Summary
Here, we provide a protocol to isolate mitochondria from cultured cells and extract differently located mitochondrial proteins. We detail steps to separate both integral and peripheral membrane proteins from soluble proteins using sonication. We describe the separation of integral membrane proteins from the peripheral membrane and soluble proteins using sodium carbonate extraction. Furthermore, we detail the use of proteinase K and Triton X-100 to distinguish outer membrane proteins from mitochondrial proteins.
Subject areas: Cell culture, Cell Membrane, Cell separation/fractionation, Protein Biochemistry
Graphical abstract
Highlights
-
•
Protocol for mitochondrial isolation and mitochondrial proteins localization
-
•
Analysis of mitochondrial proteins sub-cellular location by western blot
-
•
Isolating mitochondria from cells for various use, such as enzyme activity detection
-
•
Sub-cellular localization provides insights into the role of mitochondrial proteins
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Here, we provide a protocol to isolate mitochondria from cultured cells and extract differently located mitochondrial proteins. We detail steps to separate both integral and peripheral membrane proteins from soluble proteins using sonication. We describe the separation of integral membrane proteins from the peripheral membrane and soluble proteins using sodium carbonate extraction. Furthermore, we detail the use of proteinase K and Triton X-100 to distinguish outer membrane proteins from mitochondrial proteins.
Before you begin
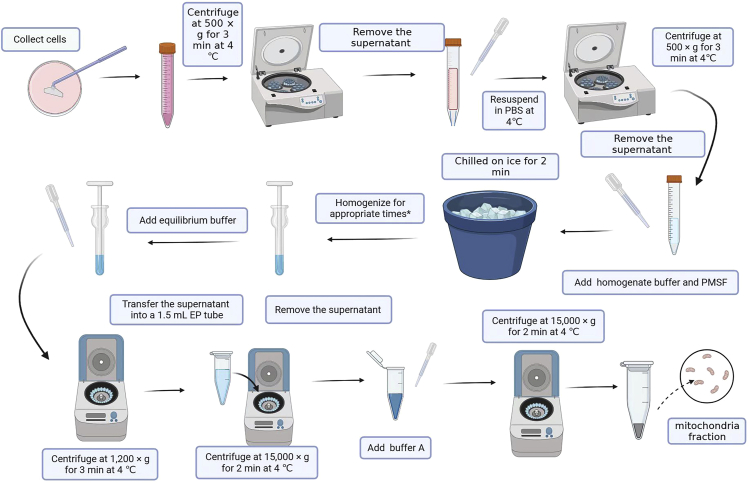
Mitochondria can be isolated from various types of cultured cells, and in this protocol, we choose HEK293T cells to demonstrate all the processes (Figure 1).
Figure 1.
Schematic workflow of mitochondrial isolation from cultured cells
The main materials needed are listed in the “key resources table”; more detailed information on fabricating reagents for the following steps is shown in “materials and equipment.”
Isolation of mitochondria from cultured cells
Timing: 60 min
-
1.
Prepare the buffers and tubes needed for the procedure (see materials and equipment).
CRITICAL: Precool all the materials needed on ice 10 min before starting the procedure. Mix the reagents taken from the refrigerator upside down before using them.
-
2.
Collect cultured cells from several 100 cm2 cell culture dishes with approximately 80% density of cells. After removing the culture medium, wash it twice with phosphate buffer saline (PBS).
Note: The number of cultured cells needed varies. The bigger the cell, the more dishes are needed. For example, HEK293T cells in a total of 1 × 108 from cell culture dishes (ten 100 cm2 cell culture dishes at approximately 80% density of cells) are needed in mitochondrial isolation.
CRITICAL: From this step onward, all activities should be performed on ice to minimize the activation of destructive proteinases.
-
3.
Remove PBS and use a cell lifter to scrape off the adherent cells. Subsequently, transfer the suspension to a 15 mL tube using a pipette, and centrifuge the tube at 500 × g for 3 min at 4°C. Remove the supernatant, then add PBS, and repeat the centrifugation process.
Note: Make sure that all the cells are collected and adherent cells are scraped off in the same direction in each dish.
-
4.
Remove the supernatant, add 2 mL of homogenate buffer and 20 μL of phenylmethylsulfonyl fluoride (PMSF) (100 mM), resuspend the pellet, and chill it on ice for 2 min.
-
5.
Homogenize the cells for appropriate times by using a Dounce tissue grinder.
