Abstract
This study aimed to evaluate the validity of a deep learning-based convolutional neural network (CNN) for detecting proximal caries lesions on bitewing radiographs. A total of 978 bitewing radiographs, 10,899 proximal surfaces, were evaluated by two endodontists and a radiologist, of which 2,719 surfaces were diagnosed and annotated with proximal caries and 8,180 surfaces were sound. The data were randomly divided into two datasets, with 818 bitewings in the training and validation dataset and 160 bitewings in the test dataset. Each annotation in the test set was then classified into 5 stages according to the extent of the lesion (E1, E2, D1, D2, D3). Faster R-CNN, a deep learning-based object detection method, was trained to detect proximal caries in the training and validation dataset and then was assessed on the test dataset. The diagnostic accuracy, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and receiver operating characteristic curve were calculated. The performance of the network in the overall and different stages of lesions was compared with that of postgraduate students on the test dataset. A total of 388 carious lesions and 1,435 sound surfaces were correctly identified by the neural network; hence, the accuracy was 0.87. Furthermore, 27.6% of lesions went undetected, and 7% of sound surfaces were misdiagnosed by the neural network. The sensitivity, specificity, PPV, and NPV of the neural network were 0.72, 0.93, 0.77, and 0.91, respectively. In contrast with the network, 52.8% of lesions went undetected by the students, yielding a sensitivity of only 0.47. The F1-score of the students was 0.57, while the F1-score of the network was 0.74 despite the accuracy of 0.82. A significant difference in the sensitivity was found between the model and the postgraduate students when detecting different stages of lesions (p < 0.05). For early lesions which limited in enamel and the outer third of dentin, the neural network had sensitivities all above or at 0.65, while students showed sensitivities below 0.40. From our results, we conclude that the CNN may be an assistant in detecting proximal caries on bitewings.
Keywords: Artificial intelligence, Bitewings, Proximal caries, Caries detection
Introduction
Dental caries is a common chronic disease that places a considerable burden on both individuals' quality of life and healthcare systems [Peres et al., 2019]. Bitewing radiography is commonly used to detect proximal caries, which require accurate diagnoses and early management and cannot be detected clinically due to tight contact surfaces [Gimenez et al., 2015]. Many factors, such as fatigue, emotions [Stec et al., 2018], and complex clinical environments [Hellén-Halme et al., 2008], could affect the accuracy of image interpretation. As reported by Tewary, the experience of clinical practice is the most influential factor [Tewary et al., 2011]. Compared with experienced examiners, low-experienced examiners were nearly four times as likely to make incorrect assessments when diagnosing proximal caries [Geibel et al., 2017].
An automated assistance system for dental radiography images may help to address these shortcomings by providing a reliable and stable diagnostic result, especially for less-experienced examiners. Additionally, this system may save diagnostic time and improve treatment efficiency.
Artificial intelligence (AI) is a branch of applied computer science that uses computer technology to simulate human behavior, such as intelligent activity, critical thinking, and decision-making [Shan et al., 2021]. Machine learning (ML), a subfield of AI, has been widely used in computer-aided diagnostic projects because it can identify meaningful patterns and structures in data and predict unknown data based on what it has learned [Chartrand et al., 2017]. However, deep learning (DL), a popular ML method characterized by multilayer neural networks and automatic feature extraction, has replaced traditional ML methods and emerged as the dominant method in healthcare. Convolutional neural networks (CNNs), a type of DL algorithm, have achieved favorable results and have become a state-of-the-art method [Esteva et al., 2019] in medical radiological diagnostic tasks, such as thoracic disease [Wang et al., 2017], mammographies [Teare et al., 2017], brain imaging [Puente-castro et al., 2020], and retinal fundus photographs [Gulshan et al., 2016]. To date, as a relatively novel method in the dental field, CNNs have been used in dentistry for a variety of purposes, such as determining periodontal bone loss [Lee et al., 2018a; Chang et al., 2020], detecting carious lesions [Srivastava et al., 2017; Tripathi et al., 2019; Cantu et al., 2020; Megalan Leo and Kalpalatha Reddy, 2020; Megalan Leo and Kalpalatha Reddy, 2021; Bayraktar and Ayan, 2022], segmenting apical lesions [Setzer et al., 2020], and identifying dental plaque [You et al., 2020].
However, only one study [Cantu et al., 2021] compared the results of interpreting bitewings with those of dentists, and one randomized trial [Mertens et al., 2021] assessed the impact of AI assistance on dentists. Comparisons of the performance between different neural networks and dentists are insufficient.
The present study aimed to evaluate the performance of the deep neural network for detecting proximal carious lesions on bitewings. Faster R-CNN was chosen, sampling strategy, anchor size setting, and many other parameters were adjusted to adapt to the caries detection task based on the training result. Then its diagnostic effectiveness was compared with that of postgraduate students.
