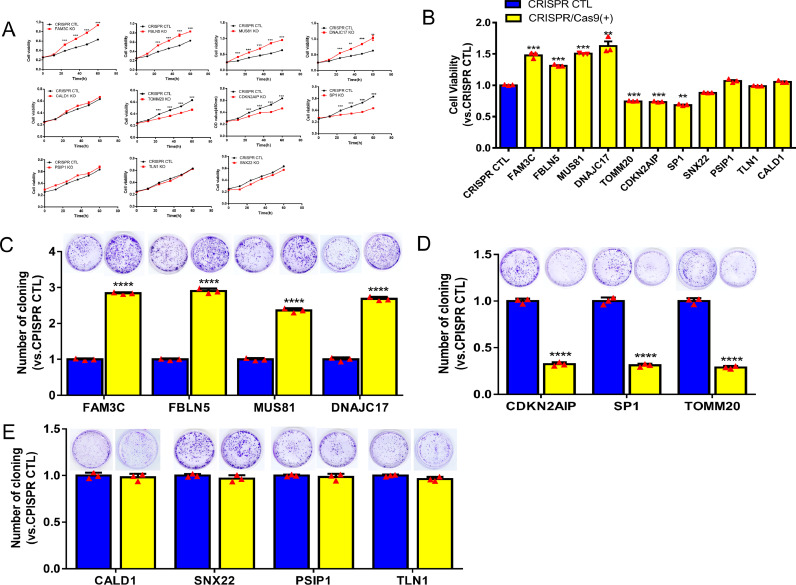
Compared with the control cells, the radioresistant ability of the cells decreased after radioresistant gene knockout, more cells died and the proliferation ability of the cells was decreased. In contrast, the cell proliferation increased when the radiosensitive gene was knocked out. The results of the CCK8 experiment and colony formation showed that the growth of C666-1 and CNE1 cells with low expression of FBLN5, FAM3C, MUS81, or DNAJC17 were significantly promoted (p < 0.05, Figs. 3A-C, S8) compared to the control group. The growth of C666-1 cells with low expression of TOMM20, CDKN2AIP or SP1 in NPC was significantly lower than that in the control group (p < 0.05, Fig. 3A-B, D). While the growth of C666-1 cells with low expression of CALD1, SNX22, PSIP1or TLN1 in NPC were no statistical difference (Fig. 3A-B, E). Compared to the control group, the growth of CNE1 cells with low expression of CDKN2AIP or SP1 were significantly inhibited (p < 0.05, Fig. S8). TOMM20 can promote the growth of C666-1 cells, but has no significant effect on CNE1, which may be related to different cell types. In conclusion, knockout of FBLN5, FAM3C, MUS81 and DNAJC17 could significantly promote the cell proliferation and increase the radiosensitivity of C666-1 and CNE1 cells, indicating that these genes may be radiosensitive genes. Knockout of CDKN2AIP and SP1 can significantly inhibit the proliferation and increase the radioresistance of C666-1 and CNE1 cells, indicating that these genes may be radioresistant genes. In addition, more activated radioresistant genes accumulate in radioresistant NPC cells compared with parental cells. RT-qPCR was used to determine the expression levels of eleven genes in C666-1R and 5-8FR cells to confirm their radioresistance or radiosensitive functions. The mRNA expression level of five genes (FBLN5, FAM3C, MUS81, DNAJC17, and CALD1) in C666-1R was significantly lower (p < 0.05), and the mRNA expression level of four genes (TOMM20, CDKN2AIP, SNX22, and SP1) in C666-1R was significantly higher (p < 0.05, Fig. 4C) compared to C666-1 cells. In 5-8FR, we found that the expression levels of FBLN5, FAM3C, MUS81 and DNAJC17 were significantly lower than those in 5-8F cells. Meanwhile, the expression levels of TOMM20, CDKN2AIP, SNX22, and SP1 were significantly higher than those in 5-8F cells (p < 0.05, Fig. S10). CALD1 was highly expressed in C666-1R, but no significant difference in 5-8FR, which may be related to different cell types. The results of the radioresistant cells indicated that, FBLN5, FAM3C, MUS81 and DNAJC17 were associated with radiosensitivity, while TOMM20, CDKN2AIP, SNX22 and SP1 were relative to radioresistance. In addition, our result suggested that CALD1 was a potential gene for radiosensitivity in C666-1R and 5-8FR.