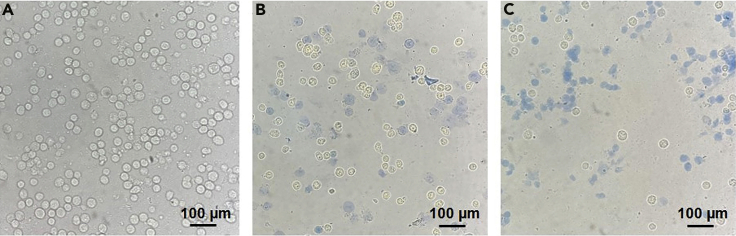
Note: Different cells have different homogenization times. They need to be determined by Trypan Blue staining, in which 80% of cells stained in the field of vision observed under a microscope is the best. For HEK293T cells, about 20 times of homogenization is usually adopted (Figure 2).
-
6.
Add 200 μL of equilibrium buffer to the Dounce tissue grinder at the end of homogenization.
Note: After centrifugation, the fraction of cells disrupted in homogenization is in the pellet, while the mitochondria needed are in the suspension. Therefore, remove the pellet and retain the suspension.
-
7.
Centrifuge at 1,200 × g for 3 min at 4°C and transfer the supernatant to a 1.5 mL tube. Repeat the centrifugation process until no sediment can be observed in order to remove the whole cells.
-
8.
Transfer the supernatant to a fresh 1.5 mL tube and centrifuge at 15,000 × g for 2 min at 4°C.
-
9.
Remove the supernatant, then resuspend the pellet in 500 μL of buffer A, and centrifuge at 15,000 × g for 2 min at 4°C.
-
10.
At this stage, the pellet is the crude mitochondria (mitochondrial enriched fractions) and can be used for electrophoresis and subsequent experiments.
Note: If not used right away, crude mitochondria should be stored frozen at −80°C for at most 1 week.
Figure 2.
Evaluation of cell homogenizing effect using trypan blue staining
(A–C) Cells before homogenization (A), cells stained by trypan blue before homogenization (B), and cells stained by trypan blue after homogenization (C).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| 0.01 M PBS (powder, pH 7.2–7.4) | Solarbio Life Science | Cat#P1010 |
| Phenylmethylsulfonyl fluoride (PMSF) | Sigma-Aldrich | Cat#P7626 |
| Dehydrated alcohol (CH3CH2OH) | ANTE | N/A |
| Hydrochloric acid (HCl) | http://www.zh-chem.com | N/A |
| Trizma base | Sigma-Aldrich | Cat#V900483 |
| Sodium chloride (NaCl) | Sigma-Aldrich | Cat#V900058 |
| Magnesium chloride (MgCl2) | Sigma-Aldrich | Cat#208337 |
| Trypan blue stain (0.4%) | Gibco | Cat#15250061 |
| Sucrose | Sigma-Aldrich | Cat#V900116 |
| Ethylenedinitrilotetraacetic acid (EDTA) | Sigma-Aldrich | Cat#V900106 |
| Potassium chloride (KCl) | Sigma-Aldrich | Cat#P5405 |
| Sodium phosphate dibasic (Na2HPO4) | Sigma-Aldrich | Cat#V900061 |
| Sodium carbonate (Na2CO3) | Sigma-Aldrich | Cat#S7795 |
| Proteinase K | Beyotime Biotechnology | Cat#ST533 |
| Triton X-100 | Sigma-Aldrich | Cat#93443 |
| Experimental models: Cell lines | ||
| Cultured cells, e.g., HEK293T cells | Stem Cell Bank, Chinese Academy of Sciences | Cat#BFN60700191 |
| Software and algorithms | ||
| BioRender | BioRender.com | N/A |
| Other | ||
| Cell culture dish | Sigma-Aldrich | Cat#CLS430167 |
| Cell lifer | Sigma-Aldrich | Cat#CLS3008 |
| 15 mL centrifuge tube | Sigma-Aldrich | Cat#430791 |
| Eppendorf 5424R Microcentrifuge | Eppendorf | Cat#5406000119 |
| Vent filter for reservoir | Pall Life Sciences | Cat#LWFS32501 |
| 15 mL Dounce tissue grinder | Duran Wheaton Kimble Life Science | Cat#357544 |
| Nikon Eclipse 80i fluorescence microscope | Nikon | N/A |
| 1.5 mL centrifuge tube | Maisinuo | Cat#HZX018-2 |
| Scientz-IID ultrasonic homogenizer | Ningbo Scientz Biotechnology | N/A |
| Type 42.2 Ti Rotor | Beckman Coulter | Cat#343007 |
| Open-top thickwall polypropylene tube | Beckman Coulter | Cat#343621 |
| Beckman Coulter Optima XPN-100 ultracentrifuge | Beckman Coulter | Cat#A94469 |
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23225 |
| SpectraMax iD3 Multi-Mode Microplate Reader | Molecular Devices | N/A |
| PowerPac Universal power supply | Bio-Rad | Cat#1645070 |
| Protec OptiMax 2010-NDT tabletop film processor | Protec | N/A |
| X-OMAT BT Film (5 × 7 in.) | Carestream | N/A |
| Epson Perfection V850 Pro photo scanner | Epson | Cat#B11B224201 |
Materials and equipment
Isolation of mitochondria from cultured cells
100 mM PMSF
| Reagent | Final concentration | Amount |
|---|---|---|
| PMSF | 100 mM | 870.95 mg |
| 100% dehydrated alcohol | N/A | 50 mL |
| Total | N/A | 50 mL |
Store at −20°C with a one-year shelf life.
