Abstract
Deletion of O-GlcNAc transferase (Ogt) in pancreatic epithelial progenitor cells results in pancreatic hypoplasia at birth, partly due to increased apoptosis during embryonic development. Constitutive loss of Ogt in β-cells results in increased ER stress and apoptosis, and in the Ogt-deficient pancreas, transcriptomic data previously revealed both tumor suppressor protein p53 and pancreatic duodenal homeobox 1 (Pdx1), key cell survival proteins in the developing pancreas, as upstream regulators of differentially expressed genes. However, the specific roles of these genes in pancreatic hypoplasia are unclear. In this study, we explored the independent roles of p53, ER stress protein CHOP, and Pdx1 in pancreas development and their use in the functional rescue of pancreatic hypoplasia in the context of Ogt loss. Using in vivo genetic manipulation and morphometric analysis, we show that Ogt plays a key regulatory role in pancreas development. Heterozygous, but not homozygous, loss of pancreatic p53 afforded a partial rescue of β-cell, α-cell, and exocrine cell masses, while whole body loss of CHOP afforded a partial rescue in pancreas weight and a full rescue in exocrine cell mass. However, neither was sufficient to fully mitigate pancreatic hypoplasia at birth in the Ogt-deficient pancreas. Furthermore, overexpression of Pdx1 in the pancreatic epithelium resulted in partial rescues in pancreas weight and β-cell mass in the Ogt loss background. These findings highlight the requirement of Ogt in pancreas development by targeting multiple proteins such as transcription factor Pdx1 and p53 in the developing pancreas.
Keywords: pancreas development, O-GlcNAc transferase, O-GlcNAcylation, Pdx1, p53, CHOP, ER stress, transcription factors, islet, beta-cell
Abbreviations: CHOP, C/EBP homologous protein; DEG, differentially expressed gene; ER, endoplasmic reticulum; Ogt, O-GlcNAc transferase; UPR, unfolded protein response
Protein O-GlcNAcylation is a nutrient and stress-sensitive protein posttranslational modification. The addition of an O-GlcNAc molecule to proteins is catalyzed by O-GlcNAc transferase (Ogt), and the enzyme O-GlcNAcase (OGA) is responsible for removal of this posttranslational modification. Ogt's substrate, UDP-GlcNAc, is synthesized through the hexosamine biosynthetic pathway, which receives 2 to 5% of glucose, as well as other nutrients such as amino acids and lipids, that enter the cell, making Ogt a nutrient sensor (1, 2). Ogt has been implicated in a variety of cellular processes related to growth, survival, and stress, including the β-cell endoplasmic reticulum (ER) and mitochondrial stress responses in a hyper-nutrient environment (3, 4, 5, 6).
The pancreatic epithelium receives nutrient signals from the surrounding tissue. As a nutrient sensor, Ogt is poised to fine tune multiple signaling pathways in response to nutrient and growth factors. Many transcription factors that are important for pancreatic β-cell health and development, such as NeuroD1 and Pdx1, are modified by Ogt (7, 8, 9, 10). The pancreas is very sensitive to nutrient levels in utero, and nutrients have been shown to regulate the differentiation and development of specific pancreatic cells such as the β-cells. We previously showed that the levels of O-GlcNAcylation in pancreatic proteins are high. Indeed, expression of Ogt in Pdx1+ pancreatic epithelial progenitors is required for pancreatic development, and its absence results in pancreatic hypoplasia driven by increased apoptosis, suggesting a role of Ogt in cellular survival (9).
Suppression of proapoptotic protein p53 by DNA methyltransferase 1 (DNMT1) is required in pancreatic development by allowing the survival of pancreatic progenitors (11). Much like the deletion of Ogt in the pancreatic progenitors (OgtKOPanc), derepression of p53 in the developing pancreas results in pancreatic atrophy through depletion of the pancreatic progenitor pool (11). RNA sequencing has implicated p53, a target of Ogt, as a key upstream regulator of differentially expressed genes (DEGs) in the OgtKOPanc model (9).
The C/EBP homologous protein (CHOP), as a transcription factor, mediates ER stress-induced apoptosis by regulating BCL2-family proteins, ultimately orchestrating the release of proapoptotic factors from the mitochondria and leading to cell death (12, 13). In cardiac tissue, CHOP deficiency confers resistance to ER stress–mediated apoptosis (14). However, its role in the developing pancreas is unknown. Studies in the mature islet suggest that pancreatic β-cells are highly susceptible to ER stress, which lead to secretory defects and cell death (15, 16). Deletion of Ogt in the β-cells (β-OgtKO) results in increased CHOP protein in the islets, and secretory defects in insulin are partially rescued by the deletion of CHOP, suggesting a role of Ogt in the regulation of ER stress in the β-cell (17).
Pancreatic Duodenal Homeobox 1 (Pdx1) is a transcription factor known to be a master regulator of pancreas development as well as β-cell function and survival (18). Pdx1-expressing pancreatic progenitors give rise to both the endocrine and exocrine pancreas compartments during pancreas development (19). Pdx1 null mice do not develop a pancreas. We and others have demonstrated that Pdx1 is O-GlcNAc modified (8). In both β-OgtKO or OgtKOPanc, Pdx1 protein levels are reduced (9, 17). O-GlcNAc modification on Pdx1 may impact DNA-binding affinity (8). In the developing pancreas, the O-GlcNAc modification is correlated with the survival of Pdx1+ pancreatic progenitors (9). Ablation of Ogt reduced the size of the Pdx1+ progenitor pool in the embryonic pancreas, and DEGs associated with Ogt loss were, in part, correlated with cellular death and survival (9).
Given that the size of the pancreatic organ is limited by the number of embryonic progenitor cells available in the pancreas, it follows that the survival of these progenitors is imperative to pancreatic development (20). In this study, we sought to determine whether reducing proapoptotic proteins p53 or ER-stress mediator CHOP in the background of Ogt loss could ameliorate the pancreatic hypoplasia in the OgtKOPanc using a mouse model.
