Abstract
A variety of walnut known as Tetracarpidium conophorum is widely cultivated in several parts of Africa for its edible nuts. These nuts have been reported for their huge antioxidant, anti-obesity, and anti-depressant potentials, but remain underutilized due to their poor storage and preservation. This is why the nuts are mostly cooked and consumed as snacks whenever in season. This data article reports the untargeted metabolite profile of boiled and dried African walnut extracted using two different mixtures of solvents. The raw nuts obtained from a local market in Osun State, Nigeria, were processed by cooking for 20 min, deshelled, diced, dried at 60 ± 2 °C for 6 h, and stored until further analysis. The dried walnut samples were extracted with acetonitrile/methanol/water (40:40:20 v/v/v) and methanol/water (80:20 v/v) as solvents, before being analysed by gas chromatography high-resolution time of flight mass spectrometry (GC-HRTOF-MS) system. Data obtained from the analysis were further classified into different compounds, including alcohols, esters, hydrocarbons, phytosterols, vitamins, and many more. Their retention time, observed ion mass-to-charge ratio, molecular formula, and average peak areas were also reported. These data thus serve as a source of metabolites comparison for other walnuts, may be useful for the identification of functional compounds available in this neglected food crop, and encourage its utilization in developing functional foods.
Keywords: Metabolite profiling, Untargeted metabolites, GC-HRTOF-MS, Boiled walnut, Conophor nut
Specifications Table
| Subject | Food Science: Food Chemistry |
| Specific subject area | Processing; Food composition and analysis; Metabolomics |
| Type of data | Table Figure Spectra |
| How the data were acquired | Raw walnuts were boiled under pressure for 20 min, sliced and dried at 60 ± 2 °C for 6 h. The dried cooked nuts were further grounded using laboratory mortar & pestle, and then extracted using two different combinations of organic solvents, acetonitrile/methanol/water (40:40:20 v/v/v), and methanol/water (80:20 v/v). The extracts were analyzed using the GC-HRTOF-MS system (LECO Pegasus, St Joseph, USA). This system featured 50,0 0 0 FWMH resolution (full peak with at one-half maximum), mass accuracies/errors of <1 ppm with acquisition rates up to 200 spectra/s, and was equipped with an Agilent 7890A gas chromatograph (Agilent Technologies, Inc., Wilmington, DE, USA). This GC-HRTOF-MS operates at high resolution and is equipped with a Gerstel MPS multipurpose autosampler (Gerstel Inc., Mülheim an der Ruhr, Germany) and a Rxi ®-5ms column (30 m × 0.25 mm ID × 0.25 µm) (Restek, Bellefonte, United States). |
| Data format | Raw data Analyzed data Filtered data Spectra of commonly identified compounds |
| Description of data collection | The already processed walnut (1 g) in its ground form was weighed and metabolites were extracted using the 10 mL mixture of different solvents (acetonitrile/methanol/water (40:40:20 v/v/v), and methanol/water (80:20 v/v) in each case. Thereafter, each sample was vacuum concentrated and reconstituted in chromatography-grade methanol (1 mL), then filtered with a 0.22 µm syringe into amber vials. Each sample (1 µL) was auto-injected into the GC-HRTOF-MS machine in triplicates and analyzed. The identities of the metabolite obtained were determined using NIST, Mainlib and Feihn metabolomics databases. |
| Data source location | Raw African walnuts were sourced from a local market in Modakeke, Osun State Nigeria (N 7°22’ 54.848” E 4°16’3.737”) on the 27th June, 2021 and processed within 24 h after collection. Thereafter, the extraction and analyses were carried out at the University of Johannesburg, Doornfontein, Johannesburg, Gauteng, South Africa (S 26°11’ 32.6” E 28°03’ 28.9”). |
| Data accessibility | Raw & processed dataset, and mass spectra of the metabolites have been deposited in the Mendeley repository. It is accessible using the details below: Repository name: Mendeley data. DOI: 10.17632/s9vrhj8tsk.1 Direct URL to data: https://data.mendeley.com/datasets/s9vrhj8tsk |
Value of the Data
-
•
The data contributed to the identification of metabolites in African walnuts and provided information on the versatility of different solvent mixtures in metabolite extraction.
