Abstract
Tuberculosis (TB) in one of the dreadful diseases present globally. This is caused by Mycobacterium tuberculosis. Mycobacterium tuberculosis dethiobiotin synthetase (MtDTBS) is an essential enzyme in biotin biosynthesis and is an ideal target to design and develop novel inhibitors. In order to effectively combat this disease six natural compound (butein) analogues were subjected to molecular docking to determine their binding mode and the binding affinities. The resultant complex structures were subjected to 500 ns simulation run to estimate their binding stabilities using GROMACS. The molecular dynamics simulation studies provided essential evidence that the systems were stable during the progression of 500 ns simulation run. The root mean square deviation (RMSD) of all the systems was found to be below 0.3 nm stating that the systems are well converged. The radius of gyration (Rg) profiles indicated that the systems were highly compact without any major fluctuations. The principle component analysis (PCA) and Gibbs energy landscape studies have revealed that the comp3, comp5 and comp11 systems navigated marginally through the PC2. The intermolecular interactions have further demonstrated that all the compounds have displayed key residue interactions, firmly holding the ligands at the binding pocket. The residue Lys37 was found consistently to interact with all the ligands highlighting its potential role in inhibiting the MtDTBS. Our investigation further put forth two novel compounds (comp10 and comp11) as putative antituberculosis agents. Collectively, we propose six compounds has plausible inhibitors to curtail TB and further can act as scaffolds in designing new compounds.
Keywords: Tuberculosis, Natural compounds, Molecular dynamics simulation, Mycobacterium tuberculosis dethiobiotin Synthetase, Butein analogues
1. Introduction
One of the ancient infections that has been invading the mankind is the tuberculosis (TB) for nearly about 4000 years. Characteristically, the primary site of TB infection is lungs but can also be seen to infect brain, intestines, kidneys, or the spine [1]. Mycobacterium tuberculosis is the causative agent of this disease and spreads through air [1]. According to world health organization (WHO), 1.5 million deaths have occurred due to TB in 2020. This is second to COVID-19 in infectious deaths and 13th leading causes of mortalities (https://www.who.int/news-room/fact-sheets/detail/tuberculosis). These facts direct towards the urgency of finding potential drugs that could reverse the condition.
Due to the slow growth and dormancy of some of the bacilli, multitude of reports illuminate on treatments that should be a prolonged or the multidrug therapeutic approach to get rid of the bacteria from the pulmonary and extrapulmonary sites [2]. Besides this, the chemical features also contribute to the success of the treatment regime. It is reported that the drugs in combination can curtail the likelihood of failure and relapse [2]. Furthermore, two effective drugs are to be prescribed to counter the appearance of resistant strains [2].
Several drug have been reported to counter TB and WHO has classified the antiTB drugs into five classes [2,3]. The drug-susceptible tuberculosis agents are grouped into first class and the drugs with uncertain or limited efficacy are grouped into fifth class [2,3], with a possibility of a new group [4]. The drugs namely rifampicin was licensed to administer for humans in 1963. Later bedaquiline and delamanid, have received approval to be used against multi-drug resistant (MDR)-TB [5]. Due to their association with side effects, these drugs are prescribed to be used when no other treatment option is available [5].
In the current study, for discovering the potential inhibitors against MtDTBS, we have used various in silico methods such as molecular docking, molecular dynamics simulation (MDS) and essential dynamics. These studies have proven to be crucial in swiftly identifying the plausible inhibitors [[6], [7], [8], [9], [10], [11], [12]]. Particularly, a few studies report the use of in silco methods in identifying potential inhibitors for Mt. Mallavarapu et al., identified the inhibitors for Mt MraY (Rv2156c). The target model was built using the homology modelling technique and was then complexed with antibiotic muraymycin D2 (MD2) [13]. Computational methods were applied to discover potential candidates from Asinex database depending on the binding modes and interactions with Mt MraY-MD2 [13]. Singh et al., used pharmacophore-based screening, molecular docking and MDS methods to retrieve potential hits against MtPknG [14]. In vitro evaluation was performed on the identified hits and eventually recommended compound NRB04248 as the potential inhibitor [14]. Sundar et al., used computational drug repurposing approaches to retrieve plausible small molecules against Mt Protein kinase A (PknA) [15]. Pitaloka et al., discovered possible drugs against Enoyl-[acyl-carrier-protein] reductase (InhA) of Mt adapting computational techniques [16]. Another group has identified prospective inhibitors via in silico methods that included structure-based drug design [17].
Nature products have been pivotal in treating TB that includes peptide actinomycin and rapamycin. Several natural products have displayed superior pharmacokinetic properties than the synthetic compounds [18]. In general, there has been a great impact of natural products in the field of medicine and have served as the only means to treat injuries and diseases [19]. A research group has identified natural products isobavachalcone and isoneorautenol and a synthetic chromene as antimycobacterial agents [20]. Another study has reported flavonoids as potential agents [21]. A study has reported the use of marine natural products to selectively kill dormant Mycobacterium tuberculosis [22].
A well-known flavonoid, butein (2′,3,4,4′-tetrahydroxychalcone) is highly regarded for its wide therapeutic potential [23] and widely studied as an anticancer agent [[24], [25], [26], [27]]. In a study, butein is reported to inhibit Mycobacterium bovis BCG [28] and has demonstrated anti-mycobacterial activity [29], however no findings are yet published with respect to the butein analogues for MtDTBS target. Therefore, in the current study, we have used the butein analogues that are available in the lab to determine its computational efficacy against Mycobacterium tuberculosis dethiobiotin synthetase (MtDTBS). This enzyme participates in the biosynthesis of biotin [30] and is a promising target to design new drug candidates [30].
2. Materials and methods
2.1. Selection of the protein
The protein for the current study is MtDTBS (PDB 6CZD). This is an X-ray crystal structure with 2.40 Å resolution [31]. The structure was prepared by using ‘prepare protein’ module obtainable in the discovery studio v16 (DS). This protocol prepares the input proteins and executes actions such as inserting missing atoms in the incomplete residues [32,33], modelling the missing loop regions, deletion of the alternate conformations, dislodges the water molecules, standardizing the atom names, and protonating titratable residues using predicted pKs. The active site of the target was selected around the cocrystallised ligand for all the residue atoms that fall in the radius of 7.9 Å that includes, Thr11, Gly12, Val13, Gly14, Lys15, Thr16, Lys37, Thr41, Asp49, Pro71, Met72, Ala73, Pro74, Glu108, Ala110, Gly111 and Val115. The XYZ coordinates for the binding site sphere are taken as 33.497189 Å, 22.149442 Å, and 33.777841 Å, respectively. To evaluate and validate the docking algorithm, the inbound cocrystallised ligand was docked into the X-ray structure of the target. The redocked binding mode and the native cocrystallised ligand binding mode were found to be similar. The RMSD between the native ligand and the redocked pose was observed to be 0.81 Å (Supplementary Figure 1).