Fig. 3.
GeCKO screening results regarding the radiosensitivity and radioresistance of candidate genes in C666-1 cells. (A-B) CCK8 assay for C666-1 cells with single gene knockout. (C) Four genes for radiosensitivity: FBLN5, FAM3C, MUS81, and DNAJC17; (D) three genes for radioresistance: CDKN2AIP, SP1, and TOMM20; (E) genes that in no statistically significant difference: CALD1, SNX22, PSIP1, and TLNI. Reported values were mean ± SD from three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001; **** p < 0.0001.
The genes be considered as radioresistant or radiosensitive genes only if the results of the single knockout cell line and the radioresistant cell line are consistent. Besides, if only one result is significant, the candidate genes would be considered as potential genes . We found nine genes involved in the radiosensitivity or radioresistance of NPC: four genes for radiosensitivity (FBLN5, FAM3C, MUS81, and DNAJC17), two genes for radioresistance (CDKN2AIP, SP1), two potential radioresistant genes (TOMM20, SNX22), and a potential radiosensitive gene (CALD1).
Based on the RT-qPCR results of C666-1R and 5-8FR and cell proliferation and clonal formation abilities of stable cell lines after single gene knockout, we concluded that there were four genes of radiosensitivity, FBLN5, FAM3C, MUS81, and DNAJC17, and two genes of radioresistance, CDKN2AIP and SP1. And CALD1 is a potential radiosensitivity gene, TOMM20 and SNX22 are potential radioresistance genes, while PSIP1 and TLNI are still uncertain.
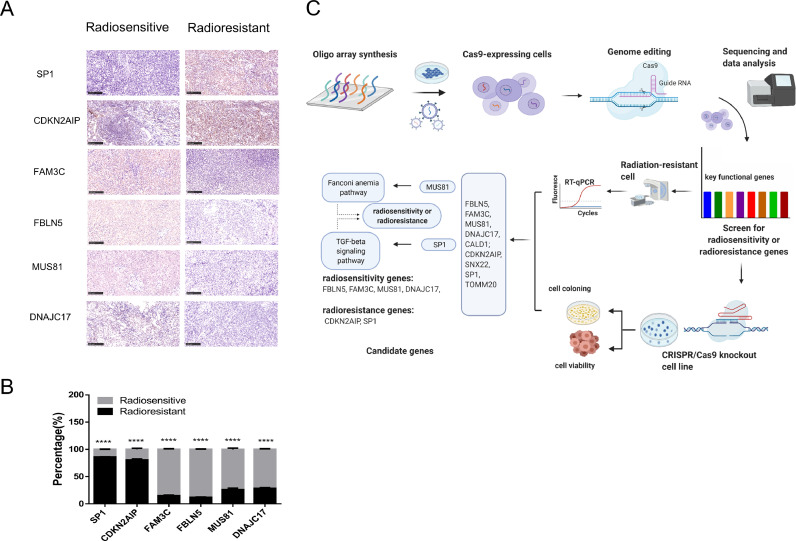
In addition, IHC was performed to confirm the expression of candidate genes in NPC patients with radiosensitivity and radioresistance. The results revealed that SP1 and CDKN2AIP were overexpressed in radioresistant tumors, while the expression of FAM3C, FBLN5, MUS81, and DNAJC17 were higher in radiosensitive tumors (p < 0.05) (Fig. 6A, B). The results of the IHC showed that FAM3C, FBLN5, MUS81, and DNAJC17 were associated with radiosensitivity, while SP1 and CDKN2AIP were associated with radioresistance. Overall, these results are consistent with the results obtained using radioresistant cells and the single gene knockout stable cell lines.