The remainder of this paper is organized as follows. Section 2 gives an overview of the material and methods. The image datasets, the training process, and evaluation approaches are described in this section. Section 3 presents the results of our experiments. Then, Section 4 discusses and analyzes the obtained results, and Section 5 concludes the paper.
Materials and Methods
In the present study, we used Faster R-CNN, a type of CNN that has been widely applied for object detection in medical imaging. The detection network has three stages: feature extraction, lesion detection with region proposals, and generation of boxes for outputting the location of caries lesions.
Sample Size Estimation
Fisher's exact test was used to estimate the sample size of the test dataset. A clustered design was used since every bitewing contains multiple surfaces (about 11 surfaces on average in this study) which was calculated by design effect (DE) with the formula DE = 1 + (m − 1) × ICC, in which m means cluster size, and ICC is the intraclass correlation coefficient. ICC of 0.2 was assumed based on the previous study [Meinhold et al., 2020]. According to the previously published studies, we assumed the accuracy of 0.85 for AI and 0.75 for the postgraduate students [Schwendicke et al., 2015], a study powered at 1-beta = 0.8 and with alpha = 0.05 would require 534 units in the test set. When DE = 1 + (11 − 1) × 0.2, i.e., 3.0, the total surfaces required should be 1,602, or 146 bitewings. The final test dataset included in this study consisted of 160 radiographs.
Dataset
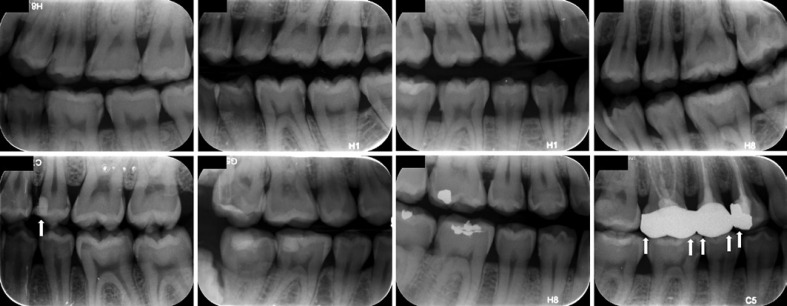
The dataset included 978 bitewing radiographs that were obtained during routine care at the Peking University School and Hospital of Stomatology in Beijing, China. The data were collected between November 2018 and July 2020. All the radiographs were obtained with the Gendex expert DC system (Gendex Dental System, Hatfield, PA, USA) at exposure settings of 65 kV, 7 mA, and an exposure time of 0.125–0.32 s. Proprietary storage phosphor plates were used to record the image. Each bitewing was exported with a resolution of 72 dpi at a size of approximately (960–980) × (750–755) pixels and saved as an 8-bit “JPG” format image file with a unique identification code. Only bitewings of permanent teeth with the crowns of at least one dental arch and at least one proximal carious lesion were included. Images of primary teeth with no proximal lesions were discarded, as were those with severe noise, haziness, distortion, and shadows. Some samples from the dataset were shown in Figure 1. Since the goal of this work was to detect proximal carious lesions, surface-level statistics were required; the mesial and distal surfaces of crowns without restorations and full crowns were calculated independently. A total of 978 bitewing radiographs were then randomly divided into the training and validation (n = 818) dataset and the testing (n = 160) dataset.
Fig. 1.
Samples of bitewing radiographs from the dataset. Surfaces with restorations or crowns were excluded (white arrows).
Reference Standard
Carious lesions in the bitewing radiographs were labeled by two endodontic experts and one radiologist with at least 5 years of relevant clinical experience to establish a fuzzy gold standard (reference standard). Prior to the labeling, each expert would receive a handbook containing diagnostic criteria and instructions of the software.
The proximal caries was defined as the radiolucent area between two adjacent contacts, visible as a notch in the enamel, a triangular shape with the apex pointing toward the enamel-dentinal junction, or the area at the dentinal junction penetrating toward the pulp. A bounding box was drawn by experts around each lesion to provide the Faster R-CNN for training and testing.
The labeling process was carried out using LabelMe (MIT, Cambridge, MA, USA) software, an open-source tool in computer vision research. In the present study, functions to adjust the brightness and contrast were added to mimic clinical diagnostic conditions. The screen resolution was 2,560 × 1,600, and the display ratio was 1:1. The images were labeled in a dimly lit room. For analytic purposes, each annotation in the test set was then classified into 5 stages: E1, E2, D1, D2, and D3. For a better fit with the real clinical situation, 5 stages were merged into three categories, “E1/E2” caries radiolucency in the outer or inner half of enamel; “D1” radiolucency in the outer third of dentin; “D2/D3” caries radiolucency in the middle and inner third of dentin.
Each expert labeled independently and was checked by others. When disagreement arose, consensus should be reached among three experts by discussion. Before the images were labeled, three observers were calibrated on diagnosing proximal caries. To evaluate consistency, each expert was asked to label the lesions again 1 week later. Fleiss kappa values were calculated to assess the Inter- and intra-observer agreement.