2.2. Selection of the ligands
The six small molecules used for the current study are available in the lab. The three compounds are earlier published from our group [7]. These small molecules were prepared by using the ‘full minimization’ module available with the DS. The CHARMm forcefield was applied with the Smart Minimizer algorithm. This is performed by 1000 steps of steepest descent with an RMS gradient tolerance of 3, subsequently followed by conjugate gradient minimization. The well-prepared protein and ligands were upgraded to molecular docking to understand the binding affinities between them.
2.3. Molecular docking to assess the binding affinity
To elucidate the predictive binding mode and to assess the binding affinity of the selected compounds towards the Mycobacterium tuberculosis, the CDOCKER available with the discovery studio v18 (DS) was used. In the current study six compounds, three from the previous study were also included [7]. The CDOCKER docking method used here is a grid-based molecular docking method that uses CHARMm. In here, the receptor is fixed in contrast to the ligands that are allowed to move during the refinement. Random conformations of the ligands are then generated. The conformations are correspondingly translated into the binding site. The poses are then generated using random rigid-body rotations followed by simulated annealing. A final minimization is then used to refine the ligand poses. Finally, the molecular docking score is evaluated according to the –CDOCKER interaction energy expressed in kcal/mol. Each ligand was allowed to generate 50 poses. Each pose was ranked from the largest cluster, with good dock score and interactions with the key residues. These complexes were upgraded to molecular dynamics simulation (MDS) to gain insights on the complex stabilities and intermolecular interactions.
2.4. Molecular dynamics simulation studies
To vividly understand the nature of small molecules at the binding pocket of the protein, the molecular dynamics simulations (MDS) were performed using the GROMACS v2016.6 [34]. The main reason for adapting the MDS is to evaluate the binding stabilities of the protein-ligand complex and to understand the interactions at the atomistic level as reported earlier [35,36]. To accomplish this, the topologies of the ligand were generated from SwissParam [37] while using the CHARMM27 all atom force field. The dodecahedron water box was generated and solvated with TIP3P water model and subsequently, the counter ions (Na+) were supplemented (Supplementary Table 1). The minimization of the system was done. The protein and the ligand were coupled and two step equilibration was done with conserved number of particles (N), system volume (V) and temperature (T) (NVT) and the constant number of particles (N), system pressure (P) and temperature (T) (NPT) for 1 ns each with V-rescale thermostat. The NPT ensembles were upgraded to MDS for 500 ns. During the equilibration process, the protein backbone was restrained. The Parrinello-Rahman barostat [38] was used to monitor the pressure of the system. The LINCS algorithm was used to maintain the geometry of the molecules that constraints the bond length [39]. Long-range electrostatic interactions were computed with Particle Mesh Ewald (PME) technique [40]. The results were analyzed using the visual molecular dynamics (VMD) [41] and DS and studied according to the root mean square deviation (RMSD), root mean square fluctuations (RMSF), radius of gyration (Rg) [42], number of hydrogen bonds, and the mode of ligand binding. Here, the protein backbone was analyzed. Furthermore, the PCA analysis and essential dynamics (ED) was conducted using the gmx covar and gmx sham to understand the dynamic motion of the protein.
3. Results
3.1. Binding affinity studies
The binding affinity of the selected molecules was conducted adapting the CDOCKER module available with the DS. The results have shown that all the molecules have occupied the defined binding pocket by good dock score (Table 1). It was also noted that several key residues have adhered with the compounds to accommodate them at the binding pocket firmly.
Table 1.
Binding affinity and intermolecular interactions.
| Comp no | Dock scorea | Hydrogen | π interactions | Carbon hydrogen | Attractive charges | van der Waals |
|---|---|---|---|---|---|---|
| 1 | 51.4398 | Lys15, Glu52 | – | – | Lys15, Asp49 | Thr16, Lys37, Glu108, Gly109, Gly111 |
| 2 | 47.7706 | Glu52,Glu108 | Asp48 | – | – | Lys15, Thr16, Lys37,Asp47, Asp49, Ala110 |
| 3 | 44.6003 | Thr16, Asp48 | – | Asp47 | – | Lys15, Lys37, Gln40, Asp49, Ala110, Gly111 |
| 5 | 40.0093 | Thr41 | Met72, Ala73, Pro74, Ala110 | Pro71 | – | Thr11, Lys37, Gln40, Gly42, Asp47, Gly111,Val115 |
| 10 | 45.6705 | Thr16, Asp47, Glu52 | Asp48 | – | – | Lys37,Thr41, Gly42, Asp49, Arg55,Pro71 |
| 11 | 42.5373 | Thr16, Asp49, Glu52, Glu108 | Ala110 | – | – | Thr11, Lys37,Gln40, Thr41, Asp47, Asp48, Ala73, Pro74 |
-CDOCKER interaction energy expressed in kcal/mol.
3.2. Molecular dynamics simulation studies
The MDS studies elaborate on the stability of the protein ligand complexes formed during the molecular docking. The top ranked poses from the molecular docking were forwarded to MDS and the results were analyzed according to RMSD, RMFS, Rg, binding mode and hydrogen bond number.
3.3. Stability analysis by RMSD
RMSD records the deviations present between the initial structure and the final structure of the simulation run. Generally, a smaller deviation denotes a relatively stable structure [43,44]. In the current study, the RMSD for the six systems has projected to be stable below 0.3 nm. During the evolution of the simulation, no abnormal behavior was noticed among the six systems. Comp1 and comp11 demonstrated marginal deviations in the plots. Comp1 showed a minor surge at around 300 ns with an average of 0.15 nm, while the comp11 displayed a surge between 160 ns and 230 ns and has remained stable thereafter, with an average of 0.13 nm. The average RMSD for Comp2 is 0.14 nm, comp3 is 0.14 nm, comp5 is 0.11 nm and comp10 is 0.13 nm, respectively. These results state that the systems were stable during 500 ns simulation run representing the RMSD values below 0.3 nm (Fig. 1A).
