Abstract
Isovaleric acidemia (IVA), due to isovaleryl-CoA dehydrogenase (IVD) deficiency, results in the accumulation of isovaleryl-CoA, isovaleric acid and secondary metabolites. The increase in these metabolites decreases mitochondrial energy production and increases oxidative stress. This contributes to the neuropathological features of IVA. A general assumption in the literature exists that glycine N-acyltransferase (GLYAT) plays a role in alleviating the symptoms experienced by IVA patients through the formation of N-isovalerylglycine. GLYAT forms part of the phase II glycine conjugation pathway in the liver and detoxifies excess acyl-CoA’s namely benzoyl-CoA. However, very few studies support GLYAT as the enzyme that conjugates isovaleryl-CoA to glycine. Furthermore, GLYATL1, a paralogue of GLYAT, conjugates phenylacetyl-CoA to glutamine. Therefore, GLYATL1 might also be a candidate for the formation of N-isovalerylglycine. Based on the findings from the literature review, we proposed that GLYAT or GLYATL1 can form N-isovalerylglycine in IVA patients. To test this hypothesis, we performed an in-silico analysis to determine which enzyme is more likely to conjugate isovaleryl-CoA with glycine using AutoDock Vina. Thereafter, we performed in vitro validation using purified enzyme preparations. The in-silico and in vitro findings suggested that both enzymes could form N-isovaleryglycine albeit at lower affinities than their preferred substrates. Furthermore, an increase in glycine concentration does not result in an increase in N-isovalerylglycine formation. The results from the critical literature appraisal, in-silico, and in vitro validation, suggest the importance of further investigating the reaction kinetics and binding behaviors between these substrates and enzymes in understanding the pathophysiology of IVA.
Keywords: Isovaleric acidemia, Glycine N-acyltransferase, Glycine conjugation, N-isovalerylglycine
Graphical Abstract
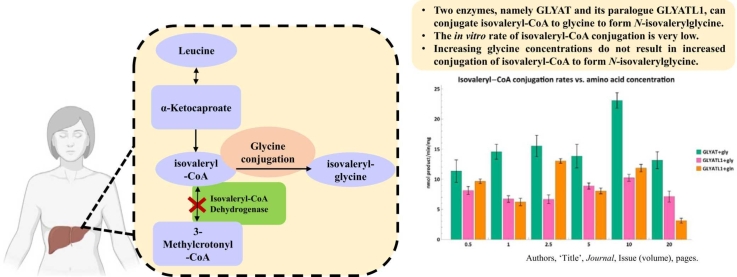
The glycine N-acyltransferases, GLYAT and GLYATL1, contribute to the detoxification of isovaleryl-CoA - an in-silico and in vitro validation.
1. Introduction
Isovaleric acidemia (IVA, OMIM: 243500) is a rare autosomal inherited recessive disorder first identified by Tanaka and Isselbacher (1967). The incidence of IVA is 1 in 250 000 in the USA [1], [2] and 1 in 526 000 in other Western populations [3]. In Germany, newborn screening (NBS) data collected between 2004 and 2017 showed a birth prevalence for IVA of 1 in 94 000 newborns [4]. In South Africa (ZA), no official statistics on the prevalence or incidence have been published. However, 15 patients have been diagnosed by the Centre for Human Metabolomics (CHM) at North-West University (NWU). In developing countries, such as ZA, NBS are typically accessed by the private sector, which accounts for the low incidence of IVA in these countries as only 6000 out of 1 000 000 births are screened [5].
IVA arises from a defect in isovaleryl-CoA dehydrogenase (IVD, E.C. 1.3.8.4) that leads to a defective catabolism of the branched chain amino acid, leucine [6]. IVD is mainly expressed in the thyroid, liver, and kidney (Data taken from NCBI – accessed 24 April 2022) and catalyzes the conversion of isovaleryl-CoA to 3-methylcrotonyl-CoA. An IVD enzyme defect results in the accumulation of various metabolites which are then shunted towards several secondary metabolic pathways including glycine conjugation. 3-Hydroxyisovaleric acid, N-isovalerylglycine and N-isovalerylcarnitine are utilized as primary biomarkers [7] in the diagnosis of IVA. Additionally, the accumulation of isovaleryl-CoA results in sequestration of CoA and formation of large amounts of isovaleric acid [8].
According to the proposed standard classification, IVA patients may fall within two phenotypic groups namely: the acute neonatal form and chronic intermittent form [9]. The acute neonatal form is characterized by encephalopathy, developmental delay, lethargy, and a sweaty feet odor. Mild neurocognitive and motor deficits are characteristic of the intermittent IVA form and is typically triggered by stresses such as infection, fever, a high-protein diet or extensive fasting [10]. With the expansion of NBS programs, an asymptomatic to mild phenotype of IVA has also been identified [11]. This emphasizes the importance of biochemical characterization and monitoring of patients to determine the degree of intervention required to assure optimal outcome [9], [10], [12], [13].
The underlying pathophysiology that leads to cerebral damage in these patients, is still not fully elucidated. Studies in rats indicate that metabolites which accumulate during IVA can induce oxidative stress in the cerebral cortex and that oxidative damage may be at least in part involved in the neuropathology of IVA [14]. Additionally, free isovaleric acid can reduce Na+ , K+ -ATPase activity in the synaptic membranes of cerebral cortexes of young rats, possibly via mechanisms involving lipid peroxidation [15]. Furthermore. hyperammonemic encephalopathy is a common feature in untreated/poorly managed patients as isovaleryl-CoA has an inhibitory effect on N-acetylglutamate synthase activity resulting in poor ammonia clearance via the urea cycle pathway [16]. The repercussion of this is irreversible central nervous damage because of accumulating ammonia acting as a neurotoxin. N-carbamylglutamate is an approved pharmacological agent for the treatment of acute and chronic hyperammonemia in IVA [17].
The haplotype frequency data for IVD available on the Ensembl database show that the reference haplotype is found at a frequency of 98.3% in the worldwide population while the remainder of the haplotypes have haplotype frequencies ranging from 0.02% to 1% (Data taken from Ensembl.org; accessed 19 Apr 2022 for transcript ENST00000487418.8 - Canonical transcript with TSL 1 score). This indicates that the IVD gene is highly conserved and that mutations are found at very low frequencies. Molecular analysis of the IVD gene has allowed for the identification of different types of pathogenic variants, with no straightforward phenotype–genotype correlation in most cases [10], [11]. Although a recent study recommends that additional investigations may be required to conclusively define if genotype-phenotype correlations can be made in some IVA populations [18]. The most common IVD mutation (c.932 C>T, p.A282V) results in a lower affinity for isovaleryl-CoA as well as a reduced catalytic efficiency and is associated with a mild biochemical, but clinical asymptomatic phenotype [4], [11]. The ZA Caucasian cohort are all homozygous for the same (c.367 G>A, p.G123R) mutation [13], which results in no protein expression or detectable IVD activity.
1.1. Treatment strategies for IVA
Presently there are three modes of treatment for IVA: prevention of metabolic decompensation by careful clinical observation, long term reduction of the production of isovaleryl-CoA through dietary manipulation, and activation of secondary metabolism by shunting isovaleryl-CoA towards reactions that produce non-toxic metabolites [7], [19]. These treatments, if applied timely, generally results in normal neurological development [13], [20], [21], [22], while motor dysfunction and neurological deficits have mostly been observed with delayed therapeutic intervention [9]. Interestingly, the same group emphasized that the neurocognitive outcome is not related to the catabolic episodes, and that further investigations are required to assess clinical heterogeneity. For this study, the focus will be on one of the secondary metabolic pathways involved in the third mode of treatment, namely glycine conjugation.
N-isovalerylglycine was identified in the urine of IVA patients by Tanaka and Isselbacher (1967). Isovaleryl-CoA, produced by oxidation of leucine through α-ketoisocaproic acid, cannot be utilized by primary metabolic pathways such as the citric acid cycle or β-oxidation. Under these circumstances, the conjugation of isovaleryl-CoA with glycine permits detoxication and elimination of accumulated isovaleryl-CoA as N-isovalerylglycine. This mechanism might be adequate to protect patients from the accumulation of isovaleric acid in the blood and other body fluids, which led to the use of glycine as treatment [23]. As a result, a clear clinical improvement was observed in IVA patients [24]. Unfortunately, as a result of the clinical variation of IVA, its rarity, and the lack of extensive, multi-center, longitudinal studies on patient outcomes, there is no consensus regarding optimal dietary management and the combined or singular use of glycine and L-carnitine for detoxification purposes [9], [25]. L-carnitine treatment decreases free isovaleric acid levels during acute metabolic decompensation [26].
Even though glycine is recommended as a treatment for IVA, several studies have reported adverse events in some patients [27], [28], [29]. A recent publication by Vliet et al. indicated that glycine supplementation should be carefully considered in a patient-to-patient basis [25]. This is since hyperglycinemia (in case of excess systemic glycine moving over the blood brain barrier) may result in acquired encephalopathy detrimental to the patient.
Tanaka and Isselbacher were the first to suggest that glycine N-acyltransferase (GLYAT, EC 2.3.1.13) might be the enzyme responsible for the formation of N-isovalerylglycine [23]. Even though Tanaka and Isselbacher referred to a study on bovine GLYAT [30], the literature states that human GLYAT can conjugate isovaleryl-CoA to glycine [13], [31], [32]. However, is there any evidence that human GLYAT is indeed responsible for the formation of N-isovalerylglycine?
1.2. The glycine conjugation pathway
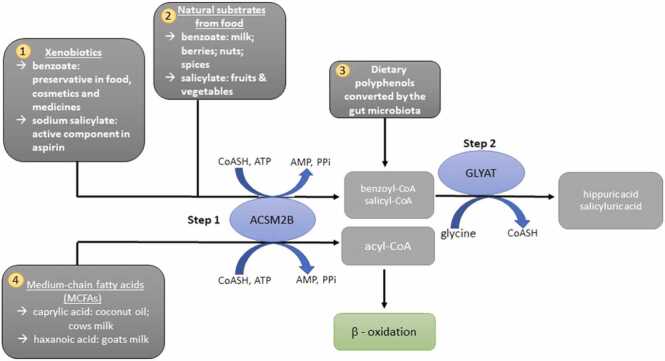
The glycine conjugation pathway is a two-step enzymatic reaction responsible for the detoxification of various substrates (Fig. 1) [23], [32], [33], [34], [35], [36]. The mitochondrial xenobiotic/medium chain fatty acid: CoA ligases (ACSM2B, EC 6.2.1.2) activate substrates, such as benzoate, to an acyl-CoA [37], [38]. The acyl-CoA’s are then subsequently conjugated to glycine by GLYAT [23], [30], [39]. The glycine conjugation pathway also plays a fundamental role in the homeostatic energy balance within the mitochondria by regenerating the CoA moiety [40].
Fig. 1.
An overview of the glycine conjugation pathway and the substrates that it detoxifies. 1 & 2: Benzoate and salicylate (food, medicine and preservatives) are activated to an acyl-CoA by the mitochondrial xenobiotic/medium chain fatty acid: CoA ligase (ACSM2B) and subsequently conjugated to glycine by glycine N-acyltransferase (GLYAT). 3. Gut microorganisms convert dietary polyphenols to benzoyl-CoA, which is a substrate for glycine conjugation. 4. MCFAs, example caprylic acid, are activated by ACSM2B ligase in the liver before entering the mitochondrial beta-oxidation cycle.
1.2.1. Glycine conjugation in IVA
Only three experimental studies (Table 1) are available that attempted to show that human GLYAT can conjugate isovaleryl-CoA with glycine [41], [42], [43].
