Summary
Phenotypic associations have been reported between blood cell traits (BCTs) and a range of neurological and psychiatric disorders (NPDs), but in most cases, it remains unclear whether these associations have a genetic basis and, if so, to what extent genetic correlations reflect causality. Here, we report genetic correlations and Mendelian randomization analyses between 11 NPDs and 29 BCTs, using genome-wide association study summary statistics. We found significant genetic correlations for four BCT-NPD pairs, all of which have prior evidence for a phenotypic correlation. We identified a previously unreported causal effect of increased platelet distribution width on susceptibility to Parkinson’s disease. We identified multiple functional genes and regulatory elements for specific BCT-NPD pairs, some of which are targets of known drugs. These results enrich our understanding of the shared genetic landscape underlying BCTs and NPDs and provide a robust foundation for future work to improve prognosis and treatment of common NPDs.
Keywords: blood cell traits, neurological and psychiatric disorders, genetic correlation, Mendelian randomization, blood-based biomarkers, platelets, Parkinson’s disease
Graphical abstract
Highlights
-
•
Extensive genome-wide and local genetic correlations between BCTs and NPDs
-
•
Evidence for a causal effect of platelet distribution width on Parkinson’s disease
-
•
Multiple functional genes and regulatory elements shared by specific BCT-NPD pairs
-
•
Genetic evidence for potential drug re-purposing candidates for common NPDs
Yang et al. report a broad landscape of genetic overlap between 29 blood cell traits (BCTs) and 11 neurological and psychiatric disorders (NPDs), including previously unreported genome-wide and local genetic correlations, a causal effect of platelet distribution width on Parkinson’s disease, and numerous functional genes and regulatory elements shared by specific BCT-NPD pairs. These results shine light on the close relationship of BCTs with risk for NPDs and provide a foundation for improving the prognosis and treatment of common NPDs.
Introduction
Variation in the functional and physiological properties of blood cells has been associated with a range of neurological and psychiatric disorders (NPDs), including major depressive disorder (MDD),1 schizophrenia (SCZ),2 multiple sclerosis (MS),3 stroke,4 and Parkinson’s disease (PD).5 Some phenotypic associations between blood cell traits (BCTs) and NPD diagnoses have been shown to have a genetic basis,6 in some cases consistent with the presence of a causal effect of hematological indices on disease. For example, Astle et al.7 reported that elevated lymphocyte count (LYMPH#) causally increases the risk for MS and SCZ, Harshfield et al.8 reported a causal role of higher plateletcrit (PCT) and eosinophil percentage of white blood cells (EO%) in susceptibility to ischemic stroke and its subtypes (i.e., cardioembolic stroke, large-artery atherosclerotic stroke), and Sealock et al.1 reported a causal effect of increased white blood cell count (WBC) on risk for depression. Previous studies have also identified specific genes shared by pairs of BCTs and NPDs, such as tyrosine kinase 2 (TYK2) underlying MS and T lymphocyte polarization,9 and the well-established PD risk gene SNCA (encoding the protein alpha-synuclein), which also plays a role in development of red blood cells.10 These discoveries improve understanding of disease etiology and potential points of intervention through re-purposing of drugs.
Despite this progress, the genetic relationships between BCTs and many NPDs remain unclear. There is also uncertainty in relation to whether genetic correlations between BCTs and NPDs predominantly reflect horizontal pleiotropy, whereby genetic variants have effects on both members of a trait pair via one (correlated horizontal pleiotropy) or more (uncorrelated horizontal pleiotropy) independent pathways. Alternatively, a causal relationship may exist between BCT and NPD, potentially involving other traits downstream of the exposure but on the same causal pathway linking the exposure to the outcome (referred to as “vertical pleiotropy”).11 A more comprehensive understanding of genetic overlap and causal relationships between BCTs and NPDs is needed to determine whether hematological measures (which are more accessible and well established than brain-based markers) represent meaningful predictive or prognostic biomarkers for risk of common brain disorders,12,13,14,15 and if hematopoietic pathways may even represent legitimate targets for development of disease-modifying treatments.
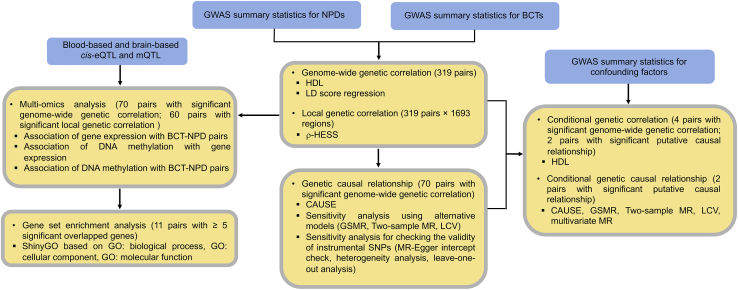
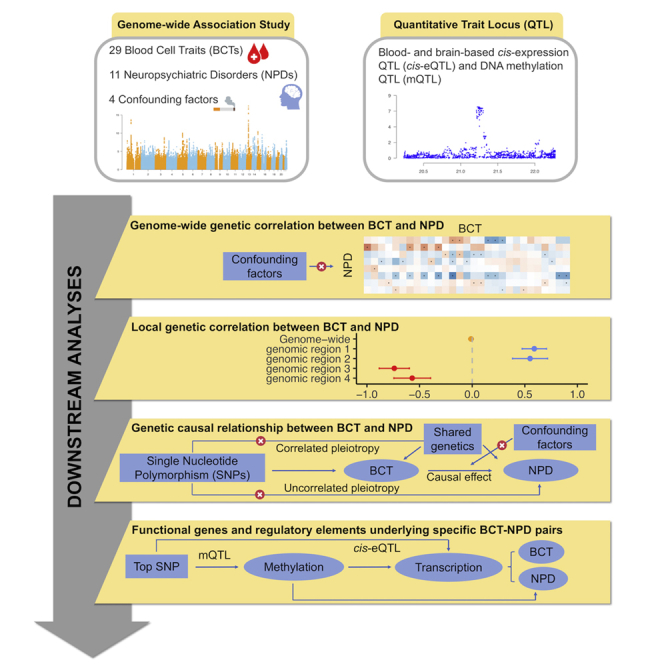
In this study, we used large-scale genome-wide association study (GWAS) summary statistics for 29 BCTs and 11 common NPDs (Tables 1 and 2) to estimate global and local genetic correlations between BCT-NPD pairs using high-definition likelihood (HDL)16 and heritability estimation from summary statistics (ρ-HESS),17 respectively. We then performed multiple Mendelian randomization (MR) analyses18,19,20,21,22,23,24 to explore evidence for causality between BCTs and NPDs, and we applied summary data-based MR (SMR)25 to identify putatively functional genes and regulatory elements shared between pairs of BCTs and NPDs. A flowchart of the main analytic steps is provided in Figure 1.
Table 1.
GWAS datasets for BCTs used in the study
| BCT | Class | Abbreviation |
|---|---|---|
| Basophil count | White cells | BASO# |
| Basophil percentage of white cells | White cells | BASO% |
| Eosinophil count | White cells | EO# |
| Eosinophil percentage of white cells | White cells | EO% |
| Lymphocyte count | White cells | LYMPH# |
| Lymphocyte percentage of white cells | White cells | LYMPH% |
| Monocyte count | White cells | MONO# |
| Monocyte percentage of white cells | White cells | MONO% |
| Neutrophil count | White cells | NEUT# |
| Neutrophil percentage of white cells | White cells | NEUT% |
| White blood cell count | White cells | WBC |
| Mean platelet volume | Platelets | MPV |
| Plateletcrit | Platelets | PCT |
| Platelet distribution width | Platelets | PDW |
| Platelet count | Platelets | PLT# |
| Hematocrit | Red cells | HCT |
| Hemoglobin | Red cells | HGB |
| High light scatter reticulocyte count | Red cells | HLSR# |
| High light scatter percentage of red cells | Red cells | HLSR% |
| Immature fraction of reticulocytes | Red cells | IRF |
| Mean corpuscular hemoglobin | Red cells | MCH |
| Mean corpuscular hemoglobin concentration | Red cells | MCHC |
| Mean corpuscular volume | Red cells | MCV |
| Mean reticulocyte volume | Red cells | MRV |
| Mean spheric corpuscular volume | Red cells | MSCV |
| Red blood cell count | Red cells | RBC# |
| Red cell distribution width | Red cells | RDW |
| Reticulocyte count | Red cells | RET# |
| Reticulocyte fraction of red cells | Red cells | RET% |
Table 2.