Fig. 1.
MDS analysis of six systems conducted for 500 ns. A) RMSD profiles. B) Rg plots. C) RMSF profiles. The residues that showed fluctuations are marked in orange box. D) number of hydrogen bonds during the MD run.
3.4. The protein backbones were compact during the simulations
The Rg plots elucidates on the compactness of the protein backbone. The Rg plots for all the systems were found to between 1.64–1.74 nm. This implies that smaller the reading highly compact is the structure. Similar to the RMSD profiles, the comp11 has shown a negligible peak between 160 ns and 230 ns and a clear plateau was noticed later. The other systems were highly compact with an average of 1.68 nm, 1.66 nm, 1.68 nm, 1.66 nm, 1.67 nm and 1.67 nm, respectively for comp1, comp2, comp3, comp5, comp 10 and comp11. These results suggest that the compounds were highly compact during the evolution of the simulation run (Fig. 1B).
3.5. Residue specific fluctuations
The residue specific fluctuations were determined according to the RMSF. The results have shown that the protein backbone has displayed minimum fluctuations except for Arg45 and Asp173. These residues are at a distance from the binding pocket and therefore we speculate that their impact might be negligible on the binding potential. While the key residues that interact with the ligands were relatively stable (Fig. 1C).
3.6. Hydrogen bond interactions
During the course of the simulation run, the presence of the hydrogen bonds were meticulously monitored. It was noticed that the hydrogen bonds were formed during the entire simulation run. Notably, the number was higher until 250 ns for all the complexes (Fig. 1D). At least one hydrogen bond was present during the entire simulation (Fig. 1D).
3.7. Binding mode analysis
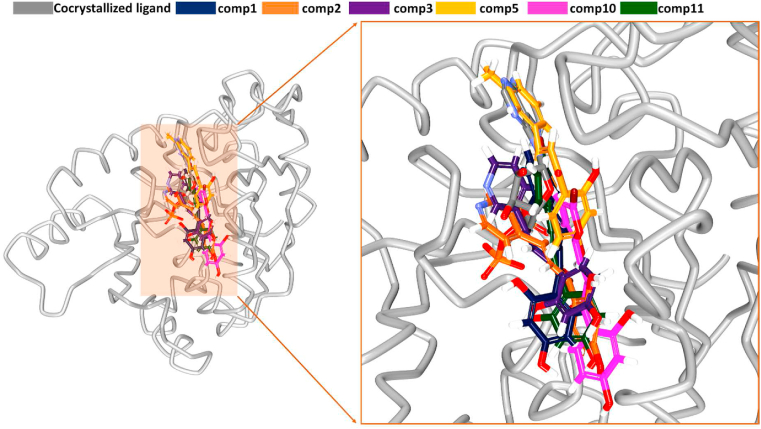
From the stable RMSD, the representative structures from last 50,000 ps were extracted and were superimposed onto the X-ray structure using the ‘align structures’ module available on DS. The results have shown that the small molecules have occupied the same position as that of the cocrystallised ligand (Fig. 2). Several key residues have positioned the ligands at the binding pocket of the target.
Fig. 2.
Accommodation of the small molecules at the binding pocket of the protein. Left panel indicates the protein and the ligands at the binding pocket and the right panel indicates its zoomed version.
3.8. Intermolecular interactions
Comp1 has formed hydrogen bond interactions with Lys15 and Glu52 residues by an acceptable bond length. Additionally, the residues Lys15, Asp49 have bound to the ligand via the attractive charges. The residues Thr16, Lys37, Glu108, Gly109 and Gly111 have interacted by van der Waals interactions holding the ligand at the binding pocket of the target (Fig. 3A, Fig. 4A and Table 1).
Fig. 3.
Intermolecular interactions between the protein and the ligands. A) comp1, B) comp2, C) comp3, D) comp5, E) comp10, F) comp11.
Fig. 4.
Intermolecular 2D interactions between the protein and the ligands. A) comp1, B) comp2, C) comp3, D) comp5, E) comp10, F) comp11.
Comp2 compound has generated hydrogen bond interactions with Glu52, Glu108 and a π-alkyl interaction with the residue Asp48. The residues Lys15, Thr16, Lys37, Asp47, Asp49 and Ala110 have prompted van der Waals interactions accommodating the ligand at the binding pocket of the target (Figs. 3B and 4B and Table 1).
Comp3 has formed hydrogen bond interactions with the residues Thr16, Asp48 and a carbon hydrogen bond with the residue Asp47. The residues Lys15, Lys37, Gln40, Asp49, Ala110 and Gly111 have formed van der Waals interactions accommodating the ligand at the binding pocket of the target protein (Figs. 3C and 4C and Table 1).
Comp5 has formed a two hydrogen bond interaction Thr41. The residues Ala73, Pro74 and Ala110 have formed π-alkyl interaction and the residue Met72 has prompted an amide π stacked interaction with the ligand. Additionally a carbon hydrogen bond was noticed with Pro71. The residues Thr11, Lys37, Gln40, Gly42, Asp47, Gly111 and Val115 have formed van der Waals interactions adhering the ligand at the binding pocket (Figs. 3D and 4D and Table 1).
Comp10 has generated hydrogen bond interactions with Thr16, Asp47 and Glu52 residues. An amide π stacked interaction was noticed with Asp48 residue. The residues Lys37, Thr41, Gly42, Asp49, Arg55 and Pro71 have held the ligand firmly by van der Waals interactions (Figs. 3E and 4E and Table 1).
Comp11 has formed hydrogen bonds with the residues Thr16, Asp49, Glu52, Glu108 and a π alkyl interaction with the residue Ala110. The residues Thr11, Lys37, Gln40, Thr41, Asp47, Asp48, Ala73 and Pro74 have communicated with the ligand via the van der Waals interaction to efficiently secure the ligand at the binding pocket of the protein (Figs. 3F and 4F and Table 1). The comprehensive 2D interactions between the target and the ligands are given in Fig. 4.
