Summary
Visualizing the nano-organization of the synapse is fundamental to elucidating the structure-function relationship of the nervous system. The advent of super-resolution microscopy provides a tool to assess and quantify the dynamic organization of numerous proteins at the synapse. Here we present a protocol assessing inhibitory synapse scaffold protein, gephyrin, in rat primary hippocampal cultures using dSTORM microscopy. We delineate the steps for artemisinin treatment, immunocytochemistry, dSTORM image acquisition, single-molecule localization, and the analysis of synaptic scaffold protein dynamics.
For complete details on the use and execution of this protocol, please refer to Guzikowski and Kavalali (2022).1
Subject areas: Microscopy, Neuroscience
Graphical abstract

Highlights
-
•
Protocol to assess inhibitory synapse scaffold protein gephyrin with dSTORM microscopy
-
•
Immunocytochemistry steps for staining primary hippocampal cultures
-
•
dSTORM imaging and single-molecule localization of pre- and post-synaptic proteins
-
•
Image analysis to investigate synaptic clusters and their spatial distribution
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Visualizing the nano-organization of the synapse is fundamental to elucidating the structure-function relationship of the nervous system. The advent of super-resolution microscopy provides a tool to assess and quantify the dynamic organization of numerous proteins at the synapse. Here we present a protocol assessing inhibitory synapse scaffold protein, gephyrin, in rat primary hippocampal cultures using dSTORM microscopy. We delineate the steps for artemisinin treatment, immunocytochemistry, dSTORM image acquisition, single-molecule localization, and the analysis of synaptic scaffold protein dynamics.
Before you begin
The protocol detailed below describes the specific steps for staining and dSTORM imaging in primary hippocampal cultures. Parental Artemisinin and its’ two semisynthetic derivatives (Artemether and Artesunate) bind in the gephyrin universal receptor binding pocket, disrupting GABAAR-gephyrin binding at central inhibitory synapses.2 In Guzikowski and Kavalali (2022)1 this protocol was used to understand how Artemisinin treatment disrupted the structural organization of the synapse and how that corresponded to subsequent synaptic signaling deficits with electrophysiology. The pharmacological manipulation of inhibitory synapses with Artemisinin can be expanded to probe other nano-organizational, functional, and singling pathways downstream of GABAAR mediated neurotransmission. In addition, the staining, image acquisition, and image analysis portion of this protocol has also been used to investigate excitatory synapses in hippocampal culture and slices including staining for vGlut, Homer, Bassoon, GluA1, and GluA2.
Institutional permissions
All animal procedures were performed in accordance with the guide for the care and use of laboratory animals and were approved by the Institutional Animal Care and Use Committee at Vanderbilt University. If you are conducting this protocol institutional permissions from relevant bodies are necessary for the use of laboratory animals.
Prepare rat hippocampal cultures
Timing: 2 weeks
-
1.Prepare primary hippocampal cultures on #1.5 thickness 12 mm diameter coverslips.
-
a.To generate dissociated hippocampal cultures, postnatal pups (P1-2 rats) are decapitated at the head/neck junction with surgical scissors. The entire brain is then removed from the skull using a surgical scalpel and put into cold 20% FBS containing Hank’s balanced salt solution. With a dissection microscope the cerebellum is removed, the two brain hemispheres are split along the longitudinal fissure, and midbrain tissue is removed. The hippocampus is subsequently removed from each hemisphere and placed in cold 20% FBS containing Hank’s balanced salt solution. Tissue is then washed and treated with 10 mg/mL of trypsin and 0.5 mg/mL DNase for 10 min at 37°C. Following trypsinization tissue is washed again and mechanically dissociated. Cells are then plated on 1:50 Matrigel:MEM coated glass coverslips. Hippocampal tissue from one rat pup is used to plate 10 coverslips. Neurons are incubated for the first 24 h in media containing: MEM (no phenol red), 5 g/l D-glucose, 0.2 g/l NaHCO3, 0.1 g/l transferrin, 10% FBS, 2 mM L-glutamine and 20 mg/l insulin. On DIV 1 media is changed to growth media with 5% FBS, 0.5 mM L-glutamine, no insulin, and the addition of B27 supplement and 4 μM cytosine arabinoside. On DIV 4 growth media is changed to contain only 2 μM cytosine arabinoside. Cells are maintained at 37°C in a 5% CO2 atmosphere until DIV 14–18 for experiments3 (Figure 1).
-
a.
Note: We have found 35 mm MatTek dishes with #1.5 thickness 12 mm coverslips are optimal for all three major steps of the protocol: culturing, staining, and imaging.
Figure 1.