Results
Deletion of one allele of p53 partially rescues β-cell, α-cell, and exocrine cell mass in OgtKOPanc
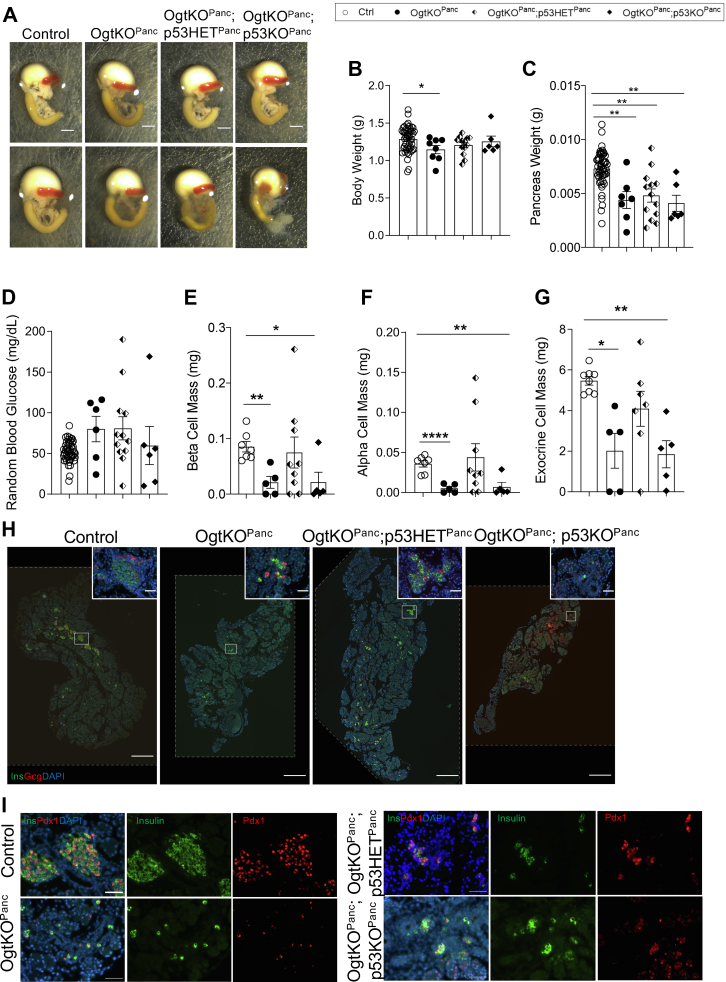
We previously demonstrated that ablation of Ogt in the pancreatic progenitors (OgtKOPanc) results in pancreatic hypoplasia at birth, in part due to increased apoptosis in utero at embryonic day 14.5. Ingenuity Pathway Analysis of single cell RNA sequencing data implicated p53, an O-GlcNAc target, as an upstream regulator of DEGs in the OgtKOPanc (9, 21), and a gene ontology analysis of these upstream regulators revealed the regulation of apoptosis among the top 30 enriched terms (Fig. S1A). In the current study, we tested whether ablation of p53 could rescue pancreatic hypoplasia in the background of Ogt loss. Contrary to deletion of p53 alone, pancreas-specific concomitant deletion of p53 and Ogt resulted in pancreatic hypoplasia like the OgtKOPanc alone (Fig. 1A). As we previously reported, OgtKOPanc displayed lower body weight than littermate controls (Fig. 1B). Interestingly, full deletion of p53 in the background of Ogt loss (OgtKOPanc;p53KOPanc) resulted in an intermediate body weight between control and OgtKOPanc, while mice with a heterozygous loss of p53 in the background of Ogt loss (OgtKOPanc;p53HETPanc) displayed a downward trend in body weight compared to littermate controls (Fig. 1B). OgtKOPanc;p53HETPanc and OgtKOPanc;p53KOPanc mice displayed similarly reduced pancreas weight as the OgtKOPanc, indicating that deletion of p53 was insufficient to rescue pancreas weight reduction in OgtKOPanc mice (Figs 1C and S2A [pancreas weight corrected by body weight]).
Figure 1.
Heterozygous deletion of p53 partially rescues endocrine and exocrine cell mass in Ogt-deficient pancreas. Representative images of pancreas, stomach, spleen, and duodenum at birth (A, scale bar represents 250 pixels). Body weight (B, n = 6–51), pancreas weight (C, n = 6–51), random blood glucose (D, n = 6–51), β-cell mass (E, n = 5–9), α-cell mass (F, n = 5–9), and exocrine cell mass (G, n=5–8) at birth. Representative images of whole pancreas (H, large panel 10× magnification, scale bar represents 500 μm; inset 40× magnification, scale bar represents 50 μm; areas outside dotted lines indicate space added from canvas size adjustment) and islets showing normal Pdx1 localization (I, 40× magnification, scale bar represents 50 μm). Statistics conducted using one-way ANOVA followed by unpaired Student t test with Welch’s correction, p ≤ 0.05 deemed significant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001. Ogt, O-GlcNAc transferase.
Random blood glucose was similar among all neonatal mice, but OgtKOPanc;p53HETPanc mice displayed a trend toward elevated blood glucose (Fig. 1D). As expected, we observed reduced β-cell mass between Ctrl and OgtKOPanc as previously published (9). While OgtKOPanc;p53KOPanc mice displayed a lower β-cell mass and α-cell mass than control, concomitant heterozygous deletion of p53 in mice lacking Ogt resulted in a partial rescue in both β-cell mass and α-cell mass (Fig. 1, E and F). Similarly, while we observed lower exocrine cell mass in the OgtKOPanc, heterozygous deletion of p53 resulted in an intermediate exocrine cell mass between OgtKOPanc and control, suggesting a partial rescue (Fig. 1, G and H). Interestingly, homozygous deletion of p53 with Ogt resulted in an exocrine mass like the OgtKOPanc alone (Fig. 1G). Immunofluorescence (IF) staining of pancreas sections revealed that although Ogt deletion resulted in lower β-cell mass, remaining β-cells in the pancreas express nuclear Pdx1 (Fig. 1I), which suggest those that survived without Ogt developed normally.