CRITICAL: PMSF is harmful to the mucosa of the respiratory tract, eyes, and skin and can be lethal if inhaled, ingested, or absorbed through the skin. In case of eye or skin contact with PMSF, rinse immediately with plenty of water. Clothing contaminated with PMSF should be discarded.
Homogenate buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl (pH 7.8) | 350 mM | 2,120 mg |
| NaCl | 250 mM | 730.50 mg |
| MgCl2 | 50 mM | 238.03 mg |
| ddH2O | N/A | 50 mL |
| Total | N/A | 50 mL |
This buffer needs to prepare fresh the day before or on the day of experiment and precooled at 4°C.
Equilibrium buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl (pH 7.8) | 3.5 mM | 21.20 mg |
| NaCl | 2.5 mM | 7.31 mg |
| MgCl2 | 0.5 mM | 2.38 mg |
| ddH2O | N/A | 50 mL |
| Total | N/A | 50 mL |
This buffer needs to prepare fresh the day before or on the day of experiment and precooled at 4°C.
Buffer A
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl (pH 7.4) | 10 mM | 60.57 mg |
| EDTA | 1 mM | 14.61 mg |
| Sucrose | 320 mM | 5476.80 mg |
| ddH2O | N/A | 50 mL |
| Total | N/A | 50 mL |
This buffer needs to prepare fresh the day before or on the day of experiment and precooled at 4°C.
Localization of membrane-associated proteins by sonication
1 × TD
| Reagent | Final concentration | Amount |
|---|---|---|
| Trizma base | 49.99 mM | 302.80 mg |
| NaCl | 274.13 mM | 801 mg |
| KCl | 20.12 mM | 75 mg |
| Na2HPO4 | 13.95 mM | 99 mg |
| ddH2O | N/A | 50 mL |
| Total | N/A | 50 mL |
Store at 4°C with a six-month shelf life.
Localization of membrane-associated proteins by sodium carbonate
100 mM Na2CO3
| Reagent | Final concentration | Amount |
|---|---|---|
| Na2CO3 | 100 mM | 529.95 mg |
| ddH2O | N/A | 50 mL |
| Total | N/A | 50 mL |
Store at 4°C with a six-month shelf life.
100 mM PMSF and 1 × TD buffer formulations are described above.
Outer membrane proteins localization using proteinase K and Triton X-100
100 mM PMSF and 1 × TD buffer formulations are described above.