Faster R-CNN
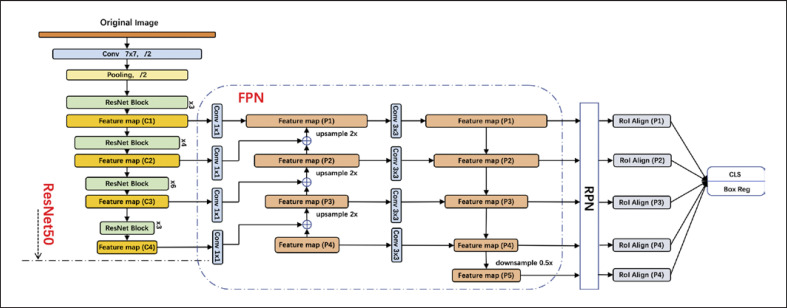
The Faster R-CNN model included two networks: a regional proposal network (RPN) for generating region proposals and an object detector network that uses these proposals to locate and classify objects, such as proximal caries lesions in this study. The two modules share convolution layers that produce representations of the raw image known as the feature map.
The raw image was input into the basic feature extraction network for convolution and pooling operations to build the feature map, which was then transmitted to the RPN to generate regional proposals. The RPN slid the window across the feature map, ranked the region boxes or candidate frames, which are known as “anchors,” and identified the regions most likely to contain objects. The RPN initially determined the position and score of the object to be detected. The output of the RPN, which consisted of a set of boxes linked by a special regressor, was then fed into the object detection network, which refined the score of the boxes to estimate whether the objects were present, and the final bounding box coordinates were generated. The architecture details are shown in Figure 2.
Fig. 2.
Overall architecture of the Faster R-CNN model.
Model Training
The Faster R-CNN model was trained on 818 labeled bitewing radiographs. The bitewings were scaled to 800 px on the shorter side and then augmented by applying random transformations, such as flipping the image, center cropping, rotating, Gaussian blurring, sharpening, and adjusting the contrast and brightness.
The network was pretrained on ImageNet to determine the initial weights. During the training process, the model weights were updated to minimize a binary-focal loss function using an Adam optimizer. The Faster R-CNN model was trained over 200 epochs (24 h) with 8 batch sizes and a learning rate of 1e−3. The entire process was carried out on an NVIDIA Tesla P40 GPU using CUDA 10.2. The model architecture and optimization process were carried out on the DL framework PaddlePaddle with Python.
A total of 160 bitewing radiographs from the test dataset were used to estimate the optimal Faster R-CNN algorithm weight factors. The results of two dental postgraduate students with less than 3 years of clinical experience were compared to the results of the neural network to assess the performance. The diagnosis standard and the calibration, consistency testing, and labeling procedures were the same as those of the experts.
Evaluation Metrics
The Fleiss kappa values were calculated in SPSS Statistics 19.0 software to determine the intra- and inter-observer agreement. The evaluation metrics of both the Faster R-CNN model and the postgraduate students were on the proximal surface level. The evaluation metrics were compared with the reference standards of the experts. The receiver operating characteristic curve (ROC) of the model was drawn. The diagnostic accuracy, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and F1-score were calculated. The F1-score was the harmonic mean of the PPV and the sensitivity. The diagnostic sensitivity and PPV at different stages were also calculated. The parametric data were analyzed using a McNemar's χ2 test to evaluate the differences between the network and the students, and a McNemar Bowker test was used to evaluate the difference in sensitivity between them. A value of p < 0.05 was considered statistically significant.
Results
The intra-observer and inter-observer Fleiss Kappa values were both greater than 0.75 (0.795–0.843) (Table 1), suggesting excellent agreement. In the present study, the reference standard established by the experts included 2,719 carious surfaces and 8,180 sound surfaces (Table 2).
Table 1.
Fleiss Kappa values for inter- and intra-observer agreement among experts
| Intra-observer | Inter-observer | |
|---|---|---|
| Kappa value | Kappa value | |
| Expert A | 0.843 | |
| Expert B | 0.842 | 0.796 |
| Expert C | 0.827 |
Table 2.
The distribution of carious and sound surfaces in the dataset
| Caries surfaces | Sound surfaces | Total | |
|---|---|---|---|
| Training and validation dataset | 2,183 | 6,629 | 8,812 |
| Test dataset | 536 | 1,551 | 2,087 |
| Total | 2,719 | 8,180 | 10,899 |
The Faster R-CNN model was evaluated with the test dataset, which included 536 carious surfaces and 1,551 sound surfaces. The accuracy, sensitivity, specificity, PPV and NPV, and F1-score are shown in Table 3. The neural network identified 388 of the 536 carious lesions, missing 27.6% and thus yielding a sensitivity of 0.72. The network misdiagnosed 116 of the 1,551 (0.7%) sound surfaces as caries, yielding a specificity of 0.93. In total, 388 carious surfaces and 1,435 sound surfaces were correctly diagnosed, yielding an overall accuracy of 0.87. In addition, the NPV, which reflects that the probability of a test is negative in the absence of a disease, was 0.91, while the PPV, which reflects the probability of a test is positive if a disease is present, was 0.77 (shown in Fig. 3).