Table 1.
Summary of studies that evaluated the ability of human GLYAT to conjugate isovaleryl-CoA to glycine.
| Enzyme preparation | CoA substrates evaluated | Amino acid substrates evaluated | Formation ofN-isovalerylglycine | Reference |
|---|---|---|---|---|
| Enzyme extract from human liver and kidney tissue | phenylacetyl-CoA benzoyl-CoA isobutyryl-CoA isovaleryl-CoA indolyl acetyl-CoA p-hydroxyphenylacetyl-CoA |
glycine glutamine |
No | [41] |
| Human liver lysate | 2-methyl butyryl-CoA isobutyryl-CoA butyryl-CoA hexanoyl-CoA octanoyl-CoA decanoyl-CoA isovaleryl-CoA |
glycine | N-isovalerylglycine formed under experimental conditions not comparable to physiological conditions | [42] |
| Enzyme preparation isolated from human liver | benzoyl-CoA butyryl-CoA salicylyl-CoA heptanoyl-CoA isovaleryl-CoA |
glycine | isovaleryl-CoA had the lowest substrate specificity | [43] |
In the first study, an enzyme extract from human liver and kidney tissue was used to test the conjugation of phenylacetyl-CoA, benzoyl-CoA, isobutyryl-CoA, isovaleryl-CoA, indolyl acetyl-CoA and p-hydroxyphenylacetyl-CoA with either glutamine or glycine. Although this enzyme preparation catalyzed the formation of hippuric acid (benzoylglycine), no evidence for the conjugation of isovaleryl-CoA to glycine could be found in either the liver or kidney [41].
Subsequently, Gregersen et al., prepared a human liver lysate and developed a GC/MS assay to determine GLYAT affinity for 2-methyl butyryl-CoA, isobutyryl-CoA, butyryl-CoA, hexanoyl-CoA, octanoyl-CoA, decanoyl-CoA and isovaleryl-CoA [42]. N-isovalerylglycine was quantified in this study, but the substrate concentrations used for isovaleryl-CoA (672 µM) and glycine (523 mM) were very high and are not comparable to intracellular substrate concentrations. For example, in nonketotic hyperglycinemia, intracellular glycine concentration in brain tissue do not exceed 30 mM, while the glycine concentration in the liver is approximately 5 mM [44], [45].
The last study determined the substrate specificity of an enzyme preparation isolated from human liver. The highest activity was obtained with benzoyl-CoA which was set as the maximum relative activity of 100%. This was followed by butyryl-CoA (28%), salicylyl-CoA (22%), heptanoyl-CoA (9.5%) and isovaleryl-CoA (9.1%) [43].
While all three studies could show conjugation between acyl-CoA substrates and amino acids, the enzyme preparations were neither homogenous nor purified. In other words, the extracts contained other mitochondrial enzymes. None of the studies could show that isovaleryl-CoA is a good substrate for GLYAT. This is puzzling when one considers that during an IVA episode, patients can excrete up to 1700 mg of N-isovalerylglycine per day, while [23]. hippurate (the conjugate formed by the preferred substrates benzoyl-CoA and glycine), is only excreted to a maximum of 716 mg per day [46]. Therefore, if isovaleryl-CoA is indeed a poor substrate for GLYAT, how is N-isovalerylglycine then excreted at significantly greater quantities than the preferred substrate? We propose that a greater understanding of the kinetic and docking behavior of the GLYAT enzymes may yield an answer to this question.
Although these three studies (Table 1) could not show that isovaleryl-CoA is a good substrate for human GLYAT, the use of crude preparations may confound these findings. Kelley and Vessey (1994) for example analyzed GLYAT substrate specificity using an enzyme preparation with three distinct proteins. Furthermore, a homologous gene, GLYATL1, is located on human 11q12.1 which [47] has 39% nucleotide identity to GLYAT. GLYATL1 is a glutamine N-acyltransferase enzyme present in the mitochondrial fractions from human kidney and liver. [41], [48]. This then raises the question whether an enzyme with a preference for L-glutamine can also conjugate isovaleryl-CoA to glycine? Previous studies have shown that GLYATL1 can conjugate benzoyl-CoA (0.027 U/mg protein) and phenylacetyl-CoA (0.085 U/mg protein) to glycine, albeit at lower rates, when compared to the glutamine conjugation rates (0.101 and 0.603 U/mg enzyme respectively) [41], [48]. Moldave and Meister, however, could not obtain evidence for GLYATL1 of N-isovalerylglycine formation from isovaleryl-CoA and glycine [41].
It is also important to distinguish between the studies done on human GLYAT [41], [42], [43] and those done on bovine GLYAT [30], [32] as the kinetic mechanisms of these two enzymes are different. Human GLYAT exhibits sigmoidal kinetics [49] whereas bovine GLYAT follow Michaelis-Menten kinetics [50]. Sigmoidal enzymes do not follow a hyperbolic curve, but a sigmoidal one i.e. the observed kinetic behaviour is a consequence of co-operativity in the substrates binding [51]. In the case of human GLYAT, the cooperativity is stronger for benzoyl–CoA than for glycine [52]. If the preferred substrate for GLYAT, benzoyl-CoA, outcompetes isovaleryl-CoA for conjugation to glycine this can impact the efficacy of treating IVA with glycine supplementation. However, this would require further study.
To date, no study has shown that a purified GLYAT enzyme can conjugate isovaleryl-CoA to glycine. It is important to verify whether the assumption that GLYAT can conjugate isovaleryl-CoA to glycine is indeed correct. Especially since in the ZA cohort of IVA patients who are all homozygous for the same IVD mutation, variation in the responsiveness to glycine supplementation was observed [13]. This variation might be due to several factors. Firstly, variants in GLYAT as well as transcriptional regulation of GLYAT, may affect enzyme activity. Secondly, substrate competition between isovaleryl-CoA and other, more preferred substrates might result in the preferred substrate outcompeting isovaleryl-CoA. Thirdly, there may be inhibitors, regulators, or interactions with other enzymes that can affect the variation in N-isovalerylglycine formation.
No studies are available on the interaction or putative competition between the various acyl-CoA substrates involved in the glycine conjugation pathway. One study did show that L-carnitine conjugation to isovaleryl-CoA occurs earlier than isovaleryl-CoA conjugation to glycine [26], which might indicate that the conjugation of isovaleryl-CoA is delayed by the competition of other substrates.
Improving our understanding of the pathophysiology of IVA, specifically the activation of secondary metabolic pathways, such as glycine conjugation, in response to increased isovaleryl-CoA accumulation, is required to optimize the monitoring of diagnosed IVA patients and to potentially improve, even personalise current treatment strategies. The potential of an acquired hyperglycinemia makes this study even more important as an understanding of the capacity of the glycine conjugation pathway to form N-isovalerylglycine may further contribute to realistic treatment recommendations. Therefore, the aim of this study is to test the hypothesis whether GLYAT and/or GLYATL1 can conjugate isovaleryl-CoA to glycine. Firstly, molecular modelling and binding interactions of the proposed enzymes (GLYAT and GLYATL1) and substrates (isovaleryl-CoA and glycine) were done to establish which of the enzymes are more likely to bind isovaleryl-CoA to glycine. The results of the in-silico analyses were then validated by analyzing various enzyme reactions using recombinantly expressed purified enzymes.
2. Methods
2.1. Retrieval of GLYAT and GLYATL1 enzyme sequences and substrate structures
The GLYAT (S156 GLYAT variant; WT Ref seq NM_201648.3) and GLYATL1 (WT ref Seq NM_001389712.2) sequences were obtained from Ensembl (ensemble.org/). The 3D structures of the substrates were retrieved from Protein Data Bank (PDB; https://www.rcsb.org/). Benzoyl-CoA (PDB: 4Z3Y), glycine (PDB: 3OWW), phenylacetyl-CoA (PDB: 4IIT), glutamine (PDB: 6QN3) and isovaleryl-CoA (PDB: 5K7H) were extracted from experimentally solved 3D structures (X-ray diffraction).
2.2. Secondary structure prediction
Secondary structure elements consisting of alpha-helices, beta-sheets, and random coils were predicted for GLYAT and GLYATL1 using the PSIPRED secondary structure prediction server3 (http://bioinf.cs.ucl.ac.uk/psipred) [53].
2.3. Prediction of disordered state of GLYAT and GLYATL1
The DISOPRED3 program was used for protein disorder prediction and protein-binding site annotation within disordered regions (http://bioinf.cs.ucl.ac.uk/disopred) [54]. The server allows users to submit a protein sequence and returns a probability estimate of each residue in the sequence being disordered. Briefly, the GLYAT and GLYATL1 protein sequences were uploaded to the database for residue disorder prediction. Here, we wanted to predict the prevalence of disordered states within GLYAT and GLYATL1 to infer their potential function. Proteins with a higher prevalence of disordered states do not always conform to the traditional “sequence-structure-function” paradigm [55] and therefore, the prediction of disordered states assists in the interpretation of findings from docking studies.
2.4. 3D structure prediction of GLYAT and GLYATL1 using SWISS-MODEL
SWISS-MODEL (https://swissmodel.expasy.org/) is a fully automated protein structure homology-modelling server [56], [57] which was used to model GLYAT and GLYATL1. The GLYAT and GLYATL1 protein sequences were used as input to the SWISS-MODEL web interface. From the input sequence, SWISS-MODEL first performs a template search, then it selects and aligns the template. The aligned template is then used to build a model and lastly the model quality is assessed [56], [57]. For the prediction of GLYAT, the template ID: 7pk1.1. A (sequence identity: 75.93% and coverage: 1.00) and for GLYATL1, the template ID: 7pk1.1. A (sequence identity 41.30% and coverage: 0.97) were selected. The templates with the highest sequence identity and coverage to the target sequences were selected for model building. The SWISS-MODEL webserver reports inbuilt quality assessment scores for protein models predicted using the webserver, such as the Global Model Quality Estimate (GMQE) score [58]. The GMQE score gives an overall model quality measurement between 0 and 1, with higher numbers indicating higher accuracy of the model built with that specific alignment and template [58]. Where predicted models were missing residues, these were remodeled using previous modelled structures from the Alpha Fold protein structure database as templates (GLYA T: Q6IB77 and GLYATL1: Q969I3). The final predicted structures were subjected to protein 3D structure quality assessment.
2.5. 3D structure quality assessment
To assess the quality of the predicted 3D structures, a variety of structural parameters were tested within each model. Procheck, VERIFY3D and ERRAT from the Structural Analysis and Verification Server (SAVES)5 were used to evaluate the quality of the predicted structures. Procheck was used to determine if the predicted residues were within the allowable region of the Ramachandran plot [59]. Structures were considered reliable if more than 90% of the residues had favorable phi and psi dihedral angle distributions. The program VERIFY3D [60], [61] measures the compatibility of a protein model with its own amino acid sequence. VERIFY3D produces an averaged 3D–1D score for each residue to evaluate the quality of the homology protein structure based on the energetic and empirical methods. Protein structures where more than 80% of the residues had a score greater than 0.2 are of high quality [62]. The program ERRAT [63] analyses the relative frequencies of noncovalent interactions between atoms of various types. ERRAT is considered the “overall quality factor” for non-bonded atomic interactions, with higher scores indicating higher quality. The generally accepted range is over 50 for a high-quality model [64]. ProSA-web-Protein Structure Analysis was used for the recognition of errors in three-dimensional structures of proteins and to measure total energy deviation within the protein structure [65]. This was used to determine whether the z-score of the input structure is within the range of scores typically found for native proteins of similar size. Lastly, the root mean square deviation (RMSD) values were calculated between the predicted structure and the homologous template structure using PYMOL/Maestro molecular visualizing software to compare backbone structural similarity to the experimentally solved template structure. Highly similar structures are considered when the RMSD is below 2 Å suggesting homology [66] whereas higher RMSD indicates that predicted structures and templates are not structurally similar. As a comparative analysis, we have also compared our structures to available structures on the Alpha Fold protein structure database for GLYAT (Q6IB77) and GLYATL1 (Q969I3). Protein structures that satisfy most or all of the quality parameter tests were considered reliable for subsequent docking studies.