On focusing on the interactions between Mg ion and the ligands, it was observed that all the ligands have formed interaction with the Mg ion. Comp1 has interacted via the attractive charge. Comp2, comp3 and comp11 have formed the metal-acceptor interaction, while comp5 and comp10 have prompted the van der Waals interaction (Fig. 4). A similar attractive charge was noticed between the Mg ion and the native inbound ligand and the docked pose (Supplementary Figure 2B and 2C). These interactions have also stabilized the ligands to be placed at the binding pocket of the protein.
3.9. Protein ligand interaction energy
We further estimated the interaction energy between the protein and ligand in terms of short-range Lennard-Jones energy (LJ-SR). The LJ-SR of all the complexes ranges to −140 kJ/mol. The average LJ-SR of comp1 is −61.80 kJ/mol, comp2 is −45.85 kJ/mol, comp3 is −54.18 kJ/mol, comp5 is −73.93 kJ/mol, comp10 is −55.36 kJ/mol and comp11 is −55.49 kJ/mol, respectively (Fig. 5). Comparatively, the cocrystallised ligand demonstrated a better interaction energy of −108.93 kJ/mol (Supplementary Figure 2A).
Fig. 5.
LJ-SR interaction energy between the target and the ligands.
4. PCA and ED
Essential dynamics plays a key role in understanding the overall motion of the proteins in the conformational spaces during the course of the simulation run [[44], [45], [46]]. Logically, a fair amount of flexibility and rigidity is necessary for a given protein to be functional [44,45]. More particularly, this may be required at the binding pocket. Here, we have studies the conformational changes of the protein backbone from the principle component analysis (PCA) [[44], [45], [46]]. The PCA findings have shown that the systems were largely stable (Fig. 6). The cocrystal, comp1, comp2 and comp10 systems was highly stable (Fig. 6A, 6B, 6C and 6F) showing that the systems were well converged. However, comp3, comp5 and comp11 systems have navigated along the PC2 before attaining the equilibrium (Fig. 6D, 6E and 6G). This pattern is in line with the RMSD and the Rg profiles (Fig. 1). Upon viewing at the Gibbs free energy landscapes a varied pattern was observed. Expect for comp2 (Fig. 7C) and comp5 (Fig. 7E) systems all the other systems (Fig. 7A, 7B, 7D, 7F and 7G) have displayed two energy minima during the process of attaining the equilibrium state (Fig. 7). From the Gibbs plots it can be revealed that the red spots represent the favourable conformations while the dark blue spots indicate the unfavourable conformations (Fig. 7).
Fig. 6.
Principle component analysis (PCA) of six systems in comparison with the cocrystallised ligand system.
Fig. 7.
Gibbs energy landscape of six systems in comparison with the cocrystallised ligand system.
5. Discussion
To find effective therapeutics against the causative agent of TB, the computational framework was undertaken. Computer-aided drug discovery/design (CADD) approaches have paved the way in the progress of discovering, expanding and retrieving the small molecules with therapeutic potential [47]. CADD methods have been instrumental in identifying inhibitors against a varied range of diseases [47].
Natural products/analogues have immensely contributed to the field of pharmacotherapy, in particular to treat cancer and infectious diseases [48]. These compounds offer several advantages and are enriched with medicinal values [48]. Flavonoids occupy a special place in providing medicinal formulation to various diseases [49]. These contain the phenolic group and are present in vegetables, bark, stems, roots, fruits, tea, flowers and wine [50]. One such flavonoid, butein has been extensively studies for its therapeutic potential against cancer [[51], [52], [53]] along with anti-inflammatory and antioxidant properties [24]. In the current study, we have designed the butein compounds computationally and have used it against TB target.
Six compounds were used against the validated target MtDTBS. Out of the six compounds, two compounds are novel and have shown anticancer activity in our previous study [7]. The target structure chosen for the current study is complexed with adenosine-5′-diphosphate and Mg ion. The binding pocket can be divided into the nucleoside-binding pocket and the phosphate-binding loop [31]. The compounds used in the study have inclined more towards the nucleoside-binding pocket. These compounds have interacted with the key residue Lys37 via the van der Waals interaction. The interaction with this residue was reported in previous literature [31]. Several other interactions present within the crystal structure were also noted with the selected compounds. The MSD studies conducted for 500 ns have shown that the systems were stable throughout the simulations displaying the stable profiles, read according to RMSD, RMSF and Rg.
Additionally, the modified Butein compounds have shown a similar MDS results as that of the cocrystallised ligand. The RMSD profiles of all the modified butein systems were below 0.3 nm, with an average between 0.11 nm and 0.15 nm (Fig. 1A), while the cocrystallised ligand has demonstrated an average RMSD of 0.09 nm without any deflection during the entire simulation run (Supplementary Figure 3A). The Rg profiles of the modified systems ranged between 1.63 and 1.72 nm (Fig. 1B) while the cocrystallised ligand was relatively compact (Supplementary Figure 3B). The RMSF plots of the modified butein systems were recorded to be ranging between 0.09 nm to 0.5 nm, respectively (Fig. 1C), while that of the cocrystallised ligand displayed an average RMSF of 0.15 nm (Supplementary Figure 3C). Interestingly, the RMSF of the cocrystallised ligand complex had shown lower fluctuations as compared to the modified compounds including the residues, Arg45, Asp173 (Supplementary Figure 3C).
A noteworthy observation was noticed in the number of hydrogen bond interactions. The cocrystallised ligand had shown higher number of hydrogen bonds (Supplementary Figure 3D) than the modified compounds (Fig. 1D). This may be due to the larger size of the cocrystallised ligand than the modified compounds. Overall, the results of the modified compounds were comparable to cocrystallised ligand.
We further analyzed to know if any deflections have occurred between the docked pose and the MD derived pose by initiating the ‘align structures’ module available on the DS. This utilizes the 3DMA program to align the protein sequences in accordance with the 3D structural similarity. This algorithm identifies the segments that are matching with the protein by comparing the C-alpha pseudo-torsion angle and subsequently completes the alignment by discriminative extension of the matching segments. Furthermore, the multiple structure alignment is performed by a progressive pairwise alignment depending on the initial guide tree derived from all-against-all pairwise comparison. Correspondingly, the sequences are clustered into alignments depending on their similarities.