Table 3.
Validity of the faster R-CNN model and the postgraduate students in diagnosing proximal caries on bitewings
| TP | FP | FN | TN | Accuracy | Sensitivity | Specificity | PPV | NPV | F1-score | |
|---|---|---|---|---|---|---|---|---|---|---|
| Neural network | 388 | 116 | 148 | 1,435 | 0.87 | 0.72 | 0.93 | 0.77 | 0.91 | 0.74 |
| Postgraduate student | 253 | 95 | 283 | 1,456 | 0.82 | 0.47 | 0.94 | 0.73 | 0.84 | 0.57 |
TP, true positive; FP, false positive; TN, true negative; FN, false negative; PPV, positive predictive value; NPV, negative predictive value.
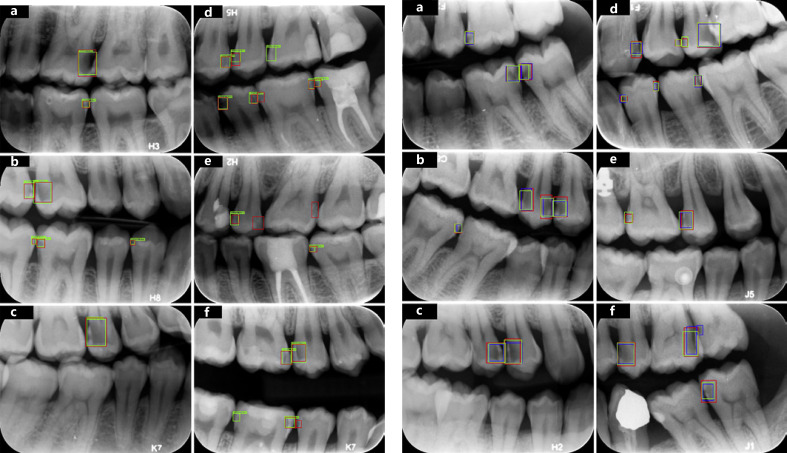
Fig. 3.
Sample results by the network. The standard proximal caries assessed by experts (red boxes) and those detected by the neural network (green boxes) are shown. Successful detections (both red and green boxes, a–f), false negative detections (red boxes only, d–f), and false positive detections (green boxes only, d–f) of the model.
Compared with the neural network, 283 of the 536 carious lesions, more than half (52.8%), went undetected by the students, nearly twice as many as the network, which resulted in a proximal caries detection sensitivity of only 0.47 for the students. However, only 95 of the 1,551 sound surfaces were misdiagnosed as caries, yielding a specificity of 0.94. The accuracy, PPV, and NPV were 0.82, 0.73, and 0.84, respectively. However, the F1-score of the network was 0.74, while the F1-score of the students was 0.57 (shown in Fig. 4). Overall, the neural network performed significantly better than the students (χ2 = 69.43642, p < 0.001).
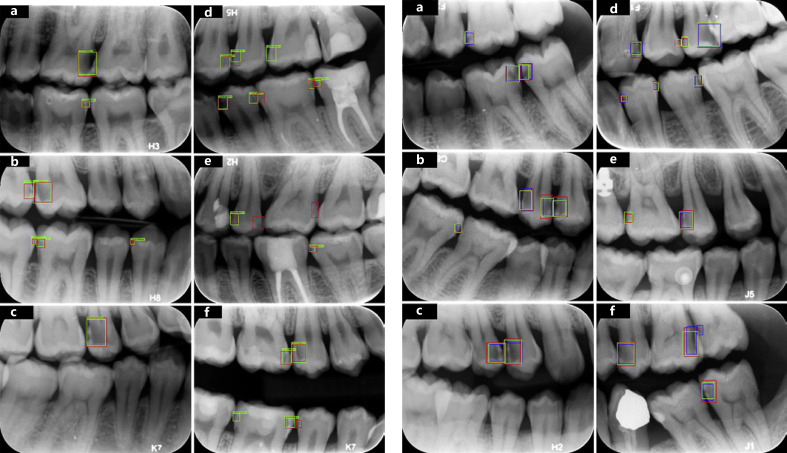
Fig. 4.
Sample cases by the network and the postgraduate students. The standard proximal caries assessed by experts (red boxes), the neural network (green boxes), and the postgraduate students (blue boxes) are shown. Successful detections (red, green, and blue boxes, a–f), false negative detections (red and green boxes only, d–f), and false positive detections (blue boxes only, d–f) of the students.