Fig. 6.
Schematic diagram of genome-wide CRISPR-Cas9 screening principle and the potential signaling pathway contributed to radiosensitivity or radioresistance in NPC.
GO and KEGG pathway analysis
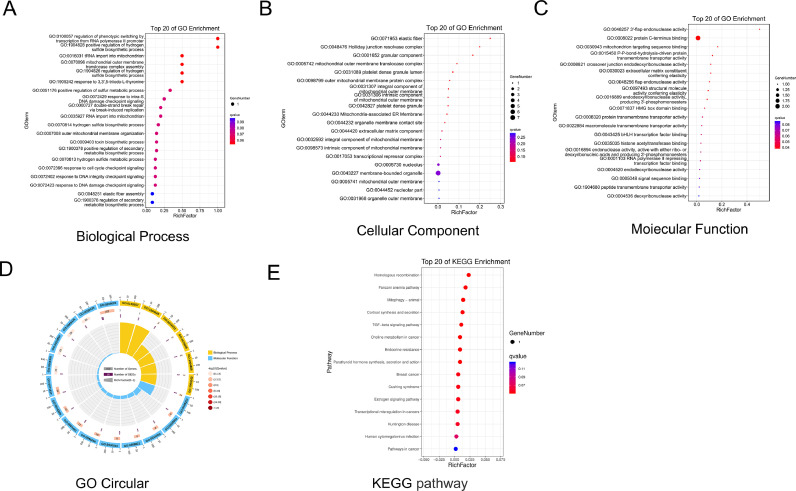
The analysis of GO and KEGG pathways would provide insight into the discovery of potential radiosensitive or radioresistant signaling pathway in NPC. GO biological process annotation revealed that the identified target genes may be enriched in the regulation of phenotypic switching by transcription from RNA polymerase II promoter, positive regulation of hydrogen sulfide biosynthetic process, and tRNA import into mitochondria. For the cellular component, the identified target genes were closely related to the elastic fiber, Holliday junction resolvase complex, and granular component. For the molecule functions, the three most enriched items were 3′-flap endonuclease activity, protein C-terminus binding, and mitochondrion-targeting sequence binding (Fig. 5A-D, Table S8). KEGG enrichment analysis showed that the potential signaling pathway contributed to radiosensitivity or radioresistance in NPC, including the Fanconi anemia pathway and the TGF-beta signaling pathway (Fig. 5E, Table S9). Fig. 6C is a schematic diagram of genome-wide CRISPR-Cas9 screening principle and the potential signaling pathway contributed to radiosensitivity or radioresistance in NPC.
Fig. 5.
Enriched annotation of GO categories and KEGG pathways for the selected potential genes. (A) Biological processes (BP). (B) Cellular components (CC). (C) Molecular factors (MF). (D) GO Circular. (E) KEGG pathway.
Discussion
Screening of radiosensitive and radioresistant genes in NPC in order to increase the sensitivity of tumor cells to radiotherapy is the basis of improving the prognosis of patients with NPC. It is believed that there are 10%–50% of the cells are hypoxic cells in solid tumors, which are radioresistant to an extent of 2.5–3.0 times compared to aerobic cells. They cannot be effectively killed and are the source of tumor recurrence and metastasis after treatment [25]. Based on gene editing technology, high-throughput functional gene screening is an important tool for analyzing gene function and exploring biological processes [26]. Compared to genome editing tools such as zinc finger nucleases (ZFNs) and Transcription activator-like effector nucleases (TALENs), the binding target of Cas9 is a gene site-specific editing technology, determined by sgRNA sequence rather than protein structure. In addition, CRISPR-Cas9 relies on the advantages such as having a simple design, strong specificity, high efficiency, and the ability to produce multiple types of editing results at the target site. The researchers used CRISPR-Cas9 genome-wide screening to obtain the target gene in mouse acute myeloid leukemia (AML) cell lines, combined with existing database analysis to narrow the gene range, and carried out a second screening in mice to obtain the gene DCPS with higher correlation with AML cell survival ability. Cap hydrolase gene at the 3 'end of mRNA (DCPS) maintains AML cell survival by influencing pre-mRNA maturation [27]. Researchers obtained a non-metastatic tumor cell line with stable expression of Cas9-EGFP from non-small-cell lung cancer (NSCLC) mice in 2015. It can induce tumorigenesis in immunodeficient mice. Cas9-EGFP cells infected with the sgRNA library were injected into mice, and the migrated tumor cells were localized according to the fluorescent protein. Shi J et al.performed high-throughput sequencing and bioinformatics analysis of transplanted tumor cells, and found 624 genes closely related to tumor metastasis [28]. This series of studies revealed that CRISPR-Cas9 genome-wide screening technology may play an important role in the life sciences.
