Abstract
In three domains of life, proteins are synthesized by large ribonucleoprotein particles called ribosomes. All ribosomes are composed of ribosomal RNAs (rRNA) and numerous ribosomal proteins (r-protein). The three-dimensional shape of ribosomes is mainly defined by a tertiary structure of rRNAs. In addition, rRNAs have a major role in decoding the information carried by messenger RNAs and catalyzing the peptide bond formation. R-proteins are essential for shaping the network of interactions that contribute to a various aspects of the protein synthesis machinery, including assembly of ribosomes and interaction of ribosomal subunits. Structural studies have revealed that many key components of ribosomes are conserved in all life domains. Besides the core structure, ribosomes contain domain-specific structural features that include additional r-proteins and extensions of rRNA and r-proteins. This review focuses specifically on those r-proteins that are found only in archaeal and eukaryotic ribosomes. The role of these archaea/eukaryote specific r-proteins in stabilizing the ribosome structure is discussed. Several examples illustrate their functions in the formation of the internal network of ribosomal subunits and interactions between the ribosomal subunits. In addition, the significance of these r-proteins in ribosome biogenesis and protein synthesis is highlighted.
Keywords: Protein synthesis, Translation, Ribosome biogenesis, Archaea, Eukaryote, Ribosomal proteins
Graphical Abstract
1. Introduction
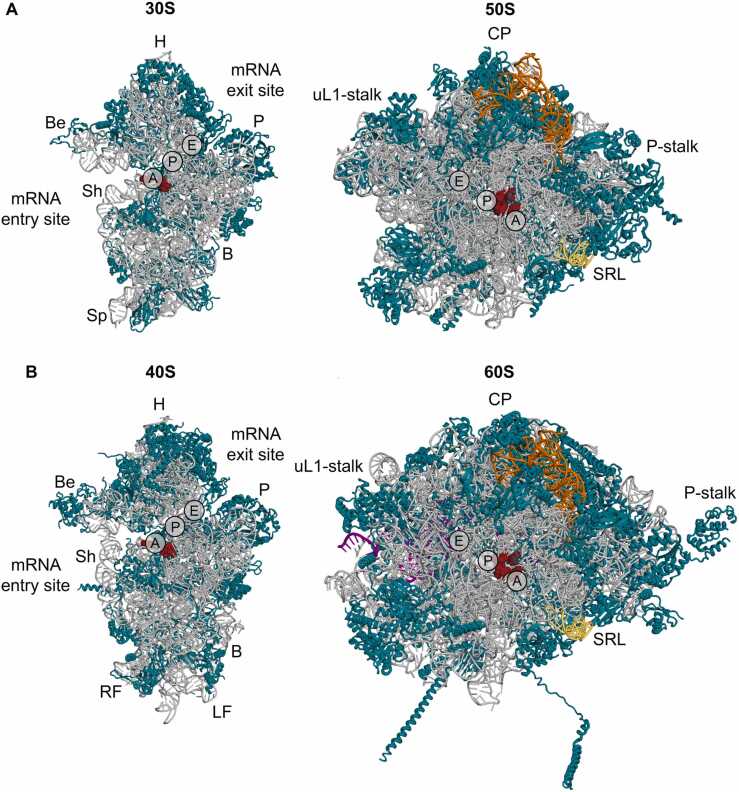
Protein synthesis, an essential process for life, is carried out by a large ribonucleoprotein complex called the ribosome. All ribosomes are composed of two subunits - the small ribosomal subunit (SSU; 30S in archaea and 40S in eukaryotes) and the large ribosomal subunit (LSU; 50S in archaea and 60S in eukaryotes) (Fig. 1). The SSU associates with mRNA during translation and mediates tRNA-mRNA interactions. Thus, SSU contains the decoding center (DC) that ensures the recognition of correct start codon during the initiation of translation and the selection of correct aminoacyl-tRNAs during the elongation of translation. The LSU catalyzes the formation of peptide bonds between the incoming amino acids and the growing peptide chain at the peptidyl transferase center (PTC). In addition, LSU stimulates the hydrolysis of factor-bound GTP molecules and drives exit of the growing peptide chain through the nascent polypeptide exit tunnel (PET). The two subunits form a translating ribosome 70S in archaea and 80S in eukaryotes) containing binding sites for three tRNAs - the aminoacyl site (A-site), the peptidyl site (P-site) and the exit site (E-site). Both subunits are made up of two types of structural components - ribosomal RNAs (rRNAs) and ribosomal proteins (r-proteins). rRNAs and r-proteins constitute the most abundant biological macromolecules in cells [1], [2]. This is illustrated by the observation that in actively dividing bacterial cells, rRNAs account for approximately 85% of the total cellular RNA and r-proteins account for 23% of the total protein [1].
Fig. 1.
Comparison of archaeal and eukaryotic ribosomal subunits. Intersubunit view of individual subunits of Pyrococcus furiosus (A) and Saccharomyces cerevisiae (B) (left – SSU (30S, 40S), right – LSU (50S, 60S). rRNA and r-proteins are shown in grey and teal, respectively. Domains of the SSU (H, head; Be, beak; P, platform; Sh, shoulder; B, body, Sp, spur; RF, right foot; and LF, left foot) and mRNA entry/exit sites are labelled. Landmarks of the LSU (uL1-stalk; P-stalk; CP, central protuberance) are shown. Sarcin-ricin loop (SRL) is colored yellow, 5S rRNA is orange, 5.8S rRNA is purple. The decoding centre in the SSU and the peptidyl transferase centre in the LSU are indicated by red spheres. In addition, approximate positions of the A-, P- and E-sites for tRNA binding are indicated. PDB coordinates 4V6U for P. furiosus [3] and 4V88 for S. cerevisiae [4] were rendered in PyMol.
High resolution atomic models have revealed that all cytoplasmic ribosomes (from bacteria to the higher eukaryotes) have a similar core structure [5], [6], [7]. This universal common core has a mass of nearly 2 MDa and comprises ∼4400 RNA bases together with 33 r-proteins. The common core contains all of the main functional centers of the ribosome - DC, PTC, GTPase-associated region, upper part of PET closer to PTC, and A-, P- and E-sites for tRNA binding. In addition to the core structure, the ribosomes from different life domains contain their own set of specific moieties: additional rRNA sequences called expansion segments (ES), domain-specific r-proteins as well as insertions and extension of r-proteins present in all cytoplasmic ribosomes [5].
In general, the r-proteins can be divided into three main groups: (1) the universally conserved r-proteins that with the rRNA form the universal common core; (2) the domain-specific r-proteins (e.g. bacteria-specific and eukaryote-specific; no archaea-specific group of r-proteins is currently recognised); and (3) the r-proteins exclusively shared between archaea and eukaryotes (also called the archaea/eukaryote-specific, A/E-specific) [8]. R-proteins are unevenly distributed in the ribosome, with most of them located on the solvent side of the subunits, leaving the subunit interface side to be dominated by rRNA [9], [10].
In bacterial ribosome, r-proteins belonging to the common core account for about 62% of all r-proteins [5]. Additional r-proteins, about 20 of them, are bacteria-specific. The exact number of these r-proteins varies between bacterial species [11]. Although the core structural components of archaeal rRNA are very similar to those of bacteria, the content of r-proteins differs markedly. In archaeal ribosomes, 71 different r-proteins have been identified [12], [13]. Only 54 of them are known to be ubiquitous across archaea [14]. In addition to the 33 universally conserved r-proteins, a set of 35 r-proteins is shared with eukaryotic ribosomes. Interestingly, archaeal ribosomes do not share any r-proteins exclusively with bacterial ribosomes. In eukaryotic ribosomes, the overall mass of r-proteins is dramatically increased. For example, unlike the bacterial SSU, where the protein-to-rRNA mass ratio is ∼1:2, the eukaryotic SSU has a mass ratio of almost 1:1 [15]. The budding yeast (S. cerevisiae) 80S ribosome contains 79 r-proteins [8], [4]. In addition to the universally conserved and A/E-specific r-proteins, 11 r-proteins are specific only to the eukaryotes.
R-proteins have an essential function in ribosome biogenesis and in ensuring optimal functioning of the ribosome. In this context, r-proteins have a role in stabilizing the intrasubunit network, in forming the subunit contacts during protein synthesis and are docking sites for translation factors and other ribosome-associated proteins during translation. This review focuses on r-proteins conserved in archaea and eukaryotes and how these A/E-specific r-proteins perform functions specific to r-proteins. In addition, a number of A/E-specific r-proteins are discussed in detail with respect to their known functions.
2. Archaea/eukaryote-specific r-proteins
A/E-specific r-proteins, like r-proteins in general, share a number of characteristics. By and large, r-proteins are among of the smallest proteins in cells. For instance, in budding yeast ribosome, the length of A/E-specific r-proteins varies from 25 aa residues in eL41 up to 261 aa residues in eS4 (Supplementary Table S1) [16], [17]. R-proteins are characterized by high isoelectric point (generally higher than 9.0) and net positiive charge compared to non-ribosomal proteins [18], [19]. In addition, they exhibit charge segregation, i.e. positively charged regions of r-proteins interact with negatively charged rRNA residues whereas negatively charged regions of r-proteins are exposed to the solvent [20], [21]. This charge segregation ensures tight binding of r-proteins to rRNA, which helps to stabilize the overall structure of ribosome. However, there are some A/E-specific r-proteins that do not share the same properties. For example, the P-proteins (archaeal P1 and eukaryotic P1 and P2) are acidic (pI ∼3.5–4.5) and are the only r-proteins present in multiple copies per ribosome [22], [23].
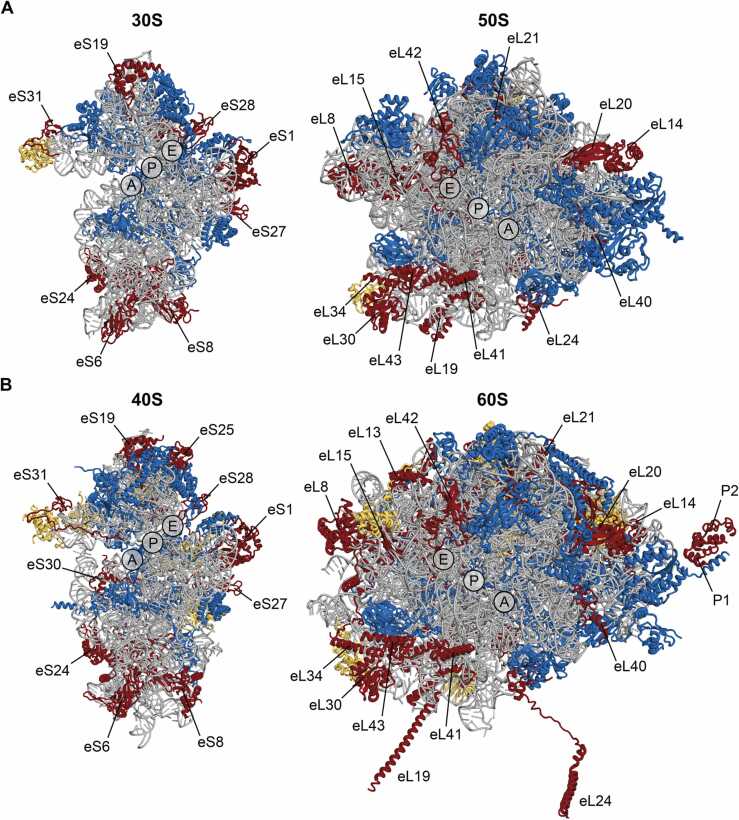
Structural analysis of archaeal and eukaryotic ribosomes has revealed that the spatial distribution of A/E-specific r-proteins in subunits is different. In the SSU, A/E-specific proteins reside at the spur/foot of the subunit, at the platform domain and at the top of the head (Fig. 2, Supplementary Fig. S1) [4]. In the LSU these r-proteins follow the cluster organization of rRNA ESs [4]. In general, the archaeal A/E-specific r-proteins consist of a single globular domain conserved in both archaea and eukaryotes (Supplementary Figs. S2-S3) [3], [24]. These globular domains contain both α-helices and β-barrels, and in some cases only α-helices (e.g. eS17, eL39, and eL41) or β-barrels (e.g. eS27 and eS28). A few of the archaeal A/E-specific r-proteins contain additional loops and/or N-terminal and/or C-terminal extensions (e.g. eS8, eS27, eL21, and eL39). In contrast, the eukaryotic A/E-specific r-proteins are characterized, in addition to the conserved globular domain, by the presence of long unstructured N-terminal and/or C-terminal extensions that frequently reach far from the globular domains [25], [26]. Thus, eukaryotic A/E-specific r-proteins usually consist of multiple domains (e.g. eS6, eL19, and eL24). Interestingly, the size and sequence of r-proteins is conserved within a single domain of life. For instance, 72 out of the 80 r-proteins from H. sapiens have nearly identical size and tertiary structures as their homologs from S. cerevisiae (Supplementary Table S1) [5].