-
•
The information provided herein will assist in understanding the usefulness of African walnuts and promote their cultivation to prevent crop extinction.
-
•
The data could be useful for a comparative analysis of the metabolite composition in raw and processed, domestic or foreign walnuts, and the developed products.
-
•
The data would be useful for food processors and researchers aiming to develop novel functional foods from African walnuts.
-
•
The data would be useful in identifying constituents that may be responsible to sensory, functional, and nutritional effects and concentration in developed food products.
-
•
The data would be useful resource for nutritionists, agronomists, food, and data scientists.
-
•
The data indicates that untargeted GC-HRTOF-MS analysis could facilitate the identification of compounds that may be responsible for the nut's health-promoting effects.
Objective
The African walnut (Tetracarpidium conophorum) plant has been extensively investigated for its high nutrients, anti-oxidants, anti-diabetic, anti-inflammatory, and other therapeutic benefits. The nut has also been explored in a few food products such as functional cookies [1], however, the nut has remained poorly utilized. This study identified metabolite present in ready-to-eat African walnuts that may enhance the potential exploitation of the nut as food ingredients in the development of functional food products that could solve many health issues.
1. Data Description
The dataset deposited in the repository contain two files (excel sheets and word document). The excel sheet 1 contains the raw data collected from the GC-HRTOF-MS analysis, it described the retention time (min), sample code, observed mass per charge number of ions, formula, area, name and synonym of the compounds extracted. Sheet 2 contains the data that was processed using the DataPrep solutions software and the class of the identified compounds, while sheet 3 represent the common compound that occurred at least two times in three injection in both samples analysed. The sample labelled W1 represent the walnut sample extracted with the mixture of acetonitrile/methanol/water (40:20:20 v/v/v), and W2 represent walnut sample extracted with the mixture of methanol/water (80:20 v/v). In addition, the word document deposited in the repository enclose the spectra of each compound identified in both samples. The spectrum of each compound typically shows a number of signals and the true peak at the highest mass per charge ion ratio. This will provide the scientist community with the structural information and the whole molecule identification.
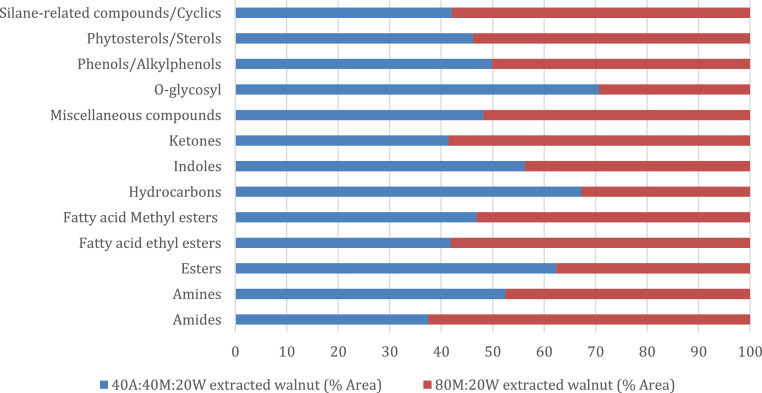
The metabolite data obtained from extracted walnut samples are presented below. Table 1 represents metabolites obtained from walnut extracted using the mixture of acetonitrile/methanol/water (40:20:20 v/v/v), and methanol/water (80:20 v/v) mixed solvent. The data in each table shows information regarding the name of each extractable compound identified, their retention time, observed ion mass-to-charge ratio, molecular formula and average peak area. These data were generated from GC-HRTOF-MS analysis and the spectra obtained were compared with NIST, Mainlib and Feihn metabolite databases. The raw and analyzed data along with the spectra of the identified compounds are available in a supplementary file deposited in the repository [2]. Figure 1 summarizes the percentage distribution of the compounds found from at least 2 out of 3 injections of each extracted sample from the extraction solvent.