GWAS datasets for neurological and psychiatric disorders used in this study
| NPDs (abbreviation) by typea | Publication | n cases | n controls | Eff. nb | n SNPsc |
|---|---|---|---|---|---|
| Alzheimer’s disease (AD) | Jansen et al.27 | 71,880 | 383,378 | 121,062 | 9.74M |
| Amyotrophic lateral sclerosis (ALS) | Nicolas et al.28 | 20,806 | 59,804 | 30,872 | 8.85M |
| Migraine (migraine) | Gormlet et al.29 | 59,674 | 316,078 | 100,394 | 8.94M |
| Multiple sclerosis (MS) | IMSGC30 | 14,802 | 26,703 | 19,046 | 6.52M |
| Parkinson’s disease (PD) | Nalls et al.31 | 37,688 cases, 18,618 proxy cases | 1,417,791 | 108,311 | 7.52M |
| Stroke (stroke) | Malik et al.32 | 40,585 | 406,111 | 73,795 | 8.26M |
| Attention-deficit/hyperactivity disorder (ADHD) | Demontis et al.33 | 19,099 | 34,194 | 24,509 | 6.91M |
| Autism spectrum disorder (ASD) | Grove et al.34 | 18,381 | 27,969 | 22,183 | 9.11M |
| Bipolar disorder (BIP) | Stahl et al.35 | 20,352 | 31,358 | 24,684 | 9.64M |
| Major depressive disorder (MDD) | Wray et al.36 | 116,404 | 314,990 | 169,989 | 9.51M |
| Schizophrenia (SCZ) | PGC37 | 51,900 | 71,675 | 60,205 | 9.55M |
GWAS, genome-wide association study; IMSGC, International Multiple Sclerosis Genetics Consortium; NPD, neurological and psychiatric disorder; PGC, Psychiatric Genomics Consortium.
NPD, neurological and psychiatric disorder
Eff n: Effective sample size calculated from RICOPILI.38
SNPs with minor allele frequency < 1%.
Figure 1.
Overview of the main analytic steps performed in the study
Results
Genetic correlations between BCTs and neurological and psychiatric disorders
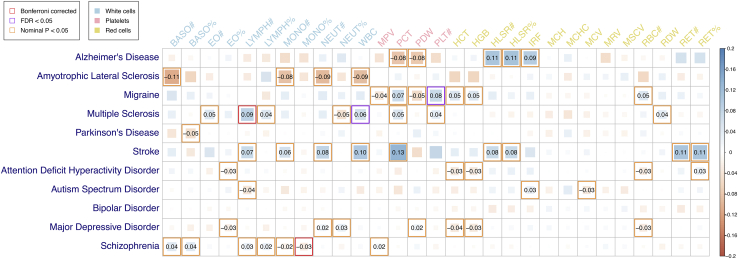
We observed Bonferroni significant (p < 1.57 × 10−4) genetic correlations (rg) for two (of 319) BCT-NPD pairs using the HDL method (MS and LYMPH#: rg = 0.09, SE = 0.02, p = 3.86 × 10−6; SCZ and monocyte percentage of white cells [MONO%]: rg = −0.03, SE = 0.01, p = 2.11 × 10−5), and a further two trait pairs (migraine and platelet count [PLT#]: rg = 0.08, SE = 0.02, p = 3.94 × 10−4; MS and WBC: rg = 0.06, SE = 0.02, p = 4.78 × 10−4) surpassed a less stringent 5% Benjamini-Hochberg false discovery rate (FDR) (Figure 2; Table S1). Notably, each of these four trait pairs had prior evidence for a significant phenotypic correlation in the same direction.39,40,41,42 We then re-estimated each of these four genetic correlations after using multi-trait-based conditional and joint analysis (mtCOJO)23 to condition each trait on each of four potential confounding factors, including cigarettes per day,43 drinks per week,43 educational attainment,44 and household income.45 The rg estimates from these conditional HDL analyses were highly consistent with our original estimates, suggesting that these confounding factors have negligible effects on the shared genetics underlying the focal pairs of BCTs and NPDs (Figure S1; Table S2).
Figure 2.
Estimated genetic correlations between BCTs and NPDs using the HDL method
Significant genetic correlations (with estimates provided) are highlighted by red, purple, or orange borders if they were Bonferroni significant (p < 1.57 × 10−4), FDR significant (p < ∼7 × 10−3), or nominally significant (p < 0.05), respectively. See Table S1 for complete details of HDL estimates.
The magnitude of rg estimates between pairs of BCTs and NPDs were weak to moderate, ranging from −0.11 to 0.13. The HDL rg estimates were highly consistent with those estimated by linkage disequilibrium (LD) score regression (LDSC; R = 0.81, 95% confidence interval [CI] = 0.76–0.84; Figures S2 and S3). Considering all nominally significant genetic correlations, HDL identified 56 pairs of BCTs and NPDs and LDSC identified 28, of which 14 were not seen with HDL (Table S3; Figure S2). We also estimated the rg among 406 BCT pairs, and whereas most BCTs were genetically distinct (322 of 406 pairs with absolute value of rg < 0.2), we identified strong positive and negative rg for a subset of BCT pairs (Figures S4 and S5; Tables S4 and S5), consistent with hematopoietic cell type classifications reported by Astle et al.7 For example, we found strong positive rg among white blood cell measures (e.g., LYMPH#, monocyte count, neutrophil count, WBC) but negative rg among ratios of these white blood cell count measures (e.g., lymphocyte percentage of white cells, MONO%, neutrophil percentage of white cells [NEUT%]).
Local genetic correlations between BCTs and neurological and psychiatric disorders
We used ρ-HESS17 to estimate local genetic correlations between 319 pairs of BCTs and NPDs at 1,693 approximately LD-independent genomic regions (excluding the major histocompatibility complex [MHC] region). The rationale was that a negligible genome-wide genetic correlation may obscure meaningful genetic correlations in defined genomic regions, and the pattern of local genetic correlations in trait-associated regions can reveal putative causal relationships between traits.17 As a validity check, we first compared the genome-wide sum of local genetic correlations per trait pair to genome-wide rg estimates (Table S6) from HDL (and LDSC), finding that these were highly correlated (R = 0.83, 95% CI = 0.79–0.86 for ρ-HESS and HDL; R = 0.74, 95% CI = 0.68–0.79 for ρ-HESS and LDSC; Figure S6), as expected.
Next, we considered if there was evidence for specific genomic regions that contribute disproportionately to trait covariance (Tables S7, S8, S9, S10, S11, S12, S13, S14, S15, S16, and S17). We identified 32 genomic regions (involving 74 trait pairs) with a Bonferroni significant (p < 9.26 × 10−8) local genetic correlation and significant local SNP heritability for both traits (Figure S7; Table S18). These included the APOE region on chromosome 19 (hg19: 44.7–46.1 Mb) contributing to Alzheimer’s disease (AD) and four BCT traits, and two regions on chromosomes 4 (0.7–1.5 Mb) and 17 (43.1–45.9 Mb) contributing to PD and a total of 20 BCT traits, the latter encompassing the highly pleiotropic 17q21 inversion region, which contains the microtubule-associated protein tau (MAPT) gene associated with AD46 and frontotemporal dementia.47 Additionally, nine and 24 regions were found to contribute to 24 MS-BCT and 26 SCZ-BCT pairs, respectively. A majority (128 of 166) of these significant local genetic correlations involved trait pairs for which there was no evidence of genome-wide rg from HDL or LDSC. However, most regions contained genome-wide significant SNPs for both traits (141 of 166), for the NPD (141 of 166) or for the BCT (165 of 166).
Finally, we explored if any BCT-NPD pairs exhibited a pattern of local genetic correlations that were consistent with a putative causal relationship between traits: that is, trait pairs for which the average local genetic correlation was significantly different in BCT-versus NPD-associated regions of the genome. We identified no single BCT-NPD trait pair satisfying this criterion after Bonferroni (p < 1.57 × 10−4) or FDR (p < ∼1.50 × 10−4) correction for multiple testing, irrespective of the p value cut-off (or SBayesR48) used to define trait-specific SNPs and genomic regions (Figures S8–S14; Tables S19, S20, S21, S22, S23, S24, and S25).