Hippocampal dissection and preparation of primary cultures
Steps to isolate hippocampi from P1/2 pups and prepare primary cultures.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Gephyrin (mouse monoclonal) | Synaptic Systems | Cat# 147 021; RRID: AB_2232546 |
| Anti-vGAT (rabbit monoclonal) | Synaptic Systems | Cat# 131 008; RRID: AB_2800534 |
| Anti-MAP2 (guinea pig polyclonal) | Synaptic Systems | Cat# 188-004 RRID: AB_2138181 |
| AF488 | Invitrogen | A11073 |
| AF568 | Invitrogen | A10042 |
| AF647 | Invitrogen | A21236 |
| Chemicals, peptides, and recombinant proteins | ||
| Glucose oxidase from Aspergillus niger | Sigma-Aldrich | Cat# G2133 |
| Cysteamine | Sigma-Aldrich | Cat#30070 |
| Catalase from bovine liver | Sigma-Aldrich | Cat#C40 |
| Artemisinin | Sigma-Aldrich | Cat# 361593 |
| Artemether | Sigma-Aldrich | Cat# A9361 |
| Artesunate | Sigma-Aldrich | Cat# A3731 |
| Paraformaldehyde | Sigma-Aldrich | Cat# P6148 |
| NaOH | Sigma-Aldrich | Cat# S8045 |
| PBS10× | Thermo Fisher | Cat# 71448-16 |
| Sucrose | Sigma-Aldrich | Cat# S0389 |
| Triton X-100 | Sigma-Aldrich | Cat# T9284 |
| Normal goat serum | Vector Laboratories | Cat# S-1000 |
| Bovine serum albumin | Sigma-Aldrich | Cat# A9647 |
| Sodium azide, 5% (w/v) aqueous solution | Thermo Fisher | Cat# 71448-16 |
| 1 mM Tris-HCl, pH 8.0 | Corning | Cat# 46-031-CM |
| NaCl | Thermo Fisher | Cat# BP358-1 |
| Glucose | Sigma-Aldrich | Cat# G7528 |
| Hydrochloric acid | Fisher Chemical | Cat# 7647-01-0 |
| Poly-D-lysine | Thermo Fisher | Cat# A3890401 |
| Experimental models: Organisms/strains | ||
| Sprague-Dawley rat pups (P1–P2) | Charles River | Strain code: 400 |
| Software and algorithms | ||
| Prism 8 | GraphPad | http://www.graphpad.com |
| Vutara VXL software | Bruker | N/A |
| Other | ||
| Vutara Comprehensive workstation | Bruker | N/A |
| MatTek glass bottom dishes #1.5 | MatTek | Cat# P35G-1.5-14-C |
| pH strips | Fisher Scientific | Cat# 13-640-516 |
| Tetraspeck beads | Thermo Fisher | T7279 |
| Silicone immersion oil | Olympus | N/A |
Materials and equipment
Equipment set-up:
Vutara VXL Comprehensive Workstation (microscope and software):
Image acquisition, single molecule localization, and analysis were completed on a Vutara comprehensive workstation. Which includes a VXL Single Molecule Localization (SML) Super-Resolution Microscope outfitted for 3D direct stochastic optical reconstruction microscopy (dSTORM) with 405 nm, 488 nm, 555 nm, and 640 nm excitation lasers with biplane illumination, a 60× objective, a sCMOS (scientific Complementary metal–oxide–semiconductor) detector, and SRX analysis software.
Artemisinins
| Reagent | Stock concentration | Working concentration | Final concentration |
|---|---|---|---|
| Artemisinin | 175 mM | 10 mM | 50 μM |
| Artemether | 50 mM | 10 mM | 50 μM |
| Artesunate | 65 mM | 10 mM | 50 μM |
Store stock solution at RT for up to one month or −20°C for up to 3 months.
Preparation: Dissolve appropriate amount of Artemisinins based on molecular weight in DMSO to reach desired stock concentration. Prepare working concentration day of experiments.
4% paraformaldehyde buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Paraformaldehyde (PFA) | 4% | 5 g |
| NaOH | 4% | 5 g |
| PBS 10× | 10% | 12.5 mL |
| Sucrose | 4% | 5 g |
| ddH2O | N/A | 112.5 mL |
| Total | N/A | 125 mL |
Store solution at 4°C for up to one month.
Preparation: Dissolve PFA and NaOH in ddH2O over medium heat in the hood. When the PFA is dissolved the solution will go from milky to clear. Adjust the pH to 7. Then add the PBS and sucrose. Filter the final solution.
CRITICAL: Paraformaldehyde is toxic and a potential carcinogen and therefore should be prepared in the hood with appropriate PPE (gloves, lab coat, safety glasses, and mask). We preferably use pH strips to test pH, in order to prevent pH meter contamination (an approximate pH is sufficient for this buffer).
0.2% Triton X-100 buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS 1× | N/A | 29.94 mL |
| Triton X-100 | 0.2% | 60 μL |
| Total | N/A | 30 mL |
Store solution at 4°C in well-sealed for up to six months.
Preparation: Dilute PBS10× (listed in key resources table) to 1× with ddH2O.
Note: Prepare 10% stock solution and before the experiment dilute to 0.2% with PBS. Due to the viscosity of Triton X it is easier to prepare a higher concentration stock solution to work from.
Blocking solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Normal goat serum | 2% | 400 μL |
| BSA | 1% | 0.2 g |
| PBS 1× | X | 20 mL |
| Total | N/A | 20 mL |
Make fresh before use.
Alternatives: If secondary antibodies are from donkey and goat, prepare blocking solution that contains 2% of each species serum.
Sample storage buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 4% PFA Buffer | 0.5% | 2.5 mL |
| Sodium azide (5%) | 0.02% | 80 μL |
| PBS 1× | N/A | 17.42 mL |
| Total | N/A | 20 mL |
Store solution at 4°C for up to one month.
Dilution buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 1 M Tris, pH 8.0 | 10 mM | 0.1 mL |
| 1 M NaCl solution | 50 mM | 0.5 mL |
| PBS 1× | N/A | 9.4 mL |
| Total | N/A | 10 mL |
Store solution at room temperature for up to 6 months.