Table 1.
Metabolites identified in the walnut sample that was extracted using the two different solvents mixture.
| Retention Time (Min) | Observed Ion m/z | Name | Molecular Formula | Average Area |
|
|---|---|---|---|---|---|
| W1 | W2 | ||||
| Acyclic alkanes | |||||
| 14.976 | 268.9873 | Eicosane | C₂₀H₄₂ | ND | 135601 |
| Alcohols/Phenols | |||||
| 2.987 | 32.0259 | Methyl Alcohol | CH₄O | 2517321 | ND |
| 12.317 | 220.1821 | Butylated Hydroxytoluene | C₁₅H₂₄O | ND | 84268 |
| Aldehydes | |||||
| 7.576 | 120.0569 | Benzaldehyde, 2-methyl- | C₈H₈O | 698094 | ND |
| 15.962 | 234.1612 | 3,5-di-tert-Butyl-4-hydroxybenzaldehyde | C₁₅H₂₂O₂ | ND | 12759 |
| Amides | |||||
| 21.457, 21.742 | 142.1226, 156.1383 | 3-Cyclopentylpropionamide, N,N-dimethyl- | C₁₀H₁₉NO | 180548 | 300932 |
| Amines | |||||
| 22.582, 22.580 | 144.1019, 144.1019 | Bis(2-(Dimethylamino)ethyl) ether | C₈H₂₀N₂O | 421813 | 382242 |
| Esters | |||||
| 21.490 | 225.4713 | 2-Propenoic acid, 3-(4-methoxyphenyl)-, 2-ethylhexyl ester | C₁₈H₂₆O₃ | ND | 19523 |
| 17.921, 17.918 | 292.2026, 292.2033 | Benzenepropanoic acid, 3,5-bis(1,1-dimethylethyl)-4-hydroxy-, methyl ester | C₁₈H₂₈O₃ | 27667 | 32997 |
| 30.801 | 530.4694 | Benzenepropanoic acid, 3,5-bis(1,1-dimethylethyl)-4-hydroxy-, octadecyl ester | C₃₅H₆₂O₃ | ND | 111822 |
| 22.581 | 219.0679 | Carbonic acid, 2-dimethylaminoethyl 2-methoxyethyl ester | C₈H₁₇NO₄ | ND | 418022 |
| 21.606, 21.654 | 170.0830, 219.1150 | Carbonic acid, 2-dimethylaminoethyl isobutyl ester | C₉H₁₉NO₃ | 316407 | 172290 |
| 25.181 | 297.2416 | Decanedioic acid, bis(2-ethylhexyl) ester | C₂₆H₅₀O₄ | ND | 196860 |
| 23.307 | 279.1594 | Dicyclohexyl phthalate | C₂₀H₂₆O₄ | ND | 67658 |
| 3.082 | 88.0519 | Ethyl Acetate | C₄H₈O₂ | ND | 8575634 |
| 18.124 | 223.5891 | Dibutyl phthalate | C₁₆H₂₂O₄ | 80025 | ND |
| 24.783, 24.780 | 279.1576, 279.1592 | Mono(2-ethylhexyl) phthalate | C₁₆H₂₂O₄ | 81688 | 48734 |
| 25.443 | 503.1072 | Phthalic acid, 8-chlorooctyl decyl ester | C₂₆H₄₁ClO₄ | 191547 | ND |
| 11.734 | 149.1072 | Succinic acid, 3-methylbut-2-en-1-yl 3-methoxyphenyl ester | C₁₆H₂₀O₅ | 111337 | ND |
| 24.557 | 328.2897 | Octadecanoic acid, 2,3-dihydroxypropyl ester | C₂₁H₄₂O₄ | ND | 646767 |
| 25.622 | 226.9907 | Phthalic acid, 4-methylhept-3-yl pentyl ester | C₂₁H₃₂O₄ | ND | 53421 |