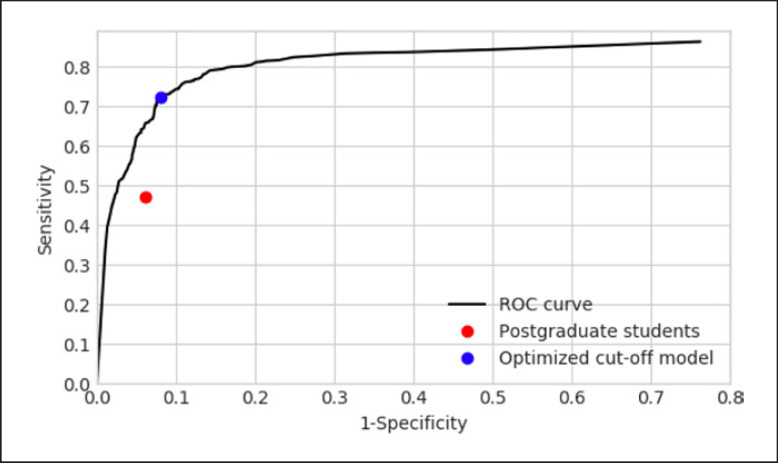
The ROC curve of the network and the students are shown in Fig. 5. The students' performance on the curve was to the lower right of the cutoff point.
Fig. 5.
Receiver operating characteristic (ROC) curve. The black line described indicates the performance of the neural network when evaluated with respect to in terms of sensitivity and specificity. The blue dot shows the sensitivity and specificity value of the network when it achieved at the optimal detection cutoff. The red dot represents the performance of the students' performance.
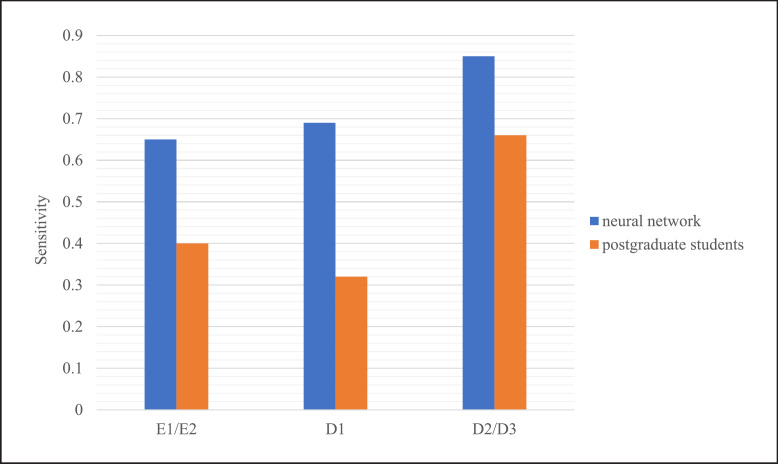
The distribution, sensitivity, and PPV of the neural network and the postgraduate students in diagnosing different stages of proximal caries are shown in Table 4 and Table 5. A significant difference in the sensitivity between the model and the students (χ2 = 93.849, p < 0.001). The model was more sensitive than the postgraduate students at different stages of the extent of lesions, all above or at 0.65. However, the students showed lower sensitivities, especially for lesions limited in enamel or outer third of dentin (0.40 and 0.32) (shown in Fig. 6).
Table 4.
Distribution of the neural network and the postgraduate students in diagnosing different stages of proximal carious lesions
| Lesions stage | Reference standard | Neural network |
Students |
||
|---|---|---|---|---|---|
| true | false | true | false | ||
| E1 | 63 | 18 | 19 | 14 | 22 |
| E2 | 188 | 145 | 53 | 87 | 44 |
| D1 | 105 | 72 | 30 | 34 | 18 |
| D2 | 100 | 80 | 13 | 58 | 9 |
| D3 | 80 | 73 | 1 | 60 | 2 |
| Total | 536 | 388 | 116 | 253 | 95 |
E1, lesions involving the outer half of enamel; E2, lesions involving the inner half of enamel; D1, lesions involving the outer third of dentin; D2, lesions involving the middle third of dentin; D3, lesions involving the inner third of dentin.
Table 5.
Sensitivity and PPV by the neural network and the postgraduate students on different stages of proximal carious lesions
| Lesions stage | Neural network |
Students |
||
|---|---|---|---|---|
| sensitivity | PPV | sensitivity | PPV | |
| E1/E2 | 0.65 | 0.69 | 0.40 | 0.60 |
| D1 | 0.69 | 0.71 | 0.32 | 0.65 |
| D2/D3 | 0.85 | 0.92 | 0.66 | 0.91 |
PPV, positive predictive value; E1/E2, lesions limited in enamel; D1, lesions involving the outer third of dentin; D2/D3, lesions involving the middle and inner third of dentin.
Fig. 6.
The sensitivities of the neural network and the postgraduate students in diagnosing different stages of proximal carious lesions.