Fig. 2.
Comparison of the location of archaea/eukaryote-specific r-proteins in archaeal and eukaryotic ribosomes. Intersubunit view of individual subunits of Pyrococcus furiosus (A) and Saccharomyces cerevisiae (B) (left – SSU (30S, 40S), right – LSU (50S, 60S) are shown. rRNA and universally conserved r-proteins are highlighted in grey and dark blue, respectively. Archaea/eukaryote-specific proteins are colored red and labelled. Domain-specific proteins are yellow. Approximate positions of the A-, P- and E-sites for tRNA binding are indicated. Protein groups and nomenclature are according to [8]. PDB coordinates 4V6U for P. furiosus [3] and 4V88 for S. cerevisiae [4] were rendered in PyMol.
Structural analysis of ribosomes has also demonstrated that several A/E-specific r-proteins are positional analogues of bacteria-specific r-proteins (Supplementary Table 2). These r-proteins occupy similar positions relative to structural elements of rRNA. In the course of evolution, the A/E-specific r-proteins have acquired additional contacts with rRNA helices. This is particularly well illustrated by structural analysis of the eukaryotic ribosome (Supplementary Table 2). In general, there is no structural homology between the positional analogues of r-proteins. An exception is eS1, whose C-terminal domain resembles the bacterial bS6, but its overall fold is different [26]. In the LSU, P-stalk proteins do not directly ineract with rRNA [27]. Instead, they are in contact with universally conserved r-proteins uL11 and uL10, which form the base and anchor the stalk to the rRNA. Although the localisation of P-stalk proteins in the ribosome is identical, the archaeal P1 and the eukaryotic P1/P2 are not homologous to bacterial stalk protein bL12.
Recent cryo-EM structures of Pyrococcus abyssi 30S initiation complex and Thermococcus celer 30S post-splitting complex identified a previously unobserved density for r-protein on the archaeal SSU platform [28], [29]. The newly identified 59 aa long r-protein occupies the same position as eS21 in the SSU of S. cerevisiae. Furthermore, these two r-proteins share a similar structural topology. However, the amino acid sequence identity of the newly discovered archaeal r-protein with the eukaryotic eS21 is low, with only 7% for the full-length protein [29]. It is therefore possible that structural homologues for other eukaryote-specific r-proteins are also present in archaea. Their low amino acid sequence similarity makes them difficult to identify.
Phylogenomic analysis of A/E-specific r-proteins has shown that the presence of these r-proteins varies across the archaeal domain of life [14], [30], [31]. In particular, ribosomes of species belonging to Crenarchaeota and Asgard superphylum (Loki-, Odin-, Thor- and Heimdallarchaeota) contain more r-proteins than those from other archaea (Table 1). There have been several probable independent losses of r-proteins in the Euryarchaeota. The A/E-specific eS25, eS30, eL13 and eL38 are not found in these species. A number of other A/E-specific r-proteins have been gradually reduced. Thus, the ribosomes of various archaea have lost several A/E-specific r-proteins, which at the same time have been preserved in eukaryotes. It should be noted that not all A/E-specific r-proteins are essential for cell viability in eukaryotes [32]. Genetic studies with budding yeast have demonstrated that eight A/E-specific r-proteins are non-essential but the loss of these r-proteins has different effect on cell growth. Cells lacking eL38 or eL41 r-protein grow similarly to wild type cells [33], [34]. At the same time, the loss of eL31, eL39, or eL25, severely limits cell growth [35], [36], [37].
Table 1.
Distribution of non-ubiquitous A/E-specific r-proteins in various archaeal phyla1.
| Archaeal phylum | r-protein2 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| eS25 | eS30 | eL13 | eL14 | eL20 | eL30 | eL34 | eL33 | eL38 | eL41 | |
| Lokiarchaeota | + | + | + | + | + | + | + | + | + | - |
| Odinarchaeota | + | + | + | + | un | + | + | un | - | - |
| Thorarchaeota | + | + | + | + | + | + | + | + | + | - |
| Heimdallarchaeota | + | + | - | + | + | + | + | un | + | - |
| Verstraetearchaeota | + | + | - | - | + | + | - | + | - | - |
| Crenarchaeota | + | + | + /- | + | + | + | + | + /- | + /- | - |
| Korarchaeota | + /- | + | + | + | - | + | + | - | - | - |
| Bathyarchaeota | + | + | + | + | + | + | + | + | + | - |
| Aigarchaeota | + | + | + /- | + | + | + | - | un | - | - |
| Thaumarchaeota | + | + | + /- | + /- | - | + | - | - | - | - |
| Nanoarchaeota | - | - | + | + | + | + | + | + | - | + |
| Euryarchaeota | - | - | - | + /- | + /- | + /- | + /- | + /- | - | + /- |
Taken together, A/E-specific r-proteins present an intriguing group of r-proteins. On the one hand, there is a structural variability within these r-proteins, in particular the presence of non-globular extensions in eukaryotic r-proteins. On the other hand, there are archaeal ribosomes that have lost some A/E-specific r-proteins. Thus, such structural variation may be significant for the functional specialization of both A/E-specific r-proteins and the ribosome as a complex. An example of such specialization is the preference of ribosomes for translating certain subsets of mRNAs. It is known that translation of mRNAs containing internal ribosome entry site (IRES) or IRES-like elements is dependent on A/E-specific r-proteins eS25 [39], [40]. As discussed above, these r-proteins belong to the group of non-ubiquitous A/E-specific r-proteins in archaea. It is therefore possible to speculate that the increased variability in the r-protein composition of archaeal ribosomes facilitates translation of a specific subset of mRNAs.
3. Functional roles of archaea/eukaryote-specific r-proteins
As already mentioned, a common feature of the r-proteins is that the vast majority of them are positively charged. It has therefore been proposed that one of the main functions of r-proteins is to stabilize the negatively charged rRNA to promote efficient protein synthesis [10]. Several studies have revealed that r-proteins also have functions in different cellular processes, e.g. cell proliferation [41], [42], differentiation [43], [44], apoptosis [45], [46], DNA repair [47], [48], as well as modulation of cell migration and invasion [49]. In addition, mutations in r-protein genes have been associated with human diseases, such as Diamond-Blackfan anemia and Schwachman-Diamond syndrome [50]. R-proteins may also participate in the development of several types of cancers [51], [52]. Although r-proteins are known to have extra-ribosomal functions, their essential role is to provide structural functionality of the ribosome. In the following sections, the role of A/E-specific r-proteins in ribosome assembly, ribosome function, and protein translation, will be discussed.
4. Role in ribosome biogenesis
Ribosome biogenesis is a highly coordinated process involving the synthesis of r-proteins, the synthesis and modification of rRNAs, and the assembly of r-proteins and rRNAs into two mature ribosomal subunits. It is estimated that exponentially growing budding yeast cell produces 2000 ribosomes per minute, making ribosome assembly an extremely fast and energy consuming process that is critical for cellular homeostasis [53], [54], [55]. In eukaryotic cells, the assembly starts in the nucleolus, where the transcription of 35S precursor rRNA (pre-rRNA) by polymerase I occurs [56], [57]. In parallel, polymerase III transcribes 5S rRNA [58], [59]. During assembly, pre-rRNAs undergo hierarchical processing, folding, chemical modification, and assembly with r-proteins and assembly factors [60], [61]. Assembly machinery proceeds through the subsequent nucleolar, nucleoplasmic and cytoplasmic steps to result in mature SSU and LSU. Recent review articles provide a detailed overview of ribosome biogenesis in eukaryotes [62], [63], [64]. Ribosome biogenesis in archaea has been less studies (reviwed in [65]).
The A/E-specific r-proteins are also involved in assembly of ribosomal subunits, as they coordinate folding of rRNAs and binding of numerous assembly factors and other r-proteins. For example, 9 out of 19 r-proteins that are associated with 35S pre-rRNA during the initial co-transcritpional assembly of the SSU belong to the A/E-specific r-protein group (Table 2) [66]. Recent cryo-EM studies of the earliest pre-LSU particles revealed that 18 r-proteins are already associated with pre-rRNAs [67]. Of these, 10 are A/E-specific r-proteins. All associated r-proteins assist in the initial folding of pre-rRNA at the solvent side of pre-LSU subunit [67], [68]. In the cytoplasm, where domains of SSU rRNA are finally folded, 3 remaining A/E-specific r-proteins (eS17, eS19 and eS25) are incorporated [66]. Cytoplasmic step of the LSU assembly involves the incorporation of the last 9 r-proteins, among them the A/E-specific r-proteins eL24, eL40, eL41, eL42, and final folding of P-stalk [62], [69], [70], [71].
Table 2.
Hierarchical assembly of archaea/eukaryote-specific r-proteins in the SSU and LSU of the budding yeast ribosome.
| Subunit | Associated in |
|||
|---|---|---|---|---|
| nucleolus early state |
nucleolus late state |
nucleoplasm | cytoplasm | |
| 40 S assembly1 | eS1, eS4, eS6, | eS17, eS19, | ||
| eS8, eS24, eS27, | eS25 | |||
| eS28, eS30, eS31 | ||||
| 60 S assembly2 | eL8, eL13, eL14, | eL19, eL21, | eL39, eL43 | eL24, eL42 |
| eL15, eL18, eL20, | eL30, eL31, | |||
| eL32, eL33, eL37 | eL34, eL38 | |||
In many cases, the ribosome assembly factors adopt a similar conformation and occupy the same rRNA-binding sites as A/E-specific r-proteins. One such example is the Sas10 protein, a component of the SSU processome complex [74]. Sas10 mimics eS30 and occupies its binding site on helix 16 of SSU rRNA. Another example is Rlp24 protein, an assembly factor that associates with pre-LSU particles during nucleolar, nuclear, and cytoplasmic steps of maturation [75]. Rlp24 and eL24 share the N-terminal domain that recognizes the same binding site on LSU [73]. Rlp24 is an important docking factor for other assembly factors and is required for the processing of the ITS2 region in pre-rRNA [76], [77]. It is removed from pre-LSU particles at the cytoplasmic step of maturation, followed by the incorporation of eL24 [78].