| Ethers | |||||
| 8.715, 12.656 | 131.1513, 131.1238 | 1,1,1,2,3,3,3-Heptafluoro-2-methoxypropane | C₄H₃F₇O | 7819 | 7624 |
| Fatty acid ethyl esters (FAEEs) | |||||
| 22.989, 22.990 | 300.2605, 311.2588 | Hexadecanoic acid, 2-hydroxy-1-(hydroxymethyl) ethyl ester | C₁₉H₃₈O₄ | 540587 | 751444 |
| Fatty Acid Methyl Esters (FAMEs) | |||||
| 19.473, 19.516 | 292.2395, 292.2398 | 9,12,15-Octadecatrienoic acid, (Z,Z,Z)-, Methyl 8,11,14-heptadecatrienoate | C₁₈H₃₀O₂ | 1476801 | 1179328 |
| 19.398, 19.398 | 294.2553, 294.2552 | 9,12-Octadecadienoic acid, methyl ester | C₁₉H₃₄O₂ | 1064918 | 1214274 |
| 21.261, 21.260 | 293.2822, 293.2820 | 9-Octadecenoic acid (Z)-, methyl ester | C₁₉H₃₆O₂ | 69858 | 151120 |
| 17.665, 17.663 | 270.2551, 270.2553 | Hexadecanoic acid, methyl ester | C₁₇H₃₄O₂ | 2729756 | 2953203 |
| 19.674, 19.673 | 298.2860, 298.2868 | Methyl stearate | C₁₉H₃₈O₂ | 1685892 | 2403367 |
| 23.094, 20.541 | 219.2037, 223.6918 | Tridecanoic acid, methyl ester | C₁₄H₂₈O₂ | 197152 | 112146 |
| 23.035 | 199.1692 | Undecanoic acid, methyl ester | C₁₂H₂₄O₂ | 118716 | ND |
| 19.451, 19.448 | 296.2707, 296.2708 | trans-13-Octadecenoic acid, methyl ester | C₁₉H₃₆O₂ | 1003628 | 812770 |
| Hydrocarbons | |||||
| 13.420 | 131.1726 | Butane, 1,1,1,2,3,3,4,4,4-nonafluoro-2-(trifluoromethyl)- | C₅F₁₂ | 6718 | ND |
| 15.032, 8.761 | 175.0624, 155.1432 | Hexadecane | C₁₆H₃₄ | 84165 | 41015 |
| 11.656 | 139.0982 | Pentadecane | C₁₅H₃₂ | ND | 79208 |
| Indoles | |||||
| 8.677, 8.680 | 117.0573, 117.0573 | Indole | C₈H₇N | 59297 | 46098 |
| Ketones | |||||
| 17.716, 17.710 | 276.1710, 267.0356 | 7,9-Di-tert-butyl-1-oxaspiro(4,5)deca-6,9-diene-2,8-dione | C₁₇H₂₄O₃ | 48189 | 94470 |
| 15.033, 15.032 | 188.1192, 219.1732 | Methanone, (1-hydroxycyclohexyl)phenyl- | C₁₃H₁₆O₂ | 219837 | 287750 |
| 9.877 | 269.0488 | 7-Chloro-1,3,4,10-tetrahydro-10-hydroxy-1-[[2-[1-pyrrolidinyl]ethyl]imino]-3-[3-(trifluoromethyl)phenyl]-9(2H)-acridinone | C₂₆H₂₅ClF₃N₃O₂ | ND |
977127 |
| Miscellaneous compounds | |||||
| 19.981 | 292.2392 | 1,2-Benzenediol, O-(2-furoyl)-O'-(pentafluoropropionyl)- | C₁₄H₇F₅O₅ | ND | 160588 |
| 20.243 | 225.0656 | 1,8,11-Heptadecatriene, (Z,Z)- | C₁₇H₃₀ | ND | 273401 |
| 20.422 | 219.1343 | 1-Acetoxynonadecane | C₂₁H₄₂O₂ | ND | 75610 |