Imaging buffer base
| Reagent | Final concentration | Amount |
|---|---|---|
| Glucose | 10% | 1 g |
| 1 M Tris, pH 8.0 | 50 mM | 0.5 mL |
| 1 mM NaCl solution | 10 mM | 0.1 mL |
| Total | N/A | 10 mL |
Store solution at room temperature for up to 6 months.
Enzyme stock solution (oxygen scavengers)
| Reagent | Final concentration | Amount |
|---|---|---|
| Glucose Oxidase | N/A | 14 mg |
| Dilution Buffer (ingredients detailed above) | N/A | 200 μL |
| Catalase [20 mg/mL] | N/A | 50 μL |
| Total | N/A | 250 μL |
Store solution at 4°C for up to 2 weeks.
Preparation: Dissolve glucose oxidase in 200 μL of dilution buffer. Dilute 20 mg of catalase in 1 mL of dilution buffer separately. Add 50 μL of catalase solution to the glucose oxidase solution.4
Cysteamine (MEA) solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Cysteamine | 1 M | 77 mg |
| 360 mM Hydrochloric acid | N/A | 1 mL |
| Total | N/A | 1 mL |
Store solution at 4°C for up to 1 month.4
CRITICAL: Store solid MEA at 4°C in desiccating conditions. Remove MEA from the fridge and let warm to room temperature before weighing.
dSTORM imaging buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Imaging buffer base | N/A | 890 μL |
| Enzyme Stock solution | 1× | 10 μL |
| MEA solution | 100 mM | 100 μL |
| Total | N/A | 1 mL |
Make solution fresh before imaging and use for up to 2 h.4
CRITICAL: Prepare solution immediately before use on ice. The buffer lasts for about two hours in an open room temperature environment, efficacy of fluorophore photo-switching will be determined by buffer conditions. Prepare fresh buffer at least every 2 h.
Alternatives: dSTORM imaging buffer with BME
dSTORM imaging buffer with βME
| Reagent | Final concentration | Amount |
|---|---|---|
| Imaging buffer base | N/A | 980 μL |
| Enzyme Stock solution | 1× | 10 μL |
| βME solution | 1× | 10 μL |
| Total | N/A | 1 mL |
Make solution fresh before imaging and use for up to 2 h.4
CRITICAL: Ensure to prepare fresh βME.
CRITICAL: Prepare solution immediately before use on ice. The buffer lasts for about two hours in an open room temperature environment, efficacy of fluorophore photo-switching will be determined by buffer conditions. Prepare fresh buffer at least every 2 h.
Step-by-step method details
Artemisinins treatment
Timing: ∼1 h
This section explains the steps to treat hippocampal neurons with Artemisinins. In Guzikowski and Kavalali (2022)1 this treatment scheme was used to understand how pharmacological manipulation with parental Artemisinin and its’ the two semisynthetic derivatives (Artemether and Artesunate) disrupts GABAergic synapse nano-organization. Therefore, the first section of this protocol is to treat cells with the Artemisinins. All three Artemisinins were used as their orientation in the Gephyrin (GephE domain) universal receptor binding pocket is different.2 An acute 1-h treatment was used to rapidly probe the functional and structural effects of distributed GABAAR-gephyrin binding however treatment length can be extended.
-
1.
The day of experiments prepare working solutions of 10 mM Artemisinin, Artemether, and Artesunate.
-
2.Treat primary hippocampal cultures with 50 μM of Artemether, Artemisinin, and Artesunate for 1 h by directly adding Artemisinins to growth media. Treat control samples with the equivalent volume of DMSO.
-
a.Recommended to have volume of DMSO < 1% of total volume.
-
a.
Note: Immediately move to step 3 following 1 h incubation to begin fixation.
Note: The above culturing and Artemisinins treatment protocol can also be used for electrophysiology whole cell voltage and current clamp recordings. It is important to also include Artemisinins in bath solution while recording.
3-color immunocytochemistry
Timing: ∼1.5 days
This section explains the steps to stain primary hippocampal cultures with desired primary and secondary antibodies to visualize protein location within the neuron. In Guzikowski and Kavalali (2022)1 the main protein of interest was post-synaptic protein, gephyrin. In addition to gephyrin staining vesicular GABA transporter (vGAT) was stained for as a pre-synaptic marker and Microtubule-associated protein 2 (MAP2) as a dendritic marker.
Day 1.
-
3.
Remove growth media from cell culture MatTek dishes and immediately replace with 4% PFA solution for 20 min at room temperature (RT) (20°C–22°C). Following fixation wash cells 3 × 5 min with PBS 1×.
-
4.
Remove PBS and permeabilize with 0.2% Triton X-100 for 30 min at RT (20°C–22°C). Following permeabilization wash cells 3 × 5 min with PBS 1×.
-
5.
Block with 1% BSA and 2% normal goat serum diluted in PBS for 2 h at RT (20°C–22°C).
-
6.Prepare primary antibodies in blocking buffer.
-
a.Prepare 120 μL of antibody solution per coverslip (optimize antibody solution volume depending on coverslip diameter, just need to cover). To investigate gephyrin, stain inhibitory presynaptic marker, vesicular GABA transporter (vGAT) (1:500), inhibitory postsynaptic scaffold protein gephyrin (1:150) and dendritic marker MAP2 (1:1000) (Refer to Table 1 for antibody dilution and concentration information).