Putative causal effects of plateletcrit on stroke and platelet distribution width on PD
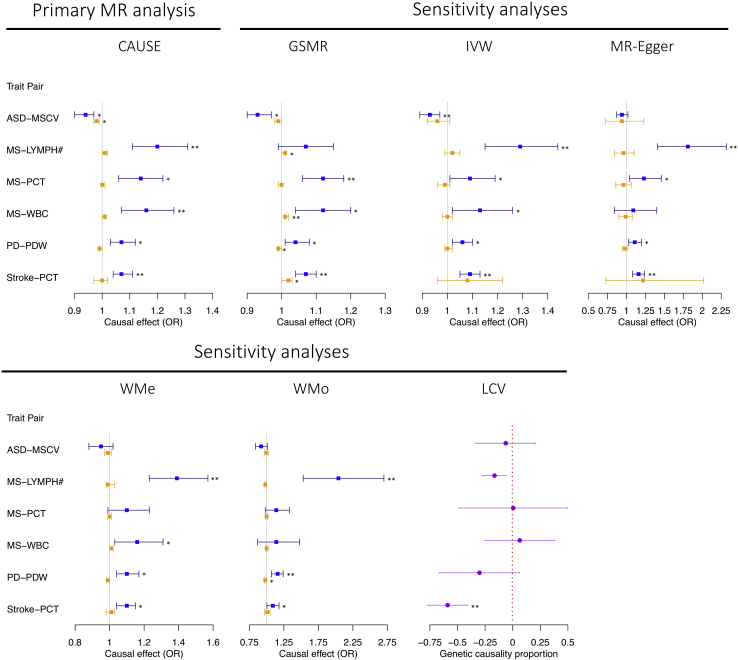
Next, we used the CAUSE (causal analysis using summary effect estimates)18 method to perform bi-directional MR analyses for pairs of BCTs and NPDs with evidence for a nominally significant genome-wide rg (from HDL or LDSC; n = 70 trait pairs; see Figures 2 and S2), recognizing that causal relationships between highly polygenic traits (such as the BCTs and NPDs included in our study) are more likely in the presence of a global genetic correlation. We identified three Bonferroni-significant (p < 3.57 × 10−4, i.e., ) causal relationships (increased LYMPH# on risk for MS: odds ratio [ORCAUSE] = 1.20 [i.e., a 1.2-fold increase in risk for MS for each SD increase in LYMPH#], 95% CI = 1.11–1.31, p = 3.94 × 10−5; increased WBC on MS: ORCAUSE = 1.16, 95% CI = 1.07–1.26, p = 2.60 × 10−4; increased PCT on stroke: ORCAUSE = 1.07, 95% CI = 1.04–1.11, p = 6.27 × 10−6), two of which remained significant after conservatively adjusting for all 319 trait pairs (i.e., LYMPH# and MS, PCT and stroke). Notably, the genetic correlation between LYMPH# and WBC (both measures of white cells) is 0.64 (SE = 0.06, p = 1.27 × 10−31), suggesting that the putative causal effects of these BCTs on MS represents a partly overlapping signal. A further three potential causal relationships were identified at a less stringent 5% FDR significance level: reduced mean spheric corpuscular volume (MSCV) on autism spectrum disorder (ASD; ORCAUSE = 0.94, 95% CI = 0.90–0.97, p = 6.45 × 10−4), increased PCT on MS (ORCAUSE = 1.14, 95% CI = 1.06–1.22, p = 4.29 × 10−4), and increased platelet distribution width (PDW) on PD (ORCAUSE = 1.07, 95% CI = 1.03–1.12, p = 6.40 × 10−4). In the reverse analyses, there was no evidence for a causal effect of any NPD on any BCT (Figure 3; Table S26), suggesting the significant genetic correlations between BCTs and NPDs are unlikely to be driven by reverse causality.
Figure 3.
Summary of significant putative causal relationships between BCTs and NPDs identified by CAUSE (primary MR analysis) and of MR sensitivity analyses (including LCV and five two-sample MR methods) for each of these relationships
Results colored in blue represent the estimated causal effect of BCTs on NPDs, while results colored in orange represent the estimated causal effect of NPDs on BCTs. Error bars for CAUSE and the five two-sample MR methods represent 95% confidence intervals, and those for LCV-based GCP point estimates represent standard errors. For LCV, a negative GCP indicates a causal effect of BCT on NPD, and vice versa. Single and double asterisks indicate results surpassing nominal significance (p < 0.05) and Bonferroni significance (p < 3.05 × 10−4), respectively. See Tables S26 and S27 for complete details of the estimates.
To further assess these potential causal relationships, we performed a series of sensitivity MR analyses, cognizant of the strong a priori expectation for pleiotropy under the plausible assumption that changes in BCTs are inherent to the disease process. Using six alternative MR methods, we identified consistent evidence for a causal effect of increased PCT on stroke, with p < 0.05 for all methods and four of six surpassing the Bonferroni significance threshold (Figure 3; Table S27). Notably, the estimated genetic causality proportion (GCP) from latent causal variable (LCV) analysis for an effect of PCT on stroke was 0.69 (SE = 0.21, p = 4.43 × 10−23), which (to put this in perspective) is roughly equivalent to the GCP estimate for the effect of high cholesterol on risk for myocardial infarction (GCP = 0.70).24 Consistent with this evidence for a causal relationship, we observed a significant (albeit modest) positive phenotypic correlation (rP) between PCT and stroke in the UK Biobank (UKB; rP = 0.01 and p = 2.86 × 10−5, assuming prevalence of 5%; see Data S1; Table S28).
We also found consistent evidence for a causal effect of elevated PDW on PD, with p < 0.05 for five of six MR methods (Figure 3; Table S27). The exception was LCV (GCP = 0.35, SE = 0.43, p = 0.17), which may be explained by the small estimated rg between PDW and PD (rg(HDL) = 0.03, SE = 0.02, p = 0.09; rg(LDSC) = 0.05, SE = 0.03, p = 0.04), given that the LCV method is known to produce conservative p values for traits with low rg.24 Again, as would be expected given a causal relationship, we observed a positive rP between PDW and PD in the UKB, although the estimate was only marginally significant (rP ∼ 0.01, p = 8.44 × 10−4 to 0.07 depending on prevalence; Data S1; Table S28), potentially because of the modest number of PD cases in the cohort (n = 1,323 compared with >5,000 for stroke).
To further evaluate the reliability of the inferred causal effects of PCT on stroke and PDW on PD, we performed several additional sensitivity analyses. We first checked the MR-Egger intercept terms in each analysis, confirming there was no evidence for non-zero estimates, and thus no indication that the MR-Egger causal estimates were confounded by pleiotropy (Table S29). Second, we performed leave-one-out analyses for each trait pair using each of the four two-sample MR methods (IVW, MR-Egger, weighted median [WMe], and weighted mode [WMo]). In all instances, there was no evidence that any single instrumental SNP was responsible for the inference of a causal relationship for either PCT-stroke or PDW-PD (Figure S15, Tables S30, S31, S32, and S33). Third, we checked for heterogeneity of instrumental SNP effects in the IVW and MR-Egger analyses, and although there was evidence for heterogeneity, after removing pleiotropic SNPs identified by generalized summary data-based MR (GSMR), the causal estimates from IVW and MR-Egger for PCT-stroke and PDW-PD remained significant and highly consistent with the original estimates (Table S29).
As a final sensitivity analysis, we explored if the causal effects of PCT on stroke and PDW on PD were influenced by common environmental factors associated with disease risk, including smoking,43 alcohol consumption,43 educational attainment,44 and socioeconomic status.45 We conditioned GWAS statistics for PCT, stroke, PDW and PD on each potential cofounder using mtCOJO and then repeated each of the MR analyses (i.e., CAUSE, GSMR, IVW, MR-Egger, WMe, and WMo) and LCV using the conditioned GWAS summary statistics (see STAR Methods). These conditional MR (and LCV) analyses were all highly consistent with our primary results (Figure S16; Table S34). We also applied multivariable MR (MVMR)49 analysis to PCT-stroke and PDW-PD, adjusting for potential pleiotropic effects of all four confounders concurrently. Again, these analyses were highly consistent with the causal inference from our primary analyses (Table S35), further supporting unidirectional causal effects of PCT on stroke and PDW on PD.
In addition to PCT-stroke and PDW-PD, we also observed suggestive support for higher LYMPH# on MS, with four of six sensitivity analyses surpassing Bonferroni significance (Figure 3; Table S27). However, effect size estimates varied widely, and contrary to expectations, there was no evidence for a phenotypic correlation between LYMPH# and MS in the UKB (rP ∼ −0.002, p > 0.50; Data S1; Table S28). These findings suggest that further research will be needed to confirm this putative causal relationship.