Our results have shown that the protein backbones have well aligned as expected. We further focussed on the ligands to understand if there is any major change from the initial pose. From the results, it can be deduced that comp1, comp5 and comp11 were relatively stable with marginal movement from the initial pose (Fig. 8A, 8D, and 8F). In case of comp2 and comp3, the linear structure present between both the peripheral rings has seemed to wobble (Fig. 8B and 8C). Comp10 has moved upwards from the initial position; however, it has still rested in the defined binding pocket (Fig. 8E).
Fig. 8.
Alignment of the docked pose and the MD pose. A) comp1, B) comp2, C) comp3, D) comp5, E) comp10, F) comp11. The linear wobbled structure in comp2, comp3, and shown in yellow oval.
We further made an analogy with the other compounds retrieved from the recently published literature [54,55]. Correspondingly 20 compounds were sketched on Biovia Draw 2017. The compounds were minimized and subjected to molecular docking using the same protocol as described before. The results show that the molecular docking score ranges from 34.30 kcal/mol to 70.21 kcal/mol (Table 2). This high dock score could be because of the size of the small molecules which are slightly bigger than our compounds. These compounds have also showed a curved pattern while accommodating at the binding pocket (Fig. 9), while our compounds of interest have rather displayed a linear form. This again could be because of the smaller size of the compounds making them to easily fit into the binding pocket.
Table 2.
Binding affinity studies of the compounds from the literature with the target of interest.
| Smiles | -CDOCKER interaction energy (kcal/mol) |
|---|---|
| O=C(CC[C@@H](NC(=O)NOCc1ccccc1)NC(=O)OCc2ccccc2)NOCc3ccccc3 | 70.2172 |
| CCCNC(=O)N[C@@H](CC(=O)NCC)NC(=O)OCc1ccccc1 | 64.7585 |
| O=C(CC[C@@H](NC(=O)OCc1ccccc1)NC(=O)N2CCCCC2)N3CCCCC3 | 64.0073 |
| CC(C)(C)C(=O)c1ccc(cc1)n2cc(CSc3nnc(Cc4ccccc4)n3\N=C\Cc5ccc(cc5)N6CCOCC6)nn2 | 61.6879 |
| O=C(N[C@H](CCNC(=O)N1CCOCC1)C(=O)N2CCOCC2)OCc3ccccc3 | 59.8071 |
| O=C(CC[C@H](NC(=O)OCc1ccccc1)NC(=O)N2CCOCC2)N3CCOCC3 | 59.5927 |
| OC(=O)c1ccc(cc1)n2cc(CSc3nnc(Cc4ccccc4)n3\N=C\Cc5cccc6OCOc56)nn2 | 57.0524 |
| OC(=O)c1ccc(cc1)n2cc(CSc3nnc(Cc4ccccc4)n3\N=C\Cc5ccc(cc5)N6CCOCC6)nn2 | 56.7373 |
| CCCNC(=O)NCC[C@H](NC(=O)OCc1ccccc1)C(=O)NCCC | 55.7313 |
| O=C(N[C@@H](CCNC(=O)N1CCCCC1)C(=O)N2CCCCC2)OCc3ccccc3 | 55.4408 |
| OC(=O)c1ccc(cc1)n2cc(CSc3nnc(Cc4ccccc4)n3\N=C\Cc5cccs5)nn2 | 53.4039 |
| CC(C)(C)C(=O)c1ccc(cc1)n2cc(CSc3nnc(Cc4ccccc4)n3\N=C\Cc5cccc6OCOc56)nn2 | 52.6891 |
| C#CCc1nnc(Cc2ccccc2)n1\N=C\C[C@@H]3CC[C@H](CC3)N4CCOCC4 | 52.2451 |
| CC(C)(C)C(=O)c1ccc(cc1)n2cc(CSc3nnc(Cc4ccccc4)n3\N=C\Cc5cccs5)nn2 | 51.9265 |
| O=C(NC1=CC(=O)NC1=O)OCc2ccccc2 | 44.3654 |
| Sc1nnc(Cc2ccccc2)n1\N=C\Cc3ccc(cc3)N4CCOCC4 | 43.6639 |
| Sc1nnc(Cc2ccccc2)n1\N=C\Cc3cccc4OCOc34 | 42.8059 |
| C#CCc1nnc(Cc2ccccc2)n1\N=C\Cc3cccc4OCOc34 | 39.651 |
| C#CCc1nnc(Cc2ccccc2)n1\N=C\CC3=CC=CC3 | 36.7983 |
| Sc1nnc(Cc2ccccc2)n1\N=C\Cc3cccs3 | 34.3048 |
Fig. 9.
Accommodation of the compounds at the binding pocket of the target.
Recent report discovered two inhibitors towards the Mycobacterium tuberculosis dethiobiotin synthase (MtDTBS) [30]. One compound, cyclopentylacetic acid (compound 2) has demonstrated a KD of 3.4 ± 0.4 mM and compound 7a, an optimized compound of compound 2 has shown a KD of 57 ± 5 nM [30]. Structurally, compound 2 appears to be similar as that of our compounds while compound 7a has a slightly bigger structure. Here, we performed a binding energy comparison after the molecular docking studies. The docking results of compound 2 and compound 7a are comparable with our compounds while the binding energies are better for our compounds except for comp 5. Interestingly, the dock score of comp1 and compound 2 are highly similar, while the binding energy of comp1 is better. From this analysis we spectulate that the KD of our compounds might also be better than compound 2 and compound 7a (Table 3).
Table 3.
Binding energy comparison between the reported Mt inhibitors and six compounds.
| Compound name | -CDOCKER interaction (kcal/mol) | Binding energy (kcal/mol) |
|---|---|---|
| comp1 | 51.4398 | −138.971 |
| comp2 | 47.7706 | −150.121 |
| comp3 | 44.6003 | −135.839 |
| comp5 | 40.0093 | −83.1029 |
| comp10 | 45.6705 | −153.886 |
| comp11 | 42.5373 | −149.82 |
| Compound 2 | 50.1015 | −127.768 |
| Compound 7a | 45.6095 | −119.457 |
The findings from the study are an elegant evidence that supports our objective that the selected butein analogues can act against TB. Taken together, our results present six compounds as prospective candidate inhibitors that could inhibit TB. Although our computational results demonstrate that the butein analogues could be potential MtDTBS inhibitors, it is essential to test these compounds in vitro and also to specifically target MtDTBS to understand is inhibitory effect.