-
a.
-
7.
Incubate cells in primary antibody diluted in blocking buffer overnight (15–20 h) at 4°C. Place dishes into a humid chamber for incubation to ensure primary antibody does not evaporate.
Note: Humid chambers can be made simply from supplies around the lab – an empty tip box padded with a layer of moist tissues will ensure the primary antibody solution does not evaporate during overnight incubation inside the tip box.
Note: For optimization of primary antibody concentration start at a 1:500 dilution or concentration recommended by the supplier (Refer to troubleshooting problems 1 and 2). In tissue increasing staining incubation length can increase antibody penetrance however for staining in cell culture, concentration should initially be adjusted before incubation length.
Note: Always vortex and spin down antibodies before use.
Table 1.
Antibody concentrations and dilutions
| Antibody | Stock concentration | Dilution |
|---|---|---|
| Anti-Gephyrin (mouse monoclonal) | 1 μL/μL Hybridoma supernatant | 1:150 |
| Anti-vGAT (rabbit monoclonal) | 1 mg/mL | 1:500 |
| Anti-MAP2 (guinea pig polyclonal) | 1 μL/μL Antiserum | 1:1000 |
| AF488 | 2 mg/mL | 1:500 |
| AF568 | 2 mg/mL | 1:1000 |
| AF647 | 2 mg/mL | 1:350 |
Manufactures instructions were followed for all antibody dilutions.
Day 2.
-
8.
Wash cells 3 × 5 min with PBS 1×.
-
9.Prepare secondary antibodies in blocking buffer.
-
a.Prepare 120 μL of antibody solution per coverslip (optimize antibody solution volume depending on coverslip diameter, just need to cover). To tag the primary antibodies listed above use AF647 (1:350), AF568 (1:1000), and AF488 (1:500). Gephyrin is the primary protein of interest and was therefore tagged with the AF647 antibody and vGAT with AF568.
-
a.
Note: AF647 is an excellent fluorophore for dSTORM as it is extremely bright (high photon yield per switching event) and has a low on-off duty cycle. Therefore, if performing single color dSTORM, AF647 should be used. The quality of super resolution images is directly dependent on the photo-switchable probe’s photon number, duty cycle, survival fraction, and the number of switching cycles,5 therefore the selection of fluorophores is of utmost importance. Although in our imaging we used AF568, CF568 is a better fluorophore in the orange channel. Additionally, MAP2 was only used as a reference under widefield and therefore a dSTORM compatible antibody was not used. If you would like to conduct 3-color dSTORM use Atto488.
Note: For optimization of primary antibody concentration start at a 1:500 dilution or concentration recommended by the supplier (Refer to troubleshooting problems 1 and 2).
Note: Always vortex and spin down antibodies before use.
-
10.
Incubate cells in secondary antibody diluted in blocking buffer for 90 min at RT (20°C–22°C). Place dishes back into the humid chamber for incubation to ensure secondary antibody does not evaporate. Cover humid chamber in foil to prevent photobleaching of secondary antibodies.
-
11.
Complete a post staining fixation by incubating cells in 4% PFA buffer for 20 min at RT (20°C–22°C).
-
12.
Store samples in storage buffer in dark at 4°C.
Note: Fixation time can be optimized to mitigate potential side effects.
Pause point: Samples can be stored for at least one month before imaging (have not used samples beyond that timepoint).
dSTORM image acquisition and SML localization
Timing: Typically, it takes ∼10 min per image (50 fps), requiring multiple hours-days to image an entire experiment
This section describes the steps to acquire images and localize single molecules of inhibitory presynaptic protein vGAT and post-synaptic scaffold protein gephyrin. The imaging acquisition and analysis portion of this protocol can be expanded to a variety of other synaptic proteins to address the same fundamental question - how do different proteins cluster and organize at the synapse. This protocol is robust and applicable to other synaptic proteins with basic adjustments in antibody concentrations and the optimization of image acquisition settings (please see troubleshooting section). In our laboratory this protocol has be used to investigate excitatory synapses in primary hippocampal culture, visualizing vGlut, PSD-95, Bassoon and in hippocampal slices visualizing Homer, Bassoon, GluA1, and GluA2.
Note: The microscopy set-up used had these essential components: biplane illumination, lasers: 405 nm, 488 nm, 555 nm, and 640 nm, sCMOS detector, multi-band dichroic mirror, pentaband emission filter, and standard emission single bandpass filters (405, 488, 555, 640). Lasers that are compatible with 647, 568, and 488 (red, orange, and green respectively) fluorophores must be used.
-
13.
Optical Calibration: For accurate single molecule localization (SML) the focal planes and color channels of the optical system need to be aligned. Furthermore, to generate 3D measured point spread functions for sub-diffraction limit localization the point spread function (PSF) of a point source must be determined. 100 nm Tetraspeck bead samples that emit at blue, green, orange, and red were prepared on poly-d-lysine coated MatTek dishes [1:500] to calibrate the channels and planes. Resolution: X-Y: 20 nm, Z: 50 nm.
-
14.
Take dSTORM images with a 60× objective (1.3 NA, silicone oil).
-
15.
Apply one drop of silicone immersion oil to the center of 60× objective (ensure no air bubbles during application).