Ablation of p53 in pancreatic Pdx1+ progenitors does not alter pancreas development and islet composition at birth
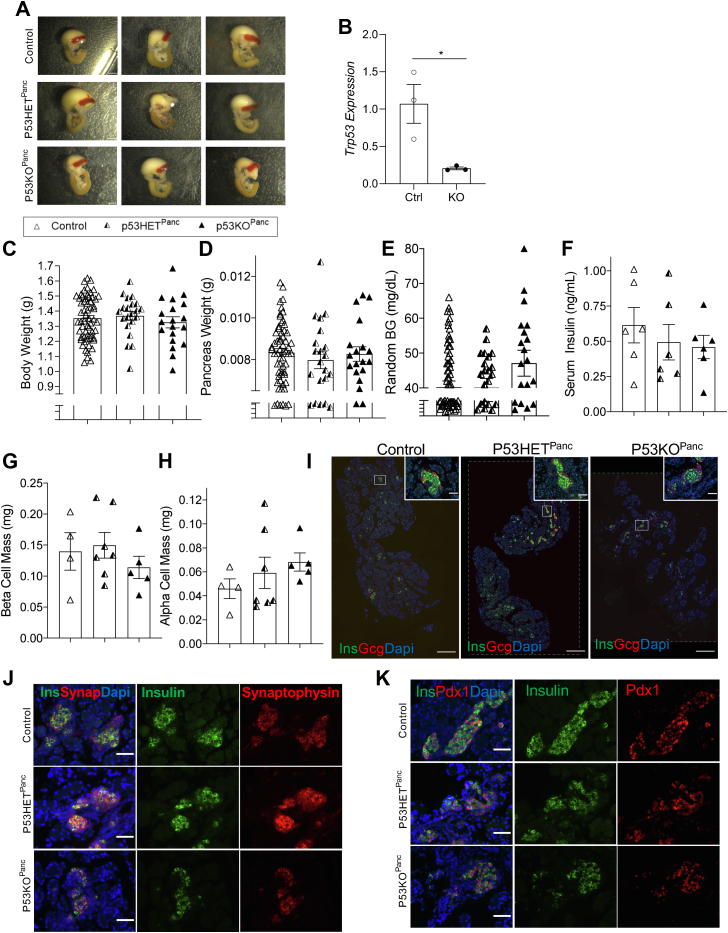
Since partial deletion of p53 in the context of pancreatic Ogt loss resulted in a partial rescue, we sought to determine the effect of genetic ablation of p53, alone, in the pancreas. Suppression of p53 via DNMT1 has previously been shown to be required for pancreatic organogenesis (11). To explore the effect of the absence of p53 in pancreas development, we studied the pancreas of animals with pancreas-specific genetic ablation of p53 using Cre-recombinase driven by the Pancreatic Duodenal Homeobox 1 (Pdx1) promotor (p53KOPanc) (22). At postnatal day 0 (p0), pancreases of p53HETPanc and p53KOPanc offspring were morphologically comparable with littermate controls (Fig. 2A). mRNA levels of p53 in the whole pancreas measured by qPCR verify that p53 expression was diminished in the pancreas (Fig. 2B). No differences were observed in body weight and pancreas weight among the p53HETPanc, p53KOPanc, and littermate controls at birth (Figs. 2, C and D and S2B [pancreas weight corrected by body weight]). Additionally, both p53HETPanc and p53KOPanc offspring displayed normal random blood glucose and serum insulin levels (Fig. 2, E and F). Analysis of stained pancreas sections showed no differences in β-cell mass or α-cell mass among the three groups (Fig. 2, G–I). To explore the morphology of the pancreatic islets, pancreas sections were immunostained against insulin and synaptophysin (a marker for all endocrine cells), and no differences in staining pattern were observed (Fig. 2J). Since Pdx1 expression is known to be important in pancreatic organogenesis, as well as β-cell identity and function, we assessed Pdx1 expression patterns in pancreas sections of these mice and observed nuclear expression in p53HETPanc, p53KOPanc, and littermate controls (Fig. 2K), suggesting normal β-cell development.
Figure 2.
P53 is dispensable in pancreas organogenesis. Representative images of pancreas, stomach, spleen, and duodenum at birth (A, scale bar represents 250 pixels). Quantitative PCR of neonatal pancreas showing reduced p53 mRNA expression in the p53 knockout (B, n = 3). Body weight (C, n = 19–60), pancreas weight (D, n = 19–60), random blood glucose (E, n = 19–60), and serum insulin (F, n = 6), β-cell mass (G, n = 4–7), and α-cell mass (H, n = 4–7) at birth. Representative images of whole pancreas (I, large panel 10× magnification, scale bar represents 500 μm; inset 40× magnification, scale bar represents 50 μm; areas outside dotted lines indicate space added from canvas size adjustment) and islets showing normal morphology (J, 40× magnification, scale bar represents 50 μm) and Pdx1 localization (K, 40× magnification, scale bar represents 50 μm). Statistics conducted using one-way ANOVA followed by unpaired Student t test with Welch’s correction, p ≤ 0.05 deemed significant. ∗p < 0.05.