The role of the universally conserved r-proteins in ribosome biogenesis has been extensively studied [79], [80], [81], [82], [83], [84]. For example, many extensions of these r-proteins provide the binding site for chaperons or harbour nuclear localization signals and thus promote active nuclear import of r-proteins. In addition, some extensions are important for specific pre-rRNA processing steps [79], [80], [85], [86], [87], [88]. The role of only a few of the A/E-specific r-proteins in ribosome biogenesis has been studies in detail. The examples include eL14, eL8, and eL19 [89], [90], [91].
eL8 contains a conserved globular domain and two eukaryote-specific extensions - at the N- and C-terminus (Supplementary Fig. S3). Contacts of the eL8 globular domain with LSU pre-rRNA are stabilized early, during the subunit assembly, while interactions between N-terminal extension and pre-rRNA are stabilized at late stages [92]. The depletion of eL8 in budding yeast cells causes the pre-RNA processing to be blocked at the nucleolar steps [93]. The deletion of the N-terminal extension of eL8 affects the late nuclear steps of pre-rRNA processing by blocking ITS2 processing and rearrangements of the central protuberance [89]. In contrast, the eukaryote-specific extension at the C-terminus is not essential for LSU biogenesis [89].
eL14 assembles into pre-LSU particles already in the nucleolus [90]. It has been demonstrated that eL14 binding to the pre-LSU is required for the stable assembly of a subset of r-proteins and a few late-acting assembly factors, most of which are essential for ITS2 processing. The eukaryotic eL14 contains a specific C-terminal extension absent in the archaeal homologs (Supplementary Fig. S3) [86]. This helix interacts with the C-terminal extension of uL13 already in the pre-LSU particles [67], [73]. It has been shown that the expression of eL14 variant lacking the last 16 amino acids, confers a mild LSU biogenesis defect [86]. The expression of eL14 variants with longer truncations is lethal [90].
eL19 is composed of N-terminal globular domain, a middle region and C-terminal α-helix (Supplementary Fig. S3). The N-terminal domain and middle region, two domains that are also present in archaeal homologs, are embedded in rRNA of the LSU. The depletion of eL19 in budding yeast cells leads to a delay in cleavage in ITS2 region, resulting the impaired pre-rRNA processing [91], [94]. However, the expression of eL19 variant lacking most of the C-terminal helix did not show any LSU assembly defects [91]. Thus, for some A/E-specific r-proteins, it is the conserved domains rather than eukaryote-specific extensions that are important in ribosome biogenesis.
Altogether, the functional analysis of A/E-specific r-proteins has expanded our knowledge of the multiple roles of r-proteins in ribosome biogenesis. It has been suggested that r-proteins establish several different stepwise interactions with pre-rRNA/rRNA during the assembly of subunits [95], [96], [97]. Since eukaryote-specific extensions of r-proteins are elongated and partially disordered, these structurally flexible extensions may facilitate interactions with multiple partners thereby allowing the dynamic rearrangement of pre-ribosome particles.
5. Stabilization of ribosome structure - participation in intrasubunit network
The structural organization of r-proteins allows the formation of an extensive neuron-like network of protein-protein interactions [25], [98], [99]. A/E-specific r-proteins play a central role in creating such a network. In archaeal ribosomes, most A/E-specific r-proteins interact with 2 partners (Table 3), which are mainly universally conserved r-proteins. Contacts between globular domains, between globular domains and protein extensions, and between extensions occur with the same frequency [99]. Interestingly, four A/E-specific r-proteins are not in contact with other r-proteins. All of these proteins are composed of a single domain and do not have any N-terminal and/or C-terminal extensions. Archaeal eS6 and eL41 localize at the interface side of the subunits (Fig. 2, Supplementary Fig. S1). In 70S ribosomes, eS6 forms the contact with the LSU r-protein eL24 and eL41 interacts extensively with SSU rRNA [4], [99]. Thus, these r-proteins are specifically involved in the formation of intersubunit contacts and do not participate in the intrasubunit network.
Table 3.
Number of intrasubunit protein partners for A/E-specific r-proteins in archaeal and eukaryotic ribosomes1.
| Subunit | Number of partners |
In archaea | In eukaryotes2 |
|---|---|---|---|
| SSU | 0 | eS6, eS31 | |
| 1 | eS1 | eS31 | |
| 2 | eS8, eS17, eS19, eS24, eS27, eS28 |
eS6, eS8, eS17, eS19, eS30 |
|
| 3 | eS21, | eS1, eS24, eS25, eS28 |
|
| 4 | eS4 | eS4, eS21, eS27 | |
| LSU | 0 | eL31, eL41 | eL31, eL38, eL41 |
| 1 | eL8, eL19, eL21, eL40 |
eL19, eL40 | |
| 2 | eL14, eL15, eL18, eL24, eL32, eL33, eL37, eL43 |
eL24, eL43 | |
| 3 | eL20, eL30, eL34, eL39 |
eL30, eL32, eL33, eL34, eL42 |
|
| 4 | eL14, eL18, eL37, eL39 |
||
| 5 | eL8 | ||
| 6 | eL13, eL21 | ||
| 7 | eL15, eL20 |
Upon transitioning from archaeal to eukaryotic ribosomes, the number of intrasubunit protein contacts increases greatly [98]. In eukaryotic ribosomes, A/E-specific r-proteins mostly interact with 2–3 partners in SSU and 3–4 partners in LSU (Table 3). On the other hand, eL15 and eL20 connect 7 partners each, making them most connected r-proteins. These r-proteins, as well as eL21, link together three functional centres of the ribosome: PTC, PET, and tRNA binding sites. This bridging has only been observed in eukaryotes [99]. Although in eukaryotic ribosome most of the A/E-specific r-proteins form many contacts with one another, there are 3 r-proteins that do not interact with any r-protein in LSU: eL31, eL38, and eL41. All these r-proteins consist of a single domain and do not contain any extensions.
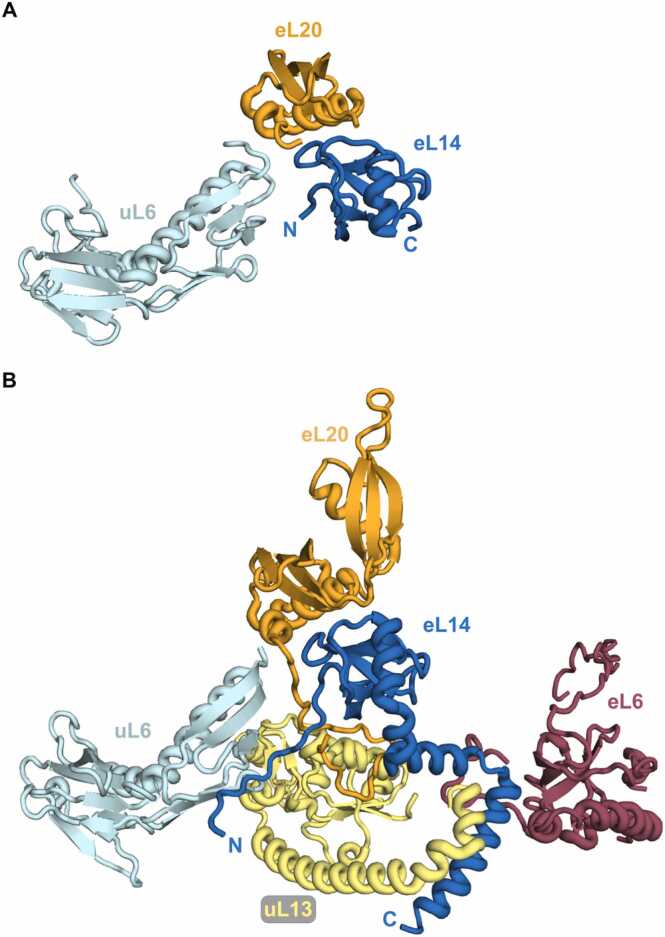
Analysis of the interaction types in the eukaryotic ribosome revealed that direct contacts between the globular domains are less frequent [98]. Most of the protein-protein ineractions are mediated by extensions. Among them, the majority of the contacts involve C-terminal extensions. One such example of the diversity of contacts formed is the eL14 hub. In archaeal LSU, eL14 forms contacts with globular domains of uL6 and eL20 (Fig. 3). In eukaryotic LSU, four additional interactions have been added. These include interactions of the eL14 N-terminal extension with the uL6 globular domain and the eL20 C-terminal helix. The eL14 C-terminal helix interacts with the uL13 C-terminal helix as well as the eL6 C-terminal extension and globular domain.
Fig. 3.
eL14 and its interacting partners in archaeal (A) and eukaryotic (B) LSU. The r-proteins conserved in archaea and eukaryote are shown in dark blue (eL14), light blue (uL6), orange (eL20), and yellow (uL13). The eukaryote-specific eL6 is colored dark red. Positions of N and C termini of eL14 are indicated. PDB coordinates 4V6U for P. furiosus [3] and 4V88 for S. cerevisiae [4] were rendered in PyMol.
Thus, eukaryote-specific extensions of r-proteins, including A/E-specific r-proteins, ensure the stability of ribosome structure and provide communication between different regions of the ribosome.
6. Stabilization of ribosome structure - structural components of intersubunit bridges
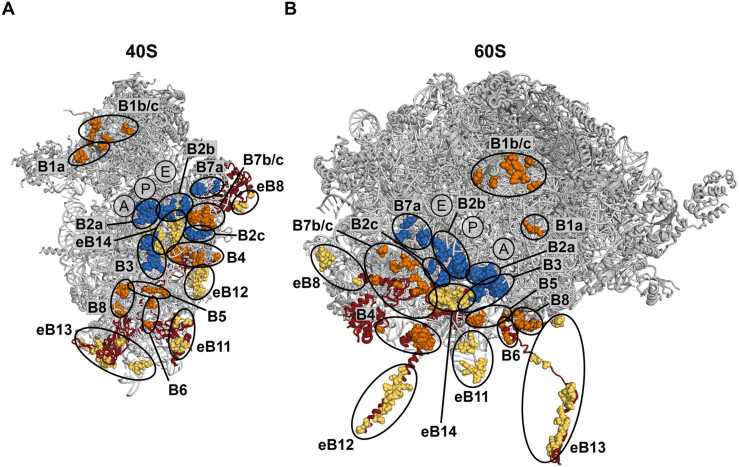
The overall conformation and functionality of the ribosome depends not only on the intrasubunit interactions, but also on the communication between ribosomal subunits provided by intersubunit contacts called bridges. Comparison of available structural models of bacterial and eukaryotic ribosomes has led to the division of intersubunit bridges into two groups. Twelve intersubunit bridges are conserved, as they have very similar composition and location in the conserved structural core of both Bacteria and Eukarya ribosomes (Fig. 4) [4]. Three A/E-specific r-proteins are involved in the conserved bridges (Table 4). Bridge B6, together with B5 and B8, forms a cluster of intersubunit bridges near the binding sites for translational GTPases. The structure of B6 varies significantly among different species and becomes less complex with increasing complexity of the ribosome. In E. coli ribosome, B6 contains numerous contacts between bacteria-specific bL19 and helix 44 of SSU rRNA [100]. In eukaryotic ribosomes, this bridge consists of two (in budding yeast) or only one (in human) interaction between the N-terminal domain of eL24 and helix 44 of SSU rRNA [4], [101]. Mutational analysis has demonstrated that bridge B6 has no apparent role in the budding yeast ribosomes. Cells expressing an eL24 variant that is not able to form this bridge showed growth rates and translation levels similar to wild type cells [102]. Interestingly, r-proteins associated with conserved bridges are tightly connected to other intersubunit bridges. Specifically, eL43 is also involved in the formation of bridge eB8 and uS15 interacts with eS7, which is a component of bridge eB12 (Table 4).