2.6. Refinement and energy minimization
The predicted 3D structures were subsequently energy minimized using Modrefiner available at https://zhanggroup.org/ModRefiner/. Modrefiner refines protein structures from Cα traces based on a two-step, atomic-level energy minimization [67]. Briefly, the predicted 3D structures for GLYAT and GLYATL1 were uploaded to the Modrefiner refinement interface. The final energy-minimized model was the lowest energy minima conformation of the protein structures, and these were utilized in molecular docking.
2.7. Molecular docking: AutoDock Vina
The molecular docking of the bi-substrates to GLYAT/GYATL1 was carried out using AutoDock Vina [68], which is an open-source program for doing molecular docking. AutoDock Vina uses a sophisticated gradient optimization method in its local optimization procedure [68]. Autodock vina was used to determine binding affinities for several ligands to either GLYAT or GLYATL1. Preparatory steps, including preparation of pdbqt files for protein and ligands and grid box creation were completed using the Graphical User Interface program AutoDock Tools (ADT). Protein preparation included the removal of water molecules, the addition of polar hydrogens and united Kollman chargers. The grid size was set to 40 × 40 × 40 xyz points with grid spacing of 0.375 Å. The grid center designated at dimensions (x, y, and z) differed with respect to each docking interaction. AutoDock Vina was employed for docking using protein and ligand information along with grid box properties in the configuration file. During the docking procedure, both the protein and ligands are considered rigid. AutoDock Vina generated 8 different docking poses. The pose with the lowest energy of binding or binding affinity was extracted and aligned with the receptor structure for further analysis. The preferred substrates for GLYAT are benzoyl-CoA and glycine [52], [69] and for GLYATL1 it is phenylacetyl-CoA and glutamine [41]. Therefore, these were used as positive controls for our computational docking. Benzoyl-CoA binds to GLYAT first, followed by glycine [70]. Therefore, we docked GLYAT-benzoyl-CoA first, and this was sequentially followed by a second docking step to bind glycine. Similarly, we docked GLYATL1-phenylacetyl-CoA first, and this was sequentially followed by a second docking step to bind glutamine. Lastly, we docked isovaleryl-CoA to GLYAT and GLYATL1 respectively, and this was sequentially followed by a second docking step to bind glycine.
2.8. Protein–ligand interaction profiler
Protein–substrate interaction analysis was done using the protein–ligand interaction profiler (PLIP) [71]. Briefly, the docked complexes of GLYAT/GLYAT1 and substrates were uploaded to the PLIP webserver (https://plip-tool.biotec.tu-dresden.de/plip-web/plip/index). PLIP detects hydrogen bonds, hydrophobic contacts, π-stacking, π-cation interactions, salt bridges, water bridges, metal complexes, and halogen bonds between ligands and targets. Cut off for interactions formed were 4.1 Å for hydrogen bonds, 4.0 Å for hydrophobic contacts, 5.5 Å for π-stacking, 6.0 Å for π-cation interactions, 5.5 Å for salt bridges, 4.1 Å for water bridges, 3.0 Å for metal complexes, and 4.0 Å for halogen bonds.
2.9. Expression and purification of recombinant human GLYAT and GLYATL1
Glycerol stocks of C41(DE3)pLysS E.coli cells which contained either the pHis17_GLYAT or pET23a(+)_GLYATL1 plasmid were incubated in 20 mL of sterile media (2% m/v bacto-tryptone, 1.25% m/v yeast extract, 0.625% NaCl, 0.5% Na2HPO4, 0.1% KH2PO4, 0.2% m/v glucose, 100 µg/mL carbenicillin, and 35 µg/mL chloramphenicol) (Sigma Merck) at 37 °C and 250 RPM (New Brunswick Innova 40 shaking incubator), overnight. The following day, the starter culture was inoculated in 100 mL of fresh media at a ratio of 1:100 and grown at 37 °C and 250 RPM until the OD600 was between 0.5 and 0.6. The expression of recombinant GLYAT or GLYATL1 was then induced by supplementing the media with 0.05 mM IPTG, 5 g/L arabinose, and 0.2% m/v glycine or 0.2% m/v glutamine (Sigma Merck) (for GLYAT or GLYATL1, respectively), then lowering the temperature to 25 °C and leaving the cells for 24 h at 250 RPM (New Brunswick Innova 40 shaking incubator).
Thereafter, the cells were pelleted at 4,500 x g and 4 °C for 10 min, the clarified supernatant was discarded, and the cells were resuspended in 10 mL of a modified RIPA buffer (0.29% m/v NaCl, 3.6% m/v NaH2PO4, 1% v/v Triton X-100)(Sigma Merck) which was supplemented to 1 U/10 mL of Pierce Universal Nuclease (Thermo Fisher Scientific). Each suspension was left on ice for 15 min before being passed through at 22-G needle 10 times. The lysate was centrifuged at 14500 x g and 4 °C for 30 min whereafter 9 mL of the clarified supernatant was removed for purification.
Protino Ni-TED 2000 (Machery-Nagel) columns were used to purify the recombinant enzymes as per the manufacturer’s instructions which resulted in three 3 mL fractions. The protein concentration of each fraction was determined using a Qubit 2.0 fluorometer and the BR protein assay kit from Invitrogen according to the manufacturer’s instructions. The proteins could be stored for at least four weeks at 4 °C without noticeable loss of protein or activity.
2.10. Relative enzyme activity
Microplate assays were all performed in a BioTek Synergy HT plate reader at 37 °C, 25 mM TRIS-HCl (final concentration, pH 8.13), with 0.5 µg of recombinant protein (from 10 µg/mL stock), at 412 nm, in 100 µL volumes. Relative activities of the enzymes, in nmol/min/mg, were calculated over the first 10 min period where the reaction proceeded linearly. Stock solutions of 50 mM of benzoyl-CoA, phenylacetyl-CoA, and isovaleryl-CoA were prepared in milliQ water and stored at − 20 °C until needed. Dilution ranges were prepared with milliQ water as necessary to ensure that only 5 µL of the relevant acyl-substrate were added to the reaction mix. Similarly, 100 mM stock solutions of glycine and glutamine were prepared in milliQ water, filtered through a 22 nm CA membrane, stored at 4 °C, and diluted as necessary to ensure that only 20 µL of the relevant amino acid substrate were added. A fresh 10 mM stock solution of DTNB was prepared in absolute ethanol of which 2 µL were added to the reaction mix for a final concentration of 200 µM. The volume was adjusted to 95 µL with milliQ water and the reactions were initiated with 5 µL of the recombinant enzyme.
Each assay was performed in a technical triplicate, the data were exported from the BioTek Gen5 1.11 software into an MS Excel spreadsheet, where initial data wrangling was performed before being analyzed and graphed in Wolfram™ Mathematica™ 13.0 [72]. Negative controls were set up similarly either excluding enzyme or excluding substrate. The enzyme did not meaningfully absorb at 412 nm, the acyl-CoA substrates did not spontaneously conjugate with the amino acid substrates, nor did the DTNB degrade or spontaneously conjugate with acyl-CoA or any free CoA in a manner detectable by the plate reader. Negative controls were used to blank the data. A variety of conditions were tested, and these are summarized in Table 2. The concentrations for the substrates which were kept constant were selected based on previous kinetic studies [52], [73].
Table 2.
Experimental design to test the activities of GLYAT and GLYATL1 with varying substrate concentrations.
| Purpose | Constant Substrate | Variable Substrate |
|---|---|---|
| Evaluate the baseline enzyme activity of GLYAT and GLYATL1 | Preferred amino acid | Preferred acyl-CoA |
| Evaluate the ability of GLYAT and GLYATL1 to conjugate isovaleryl-CoA as a substrate | Preferred amino acid | Isovaleryl-CoA |
| Determine the correlation between amino acid concentration and GLYAT and GLYATL1 reaction rate | Preferred acyl-CoA | Preferred amino acid |
| Determine the correlation between amino acid concentration and GLYAT and GLYATL1 reaction rate | Isovaleryl-CoA | Preferred amino acid |
Amino acid substrates, when constant, were 20 mM whereas the preferred acyl-CoA substrates (benzoyl-CoA or phenylacetyl-CoA) were 250 µM and isovaleryl-CoA was 500 µM. Concentration variables for the acyl-CoA substrates were: 50, 100, 250, and 500 µM. Amino acid substrate concentrations were varied as follows: 0.5, 1, 2, 5, 10, and 20 mM.
3. Results and discussion
In this study we proposed that GLYAT and/or GLYATL1 might be responsible for the formation of N-isovalerylglycine in IVA patients. To test this hypothesis, we performed an in-silico analysis to determine which enzyme is more likely to conjugate isovaleryl-CoA with glycine followed by an in vitro validation using purified enzyme preparations.
Genetic variants can affect enzyme activity. Therefore, we first analysed the variant data available on the Ensembl database (ensemble.org/) to determine the haplotype of GLYAT and GLYATL1 with the highest frequencies in the worldwide population. This was done to enzymatically characterize relevant variants of the enzymes. Previous studies have shown that for GLYAT, the S156 haplotype has the highest haplotype frequency and enzyme activity [52], [73]. GLYATL1 has not been previously characterized and therefore, the wildtype reference haplotype (NM_001389712.2) with the highest haplotype frequency (97.9%) was characterized in this study.
3.1. Molecular modelling and docking
For the in-silico analyses, we have created three-dimensional structures of the GLYAT and GLYATL1 enzymes using SWISS-MODEL. We considered these structures reliable for docking as they satisfied most, or all of the quality parameter tests. Thereafter, we performed a molecular docking of the bi-substrate enzymes GLYAT/GLYATL1 with several substrates using AutoDock Vina. As positive controls and as known substrates for GLYAT/GLYATL1, we performed molecular docking of GLYAT to benzoyl-CoA and glycine and GLYATL1 to phenylacetyl-CoA and glutamine. We, subsequently, tested the hypothesis whether GLYAT and/or GLYATL1 can bind isovaleryl-CoA and glycine.
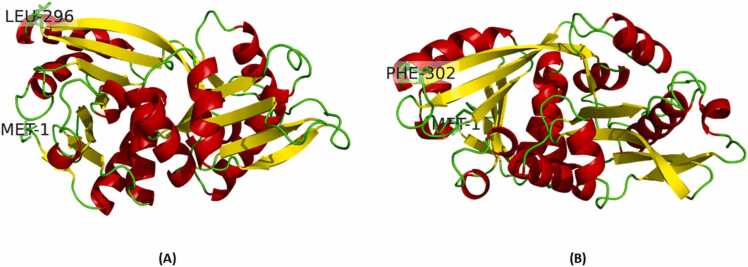
Secondary structure prediction from PSIPRED for GLYAT indicated 12 α-helices, 14 β-strands connected with random coils (Supplementary Figure 1). Secondary structure prediction from PSIPRED for GLYATL1 indicated 11 α-helices and 14 β-strands and connected with random coils (Supplementary Figure 2). None of the proteins had predicted disordered states (Supplementary Figures 3 and 4). However, findings from the 3D structures for GLYAT indicated 12 α-helices and 13 β-strands (Fig. 2A) and GLYATL1 indicated 13 α-helices and 12 β-strands (Fig. 2B).