Whole body deletion of CHOP results in larger body weight and total pancreas weight
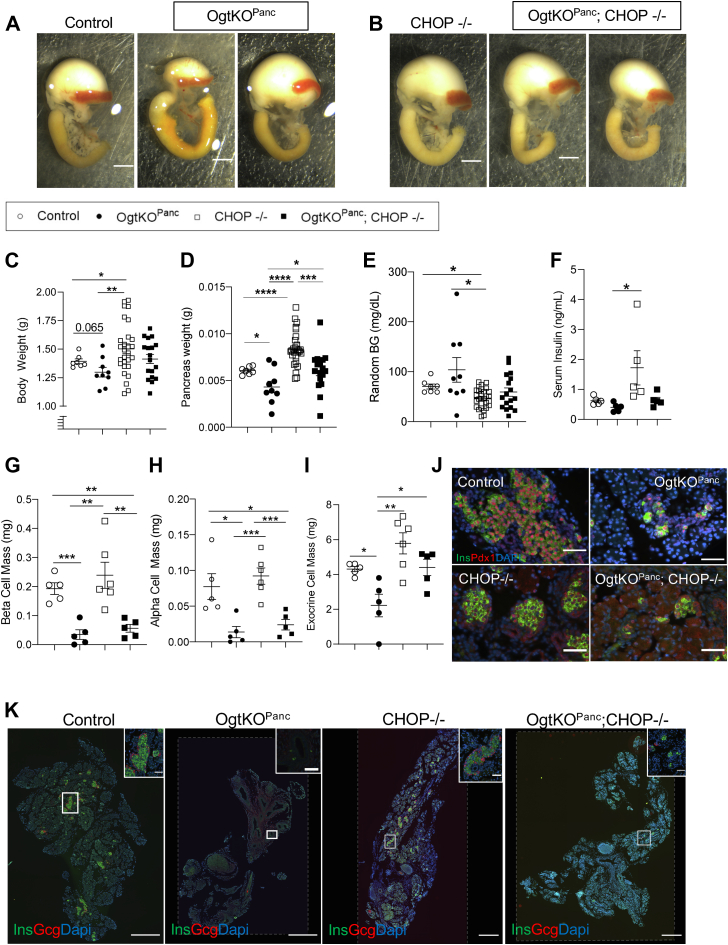
As previously shown, OgtKOPanc display pancreatic hypoplasia at birth (Fig. 3, A and K). We have previously demonstrated that pancreatic hypoplasia in the Ogt loss model is due, in part, to increased apoptosis in the developing pancreatic epithelium. ER stress was previously implicated in the apoptosis of Ogt-deficient β-cells, where CHOP protein levels are increased (17). Furthermore, single cell RNA sequencing of embryonic pancreas has implicated the ER unfolded protein sensor ERN1 (also known as IRE1) as a putative O-GlcNAc target and upstream regulator of DEGs in the Ogt-deficient pancreas (9). This provides a strong rationale to examine ER stress. To assess the role of ER-stress in the Ogt-deficient pancreatic epithelium, we generated a mouse model conferring a full body loss of CHOP (CHOP−/−). Full body deletion of CHOP does not change pancreas morphology, but in the absence of Ogt (OgtKOPanc;CHOP−/−), mice are born with varying degrees of pancreatic hypoplasia (Fig. 3, B and K). Deletion of CHOP, by itself, resulted in increased body weight and pancreas weight compared to controls (Figs. 3, C and D and S2C (pancreas weight corrected by body weight)), and deletion of CHOP in the absence of Ogt afforded a rescue in pancreas weight (Figs. 3, D and S2C). Random blood glucose at birth was lower in the CHOP−/−than OgtKOPanc and controls (Fig. 3E). Whole body deletion of CHOP resulted in a significant increase in serum insulin compared to the OgtKOPanc and a trending increase compared to controls (Fig. 3F).
Figure 3.
Whole body deletion of CHOP partially rescues pancreas weight through ameliorating exocrine cell mass. Representative images of pancreas, stomach, spleen, and duodenum at birth showing varying degrees of pancreatic hypoplasia in OgtKOPanc (A, scale bar represents 250 pixels) and OgtKOPanc;CHOP−/− (B). Body weight (C, n = 7–28), pancreas weight (D, n = 7–28), random blood glucose (E, n = 7–29), serum insulin (F, n = 5), β-cell mass (G, n = 5–6), α-cell mass (H, n = 5–6), and exocrine cell mass (I, n = 5–6) at birth. Representative images of islets showing normal Pdx1 localization (J, 40× magnification, scale bar represents 50 μm) and whole pancreas (K, large panel 10× magnification, scale bar represents 500 μm; inset 40× magnification, scale bar represents 50 μm; areas outside dotted lines indicate space added from canvas size adjustment). Statistics conducted using one-way ANOVA followed by unpaired Student t test with Welch’s correction, p ≤ 0.05 deemed significant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. CHOP, C/EBP homologous protein.
Deletion of CHOP rescues exocrine cell mass in the OgtKOPanc pancreas
While CHOP−/− mice displayed elevated α-cell mass and β-cell mass compared to OgtKOPanc, homozygous deletion of CHOP in the absence of Ogt resulted in reduced β-cell mass and α-cell mass, suggesting that CHOP-dependent ER stress is not a major contributor to the loss of mass in the endocrine compartment in the OgtKOPanc (Fig. 3, G and H). As expected, deletion of Ogt resulted in decreased exocrine cell mass (Fig. 3I). However, concomitant deletion of CHOP normalized exocrine cell mass, suggesting the rescue in pancreas weight originates only from the exocrine compartment (Fig. 3I). Mice from all groups display nuclear Pdx1 staining in β-cells (Fig. 3J).
Overexpression of Pdx1 in OgtKOPanc pancreas partially rescues pancreas weight and β-cell mass
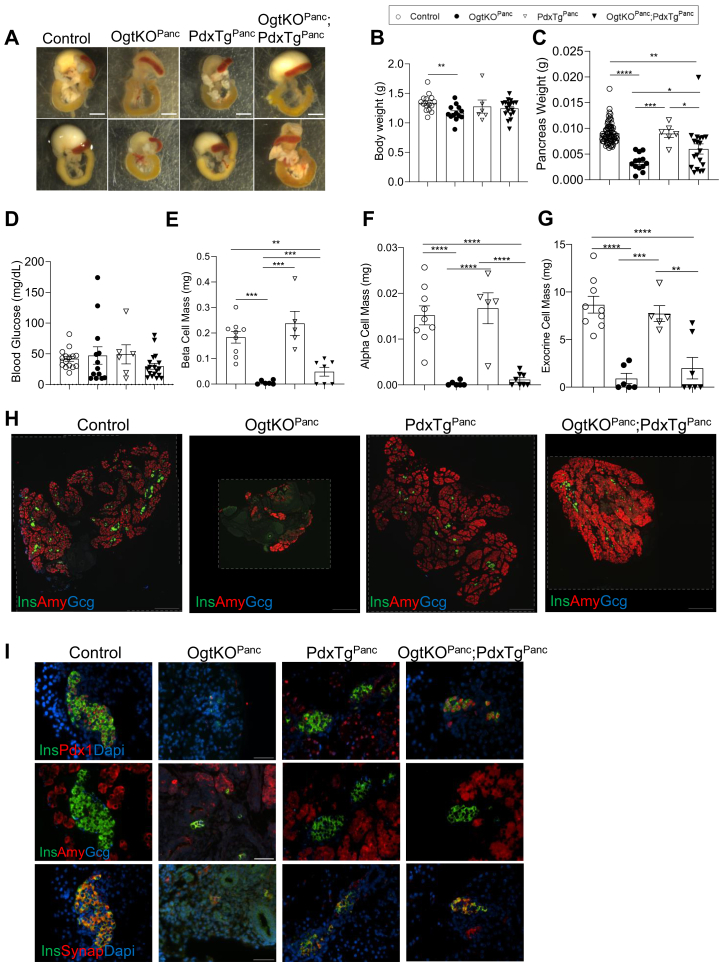
Pdx1 protein is reduced in both the β-cell–specific and pancreas-specific Ogt loss models (9, 17). During embryonic development, OgtKOPanc exhibit a reduced number of Pdx1-expressing pancreatic progenitor cells (9). The size of the pancreatic progenitor pool is known to limit ultimate pancreas size (20). Thus, we tested whether genetically reconstituting Pdx1 will rescue pancreas hypoplasia in the OgtKOPanc animals. PdxTgPanc mice express Pdx1 in the acinar tissue, confirming efficient expression of the transgene by Pdx1Cre (Fig. S2D). Overexpression of Pdx1, alone, does not affect pancreas morphology at birth (Fig. 4, A and H). Body weight, pancreas weight, and blood glucose, as well as endocrine and exocrine pancreas compartments were not different between Pdx1Tg compared to littermate controls (Fig. 4, B–I). Pancreatic Ogt loss results in reduced neonatal body weight, and concomitant overexpression of Pdx1 in the context Ogt loss did not rescue the reduction in body weight (Fig. 4B). However, overexpression of Pdx1 afforded a partial rescue in pancreas weight in the Ogt-deficient model (Figs 4C and S2E). Blood glucose levels were comparable among all groups (Fig. 4D). Overexpression of Pdx1 partially rescued β-cell mass in the Ogt-deficient pancreas but did not mitigate the reduction of α-cell mass or exocrine cell mass (Fig. 4, E–I).