Step-by-step method details
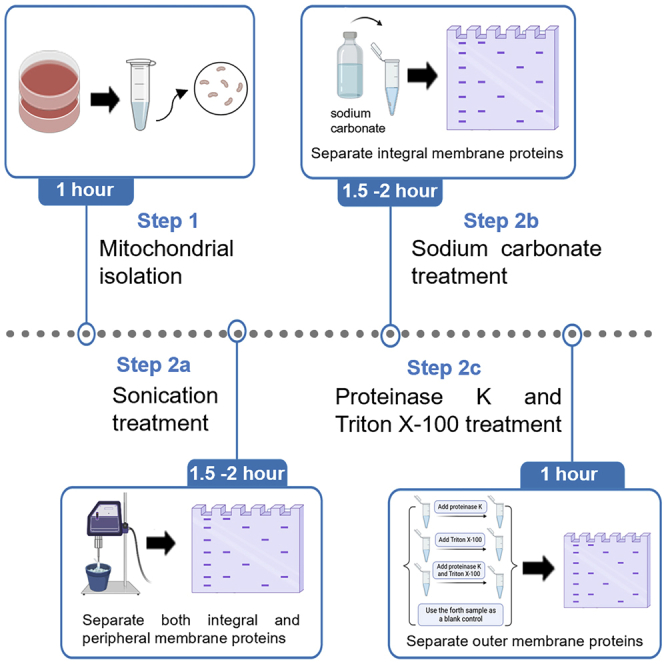
Although six discrete mitochondrial compartments can be recognized in some articles,1 to simplify the structure, we roughly divide a mitochondrion into four compartments: outer mitochondrial membrane, intermembrane space, inner mitochondrial membrane, and matrix. Mitochondrial proteins can be categorized into soluble and membrane proteins (MPs). Soluble proteins are localized in intermembrane space and mitochondrial matrix, whereas membrane proteins are associated with inner or outer mitochondrial membranes. Moreover, membrane proteins can be either peripheral or integral. Peripheral MPs crowd the surface of the membrane through electrostatic, hydrogen-bonding, or hydrophobic interactions with lipid head groups or other integral MPs and can be detached from the membrane by relatively mild treatments such as changing the ionic strength or pH in the buffer. Integral MPs are firmly embedded in the lipid bilayer by hydrophobic interactions between the hydrocarbon chains of lipids and the hydrophobic domains of proteins and can only be removed by detergent solubilization.2,3,4,5,6 In addition, isotopes are also used in localization by labeling proteins in mitochondria.7 The three methods to extract and localize proteins from mitochondria are discussed subsequently.
Localization of membrane-associated proteins by sonication
Timing: between 90 and 120 min
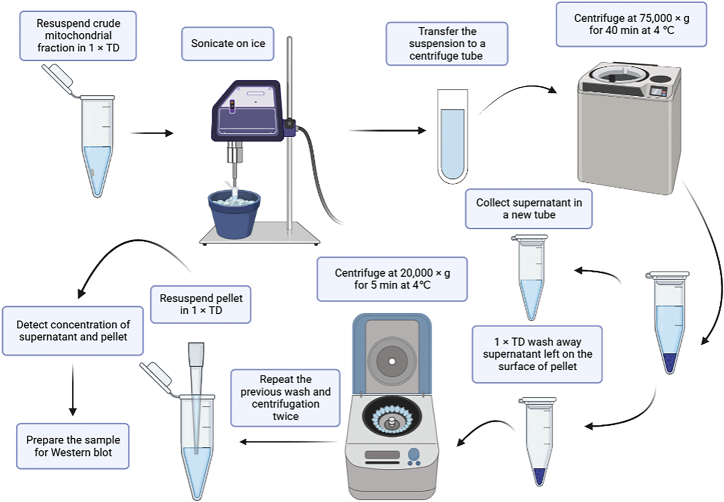
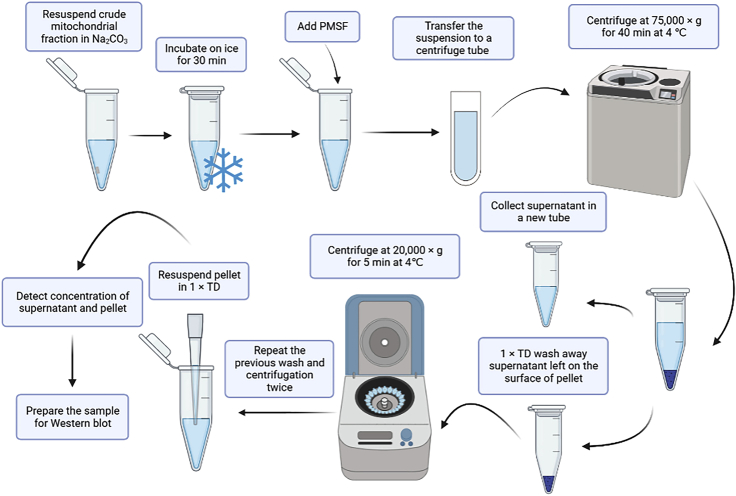
Sonication can break mitochondrial through high frequency ultrasonic vibration energy generated by ultrasonic generator. Thus, sonication can distinguish between the membrane proteins (integral and peripheral) in the pellet (P) and soluble proteins in the supernatant (S) (Figure 3).
-
1.
Obtain cultured cells from several 100 cm2 cell culture dishes with approximately 80% density of cells, according to the aforementioned corresponding steps to collect the crude mitochondrial fraction.
Note: For HEK293T cells, a total of 3 × 107 (three 100 cm2 cell culture dishes at approximately 80% density of cells) are needed in mitochondrial isolation to perform sonication.
-
2.