6. Conclusion
In order to find effective therapeutics towards the infectious TB, we have used several computational methods such as molecular docking and molecular dynamics simulations. The ligands used are the butein analogues that are present in the lab. These compounds have shown good binding affinity potential and have established stable RMSD profiles. The key residue Lys37 was noticed with all compounds. In conclusion, we put forth six compounds as potential antiTB drugs. These compounds can also act as starting structures for designing and developing new candidate drugs.
Author contribution statement
Shailima Rampogu, Baji Shaik, Ju Hyun Kim, Tae Sung Jung, Min Woo Ha, Keun Woo Lee: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by two National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIT) (Grant Nos. NRF-2021R1F1A1059797 and NRF-2021R1A2B5B02002220).
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interest's statement
The authors declare no conflict of interest.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e13324.
Contributor Information
Shailima Rampogu, Email: shailima.rampogu@gmail.com.
Min Woo Ha, Email: minuha@jejunu.ac.kr.
Keun Woo Lee, Email: kwlee@gnu.ac.kr.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Zaman K. Tuberculosis: a global health problem. J. Health Popul. Nutr. 2010;28:111–113. doi: 10.3329/jhpn.v28i2.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sotgiu G., Centis R., D’ambrosio L., Migliori G.B. Tuberculosis treatment and drug regimens. Cold Spring Harb. Perspect. Med. 2015;5:a017822. doi: 10.1101/cshperspect.a017822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tiberi S., Scardigli A., Centis R., D'Ambrosio L., Muñoz-Torrico M., Salazar-Lezama M.Á., Spanevello A., Visca D., Zumla A., Migliori G.B., Caminero Luna J.A. Classifying new anti-tuberculosis drugs: rationale and future perspectives. Int. J. Infect. Dis. 2017;56:181–184. doi: 10.1016/j.ijid.2016.10.026. [DOI] [PubMed] [Google Scholar]
- 4.Caminero J.A., Scardigli A. Classification of antituberculosis drugs: a new proposal based on the most recent evidence. Eur. Respir. J. 2015;46:887. doi: 10.1183/13993003.00432-2015. LP – 893. [DOI] [PubMed] [Google Scholar]
- 5.Quan D., Nagalingam G., Payne R., Triccas J.A. New tuberculosis drug leads from naturally occurring compounds. Int. J. Infect. Dis. 2017;56:212–220. doi: 10.1016/j.ijid.2016.12.024. [DOI] [PubMed] [Google Scholar]
- 6.Alnajjar R., Mostafa A., Kandeil A., Al-Karmalawy A.A. Molecular docking, molecular dynamics, and in vitro studies reveal the potential of angiotensin II receptor blockers to inhibit the COVID-19 main protease. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e05641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rampogu S., Kim S.M., Shaik B., Lee G., Kim J.H., Kim G.S., Lee K.W., Kim M.O. Novel butein derivatives repress DDX3 expression by inhibiting PI3K/AKT signaling pathway in MCF-7 and MDA-MB-231 cell lines. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.712824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rampogu S., Kim S.M., Son M., Baek A., Park C., Lee G., Kim Y., Kim G.S., Kim J.H., Lee K.W. A computational approach with biological evaluation: combinatorial treatment of curcumin and exemestane synergistically regulates ddx3 expression in cancer cell lines. Biomolecules. 2020 doi: 10.3390/biom10060857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ezugwu J.A., Okoro U.C., Ezeokonkwo M.A., Hariprasad K.S., Rudrapal M., Ugwu D.I., Gogoi N., Chetia D., Celik I., Ekoh O.C. Design, synthesis, molecular docking, molecular dynamics and in vivo antimalarial activity of new dipeptide-sulfonamides. ChemistrySelect. 2022;7 doi: 10.1002/slct.202103908. [DOI] [Google Scholar]
- 10.Cerqueira A.P.M., Santana I.B., Araújo J.S.C., Lima H.G., Batatinha M.J.M., Branco A., dos Santos Junior M.C., Botura M.B. Homology modeling, docking, molecular dynamics and in vitro studies to identify Rhipicephalus microplus acetylcholinesterase inhibitors. J. Biomol. Struct. Dyn. 2022;40:6787–6797. doi: 10.1080/07391102.2021.1889666. [DOI] [PubMed] [Google Scholar]
- 11.Askarzadeh M., Azizian H., Adib M., Mohammadi-Khanaposhtani M., Mojtabavi S., Faramarzi M.A., Sajjadi-Jazi S.M., Larijani B., Hamedifar H., Mahdavi M. Design, synthesis, in vitro α-glucosidase inhibition, docking, and molecular dynamics of new phthalimide-benzenesulfonamide hybrids for targeting type 2 diabetes. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-14896-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandra Babu T.M., Rajesh S.S., Bhaskar B.V., Devi S., Rammohan A., Sivaraman T., Rajendra W. Molecular docking, molecular dynamics simulation, biological evaluation and 2D QSAR analysis of flavonoids from Syzygium alternifolium as potent anti-Helicobacter pylori agents. RSC Adv. 2017;7:18277–18292. doi: 10.1039/C6RA27872H. [DOI] [Google Scholar]
- 13.Mallavarapu B.D., Abdullah M., Saxena S., Guruprasad L. Inhibitor binding studies of Mycobacterium tuberculosis MraY (Rv21 56c): insights from molecular modeling, docking, and simulation studies. J. Biomol. Struct. Dyn. 2019;37:3751–3763. doi: 10.1080/07391102.2018.1526715. [DOI] [PubMed] [Google Scholar]
- 14.Singh N., Tiwari S., Srivastava K.K., Siddiqi M.I. Identification of novel inhibitors of Mycobacterium tuberculosis PknG using pharmacophore based virtual screening, docking, molecular dynamics simulation, and their biological evaluation. J. Chem. Inf. Model. 2015;55:1120–1129. doi: 10.1021/acs.jcim.5b00150. [DOI] [PubMed] [Google Scholar]