-
16.
Mount MatTek dish in chamber and replace storage solution with imaging buffer (∼200 μL to only cover the coverslip). Note the time to ensure image acquisition is within the buffer’s lifetime.
-
17.
Mount the chamber into the stage-holder.
-
18.
Using the 488 channel (exciting fluorophores tagged to MAP2) find your region of interest. Conduct search on a low laser power to prevent photo-switching and photobleaching. Ideal region is not too dense to allow for the isolation of pre and post synaptic localizations.
-
19.Perform sequential imaging of the 640 channel and then the 555 channel. Begin image acquisition with constant illumination with the 640 nm laser at 463.4 mW while gradually increasing the 405 laser power from 1–10% of max power (49.7 mW) over imaging period. Take 20,000 frames of the 647 channel. Then illuminate the 555 nm laser at constant 555 mW for 10,000 frames. Generally, 3 regions of interest were imaged per coverslip.
-
a.Take single z-plane images (z-distance = 2 μm due to biplane illumination).
-
b.Exposure time: 20 ms, 50 Frames per second.
-
c.Widefield field of view of 200 μm × 200 μm and super resolution field of view 50 μm × 50 μm.
-
a.
Note: Upon excitation of the sample there should be robust photo-switching of the fluorophore, appearing as blinking. Following initiation, photo-switching should reach a steady state level. Adequate blinking is fundamental to SML therefore antibody concentrations, buffer conditions, and laser power should be optimized for experiments (please refer to troubleshooting problems 1, 2, and 3).
Note: Silicone oil closely matches the refractive index of cells decreasing spherical aberration while imaging and therefore increasing the quality of super resolution images; neurons (n = ∼1.38), silicone oil (n = 1.406).6
-
20.Identify individual molecules by their brightness above the background in a frame-by-frame process (Figure 2). Generate a molecule’s coordinates in space by fitting the imaged PSF with the 3D measured PSF (step 13). As the raw data comes from the camera the SRX software works in parallel to conduct single molecule localizations. Images are filtered, particles are defined, and fluorophores localized. Analysis and filtering settings7:
-
a.Particle Detection: Parameters for PSF identification and extraction (filtering) to define a particle:
-
i.Cutout width: 15 px (1px = 100 nm) - Size around an ROI’s brightest pixel that is used for localization routines.
-
ii.ROI border: 0 px - How large of a region around the field of view border is discarded.
-
iii.Max frame accumulation: 1 frame - Depending how long a fluorophore is in its “on” state (fluorescing) will determine how many frames it is visible. If a fluorophore is active for a longer period of time max frame accumulation should be increased so the same fluorophore will not be detected as multiple blinking events.
-
iv.Max accumulation offset: 1px (100 nm) - Degree of fluorophore shift across sequential frames to be considered the same fluorophore.
-
v.Max particles per frame: no max - Number of particles to be localized per frame.
-
vi.Background slope removal threshold: 20 - The threshold of heterogeneity across the background of fluorophore’s cutout that is admissible for localization.
-
vii.Cutout/background removal threshold: 0.2 - A signal to noise threshold ((total photon count of cutout – background photon count)/background photon count) that determines if a fluorophore is included for localization.
-
viii.Detection bandpass low frequency cutoff: 7.07px, Detection bandpass high frequency cutoff: 1.41px - 1st step of localization is the application of a smoothing filter to reduce noise for fluorophore identification.
-
i.
-
b.Parameters for fluorophore localization:
-
i.3D measured point spread functions are used for localization (experimental PSF is collected from bead calibration step 13). This is the optimal routine to use as calibration is specific to your system and imaging set-up.
-
ii.Max iterations: 40 – maximum number of iterations for one candidate particle.
-
iii.PSF Interpolation Spline method: B-Spline – method for PSF fitting for localization.
-
iv.Temporal Median Filter: 0 frames – method for the estimation of background signal.8
-
v.Background threshold 647 channel: 50, 568 channel: 200 - Fluorescence values above the mean background that the software will treat as a localization.
-
i.
-
a.
Note: Refer to troubleshooting problem 4 for suggestions on how to optimize localization parameters.
Note: Single molecule localizations can generate large data sets, ensure your computer or external hard drive have adequate storage space (TB range).
Figure 2.
Representative images during particle detection and fluorophore localization
(A) Representative images of the experimental point spread function (PSF) in the XY and YZ views for the two imaging colors (red 647, orange 568) collected in step 13.
(B) Particle accumulation of single molecule localizations summated from multiple frames of a neuronal projection.
(C) Representative image of a single frame during particle detection with detected particles in pink boxes.
dSTORM image analysis
Timing: Typically, it takes ∼10 min per image, requiring multiple hours to days to analyze an entire experiment
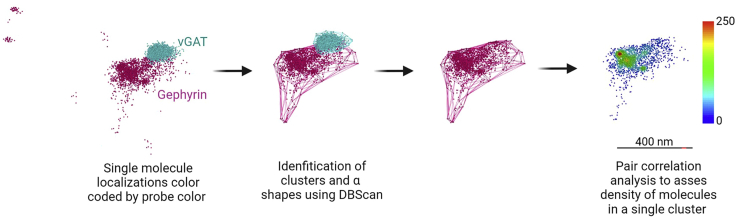
This section describes the steps to analyze single molecule localizations of inhibitory presynaptic marker vGAT and post-synaptic scaffold protein gephyrin. After single molecules have been localized to a X,Y, and Z position in space based on their PSF, they are then grouped and clustered; single molecules are localized by their PSFs and clusters defined based on their spatial proximity to other molecules. To define synaptic clusters a clustering algorithm Density-Based Spatial Clustering of Applications with Noise (DBScan) is used as it can define arbitrarily shaped clusters from a spatial database.9 Stochastic optical reconstruction microscopy-based relative localization analysis (Storm-RLA) is used to define a pre- and post-synaptic pair.