| 17.249 | 131.1923 | 1H-1,3-Benzimidazole-1-ethanol, a-(4-morpholinylmethyl)- | C₁₄H₁₉N₃O₂ | ND | 30635 |
| 24.268 | 131.0521 | 1H-Indole, 4-methyl- | C₉H₉N | ND | 50238 |
| 13.318, 11.381 | 69.1009, 69.0252 | 2-Propynenitrile, 3-fluoro- | C₃FN | 56110 | 10723 |
| 20.441 | 322.2496 | 3,4-Dimethoxybenzoic anhydride | C₁₈H₁₈O₇ | ND | 68270 |
| 11.937 | 503.1066 | 3-Isopropoxy-1,1,1,7,7,7-hexamethyl-3,5,5-tris(trimethylsiloxy)tetrasiloxane | C₁₈H₅₂O₇Si₇ | ND | 284860 |
| 13.434, 13.433 | 157.0884, 157.0882 | 3-Methyl-4-phenyl-1H-pyrrole | C₁₁H₁₁N | 62006 | 50364 |
| 20.423 | 265.1814 | Heneicosyl acetate | C₂₃H₄₆O₂ | 98265 | ND |
| 6.115 | 144.0419 | Acetic acid, trifluoro-, ethyl ester | C₄H₅F₃O₂ | ND | 43042 |
| 23.999 | 269.0457 | Anthranilic acid, 2TMS derivative | C₁₃H₂₃NO₂Si₂ | ND | 9488 |
| 6.517 | 357.0670 | Cyclopentasiloxane, decamethyl- | C₁₀H₃₀O₅Si₅ | ND | 126744 |
| 19.172 | 219.1294 | Dimethylmalonic acid, di(2-formylphenyl) ester | C₁₉H₁₆O₆ | ND | 3508 |
| 22.124 | 504.1076 | Heptasiloxane, hexadecamethyl- | C₁₆H₄₈O₆Si₇ | ND | 176174 |
| 21.455 | 170.1539 | Octanamide, N,N-dimethyl- | C₁₀H₂₁NO | ND | 319657 |
| 14.843 | 210.0891 | Methyl 3-(4-hydroxy-3-methoxyphenyl)propanoate | C₁₁H₁₄O₄ | 29002 | ND |
| 5.246 | 141.0699 | Methyl 3-O-benzyl-alpha-d-glucofuranoside 5,6-carbonate | C₁₅H₁₈O₇ | 156833 | ND |
| 19.564, 19.562 | 503.1055, 505.1041 | Octasiloxane, 1,1,3,3,5,5,7,7,9,9,11,11,13,13,15,15-hexadecamethyl- | C₁₆H₅₀O₇Si₈ | 118565 | 166258 |
| 13.576, 18.274 | 219.1379, 219.0369 | Phosphine, tris(trifluoromethyl)- | C₃F₉P | 4708 | 6508 |
| 6.721, 6.719 | 139.0991, 139.0991 | Quinoline, decahydro- | C₉H₁₇N | 200840 | 170716 |
| 13.535, 18.732 | 131.1894, 131.0855 | Tris(trifluoromethyl) bromomethane | C₄BrF₉ | 6445 | 5364 |
| O-glycosyl | |||||
| 14.108, 14.093 | 217.1589, 137.0398 | Ethyl a-d-glucopyranoside | C₈H₁₆O₆ | 352422 | 146265 |
| Phenols/Alkylphenols | |||||
| 12.252, 12.250 | 206.1660, 206.1658 | 2,4-Di-tert-butylphenol | C₁₄H₂₂O | 537673 | 454311 |
| 22.354, 22.352 | 340.2395 | Phenol, 2,2′-methylenebis[6-(1,1-dimethylethyl)-4-methyl- | C₂₃H₃₂O₂ | 1244321 | 1333882 |
| Phenylpropanes | |||||
| 27.948 | 278.0431 | 4-tert-Octylphenol, TMS derivative | C₁₇H₃₀OSi | 1089432 | ND |
| Phytosterols/Sterols | |||||
| 29.015, 29.010 | 412.3703, 412.3677 | Stigmasta-5,24(28)-dien-3-ol, (3ß,24Z)- | C₂₉H₄₈O | 274827 | 248085 |