In this study, CRISPR-Cas9 genome-wide library lentivirus was used to screen genes associated with radiosensitivity and radioresistance in NPC. Second-generation sequencing technology identifies specific sgRNA, and combines bioinformatics analysis to screen out radiosensitive and radioresistant function genes. By constructing the radiotherapy resistant cells, single-gene knockout stable cell line and NPC patients with radiosensitivity and radioresistance, we identified that FBLN5, FAM3C, MUS81, and DNAJC17 may be genes for radiosensitivity, CDKN2AIP and SP1 may be genes for radioresistance. However, the effects of PSIP1, and TLN1 on radiosensitivity and radioresistance have not been determined. We initially screened for radiosensitive and radioresistant functional genes in NPC, which laid the foundation for new studies to explain the mechanisms of radiosensitivity in NPC, and makes some contribution to the discovery of new therapeutic targets and new drugs to increase the radiosensitivity of NPC. Of course, there are still some shortcomings in our study, and more experiments are needed to explore the molecular mechanisms of candidate genes in NPC.
MUS81 is a component of the MUS81-EME1 structure-specific endonuclease, a structure-specific nuclease that involved in ICL repair [29,30] via cutting on the 3’side of X-shaped structures [31]. MUS81, first identified by interacting with Rad54 in budding yeast [32], has been identified as an important component of the HR-mediated double-chain fracture repair pathway in mammalian cells. MUS81 has been implicated in the formation of DNA double-strand breaks (DSBs) at interstrand cross-link (ICL) -stalled replication forks [33]. A study done by John showed that a critical role for the proper biallelic expression of the mammalian Mus81 in the maintenance of genomic integrity and tumor suppression [34]. In addition, MUS81 is involved in the response to ultraviolet irradiation and methylation-induced DNA damage in cereviscera [34]. It is considered as a potential therapeutic target for multiple cancer, such as breast cancer, Gastric Cancer, Thyroid Cancer [35], [36], [37]. In this study, MUS81 was preliminarily believed to have the effect of radiotherapy sensitization in NPC cells. KEGG enrichment analysis results suggest that MUS81 may play a role through the Fanconi anemia pathway. The Fanconi anemia pathway is a critical component of the DNA damage response, regulating the repair of interstrand cross-links [38,39]. Several reports have suggested that FA proteins play important roles in the genome stability throughout the cell cycle, especially in M phase [40,41].
SP1 is a transcription factor binding to the promoter elements rich in GC/GT, to regulate the promoter activity of multiple genes involved in the cell cycle, differentiation, and tumorigenesis [42]. SP1 is considered as a candidate tumor suppressor gene, which has been shown to regulate RAS domain protein 1, vascular endothelial growth factor, and matrix metallopeptidase 9 in NPC, leading to a positive prognosis [43,44]. SP1 is overexpressed in many cancers, including rectal cancer [45], hepatocellular carcinoma [46], lung cancer [47] and so on. SP1 and C-MYC synergistically bind the BMI1 gene promoter to participate in the pathogenesis of NPC [48]. Our previous report and Jun Wang et al indicated that SP1 has a crucial role in increasing the radioresistance of NPC cells. Inhibition of Sp1 could increase the radiosensitivity of NPC cells [21,49]. The results of the present study are consistent with these reports. Therefore, SP1 may play an important role in the radiotherapy resistance of NPC and is a potential therapeutic target.