Figure 4.
Constitutive expression of Pdx1 partially rescues pancreas weight and β-cell mass. Representative images of pancreas, stomach, spleen, and duodenum at birth showing varying degrees of pancreatic hypoplasia in OgtKOPanc and OgtKOPanc;Pdx1TgPanc (A, scale bar represents 250 pixels). Body weight (B, n = 6–19), pancreas weight (C, n = 6–73), random blood glucose (D, n = 6–18), β-cell mass (E, n = 5–9), α-cell mass (F, n = 5–9), and exocrine cell mass (G, n = 5–9) at birth. Representative images of whole pancreas (H, 10× magnification, scale bar represents 500 μm; areas outside dotted lines indicate space added from canvas size adjustment) and islets (I, 40× magnification, scale bar represents 50 μm; areas outside dotted lines indicate space added from canvas size adjustment). Statistics conducted using one-way ANOVA followed by unpaired Student t test with Welch’s correction, p ≤ 0.05 deemed significant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Discussion
In this study, we demonstrate the indispensable role of nutrient-driven O-GlcNAcylation in pancreas development. While hypoplasia in the Ogt-deficient pancreas is associated with perturbations of genes regulated upstream by p53, ER stress, and Pdx1, manipulation of any single one of these regulators is insufficient to fully ameliorate the hypoplasia.
The lack of phenotype in the p53KOPanc is consistent with previous evidence showing that the suppression of p53 expression via methylation of the regulatory region of the p53 gene is necessary to maintain pancreatic progenitor survival during the formation of the pancreas (11). P53 function, though important in suppressing carcinogenesis, is not required for normal mouse pancreas development (11, 23). Yamauchi et al., also reported that p53 deletion results in no differences in pancreatic islet morphology in adult mice (24).
In the Ogt-deficient pancreas, ablation of p53, whether homozygous or heterozygous, did not fully rescue pancreatic hypoplasia. This is not entirely surprising given the diverse and opposing ways in which p53 is known to affect metabolic pathways in response to stress, such as through the activation of apoptosis and cell-proliferation inhibitor p21 or decreasing reactive oxygen species (25). It is, however, unclear why heterozygous, but not homozygous, loss of p53 confers a partial rescue in the Ogt-deficient pancreas. Based on analysis of the DEGs, both cellular survival and production of reactive oxygen species were top canonical pathways implicated in the Ogt-deficient pancreas (9). It is possible that a total loss of p53 resulted in the cancellation of both prosurvival and proapoptotic effects, thus negating any protective effects. Additionally, in cancer cell lines, overexpression of either Ogt or OGA is known to upregulate p53 protein, suggesting that p53 is sensitive to the changes in O-GlcNAc homeostasis rather than absolute O-GlcNAc levels (26). Nonetheless, p53 activity in the OgtKOPanc and levels of apoptosis and cell proliferation in OgtKOPanc with heterozygous and homozygous loss of p53 loss warrants further exploration. Moreover, additional studies to define the molecular relationship of Ogt and p53 are warranted.
Like other cell types with high secretory demand, the pancreatic acinar is equipped with a substantial network of ER, whose folding capacity changes in accordance to the flux of incoming polypeptide chains (27, 28). Perturbation of the homeostasis between incoming unfolded polypeptides and the capacity of the ER to fold these proteins activate the Unfolded Protein Response (UPR) (28), a branch of which destines the cell for apoptosis via induction of CHOP (29). Numerous studies demonstrate that inhibition of CHOP in the germline or in the β-cells, specifically, has protective effects against β-cell failure and potentiating effects on insulin secretion (17, 30, 31, 32, 33, 34). Emerging evidence in β-cells has implicated the UPR itself as a sensor of secretory demand, in which below a certain level of ER stress, cells with an active UPR tend to proliferate and expand cell mass to compensate for increased secretory demand (35). Full body deletion of CHOP did not rescue the endocrine compartment of the Ogt-deficient pancreas, possibly indicating the ER stress was beyond the threshold for which the UPR could trigger a compensatory proliferation event. However, the full rescue in the exocrine compartment in the OgtKOPanc;CHOP−/− mice suggests that ER stress–mediated apoptosis is a major contributor only to hypoplasia in the exocrine compartment. Indeed, there is abundant evidence to suggest that the pancreatic acinar is particularly susceptible to ER stress through UPR or ER overload response (36).