Fig. 4.
Position of the intersubunit bridges in the SSU (A) and LSU (B) of the budding yeast ribosome. rRNA and r-proteins are colored grey. Archaea/eukaryote-specific proteins that are involved in bridge formation are colored red. Conserved bridges that contain only rRNA-rRNA contacts or comprise also r-protein component, are shown in blue and orange spheres, respectively. Eukaryote-specific bridges are shown as yellow spheres. Approximate positions of the A-, P- and E-sites for tRNA binding are indicated. Bridge nomenclature and coordinates are from [4]. PDB coordinates 4V88 [4] were rendered in PyMol.
Table 4.
Components of the intersubunit bridges that involve the A/E-specific r-proteins in budding yeast ribosome1.
| Bridge2 | Pre-translocational |
Post-translocational |
||||
|---|---|---|---|---|---|---|
| SSU | LSU | Number of contacts | SSU | LSU | Number of contacts | |
| Conserved bridges | ||||||
| B4 | h20 | H34 | 6 | h20 | H34 | 7 |
| uS15 | H34 | 1 | uS15 | H34 | 5 | |
| uS15 | eL30 | 1 | ||||
| B6 | h44 | eL24 | 2 | h44 | eL24 | 2 |
| B7b/c | h24 | uL2 | 6 | h24 | uL2 | 4 |
| h24 | eL43 | 1 | h24 | eL43 | 4 | |
| h27 | eL43 | 1 | ||||
| Eukaryote-specific bridges | ||||||
| eB8 | eS1 | ES31L | 2 | eS1 | ES31L | 2 |
| eS1 | eL43 | 2 | ||||
| eB11 | eS8 | H63 | 1 | eS8 | ES41L | 5 |
| eS8 | ES41L | 6 | ||||
| eB12 | ES6S | eL19 | 13 | ES6S | eL19 | 14 |
| uS17 | eL19 | 3 | eS7 | eL19 | 2 | |
| eB13 | eS6 | uL3 | 3 | h6 | uL3 | 1 |
| eS6 | eL24 | 8 | ES3S | eL24 | 6 | |
| ES12S | eL24 | 3 | ||||
| eS6 | uL3 | 2 | ||||
| eS6 | eL24 | 11 | ||||
| eB14 | h27 | eL41 | 10 | h27 | eL41 | 10 |
| h44 | eL41 | 4 | h44 | eL41 | 5 | |
| h45 | eL41 | 14 | h45 | eL41 | 14 | |
The interaction surface between the two ribosome subunits has almost doubled in eukaryotes and additional five eukaryote-specific bridges have formed (Fig. 4) [4]. These bridges are predominated by A/E-specific r-proteins (Table 4). Only one of the eukaryote-specific bridges, eB14, locates in the core of the ribosome. The remaining four bridges (eB8, eB11, eB12 and eB13) are positioned at the peripheral regions, where relative motion of ribosomal subunits is most pronounced. The functional importance of eukaryote-specific bridges remains largely obscure due to limited number of studies, when compared to knowledge about conserved bridges.
Bridge eB12 is distinguished by the long C-terminal α-helix of eL19, which extends from the E-site side of the LSU (Fig. 4). This helix interacts with expansion segment ES6S of the SSU. Additional stabilizing interactions between eL19 and eS7 or eS17 of the SSU are involved, depending on the rotational state of the ribosome. Mutational analysis of eL19 has revealed that the functional integrity of bridge eB12 depends on the protein-rRNA contacts [91]. The archaeal version of eL19 is shorter (Supplementary Fig. S3) [103]. Instead of the long C-terminal helical domain, it has a ∼10 aa residues long extension after the conserved middle region.
A budding yeast mutant expressing the eL19 variant mimicking the archaeal version has been shown to be viable but displays a slow growth phenotype [91], [104]. In addition, the translation in this mutant was reduced by 1.8-fold compared to wild type cells [104].
By analogy to eB12, bridge eB13 is recognizable by the long C-terminal α-helix and the linker region of eL24 that extend from the A-site side of LSU (Fig. 4). Bulk of the bridge eB13 is formed by interactions of α-helix and linker of eL24 with eS6 of SSU. These contacts are assisted by interactions between uL3 and eS6. The additional contacts between eL24 and SSU rRNA occur depending on the rotational state of the ribosome. The N-terminal domain of the eL24 resides on the surface of the LSU and interacts with uL3 and uL14. These three interconnected proteins form a structural cluster that gives rise to the interconnected intersubunit bridges B5, B6, B8, and eB13. The archaeal homolog of eL24 is a short one-domain protein (Supplementary Fig. S3) [103], [105]. Mutational analysis of eL24 indicated that the functionality of eB13 bridge depends on the protein-protein contacts between eL24 and eS6 [102]. Analysis of yeast mutant expressing the archaeal variant of eL24 has revealed that bridge eB13 is important for subunit joining in vivo and in vitro [102]. Further analysis utilizing the cell-free translation system has demonstrated the role of this bridge in both the initiation and elongation steps of translation.
Bridge eB14 is formed by extensive interactions of the smallest r-protein, eL41, of the LSU with conserved SSU helices 27, 44, and 45. In eukaryotes, eL41 has more contacts with rRNA of the SSU than with rRNA of the LSU [4]. The eL41 encoding gene is not present in all archaeal genomes (Table 1) [3]. Furthermore, archaeal eL41 can vary in length. A protein with N-terminal extension has been annotated in several archaeal genomes [28]. eL41 is one of the few nonessential r-proteins [32], [106]. Yeast cells lacking eL41 display a similar wild type growth rate [34]. The functional importance of eL41 was revealed by studying the genetic interactions between r-proteins, which form eukaryote-specific bridges [104]. Mutant cells lacking two r-proteins, eL41 and eL24, or lacking eL41 and eB12 bridge-forming helix of eL19 displayed severely reduced cell growth and decrease in total translation.
Two budding yeast mutants with archaea-like ribosomes have been constructed [104]. Ribosomes in cells expressing the archaeal variants of eL19 and eL24 are defective in formation of eB12 and eB13 bridges. These ribosomes resemble the Euryarchaeota-like ribosomes. In the triple mutant, the ribosomes are deficient in three eukaryote-specific bridges (eB12, eB13, and eB14). This mutant carries ribosomes similar to those of Korarchaeota and Crenarchaeota. Phenotypic analysis of these mutants revealed that the formation of functional 80S ribosomes during translation initiation is severely hampered. This results in a slow growth phenotype and reduced levels of translation. Thus, the non-essential structural elements of r-proteins become highly important in the context of disturbed subunit association.
During evolution, ribosomes have increased in size. Following the splitting of eukaryotic and archaeal linages, eukaryotic ribosomes acquired additional protein and RNA sequences. The expansion of eukaryotic ribosomes also led to an enlargement of the area between SSU and LSU. To stabilize the sophisticated 80S structure, intersubunit bridges characteristic for eukaryotes have evolved. It is the A/E-specific r-proteins that play a dominant role in the formation of these bridges. In addition, structural studies indicate that also intersubunit bridges are connected to each other. Such a network of connections is also formed by A/E-specific r-proteins.
7. Involvement in translation
R-proteins and rRNA on the surface of the ribosome act as docking sites for translation factors and many other ribosome-associated proteins. Information on the interactions between translation factors and ribosome components is mainly derived from atomic models of cryo-EM and X-ray crystallographic structures. This is complemented by data from biochemical and genetic experiments. Several examples are provided below to illustrate the role of A/E-specific r-proteins at different steps of translation.
Of the four steps of translation - initiation, elongation, termination and ribosome recycling - it is the molecular mechanisms of initiation that differ most in the three domains in life. In bacteria, SSU binds upstream of the start codon during initiation complex formation through interactions between the mRNA Shine-Dalgarno sequence and the complementarty sequence in SSU rRNA [107]. In eukaryotes, translation initiation follows the canonical, 5′ m7G-cap-dependent mechanism [108], [109]. In archaea, the initiation mechanism is determined by the properties of mRNAs similar to those of bacterial mRNAs. Depending on the organism, archaeal mRNAs may contain a Shine-Dalgarno sequence or a very short 5′ untranslated region or be leaderless [110], [111], [112]. Despite differences in archaea and eukaryotes in recruitment of the preinitiation complexes on the mRNA, the selection of start codon is achieved by the common structural core [113]. Several high-resolution structures of initiation complexes formed after recognition of start codon have revealed that the set of r-proteins contacting the mRNA in the exit channel is very similar in archaeal and eukaryotic ribosomes [28], [114], [115]. In archaea, the Shine-Dalgarno helix of mRNA is positioned on one side by uS11, eS1 and helix 26 of the SSU rRNA and, on the other side, by uS7, eS28, and helices 28 and 37 of the SSU rRNA [28]. In eukaryotes, similar interactions are formed and two more are introduced. Namely, the long C-terminal extension of eS17, which is absent in archaeal orthologues, is in contact with mRNA and the eukaryote-specific r-protein eS26 stabilizes the 3′ end of the mRNA [114], [115]. In bacteria, two bacteria-specific r-proteins, bS6 and bS18, are found in place of eS1. In addition, eS28 is absent in bacteria and uS2 possesses an additional C-terminal domain at the location of eS17. Because of these differences, the archaeal and bacterial mRNA exit channels serve as two structural solutions for Shine-Dalgarno duplex binding [28]. In archaea and eukaryotes, A/E-specific r-proteins play an important role in the stabilization of mRNA in the exit channel.
Some alternative initiation mechanisms have been described in eukaryotes [116], [117]. One such mechanism, employed mainly by certain viruses, requires a specific RNA structure - IRES [118]. A number of studies have demonstrated that several structurally and functionally diverse IRESs rely on the A/E-specific eS25 [39], [40], [119], [120]. eS25 is one of the non-essential r-proteins of yeast [32]. Its globular domain resides in the E-site of SSU and its N-terminal extension stretches towards the P-site of SSU [4], [26]. Recent high-resolution structures from eukaryotic ribosome:IRES complexes have revealed the detailed information about interactions between IRESs and r-proteins. For example, Hepatitis C virus IRES is in contact with several SSU r-proteins: eS1, uS7, uS11, eS25, eS27, and eS28 [121], [122], [123], [124]. The cricket paralysis virus intergenic region (IGR) IRES has been reportered to contact also with eS25 and other r-proteins such as uS7, uL1, uL5, and uL11 [125], [126]. Of these r-proteins, it is e25 that is critical for initial binding of IGR IRES to the SSU [127].
During translation elongation, the nascent polypeptide chain passes through the PET of the ribosome, which extends from PTC to the surface of the LSU. In all ribosomes, the PET is predominantly composed of rRNA [128]. The internal loops of two r-proteins, uL4 and uL22 form the conserved construction sites. In eukaryotic and archaeal ribosomes, the loop of uL4 yields a second construction site of the tunnel that is absent in bacteria. Close to the subunit surface, the bacterial tunnel is surrounded by rRNA and uL23. In eukarotes and archaea, uL23 is also present, but the segment covering the tunnel region is replaced by the A/E-specific eL39. eL39 also extends to the tunnel exit, resulting in a narrowing of the tunnel. This difference may help to explain certain differences between eukaryotes and bacteria in their translocation modes of membrane proteins mediated by the signal recognition particle [129], [130]. In addition, reducing the size of the the tunnel exit enables the antibiotic resistance in eukaryotes [128], [131], [132].