Synaptic protein cluster analysis
The goal of this analysis is to understand the clustering dynamics of vGAT and gephyrin single molecule localizations using a proximity- and density-based analysis.
-
21.Conduct Density-Based Spatial Clustering of Applications with Noise (DBScan) to define clusters of SMLs of the same probe based on proximity (Figures 3 and 4). Parameters:
-
a.Maximum Particle distance: 0.1 μm - threshold distance to determine if a particle is in one cluster or not clustered.
-
b.Cluster minimum particle count: 6 particles - need at least 6 particles within the defined distance to be labeled a cluster.
- c.
-
a.
Note: Cluster characteristics of the least dense cluster were used as global parameters to set threshold values.9 Due to Artemisinins treatment disrupting gephyrin clustering, the cluster minimum particle count value was quite low and should be optimized based on experimental question.
-
22.
Following the determination of clusters, localizations not grouped were eliminated from analysis and considered noise (Figure 4).
-
23.
From α shapes calculate cluster volume.
Figure 3.
Image analysis pipeline
Single molecule localizations of the same probe are clustered according to the DBScan algorithm. Molecules not localized to a cluster are eliminated from further analysis. Cluster α shapes are determined and further analysis is conducted on gephyrin volume, vGAT-gephyrin centroid distance, and density. (Representative clusters obtained from data generated in Guzikowski and Kavalali,1 Images have been manipulated to represent potential outcomes of this protocol).
Figure 4.
DBScan clustering schematic
r is the distance around one molecule that is searched for another molecule of the same probe. For a molecule to form a cluster it must have 2 molecules within the distance r. This molecule is considered a core molecule. The same routine is conducted for the 2 new molecules and subsequent localizations in their radii are considered part of the original cluster. If a molecule is considered part of a cluster but does not meet the density parameter, it is considered a boundary molecule. A molecule that does not meet the density or distance parameters is considered noise.9
Pre-synaptic vGAT and post-synaptic gephyrin cluster spatial proximity
The spatial distribution of vGAT and gephyrin clusters were mapped to define which clusters composed a pre- and post-synaptic juxtaposed synapse using stochastic optical reconstruction microscopy-based relative localization analysis (Storm-RLA). Only vGAT and gephyrin clusters that were categorized as a paired pre- and post-synapse using this relative localization analysis (details in step 24) were included in further analysis.
Note: With conventional confocal microscopy often pre-post-synapse colocalization analysis is done to define synaptic puncta. However, with super resolution microscopy x-y resolution is 20 nm, therefore pre- and post-markers can be differentiated in space and a spatial proximity analysis is performed (however there can be overlap in localizations from two different probes).
-
24.Investigate the spatial organization of vGAT and gephyrin clusters using stochastic optical reconstruction microscopy-based relative localization analysis (Storm-RLA). Using the Storm—RLA algorithm cluster α shapes generated in step 21 are used to define a pre- and post-synaptic pair based on their position in space. From this eliminate non-paired clusters and calculate vGAT and gephyrin cluster centroid distance (Figure 3).11 Parameters:
-
a.Max cluster distance: 1 μm (generous threshold however the matrix created allowed the selection of the closest vGAT and gephyrin clusters as a synapse).
-
a.
Cluster density analysis
To understand the spatial distribution of gephyrin molecules.
-
25.Asses individual gephyrin clusters in 2D by condensing localizations to the X-Y plane. For each individual cluster determine a theoretical pair correlation function (PCF). A theoretical fit accounts for the density and shape of the cluster and generates a function as if the molecules were randomly organized. To compare density distributions across groups normalize individual PCFs of each individual cluster to their theoretical fit to account for shape, volume, and density.12,13 Parameters:
-
a.Search radius from center: 500 nm, 100 bins.
- b.
-
a.
Note: The pair correlation function generated represents the point pattern of individual gephyrin molecules within a single cluster; g(r) as a function of distance from reference particle. Ultimately providing insight into density variations and nanoclustering within a defined cluster.
Figure 5.
Pair correlation function density analysis
Example pair correlation functions with corresponding gephyrin clusters colored coded by heat map. The function quantifies the probability of another molecule being within a given distance, g(r), of a given molecule. With a g(r) > 1 representing a heterogenous organization (i.e., clustering), g(r) = 1 a uniform random distribution and g(r) < 1 a molecularly de-enriched area. The peaks in the function when g(r) > 1 demonstrate sub-synaptic clustering within a single cluster (Images have been manipulated to represent potential outcomes of this protocol).