Discussion
Bitewing radiography is a noninvasive radiographic procedure that has been widely used to detect and assess the extent of proximal carious lesions. In this study, the performance of CNNs for detecting proximal caries on bitewing radiographs was evaluated. As a type of DL algorithm in the field of AI, CNNs have an impressive track record in medical imaging analysis and interpretation; however, they are still a novel method in the field of dentistry. There have been some studies on using different neural networks for lesions detection in recent years [Srivastava et al., 2017; Tripathi et al., 2019; Cantu et al., 2020; Megalan Leo and Kalpalatha Reddy, 2021; Bayraktar and Ayan, 2022], but considering the practical clinical applications in the future, we taken the advantages of fast inference speed and high accuracy of Faster R-CNN to apply it to carious lesions detection task.
Faster R-CNN was developed on the basis of the R-CNN (region-based CNN) method, combining DL and traditional computer vision knowledge to achieve object detection. Faster R-CNN improved the network performance by using a neural network for classification instead of a traditional method; this model integrated extraction, judgment, and regression in one operation, thus improving the speed. The efficiency of Faster R-CNN was improved by relying on deep CNNs and running the algorithm on a GPU.
In the test set, the neural network exhibited a sensitivity and specificity of 0.72 and 0.93, respectively, in detecting proximal caries; 27.6% of lesions (388/536) were missed, while only 0.7% (116/1,551) of sound surfaces were misdiagnosed as caries lesions. This result was similar to that of Cantu [Cantu et al., 2020], in which U-Net was used to segment lesions on the bitewing radiographs, and Bayraktar [Bayraktar and Ayan, 2022], in which YOLO was used for detection. Both studies found that the specificity was greater than the sensitivity: 75% versus 83% in Cantu's study and 72.26% versus 98.19% in Bayraktar's study. However, a higher sensitivity and lower specificity (92.3% vs. 84.0%) for molars were reported by Lee [Lee et al., 2018b], who cropped periapical radiographs with one tooth in each image and did not limit the type and positions of caries.
The Faster R-CNN model showed a higher sensitivity value than the less-experienced students (0.72 vs. 0.47). The students missed more than half of the lesions, nearly doubling the risk of underestimation of proximal caries. With similar PPVs, the neural network also has a higher sensitivity than the students in diagnosing lesions at different stages. Especially for the lesions limited in enamel or outer third of dentin, the students had sensitivities as low as 0.32, while the model could achieve 0.65 and above. This was similar to the results of Cantu [Cantu et al., 2020], who found that the sensitivities of the network and dentists were 0.75 and 0.36, respectively. But for initial lesions, dentists had a very limited sensitivity of less than 0.25, compared to the model's sensitivity which is above 0.7. Cantu's interpretation of this result was that diagnosing small carious lesions was more difficult than detecting advanced lesions, especially for inexperienced dentists; however, this was not the case for the neural network. We also found that mistakes taken by the model in degree judgment also tend to occur in early lesions and the model tends to underestimate the extent of lesions. These findings suggest that neural networks could be useful in initial diagnoses and for rechecking lesions that may have been missed by less-experienced practitioners, which allows for early non- or micro-invasive treatment at an earlier stage. A randomized trial found that not only may dentists' sensitivity be improved with the help of AI in detecting early caries, but the risk of invasive treatment decisions was also improved [Mertens et al., 2021]. We remain neutral on this conclusion because radiography can only be used as an assistant method of diagnosis, and decision should not be made before a complete clinical examination. Thus, AI should work as assistance, a double check by dentist is necessary to avoid overtreatment.
The present study found that the accuracies of the neural network, and the students were similar (0.87 vs. 0.82), whereas the F1-score was 0.74 for the network and 0.57 for the students. The F1-score was used in the present study because the accuracy measures the correct cases without emphasizing false negatives and false positives, while the F1-score, defined as 2·(sensitivity·PPV)/(sensitivity + PPV), is the harmonic mean of the PPV and sensitivity, thus accounting for both false positives and false negatives. In medical cases where misdiagnoses are unacceptable, a high accuracy may not necessarily indicate a good performance. For example, if a method identifies each surface as a caries lesion, while the accuracy may be 1, but does not make any sense and may even cause irreversible damage to healthy teeth. However, a high F1-score indicates that a method can correctly identify positive cases while avoiding overestimation and underestimation.
The cutoff point on the ROC of the neural network was determined according to the maximum value of sensitivity × specificity to ensure that both the sensitivity and specificity were high, which was located near the upper left region of the curve. The neural network performed better than the students, whose cutoff point was in the lower right region of the curve.