Fig. 2.
3D structure of (A) GLYAT and (B) GLYATL1. The alpha-helical structures are indicated in red, and the beta-strands are indicated in yellow. The N-terminal Met1 and C-terminal Leu296/Phe302 are shown as stick structures.
Further, the 3D predicted GLYAT and GLYATL1 protein structures had high GMQE indicating that the models were of reliable quality and accuracy (Table 3). Both models passed all quality assessments including the Procheck assessment as more than 90% of the residues were within the allowable regions of the Ramachandran plot (Table 3), the VERIFY3D assessment as both protein structures had more than 80% of residues with a score of greater than 0.2 and the ERRAT assessment had scores higher than 50. ProSA analysis indicated that both structures z-scores were within the range of scores typically found for native proteins of a similar size. Furthermore, the RMSD scores for the GLYAT/GLYATL1 proteins were less than 2 Å when compared to the homologous templates as well as the alphafold templates suggesting high structural similarity (Table 3). In addition, we have also compared our 3D predicted GLYAT structure (S156 haplotype of the Ref seq NM_201648.3) to a previously generated structure (NM_201648) [73]. High similarity was predicted when comparing these structures (RMSD = 0.005 Å). Therefore, our predicted 3D structures of GLYAT and GLYATL1 satisfied all the quality parameter tests and were considered for subsequent docking studies.
Table 3.
Summary of the quality assessment scores for the 3D predicted structures of GLYAT and GLYAT1.
| Enzyme | Template | GMQE | Procheck (Percentage in allowed region) | VERIFY3D | ERRAT |
ProSA (z-score) |
RMSD (Å) |
Comparative RMSD (Å) |
|---|---|---|---|---|---|---|---|---|
| GLYAT | 7pk1. | 0.92 | Pass (100%) | Pass (98.65%) | 92.71 | Pass (−8.69) | 0.893 | 0.339 |
| GLYATL1 | 7pk1. | 0.80 | Pass (100%) | Pass (100%) | 86.06 | Pass (−7.85) | 1.148 | 0.488 |
VERIFY3D: % of the residues with a 3D–1D score > = 0.2.
Comparative analysis: RMSD when comparing generated structures and structures from the alphafold database.
We analyzed the molecular docking of all substrates in two steps. First, GLYAT or GLYATL1 was docked against one substrate and the output docking complex was then used to dock the second substrate. Table 4 presents the respective docking profiles for when (1) GLYAT or GLYATL1 was docked against one substrate and (2) the output docking complex was docked against the second substrate. Therefore, Table 4, represents the interaction profile as each substrate is being docked respectively. GLYAT docked with a slightly higher affinity to benzoyl-CoA (−6.6 kcal/mol) in comparison to isovaleryl-CoA (−6.3 kcal/mol), however, the docking of glycine resulted in the same affinity for both (−4.1 kcal/mol) interactions (Table 4). The same affinity for glycine may be due to interaction with the same binding pocket in both docking interactions (Fig. 3 A and B). The docking affinity of GLYAT+isovaleryl-CoA to glutamine (−6.4 kcal/mol) was higher than that of GLYAT+isovaleryl-CoA to glycine (−4.1 kcal/mol). Interestingly, when comparing the number and type of interactions between the GLYAT benzoyl-CoA+glycine complex versus the GLYAT isovaleryl-CoA+glycine complex, the latter had a greater number of interactions of 19 compared to the former of 16 (Table 4). When comparing and profiling the number and type of interactions for the entire complex (GLYAT+both substrates) as one docking reaction, GLYAT+isovaleryl-CoA/glutamine had the greatest number of interactions (29), followed by the GLYAT+isovaleryl-CoA/glycine (25) and GLYAT+benzoyl-CoA/glycine (21) (Supplementary Table 1). Supplementary Table 1 represents the interaction profile of the entire complex (GLYAT/GLYATL1 + both substrates) and therefore presents the docking interactions after both substrates have bound. These findings are also in line with the predicted binding affinities (Table 4).
Table 4.
The interaction binding affinities and the number and type of interactions for both GLYAT and GLYATL1 docked to the respective substrates.
| Protein | Substrate | Affinity (kcal/mol) | H-bonds (amino acids) |
Salt bridges (Amino acids) |
π-Cation (Amino acids) |
Hydrophobic (Amino acids) |
|---|---|---|---|---|---|---|
| GLYAT | Benzoyl-CoA | -6.6 kcal/mol | TYR72, GLN103, LYS127, ARG131, HIS186, ASN190, ASP266, ASN292 | HIS71, HIS71, HIS101, LYS127 | GLN130, HIS186, PHE187, TYR267 | |
| GLYAT+ Benzoyl-CoA | Glycine | -4.1 kcal/mol | THR75, GLN103, GLN105 | ARG229 | ||
| GLYAT | Isovaleryl-CoA | -6.3 kcal/mol | TRP185, HIS186, THR233, ARG238, ARG238, ARG238, LEU239, GLY241, LEU242, VAL243, THR244, THR244, ASN269 | HIS186, ARG238, ARG238 | HIS186, PHE187, ALA271 | |
| GLYAT + Isovaleryl-CoA | Glycine | -4.1 kcal/mol | TYR72, GLN105, GLN105, ARG229, ARG229- | |||
| GLYAT + Isovaleryl-CoA | Glutamine | -6.4 kcal/mol | 103 A, 105 A, 105 A, 224 A, 224 A, 225 A | 229 A | 75 A, 196 A | |
| GLYATL1 | Phenylacetyl-CoA | -8.6 kcal/mol | LYS187, LYS187, SER234, SER234, SER234, ARG239, ARG240, ASN270 | 239 A, 239 A | 186 A, 289 A | TRP186, ALA232, TYR233 |
| GLYATL1 + Phenylacetyl-CoA | Glutamine | -4.1 kcal/mol | GLU63, ASP66, ASP69, THR72, HIS196, ARG200, ARG200 | |||
| GLYATL1 + Phenylacetyl-CoA | Glycine | -3.6 kcal/mol | GLN104, GLN104, SER226 | LYS26, ARG76 | ||
| GLYATL1 | Isovaleryl-CoA | -8.4 kcal/mol | LYS187, ALA232, SER234, SER234, SER234, ARG239, ARG240, THR241, GLY242, ASN243, MET244, SER265, ASN270, SER273, SER273 | TRP186, TRP186, LYS187, VAL266 | ||
| GLYATL1 + Isovaleryl-CoA | Glycine | -3.5 kcal/mol | PHE40, THR72, ARG200 | |||
| GLYATL1 + Isovaleryl-CoA | Glutamine | -5.0 kcal/mol | ASP159, PHE280, GLY281 | HIS157, ARG256 | MET252 |
Fig. 3.
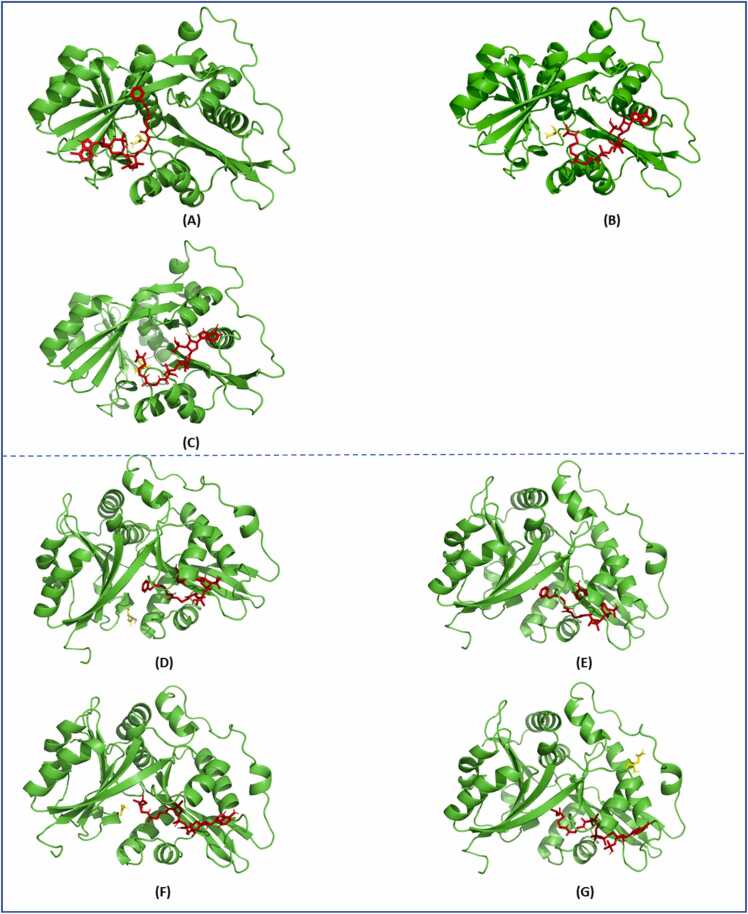
Docking pose of (A) GLYAT and benzoyl-CoA/glycine, (B) GLYAT and isovaleryl-CoA/ glycine, (C) GLYAT and isovaleryl-CoA/glutamine, (D) GLYATL1 and phenylacetyl-CoA/ glutamine and (E) GLYATL1 and phenylacetyl-CoA/glycine, (F) GLYATL1 and isovaleryl-CoA/glycine and (G) GLYATL1 and isovaleryl-CoA/glutamine.
Structurally, the binding of these substrates was very similar (Fig. 3 A, B and C). GLYATL1 docked with a slightly higher affinity to phenylacetyl-CoA (−8.6 kcal/mol) compared to isovaleryl-CoA (−8.4 kcal/mol). When comparing the number and type of interactions between GLYATL1 and phenylacetyl-CoA versus GLYATL1 and isovaleryl-CoA the latter had a greater number of interactions of 19 compared to the former of 15 (Table 4). This finding is in line with the findings for GLYAT. Further, GLYATL1 + phenylacetyl-CoA docked with a higher binding affinity to glycine (−3.6 kcal/mol) compared to glutamine (−4.1 kcal/mol). On the other end, GLYATL1 + isovaleryl-CoA docked with a higher affinity to glutamine (−5.0 kcal/mol) compared to glycine (−3.5 kcal/mol) (Table 4). When comparing the number and type of interactions for the entire complex (GLYATL1 + both substrates), GLYATL1 + isovaleryl-CoA/glutamine had the greatest number of interactions (26), followed by GLYATL1 + phenylacetyl-CoA/glutamine (24), GLYATL1 + isovaleryl-CoA/glycine (23) and GLYATL1 + phenylacetyl-CoA/glycine (22) (Supplementary Table 1). Structurally, the binding was similar for all interactions with GLYATL1 (Fig. 3D-F) besides for GLYATL1 and isovaleryl-CoA/glutamine (Fig. 3G).