Expected outcomes
Synaptic nanostructure is fundamental in directing different modes of neurotransmission and their autonomous downstream roles.1,10,14,15,16 Understanding basal synaptic structural dynamics in addition to how pharmacological or genetic manipulations disrupt synaptic architecture provides insight into how an individual protein’s organization facilitates cell signaling. Since dSTORM microscopy provides single synapse level information this staining protocol in parallel to electrophysiology is a very powerful tool; allowing you to delineate how structural changes in scaffolding proteins might lend to changes in evoked and spontaneous neurotransmission. We have demonstrated that gephyrin clusters in rat hippocampal neurons are mostly uni-domain with the center having a higher density of molecules than the periphery. Furthermore, using Artemisinins as a tool to disrupt GABAAR binding to gephyrin we establish a center surround organization of evoked and spontaneous GABAergic neurotransmission at central synapses, with the periphery of gephyrin clusters fundamental in maintaining spontaneous neurotransmission.1
Quantification and statistical analysis
The image analysis steps detailed above will produce individual data points for cluster volume, vGAT-gephyrin centroid distance, and pair correlation functions for each gephyrin cluster. For statistical analysis GraphPad Prism was used to compare groups pre and post pharmacological manipulation.1 For statistical analysis experimental replicates were pooled together and an equal number of points were selected to compare across groups, a test for normality was run, and the appropriate statistical test conducted. To analyze pair correlation functions the y = 1 intercepts of cluster functions were compared.
Limitations
The super-resolution optical analysis of synaptic scaffold proteins is a valuable methodology to visualize the synapse at the nanometer level. However, it should be noted that the use of this protocol requires multiple points of optimization for your microscopy set-up. First, sample preparation and antibody concentrations must be optimized for protein density and synapse density. Second, due to variation in labeling density and fluorophore brightness PSF identification, extraction and single molecule localization criteria need to be fine-tuned to detect signal while preventing the inclusion of noise. The first parameters to consider are max frame accumulation and background removal thresholds. Furthermore, this analysis does not account for neuronal morphology i.e., where post-synaptic clusters are located on the dendrite, spine vs. shaft. If neuronal morphology is of interest other super-resolution approaches (e.g., STED nanoscopy) or conventional light microscopy methods will likely be better suited (cell filling probes can be used to visualize morphology).
Troubleshooting
Problem 1
Weak Staining (step 7).
Potential solution
The immunoreactivity of different primary antibodies can cause weak staining, while the recommended gephyrin concentration is high, 1:150, this can be increased to boost immunoreactivity. Furthermore, to optimize antibody concentrations the dilution can be adjusted while all other variables are maintained (buffer and incubation length).
Problem 2
High background (steps 7, 10).
Potential solution
Primary or secondary antibody concentration is potentially too high. To combat this antibody concentrations can be decreased, or secondary incubation time can be decreased to 1 h.
Problem 3
Low or absent photo-switching (step 19).
Potential solution
One of the most sensitive portions of the protocol is the imaging buffer. Quality photo-switching is directly dependent on buffer quality ultimately determining the success of the experiment.
Prepare fresh solutions of Glucose Oxidase, Catalase and MEA, if any have degraded, fluorophores will not be able to photo-switch well. Ensure you are using imaging buffer for no more than 2 h at room-temperature and are keeping all stock solutions on ice. To increase the longevity of the buffer you can seal your sample. Double check the pH of your buffer solution is pH 8.
Buffer components can also be optimized for your sample and the number of colors being imaged. For single color imaging the 647 channel and a βME based imaging buffer should be used. A βME based buffer will produce a higher photon count relative to MEA. However, if your sample is densely labeled, you are labeling highly abundant proteins, or using more than one color a MEA based buffer is optimal.
Laser power must be strong enough to cause fluorophores to photo-switch. Low laser power should be used to find regions of interest (view as an epifluorescence image) and increased for image acquisition. Laser power should be optimized for photo-switching behavior.
Problem 4
Poor single molecule localization (step 20).
Potential solution
The first thing to check is that image acquisition is working correctly, and fluorophore photo-switching is robust. Following this, parameters for PSF identification, extraction and molecule localization can be adjusted to ensure the adequate detection of signal. Parameter optimization suggestions7:
-
•
Cutout width: If samples are densely labeled the cutout width should be lowered to decrease the probability that light from another particle is included in localization.
-
•
Max frame accumulation: It is important to tune this setting to the blinking behavior of individual fluorophores. Depending how long a fluorophore is in its “on” state (fluorescing) will determine how many frames it is visible. If a fluorophore is active for a longer period of time max frame accumulation should be increased so the same fluorophore will not be detected as multiple blinking events. If there is a high degree of noise adjust max frame accumulation to eliminate bright spots that aren’t blinking from your localization acquisition.
-
•
Max particles per frame: Can be optimized to eliminate dim candidate events from localization routines. If you want to only localize a subset of the brightest fluorophores per frame the max number of particles per frame can be set.
-
•
Back slope removal threshold: Adjusted based on background homogeneity of fluorophore cutouts of your sample. If your background is heterogenous the background slope threshold can be increased. For brain slices this value can be increased due to the decreased homogeneity of the imaging area relative to culture.
-
•
Cutout/background removal threshold: Adjusted based on signal to noise ratio, i.e., if the background is high this number can be lowered. For brain slices this value can be decreased due to high background signal.
-
•
Background threshold: If your signal is dim decrease background thresholds, typically CF568 will not be as bright as AF647. For brain slices these values were decreased relative to cell culture due to weaker signal in tissue.
Problem 5
Discrepancies in protein organization analysis compared to what is in the literature (steps 20, 21, 25).