The A/E-specific r-proteins that are exceptional in many ways are the P-proteins, which form the ribosomal P-stalk located on the LSU (Fig. 1) [27], [133]. The P-stalk together with the sarcin-ricin loop forms the GTPase-associated center responsible for stimulating factor-dependent GTP hydrolysis [134], [135]. P-proteins are acidic, they do not interact directly with rRNA, and they are the only ribosomal components that exist in more than one copy per ribosome. The archaeal P1 and the eukaryotic P1/P2 are closely related to each other [136], [137]. Although bacterial stalk protein bL12 is located at the identical site on the ribosome, this r-protein is not phylogenetically and structurally related to its archaeal/eukaryotic counterparts [137]. Nevertheless, P-proteins play identical roles on the ribosome, being part of the GTPase-associated region wich is directly responsible for the stimulation of translation factor-dependent GTP hydrolysis [134].
All P-proteins are organized into three functional domains [137], [138], [139]. A globular N-terminal domain is responsible for oligomerization and for the interaction with uL10. A highly conserved C-terminal domain is implicated in translation factor binding. These two domains are connected through a flexible hinge region. The interactions between P-proteins and translation factors (namely archaeal initiator factor IF5B, and archaeal elongation factors EF1A and EF2) are biochemically and structurally well documented [140], [141], [142], [143], [144]. Two mechanistic functions of the P-stalk have been proposed [145], [146], [147]. One possibility is that the P-stalk increases the local concentration of translational GTPases by recruiting them to the factor-binding center. This in turn increases the rate of GTP hydrolysis. Alternatively, P-stalk may stabilize translational GTPases when bound to the SRL of LSU rRNA, thereby increasing the rate of GTP hydrolysis. In eukaryotes, specifically in budding yeast, P1/P2 r-proteins are not required for cell viability and, consequently, for protein synthesis [148]. Ribosomes depleted of P1/P2 exhibit reduced translation fidelity [149]. Unexpectedly, the lack of P1/P2 has little effect in vivo on translocation. It has been proposed that decrease in decoding accuracy due to the P1/P2 depletion is the main factor causing the translational slowdown, which in turn affects the metabolic fitness and growth rate of yeast cells. Indeed, the mutant lacking P1/P2 exhibits a cold-sensitive phenotype and reduced cell growth [148].
Altogether, A/E-specific r-proteins play an important role in ensuring ribosome functionality during translation. As illustrated by the examples above, these r-proteins are involved both in binding of translation factors and in stabilizing the contacts between mRNA and the ribosome.
8. Summary and Outlook
In general, ribosomes are widely considered to be molecular machines that decode information carried by messenger RNAs and translate this information into proteins. They are universally composed of rRNAs and numerous r-proteins. Because ribosomes are the central sites for protein synthesis, many key components of ribosomes are conserved in all three domains of life.
Despite ribosomes carrying out the central function of protein synthesis and sharing a universally conserved common core, ribosomes have diverged considerably during evolution. Eukaryotic ribosomes are large and more complex containing rRNA and r-proteins extensions as well as additional r-proteins. The expansion of eukaryotic ribosomes also led to an increase in the surface between the two ribosomal subunits and in the formation of eukaryote-specific intersubunit contacts in which r-proteins play a major role. Archaeal ribosomes are only partially similar to eukaryotic ribosomes. While their rRNAs share similarities with bacterial rRNAs, their r-protein composition is more similar to that of eukaryotic ribosomes. The archaeal and eukaryotic ribosomes share a common set of 35 r-proteins. However, these A/E-specific r-proteins are not ubiquitously distributed across archaea. Thus, the r-protein composition of ribosomes varies between archaeal species. This opens up the possibility to investigate how the minimal set of A/E-specific r-proteins influences the structure and the functionality of the ribosome. It is possible that ribosomes with different A/E-specific r-protein composition have a bias towards translation of specific mRNAs, which in turn may be reflected in the cellular proteome. Future investigations may reveal the existence of such regulation.
The A/E-specific r-proteins are good examples of how r-proteins have evolved into multifunctional proteins. Their primary function is devoted to the assembly and stabilization of the complex structure of the ribosome. In addition, these r-proteins have acquired new functions connected with the increasing complexity of the ribosomes. Structures of ribosomes and ribosomal complexes from a wide range of organisms have provided a framework for understanding how ribosomal components are organized into a sophisticated network and how the components are interacting with translational factors. These structures have also made it possible to observe how A/E-specific r-proteins have changed during evolution. Several non-globular extensions and internal loops have been added to the r-proteins that make up the eukaryotic ribosome. It has been suggested that these structural variations may reflect the functional specialization of r-proteins. Thus, functional studies based on structural data are important to extend the understanding of the functional roles of A/E-specific r-proteins in protein synthesis. Recent advances in genetic engineering will allow more such studies to be carried out in higher eukaryotes.
CRediT authorship contribution statement
Ivan Kisly: Investigation, Data curation, Visualization, Writing – review & editing. Tiina Tamm: Conceptualization, Writing – original draft, Writing – review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank all members of the Chair of Molecular Biology, especially Dr. Margus Leppik and Dr. Rya Ero, for helpful discussions. This work was supported by the Estonian Research Council grants (grant numbers: PSG295, PRG669, PRG1179 and PRG1741).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.01.037.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Bremer H., Dennis P.P. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal. 2008;3(1) doi: 10.1128/ecosal.5.2.3. [DOI] [PubMed] [Google Scholar]
- 2.Milo R., Phillips R. xlii. Taylor & Francis Group; 2016. p. 356. (Cell biology by the numbers: Garland Science). [Google Scholar]
- 3.Armache J.P., Anger A.M., Marquez V., Franckenberg S., Frohlich T., Villa E., et al. Promiscuous behaviour of archaeal ribosomal proteins: implications for eukaryotic ribosome evolution. Nucleic Acids Res. 2013;41(2):1284–1293. doi: 10.1093/nar/gks1259. PubMed PMID: 23222135; PubMed Central PMCID: PMC3553981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 5.Melnikov S., Ben-Shem A., Garreau de Loubresse N., Jenner L., Yusupova G., Yusupov M. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol. 2012;19(6):560–567. doi: 10.1038/nsmb.2313. [DOI] [PubMed] [Google Scholar]
- 6.Bernier C.R., Petrov A.S., Kovacs N.A., Penev P.I., Williams L.D. Translation: the universal structural core of life. Mol Biol Evol. 2018;35(8):2065–2076. doi: 10.1093/molbev/msy101. PubMed PMID: 29788252; PubMed Central PMCID: PMC6063299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowman J.C., Petrov A.S., Frenkel-Pinter M., Penev P.I., Williams L.D. Root of the tree: the significance, evolution, and origins of the ribosome. Chem Rev. 2020;120(11):4848–4878. doi: 10.1021/acs.chemrev.9b00742. [DOI] [PubMed] [Google Scholar]
- 8.Ban N., Beckmann R., Cate J.H., Dinman J.D., Dragon F., Ellis S.R., et al. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 2014;24 doi: 10.1016/j.sbi.2014.01.002. PubMed PMID: 24524803; PubMed Central PMCID: PMC4358319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson D.N., Nierhaus K.H. Ribosomal proteins in the spotlight. Crit Rev Biochem Mol Biol. 2005;40(5):243–267. doi: 10.1080/10409230500256523. [DOI] [PubMed] [Google Scholar]
- 10.Nikolay R., van den Bruck D., Achenbach J., Nierhaus K.H. Ribosomal Proteins: Role in Ribosomal Functions. eLS2015. p. 1–12.
- 11.Galperin M.Y., Wolf Y.I., Garushyants S.K., Vera Alvarez R., Koonin E.V. Non-essential ribosomal proteins in bacteria and archaea identified using COGs. J Bacteriol. 2021 doi: 10.1128/JB.00058-21. PubMed PMID: 33753464; PubMed Central PMCID: PMC8117519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lecompte O., Ripp R., Thierry J.C., Moras D., Poch O. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 2002;30(24):5382–5390. doi: 10.1093/nar/gkf693. PubMed PMID: 12490706; PubMed Central PMCID: PMC140077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marquez V., Frohlich T., Armache J.P., Sohmen D., Donhofer A., Mikolajka A., et al. Proteomic characterization of archaeal ribosomes reveals the presence of novel archaeal-specific ribosomal proteins. J Mol Biol. 2011;405(5):1215–1232. doi: 10.1016/j.jmb.2010.11.055. [DOI] [PubMed] [Google Scholar]
- 14.Yutin N., Puigbo P., Koonin E.V., Wolf Y.I. Phylogenomics of prokaryotic ribosomal proteins. PloS One. 2012;7(5) doi: 10.1371/journal.pone.0036972. PubMed PMID: 22615861; PubMed Central PMCID: PMC3353972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson D.N., Doudna, Cate J.H. The structure and function of the eukaryotic ribosome. Cold Spring Harb Perspect Biol. 2012;4(5) doi: 10.1101/cshperspect.a011536. PubMed PMID: 22550233; PubMed Central PMCID: PMC3331703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Planta R.J., Mager W.H. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast. 1998;14(5):471–477. doi: 10.1002/(SICI)1097-0061(19980330)14:5<471::AID-YEA241>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 17.Warringer J., Blomberg A. Evolutionary constraints on yeast protein size. BMC Evolut Biol. 2006;6:61. doi: 10.1186/1471-2148-6-61. PubMed PMID: 16911784; PubMed Central PMCID: PMC1560397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaltschmidt E. Ribosomal proteins. XIV. Isoelectric points of ribosomal proteins of E. coli as determined by two-dimensional polyacrylamide gel electrophoresis. Anal Biochem. 1971;43(1):25–31. doi: 10.1016/0003-2697(71)90103-5. [DOI] [PubMed] [Google Scholar]