Pancreas development is governed by a hierarchy of transcriptional activation. The pool of pancreatic progenitors (Pdx1+) is known to be a significant determinant of pancreas size at birth and is not mitigated by postnatal growth compensation (20). We have previously shown that deletion of Ogt in the developing pancreatic epithelium reduces the number of Pdx1+ cells in the developing pancreas as well as overall Pdx1 protein levels (9). In the β-cell, loss of Pdx1 is associated with increased susceptibility to ER stress and autophagy-mediated cell death (18). Increasing Pdx1 protein levels in the background of Ogt loss afforded a partial rescue in both pancreas weight and β-cell mass. As Ogt loss in the pancreatic β-cell is associated with increased ER stress–induced apoptosis, it is not entirely surprising that increasing Pdx1 partially ameliorates the effects of Ogt loss on β-cell mass in the OgtKOPanc (17).
Conclusion
In summary, we show that p53 and CHOP are not necessary for pancreas and islet development and that neither protein is the main regulator of pancreatic hypoplasia resulting from Ogt loss in the pancreatic progenitors. However, we demonstrate that the overexpression of Pdx1 in the pancreas can partially mitigate pancreatic development defects associated with the loss of Ogt. Altogether, these findings underscore the essential role of Ogt in pancreas development and highlight its multiple protein targets such as transcription factor Pdx1 to promote proper pancreas development.
Experimental procedures
Animals
Late Pdx1cre line (L-Pdx1cre) was provided by Dr Pedro Herrera (37). Ogtflox/flox, p53flox/flox, CHOP−/−, and CAG-EGFP reporter transgenic animals were purchased from Jackson laboratory. Expression of Pdx1-cre begins in the pancreatic epithelium at embryonic day 11.5 (38). Pdx-cre; Ogtflox/y (OgtKOPanc) males were crossed with Ogtflox/flox females to produce Ogtflox/flox or Ogtf/y littermate controls. P53KOPanc mice were generated in a similar manner. CAG-CAT-Pdx1 mice (PdxTg) were obtained from Yoshio Fujitani and bred into the Pdx-cre line (39). Briefly, Pdx1 is constitutively expressed under the CAG promotor when the floxed STOP cassette encoding CAT ahead of Pdx1 is removed during cre-recombination (39). All mice were generated on a C57Bl/6J background and group housed on a 14:10 light-dark cycle. Controls are both Pdxcre+ only and Pdxcre-. All experiments were performed with both male and female mice, and all procedures were approved by the University of Minnesota Institutional Animal Care and Use Committee (Protocol Number# 2106A39213).
Blood and tissue collection
Neonatal mice were sacrificed by decapitation at postnatal day 0. Whole blood and blood glucose were obtained via the trunk at sacrifice. Tissues were treated in 3.7% formaldehyde at 4 °C for 6 h before being transferred to 70% EtOH in preparation for paraffin embedding.
Serum insulin analysis
Serum obtained from neonatal trunk blood was assessed using the Rat or Mouse Ultrasensitive Insulin ELISA kit from Alpco Diagnostics following manufacturer’s instructions. Control and knockout conditions for each model were run on a single plate at the same time.
Tissue preparation and immunofluorescence staining
Neonatal pancreases were paraffin embedded and sectioned from top to bottom at 5 μm thickness. Deparaffination and tissue preparation for immunofluorescence (IF) imaging were done as previously described (40). Tissues were treated with primary antibodies against guinea pig insulin (Sigma, 1:10,000), mouse glucagon (Abcam, 1:500), rabbit amylase (Sigma, 1:200), and rabbit Pdx1 (Millipore or Abcam, 1:400) and incubated at 4 °C for 12 to 16 h. Secondary antibodies conjugated to FITC (1:400) and Cy3 (1:500) were used, and nuclei were stained with DAPI dip (1:1000).
Cell counts and morphometric analysis
All morphometric analyses and cell counts were conducted with ImageJ2 as previously described (40, 41). Fluorescent images for endocrine/exocrine cell mass were captured at 10× magnification and individual islet pictures at 20× and 40×, respectively with a Nikon Eclipse NI-E (Nikon Instruments) microscope equipped with a motorized stage, a Nikon TI-E Deconvolution Scope (Nikon Instruments) equipped with a motorized stage, or with the Keyence BZ-X series all-in-one fluorescence microscope (Keyence Corporation). Cell types and areas were identified by hormone-positive IF staining and normalized to total pancreatic area as previously described (9). Endocrine and exocrine cell mass calculations were performed using five and three pancreas sections, respectively, as previously described (9).
Statistical analysis
All values were expressed as mean ± s.e.m. Analyses were conducted in Prism (v.9.12) utilizing One-way ANOVA followed by student t test with Welch’s correction. Results were deemed significant when p ≤ 0.05 and trending when 0.05<p < 0.1.
Quantitative PCR
RNA was isolated using TRIzol. cDNA was synthesized from total RNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Relative gene expression was assessed on an Applied Sciences QuantStudio Flex 6 Real Time PCR System using Power SYBR Green (Applied Biosciences), according to the ΔΔCT method, normalized to β-actin housekeeping gene.
Data availability
All data are contained within this article and provided as supporting data.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Ms Lensa Ali for technical support. We also thank Dr Mark Sanders and Nadia Kane at the University of Minnesota Imaging Centers for providing training and access to the Nikon Ti-E Deconvolution Scope. Dr Alejandro is the guarantor of this work.
Author contributions
E. U. A. conceptualization; A. W., S. P., M. M., B. A., N. A., M. B., Y. F., and E. U. A. methodology; A. W., S. P., M. M., B. A., N. A., and E. U. A. software; A. W., S. P., M. M., B. A., N. A., M. B., Y. F., and E. U. A. validation; A. W., S. P., M. M., N. A., M. B., and E. U. A. formal analysis; A. W., S. P., M. M., B. A., N. A., M. B., and E. U. A. investigation; E. U. A. resources; A. W., S. P., B. A., and M. B. data curation; A. W. and E. U. A. writing–original draft; A. W., B. A., N. A., M. B., Y. F., and E. U. A. writing–review and editing; A. W., S. P., B. A., N. A., M. B., and E. U. A. visualization; E. U. A. supervision; E. U. A. project administration; E. U. A. funding acquisition.