As expected, sensitivity analyses using stroke, PD and MS as exposures for PCT, PDW and LYMPH# (respectively), were universally (or largely) non-significant. Sensitivity analyses of the effect of MSCV on ASD, and of PCT and WBC on MS were highly inconsistent, and thus we could not further determine if their genetic correlations were due to causality or pleiotropy.
Prioritization of putatively functional genes and regulatory elements shared by BCTs and neurological and psychiatric disorders
Finally, we used SMR to identify putatively functional genes and regulatory elements shared by specific pairs of BCTs and NPDs because of causality or pleiotropy.
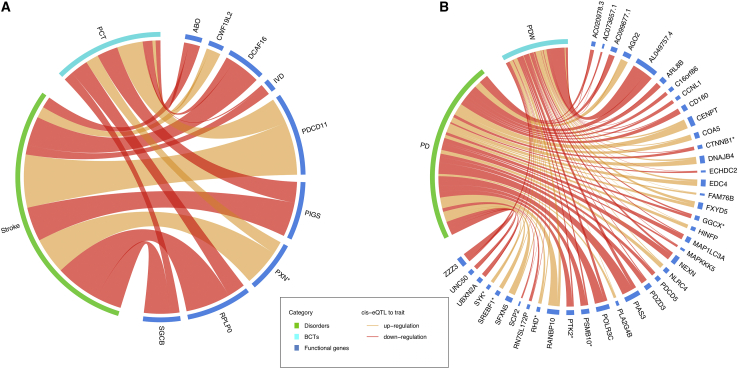
We first applied SMR to PCT, PDW, stroke and PD to identify functional genes and regulatory elements underlying the putative causal effects of elevated PCT on risk of stroke and increased PDW on risk for PD. In analyses using blood-based cis-expression quantitative trait loci (cis-eQTL) in eQTLGen50 (Table S36) or platelet cis-eQTLs from GeneSTAR51 (Table S37), no single gene survived Bonferroni correction (pSMR < 3.18 × 10−6) for both PDW and PD, or PCT and stroke. However, a total of 18 genes were Bonferroni significant for PCT (n = 5), PDW (n = 12), or PD (n = 1), in agreement with evidence for stronger genetic signals for exposures (i.e., PCT, PDW) than outcomes (i.e., stroke, PD). At a less stringent 5% FDR threshold, we identified 56 significant genes (also passing the heterogeneity in dependent instruments [HEIDI] test) for PDW-PD and 20 for PCT-stroke using blood-based cis-eQTLs in eQTLGen50 (Table S36), and a further four genes (RHD, FXYD5, MAP1LC3A, and SRSF6) for PDW-PD in analyses using platelet cis-eQTLs from GeneSTAR51 (Table S37). Among these aforementioned genes, 40 of 60 and nine of 20 had consistent direction of SMR effect for PDW-PD and PCT-stroke, respectively (Figure 4). Using brain-based cis-eQTLs, none of these genes surpassed the Bonferroni significance threshold (pSMR < 6.63 × 10−6), but a total of 17 genes were FDR significant for PD (Table S38) and three were identified for stroke (CLBA1, IVD, and RMC1), with consistent direction of SMR associations in blood and brain for a large proportion of genes (with the exception of FXYD5, MGAT3, RANBP10, RHD, and UBXN2A).
Figure 4.
Summary of FDR-significant functional genes associated with PCT-stroke and PDW-PD trait pairs, with consistent direction of SMR effects
(A and B) PCT-stroke (A) and PDW-PD (B) trait pairs. The width of each line represents the SMR association strength. Line color indicates whether upregulation (orange) or downregulation (red) of functional genes is associated with increased disorder risk or BCT values. Genes with an asterisk symbol are known drug targets (Table S49). See Tables S36 and S37 for complete details of the SMR estimates.
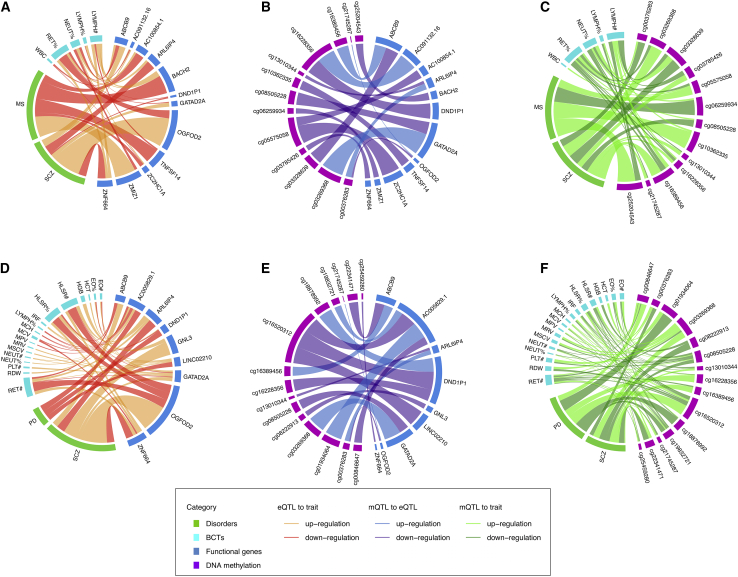
Second, we applied SMR to a further 68 BCT-NPD pairs with a nominally significant (p < 0.05) genome-wide rg (from HDL or LDSC). Using data on blood-based cis-eQTLs from eQTLGen,50 we identified 51 pleiotropic genes whose expression level was Bonferroni-significantly (pSMR < 3.18 × 10−6 and pHEIDI > 0.01 with ≥10 SNPs) associated with both members of specific BCT-NPD pairs (Table S39). We then tested for association of DNA methylation with expression of these 51 genes, using data on DNA methylation quantitative trait loci (mQTL) in blood-derived DNA from the Brisbane Systems Genetics Study (BSGS) and Lothian Birth Cohorts (LBCs).52 We identified 273 DNA methylation probes that were Bonferroni-significantly associated with expression of one or more of these genes (pSMR < 5.37 × 10−7, pHEIDI > 0.01 with ≥10 SNPs; Table S40), of which 13 independent probes, associated with a total of 12 genes, were also Bonferroni-significantly associated (pSMR < 5.37 × 10−7, pHEIDI > 0.01 with ≥10 SNPs) with both members of the respective focal BCT-NPD pair (Figure 5; Tables S41, S42, and S43). These included six genes (ABCB9, AC100854.1, BACH2, TNFSF14, ZC2HC1A, and ZMIZ1) for MS and one or more BCTs on chromosomes 6, 8, 10, and 19, and seven genes (ABCB9, AC091132.16, ARL6IP4, DND1P1, GATAD2A, OGFOD2, and ZNF664) for SCZ and one or more BCTs on chromosomes 12, 17, and 19 (including one gene [ABCB9] overlapping with MS and related BCTs). Two functional genes (ARL6IP4 and GATAD2A) and regulatory elements for SCZ and reticulocyte fraction of red cells (RET%) surpassed the experiment-wide Bonferroni-corrected thresholds (pSMR < 9.96 × 10−9 [i.e., ] using cis-eQTL as exposure and GWAS as outcome; pSMR < 1.05 × 10−8 [i.e., ] using mQTL as exposure and cis-eQTL as outcome; pSMR < 1.68 × 10−9 [i.e., ] using mQTL as exposure and GWAS as outcome; pHEIDI > 0.01 with ≥10 SNPs for each analysis) across all SMR analyses. Notably, there was evidence for involvement of regulatory elements located in both promoter and repressor regions for four genes (ABCB9, ARL6IP4, GATAD2A, and OGFOD2). For example, reduced methylation in the OGFOD2 promoter and increased methylation in the OGFOD2 repressor, in each case associated with up-regulation of this gene, was associated with elevated risk for SCZ and reduced RET% (Table S43). In addition, for all 12 genes that were Bonferroni significant for both members of specific BCT-NPD trait pairs across the three tiers of SMR analyses, the effect of expression on the two traits was concordant with their local genetic correlations (n = 13; Table S43). Additionally, a high proportion (10 of 13) of these local genetic correlations surpassed a 5% nominal significance threshold, indicating a high degree of consistency between SMR and ρ-HESS in relation to shared genetic risk factors.
Figure 5.
Summary of blood-based Bonferroni-significant functional genes and regulatory elements associated with both members of BCT-NPD trait pairs with significant genome-wide genetic correlations and significant local genetic correlations (but negligible genome-wide genetic correlations)
(A and D) Association of gene expression with each member of the focal BCT-NPD pair. (B and E) Association of DNA methylation with expression of trait-pair-associated genes. (C and F) Association of DNA methylation with BCT-NPD pairs. The width of each line represents the SMR association strength. Line color indicates whether up-regulation or down-regulation of functional genes (or DNA methylation levels) is associated with increased disorder risk or BCT values. See Tables S43 and S44 for complete details of the SMR estimates.