Potential solution
Particle detection and fluorophore localization threshold settings can cause discrepancies in results across research groups. Furthermore, parameters set for DBScan strongly effect cluster size and density measurements. The normalization of data within your experimental set-up and treatment group will greatly augment the applicability of results across research groups, ultimately aiding the consensus of how different manipulations alter structure.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ege T. Kavalali (ege.kavalali@vanderbilt.edu).
Materials availability
This study did not generate new reagents.
Acknowledgments
We thank Dr. Melike Lakadamyali (Univ. of Pennsylvania) for sharing her expertise in dSTORM imaging buffer optimization. This work was supported by the National Institute of Mental Health (grant MH066198 to E.T.K. and T32 MH064913 to N.J.G). Figure schematics were created with BioRender.com.
Author contributions
Conceptualization, discussion, writing, and editing: N.J.G., E.T.K.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Natalie J. Guzikowski, Email: natalie.j.guzikowski@vanderbilt.edu.
Ege T. Kavalali, Email: ege.kavalali@vanderbilt.edu.
Data and code availability
No original code has been generated in this study.
References
- 1.Guzikowski N.J., Kavalali E.T. Nano-organization of spontaneous GABAergic transmission directs its autonomous function in neuronal signaling. Cell Rep. 2022;40:111172. doi: 10.1016/j.celrep.2022.111172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasaragod V.B., Hausrat T.J., Schaefer N., Kuhn M., Christensen N.R., Tessmer I., Maric H.M., Madsen K.L., Sotriffer C., Villmann C., et al. Elucidating the molecular basis for inhibitory neurotransmission regulation by Artemisinins. Neuron. 2019;101:673–689.e11. doi: 10.1016/j.neuron.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Kavalali E.T., Klingauf J., Tsien R.W. Activity-dependent regulation of synaptic clustering in a hippocampal culture system. Proc. Natl. Acad. Sci. USA. 1999;96:12893–12900. doi: 10.1073/pnas.96.22.12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin L., Vicario C., Castells-García Á., Lakadamyali M., Neguembor M.V., Cosma M.P. A protocol to quantify chromatin compaction with confocal and super-resolution microscopy in cultured cells. STAR Protoc. 2021;2:100865. doi: 10.1016/j.xpro.2021.100865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dempsey G.T., Vaughan J.C., Chen K.H., Bates M., Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gul B., Ashraf S., Khan S., Nisar H., Ahmad I. Cell refractive index: models, insights, applications and future perspectives. Photodiagnosis Photodyn. Ther. 2021;33:102096. doi: 10.1016/j.pdpdt.2020.102096. [DOI] [PubMed] [Google Scholar]
- 7.Ebeling C. SRX full system manual | SRX manual. 2020. https://guide.vutara.bruker.com/m/11201
- 8.Hoogendoorn E., Crosby K.C., Leyton-Puig D., Breedijk R.M.P., Jalink K., Gadella T.W.J., Postma M. The fidelity of stochastic single-molecule super-resolution reconstructions critically depends upon robust background estimation. Sci. Rep. 2014;4:3854. doi: 10.1038/srep03854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ester M., Kriegel H.-P., Sander J., Xu X. Proceedings of the Second International Conference on Knowledge Discovery and Data Mining KDD’96. AAAI Press; 1996. A density-based algorithm for discovering clusters in large spatial databases with noise; pp. 226–231. [Google Scholar]
- 10.Tang A.-H., Chen H., Li T.P., Metzbower S.R., MacGillavry H.D., Blanpied T.A. A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature. 2016;536:210–214. doi: 10.1038/nature19058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veeraraghavan R., Gourdie R.G. Stochastic optical reconstruction microscopy–based relative localization analysis (STORM-RLA) for quantitative nanoscale assessment of spatial protein organization. Mol. Biol. Cell. 2016;27:3583–3590. doi: 10.1091/mbc.E16-02-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sengupta P., Jovanovic-Talisman T., Lippincott-Schwartz J. Quantifying spatial organization in point-localization superresolution images using pair correlation analysis. Nat. Protoc. 2013;8:345–354. doi: 10.1038/nprot.2013.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartley J.M., Chu T.-W., Peterson E.M., Zhang R., Yang J., Harris J., Kopeček J. Super-resolution imaging and quantitative analysis of membrane protein/lipid raft clustering mediated by cell-surface self-assembly of hybrid nanoconjugates. Chembiochem. 2015;16:1725–1729. doi: 10.1002/cbic.201500278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzikowski N.J., Kavalali E.T. Nano-organization at the synapse: segregation of distinct forms of neurotransmission. Front. Synaptic Neurosci. 2021;13:796498. doi: 10.3389/fnsyn.2021.796498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S., Raychaudhuri S., Lee S.A., Brockmann M.M., Wang J., Kusick G., Prater C., Syed S., Falahati H., Ramos R., et al. Asynchronous release sites align with NMDA receptors in mouse hippocampal synapses. Nat. Commun. 2021;12:677. doi: 10.1038/s41467-021-21004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramsey A.M., Tang A.-H., LeGates T.A., Gou X.-Z., Carbone B.E., Thompson S.M., Biederer T., Blanpied T.A. Subsynaptic positioning of AMPARs by LRRTM2 controls synaptic strength. Sci. Adv. 2021;7:eabf3126. doi: 10.1126/sciadv.abf3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No original code has been generated in this study.