- 19.Kaltschmidt E., Wittmann H.G. Ribosomal proteins. XII. Number of proteins in small and large ribosomal subunits of Escherichia coli as determined by two-dimensional gel electrophoresis. Proc Natl Acad Sci USA. 1970;67(3):1276–1282. doi: 10.1073/pnas.67.3.1276. PubMed PMID: 4922286; PubMed Central PMCID: PMC283348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein D.J., Moore P.B., Steitz T.A. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol. 2004;340(1):141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 21.Fedyukina D.V., Jennaro T.S., Cavagnero S. Charge segregation and low hydrophobicity are key features of ribosomal proteins from different organisms. J Biol Chem. 2014;289(10):6740–6750. doi: 10.1074/jbc.M113.507707. PubMed PMID: 24398678; PubMed Central PMCID: PMC3945335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newton C.H., Shimmin L.C., Yee J., Dennis P.P. A family of genes encode the multiple forms of the Saccharomyces cerevisiae ribosomal proteins equivalent to the Escherichia coli L12 protein and a single form of the L10-equivalent ribosomal protein. J Bacteriol. 1990;172(2):579–588. doi: 10.1128/jb.172.2.579-588.1990. PubMed PMID: 2404943; PubMed Central PMCID: PMC208480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Remacha M., Jimenez-Diaz A., Santos C., Briones E., Zambrano R., Rodriguez Gabriel M.A., et al. Proteins P1, P2, and P0, components of the eukaryotic ribosome stalk. New structural and functional aspects. Biochem Cell Biol = Biochim Et Biol Cell. 1995;73(11–12):959–968. doi: 10.1139/o95-103. [DOI] [PubMed] [Google Scholar]
- 24.Sas-Chen A., Thomas J.M., Matzov D., Taoka M., Nance K.D., Nir R., et al. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature. 2020;583(7817):638–643. doi: 10.1038/s41586-020-2418-2. PubMed PMID: 32555463; PubMed Central PMCID: PMC8130014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klinge S., Voigts-Hoffmann F., Leibundgut M., Arpagaus S., Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334(6058):941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 26.Rabl J., Leibundgut M., Ataide S.F., Haag A., Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331(6018):730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 27.Wahl M.C., Moller W. Structure and function of the acidic ribosomal stalk proteins. Curr Protein Pept Sci. 2002;3(1):93–106. doi: 10.2174/1389203023380756. [DOI] [PubMed] [Google Scholar]
- 28.Coureux P.D., Lazennec-Schurdevin C., Bourcier S., Mechulam Y., Schmitt E. Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation. Commun Biol. 2020;3(1):58. doi: 10.1038/s42003-020-0780-0. PubMed PMID: 32029867; PubMed Central PMCID: PMC7005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nurenberg-Goloub E., Kratzat H., Heinemann H., Heuer A., Kotter P., Berninghausen O., et al. Molecular analysis of the ribosome recycling factor ABCE1 bound to the 30S post-splitting complex. EMBO J. 2020;39(9) doi: 10.15252/embj.2019103788. PubMed PMID: 32064661; PubMed Central PMCID: PMC7196836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spang A., Saw J.H., Jorgensen S.L., Zaremba-Niedzwiedzka K., Martijn J., Lind A.E., et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521(7551):173–179. doi: 10.1038/nature14447. PubMed PMID: 25945739; PubMed Central PMCID: PMC4444528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaremba-Niedzwiedzka K., Caceres E.F., Saw J.H., Backstrom D., Juzokaite L., Vancaester E., et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541(7637):353–358. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 32.Steffen K.K., McCormick M.A., Pham K.M., MacKay V.L., Delaney J.R., Murakami C.J., et al. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics. 2012;191(1):107–118. doi: 10.1534/genetics.111.136549. PubMed PMID: 22377630; PubMed Central PMCID: PMC3338253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Veronneau S., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 34.Yu X., Warner J.R. Expression of a micro-protein. J Biol Chem. 2001;276(36):33821–33825. doi: 10.1074/jbc.M103772200. [DOI] [PubMed] [Google Scholar]
- 35.Peisker K., Braun D., Wolfle T., Hentschel J., Funfschilling U., Fischer G., et al. Ribosome-associated complex binds to ribosomes in close proximity of Rpl31 at the exit of the polypeptide tunnel in yeast. Mol Biol Cell. 2008;19(12):5279–5288. doi: 10.1091/mbc.E08-06-0661. PubMed PMID: 18829863; PubMed Central PMCID: PMC2592665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sachs A.B., Davis R.W. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990;247(4946):1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- 37.Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E.D., Sevier C.S., et al. The genetic landscape of a cell. Science. 2010;327(5964):425–431. doi: 10.1126/science.1180823. PubMed PMID: 20093466; PubMed Central PMCID: PMC5600254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.UniProt C. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–D489. doi: 10.1093/nar/gkaa1100. PubMed PMID: 33237286; PubMed Central PMCID: PMC7778908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landry D.M., Hertz M.I., Thompson S.R. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23(23):2753–2764. doi: 10.1101/gad.1832209. PubMed PMID: 19952110; PubMed Central PMCID: PMC2788332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hertz M.I., Landry D.M., Willis A.E., Luo G., Thompson S.R. Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol Cell Biol. 2013;33(5):1016–1026. doi: 10.1128/MCB.00879-12. PubMed PMID: 23275440; PubMed Central PMCID: PMC3623076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volarevic S., Stewart M.J., Ledermann B., Zilberman F., Terracciano L., Montini E., et al. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288(5473):2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 42.Wang H., Zhao L.N., Li K.Z., Ling R., Li X.J., Wang L. Overexpression of ribosomal protein L15 is associated with cell proliferation in gastric cancer. BMC Cancer. 2006;6(91) doi: 10.1186/1471-2407-6-91. PubMed PMID: 16608517; PubMed Central PMCID: PMC1459873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhan Y., Melian N.Y., Pantoja M., Haines N., Ruohola-Baker H., Bourque C.W., et al. Dystroglycan and mitochondrial ribosomal protein L34 regulate differentiation in the Drosophila eye. PloS One. 2010;5(5) doi: 10.1371/journal.pone.0010488. PubMed PMID: 20463973; PubMed Central PMCID: PMC2864756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Da Costa L., Narla G., Willig T.N., Peters L.L., Parra M., Fixler J., et al. Ribosomal protein S19 expression during erythroid differentiation. Blood. 2003;101(1):318–324. doi: 10.1182/blood-2002-04-1131. [DOI] [PubMed] [Google Scholar]
- 45.Jang C.-Y., Lee J.Y., Kim J. RpS3, a DNA repair endonuclease and ribosomal protein, is involved in apoptosis. FEBS Lett. 2004;560(1–3):81–85. doi: 10.1016/S0014-5793(04)00074-2. [DOI] [PubMed] [Google Scholar]
- 46.He H., Sun Y. Ribosomal protein S27L is a direct p53 target that regulates apoptosis. Oncogene. 2007;26(19):2707–2716. doi: 10.1038/sj.onc.1210073. [DOI] [PubMed] [Google Scholar]
- 47.Kim J., Chubatsu L.S., Admon A., Stahl J., Fellous R., Linn S. Implication of mammalian ribosomal protein S3 in the processing of DNA damage. The. J Biol Chem. 1995;270(23):13620–13629. doi: 10.1074/jbc.270.23.13620. [DOI] [PubMed] [Google Scholar]
- 48.Hegde V., Wang M., Deutsch W.A. Human ribosomal protein S3 interacts with DNA base excision repair proteins hAPE/Ref-1 and hOGG1. Biochemistry. 2004;43(44):14211–14217. doi: 10.1021/bi049234b. [DOI] [PubMed] [Google Scholar]
- 49.Yang Z.Y., Jiang H., Qu Y., Wei M., Yan M., Zhu Z.G., et al. Metallopanstimulin-1 regulates invasion and migration of gastric cancer cells partially through integrin beta4. Carcinogenesis. 2013;34(12):2851–2860. doi: 10.1093/carcin/bgt226. [DOI] [PubMed] [Google Scholar]
- 50.Narla A., Ebert B.L. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115(16):3196–3205. doi: 10.1182/blood-2009-10-178129. PubMed PMID: 20194897; PubMed Central PMCID: PMC2858486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou X., Liao W.J., Liao J.M., Liao P., Lu H. Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol. 2015;7(2):92–104. doi: 10.1093/jmcb/mjv014. PubMed PMID: 25735597; PubMed Central PMCID: PMC4481666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie X., Guo P., Yu H., Wang Y., Chen G. Ribosomal proteins: insight into molecular roles and functions in hepatocellular carcinoma. Oncogene. 2018;37(3):277–285. doi: 10.1038/onc.2017.343. [DOI] [PubMed] [Google Scholar]
- 53.Kief D.R., Warner J.R. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol Cell Biol. 1981;1(11):1007–1015. doi: 10.1128/mcb.1.11.1007-1015.1981. PubMed PMID: 7050661; PubMed Central PMCID: PMC369722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24(11):437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 55.Strunk B.S., Karbstein K. Powering through ribosome assembly. Rna. 2009;15(12):2083–2104. doi: 10.1261/rna.1792109. PubMed PMID: 19850913; PubMed Central PMCID: PMC2779681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller O.L., Jr., Beatty B.R. Visualization of nucleolar genes. Science. 1969;164(3882):955–957. doi: 10.1126/science.164.3882.955. [DOI] [PubMed] [Google Scholar]
- 57.Neyer S., Kunz M., Geiss C., Hantsche M., Hodirnau V.V., Seybert A., et al. Structure of RNA polymerase I transcribing ribosomal DNA genes. Nature. 2016;540(7634):607–610. doi: 10.1038/nature20561. [DOI] [PubMed] [Google Scholar]
- 58.Dieci G., Fiorino G., Castelnuovo M., Teichmann M., Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet: Tig. 2007;23(12):614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 59.Han Y., Yan C., Fishbain S., Ivanov I., He Y. Structural visualization of RNA polymerase III transcription machineries. Cell Discov. 2018;4:40. doi: 10.1038/s41421-018-0044-z. PubMed PMID: 30083386; PubMed Central PMCID: PMC6066478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernandez-Pevida A., Kressler D., de la Cruz J. Processing of preribosomal RNA in Saccharomyces cerevisiae. Wiley interdisciplinary reviews. RNA. 2015;6(2):191–209. doi: 10.1002/wrna.1267. [DOI] [PubMed] [Google Scholar]
- 61.Henras A.K., Plisson-Chastang C., O'Donohue M.F., Chakraborty A., Gleizes P.E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley interdisciplinary reviews. RNA. 2015;6(2):225–242. doi: 10.1002/wrna.1269. PubMed PMID: 25346433; PubMed Central PMCID: PMC4361047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Konikkat S., Woolford J.L., Jr. Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast. Biochem J. 2017;474(2):195–214. doi: 10.1042/BCJ20160516. PubMed PMID: 28062837; PubMed Central PMCID: PMC5555582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klinge S., Woolford J.L., Jr. Ribosome assembly coming into focus. Nat Rev Mol Cell Biol. 2019;20(2):116–131. doi: 10.1038/s41580-018-0078-y. PubMed PMID: 30467428; PubMed Central PMCID: PMC7725133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bassler J., Hurt E. Eukaryotic ribosome assembly. Annu Rev Biochem. 2019;88:281–306. doi: 10.1146/annurev-biochem-013118-110817. [DOI] [PubMed] [Google Scholar]