Third, we performed SMR analyses in regions of Bonferroni-significant local genetic correlation, focusing on the subset of BCT-NPD pairs with negligible genome-wide rg (p > 0.05). On the basis of these criteria, a total of 60 trait pairs and 31 genomic regions were selected for investigation. Using the same multi-step SMR analytic strategy described above, we identified a total of nine Bonferroni-significant genes (ABCB9, ARL6IP4, DND1P1, GATAD2A, OGFOD2, ZNF664, GNL3, LINC02210, and AC005829.1) and regulatory elements for SCZ and PD and one or more BCTs (Figure 5; Tables S39, S40, S41, S42, and S44), two of which (GNL3 and GATAD2A; the latter was also reported in pairs of SCZ and RET% of nominally significant genome-wide rg; Tables S45–S48) were also Bonferroni-significantly associated with their specific NPDs using brain cis-eQTLs53 and mQTLs.53 In each case, the expression effects of these genes on specific pairs of BCTs and NPDs were consistent with their respective local genetic correlations (Table S44).
Last, we conducted gene set enrichment analysis (GSEA) using ShinyGO54 to identify biological pathways shared between specific pairs of BCTs and NPDs. We focused on 11 trait pairs with ≥5 shared genes identified using SMR applied to blood-based or brain-based cis-eQTLs (Table S49). We identified 31 pathways containing ≥2 gene set members at an FDR < 0.05, including n = 5 for MS-EO#, n = 2 for MS-RDW, n = 5 for SCZ-LYMPH#, n = 12 for SCZ-MONO%, n = 3 for SCZ-NEUT%, n = 1 for SCZ-RET#, and n = 3 for SCZ-RET% (Table S50). However, whereas the minimum gene set enrichment for these pathway terms was >5-fold, the highest proportion of pathway-specific genes was only 0.04, for tau protein binding in SCZ and MONO% (2 of 45 pathway genes, 338-fold enrichment, FDR p = 4.68 × 10−4).
Discussion
We generated new insights into the shared genetics of BCTs and neurological and psychiatric disorders using large-scale GWAS summary statistics, and through integration of these data with blood- and brain-based gene expression and DNA methylation QTLs.
We identified a broad landscape of genetic correlations between BCTs and NPDs, including Bonferroni-significant genetic correlations between two BCT-NPD pairs (MS-LYMPH#, SCZ-MONO%), each of which has prior evidence for a significant phenotypic correlation40,42; our results provide evidence for a genetic contribution to these previously reported correlations. Overall, the magnitude of HDL-based rg estimates between BCTs and neurological diseases were greater than those between BCTs and psychiatric disorders (T statistic = 2.68, ptwo-sample t test = 7.87 × 10−3); this suggests that neurological diseases (in aggregate) have greater genetic overlap with BCTs than psychiatric disorders, although we acknowledge that not all NPDs are represented in our study. Notwithstanding this caveat, our findings are consistent with evidence for a crucial role of the peripheral immune system in regulating some neurological diseases (e.g., MS, stroke).55 Interestingly, we failed to replicate some previously reported positive genetic correlations between attention-deficit/hyperactivity disorder (ADHD)-RET%, MDD-WBC, and SCZ-LYMPH#,6 potentially due to the use of different GWAS summary statistics for some psychiatric disorders (e.g., a subset of the European-based PGC3 SCZ GWAS [n = 123,575] used in this study versus complete PGC3 SCZ GWAS [n = 130,644] in Reay et al.6) and BCTs (i.e., our BCT GWAS were obtained from Vuckovic et al.,26 whereas the previous study used the publicly available blood-based biomarker GWAS from http://www.nealelab.is/uk-biobank), and the use of different LDSC-based SNP sets (i.e., our study focused on SNP with minor allele frequency [MAF] ≥ 0.01, whereas Reay et al.6 applied MAF ≥ 0.05). Further studies will be required to establish these findings conclusively.
Investigation of local genetic correlations between BCTs and NPDs detected 32 Bonferroni-significant local genetic correlations for 74 trait pairs. Interestingly, the majority of these trait pairs (n = 60) did not have a significant genome-wide HDL- or LDSC-based rg, suggesting that meaningful local genetic correlations are common between traits with negligible genome-wide rg, and in most cases, a significant genome-wide rg reflects the presence of moderately significant local genetic correlations across the genome rather than highly significant local genetic correlations in specific genomic regions.
MR analyses identified compelling evidence for two putative causal relationships between BCTs and NPDs: a causal effect of increased PCT on stroke, which has been previously reported by Harshfield et al.,8 and a causal effect of increased PDW on PD, which to our knowledge has not been previously reported. MR analyses also revealed suggestive evidence for a causal effect of elevated LYMPH# on MS, previously reported by Astle et al.,7 although the evidence for this relationship remains inconclusive. In relation to increased PCT and stroke, our analyses strengthened the evidence for a causal effect through use of more powerful GWAS summary statistics for BCTs (sample size of 408,000 compared with 173,000 in Harshfield et al.8), and the application of additional, and arguably more sophisticated MR methods, including GSMR, LCV, and CAUSE, the latter of which is capable of differentiating causality from both correlated and uncorrelated pleiotropy. High PCT levels may induce stroke via a reduction in blood flow and vessel patency,56 which have been associated with stroke severity.57
In relation to increased PDW and risk for PD, we found consistent evidence for a causal relationship using six of seven MR methods. Weak genetic correlation has been previously reported between PDW and PD,58 but our results suggest this is due (at least in part) to a causal effect of PDW on PD. Increased PDW is an indicator of platelet activation, which is known to occur as part of the inflammatory response.59,60,61 Platelet activation has also been specifically implicated in neuroinflammation,62 which is hypothesized to be a core pathogenic mechanism in PD.63,64 Our findings are consistent with recent evidence that regular use of non-steroidal anti-inflammatory drugs (i.e., anti-platelet medications such as ibuprofen and aspirin) is protective for PD in carriers of mutations in the established PD gene LRRK2.65
To explore if the inferred causal effect of increased PDW on risk for PD is a signature of underlying inflammation, we adjusted SNP effects for PDW (and PD) using summary data from a large GWAS of C-reactive protein66 (CRP; see Data S2). Interestingly, the magnitude of the causal effect of PDW on PD remained largely unchanged after adjusting for CRP, suggesting that the causal effect of PDW on PD may involve platelet functions independent of the inflammatory response. Notably, the protein implicated in the primary cellular pathology of PD, alpha-synuclein, is present in large quantities in platelets, where it regulates the Ca2+-dependent release of alpha granules.67 Irrespective of the mechanism, our findings highlight the potential clinical utility of these platelet parameters as potential risk markers and targets for improving prevention and prognosis of stroke and PD.
With the exception of the PCT-stroke and PDW-PD (and to a lesser extent LYMPH#-MS) associations, there was no consistent evidence for a causal relationship, using multiple MR models or ρ-HESS, for any other BCT-NPD pair, including those with significant genome-wide rg. This implies, as expected, that pleiotropy is pervasive between BCTs and NPDs, which is consistent with findings reported by previous studies of vascular-neuropsychiatric associations.7,68
Using SMR, we identified a total of 60 and 20 FDR-significant genes associated with PDW-PD and PCT-stroke, respectively (Tables S36 and S37). Given evidence for a causal effect of increased PDW on PD risk, and of the well-established role of elevated PCT on susceptibility to stroke, these genes represent especially interesting candidates for mitigating risk for PD and/or stroke via modulation of platelet activity or function. Interestingly, 11 of the 60 PDW-PD genes and two of the 20 PCT-stroke genes are known drug targets (Table S51). For example, the SYK (spleen-associated tyrosine kinase) gene, which is targeted by 30 drugs with primary indications for a range of cancers and autoimmune disorders, plays an essential role in platelet activation as part of the collagen receptor glycoprotein (GPV1)-induced signaling pathway.69 In our analyses, up-regulation of SYK was Bonferroni-significantly associated with increased PDW and FDR-significantly associated with higher risk for PD, suggesting that SYK inhibitors (e.g., cerdulatinib, apitolisib, fostamatinib, entospletinib, lanraplenib) may represent potential drug re-purposing opportunities for mitigation of PD risk. Another notable example is the catenin beta 1 gene (CTNNB1) which codes for β-catenin, a protein that plays essential roles in the Wnt/β-catenin pathway and cadherin-catenin cell adhesion, which in platelets undergoes complete proteolysis during platelet aggregation.70 Our analyses indicate that up-regulation of CTNNB1 is associated with reduced PDW and is protective for PD (both at FDR 5% significance level), suggesting that β-catenin inhibitors may be worthy of further investigation in relation to ameliorating risk for PD. Interestingly, although a number of anti-platelet and anticoagulation therapies are commonly used in stroke prevention (e.g., aspirin), the genes targeted by these drugs were not identified in our SMR analyses, an omission that may be attributable to a lack of power in the stroke GWAS and/or the fact that anticoagulation factors produced in the liver may not be captured by blood-based cis-eQTLs.