Our findings indicate that molecular modelling assisted in generating reliable GLYAT and GLYATL1 protein structures useful for molecular docking studies. Molecular docking suggested that GLYAT had a slightly higher affinity for benzoyl-CoA in comparison to isovaleryl-CoA, however, the docking of glycine resulted in the same affinity for both interactions, most likely due to glycine utilizing the same binding site. When glutamine is used as the amino acid substrate to the GLYAT- isovaleryl-CoA complex, the binding affinity was higher compared to when glycine was used as the amino acid substrate. This may suggest that glutamine may be the preferred amino acid substrate for isovaleryl-CoA interactions. Interestingly, the docking of GLYAT to isovaleryl-CoA and glycine/glutamine had a greater number of interactions (H-bonds, Salt bridges, π-Cation and Hydrophobic) compared to the docking of GLYAT to benzoyl-CoA and glycine, despite a lower binding affinity. This may be due to different binding sites utilized by isovaleryl-CoA compared to benzoyl-CoA (Fig. 3A versus Figures3B and 3 C) as in addition to the non-covalent intermolecular interactions that influence binding affinity, the dynamics of protein binding pockets are also crucial for their interaction specificity and affinity [74]. However, this warrants further investigation. GLYATL1 docked with a slightly higher affinity and a greater number of interactions with phenylacetyl-CoA in comparison to isovaleryl-CoA. Further, GLYATL1 docked with a higher affinity to phenylacetyl-CoA and glutamine compared to phenylacetyl-CoA and glycine. These findings support the notion that phenylacetyl-CoA and glutamine are the preferred substrates for GLYATL1 [41], [48]. Similar to the findings reported for GLYAT, GLYATL1 + isovaleryl-CoA docked with a higher affinity to glutamine compared to glycine, which again questions whether glutamine may be the preferred amino acid substrate for isovaleryl-CoA interactions.
The focus of the current study was to determine whether GLYAT or GLYATL1 can conjugate isovaleryl-CoA to glycine. Therefore, the relative enzyme activity of both enzymes for their preferred substrates were included as control reactions and compared to the ability of the enzymes to form isovalerylglycine.
3.2. Evaluation of the relative enzyme activity of GLYAT and GLYATL1 using different substrate combinations
Previous studies have shown that the preferred substrates for GLYAT is benzoyl-CoA and glycine [41], [43] and for GLYATL1 it is phenylacetyl-CoA and glutamine [41], [48]. We expanded on their work by testing three hypotheses. Firstly, whether GLYAT or GLYATL1 can conjugate isovaleryl-CoA to their preferred amino acid substrates (glycine and glutamine respectively). Secondly, we tested whether increasing the amino acid (glycine or glutamine) concentration increased the rate of acyl-CoA conjugation by GLYAT or GLYATL1. Thirdly, whether GLYATL1 can conjugate isovaleryl-CoA with glycine. In order to test our hypotheses, we expressed and purified human GLYAT and GLYATL1 and we performed a series of DNTB-based spectrophotometric kinetic assays as outlined in Table 2.
In this study we determined the relative enzyme activity between the ability of GLYAT to conjugate benzoyl-CoA and isovaleryl-CoA to glycine and GLYATL1 to conjugate phenylacetyl-CoA and isovaleryl-CoA to glutamine. The concentration range for benzoyl-CoA and the glycine concentration used, was chosen according to previously determined enzyme kinetic values [42], [43], [48], [49], [52], [73], [75], [76], [77]. The kinetic parameters for GLYATL1 have not been determined previously and therefore similar concentration ranges to those chosen for GLYAT, was used.
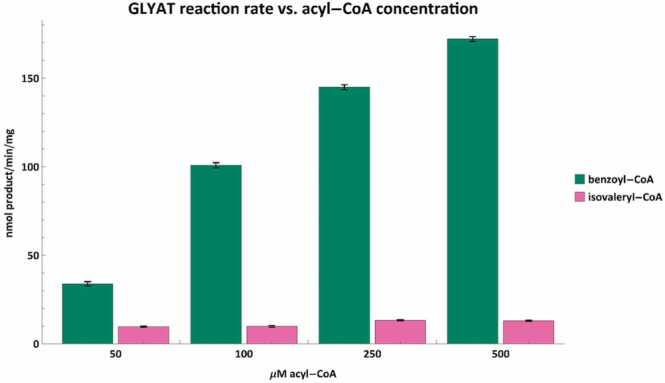
The benzoyl-CoA conjugation to glycine by GLYAT (Fig. 4) corresponds with previous in vivo and in vitro studies which showed that as the amount of benzoic acid (benzoyl-CoA as substrate) is increased, the formation and excretion of hippuric acid will also increase [52], [78], [79], [80]. Confirming these results supports the notion that GLYAT is indeed responsible for the detoxification of benzoyl-CoA.
Fig. 4.
The acyl-CoA conjugation rate of GLYAT was tested with varying concentrations of benzoyl-CoA (green) and isovaleryl-CoA (pink) using 20 mM glycine. The reaction rate of GLYAT correlates with increases in benzoyl-CoA concentration. While there are minor fluctuations in the conjugation rate of isovaleryl-CoA, overall, it appears as if the conjugation rate is largely independent of isovaleryl concentrations at or below 500 µM. Error bars indicate the mean ± standard deviation for triplicate assays.
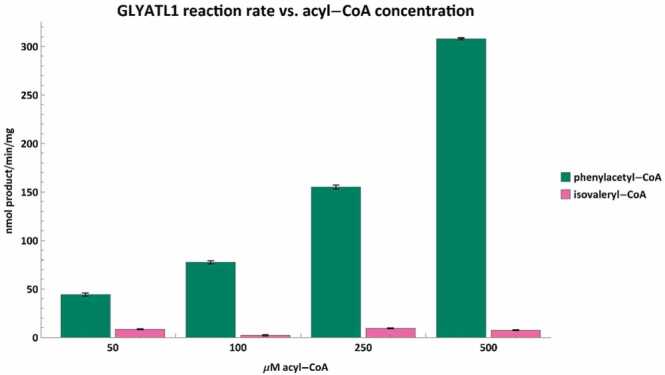
This is the first study to show a dose-dependent increase in reaction rate between the concentration of phenylacetyl-CoA and that of phenylacetylglutamine formed by GLYATL1 (Fig. 5). Furthermore, both GLYAT and GLYATL1 can conjugate isovaleryl-CoA with their preferred amino acid substrate (Fig. 4, Fig. 5). However, there is no dose-dependent increase in reaction rate when isovaleryl-CoA is used as the acyl-CoA substrate. The difference in reaction rate between 500 µM benzoyl-CoA and 500 µM isovaleryl-CoA was more than 13-fold whereas it was nearly 42-fold with the same concentration of phenylacetyl-CoA and isovaleryl-CoA for GLYATL1. Excretion of N-isovalerylglutamate has been mentioned in various IVA studies [81], [82], [83] with one study reporting it as 122 ± 87 mmol/mol creatinine in urine of untreated IVA patients [82].
Fig. 5.
The conjugation rates of GLYATL1 were tested with varying phenylacetyl-CoA (green) and isovaleryl-CoA (pink) concentrations using 20 mM glutamine. The reaction rate correlates strongly with phenylacetyl-CoA concentration. Similar to GLYAT however, the isovaleryl-CoA conjugation rate appears largely insensitive to concentrations below 500 µM. Error bars indicate the mean ± standard deviation for triplicate assays.
Therefore, while both enzymes convert isovaleryl-CoA to an isovaleryl-amino acid conjugate, the rate at which they conjugate isovaleryl-CoA is not only significantly lower than for the preferred acyl-CoA substrate, but they are also less responsive to increases in isovaleryl-CoA concentration. Additionally, these results align with the molecular docking study which indicates that the affinities of GLYAT and GLYATL1 for isovaleryl-CoA are lower than for the preferred acyl-CoA substrates (Table 4). However, the significant difference in conjugation rates does conflict with the predicted affinities which are only slightly lower for isovaleryl-CoA than for the preferred acyl-CoA.
Although several case studies have investigated the appropriate glycine dose to use in the treatment of IVA patients [19], [26], [29], [84], [85], [86], no consensus has been reached due to the large interindividual variation observed. Therefore, we investigated whether increasing the amino acid (glycine or glutamine) concentration increased the rate of acyl-CoA conjugation by GLYAT or GLYATL1 (Fig. 6, Fig. 7). The preferred acyl-CoA substrate was evaluated at 250 µM and isovaleryl-CoA at 500 µM. We selected a higher concentration of isovaleryl-CoA than preferred acyl-CoA given the relatively low isovaleryl-CoA conjugation rates seen in Fig. 4, Fig. 5.
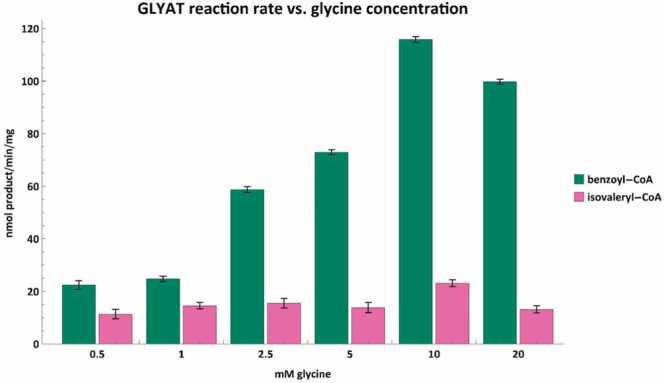
Fig. 6.
Acyl-CoA conjugation rates for benzoyl-CoA (green) and isovaleryl-CoA (pink) against varying glycine concentrations. Assays were performed with either 250 µM benzoyl-CoA or 500 µM isovaleryl-CoA. Error bars indicate the mean ± standard deviation for triplicate assays.
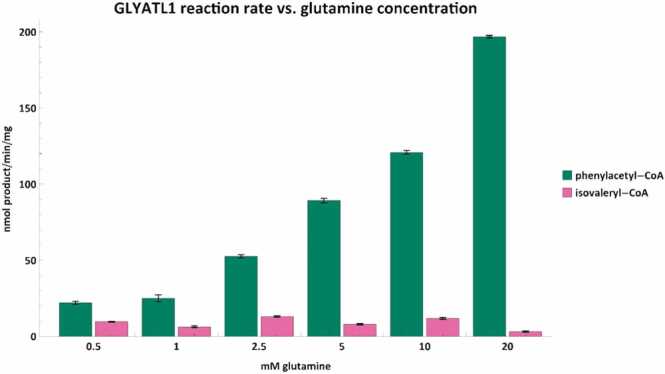
Fig. 7.
Phenylacetyl-CoA (green) and isovaleryl-CoA (pink) conjugation rates vs. glutamine concentration. Assays were performed with either 250 µM phenylacetyl-CoA or 500 µM isovaleryl-CoA. Error bars indicate the mean ± standard deviation for triplicate assays.
As was the case for the preferred acyl-CoA substrates, there is a dose-dependent increase in reaction rate when the preferred amino acid is used. However, in the case of GLYAT, glycine concentrations above 10 mM inhibit the reaction rate when 250 µM of benzoyl-CoA is used (Fig. 6). Previous studies have also shown that as the glycine concentration increased, the affinity of GLYAT for benzoyl-CoA decreased [49], [87]. This indicates that there is a glycine concentration range at which GLYAT functions optimally and therefore increasing the glycine concentration above this range will not result in more product being formed.
It is important to remember that GLYAT is a sigmoidal enzyme [52], which means that reaction kinetics are influenced by the concentration of both substrates used [88]. In the case of sulfotransferases, classic Michaelis–Menten kinetics are followed over a narrow range of substrate concentrations, while some sulfotransferases exhibit positive or negative cooperativity (allosteric sigmoidal enzyme kinetics) when studied over a wide range of substrate concentrations. In vivo studies have shown that the formation of hippuric acid from benzoic acid is limited by the availability of glycine [89] and the administration of exogenous glycine to rats increases the amount of hippuric acid formed [90]. However, the sigmoidal nature of GLYAT makes it extremely difficult to interpret in vivo studies where the concentrations of the substrates or enzyme are not known.