Hwang et al. reported that the high expression of FBLN5 in NPC cells led to high expression of FLJ10540 and the activity of AKT, thereby inducing the migration and invasion of NPC cells, which may be related to the progression and poor prognosis of NPC [50]. FAM3C can promote the occurrence and development of oral squamous cell carcinoma and breast cancer. For example, FAM3C high expression is associated with poor prognosis of oral squamous carcinoma [51], FAM3C YY1 - HSF1 pathway are pivotal for the proliferation and migration in breast cancer MDA-MB-231 cells [52]. CDKN2AIP (also known as CARF or FLJ20036) has been shown to be an emerging regulator of tumor inhibition [53]. It is a new p53 regulatory protein that is upregulated during replication, carcinogenesis, and stress-induced aging. It induces senescence (overexpression) and apoptosis (downregulation) in a dose-dependent manner [54]. Cheung et al. demonstrated that CDKN2AIP plays an important role in genome preservation and tumor inhibition, CDKN2AIP silencing induces apoptosis in human cancer cells, and CDKN2AIP siRNA is a new and effective cancer therapy drug. At present, there is few reports of CLAD1 and DNAJC17 in the development of NPC. FBLN5, FAM3C, DNAJC17 and CDKN2AIP were considered for the first time as genes related to radiaosensitivity or radioresistance of NPC in this study.
Conclusions
Nine genes involved in the radiosensitivity or radioresistance of NPC: four genes for radiosensitivity (FBLN5, FAM3C, MUS81, and DNAJC17), two genes for radioresistance (CDKN2AIP, SP1), two potential radioresistant genes (TOMM20, SNX22), and a potential radiosensitive gene (CALD1). Genome-scale CRISPR-Cas9 knockout screening for radiosensitive and radioresistant genes in NPC may provide new insights into the mechanisms underlying clinical radioresistance to improve the efficacy of radiotherapy for NPC.
Ethics approval and consent to participate
The study was approved by the Research Ethics Committee of Guangxi Medical University (Nanning, China, approval number:KY-E-232). Written informed consent was obtained from all patients. All methods were performed in accordance with the relevant guidelines and regulations. The study was performed in accordance with the Declaration of Helsinki.
Consent to publish
All authors read and consented to publication of the paper.
Availability of data and material
All data generated and analyzed during this study are included in this published article (and its supplementary information files).
CRediT authorship contribution statement
Ziyan Zhou: Conceptualization, Methodology, Validation, Formal analysis, Writing – original draft. Gang Chen: Conceptualization, Methodology, Validation, Formal analysis, Writing – original draft. Mingjun Shen: Software, Validation, Formal analysis. Jixi Li: Software, Validation, Formal analysis. Kang Liu: Software, Validation, Formal analysis. Ming Liu: Resources. Shuo Shi: Methodology, Investigation. Dong Yang: Investigation. Wei Chen: Investigation. Sixia Chen: Investigation. Yuanxiu Yin: Investigation. Yating Qin: Investigation. Xuejin Su: Investigation. Weimin Chen: Investigation. Min Kang: Supervision, Conceptualization, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81460460,81760542), The Research Foundation of the Science and Technology Department of Guangxi Province, China (grant No. 2016GXNSFAA380252, 2018AB61001 and 2014GXNSFBA118114), the Research Foundation of the Health Department of Guangxi Province, China (No. S2018087), Guangxi Medical University Training Program for Distinguished Young Scholars (2017), Medical Excellence Award Funded by the Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University(2016). Guangxi Medical High-level Talents Training Program. The central government guide local science and technology development projects (ZY18057006).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101625.