Funding and additional information
The NIH funding was provided for E. U. A. (R01DK115720 and Regenerative Medicine Minnesota), University of Minnesota Foundation, McKnight Foundation, and the UMN Genomics Center. A. W. was supported by T32GM140936, S. P. and M. B. were supported by T32DK007203, and M. M. was supported by T32DK108733. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Robert Haltiwanger
Supporting information
S. Fig 1: Programmed Cell Death Pathways Associated with Upstream Regulators of Differentially Expressed Genes in OgtKOPanc. Top 30 regulatory pathways identified by gene ontology analysis of upstream regulators of differentially expressed genes in OgtKOPanc (A). Analysis performed using GOTERM BP FAT in David Knowledgebase (42).
S. Fig 2: Pancreas Weight Differences Among Genotypes is Similar when Pancreas Weight is Corrected Over Body weight. Pancreas weight corrected over body weight for pancreas Ogt and p53 co-deficient (A, n = 5-63), pancreas p53-deficient (B, n = 10-32), and pancreas Ogt and whole body CHOP-deficient mice (C, n = 7-27). Representative images of islets and acinar in control and PdxTgPanc showing constitutive expression of Pdx1 in the acinar tissue (D, 40x magnification, scale = 50 μm). Pancreas weight corrected over body weight for pancreas Ogt and PdxTg (E, n = 4-20). Statistics conducted using one-way ANOVA followed by unpaired Student t-test with Welch’s correction, p ≤ 0.05 deemed significant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
References
- 1.Marshall S., Bacote V., Traxinger R.R. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J. Biol. Chem. 1991;266:4706–4712. [PubMed] [Google Scholar]
- 2.Akella N.M., Ciraku L., Reginato M.J. Fueling the fire: emerging role of the hexosamine biosynthetic pathway in cancer. BMC Biol. 2019;17:52. doi: 10.1186/s12915-019-0671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bond M.R., Hanover J.A. A little sugar goes a long way: the cell biology of O-GlcNAc. J. Cell Biol. 2015;208:869–880. doi: 10.1083/jcb.201501101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varki A., Cummings R.D., Esko J.D., Freeze H.H., Stanley P., Bertozzi C.R., et al. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2009. Essentials of Glycobiology. [PubMed] [Google Scholar]
- 5.Ruan H.B., Singh J.P., Li M.D., Wu J., Yang X. Cracking the O-GlcNAc code in metabolism. Trends Endocrinol. Metab. 2013;24:301–309. doi: 10.1016/j.tem.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohan R., Jo S., Lockridge A., Ferrington D.A., Murray K., Eschenlauer A., et al. OGT regulates mitochondrial biogenesis and function via diabetes susceptibility gene Pdx1. Diabetes. 2021;70:2608–2625. doi: 10.2337/db21-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrali S.S., Qian Q., Ozcan S. Glucose mediates the translocation of NeuroD1 by O-linked glycosylation. J. Biol. Chem. 2007;282:15589–15596. doi: 10.1074/jbc.M701762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Y., Miyazaki J., Hart G.W. The transcription factor PDX-1 is post-translationally modified by O-linked N-acetylglucosamine and this modification is correlated with its DNA binding activity and insulin secretion in min6 beta-cells. Arch. Biochem. Biophys. 2003;415:155–163. doi: 10.1016/s0003-9861(03)00234-0. [DOI] [PubMed] [Google Scholar]
- 9.Baumann D., Wong A., Akhaphong B., Jo S., Pritchard S., Mohan R., et al. Role of nutrient-driven O-GlcNAc-post-translational modification in pancreatic exocrine and endocrine islet development. Development. 2020;147 doi: 10.1242/dev.186643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akimoto Y., Hart G.W., Wells L., Vosseller K., Yamamoto K., Munetomo E., et al. Elevation of the post-translational modification of proteins by O-linked N-acetylglucosamine leads to deterioration of the glucose-stimulated insulin secretion in the pancreas of diabetic Goto-Kakizaki rats. Glycobiology. 2007;17:127–140. doi: 10.1093/glycob/cwl067. [DOI] [PubMed] [Google Scholar]
- 11.Georgia S., Kanji M., Bhushan A. DNMT1 represses p53 to maintain progenitor cell survival during pancreatic organogenesis. Genes Dev. 2013;27:372–377. doi: 10.1101/gad.207001.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oyadomari S., Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11:381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 13.Hu H., Tian M., Ding C., Yu S. The C/EBP homologous protein (CHOP) transcription factor functions in endoplasmic reticulum stress-induced apoptosis and microbial infection. Front. Immunol. 2018;9:3083. doi: 10.3389/fimmu.2018.03083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nam D.H., Han J.H., Lee T.J., Shishido T., Lim J.H., Kim G.Y., et al. CHOP deficiency prevents methylglyoxal-induced myocyte apoptosis and cardiac dysfunction. J. Mol. Cell Cardiol. 2015;85:168–177. doi: 10.1016/j.yjmcc.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 15.Araki E., Oyadomari S., Mori M. Impact of endoplasmic reticulum stress pathway on pancreatic beta-cells and diabetes mellitus. Exp. Biol. Med. (Maywood) 2003;228:1213–1217. doi: 10.1177/153537020322801018. [DOI] [PubMed] [Google Scholar]
- 16.Fonseca S.G., Gromada J., Urano F. Endoplasmic reticulum stress and pancreatic β-cell death. Trends Endocrinol. Metab. 2011;22:266–274. doi: 10.1016/j.tem.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alejandro E.U., Bozadjieva N., Kumusoglu D., Abdulhamid S., Levine H., Haataja L., et al. Disruption of O-linked N-acetylglucosamine signaling induces ER stress and β cell failure. Cell Rep. 2015;13:2527–2538. doi: 10.1016/j.celrep.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujimoto K., Polonsky K.S. Pdx1 and other factors that regulate pancreatic beta-cell survival. Diabetes Obes. Metab. 2009;11:30–37. doi: 10.1111/j.1463-1326.2009.01121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Y., Liu Q., Zhou Z., Ikeda Y. PDX1, neurogenin-3, and MAFA: critical transcription regulators for beta cell development and regeneration. Stem Cell Res. Ther. 2017;8:240. doi: 10.1186/s13287-017-0694-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanger B.Z., Tanaka A.J., Melton D.A. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature. 2007;445:886–891. doi: 10.1038/nature05537. [DOI] [PubMed] [Google Scholar]