Neural networks continuously improve by learning large amounts of data; thus, high-quality, consistent labels were prioritized. Three experts with more than 5 years of experience were involved in the present study. Together with strict diagnostic criteria, detailed calibration, double check, and consensus on the labeling process provided a high-quality, reliable “ground truth” establishment [Mohammad-Rahimi et al., 2022]. The distribution of the lesions, which is essential for estimating the diagnostic effectiveness, was similar in the training (24.8%) and testing set (25.7%) in this study. Therefore, under the same clinical conditions and equipment, the model is robust and did not run the risk of over-fitting. The model needs to be explored in randomized, practice-based settings [Mohammad-Rahimi et al., 2022] since class imbalance is the actual situation in real clinical radiographs. Furthermore, in the present study, the performance of the neural network was compared with that of postgraduate students on a hold-out test dataset, which was the data that the model has never seen during the training process and will only be used once the training has been finished. Since experience is found to be the most important factor in diagnostics and DL is well-known for its identification ability, it is necessary to compare the network with less-experienced practitioners to evaluate its performance. The results of this study showed that neural networks could be useful to assist dentists or in dental education, especially for early caries which limited in enamel and outer third of dentin. In addition, functions for adjusting the brightness and contrast were added to the labeling software to mimic real clinical environments, which was necessary considering that most diagnostic tasks are performed chairside. However, no testing was performed on an external dataset, such as bitewings from other hospitals. The performance of the neural network on more complicated and larger datasets needs further exploration.
Conclusion
The present study evaluated the performance of the neural network in detecting proximal caries in bitewing radiographs; this network may be useful in standardizing practitioners' imaging diagnoses and improving efficiency. However, the data source used in either this study or most of the previous ones was relatively homogenous; the generalizability of the model needs to be well evidenced in future studies.
Statement of Ethics
This study was approved by the Biomedical Institutional Review Board of Peking University School of Stomatology, document number PKUSSIRB-202274064. The consent protocol was reviewed, and the need for written and informed consent was waived by the Institutional Review Board of Peking University School of Stomatology, as this study had a noninterventional retrospective design with extremely low risk to the subjects, and all the data were anonymized prior to analysis.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
No funding sources contributed to this research.
Author Contributions
Xiaotong Chen, Jiachang Guo, and Yuhong Liang contributed to the study conception and idea. The design of methodology and data analysis was performed by Xiaotong Chen and Jiachang Guo. Data preparation and collection were carried out by Xiaotong Chen and Jiaxue Ye. The recheck of the data was accomplished by Mingming Zhang. The paper was written by Xiaotong Chen, Jiachang Guo, and Yuhong Liang contributed to interpretation and analysis of the results. All the authors read and edited this paper.
Data Availability Statement
The data that support the findings of this study are not publicly available due to their containing private information of research participants but are available from the corresponding author, Y.H. Liang, upon reasonable request.
Funding Statement
No funding sources contributed to this research.
References
- 1.Bayraktar Y, Ayan E. Diagnosis of interproximal caries lesions with deep convolutional neural network in digital bitewing radiographs. Clin Oral Investig. 2022;26((1)):623–632. doi: 10.1007/s00784-021-04040-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantu AG, Gehrung S, Krois J, Chaurasia A, Rossi JG, Gaudin R, et al. Detecting caries lesions of different radiographic extension on bitewings using deep learning. J Dent. 2020;100:103425. doi: 10.1016/j.jdent.2020.103425. [DOI] [PubMed] [Google Scholar]
- 3.Chang HJ, Lee SJ, Yong TH, Shin NY, Jang BG, Kim JE, et al. Deep learning hybrid method to automatically diagnose periodontal bone loss and stage periodontitis. Sci Rep. 2020;10((1)):1–8. doi: 10.1038/s41598-020-64509-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, et al. Deep learning: a primer for radiologists. Radiographics. 2017;37((7)):2113–2131. doi: 10.1148/rg.2017170077. [DOI] [PubMed] [Google Scholar]
- 5.Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, et al. A guide to deep learning in healthcare. Nat Med. 2019;25((1)):24–29. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 6.Geibel MA, Carstens S, Braisch U, Rahman A, Herz M, Jablonski-Momeni A. Radiographic diagnosis of proximal caries: influence of experience and gender of the dental staff. Clin Oral Investig. 2017;21((9)):2761–2770. doi: 10.1007/s00784-017-2078-2. [DOI] [PubMed] [Google Scholar]