- 65.Londei P., Ferreira-Cerca S. Ribosome biogenesis in archaea. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.686977. PubMed PMID: 34367089; PubMed Central PMCID: PMC8339473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun Q., Zhu X., Qi J., An W., Lan P., Tan D., et al. Molecular architecture of the 90S small subunit pre-ribosome. eLife. 2017;6 doi: 10.7554/eLife.22086. PubMed PMID: 28244370; PubMed Central PMCID: PMC5354517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kater L., Thoms M., Barrio-Garcia C., Cheng J., Ismail S., Ahmed Y.L., et al. Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes. Cell. 2017;171(7):1599–1610. doi: 10.1016/j.cell.2017.11.039. PubMed PMID: 29245012; PubMed Central PMCID: PMC5745149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanghai Z.A., Miller L., Molloy K.R., Barandun J., Hunziker M., Chaker-Margot M., et al. Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature. 2018;556(7699):126–129. doi: 10.1038/nature26156. PubMed PMID: 29512650; PubMed Central PMCID: PMC6118127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lo K.Y., Li Z., Wang F., Marcotte E.M., Johnson A.W. Ribosome stalk assembly requires the dual-specificity phosphatase Yvh1 for the exchange of Mrt4 with P0. J Cell Biol. 2009;186(6):849–862. doi: 10.1083/jcb.200904110. PubMed PMID: 19797078; PubMed Central PMCID: PMC2753163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lo K.Y., Li Z., Bussiere C., Bresson S., Marcotte E.M., Johnson A.W. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell. 2010;39(2):196–208. doi: 10.1016/j.molcel.2010.06.018. PubMed PMID: 20670889; PubMed Central PMCID: PMC2925414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma C., Wu S., Li N., Chen Y., Yan K., Li Z., et al. Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1. Nat Struct Mol Biol. 2017;24(3):214–220. doi: 10.1038/nsmb.3364. PubMed PMID: 28112732; PubMed Central PMCID: PMC5555584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scaiola A., Pena C., Weisser M., Bohringer D., Leibundgut M., Klingauf-Nerurkar P., et al. Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit. EMBO J. 2018;37(7) doi: 10.15252/embj.201798499. PubMed PMID: 29459436; PubMed Central PMCID: PMC5881545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu S., Tutuncuoglu B., Yan K., Brown H., Zhang Y., Tan D., et al. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature. 2016;534(7605):133–137. doi: 10.1038/nature17942. PubMed PMID: 27251291; PubMed Central PMCID: PMC5237361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barandun J., Chaker-Margot M., Hunziker M., Molloy K.R., Chait B.T., Klinge S. The complete structure of the small-subunit processome. Nat Struct Mol Biol. 2017;24(11):944–953. doi: 10.1038/nsmb.3472. [DOI] [PubMed] [Google Scholar]
- 75.Saveanu C., Namane A., Gleizes P.E., Lebreton A., Rousselle J.C., Noaillac-Depeyre J., et al. Sequential protein association with nascent 60S ribosomal particles. Mol Cell Biol. 2003;23(13):4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. PubMed PMID: 12808088; PubMed Central PMCID: PMC164837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kappel L., Loibl M., Zisser G., Klein I., Fruhmann G., Gruber C., et al. Rlp24 activates the AAA-ATPase Drg1 to initiate cytoplasmic pre-60S maturation. The. J Cell Biol. 2012;199(5):771–782. doi: 10.1083/jcb.201205021. PubMed PMID: 23185031; PubMed Central PMCID: PMC3514788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Talkish J., Zhang J., Jakovljevic J., Horsey E.W., Woolford J.L., Jr. Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40(17):8646–8661. doi: 10.1093/nar/gks609. PubMed PMID: 22735702; PubMed Central PMCID: PMC3458554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pertschy B., Saveanu C., Zisser G., Lebreton A., Tengg M., Jacquier A., et al. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol Cell Biol. 2007;27(19):6581–6592. doi: 10.1128/MCB.00668-07. PubMed PMID: 17646390; PubMed Central PMCID: PMC2099225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jakovljevic J., de Mayolo P.A., Miles T.D., Nguyen T.M., Leger-Silvestre I., Gas N., et al. The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol Cell. 2004;14(3):331–342. doi: 10.1016/s1097-2765(04)00215-1. [DOI] [PubMed] [Google Scholar]
- 80.Stelter P., Huber F.M., Kunze R., Flemming D., Hoelz A., Hurt E. Coordinated ribosomal L4 protein assembly into the pre-ribosome is regulated by its eukaryote-specific extension. Mol Cell. 2015;58(5):854–862. doi: 10.1016/j.molcel.2015.03.029. PubMed PMID: 25936803; PubMed Central PMCID: PMC6742479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moradi H., Simoff I., Bartish G., Nygard O. Functional features of the C-terminal region of yeast ribosomal protein L5. Mol Genet Genom: Mgg. 2008;280(4):337–350. doi: 10.1007/s00438-008-0369-7. [DOI] [PubMed] [Google Scholar]
- 82.Deshmukh M., Stark J., Yeh L.C., Lee J.C., Woolford J.L., Jr. Multiple regions of yeast ribosomal protein L1 are important for its interaction with 5 S rRNA and assembly into ribosomes. J Biol Chem. 1995;270(50):30148–30156. doi: 10.1074/jbc.270.50.30148. [DOI] [PubMed] [Google Scholar]
- 83.Sulima S.O., Gulay S.P., Anjos M., Patchett S., Meskauskas A., Johnson A.W., et al. Eukaryotic rpL10 drives ribosomal rotation. Nucleic Acids Res. 2014;42(3):2049–2063. doi: 10.1093/nar/gkt1107. PubMed PMID: 24214990; PubMed Central PMCID: PMC3919601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schaap P.J. van't Riet J, Woldringh CL, Raue HA. Identification and functional analysis of the nuclear localization signals of ribosomal protein L25 from Saccharomyces cerevisiae. J Mol Biol. 1991;221(1):225–237. doi: 10.1016/0022-2836(91)80216-h. [DOI] [PubMed] [Google Scholar]
- 85.Bussiere C., Hashem Y., Arora S., Frank J., Johnson A.W. Integrity of the P-site is probed during maturation of the 60S ribosomal subunit. J Cell Biol. 2012;197(6):747–759. doi: 10.1083/jcb.201112131. PubMed PMID: 22689654; PubMed Central PMCID: PMC3373404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pratte D., Singh U., Murat G., Kressler D. Mak5 and Ebp2 act together on early pre-60S particles and their reduced functionality bypasses the requirement for the essential pre-60S factor Nsa1. PloS One. 2013;8(12) doi: 10.1371/journal.pone.0082741. PubMed PMID: 24312670; PubMed Central PMCID: PMC3846774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Francisco-Velilla R., Remacha M., Ballesta J.P. Carboxy terminal modifications of the P0 protein reveal alternative mechanisms of nuclear ribosomal stalk assembly. Nucleic Acids Res. 2013;41(18):):8628–):8636. doi: 10.1093/nar/gkt637. PubMed PMID: 23880660; PubMed Central PMCID: PMC3794597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Garcia-Gomez J.J., Fernandez-Pevida A., Lebaron S. Rosado IV, Tollervey D, Kressler D, et al. Final pre-40S maturation depends on the functional integrity of the 60S subunit ribosomal protein L3. PLoS Genet. 2014;10(3) doi: 10.1371/journal.pgen.1004205. PubMed PMID: 24603549; PubMed Central PMCID: PMC3945201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tutuncuoglu B., Jakovljevic J., Wu S., Gao N., Woolford J.L., Jr. The N-terminal extension of yeast ribosomal protein L8 is involved in two major remodeling events during late nuclear stages of 60S ribosomal subunit assembly. Rna. 2016;22(9):1386–1399. doi: 10.1261/rna.055798.115. PubMed PMID: 27390266; PubMed Central PMCID: PMC4986894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Espinar-Marchena F., Rodriguez-Galan O., Fernandez-Fernandez J., Linnemann J., de la Cruz J. Ribosomal protein L14 contributes to the early assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. Nucleic Acids Res. 2018;46(9):4715–4732. doi: 10.1093/nar/gky123. PubMed PMID: 29788267; PubMed Central PMCID: PMC5961077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kisly I., Gulay S.P., Maeorg U., Dinman J.D., Remme J., Tamm T. The Functional Role of eL19 and eB12 Intersubunit Bridge in the Eukaryotic Ribosome. J Mol Biol. 2016;428(10 Pt B):2203–2216. doi: 10.1016/j.jmb.2016.03.023. PubMed PMID: 27038511; PubMed Central PMCID: PMC4884501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gamalinda M., Ohmayer U., Jakovljevic J., Kumcuoglu B., Woolford J., Mbom B., et al. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev. 2014;28(2):198–210. doi: 10.1101/gad.228825.113. PubMed PMID: 24449272; PubMed Central PMCID: PMC3909792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jakovljevic J., Ohmayer U., Gamalinda M., Talkish J., Alexander L., Linnemann J., et al. Ribosomal proteins L7 and L8 function in concert with six A(3) assembly factors to propagate assembly of domains I and II of 25S rRNA in yeast 60S ribosomal subunits. Rna. 2012;18(10):1805–1822. doi: 10.1261/rna.032540.112. PubMed PMID: 22893726; PubMed Central PMCID: PMC3446705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Poll G., Braun T., Jakovljevic J., Neueder A., Jakob S., Woolford J.L., Jr., et al. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PloS One. 2009;4(12) doi: 10.1371/journal.pone.0008249. PubMed PMID: 20011513; PubMed Central PMCID: PMC2788216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim H., Abeysirigunawarden S.C., Chen K., Mayerle M., Ragunathan K., Luthey-Schulten Z., et al. Protein-guided RNA dynamics during early ribosome assembly. Nature. 2014;506(7488):334–338. doi: 10.1038/nature13039. PubMed PMID: 24522531; PubMed Central PMCID: PMC3968076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Abeysirigunawardena S.C., Kim H., Lai J., Ragunathan K., Rappe M.C., Luthey-Schulten Z., et al. Evolution of protein-coupled RNA dynamics during hierarchical assembly of ribosomal complexes. Nat Commun. 2017;8(1):492. doi: 10.1038/s41467-017-00536-1. PubMed PMID: 28887451; PubMed Central PMCID: PMC5591316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Duss O., Stepanyuk G.A., Puglisi J.D., Williamson J.R. Transient protein-RNA interactions guide nascent ribosomal RNA folding. Cell. 2019;179(6):1357–1369. doi: 10.1016/j.cell.2019.10.035. e16. PubMed PMID: 31761533; PubMed Central PMCID: PMC7006226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Poirot O., Timsit Y. Neuron-like networks between ribosomal proteins within the ribosome. Sci Rep. 2016;6:26485. doi: 10.1038/srep26485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Timsit Y., Sergeant-Perthuis G., Bennequin D. Evolution of ribosomal protein network architectures. Scientific reports. 2021;11(1):625. doi: 10.1038/s41598-020-80194-4. PubMed PMID: 33436806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu Q., Fredrick K. Intersubunit bridges of the bacterial ribosome. J Mol Biol. 2016;428(10 Pt B):2146–2164. doi: 10.1016/j.jmb.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Khatter H., Myasnikov A.G., Natchiar S.K., Klaholz B.P. Structure of the human 80S ribosome. Nature. 2015;520(7549):640–645. doi: 10.1038/nature14427. [DOI] [PubMed] [Google Scholar]
- 102.Kisly I., Remme J., Tamm T. Ribosomal protein eL24, involved in two intersubunit bridges, stimulates translation initiation and elongation. Nucleic Acids Res. 2019;47(1) doi: 10.1093/nar/gky1083. 406-20. PubMed PMID: 30407570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gabdulkhakov A., Nikonov S., Garber M. Revisiting the Haloarcula marismortui 50S ribosomal subunit model. Acta Crystallogr Sect D, Biol Crystallogr. 2013;69(Pt 6):997–1004. doi: 10.1107/S0907444913004745. [DOI] [PubMed] [Google Scholar]
- 104.Tamm T., Kisly I., Remme J. Functional interactions of ribosomal intersubunit bridges in Saccharomyces cerevisiae. Genetics. 2019;213(4):1329–1339. doi: 10.1534/genetics.119.302777. PubMed PMID: 31649153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289(5481):905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 106.Dresios J., Panopoulos P., Suzuki K., Synetos D. A dispensable yeast ribosomal protein optimizes peptidyltransferase activity and affects translocation. J Biol Chem. 2003;278(5):3314–3322. doi: 10.1074/jbc.M207533200. [DOI] [PubMed] [Google Scholar]