Appendix. Supplementary materials
References
- 1.Ge Y, Long Y, Xiao S, et al. CD38 affects the biological behavior and energy metabolism of nasopharyngeal carcinoma cells. Int. J. Oncol. 2019;54(2):585–599. doi: 10.3892/ijo.2018.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mao C, Tao Z, Chen Z, et al. In vivoIndole-3-carbinol inhibits nasopharyngeal carcinoma cell growth and through inhibition of the PI3K/Akt pathway. Exp. Ther. Med. 2014;8(1):207–212. doi: 10.3892/etm.2014.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tao Q, Chan A. Nasopharyngeal carcinoma: molecular pathogenesis and therapeutic developments. Expert Rev. Mol. Med. 2007;9(12):1–24. doi: 10.1017/s1462399407000312. [DOI] [PubMed] [Google Scholar]
- 4.Jin Y, Yao J, Zhang F, et al. Is pretreatment Epstein-Barr virus DNA still associated with 6-year survival outcomes in locoregionally advanced nasopharyngeal carcinoma? J. Cancer. 2017;8(6):976–982. doi: 10.7150/jca.18124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang M, Zhou P, Liao X, et al. Prognostic value of masticatory muscle involvement in nasopharyngeal carcinoma patients treated with intensity-modulated radiation therapy. Oral Oncol. 2017;75:100–105. doi: 10.1016/j.oraloncology.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Hong R, Hsiao C, Ting L, et al. Final results of a randomized phase III trial of induction chemotherapy followed by concurrent chemoradiotherapy versus concurrent chemoradiotherapy alone in patients with stage IVA and IVB nasopharyngeal carcinoma-Taiwan Cooperative Oncology Group (TCOG) 1303 Study. Ann. Oncol. 2018;29(9):1972–1979. doi: 10.1093/annonc/mdy249. [DOI] [PubMed] [Google Scholar]
- 7.Sun X, Liu S, Luo M, et al. The association between the development of radiation therapy, image technology, and chemotherapy, and the survival of patients with nasopharyngeal carcinoma: a Cohort Study From 1990 to 2012. Int. J. Radiat. Oncol. Biol. Phys. 2019;105(3):581–590. doi: 10.1016/j.ijrobp.2019.06.2549. [DOI] [PubMed] [Google Scholar]
- 8.Prawira A, Oosting S, Chen T, et al. Systemic therapies for recurrent or metastatic nasopharyngeal carcinoma: a systematic review. Br. J. Cancer. 2017;117(12):1743–1752. doi: 10.1038/bjc.2017.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan X, Zhang H, Qin H, et al. CRISPR-Cas9-mediated whole genomic wide knockout screening identifies mitochondrial ribosomal proteins involving in oxygen-glucose deprivation/reperfusion resistance. J. Cell. Mol. Med. 2020 doi: 10.1111/jcmm.15580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye J, Gu Y, Zhang F, et al. IDH1 deficiency attenuates gluconeogenesis in mouse liver by impairing amino acid utilization. Proc. Nat. Acad. Sci. U.S.A. 2017;114(2):292–297. doi: 10.1073/pnas.1618605114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamauchi T, Masuda T, Canver M, et al. Genome-wide CRISPR-Cas9 screen identifies leukemia-specific dependence on a Pre-mrna metabolic pathway regulated by DCPS. Cancer Cell. 2018;33(3):386–400. doi: 10.1016/j.ccell.2018.01.012. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manguso R, Pope H, Zimmer M, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547(7664):413–418. doi: 10.1038/nature23270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cong L, Ran F, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiCarlo J, Norville J, Mali P, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41(7):4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Z, Ren M, Wang Z, et al. Highly efficient genome modifications mediated by CRISPR-Cas9 in Drosophila. Genetics. 2013;195(1):289–291. doi: 10.1534/genetics.113.153825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Yang H, Shivalila C, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Li J, Suzuki K, et al. Genetic enhancement in cultured human adult stem cells conferred by a single nucleotide recoding. Cell Res. 2017;27(9):1178–1181. doi: 10.1038/cr.