Resuspend the crude mitochondria fraction obtained in step 1 in 150 μL of 1 × TD and collect in a 1.5 mL precooled tube. Clean the ultrasonic homogenizer and set the power to 150 W and amplitude to 30% before homogenizing the samples on ice for 1 min with a pause of 1 s every 1.5 s.
Note: Typically, before drying the probe of the ultrasonic homogenizer, wash the probe with 15 mL of 75% ethanol and subsequently with water at high pressure. Do not forget to keep a portion as the control group.
-
3.
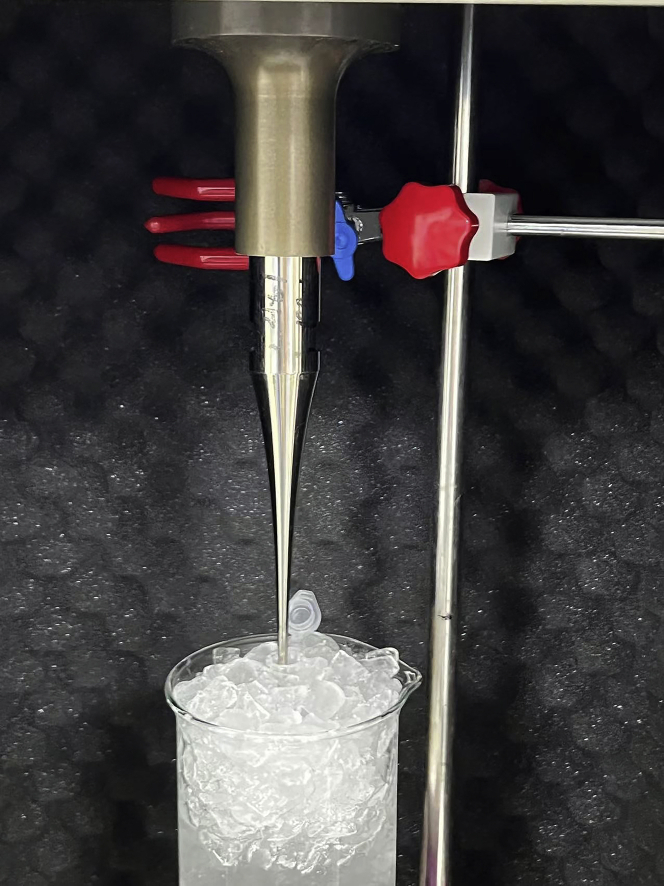
Allow the 1.5 mL tube containing mitochondrial suspension collected in step 2 to stand on the ice and turn on the ultrasonic homogenizer to sonicate the suspension (Figure 4).
CRITICAL: The probe is close to the bottom of the liquid and don’t touch the tube wall to avoid foam in the process of ultrasound. This entire step is performed on ice to prevent overheating caused by sonication. Be cautious during the procedure to avoid splashes or extremely high temperatures.
-
4.
After sonication, transfer the suspension to the special centrifuge tube of the ultra-speed Type 42.2 Ti Rotor and centrifuge for 40 min at 75,000 × g in Beckman Coulter Optima XPN-100 ultracentrifuge at 4°C (Figure 5). Then collect the supernatant (S) in a fresh 1.5 mL tube.
-
5.
Use 100 μL 1 × TD to gently rinse off the remaining supernatant from the surface of the pellet; subsequently, centrifuge at 20,000 × g for 5 min at 4°C and pipette off the supernatant.
Note: Gently add 1 × TD against the wall to avoid dispersing the pellet.
CRITICAL: In case of accidental dispersion and resuspension of the pellet into the supernatant, centrifuge further to maximize the purity of the pellet.
-
6.
Repeat step 5 twice for a total of three times.
-
7.
Fully resuspend the pellet (P) in 20 μL of 1 × TD.
-
8.
Identify the concentration of the supernatant (S) and pellet (P) using the Pierce BCA Protein Assay Kit.
-
9.
Prepare the sample for the Western blot.
Figure 3.
Localization of membrane-associated proteins by sonication schematic workflow
Figure 4.
Sonication of the sample on ice
Figure 5.
Beckman Coulter centrifugal machine and Type 42.2 Ti Rotor
(A) Beckman Coulter centrifugal machine.
(B) Type 42.2 Ti Rotor.
Localization of membrane-associated proteins by sodium carbonate
Timing: between 90 and 120 min
The sodium carbonate can break mitochondrial membranes through low surface tension based on the principle of osmotic pressure. Thus, this experiment can separate the integral proteins in the pellet (P) from peripheral and soluble proteins in the supernatant (S) (Figure 6).