- 15.Sundar S., Thangamani L., Manivel G., Kumar P., Piramanayagam S. Molecular docking, molecular dynamics and MM/PBSA studies of FDA approved drugs for protein kinase a of Mycobacterium tuberculosis; application insights of drug repurposing. Inform. Med. Unlocked. 2019;16 doi: 10.1016/j.imu.2019.100210. [DOI] [Google Scholar]
- 16.Pitaloka D.A.E., Ramadhan D.S.F., Chaidir L., Fakih T.M. Docking-based virtual screening and molecular dynamics simulations of quercetin analogs as enoyl-acyl carrier protein reductase (Inha) inhibitors of mycobacterium tuberculosis. Sci. Pharm. 2021;89:20. [Google Scholar]
- 17.Rajasekhar S., Karuppasamy R., Chanda K. Exploration of potential inhibitors for tuberculosis via structure-based drug design, molecular docking, and molecular dynamics simulation studies. J. Comput. Chem. 2021;42:1736–1749. doi: 10.1002/jcc.26712. [DOI] [PubMed] [Google Scholar]
- 18.Maiolini M., Gause S., Taylor J., Steakin T., Shipp G., Lamichhane P., Deshmukh B., Shinde V., Bishayee A., Deshmukh R.R. The war against tuberculosis: a review of natural compounds and their derivatives. Molecules. 2020;25:3011. doi: 10.3390/molecules25133011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji H.-F., Li X.-J., Zhang H.-Y. Natural products and drug discovery. Can thousands of years of ancient medical knowledge lead us to new and powerful drug combinations in the fight against cancer and dementia? EMBO Rep. 2009;10:194–200. doi: 10.1038/embor.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollo L.A.E., Martin E.F., Machado V.R., Cantillon D., Wildner L.M., Bazzo M.L., Waddell S.J., Biavatti M.W., Sandjo L.P. Search for antimicrobial activity among fifty-two natural and synthetic compounds identifies anthraquinone and polyacetylene classes that inhibit Mycobacterium tuberculosis. Front. Microbiol. 2021;11:3619. doi: 10.3389/fmicb.2020.622629. https://www.frontiersin.org/article/10.3389/fmicb.2020.622629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pawar A., Jha P., Chopra M., Chaudhry U., Saluja D. Screening of natural compounds that targets glutamate racemase of Mycobacterium tuberculosis reveals the anti-tubercular potential of flavonoids. Sci. Rep. 2020;10:949. doi: 10.1038/s41598-020-57658-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodrigues Felix C., Gupta R., Geden S., Roberts J., Winder P., Pomponi S.A., Diaz M.C., Reed J.K., Wright A.E., Rohde K.H. Selective killing of dormant Mycobacterium tuberculosis by marine natural products. Antimicrob. Agents Chemother. 2017;61:e00743. doi: 10.1128/AAC.00743-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Semwal R.B., Semwal D.K., Combrinck S., Viljoen A. Butein: from ancient traditional remedy to modern nutraceutical. Phytochem. Lett. 2015 doi: 10.1016/j.phytol.2014.12.014. [DOI] [Google Scholar]
- 24.Bordoloi D., Monisha J., Roy N.K., Padmavathi G., Banik K., Harsha C., Wang H., Kumar A.P., Arfuso F., Kunnumakkara A.B. An investigation on the therapeutic potential of butein, a tretrahydroxychalcone against human oral squamous cell carcinoma. Asian Pac. J. Cancer Prev. APJCP. 2019 doi: 10.31557/APJCP.2019.20.11.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho S.G., Woo S.M., Ko S.G. Butein suppresses breast cancer growth by reducing a production of intracellular reactive oxygen species. J. Exp. Clin. Cancer Res. 2014;33 doi: 10.1186/1756-9966-33-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Padmavathi G., Rathnakaram S.R., Monisha J., Bordoloi D., Roy N.K., Kunnumakkara A.B. Potential of butein, a tetrahydroxychalcone to obliterate cancer. Phytomedicine. 2015;22:1163–1171. doi: 10.1016/j.phymed.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 27.Yang L.-H., Ho Y.-J., Lin J.-F., Yeh C.-W., Kao S.-H., Hsu L.-S. Butein inhibits the proliferation of breast cancer cells through generation of reactive oxygen species and modulation of ERK and p38 activities. Mol. Med. Rep. 2012;6:1126–1132. doi: 10.3892/mmr.2012.1023. [DOI] [PubMed] [Google Scholar]
- 28.Brown A.K., Papaemmanouil A., Bhowruth V., Bhatt A., Dover L.G., Besra G.S. Flavonoid inhibitors as novel antimycobacterial agents targeting Rv0636, a putative dehydratase enzyme involved in Mycobacterium tuberculosis fatty acid synthase II. Microbiology. 2007 doi: 10.1099/mic.0.2007/009936-0. [DOI] [PubMed] [Google Scholar]
- 29.Sundarrajan S., Lulu S., Arumugam M. Computational evaluation of phytocompounds for combating drug resistant tuberculosis by multi-targeted therapy. J. Mol. Model. 2015;21:247. doi: 10.1007/s00894-015-2785-z. [DOI] [PubMed] [Google Scholar]
- 30.Schumann N.C., Lee K.J., Thompson A.P., Salaemae W., Pederick J.L., Avery T., Gaiser B.I., Hodgkinson-Bean J., Booker G.W., Polyak S.W., Bruning J.B., Wegener K.L., Abell A.D. Inhibition of Mycobacterium tuberculosis dethiobiotin synthase (MtDTBS): toward next-generation antituberculosis agents. ACS Chem. Biol. 2021;16:2339–2347. doi: 10.1021/acschembio.1c00491. [DOI] [PubMed] [Google Scholar]
- 31.Thompson A.P., Wegener K.L., Booker G.W., Polyak S.W., Bruning J.B. Precipitant–ligand exchange technique reveals the ADP binding mode in Mycobacterium tuberculosis dethiobiotin synthetase. Acta Crystallogr. Sect. D Struct. Biol. 2018;74:965–972. doi: 10.1107/S2059798318010136. [DOI] [PubMed] [Google Scholar]
- 32.Spassov V.Z., Flook P.K., Yan L. LOOPER: a molecular mechanics-based algorithm for protein loop prediction. Protein Eng. Des. Sel. 2008;21:91–100. doi: 10.1093/protein/gzm083. [DOI] [PubMed] [Google Scholar]