The glycine conjugation pathway maintains a delicate balance in CoA levels within the mitochondria [91] and glycine concentration within the mitochondria is also tightly regulated [92]. Allosteric regulation of certain enzymes evolved in order to control metabolic flow [93] by, for example, preventing the depletion of critical substrates in the mitochondria such as glycine and acyl-CoA [94]. Support for this idea may also be found in the lower binding affinities that GLYAT and GLYATL1 exhibit for their amino acid substrates (Table 4). This may be a mechanism by which acyl-CoA and amino acid homeostasis are preserved. However, a more detailed investigation of the GLYAT and GLYATL1 kinetics would be needed to better understand this.
While the conjugation of benzoyl-CoA by GLYAT appears to be slightly inhibited at 20 mM of glycine, GLYATL1 still appears responsive to glutamine concentrations of up to at least 20 mM (Fig. 7). It is difficult to interpret these values as the kinetic parameters and mechanism for GLYATL1 still needs to be elucidated. Studies have shown that liver tissue can contain up to 11 mmol/kg and 15 mmol/kg of glycine or glutamine, respectively [95]. A recent in vitro study has shown that cytosolic concentrations of glycine or glutamine is approximately 5 mM and 12 mM respectively [96], which are well below the amino acid concentration at which GLYAT and GLYATL1 function optimally.
Interestingly, the isovaleryl-CoA conjugation rates for GLYAT and GLYATL1 seemed largely independent of glycine and glutamine concentrations, respectively. Although GLYAT appeared to have a significant (p < 0.05) increase in reaction rate at 10 mM glycine (Fig. 6), GLYATL1 showed a significant (p < 0.05) decrease in the isovaleryl-CoA conjugation rate at 20 mM of glutamine (Fig. 7). This suggests that there might be an optimal concentration of glycine that should be used as a supplement for IVA patients. However, further in vivo studies will be required to confirm this. Furthermore, the reaction rates seen here using 500 µM isovaleryl-CoA will differ when the isovaleryl-CoA concentration is changed. This apparent variability in N-isovalerylglycine formation rates supports previous case studies, which showed that there is a limit to the patient’s capability to form N-isovalerylglycine even when plasma glycine concentrations are high [19], [27], [28], [29], [84]. This indicates that the ability of an IVA patient to metabolize isovaleryl-CoA depends on the interplay of metabolite concentrations, enzyme isoforms, and treatment regimen.
Both the variability of isovaleryl-CoA conjugation rates and the apparent insensitivity to amino acid concentrations further complicate possible treatments for IVA patients. High glycine concentrations, for example during glycine supplementation or inefficient glycine clearance, could be toxic for humans [97], [98]. Glycine supplementation in some IVA patients had to be stopped because of side-effects due to hyperglycinemia [28], [99]. A more recent long-term study on the neurological outcome of 80 patients with classical organic acidurias (including isovaleric acidemia) promoted glycine treatment in IVA and indicated that they did not observe any correlation between plasma glycine and neurological outcome. No precise therapeutic dose were however mentioned in this publication [100]. van Vliet and co-workers (2014) clearly state that a plasma glycine concentration over 1000 µmol/L should be avoided as historical studies indicated that encephalopathy side effects were observed in dosages of 300–600 mg/kg/day [25]. For some patients, monotherapy with L-carnitine may be more advantageous [9]. However, assessing the glycine conjugation capacity in patients on a combined or mono-therapeutic trial may be required in order to optimise treatment.
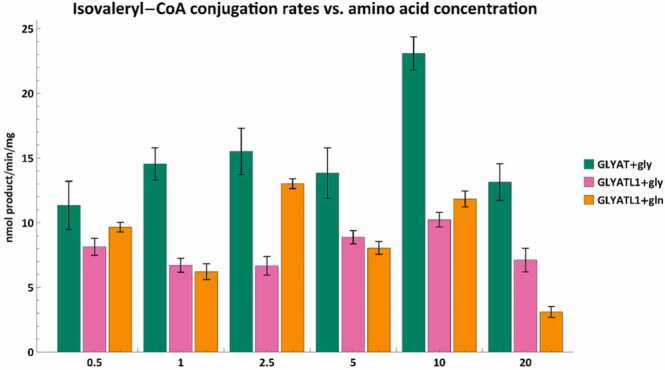
Finally, we tested whether GLYATL1 was able to conjugate isovaleryl-CoA with glycine because glycine is not the preferred amino acid for GLYATL1 [41]. These isovaleryl-CoA conjugation rates where then compared to the conjugation rates of GLYAT using glycine and GLYATL1 using glutamine (Fig. 8).
Fig. 8.
Isovaleryl-CoA conjugation rates between GLYAT and glycine (green), GLYATL1 and glycine (pink) and GLYATL1 and glutamine (orange). The amino acid concentrations were varied while the isovaleryl-CoA concentration was 500 µM. Error bars indicate the mean ± standard deviation for triplicate assays.
In Fig. 8 we can see that GLYATL1 can conjugate isovaleryl-CoA with both amino acids, albeit at a lower rate than GLYAT. The isovaleryl conjugation rates for GLYAT and GLYATL1 appear to be somewhat independent of glycine or glutamine concentrations as opposed to when the preferred acyl-CoA substrate is used. GLYATL1 is seemingly unique in its ability to conjugate benzoyl-CoA and isovaleryl-CoA with glycine whereas GLYAT appears unable to conjugate isovaleryl-CoA with glutamine (data not shown).
Given that GLYAT and GLYATL1 are co-expressed in vivo, it seems likely that the excreted N-isovaleryglycine observed in IVA patients is a combination of N-isovalerylglycine formed by both GLYAT and GLYATL1. One case study showed that urinary N-isovalerylglycine excretion remained at a plateau for 4 days after the highest level of plasma isovaleric acid was observed [101]. This further supports the results observed in this study as it was shown that although N-isovalerylglycine is formed, it is at a very low rate when compared to the preferred substrates. The elimination of isovaleryl-CoA in an IVA patient, may take several days especially in patients experiencing an ongoing attack.
4. Conclusion
Evidence from our molecular docking and enzyme kinetic studies confirm previously accepted evidence that benzoyl-CoA and glycine and phenylacetyl-CoA and glutamine are the preferred substrate pairs for GLYAT and GLYATL1 respectively. The docking evaluation of GLYAT to isovaleryl-CoA had a greater number of interactions compared to the docking of GLYAT to natural substrate, benzoyl-CoA. Additionally, we were able to show that the affinities for the amino acid substrates are moderately lower than for the acyl-CoA substrates – including the non-preferred substrate isovaleryl-CoA. Using computational docking studies in understanding enzymatic reactions remains complex and therefore validation steps such as in vitro enzyme activity assays were also evaluated.
The relative enzyme activity studies showed that the rate of product formation was dose dependent when the preferred substrates for GLYAT and GLYATL1 was used. GLYATL1 can form N-phenylacetylglutamine, N-isovalerylglutamine, N-isovalerylglycine and N-benzoylglycine (hippurate) while GLYAT can only form the latter two compounds. Contrary to established treatment methodologies [26], [102], [103], increasing glycine concentration does not appear to increase the rate at which GLYAT and GLYATL1 conjugate isovaleryl-CoA. The complex interplay between genetic variations, metabolite concentrations, dietary supplementation, and the kinetics of GLYAT and GLYATL1 likely account for the observed interindividual variation in the amount of N-isovalerylglycine excreted by IVA patients.
This study furthered our understanding of the role GLYAT and GLYATL1 play in the phase II detoxification of acyl-esters of CoA. We highlight the need for activity studies of purified glycine-N-acyltransferase family enzymes with a variety of acyl-CoA and amino acid substrates. Additionally, we demonstrate the value of using molecular docking studies to guide and interpret such kinetic experiments. This study contributes to not only the treatment strategy for IVA but also other branched chain amino acidurias. Limiting the other sources of substrates for GLYAT (Fig. 1), for example benzoate that results in benzoyl-CoA or benzoyl-CoA that is formed by the gut microbes, may even play a role to reduce the primary substrates of GLYAT and GLYATL1 and promote the formation of N-isovalerylglycine.
Role of the funding source
Funding was provided by the North-West University, South Africa. They played no role in the study design, execution of the study or the writing of this article.
CRediT authorship contribution statement
Stefan Kühn: Data curation; Formal analysis; Methodology; Validation; Visualization; Writing - original draft; Writing - review & editing. Monray E. Williams: Data curation; Formal analysis; Methodology; Validation; Visualization; Writing - original draft; Writing - review & editing. Marli Derksen: Writing - original draft. Jörn Oliver Sass: Writing - review & editing. Rencia van der Sluis: Conceptualization; Data curation; Formal analysis; Methodology; Project administration; Resources; Supervision; Writing - original draft; Writing - review & editing.
Declaration of Competing Interest
The authors declare that there are no conflict of interest.
Acknowledgement
Research support by Heinz und Heide Dürr Stiftung (Berlin, Germany) to J.O. Sass is gratefully acknowledged. MW was funded by DSI-NRF Research Development Grants for new Generation of Academics Programme (nGAP) Scholars.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.01.041.
Contributor Information
Stefan Kühn, Email: 44926324@mynwu.ac.za.
Monray E. Williams, Email: Monray.Williams@nwu.ac.za.
Marli Dercksen, Email: Marli.Dercksen@nwu.ac.za.
Jörn Oliver Sass, Email: joern.oliver.sass@h-brs.de.