-
10.
Obtain cultured cells from several 100 cm2 cell culture dishes with approximately 80% density of cells, according to the aforementioned corresponding steps to collect the crude mitochondrial fraction.
Note: For HEK293T cells, a total of 3 × 107 (three 100 cm2 cell culture dishes at approximately 80% density of cells) are needed in mitochondrial isolation for the sodium carbonate extraction experiment performed at one pH gradient.
-
11.
Resuspend crude mitochondrial fraction obtained in step 10 in 30 μL of Na2CO3 (100 mM).
Note: Do not forget to keep a portion as the control group. In addition, the volume mentioned here is just sufficient for one pH gradient; in case the sodium carbonate extraction experiment is performed at different pH values, the volume of reagents needed should be prepared after calculation.
-
12.
Incubate on ice for 30 min. After 1 μL of PMSF (100 mM) is added, mix the sample well.
-
13.
Transfer the suspension to the special centrifuge tube of the ultra-speed Type 42.2 Ti Rotor and centrifuge for 40 min at 75,000 × g in Beckman Coulter Optima XPN-100 ultracentrifuge at 4°C (Figure 5). Collect the supernatant (S) in a fresh 1.5 mL tube.
-
14.
Gently wash away the supernatant left on the surface of the pellet using 100 μL 1 × TD and then centrifuge at 20,000 × g for 5 min at 4°C.
Note: Gently add 1 × TD against the wall to avoid dispersing the pellet.
CRITICAL: In case of accidental dispersion and resuspension of the pellet into the supernatant, centrifuge further to maximize the purity of the pellet.
-
15.
Repeat step 14 twice for a total of three times.
-
16.
Thoroughly resuspend the pellet (P) in 20 μL of 1 × TD.
-
17.
Identify the concentration of the supernatant (S) and pellet (P) using the Pierce BCA Protein Assay Kit.
-
18.
Prepare the sample for the Western blot.
Figure 6.
Localization of membrane-associated proteins by sodium carbonate schematic workflow
Outer membrane proteins localization using proteinase K and Triton X-100
Timing: 60 min
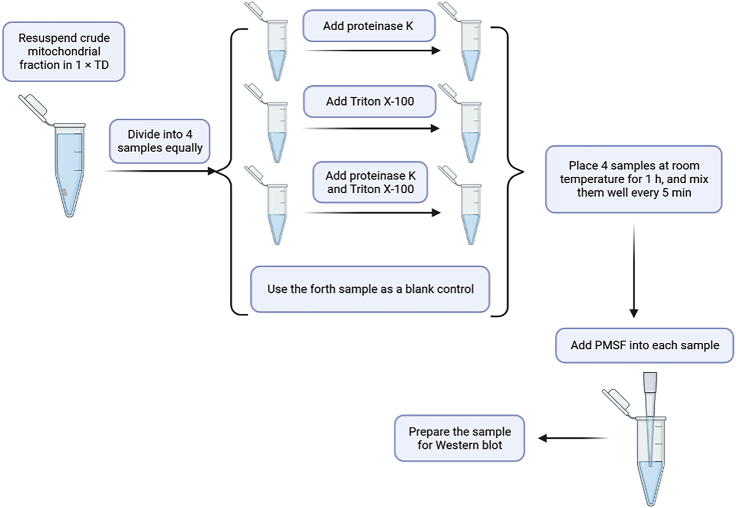
Proteinase K can only digest outer membrane proteins, but after adding Triton X-100 to dissolve mitochondrial membranes, proteinase K can pass through all compartments to digest all mitochondrial proteins. Therefore, proteinase K and Triton X-100 are used to distinguish between outer membrane proteins and other mitochondrial proteins (Figure 7).
-
19.
Obtain cultured cells from several 100 cm2 cell culture dishes with approximately 80% density of cells, according to the aforementioned corresponding steps to collect the crude mitochondrial fraction.
Note: For HEK293T cells, a total of 5 × 107 (five 100 cm2 cell culture dishes at approximately 80% density of cells) are needed in mitochondrial isolation to perform this experiment.
-
20.
Fully resuspend crude mitochondrial fraction obtained in step 19 in 120 μL of 1 × TD and equally divide them into four samples.
-
21.
Add 3.33 μL of proteinase K solution (1 mg/mL) (10 μg of proteinase K/mg of mitochondrial protein) in the first sample, 3.33 μL of 20% Triton X-100 (2 mg of Triton X-100/mg of mitochondrial protein) in the second sample, and 3.33 μL of proteinase K solution (1 mg/mL) and 3.33 μL of 20% Triton X-100 in the third sample, and use the fourth sample as a blank control.