- 7.Gimenez T, Piovesan C, Braga MM, Raggio DP, Deery C, Ricketts DN, et al. Clinical relevance of studies on the accuracy of visual inspection for detecting caries lesions: a systematic review. Caries Res. 2015;49((2)):91–98. doi: 10.1159/000365948. [DOI] [PubMed] [Google Scholar]
- 8.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316((22)):2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 9.Hellén-Halme K, Petersson A, Warfvinge G, Nilsson M. Effect of ambient light and monitor brightness and contrast settings on the detection of approximal caries in digital radiographs: an in vitro study. Dentomaxillofac Radiol. 2008;37((7)):380–384. doi: 10.1259/dmfr/26038913. [DOI] [PubMed] [Google Scholar]
- 10.Lee JH, Kim DH, Jeong SN, Choi SH. Diagnosis and prediction of periodontally compromised teeth using a deep learning-based convolutional neural network algorithm. J Periodontal Implant Sci. 2018a;48((2)):114–123. doi: 10.5051/jpis.2018.48.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JH, Kim DH, Jeong SN, Choi SH. Detection and diagnosis of dental caries using a deep learning-based convolutional neural network algorithm. J Dent. 2018b;77:106–111. doi: 10.1016/j.jdent.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 12.Megalan Leo L, Kalpalatha Reddy T. Dental caries classification system using deep learning based convolutional neural network. J Comput Theor Nanosci. 2020;17((9–10)):4660–4665. [Google Scholar]
- 13.Megalan Leo L, Kalpalatha Reddy T. Learning compact and discriminative hybrid neural network for dental caries classification. Microprocess Microsyst. 2021;82:103836. [Google Scholar]
- 14.Meinhold L, Krois J, Jordan R, Nestler N, Schwendicke F. Clustering effects of oral conditions based on clinical and radiographic examinations. Clin Oral Investig. 2020;24((9)):3001–3008. doi: 10.1007/s00784-019-03164-9. [DOI] [PubMed] [Google Scholar]
- 15.Mertens S, Krois J, Cantu AG, Arsiwala LT, Schwendicke F. Artificial intelligence for caries detection: Randomized trial. J Dent. 2021;115:103849. doi: 10.1016/j.jdent.2021.103849. [DOI] [PubMed] [Google Scholar]
- 16.Mohammad-Rahimi H, Motamedian SR, Rohban MH, Krois J, Uribe S, Nia EM, et al. Deep learning for caries detection: A systematic review: DL for caries detection. J Dent. 2022;112:104115. doi: 10.1016/j.jdent.2022.104115. [DOI] [PubMed] [Google Scholar]
- 17.Peres MA, Macpherson LM, Weyant RJ, Daly B, Venturelli R, et al. Oral diseases: A global public health challenge. Lancet. 2019;394((10194)):249–260. doi: 10.1016/S0140-6736(19)31146-8. [DOI] [PubMed] [Google Scholar]
- 18.Puente-Castro A, Fernandez-Blanco E, Pazos A, Munteanu CR. Automatic assessment of Alzheimer's disease diagnosis based on deep learning techniques. Comput Biol Med. 2020;120:103764. doi: 10.1016/j.compbiomed.2020.103764. [DOI] [PubMed] [Google Scholar]
- 19.Schwendicke F, Tzschoppe M, Paris S. Radiographic caries detection: A systematic review and meta-analysis. J Dent. 2015;43((8)):924–933. doi: 10.1016/j.jdent.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Setzer FC, Shi KJ, Zhang Z, Yan H, Yoon H, Mupparapu M, et al. Artificial intelligence for the computer-aided detection of periapical lesions in cone-beam computed tomographic images. J Endod. 2020;46((7)):987–993. doi: 10.1016/j.joen.2020.03.025. [DOI] [PubMed] [Google Scholar]
- 21.Shan T, Tay FR, Gu L. Application of artificial intelligence in dentistry. J Dental Res. 2021;100((3)):232–244. doi: 10.1177/0022034520969115. [DOI] [PubMed] [Google Scholar]
- 22.Srivastava MM, Kumar P, Pradhan L, Varadarajan S. Detection of tooth caries in bitewing radiographs using deep learning. arXiv. 2017 Epub ahead of print. [Google Scholar]
- 23.Stec N, Arje D, Moody AR, Krupinski EA, Tyrrell PN. A systematic review of fatigue in radiology: is it a problem? AJR Am J Roentgenol. 2018;210((4)):799–806. doi: 10.2214/AJR.17.18613. [DOI] [PubMed] [Google Scholar]
- 24.Teare P, Fishman M, Benzaquen O, Toledano E, Elnekave E. Malignancy detection on mammography using dual deep convolutional neural networks and genetically discovered false color input enhancement. J Digit Imaging. 2017;30((4)):499–505. doi: 10.1007/s10278-017-9993-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tewary S, Luzzo J, Hartwell G. Endodontic radiography: who is reading the digital radiograph? J Endod. 2011;37((7)):919–921. doi: 10.1016/j.joen.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 26.Tripathi P, Malathy C, Prabhakaran M. Genetic algorithms based approach for dental caries detection using back propagation neural network. Int J Recent Technol Eng. 2019;8((1)):317–319. [Google Scholar]
- 27.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases; In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2017. pp. p. 2097–106. [Google Scholar]
- 28.You W, Hao A, Li S, Wang Y, Xia B. Deep learning-based dental plaque detection on primary teeth: a comparison with clinical assessments. BMC Oral Health. 2020;20((1)):1–7. doi: 10.1186/s12903-020-01114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are not publicly available due to their containing private information of research participants but are available from the corresponding author, Y.H. Liang, upon reasonable request.