| 28.506, 28.504 | 412.3700, 412.3699 | Stigmasterol | C₂₉H₄₈O | 452029 | 599436 |
| Pyrazine/Pyridines | |||||
| 4.866 | 123.0679 | 4(H)-Pyridine, N-acetyl- | C₇H₉NO | 593145 | ND |
| 19.154 | 116.0705 | 1,4-Di(methyl-d3)benzene-d4 | C₈D₁₀ | ND | 72152 |
| Sesquiterpenoids | |||||
| 12.023 | 204.1864 | (1R,5R)-2-Methyl-5-((R)-6-methylhept-5-en-2-yl)bicyclo[3.1.0]hex-2-ene | C₁₅H₂₄ | ND | 52683 |
| 10.838 | 199.9870 | Bicyclo[7.2.0]undec-4-ene, 4,11,11-trimethyl-8-methylene-,[1R-(1R*,4Z,9S*)]- | C₁₅H₂₄ | ND | 28580 |
| Silane-related compounds/Cyclics | |||||
| 21.020 | 432.0861 | 1,1,1,5,7,7,7-Heptamethyl-3,3-bis(trimethylsiloxy)tetrasiloxane | C₁₃H₄₀O₅Si₆ | ND | 184425 |
| 18.940, 14.547 | 504.1078, 415.0368 | Cyclooctasiloxane, hexadecamethyl- | C₁₆H₄₈O₈Si₈ | 132164 | 214475 |
| 28.287 | 221.0456 | Cyclotrisiloxane, hexamethyl- | C₆H₁₈O₃Si₃ | 3083849 | ND |
| 8.916, 8.915 | 432.0872, 432.0848 | Cyclohexasiloxane, dodecamethyl- | C₁₂H₃₆O₆Si₆ | 315246 | 401864 |
| Trialkylheterosilanes | |||||
| 11.938 | 503.1089 | 3-Isopropoxy-1,1,1,7,7,7-hexamethyl-3,5,5-tris(trimethylsiloxy)tetrasiloxane | C₁₈H₅₂O₇Si₇ | 198812 | ND |
| Vitamins | |||||
| 26.254, 26.253 | 402.3488, 402.3486 | d-Tocopherol | C₂₇H₄₆O₂ | 1045910 | 686038 |
ND: Not detected; m/z: mass-to-charge ratio.
W1: Walnut sample extracted with acetonitrile/methanol/water (40:20:20 v/v/v); W2: Walnut sample extracted with methanol/water (80:20 v/v).
Fig. 1.
Percentage distribution of compounds common to both extracted walnut samples.
2. Experimental Design, Materials and Methods
2.1. Walnut collection and processing
Matured raw walnuts (Tetracarpidium conophorum) were sourced from a local market in Nigeria (N 7°22’ 54.848” E 4°16’3.737”) on the 27th June, 2021. They were physically cleaned and washed under running water to remove extraneous materials. The nuts were cooked (Pressure Pot, Master Chef, 12L) for about 20 minutes after pressure has been built within the system. The nuts were allowed to cool, de-shelled, and shredded into smaller sizes using a hand grater. The grated walnuts were dried in a food dehydrator (Bosch BS-6605, Germany) set at a temperature of 60 ± 2 °C for 6 h. The dried walnut was allowed to stand at room temperature before milling (Perten 3600, Sweden) to a coarse powder.