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shalem O, Sanjana N, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 2015;16(5):299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin L, Li X, Lin Z, et al. EBV-LMP1 regulating AKT/mTOR signaling pathway and WWOX in nasopharyngeal carcinoma. Int. J. Clin. Exp. Pathol. 2017;10(8):8619–8625. [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Hsieh M, Lin J, et al. Erianin induces cell apoptosis through ERK pathway in human nasopharyngeal carcinoma. Biomed. Pharmacother. 2019;111:262–269. doi: 10.1016/j.biopha.2018.12.081. [DOI] [PubMed] [Google Scholar]
- 21.Kang M, Xiao J, Wang J, et al. MiR-24 enhances radiosensitivity in nasopharyngeal carcinoma by targeting SP1. Cancer Med. 2016;5(6):1163–1173. doi: 10.1002/cam4.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang M, Tang B, Li J, et al. Identification of miPEP133 as a novel tumor-suppressor microprotein encoded by miR-34a pri-miRNA. Mol. Cancer. 2020;19(1):143. doi: 10.1186/s12943-020-01248-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shalem O, Sanjana N, Hartenian E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanjana N, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borchiellini D, Etienne-Grimaldi M, Thariat J, et al. The impact of pharmacogenetics on radiation therapy outcome in cancer patients. A focus on DNA damage response genes. Cancer Treat. Rev. 2012;38(6):737–759. doi: 10.1016/j.ctrv.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Allen F, Behan F, Khodak A, et al. JACKS: joint analysis of CRISPR-Cas9 knockout screens. Genome Res. 2019;29(3):464–471. doi: 10.1101/gr.238923.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blondel C, Park J, Hubbard T, et al. CRISPR-Cas9 Screens reveal requirements for host cell sulfation and fucosylation in bacterial type III secretion system-mediated cytotoxicity. Cell Host Microbe. 2016;20(2):226–237. doi: 10.1016/j.chom.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi J, Wang E, Milazzo J, et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 2015;33(6):661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crossan G, van der Weyden L, Rosado I, et al. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat. Genet. 2011;43(2):147–152. doi: 10.1038/ng.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang A, Sengerová B, Cattell E, et al. Human SNM1A and XPF-ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes Dev. 2011;25(17):1859–1870. doi: 10.1101/gad.15699211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Walter J. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair. 2014;19:135–142. doi: 10.1016/j.dnarep.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Interthal H, Heyer W. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2000;263(5):812–827. doi: 10.1007/s004380000241. [DOI] [PubMed] [Google Scholar]
- 33.Hanada K, Budzowska M, Modesti M, et al. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006;25(20):4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McPherson J, Lemmers B, Chahwan R, et al. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science. 2004;304(5678):1822–1826. doi: 10.1126/science.1094557. [DOI] [PubMed] [Google Scholar]
- 35.Yin Y, Liu W, Shen Q, et al. The DNA endonuclease Mus81 Regulates ZEB1 expression and serves as a target of BET4 inhibitors in gastric cancer. Mol. Cancer Ther. 2019;18(8):1439–1450. doi: 10.1158/1535-7163.mct-18-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinheiro M, Lupinacci F, Santiago K, et al. mus81germline mutation in resulting in impaired protein stability is associated with familial breast and thyroid cancer. Cancers. 2020;12(5) doi: 10.3390/cancers12051289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemaçon D, Jackson J, Quinet A, et al. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 2017;8(1):860. doi: 10.1038/s41467-017-01180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niedernhofer L, Lalai A, Hoeijmakers J. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123(7):1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Sejas D, Qiu Y, et al. Inflammatory ROS promote and cooperate with the Fanconi anemia mutation for hematopoietic senescence. J. Cell Sci. 2007;120:1572–1583. doi: 10.1242/jcs.003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nalepa G, Clapp D. Fanconi anemia and the cell cycle: new perspectives on aneuploidy. F1000Prime Rep. 2014;6:23. doi: 10.12703/p6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat. Cell Biol. 2009;11(6):761–768. doi: 10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- 42.Davie J, He S, Li L, et al. Nuclear organization and chromatin dynamics–Sp1, Sp3 and histone deacetylases. Adv. Enzyme. Regul. 2008;48:189–208. doi: 10.1016/j.advenzreg.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 43.Su B, Xiang B, Wang L, et al. Profiling and comparing transcription factors activated in non-metastatic and metastatic nasopharyngeal carcinoma cells. J. Cell. Biochem. 2010;109(1):173–183. doi: 10.1002/jcb.22395. [DOI] [PubMed] [Google Scholar]
- 44.Beishline K, Azizkhan-Clifford J. Sp1 and the 'hallmarks of cancer. FEBS J. 2015;282(2):224–258. doi: 10.1111/febs.13148. [DOI] [PubMed] [Google Scholar]
- 45.Spindler K, Nielsen J, Lindebjerg J, et al. Prediction of response to chemoradiation in rectal cancer by a gene polymorphism in the epidermal growth factor receptor promoter region. Int. J. Radiat. Oncol. Biol. Phys. 2006;66(2):500–504. doi: 10.1016/j.ijrobp.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 46.Rhee H, Kim H, Choi J, et al. Keratin 19 expression in hepatocellular carcinoma is regulated by fibroblast-derived HGF via a MET-ERK1/2-AP1 and SP1 Axis. Cancer Res. 2018;78(7):1619–1631. doi: 10.1158/0008-5472.can-17-0988. [DOI] [PubMed] [Google Scholar]
- 47.Wang R, Xu J, Xu J, et al. MiR-326/Sp1/KLF3: a novel regulatory axis in lung cancer progression. Cell Prolif. 2019;52(2):e12551. doi: 10.1111/cpr.12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, Liu G, Zhang H, et al. Sp1 and c-Myc regulate transcription of BMI1 in nasopharyngeal carcinoma. FEBS J. 2013;280(12):2929–2944. doi: 10.1111/febs.12299. [DOI] [PubMed] [Google Scholar]
- 49.Wang J, Kang M, Wen Q, et al. Berberine sensitizes nasopharyngeal carcinoma cells to radiation through inhibition of Sp1 and EMT. Oncol. Rep. 2017;37(4):2425–2432. doi: 10.3892/or.2017.5499. [DOI] [PubMed] [Google Scholar]
- 50.Hwang C, Shiu L, Su L, et al. Oncogenic fibulin-5 promotes nasopharyngeal carcinoma cell metastasis through the FLJ10540/AKT pathway and correlates with poor prognosis. PLoS One. 2013;8(12):e84218. doi: 10.1371/journal.pone.0084218. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Wu C, Xiao Y, Li H, et al. Overexpression of FAM3C is associated with poor prognosis in oral squamous cell carcinoma. Pathol. Res. Pract. 2019;215(4):772–778. doi: 10.1016/j.prp.2019.01.019. [DOI] [PubMed] [Google Scholar]
- 52.Yang W, Feng B, Meng Y, et al. FAM3C-YY1 axis is essential for TGFβ-promoted proliferation and migration of human breast cancer MDA-MB-231 cells via the activation of HSF1. J. Cell. Mol. Med. 2019;23(5):3464–3475. doi: 10.1111/jcmm.14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheung C, Hasan M, Widodo N, et al. CARF: an emerging regulator of p53 tumor suppressor and senescence pathway. Mech. Ageing Dev. 2009;130:18–23. doi: 10.1016/j.mad.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 54.Cheung C, Kaul S, Wadhwa R. Molecular bridging of aging and cancer: a CARF link. Ann. N.Y. Acad. Sci. 2010;1197:129–133. doi: 10.1111/j.1749-6632.2009.05392.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and analyzed during this study are included in this published article (and its supplementary information files).