- 21.Yang W.H., Kim J.E., Nam H.W., Ju J.W., Kim H.S., Kim Y.S., et al. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 2006;8:1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- 22.Gannon M., Herrera P.L., Wright C.V. Mosaic Cre-mediated recombination in pancreas using the pdx-1 enhancer/promoter. Genesis. 2000;26:143–144. doi: 10.1002/(sici)1526-968x(200002)26:2<143::aid-gene13>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 23.Jacks T., Remington L., Williams B.O., Schmitt E.M., Halachmi S., Bronson R.T., et al. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 24.Yamauchi Y., Kodama Y., Shiokawa M., Kakiuchi N., Marui S., Kuwada T., et al. Rb and p53 execute distinct roles in the development of pancreatic neuroendocrine tumors. Cancer Res. 2020;80:3620–3630. doi: 10.1158/0008-5472.CAN-19-2232. [DOI] [PubMed] [Google Scholar]
- 25.Vousden K.H., Lane D.P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 26.de Queiroz R.M., Madan R., Chien J., Dias W.B., Slawson C. Changes in O-linked N-acetylglucosamine (O-GlcNAc) homeostasis activate the p53 pathway in ovarian cancer cells. J. Biol. Chem. 2016;291:18897–18914. doi: 10.1074/jbc.M116.734533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Case R.M. Synthesis, intracellular transport and discharge of exportable proteins in the pancreatic acinar cell and other cells. Biol. Rev. Camb Philos. Soc. 1978;53:211–354. doi: 10.1111/j.1469-185x.1978.tb01437.x. [DOI] [PubMed] [Google Scholar]
- 28.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 29.Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R.T., Remotti H., et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song B., Scheuner D., Ron D., Pennathur S., Kaufman R.J. Chop deletion reduces oxidative stress, improves beta cell function, and promotes cell survival in multiple mouse models of diabetes. J. Clin. Invest. 2008;118:3378–3389. doi: 10.1172/JCI34587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu H., Peng L., Jiang H., Shen H., Zhou P., Gao Y. Silenced CHOP protects pancreatic B-cell function by targeting peroxisome proliferator-activated receptor-γ coactivator-1α through nuclear factor-κB signaling pathway in diabetes mellitus. J. Cell Biochem. 2019;120:12595–12603. doi: 10.1002/jcb.28526. [DOI] [PubMed] [Google Scholar]
- 32.Oyadomari S., Koizumi A., Takeda K., Gotoh T., Akira S., Araki E., et al. Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J. Clin. Invest. 2002;109:525–532. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maris M., Overbergh L., Gysemans C., Waget A., Cardozo A.K., Verdrengh E., et al. Deletion of C/EBP homologous protein (Chop) in C57Bl/6 mice dissociates obesity from insulin resistance. Diabetologia. 2012;55:1167–1178. doi: 10.1007/s00125-011-2427-7. [DOI] [PubMed] [Google Scholar]
- 34.Yong J., Parekh V.S., Reilly S.M., Nayak J., Chen Z., Lebeaupin C., et al. Chop/Ddit3 depletion in β cells alleviates ER stress and corrects hepatic steatosis in mice. Sci. Transl. Med. 2021;13:604. doi: 10.1126/scitranslmed.aba9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma R.B., O'Donnell A.C., Stamateris R.E., Ha B., McCloskey K.M., Reynolds P.R., et al. Insulin demand regulates β cell number via the unfolded protein response. J. Clin. Invest. 2015;125:3831–3846. doi: 10.1172/JCI79264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Logsdon C.D., Ji B. The role of protein synthesis and digestive enzymes in acinar cell injury. Nat. Rev. Gastroenterol. Hepatol. 2013;10:362–370. doi: 10.1038/nrgastro.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herrera P.L. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127:2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- 38.Heiser P.W., Lau J., Taketo M.M., Herrera P.L., Hebrok M. Stabilization of beta-catenin impacts pancreas growth. Development. 2006;133:2023–2032. doi: 10.1242/dev.02366. [DOI] [PubMed] [Google Scholar]
- 39.Miyatsuka T., Kaneto H., Kajimoto Y., Hirota S., Arakawa Y., Fujitani Y., et al. Ectopically expressed PDX-1 in liver initiates endocrine and exocrine pancreas differentiation but causes dysmorphogenesis. Biochem. Biophys. Res. Commun. 2003;310:1017–1025. doi: 10.1016/j.bbrc.2003.09.108. [DOI] [PubMed] [Google Scholar]
- 40.Akhaphong B., Lockridge A., Jo S., Mohan R., Wilcox J.A., Wing C.R., et al. Reduced uterine perfusion pressure causes loss of pancreatic β-cell area but normal function in fetal rat offspring. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2018;315:R1220–R1231. doi: 10.1152/ajpregu.00458.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rueden C.T., Schindelin J., Hiner M.C., DeZonia B.E., Walter A.E., Arena E.T., et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017;18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherman B.T., Hao M., Qiu J., Jiao X., Baseler M.W., Lane H.C., et al. David: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucl. Acids Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S. Fig 1: Programmed Cell Death Pathways Associated with Upstream Regulators of Differentially Expressed Genes in OgtKOPanc. Top 30 regulatory pathways identified by gene ontology analysis of upstream regulators of differentially expressed genes in OgtKOPanc (A). Analysis performed using GOTERM BP FAT in David Knowledgebase (42).
S. Fig 2: Pancreas Weight Differences Among Genotypes is Similar when Pancreas Weight is Corrected Over Body weight. Pancreas weight corrected over body weight for pancreas Ogt and p53 co-deficient (A, n = 5-63), pancreas p53-deficient (B, n = 10-32), and pancreas Ogt and whole body CHOP-deficient mice (C, n = 7-27). Representative images of islets and acinar in control and PdxTgPanc showing constitutive expression of Pdx1 in the acinar tissue (D, 40x magnification, scale = 50 μm). Pancreas weight corrected over body weight for pancreas Ogt and PdxTg (E, n = 4-20). Statistics conducted using one-way ANOVA followed by unpaired Student t-test with Welch’s correction, p ≤ 0.05 deemed significant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Data Availability Statement
All data are contained within this article and provided as supporting data.