- 107.Rodnina M.V. Translation in Prokaryotes. Cold Spring Harbor Perspect. Biol. 2018;10(9) doi: 10.1101/cshperspect.a032664. PubMed PMID: 29661790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hinnebusch A.G. Structural Insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem Sci. 2017;42(8):589–611. doi: 10.1016/j.tibs.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 109.Shirokikh N.E., Preiss T. Translation initiation by cap-dependent ribosome recruitment: recent insights and open questions. Wiley interdisciplinary reviews RNA. 2018;9(4) doi: 10.1002/wrna.1473. [DOI] [PubMed] [Google Scholar]
- 110.Dennis P.P. Ancient ciphers: translation in Archaea. Cell. 1997;89(7):1007–1010. doi: 10.1016/s0092-8674(00)80288-3. [DOI] [PubMed] [Google Scholar]
- 111.Ma J., Campbell A., Karlin S. Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J Bacteriol. 2002;184(20):5733–5745. doi: 10.1128/JB.184.20.5733-5745.2002. PubMed PMID: 12270832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Benelli D., Londei P. Translation initiation in Archaea: conserved and domain-specific features. Biochem Soc Trans. 2011;39(1):89–93. doi: 10.1042/BST0390089. [DOI] [PubMed] [Google Scholar]
- 113.Schmitt E., Coureux P.D., Monestier A., Dubiez E., Mechulam Y. Start codon recognition in eukaryotic and archaeal translation initiation: a common structural core. Int J Mol Sci. 2019;20(4) doi: 10.3390/ijms20040939. PubMed PMID: 30795538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Llacer J.L., Hussain T., Saini A.K., Nanda J.S., Kaur S., Gordiyenko Y., et al. Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition. eLife. 2018;7 doi: 10.7554/eLife.39273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Simonetti A., Guca E., Bochler A., Kuhn L., Hashem Y. Structural insights into the mammalian late-stage initiation complexes. Cell Rep. 2020;31(1) doi: 10.1016/j.celrep.2020.03.061. PubMed PMID: 32268096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kwan T., Thompson S.R. Noncanonical translation initiation in eukaryotes. cold spring harbor perspectives in biology. 2019;11(4) doi: 10.1101/cshperspect.a032672. PubMed PMID: 29959190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shatsky I.N., Terenin I.M., Smirnova V.V., Andreev D.E. Cap-independent translation: what's in a name? Trends Biochem Sci. 2018;43(11):882–895. doi: 10.1016/j.tibs.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 118.Mailliot J., Martin F. Viral internal ribosomal entry sites: four classes for one goal. Wiley Interdiscip Rev RNA. 2018;9(2) doi: 10.1002/wrna.1458. [DOI] [PubMed] [Google Scholar]
- 119.Carvajal F., Vallejos M., Walters B., Contreras N., Hertz M.I., Olivares E., et al. Structural domains within the HIV-1 mRNA and the ribosomal protein S25 influence cap-independent translation initiation. FEBS J. 2016;283(13):2508–2527. doi: 10.1111/febs.13756. PubMed PMID: 27191820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Olivares E., Landry D.M., Caceres C.J., Pino K., Rossi F., Navarrete C., et al. The 5′ untranslated region of the human T-cell lymphotropic virus type 1 mRNA enables cap-independent translation initiation. J Virol. 2014;88(11):5936–5955. doi: 10.1128/JVI.00279-14. PubMed PMID: 24623421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Quade N., Boehringer D., Leibundgut M., van den Heuvel J., Ban N. Cryo-EM structure of Hepatitis C virus IRES bound to the human ribosome at 3.9-A resolution. Nat Commun. 2015;6:7646. doi: 10.1038/ncomms8646. PubMed PMID: 26155016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yamamoto H., Collier M., Loerke J., Ismer J., Schmidt A., Hilal T., et al. Molecular architecture of the ribosome-bound Hepatitis C Virus internal ribosomal entry site RNA. EMBO J. 2015;34(24):3042–3058. doi: 10.15252/embj.201592469. PubMed PMID: 26604301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yokoyama T., Machida K., Iwasaki W., Shigeta T., Nishimoto M., Takahashi M., et al. HCV IRES Captures an Actively Translating 80S Ribosome. Mol Cell. 2019;74(6):1205–1214. doi: 10.1016/j.molcel.2019.04.022. e8. [DOI] [PubMed] [Google Scholar]
- 124.Brown Z.P., Abaeva I.S., De S., Hellen C.U.T., Pestova T.V., Frank J. Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES. EMBO J. 2022;41(16) doi: 10.15252/embj.2022110581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schuler M., Connell S.R., Lescoute A., Giesebrecht J., Dabrowski M., Schroeer B., et al. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol. 2006;13(12):1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 126.Fernandez I.S., Bai X.C., Murshudov G., Scheres S.H., Ramakrishnan V. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell. 2014;157(4):823–831. doi: 10.1016/j.cell.2014.04.015. PubMed PMID: 24792965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Walters B., Axhemi A., Jankowsky E., Thompson S.R. Binding of a viral IRES to the 40S subunit occurs in two successive steps mediated by eS25. Nucleic Acids Res. 2020;48(14):8063–8073. doi: 10.1093/nar/gkaa547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dao Duc K., Batra S.S., Bhattacharya N., Cate J.H.D., Song Y.S. Differences in the path to exit the ribosome across the three domains of life. Nucleic Acids Res. 2019;47(8):4198–4210. doi: 10.1093/nar/gkz106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bornemann T., Jockel J., Rodnina M.V., Wintermeyer W. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat Struct Mol Biol. 2008;15(5):494–499. doi: 10.1038/nsmb.1402. [DOI] [PubMed] [Google Scholar]
- 130.Berndt U., Oellerer S., Zhang Y., Johnson A.E., Rospert S. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc Natl Acad Sci USA. 2009;106(5):1398–1403. doi: 10.1073/pnas.0808584106. PubMed PMID: 19164516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zaman S., Fitzpatrick M., Lindahl L., Zengel J. Novel mutations in ribosomal proteins L4 and L22 that confer erythromycin resistance in Escherichia coli. Mol Microbiol. 2007;66(4):1039–1050. doi: 10.1111/j.1365-2958.2007.05975.x. PubMed PMID: 17956547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tu D., Blaha G., Moore P.B., Steitz T.A. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121(2):257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 133.Tchorzewski M. The acidic ribosomal P proteins. Int J Biochem Cell Biol. 2002;34(8):911–915. doi: 10.1016/s1357-2725(02)00012-2. [DOI] [PubMed] [Google Scholar]
- 134.Rodnina M.V., Stark H., Savelsbergh A., Wieden H.J., Mohr D., Matassova N.B., et al. GTPases mechanisms and functions of translation factors on the ribosome. Biol Chem. 2000;381(5–6):377–387. doi: 10.1515/BC.2000.050. [DOI] [PubMed] [Google Scholar]
- 135.Diaconu M., Kothe U., Schlunzen F., Fischer N., Harms J.M., Tonevitsky A.G., et al. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121(7):991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 136.Shimmin L.C., Ramirez C., Matheson A.T., Dennis P.P. Sequence alignment and evolutionary comparison of the L10 equivalent and L12 equivalent ribosomal proteins from archaebacteria, eubacteria, and eucaryotes. J Mol Evol. 1989;29(5):448–462. doi: 10.1007/BF02602915. [DOI] [PubMed] [Google Scholar]
- 137.Grela P., Bernado P., Svergun D., Kwiatowski J., Abramczyk D., Grankowski N., et al. Structural relationships among the ribosomal stalk proteins from the three domains of life. J Mol Evol. 2008;67(2):154–167. doi: 10.1007/s00239-008-9132-2. [DOI] [PubMed] [Google Scholar]
- 138.Mulder F.A., Bouakaz L., Lundell A., Venkataramana M., Liljas A., Akke M., et al. Conformation and dynamics of ribosomal stalk protein L12 in solution and on the ribosome. Biochemistry. 2004;43(20):5930–5936. doi: 10.1021/bi0495331. [DOI] [PubMed] [Google Scholar]
- 139.Bernado P., Modig K., Grela P., Svergun D.I., Tchorzewski M., Pons M., et al. Structure and dynamics of ribosomal protein L12: an ensemble model based on SAXS and NMR relaxation. Biophys J. 2010;98(10):2374–2382. doi: 10.1016/j.bpj.2010.02.012. PubMed PMID: 20483347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nomura N., Honda T., Baba K., Naganuma T., Tanzawa T., Arisaka F., et al. Archaeal ribosomal stalk protein interacts with translation factors in a nucleotide-independent manner via its conserved C terminus. Proc Natl Acad Sci USA. 2012;109(10):3748–3753. doi: 10.1073/pnas.1112934109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ito K., Honda T., Suzuki T., Miyoshi T., Murakami R., Yao M., et al. Molecular insights into the interaction of the ribosomal stalk protein with elongation factor 1alpha. Nucleic Acids Res. 2014;42(22):14042–14052. doi: 10.1093/nar/gku1248. PubMed PMID: 25428348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Murakami R., Singh C.R., Morris J., Tang L., Harmon I., Takasu A., et al. The interaction between the ribosomal stalk proteins and translation initiation factor 5B promotes translation initiation. Mol Cell Biol. 2018;38(16) doi: 10.1128/MCB.00067-18. PubMed PMID: 29844065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tanzawa T., Kato K., Girodat D., Ose T., Kumakura Y., Wieden H.J., et al. The C-terminal helix of ribosomal P stalk recognizes a hydrophobic groove of elongation factor 2 in a novel fashion. Nucleic Acids Res. 2018;46(6):3232–3244. doi: 10.1093/nar/gky115. PubMed PMID: 29471537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Imai H., Uchiumi T., Kodera N. Direct visualization of translational GTPase factor pool formed around the archaeal ribosomal P-stalk by high-speed AFM. Proc Natl Acad Sci USA. 2020;117(51):32386–32394. doi: 10.1073/pnas.2018975117. PubMed PMID: 33288716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liljas A., Sanyal S. The enigmatic ribosomal stalk. Q Rev Biophys. 2018;51 doi: 10.1017/S0033583518000100. e12. [DOI] [PubMed] [Google Scholar]
- 146.Naganuma T., Nomura N., Yao M., Mochizuki M., Uchiumi T., Tanaka I. Structural basis for translation factor recruitment to the eukaryotic/archaeal ribosomes. J Biol Chem. 2010;285(7):4747–4756. doi: 10.1074/jbc.M109.068098. PubMed PMID: 20007716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Carlson M.A., Haddad B.G., Weis A.J., Blackwood C.S., Shelton C.D., Wuerth M.E., et al. Ribosomal protein L7/L12 is required for GTPase translation factors EF-G, RF3, and IF2 to bind in their GTP state to 70S ribosomes. FEBS J. 2017;284(11):1631–1643. doi: 10.1111/febs.14067. PubMed PMID: 28342293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Remacha M., Jimenez-Diaz A., Bermejo B., Rodriguez-Gabriel M.A., Guarinos E., Ballesta J.P. Ribosomal acidic phosphoproteins P1 and P2 are not required for cell viability but regulate the pattern of protein expression in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15(9):4754–4762. doi: 10.1128/MCB.15.9.4754. PubMed PMID: 7651393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wawiorka L., Molestak E., Szajwaj M., Michalec-Wawiorka B., Molon M., Borkiewicz L., et al. Multiplication of ribosomal p-stalk proteins contributes to the fidelity of translation. Mol Cell Biol. 2017;37(17) doi: 10.1128/MCB.00060-17. PubMed PMID: 28606931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material