Rencia van der Sluis, Email: 21224919@nwu.ac.za.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Zytkovicz T.H., Fitzgerald E.F., Marsden D., et al. Tandem mass spectrometric analysis for amino, organic, and fatty acid disorders in newborn dried blood spots: a two-year summary from the New England Newborn Screening Program. Clin Chem. 2001;47(11):1945–1955. [PubMed] [Google Scholar]
- 2.Chace D.H., Kalas T.A., Naylor E.W. Use of tandem mass spectrometry for multianalyte screening of dried blood specimens from newborns. Clin Chem. 2003;49(11):1797–1817. doi: 10.1373/clinchem.2003.022178. [DOI] [PubMed] [Google Scholar]
- 3.Ensenauer R., Vockley J., Willard J.-M., et al. A common mutation is associated with a mild, potentially asymptomatic phenotype in patients with isovaleric acidemia diagnosed by newborn screening. Am J Hum Genet. 2004;75(6):1136–1142. doi: 10.1086/426318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mütze U., Henze L., Gleich F., et al. Newborn screening and disease variants predict neurological outcome in isovaleric aciduria. J Inherit Metab Dis. 2021;44(4):857–870. doi: 10.1002/jimd.12364. [DOI] [PubMed] [Google Scholar]
- 5.Conradie E.H., Malherbe H., Hendriksz C.J., Dercksen M., Vorster B.C. An overview of benefits and challenges of rare disease biobanking in Africa, Focusing on South Africa. Biopreservation Biobanking. 2021;19(2):143–150. doi: 10.1089/bio.2020.0108. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka K., Budd M.A., Efron M.L., Isselbacher K.J. Isovaleric acidemia: a new genetic defect of leucine metabolism. Proc Natl Acad Sci USA. 1966;56(1):236–242. doi: 10.1073/pnas.56.1.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vockley J., Ensenauer R. Isovaleric acidemia: new aspects of genetic and phenotypic heterogeneity. Wiley Online Libr. 2006:95–103. doi: 10.1002/ajmg.c.30089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trost L.C., Lemasters J.J. The mitochondrial permeability transition: a new pathophysiological mechanism for Reye's syndrome and toxic liver injury. J Pharmacol Exp Ther. 1996;278(3):1000–1005. [PubMed] [Google Scholar]
- 9.Grünert S.C., Wendel U., Lindner M., et al. Clinical and neurocognitive outcome in symptomatic isovaleric acidemia. Orphanet J Rare Dis. 2012;7(1):1–9. doi: 10.1186/1750-1172-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vockley J., Ensenauer R. Isovaleric acidemia: new aspects of genetic and phenotypic heterogeneity. Am J Med Genet C Semin Med Genet. 2006;142c(2):95–103. doi: 10.1002/ajmg.c.30089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ensenauer R., Vockley J., Willard J.M., et al. A common mutation is associated with a mild, potentially asymptomatic phenotype in patients with isovaleric acidemia diagnosed by newborn screening. Am J Hum Genet. 2004;75(6):1136–1142. doi: 10.1086/426318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlune A., Riederer A., Mayatepek E., Ensenauer R. Aspects of newborn screening in isovaleric acidemia. Int J Neonatal Screen. 2018;4(1):7. doi: 10.3390/ijns4010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dercksen M., Duran M., Ijlst L., et al. Clinical variability of isovaleric acidemia in a genetically homogeneous population. J Inherit Metab Dis Feb. 2012;17(17):2012. doi: 10.1007/s10545-012-9457-2. [DOI] [PubMed] [Google Scholar]
- 14.Solano A.F., Leipnitz G., De Bortoli G.M., et al. Induction of oxidative stress by the metabolites accumulating in isovaleric acidemia in brain cortex of young rats. Free Radic Res. 2008;42(8):707–715. doi: 10.1080/10715760802311179. [DOI] [PubMed] [Google Scholar]
- 15.Ribeiro C.A., Balestro F., Grando V., Wajner M. Isovaleric acid reduces Na+, K+-ATPase activity in synaptic membranes from cerebral cortex of young rats. Cell Mol Neurobiol. 2007;27(4):529–540. doi: 10.1007/s10571-007-9143-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dercksen M., IJlst L., Duran M., et al. Inhibition of N-acetylglutamate synthase by various monocarboxylic and dicarboxylic short-chain coenzyme A esters and the production of alternative glutamate esters. Biochim Et Biophys Acta (BBA)-Mol Basis Dis. 2014;1842(12):2510–2516. doi: 10.1016/j.bbadis.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 17.Kasapkara C.S., Ezgu F.S., Okur I., Tumer L., Biberoglu G., Hasanoglu A. N-carbamylglutamate treatment for acute neonatal hyperammonemia in isovaleric acidemia. Eur J Pediatr. 2011;170(6):799–801. doi: 10.1007/s00431-010-1362-9. [DOI] [PubMed] [Google Scholar]
- 18.Szymańska E., Jezela-Stanek A., Bogdańska A., et al. Long term follow-up of polish patients with isovaleric aciduria. Clinical and molecular delineation of isovaleric aciduria. Diagnostics. 2020;10(10):738. doi: 10.3390/diagnostics10100738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naglak M.C., Madsen K.M., Dembure P.P., Salvo R., Elsas L.J. Treatment of isovaleric acidemia with glycine supplements. Pediatr Res. 1987;21(4) doi: 10.1203/00006450-198807000-00004. 293-293. [DOI] [PubMed] [Google Scholar]
- 20.Itoh T., Ito T., Ohba S., et al. Effect of carnitine administration on glycine metabolism in patients with isovaleric acidemia: significance of acetylcarnitine determination to estimate the proper carnitine dose. Tohoku J Exp Med. 1996;179(2):101–109. doi: 10.1620/tjem.179.101. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka K., Isselbacher K.J. The isolation and identification of N-isovalerylglycine from urine of patients with isovaleric acidemia. J Biol Chem. 1967;242(12):2966–2972. [PubMed] [Google Scholar]
- 22.Sweetman L., Williams J.C. Branched Chain Organic Acidurias. McGraw Hill,; 2013. Part 9: Organic acids.〈http://www.ommbid.com/〉 [Google Scholar]
- 23.Tanaka K., Isselbacher K.J. The isolation and identification of N-isovalerylglycine from urine of patients with isovaleric acidemia. JBiolChem. 1967;242(12):2966–2972. [PubMed] [Google Scholar]
- 24.Sweetman L., Williams J.C. Metabolic and Molecular Bases of Inherited Diseases. 8th ed. McGraw-Hill; 2001. Branched Chain Organic Acidurias. [Google Scholar]
- 25.van Vliet D., Derks T.G., van Rijn M., et al. Single amino acid supplementation in aminoacidopathies: a systematic review. Orphanet J Rare Dis. 2014;9(1):1–14. doi: 10.1186/1750-1172-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chinen Y., Nakamura S., Tamashiro K., et al. Isovaleric acidemia: Therapeutic response to supplementation with glycine, l-carnitine, or both in combination and a 10-year follow-up case study. Mol Genet Metab Rep. 2017;11:2–5. doi: 10.1016/j.ymgmr.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Childs B., Nyhan W.L., Borden M., Bard L., Cooke R.E. Idiopathic hyperglycinemia and hyperglycinuria: a new disorder of amino acid metabolism. 1. Pediatrics. 1961;27:522–538. [PubMed] [Google Scholar]
- 28.De Sousa C., Chalmers R., Stacey T., Tracey B., Weaver C., Bradley D. The response to L-carnitine and glycine therapy in isovaleric acidaemia. Eur J Pediatr. 1986;144(5):451–456. doi: 10.1007/BF00441737. [DOI] [PubMed] [Google Scholar]
- 29.Krieger I., Tanaka K. Therapeutic effects of glycine in isovaleric acidemia. Pediatr Res. 1976;10(1):25–29. doi: 10.1203/00006450-197601000-00005. [DOI] [PubMed] [Google Scholar]
- 30.Schachter D., Taggart J.V. Glycine N-acylase: purification and properties. JBiolChem. 1954;208(1):263–275. [PubMed] [Google Scholar]
- 31.Naglak M., Salvo R., Madsen K., Dembure P., Elsas L. The treatment of isovaleric acidemia with glycine supplement. Pediatr Res 1988/07/01. 1988;24(1):9–13. doi: 10.1203/00006450-198807000-00004. [DOI] [PubMed] [Google Scholar]
- 32.Bartlett K., Gompertz D. The specificity of glycine-N-acylase and acylglycine excretion in the organicacidaemias. BiochemMed. 1974;10(1):15–23. doi: 10.1016/0006-2944(74)90004-0. [DOI] [PubMed] [Google Scholar]
- 33.Del Olmo A., Calzada J., Nunez M. Benzoic acid and its derivatives as naturally occurring compounds in foods and as additives: Uses, exposure, and controversy. Crit Rev Food Sci Nutr. 2017;57(14):3084–3103. doi: 10.1080/10408398.2015.1087964. [DOI] [PubMed] [Google Scholar]
- 34.van der Sluis R. Analyses of the genetic diversity and protein expression variation of the acyl: CoA medium-chain ligases, ACSM2A and ACSM2B. Mol Genet Genom: MGG. 2018;293(5):1279–1292. doi: 10.1007/s00438-018-1460-3. [DOI] [PubMed] [Google Scholar]
- 35.Rechner A.R., Kuhnle G., Bremner P., Hubbard G.P., Moore K.P., Rice-Evans C.A. The metabolic fate of dietary polyphenols in humans. Free Radic Biol Med. 2002;33(2):220–235. doi: 10.1016/s0891-5849(02)00877-8. [DOI] [PubMed] [Google Scholar]
- 36.Lemarie F., Beauchamp E., Legrand P., Rioux V. Revisiting the metabolism and physiological functions of caprylic acid (C8:0) with special focus on ghrelin octanoylation. Biochimie. 2016;120:40–48. doi: 10.1016/j.biochi.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Killinberg P.G., Davidson E.D., Webster L.T. Evidence for a medium-chain fatty acid: coenzyme A ligase (adenosine monophosphate) that activates salicylate. Mol Pharm. 1971;7:260–268. [PubMed] [Google Scholar]
- 38.Knights K.M. Role of hepatic fatty acid:coenzyme A ligases in the metabolism of xenobiotic carboxylic acids. Clin. Exp Pharmacol Physiol. 1998;25:776–782. doi: 10.1111/j.1440-1681.1998.tb02152.x. [DOI] [PubMed] [Google Scholar]
- 39.Nandi D.L., Lucas S.V., Webster L.T., Jr. Benzoyl-coenzyme A:glycine N-acyltransferase and phenylacetyl-coenzyme A:glycine N-acyltransferase from bovine liver mitochondria. Purification and characterization. JBiolChem. 1979;254(15):7230–7237. [PubMed] [Google Scholar]
- 40.Mitchell G.A., Gauthier N., Lesimple A., Wang S.P., Mamer O., Qureshi I. Hereditary and acquired diseases of acyl-coenzyme A metabolism. Mol Genet Metab. 2008;94(1):4–15. doi: 10.1016/j.ymgme.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 41.Moldave K., Meister A. Synthesis of phenylacetylglutamine by human tissue. J Biol Chem. 1957;229(1):463–476. [PubMed] [Google Scholar]
- 42.Gregersen N., Kolvraa S., Mortensen P.B. Acyl-CoA: glycine N-acyltransferase: in vitro studies on the glycine conjugation of straight- and branched-chained acyl-CoA esters in human liver. Biochem Biol. 1986;35(2):210–218. doi: 10.1016/0885-4505(86)90076-9. [DOI] [PubMed] [Google Scholar]
- 43.Kelley M., Vessey D. Characterization of the acyl-CoA: Amino acid N-acyltransferases from primate liver mitochondria. J Biochem Toxicol. 1994;9:153–158. doi: 10.1002/jbt.2570090307. [DOI] [PubMed] [Google Scholar]
- 44.Nilsson A., Haanstra J.R., Engqvist M., et al. Quantitative analysis of amino acid metabolism in liver cancer links glutamate excretion to nucleotide synthesis. Proc Natl Acad Sci. 2020;117(19):10294–10304. doi: 10.1073/pnas.1919250117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker P.R., 2nd, Friederich M.W., Swanson M.A., et al. Variant non ketotic hyperglycinemia is caused by mutations in LIAS, BOLA3 and the novel gene GLRX5. Brain. 2014;137(Pt 2):366–379. doi: 10.1093/brain/awt328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pero R.W. Health consequences of catabolic synthesis of hippuric acid in humans. Curr Clin Pharm. 2010;5(1):67–73. doi: 10.2174/157488410790410588. [DOI] [PubMed] [Google Scholar]
- 47.Zhang H., Lang Q., Li J., et al. Molecular cloning and characterization of a novel human glycine-n-acyltransferase gene GLYATL1, which activates transcriptional activity of HSE pathway. Int J Mol Sci. 2007;8:433–444. [Google Scholar]
- 48.Matsuo M., Terai K., Kameda N., et al. Designation of enzyme activity of glycine-N-acyltransferase family genes and depression of glycine-N-acyltransferase in human hepatocellular carcinoma. BiochemBiophysResCommun. 2012;420(4):901–906. doi: 10.1016/j.bbrc.2012.03.099. [DOI] [PubMed] [Google Scholar]
- 49.van der Sluis R., Ungerer V., Nortje C., AvD A., Erasmus E. New insights into the catalytic mechanism of human glycine N-acyltransferase. J Biochem Mol Toxicol. 2017;31(11) doi: 10.1002/jbt.21963. [DOI] [PubMed] [Google Scholar]
- 50.Badenhorst C.P.S., Jooste M., van Dijk A.A. Enzymatic characterization and elucidation of the catalytic mechanism of a recombinant bovine glycine N-acyltransferase. Drug Metab Dispos. 2012;40(2):346–352. doi: 10.1124/dmd.111.041657. [DOI] [PubMed] [Google Scholar]
- 51.Cárdenas M.L. Understanding mechanisms of enzyme co-operativity: The importance of not being at equilibrium. Perspect Sci 2015/03/01/ 2015;4:10–16. doi: 10.1016/j.pisc.2014.12.003. [DOI] [Google Scholar]
- 52.Rohwer J.M., Schutte C., van der Sluis R. Functional characterisation of three glycine n-acyltransferase variants and the effect on glycine conjugation to Benzoyl-CoA. Int J Mol Sci. 2021;22(6) doi: 10.3390/ijms22063129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGuffin L.J., Bryson K., Jones D.T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16(4):404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 54.Jones D.T., Cozzetto D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics. 2015;31(6):857–863. doi: 10.1093/bioinformatics/btu744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He B., Wang K., Liu Y., Xue B., Uversky V.N., Dunker A.K. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009;19(8):929–949. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- 56.Schwede T., Kopp J., Guex N., Peitsch M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31(13):3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waterhouse A., Bertoni M., Bienert S., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–w303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biasini M., Bienert S., Waterhouse A., et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. Jul 2014;42(Web Server issue):W252–8. doi:10.1093/nar/gku340. [DOI] [PMC free article] [PubMed]
- 59.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26(2):283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 60.Bowie J.U., Lüthy R., Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253(5016):164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 61.Lüthy R., Bowie J.U., Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356(6364):83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 62.Jain C.K., Gupta M., Prasad Y., Wadhwa G., Sharma S.K. Homology modeling and protein engineering of alkane monooxygenase in Burkholderia thailandensis MSMB121: in silico insights. J Mol Model. 2014;20(7):2340. doi: 10.1007/s00894-014-2340-3. [DOI] [PubMed] [Google Scholar]
- 63.Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9):1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Messaoudi A., Belguith H., Ben Hamida J. Homology modeling and virtual screening approaches to identify potent inhibitors of VEB-1 β-lactamase. Theor Biol Med Model. 2013;10:22. doi: 10.1186/1742-4682-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(Web Server issue):W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chothia C., Lesk A.M. The relation between the divergence of sequence and structure in proteins. Embo J. 1986;5(4):823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu D., Zhang Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J. 2011;101(10):2525–2534. doi: 10.1016/j.bpj.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mawal Y.R., Qureshi I.A. Purification to homogeneity of mitochondrial acyl coa:glycine n-acyltransferase from human liver. Biochem Biophys Res Commun. 1994;205(2):1373–1379. doi: 10.1006/bbrc.1994.2817. [DOI] [PubMed] [Google Scholar]
- 70.Schachter D., Taggart J.V. Benzoyl coenzyme A and hippurate synthesis. J Biol Chem. 1953;203:925–934. [PubMed] [Google Scholar]
- 71.Salentin S., Schreiber S., Haupt V.J., Adasme M.F., Schroeder M. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015;43(W1):W443–W447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mathematica. 2022. https://www.wolfram.com/mathematica.