-
22.
Store the four samples at room temperature for 1 h and mix them well every 5 min by using a pipette.
Note: The room temperature mentioned here maintains between 20°C and 27°C.
-
23.
Add 1 μL of 100 mM PMSF into the four samples and mix well.
-
24.
Prepare the sample for the Western blot.
Figure 7.
Schematic workflow of outer membrane protein localization using proteinase K and Triton X-100
Expected outcomes
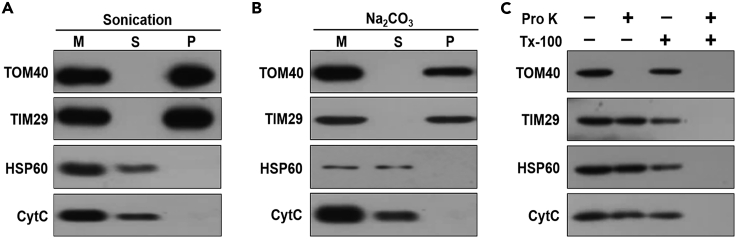
Sonication can distinguish between the membrane proteins (integral and peripheral) in the pellet (P) and soluble proteins in the supernatant (S). In addition, the sodium carbonate extraction experiment can separate the integral proteins in the pellet (P) from peripheral and soluble proteins in the supernatant (S). Combining the two methods, mitochondrial proteins theoretically can be divided into integral and peripheral membrane proteins and soluble proteins. In contrast, proteinase K and Triton X-100 are used to distinguish between outer membrane proteins and other mitochondrial proteins (Figure 8). In our recent work, we have performed sub-cellular localization of SERAC1 using proteinase K and Triton X-100, and it is easy to conclude that SERAC1 is both soluble and outer membrane protein and is supposed to be located at more than just the outer mitochondrial membrane.8
Figure 8.
Expected outcomes of protein localization using sonication, sodium carbonate or proteinase K and Triton X-100
(A–C) Mitochondria from HEK293T cells were sonicated (A) or treated with Na2CO3 (B), proteinase K or/and Triton X-100 (C), and then left untreated (M) or separated into supernatant (S) and pellet (P) and analyzed by Western blot. TOM40, TIM29, HSP60 and CytC are used as markers representing outer membrane, inner membrane, mitochondrial matrix and mitochondrial intermembrane proteins, respectively.
Limitations
There are various experiments performed on sodium carbonate extraction at pH 11.5. However, the strength of proteins connected to the membrane and the environment can vary significantly. Performing the sodium carbonate extraction experiment at different pH gradients favors both finding the optimum pH for your experiment and avoiding interpreting some peripheral proteins with looser connections to the membrane as soluble proteins or with tighter connections as integral proteins. Although Triton X-100 exerts the function of solubilization, mitochondrial inner and outer membranes differ in various aspects, leading to selective solubilization in some components and quite different behaviors toward Triton X-100.9 Mitochondrial fractions were incubated with 0.38 mg, 2 mg, 2 mg and 5 mg Triton X-100/mg of protein in pig liver, yeast, cell lines and beef heart, respectively.10,11,12,13 A suitable concentration of Triton X-100 applied to different samples needs to be tested.
Troubleshooting
Problem 1
Cells are in critical condition when isolating mitochondria (step 2).
Potential solution
Control the density of cells to at least reach 80%, and adherent cells are the desired culture condition, but cells are not supposed to be dense.
Problem 2
The number of cells scraped off from dishes is less than expected (step 3).
Potential solution
Flush the dish several times with PBS to maximize the cells collected. The osmotic pressure of PBS is similar to that of cells; thus, pipetting PBS surrounding cells while transferring the cells to a tube is reasonable and acceptable.
Problem 3
Overly homogenize when isolating mitochondria (step 5).
Potential solution
Extract the supernatant every several rounds (depending on the strength used) during homogenization, stain it with Trypan Blue, and observe under a microscope.
Problem 4
Obtain less sample than expected after ultracentrifugation (steps 4 and 13).
Potential solution
Please try your best to shorten the time in this process, because the special centrifuge tube is open-top, and practicing longer means evaporating more.
Problem 5
Acquire impure substances (supernatant or pellet) after ultracentrifugation in the sonication or sodium carbonate extraction experiment (steps 4 and 13).