Among the remaining FDR-significant genes (i.e., excluding drug targets), a particularly notable gene for PD is COA5 (cytochrome c oxidase assembly factor 5), which is known to be associated with mitochondrial complex IV deficiency.71 This is significant given the broad body of evidence implicating mitochondrial dysfunction in degeneration of dopaminergic neurons in PD patients,72 and the intimate role of mitochondria in platelet activation.73 Further studies will be required to evaluate the clinical significance of this and other putative functional genes and regulatory elements for PD and stroke. Clarification of the specific genes and molecular mechanism(s) linking PDW to PD may present opportunities for mitigating risk for PD via platelet-targeting therapeutics, as is currently the case for stroke.
Looking beyond PCT-stroke and PDW-PD, the blood-based SMR analyses identified 51 genes for BCT-NPD pairs with at least nominally significant genome-wide rg (Table S39). Among these, a total of 12 genes (Table S43)—all involving associations between MS and/or SCZ and one or more BCTs—showed Bonferroni-significant evidence for a regulatory pathway linking specific DNA methylation probe(s) to gene expression, gene expression to both members of specific BCT-NPD pairs, and for direct and consistent effects of DNA methylation on the same BCT-NPD pairs. Several of these functional genes are worthy of mention:
GATAD2A (GATA zinc finger domain containing 2A) is a crucial subunit of the nucleosome remodeling and histone deacetylation (NuRD) complex, which is one of the primary chromatin remodeling complexes in mammalian cells.74 GATAD2A has been reported to be implicated in elevated risk for SCZ, potentially via its role as a regulator of gene expression during neurodevelopment.75,76,77 The NuRD complex is also centrally involved in hematopoiesis,78 although the specific role of GATAD2A in relation to the seven BCTs associated with this gene in our SMR analyses remains unclear. Interestingly, whereas the SCZ risk allele at GATAD2A was associated with up-regulation of this gene in blood, it was associated with down-regulation in brain, consistent with prior reports79 and the idea that a proportion of risk alleles have different effects on gene expression in different organs.
Another notable gene is MAPK3 (mitogen-activated protein kinase 3), a key member of the extra-cellular signal-regulated kinase pathway, which plays a central role in cell growth, differentiation and survival via regulation of transcription and translation. In our analyses, up-regulation of MAPK3 was Bonferroni-significantly associated with increased risk for SCZ and decreased NEUT%, the latter of which is consistent with neutropenia, a potentially life-threatening condition characterized by reduced neutrophil count and correspondingly higher risk for infection. This is significant, because whereas neutropenia (and the related condition of agranulocytosis) is a well-known side effect of some second-generation anti-psychotics, such as clozapine, our results suggest that some genetic variants associated with increased risk for SCZ also directly increase susceptibility to neutropenia.80,81 There was no evidence for a causal effect of NEUT% on risk for SCZ in the MR analyses, suggesting that the association of MAPK3 with SCZ and NEUT% is due to pleiotropy. We also found no evidence for an association of DNA methylation with expression of MAPK3, or with risk for schizophrenia or variation in NEUT%. Further study will be needed to decipher the pleiotropic mechanism(s) via which MAPK3 influences NEUT% and susceptibility to SCZ.
Another gene associated with increased risk for SCZ and reduced NEUT% was ABCB9 (ATP-binding cassette subfamily B member 9), which is a member of the MDR/TAP subfamily of ABC transporters, responsible for translocation of MHC peptides into lysosomes. Up-regulation of ABCB9 was also associated with increased risk for MS, increased LYMPH#, and reduced RET%. The association of ABCB9 with SCZ (among other traits such as cardiometabolic disease) has been previously reported,76,82,83 but here we provide evidence for a role of this gene in MS, LYMPH#, NEUT%, and RET%. The association of ABCB9 with both MS and LYMPH# indicates that this is one of the genes underlying the positive genetic correlation between these traits.
The application of gene set enrichment analysis to Bonferroni-significant genes identified through SMR identified multiple FDR-significant biological pathways that were shared by specific pairs of BCTs and NPDs. The top-ranked pathway was tau protein binding for SCZ-MONO%, which is notable given that aberrantly phosphorylated tau protein has been associated with risk for SCZ84; and there is evidence that monocyte-derived macrophages play an important role in phagocytosing extra-cellular oligomeric tau protein.85 Although this observation has potential therapeutic implications, an important caveat is that the proportion of pathway-specific genes was small (<5%). This was also the case for other FDR-significant pathways identified by GSEA, which suggests that larger studies will be needed for robust inference of biological pathways shared by specific pairs of BCTs and NPDs.
In conclusion, we report a broad landscape of genetic overlap between BCTs and common NPDs, finding evidence to suggest that platelet parameters may be useful biomarkers for risk stratification of primary prevention trials of PD. Additionally, we identified multiple functional genes and regulatory elements for specific pairs of BCTs and NPDs, some of which are previously unreported, including known drug targets that may present drug re-purposing opportunities for PD. Our results provide a robust genetic foundation for improving prognosis, prevention, and possibly new avenues for treatment of common NPDs on the basis of readily assayed BCTs.
Limitations of the study
We note several limitations in our analyses. First, potential sample overlap between BCT and NPD GWAS summary statistics (i.e., both AD and PD GWAS included participants of UKB that overlap with the BCT GWAS) may introduce weak instrument bias and thus decrease the power of MR methods for detecting causal relationships.86 Nevertheless, we believe the magnitude of such bias is minimal for two reasons: (1) the intercept from bivariate LDSC was <0.02 for all pairs of BCTs and NPDs, and (2) we applied multiple MR models with different instrumental SNP sets and observed consistent results. Second, the blood-based cis-eQTL summary data used in our SMR analyses were not corrected for blood cell proportions. This has the potential to influence the SMR results for BCTs, although because the eQTLGen dataset is very large, any potential influence of blood cell heterogeneity is likely to be averaged out. Third, although we report a number of statistically significant findings, further investigation will be required to determine if they are clinically meaningful.
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yuanhao Yang (yuanhao.yang@mater.uq.edu.au).
Materials availability
This study did not generate new unique reagents.
Method details
GWAS summary data
We obtained publicly available European-ancestry GWAS summary data for 29 BCTs,26 including measures of red cells (e.g., hematocrit, hemoglobin), platelets (e.g., PCT, PDW), and white cells (e.g., BASO#, LYMPH#), and 11 common NPDs, including AD,27 ADHD,33 ALS,28 ASD,34 bipolar disorder (BIP),35 MDD,36 migraine,29 MS,30 PD,31 SCZ37 and stroke32 (Tables 1 and 2). BCTs reported by Vuckovic et al.26 were extracted from the main hematological indices of clinic-based blood samples from 408,112 European participants in the UKB. The authors implemented linear mixed regression to generate BCT GWAS summary statistics for the transformed and inversely normalized BCT residuals (i.e., regressing out effects from sex, age, age-squared, principal components, and recruitment center). GWAS of common NPDs were performed based on large-scale European samples, with the number of individuals ranging from ∼40K for MS to >1.45M for PD. Among 11 NPDs, eight (ADHD, AD, ALS, ASD, BIP, MS, SCZ and stroke) involved analysis of cases diagnosed by clinicians or physicians (or with an equivalent clinical diagnosis), and three (MDD, migraine, PD) involved a combination clinical and self-reported cases. Each GWAS summary dataset comprised ∼6-10M common SNPs after filtering out SNPs with MAF <1% (Tables 1 and 2).