- 73.van der Sluis R., Badenhorst C.P., van der Westhuizen F.H., van Dijk A.A. Characterisation of the influence of genetic variations on the enzyme activity of a recombinant human glycine N-acyltransferase. Gene. 2013;515(2):447–453. doi: 10.1016/j.gene.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 74.Stank A., Kokh D.B., Fuller J.C., Wade R.C. Protein binding pocket dynamics. Acc Chem Res. 2016;49(5):809–815. doi: 10.1021/acs.accounts.5b00516. [DOI] [PubMed] [Google Scholar]
- 75.van der Westhuizen F.H., Pretorius P.J., Erasmus E. The utilization of alanine, glutamic acid, and serine as amino acid substrates for glycine N-acyltransferase. J Biochem Mol Toxicol. 2000;14(2):102–109. doi: 10.1002/(sici)1099-0461(2000)14:2<102::aid-jbt6>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 76.Mawal Y.R., Qureshi I.A. Purification to homogeneity of mitochondrial acyl coa:glycine n-acyltransferase from human liver. BiochemBiophysResCommun. 1994;205(2):1373–1379. doi: 10.1006/bbrc.1994.2817. [DOI] [PubMed] [Google Scholar]
- 77.Schulke D., Sass J.O. Frequent sequence variants of human glycine N-acyltransferase (GLYAT) and inborn errors of metabolism. Biochimie. 2021 doi: 10.1016/j.biochi.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 78.Levy G. Pharmacokinetics of salicylate elimination in man. J Pharm Sci. 1965;54(7):959–967. doi: 10.1002/jps.2600540703. [DOI] [PubMed] [Google Scholar]
- 79.Quick A.J., Cooper M.A. The effect of liver injury on the conjugation of benzoic acid in the dog. J Biol Chem. 1932;99:119–124. [Google Scholar]
- 80.Schachter D. The chemical estimation of acyl glucuronides and its application to studies on the metabolism of benzoate and salicylate in man. J Clin Investig. 1957;36(2):297–302. doi: 10.1172/JCI103424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lehnert W. Excretion of N-isovalerylglutamic acid in isovaleric acidemia. Clin Chim Acta. 1981;116(2):249–252. doi: 10.1016/0009-8981(81)90030-9. [DOI] [PubMed] [Google Scholar]
- 82.Dercksen M., Koekemoer G., Duran M., Wanders R.J., Mienie L.J., Reinecke C.J. Organic acid profile of isovaleric acidemia: a comprehensive metabolomics approach. Metabolomics. 2013;9(4):765–777. [Google Scholar]
- 83.Loots D.T., Erasmus E., Mienie L.J. Identification of 19 new metabolites induced by abnormal amino acid conjugation in isovaleric acidemia. Clin Chem. 2005;51(8):1510–1512. doi: 10.1373/clinchem.2005.048421. [DOI] [PubMed] [Google Scholar]
- 84.Fries M.H., Rinaldo P., Schmidt-Sommerfeld E., Jurecki E., Packman S. Isovaleric acidemia: response to a leucine load after three weeks of supplementation with glycine, L-carnitine, and combined glycine-carnitine therapy. J Pediatr. 1996;129(3):449–452. doi: 10.1016/s0022-3476(96)70081-1. [DOI] [PubMed] [Google Scholar]
- 85.Cohn R.M., Yudkoff M., Rothman R., Segal S. Isovaleric acidemia: use of glycine therapy in neonates. N Engl J Med. 1978;299(18):996–999. doi: 10.1056/nejm197811022991807. [DOI] [PubMed] [Google Scholar]
- 86.Yudkoff M., Cohn R.M., Puschak R., Rothman R., Segal S. Glycine therapy in isovaleric acidemia. J Pedia. 1978;92(5):813–817. doi: 10.1016/s0022-3476(78)80164-4. [DOI] [PubMed] [Google Scholar]
- 87.Dempsey D.R., Bond J.D., Carpenter A.M., Rodriguez Ospina S., Merkler D.J. Expression, purification, and characterization of mouse glycine N-acyltransferase in Escherichia coli. Protein Expr Purif. 2014;97:23–28. doi: 10.1016/j.pep.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Palmer T. Horwood Series in Chemical Science. 5 ed. Horwood Publishing Limited; 2001. ENZYMES Biochemistry, Biotechnology, Clinical Chemistry; p. 402. [Google Scholar]
- 89.Knights K., Vessey D. Comprehensive Toxicology. Elsevier; 2010. Enzymology of amino acid conjugation reactions; pp. 459–483. [Google Scholar]
- 90.Gregus Z., Fekete T., Varga F., Klaassen C. Dependence of glycine conjugation on availability of glycine: role of the glycine cleavage system. Xenobiotica. 1993;23(2):141–153. doi: 10.3109/00498259309059370. [DOI] [PubMed] [Google Scholar]
- 91.Badenhorst C.P., van der Sluis R., Erasmus E., van Dijk A.A. Glycine conjugation: importance in metabolism, the role of glycine N-acyltransferase, and factors that influence interindividual variation. Expert Opin Drug Metab Toxicol. 2013;9(9):1139–1153. doi: 10.1517/17425255.2013.796929. [DOI] [PubMed] [Google Scholar]
- 92.Kikuchi G., Motokawa Y., Yoshida T., Hiraga K. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad, Ser B. 2008;84(7):246–263. doi: 10.2183/pjab/84.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ebrecht A.C., Solamen L., Hill B.L., Iglesias A.A., Olsen K.W., Ballicora M.A. Allosteric control of substrate specificity of the escherichia coli ADP-glucose pyrophosphorylase. Front Chem. 2017;5:41. doi: 10.3389/fchem.2017.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Badenhorst C.P., Erasmus E., van der Sluis R., Nortje C., van Dijk A.A. A new perspective on the importance of glycine conjugation in the metabolism of aromatic acids. Drug Metab Rev. 2014;46(3):343–361. doi: 10.3109/03602532.2014.908903. [DOI] [PubMed] [Google Scholar]
- 95.Barle BA H., Nyberg B., Andersson K., Essén P., Wernerman J. The concentrations of free amino acids in human liver tissue obtained during laparoscopic surgery. Clin Physiol. 1996;16(3):217–227. doi: 10.1111/j.1475-097x.1996.tb00570.x. [DOI] [PubMed] [Google Scholar]
- 96.Gauthier-Coles G., Vennitti J., Zhang Z., et al. Quantitative modelling of amino acid transport and homeostasis in mammalian cells. Nat Commun. 2021;12(1):5282. doi: 10.1038/s41467-021-25563-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sandfeldt L., Riddez L., Rajs J., Ewaldsson C., Piros D., Hahn R.G. High-dose intravenous infusion of irrigating fluids containing glycine and mannitol in the pig. J Surg Res. 2001;95(2):114–125. doi: 10.1006/jsre.2000.6028. [DOI] [PubMed] [Google Scholar]
- 98.Garlick P.J. The nature of human hazards associated with excessive intake of amino acids. J Nutr. 2004;134(6):1633S–1639S. doi: 10.1093/jn/134.6.1633S. [DOI] [PubMed] [Google Scholar]
- 99.Berry G.T., Yudkoff M., Segai S. Isovaleric acidemia: medical and neurodevelopmental effects of long-term therapy. J Pediatr. 1988;113(1):58–64. doi: 10.1016/s0022-3476(88)80528-6. [DOI] [PubMed] [Google Scholar]
- 100.Nizon M., Ottolenghi C., Valayannopoulos V., et al. Long-term neurological outcome of a cohort of 80 patients with classical organic acidurias. Orphanet J Rare Dis. 2013;8(1):1–12. doi: 10.1186/1750-1172-8-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shigematsu Y., Sudo M., Momoi T., Inoue Y., Suzuki Y., Kameyama J. Changing plasma and urinary organic acid levels in a patient with isovaleric acidemia during an attack. Pediatr Res. 1982;16(9):771–775. doi: 10.1203/00006450-198209000-00013. [DOI] [PubMed] [Google Scholar]
- 102.Naglak M.C., Madsen K.M., Dembure P.P., Salvo R., Elsas L.J. Treatment of isovaleric acidemia with glycine supplements. Pediatr Res. 1987;21:293. doi: 10.1203/00006450-198807000-00004. [DOI] [PubMed] [Google Scholar]
- 103.Fries M.H., Ronaldo P., Schmidt-Sommerfeld E., Jurecki E., Packman S. Isovaleric acidemia: response to a leucine load after three weeks of supplementation with glycine, L-carnitine, and combined glycine-carnitine therapy. J Pediatr. 1996;129(3):449–452. doi: 10.1016/s0022-3476(96)70081-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material