Potential solution
Carefully aspirate the supernatant and strictly avoid touching the pellet by leaving some supernatant on its surface. In addition, when collecting the pellet, thoroughly rinse it with 1 × TD. Repeat this process several times until the substance is pure.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ya Wang (yawang@wmu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
We thank Anran Xie (Nanjing Drum Tower Hospital) for helpful discussion. This work was supported by National Natural Science Foundation of China-excellent young scientists fund (82222043), National Natural Science Foundation of China (82172322), Zhejiang Provincial Natural Science Foundation of China (LR20H200001 and Q23H200005), and the Key Discipline of Zhejiang Province in Medical Technology (First Class, Category A). This work is partially supported by BioRender (https://biorender.com/).
Author contributions
H.F. supervised this work. S.Z., D.Z., X.H., and D.L. performed experiments and wrote the manuscript. H.F. and Y.W. edited the manuscript. All authors reviewed the manuscript.
Declaration of interests
The authors declare no competing interests.
Data and code availability
This study did not generate new data or code.
References
- 1.Logan D.C. The mitochondrial compartment. J. Exp. Bot. 2006;57:1225–1243. doi: 10.1093/jxb/erj151. [DOI] [PubMed] [Google Scholar]
- 2.Kim H., Botelho S.C., Park K., Kim H. Use of carbonate extraction in analyzing moderately hydrophobic transmembrane proteins in the mitochondrial inner membrane. Protein Sci. 2015;24:2063–2069. doi: 10.1002/pro.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.le Maire M., Champeil P., Moller J.V. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta. 2000;1508:86–111. doi: 10.1016/s0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 4.Chiu M.L., Tsang C., Grihalde N., MacWilliams M.P. Over-expression, solubilization, and purification of G protein-coupled receptors for structural biology. Comb. Chem. High Throughput Screen. 2008;11:439–462. doi: 10.2174/138620708784911456. [DOI] [PubMed] [Google Scholar]
- 5.Grisshammer R. Elsevier; 2009. Purification of Recombinant G-Protein-Coupled Receptors; pp. 631–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith S.M. Humana Press; 2011. Strategies for the Purification of Membrane Proteins; pp. 485–496. [DOI] [PubMed] [Google Scholar]
- 7.Vögtle F.N., Burkhart J.M., Gonczarowska-Jorge H., Kücükköse C., Taskin A.A., Kopczynski D., Ahrends R., Mossmann D., Sickmann A., Zahedi R.P., Meisinger C. Landscape of submitochondrial protein distribution. Nat. Commun. 2017;8:290. doi: 10.1038/s41467-017-00359-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang H., Xie A., Du M., Li X., Yang K., Fu Y., Yuan X., Fan R., Yu W., Zhou Z., et al. SERAC1 is a component of the mitochondrial serine transporter complex required for the maintenance of mitochondrial DNA. Sci. Transl. Med. 2022;14:eabl6992. doi: 10.1126/scitranslmed.abl6992. [DOI] [PubMed] [Google Scholar]
- 9.Gurtubay J.I., Goñi F.M., Gómez-Fernández J.C., Otamendi J.J., Macarulla J.M. Triton X-100 solubilization of mitochondrial inner and outer membranes. J. Bioenerg. Biomembr. 1980;12:47–70. doi: 10.1007/bf00745012. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen X.T., Pabarue H.A., Geyer R.R., Shroyer L.A., Estey L.A., Parilo M.S., Wilson K.S., Prochaska L.J. Biochemical and biophysical properties of purified phospholipid vesicles containing bovine heart cytochrome c oxidase. Protein Expr. Purif. 2002;26:122–130. doi: 10.1016/s1046-5928(02)00503-x. [DOI] [PubMed] [Google Scholar]
- 11.Rubinson K.A., Pokalsky C., Krueger S., Prochaska L.J. Structure determination of functional membrane proteins using small-angle neutron scattering (SANS) with small, mixed-lipid liposomes: native beef heart mitochondrial cytochrome c oxidase forms dimers. Protein J. 2013;32:27–38. doi: 10.1007/s10930-012-9455-0. [DOI] [PubMed] [Google Scholar]
- 12.Rubalcava-Gracia D., García-Rincón J., Pérez-Montfort R., Hamel P.P., González-Halphen D. Key within-membrane residues and precursor dosage impact the allotopic expression of yeast subunit II of cytochrome c oxidase. Mol. Biol. Cell. 2019;30:2358–2366. doi: 10.1091/mbc.E18-12-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montesinos J., Area-Gomez E. Isolation of mitochondria-associated ER membranes. Methods Cell Biol. 2020;155:33–44. doi: 10.1016/bs.mcb.2019.12.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate new data or code.