cis-eQTL and mQTL summary data
We used summary data for blood- and brain-based cis-eQTL and mQTL for SMR analyses (see below). Blood-based cis-eQTL and mQTL data were obtained from the eQTLGen consortium (nsample = 31,684; nprobe = 19,250)50 and the BSGS and LBC (1921 and 1936; nsample = 1,980; nprobe = 94,338),52 respectively. We also obtained platelet cis-eQTL data (nsample = 180; nprobe = 4,555) from the GeneSTAR Research Study.51 Brain-based cis-eQTL data (neffect sample = 1,194; nprobe = 28,538)53 were based on a meta-analysis of 10 Genotype-Tissue Expression (GTEx v6)89 brain regions, the CommonMind Consortium (CMC)90 and Religious Orders Study and Memory and Aging Project (ROSMAP)91; and the brain-based mQTL data (neffect sample = 1,160; nprobe = 436,077)53 were based on a meta-analysis of three datasets: ROSMAP,91 Hannon et al.92 and Jaffe et al.93 All these multi-omics data were publicly available and of European descent, with imputation based on the 1000 Genomes European reference panel (hg19 genome build).94
Estimation of genetic correlations using high-definition likelihood (HDL)
We used the HDL16 method to estimate the genetic correlation for each pair of BCTs and NPDs, using GWAS summary statistics. HDL is a recently developed extension of bivariate LDSC,87 which makes use of LD across the entire autosomal genome with exclusion of the MHC region (chromosome 6: 28,477,797-33,448,354 bp) via fitting an additional variance-covariance LD matrix to achieve significant reductions in variance of rg estimates, thereby improving power. Here, HDL was applied to 319 pairs of BCTs and NPDs, with the method implemented using the default UKB reference with ∼1M imputed HapMap3 autosomal SNPs, after excluding strand-ambiguous SNPs (i.e., A/T, C/G). We defined significant HDL rg estimates as those surpassing the Bonferroni corrected threshold (p < 1.57 × 10−4, i.e., ).
Estimation of genetic correlations using LD score regression (LDSC)
We also conducted bivariate LDSC87,88 for each pair of traits as a sensitivity analysis. Bivariate LDSC estimates the rg between traits using the slope from the regression of the product of the single trait GWAS test statistics (Z scores) on LD score. We applied bivariate LDSC to 319 pairs of BCTs and NPDs using the default LD scores of the 1000 Genomes European reference, again excluding SNPs if they were strand-ambiguous or located within the MHC region. As with HDL, we defined significant LDSC-based rg estimates as those surpassing the Bonferroni corrected threshold (p < 1.57 × 10−4).
Investigating potential confounding factors mediating the shared genetics underlying pairs of traits with significant genetic correlations
For BCT-NPD trait pairs with Bonferroni-corrected or FDR significant genome-wide rg (from HDL or LDSC), we investigated whether their shared genetics were driven by potential confounding factors, including smoking, drinking, educational attainment, and socioeconomic status that have been reported to be commonly associated with BCTs7,95,96 and NPDs.97,98,99,100 We investigated the roles of these potential confounding factors using a conditional approach. First, we utilised mtCOJO23 to condition GWAS summary data of BCTs and NPDs on the GWAS of cigarettes per day,43 drinks per week,43 educational attainment,44 and household income,45 respectively. We used genotype data from unrelated Europeans in the UK Biobank as a reference. We then re-estimated the genetic correlations between specific pairs of BCTs and NPDs on the basis of their conditional GWAS summary statistics, using HDL.
Estimation of local genetic correlations using ρ-HESS
Pairs of BCTs and NPDs may share genetic variance in small genomic regions even in the absence of a significant genome-wide genetic correlation. Moreover, the pattern of local genetic correlations in trait-associated regions can indicate potential causal relationships between traits. To investigate these possibilities, we used ρ-HESS17 to estimate local genetic correlations between 319 BCT-NPD pairs in 1,693 approximately independent LD regions (average width ≈1.5Mb,101 based on the hg19-based 1000 Genomes European reference), excluding those in the MHC region. We defined significant local genetic correlations as those surpassing the Bonferroni corrected threshold (p < 9.26 × 10−8; i.e., ).
We then followed the approach proposed by Shi et al.17 and classified the local genetic correlations into four groups for each pair of traits: (i) regions harboring only NPD-specific SNPs, (ii) regions harboring only BCT-specific SNPs, (iii) regions harboring SNPs shared by both members of the BCT-NPD trait pair (“Intersection”), and (iv) other regions (“Neither”). We defined trait-specific SNPs (used to classify genomic regions into each of the four groups) as those with p < 1 × 10−5, and compared these results to four other p value cut-offs (p < 5 × 10−8, p < 1 × 10−6, p < 1 × 10−4, and p < 1 × 10−3), as a means of accommodating differential GWAS power between BCTs and NPDs. We also performed a further sensitivity analysis using a Bayesian-based approach (SBayesR)48 to define trait-specific SNPs. SBayesR implements a Bayesian likelihood multiple regression procedure to refine the estimated effect sizes of trait-associated SNPs by updating the ‘prior’ SNP effects of GWAS summary statistics to ‘posterior’ SNP effects. We used a sparse LD correlation matrix generated from Europeans in the UK Biobank as a reference for SBayesR. Genomic regions were excluded if the estimated local genetic correlation was missing (e.g., because the local estimated single-trait heritability was negative), less than −1 or greater than 1 (e.g., because at least one of the local estimated single-trait heritability estimates was close to zero). For each group that comprised ≥5 local genetic correlation estimates, we calculated the mean and SE of the local genetic correlations within the group. A causal effect of BCT on NPD was suggested if the average local genetic correlation in regions harboring BCT-specific SNPs was Bonferroni significantly non-zero (p < 1.57 × 10−4, for 319 pairs) and Bonferroni significantly different from that in “Intersection” regions, “Neither” regions, and regions harboring NPD-specific SNPs (p < 1.57 × 10−4 based on a two-tailed Z-test). We used an equivalent strategy to identify patterns of local genetic correlations consistent with a potential causal effect of NPD on BCT.
Mendelian randomization analyses
We used multiple MR methods to evaluate evidence for causality of BCTs on risk for NPDs, and vice versa. We focused on a total of 70 trait pairs with nominally significant (p < 0.05) genome-wide rg (from HDL or LDSC), recognizing that a causal relationship is unlikely to exist in the absence of a global genetic correlation between highly polygenic traits like the BCTs and NPDs used in our study.
We first utilized the Bayesian multivariate linear model-based MR method CAUSE,18 that assumes the genetic effect of the exposure on the outcome is comprised of a causal effect (), correlated pleiotropy (; defined as instruments with horizontal effects on both exposure and outcome through a shared pathway), and uncorrelated pleiotropy (q; defined as instruments with horizontal effects on the exposure and outcome via separate mechanisms or pathways). This approach enables CAUSE to distinguish a causal effect from correlated pleiotropy by quantifying the joint distribution of instrumental SNP effects, under the assumption that all instrumental SNPs are influenced by a causal effect, whereas only a subset are influenced by correlated pleiotropy. The CAUSE method utilizes more (approximately independent; LD r2 < 0.10 based on the 1000 Genomes European reference) instrumental SNPs (with an arbitrary p < 1 × 10−3) than many other MR methods that rely solely on genome-wide significant SNPs, which is purported to increase power.18 CAUSE also implements an approach called expected log pointwise posterior density (ELPD) to compare the overall model fit between the null model (no causal or pleiotropy effect), sharing model (the existence of a pleiotropic effect but no causal effect), and causal model (the existence of both causal and pleiotropic effects). We applied CAUSE using the R package ‘cause’, excluding SNPs in the MHC region. We considered a relationship to be putatively causal if the causal effect estimated by the causal model surpassed the Bonferroni corrected significance threshold (p < 3.57 × 10−4, i.e., ), and the overall fitness of the causal model was significantly (p < 0.05) better than the sharing model or null model.
To strengthen evidence of causality from the CAUSE analysis, we performed sensitivity analyses using six alternative MR methods with different assumptions on pleiotropy, including inverse variance weighting (IVW),19 MR-Egger,20 WMo,21 WMe,22 and GSMR.23 Among the six alternative models, IVW19 is the basic approach that assumes no correlated pleiotropy and the presence of uncorrelated pleiotropy (with mean zero), thereby adding noise to the random-effects meta-analysis of SNP effects. MR-Egger20 is an extension of IVW that assumes no correlated pleiotropy but non-zero uncorrelated pleiotropy, which adds an extra intercept to the IVW to account for such uncorrelated pleiotropy. GSMR23 has the same assumptions as MR-Egger and utilizes the HEIDI approach to identify and exclude confounding SNPs due to uncorrelated pleiotropy. Both WMe22 and WMo21 are capable of removing partial correlated and uncorrelated pleiotropy. WMe22 is implemented using the weighted median but not weighted mean of the SNP ratio, and thus is capable of identifying true causality if the proportion of invalid instrumental SNPs of correlated and uncorrelated pleiotropy is ≤50%. WMo21 estimates the causal effect merely from the largest subset of SNPs with consistent effects. While WMo drops some instrumental SNPs and likely has reduced power, it has the ability to identify true causality, particularly when a majority of instrumental SNPs are invalid. Furthermore, we inferred putative causal relationships between BCTs and NPDs using the LCV (latent causal variable) model.24 The LCV model assumes that the genetic correlation between two traits is mediated by a latent variable, which has a causal effect on each trait and can be quantified by estimating the GCP (genetic causality proportion) using the mixed fourth moments of marginal SNP effect sizes for each trait. GCP estimates vary between 0 (no causal relationship) to 1 (fully causal relationship), with higher values implying a stronger partially causal relationship.