- 33.Spassov V.Z., Yan L. A fast and accurate computational approach to protein ionization. Protein Sci. 2008;17:1955–1970. doi: 10.1110/ps.036335.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005 doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 35.Rampogu S., Parameswaran S., Lemuel M.R., Lee K.W. Exploring the therapeutic ability of fenugreek against type 2 diabetes and breast cancer employing molecular docking and molecular dynamics simulations, evidence-based complement. Altern. Med. 2018 doi: 10.1155/2018/1943203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rampogu S., Lee K.W. Old drugs for new purpose—fast pace therapeutic identification for SARS‐CoV ‐2 infections by pharmacophore guided drug repositioning approach. Bull. Kor. Chem. Soc. 2021 doi: 10.1002/bkcs.12171. [DOI] [Google Scholar]
- 37.Zoete V., Cuendet M.A., Grosdidier A., Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J. Comput. Chem. 2011 doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 38.Parrinello M. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 39.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1002/(SICI)1096-987X. [DOI] [Google Scholar]
- 40.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981 doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 41.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996 doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 42.Rampogu S., Baek A., Son M., Park C., Yoon S., Parate S., Lee K.W. ACS Omega; 2020. Discovery of Lonafarnib-like Compounds: Pharmacophore Modeling and Molecular Dynamics Studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rampogu S., Lee K.W. Pharmacophore modelling-based drug repurposing approaches for SARS-CoV-2 therapeutics. Front. Chem. 2021;9:38. doi: 10.3389/fchem.2021.636362. https://www.frontiersin.org/article/10.3389/fchem.2021.636362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rampogu S., Lemuel M.R., Lee K.W. Virtual screening, molecular docking, molecular dynamics simulations and free energy calculations to discover potential DDX3 inhibitors. Adv. Cancer Biol. - Metastasis. 2021 doi: 10.1016/j.adcanc.2021.100022. [DOI] [Google Scholar]
- 45.Mishra C.B., Pandey P., Sharma R.D., Malik M.Z., Mongre R.K., Lynn A.M., Prasad R., Jeon R., Prakash A. Identifying the natural polyphenol catechin as a multi-targeted agent against SARS-CoV-2 for the plausible therapy of COVID-19: an integrated computational approach. Briefings Bioinf. 2020 doi: 10.1093/bib/bbaa378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Londhe A.M.H., Gadhe C.G., Lim S.M., Pae A.N. Investigation of molecular details of Keap1-Nrf2 inhibitors using molecular dynamics and umbrella sampling techniques. Molecules. 2019 doi: 10.3390/molecules24224085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sliwoski G., Kothiwale S., Meiler J., Lowe E.W., Jr. Computational methods in drug discovery. Pharmacol. Rev. 2013;66:334–395. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atanasov A.G., Zotchev S.B., Dirsch V.M., Orhan I.E., Banach M., Rollinger J.M., Barreca D., Weckwerth W., Bauer R., Bayer E.A., Majeed M., Bishayee A., Bochkov V., Bonn G.K., Braidy N., Bucar F., Cifuentes A., D'Onofrio G., Bodkin M., Diederich M., Dinkova-Kostova A.T., Efferth T., El Bairi K., Arkells N., Fan T.P., Fiebich B.L., Freissmuth M., Georgiev M.I., Gibbons S., Godfrey K.M., Gruber C.W., Heer J., Huber L.A., Ibanez E., Kijjoa A., Kiss A.K., Lu A., Macias F.A., Miller M.J.S., Mocan A., Müller R., Nicoletti F., Perry G., Pittalà V., Rastrelli L., Ristow M., Russo G.L., Silva A.S., Schuster D., Sheridan H., Skalicka-Woźniak K., Skaltsounis L., Sobarzo-Sánchez E., Bredt D.S., Stuppner H., Sureda A., Tzvetkov N.T., Vacca R.A., Aggarwal B.B., Battino M., Giampieri F., Wink M., Wolfender J.L., Xiao J., Yeung A.W.K., Lizard G., Popp M.A., Heinrich M., Berindan-Neagoe I., Stadler M., Daglia M., Verpoorte R., Supuran C.T. Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discov. 2021 doi: 10.1038/s41573-020-00114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tungmunnithum D., Thongboonyou A., Pholboon A., Yangsabai A. Flavonoids and other phenolic compounds from medicinal plants for pharmaceutical and medical aspects: an overview. Med. (Basel, Switzerland) 2018;5:93. doi: 10.3390/medicines5030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Panche A.N., Diwan A.D., Chandra S.R. Flavonoids: an overview. J. Nutr. Sci. 2016 doi: 10.1017/jns.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y., Ma C., Qian M., Wen Z., Jing H., Qian D. Butein induces cell apoptosis and inhibition of cyclooxygenase-2 expression in A549 lung cancer cells. Mol. Med. Rep. 2014;9:763–767. doi: 10.3892/mmr.2013.1850. [DOI] [PubMed] [Google Scholar]
- 52.LiRui Z., Wei C., Xu L. A novel anticancer effect of butein: inhibition of invasion through the ERK1/2 and NF‐κB signaling pathways in bladder cancer cells. FEBS Lett. 2008;582:1821–1828. doi: 10.1016/j.febslet.2008.04.046. [DOI] [PubMed] [Google Scholar]
- 53.Jayasooriya R.G.P.T., Molagoda I.M.N., Park C., Jeong J.W., Choi Y.H., Moon D.O., Kim M.O., Kim G.Y. Molecular chemotherapeutic potential of butein: a concise review. Food Chem. Toxicol. 2018 doi: 10.1016/j.fct.2017.12.028. [DOI] [PubMed] [Google Scholar]
- 54.El Sawy M.A., Elshatanofy M.M., El Kilany Y., Kandeel K., Elwakil B.H., Hagar M., Aouad M.R., Albelwi F.F., Rezki N., Jaremko M., El Ashry E.S.H. Novel hybrid 1,2,4- and 1,2,3-triazoles targeting Mycobacterium tuberculosis enoyl acyl carrier protein reductase (InhA): design, synthesis, and molecular docking. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23094706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kijewska M., Sharfalddin A.A., Jaremko Ł., Cal M., Setner B., Siczek M., Stefanowicz P., Hussien M.A., Emwas A.-H., Jaremko M. Lossen rearrangement of p-toluenesulfonates of N-oxyimides in basic condition, theoretical study, and molecular docking. Front. Chem. 2021;9 doi: 10.3389/fchem.2021.662533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.