2.2. Metabolites extraction from the samples and analysis using GC-HRTOF-MS
The cooked walnut powder sample was extracted using two different mixtures of extraction solvents, acetonitrile/methanol/water (40:40:20 v/v/v) and methanol/water (80:20 v/v), following the method previously described by Oyedeji et al. [3]. One (1) gram of each of the walnut samples was weighed separately into 50 mL centrifuge tubes, 10 mL of each extraction solvent was added, and vortexed (Vortex-Gernie K-550-GE, Bohemia USA) vigorously to ensure even mixing. The tube containing the mixture was sonicated (Ultrasonic AU-200 Argo Lab, Italia Italy) for an hour, and then centrifuged (Eppendorf 5702R, Merck, Modderfontein South Africa) for 5 min at 4°C and 3500 rpm. The supernatants from each centrifuge tube were decanted into new tubes, and allowed to dry in a vacuum concentrator (Eppendorf Plus, Merck, Modderfontein South Africa). These recovered dried extracts were reconstituted with 1 mL chromatography-grade methanol (99.9% pure), and vortexed to ensure there is even dissolution of the extracts in each tube. The extracts was filtered into dark amber vials using PTFE-L 0.22 µm. Using the Pegasus GC-HRTOF-MS system (LECO Corporation, St. Joseph, MI, USA) with a resolution of 50,0 0 0 FWMH (full peak with at one-half maximum), mass accuracies/errors of < 1 ppm and acquisition rates of up to 200 spectra/s, the samples were analysed. This analytical system was equipped with a multipurpose sampler (Gerstel Inc., Mülheim an der Ruhr Germany) and Rxi ®-5 ms column (30 m × 0.25 mm ID × 0.25 µm) (Restek, Bellefonte, USA). An aliquot of each sample was injected without spit and pumped with helium as the carrier gas at a constant flow rate of 1 mL/min. Inlet and transfer line temperatures were set at 250 and 225 respectively and the ion source temperature was at 250. The oven temperature cycle used was: 70, 0.5 min for initial temperature; then an increase form 10/min to 150 for 2 min; then ramped up to 330 °C at 10 °C/min and held for 3 min to allow the column to ‘bake-out’. The solvent blanks were also tested in parallel to monitor for potential impurities and contamination. When processing the raw data with DataPrep solutions, parameters such as a signal-to-noise ratio of 50, a similarity match of over 70 % and at least twofold occurrence of metabolites from the triplicate data were strictly considered. The properties of the metabolites were identified by matching the spectra to NIST, Mainlib, and Feihn reference library databases. Data obtained from samples extracted with acetonitrile/methanol/water (40:40:20 v/v/v) and methanol/water (80:20 v/v) are presented in Table 1, and commonly detected compounds are summarised in Fig. 1. The raw and processed data, are presented in the supplementary file along with the raw spectra of some identified compounds.
Ethics Statements
This work does not involve chemicals, procedures or equipment that have any unusual hazards inherent in their use, and it does not involve human subjects, animal experiments, or any data collected from social media platforms.
CRediT Author Statement
Beatrice Mofoluwaso Oladimeji: Conceptualization, Sample preparation, Formal data analysis, Methodology, Visualization, Validation, Writing- original draft; Oluwafemi Ayodeji Adebo: Conceptualization, Funding acquisition, Data curation, Methodology, Resources, Software, Visualization, Supervision, Writing –review & editing.
Funding
This work was supported financially by the University of Johannesburg (UJ) Global Excellence and Stature (GES 4.0) grant offered to Beatrice M. Oladimeji, and the UJ Research Committee (URC 2022) research grant awarded to Oluwafemi Ayodeji Adebo.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors wish to acknowledge the assistance received from colleagues in Food Innovation Research Group.
Data Availability
References
- 1.Fasogbon B.M., Akinwande F.F., Ademuyiwa O.H., Bamidele O.P. The influence of cooked grated African walnut on the nutritional composition, antioxidant and sensorial properties of a cookie snack. J. Cul. Sci. Tech. 2021 doi: 10.1080/15428052.2021.1955797. [DOI] [Google Scholar]
- 2.Fasogbon B.M., Adebo O.A. Supplementary data for manuscript on metabolites extracted from African walnut (Tetracarpidium conophorum) using two different solvents. Mendeley Data. 2022;V1 doi: 10.17632/s9vrhj8tsk.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oyedeji A.B., Chinma C.E., Green E., Adebo O.A. Metabolite data of germinated Bambara groundnut flour and starch extracted with two different solvents. Data Br. 2021;38 doi: 10.1016/j.dib.2021.107288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.