We considered causal relationships that were consistently identified (p < 0.05) by all alternative methods as unlikely to be false positives. We applied these alternative methods using the R packages ‘TwoSampleMR’, ‘gsmr’, and ‘LCV’, with exclusion of SNPs within the MHC region. Independent SNPs (LD clumping r2 < 0.05 within 1,000Kb windows based on the 1000 Genomes European reference) with p < 5 × 10−8 were selected as instrumental variables for all these alternative MR methods except LCV, for which all SNPs were used. For MR effect size estimates for binary exposures (i.e., NPDs), we converted the MR effects from logit-scale to liability-scale using the method of Byrne et al.102 All MR effects were then transformed into ORs for concise interpretation. The interpretation of the OR for “BCT → NPD” is that, as an example, if the estimated OR is 1.2, the NPD risk is increased by 1.2-fold for each SD increase in the BCT. Similarly, the interpretation of the OR for “NPD → BCT” at 1.2 (as an example) is that the level of BCT is increased by 1.2-fold per SD increase in NPD liability.
To evaluate the reliability of our MR results, we implemented several additional sensitivity analyses to evaluate the validity of instrumental SNPs for pairs of traits with consistent evidence for a causal relationship across all MR models.103 The sensitivity analyses included: (i) checking whether the intercept term in MR-Egger regression is significantly different from zero; (ii) checking for heterogeneity among instrumental SNPs using Cochran’s Q and I2; and (iii) performing leave-one-out analyses using each of the four two-sample MR models to evaluate if single instrumental SNPs may be responsible for the inferred causal relationship(s). MR results satisfying all three sensitivity analyses were considered robust.
Finally, we investigated the contribution of four potential confounding factors (i.e., cigarettes per day,43 drinks per week,43 educational attainment,44 household income45) on estimates of inferred causality between specific pairs of BCTs and NPDs, using two approaches. First, we re-estimated the causal effects for each pair of traits with consistent evidence for a causal relationship by applying the same MR methods (and LCV) to their conditional GWAS summary statistics, generated using mtCOJO. Second, we applied MVMR49 analysis to each BCT-NPD trait pair with consistent evidence for a causal relationship. The MVMR method is capable of estimating causal effects between an exposure and outcome whilst adjusting for the potential pleiotropic effects of multiple outcome-related confounding factors concurrently.
Multi-omics analysis of putatively functional mechanisms underlying shared genetic loci for BCTs and neurological and psychiatric disorders
Next, we utilized SMR25 to identify putatively functional genes and regulatory elements shared by pairs of BCTs and NPDs. SMR is an MR-equivalent analysis method that utilizes GWAS summary statistics to test for an association between gene expression (i.e., exposure) and a target phenotype (i.e., outcome), using genome-wide significant SNPs as instrumental variables. In our analyses, we used SMR to test for (i) association of gene expression (exposure) with pairs of BCTs and NPDs (outcomes), (ii) association of DNA methylation (exposure) with gene expression (outcome), and (iii) association of DNA methylation (exposure) with pairs of BCTs and NPDs (outcome). We also implemented the HEIDI test to distinguish linkage from a causal effect or pleiotropy, given a significant SMR association could be explained by different causal SNPs in high LD having effects on the exposure and outcome separately (i.e., linkage), rather than by a causal variant affecting outcome via changes in the exposure (i.e., causal effect) or the causal variant having a shared effect on both exposure and outcome (i.e., pleiotropy). Here, we performed SMR analyses across 70 pairs of BCTs and NPDs with evidence for a nominally significant (p < 0.05) rg (by HDL or LDSC) and/or a causal relationship on the basis of MR analyses.
We first tested for an association of blood-based gene expression (exposure) with BCT-NPD pairs (outcome traits) using blood-based cis-eQTLs from eQTLGen,50 reporting genes surpassing Bonferroni correction (pSMR < 3.18 × 10−6, based on correction for testing of 15,743 probes with cis-eQTL p < 5 × 10−8) and also the HEIDI test (pHEIDI > 0.01 with ≥10 SNPs). With respect to pairs of platelet parameters and related NPDs (e.g., PCT-stroke and PDW-PD), we further carried out SMR using platelet cis-eQTLs from GeneSTAR51 (Bonferroni-corrected pSMR < 6.31 × 10−5, i.e., , pHEIDI > 0.01 with ≥10 SNPs). For all Bonferroni-significant genes, we then used SMR to test for an association of blood-based DNA methylation (exposure) with gene expression (outcome), using mQTL summary data from BSGS and LBC and cis-eQTL data from eQTLGen, respectively. DNA methylation-gene expression associations were declared significant if they had a Bonferroni corrected pSMR < 5.37 × 10−7 (correction for testing of 93,122 probes with mQTL p < 5 × 10−8) and surpassed the HEIDI test (pHEIDI > 0.01 with ≥10 SNPs). For all Bonferroni-significant DNA methylation probes, we then performed SMR using DNA methylation as exposure and BCT-NPD pairs as outcome, to determine if DNA methylation was also directly associated (Bonferroni corrected pSMR < 5.37 × 10−7; and pHEIDI > 0.01 with ≥10 SNPs) with the same pairs of BCTs and NPDs. We considered genes and regulatory elements with consistent evidence for significant associations across all SMR analyses to be noteworthy. Additionally, we applied the same three types of SMR analyses in regions of Bonferroni-significant local genetic correlation (p < 9.23 × 10−8), focusing on the subset of BCT-NPD pairs that also had negligible (p > 0.05) genome-wide rg.
We also conducted parallel SMR analyses using brain-based cis-eQTL53 and mQTL summary data53 as sensitivity analyses, using the same analytic process as described above (but limited to the specific NPDs instead of pairs of BCTs and NPDs). We concentrated on functional genes and regulatory elements that were identified with consistent evidence for significant blood-based associations across the three types of SMR analyses. We considered brain-based SMR associations to be significant if they surpassed both a Bonferroni-corrected SMR p value threshold (pSMR < 6.63 × 10−6, i.e., , using cis-eQTL as exposure and GWAS of NPD as outcome; <5.28 × 10−7, i.e., , using mQTL as exposure and cis-eQTL as outcome, and when using mQTL as exposure and GWAS of NPD as outcome) and the HEIDI test (pHEIDI > 0.01 with ≥10 SNPs).
All SMR analyses were restricted to expression (or DNA methylation) probes with a cis-eQTL (or mQTL) p < 5 × 10−8, and probes located in the MHC region were excluded. LD was adjusted according to the 1000 Genomes European ref. 94. For SMR analysis using mQTL as exposure and GWAS as outcome, genes were matched to each DNA methylation site if they were located within a 500Kb window. For multiple DNA methylation sites associated with the same gene with consistent direction of effect, we calculated the correlations between each pair of DNA methylation sites by measuring the correlations between SMR effects of their common genes. We filtered out DNA methylation sites if they were less significant and correlated (R > 0.05) with other DNA methylation sites.
Gene set enrichment analysis (GSEA)
Finally, we performed GSEA to identify biological pathways shared by specific pairs of BCTs and NPDs. We used the ShinyGO tool54 based on the Gene Ontology (GO) annotation resource, comprising a hierarchy of biological processes, cellular components and molecular functions. To maintain power and sensitivity, we applied ShinyGO to candidate gene sets comprising five or more genes whose expression levels were Bonferroni-significant for both members of specific pairs of BCTs and NPDs, based on SMR analysis of blood-based and/or brain-based cis-eQTLs (Table S50). We defined significantly enriched pathways as those with ≥2 pathway-specific genes and an FDR <5%.
Quantification and statistical analysis
Statistical analyses were performed using R 4.0.5, HDL 1.3.8, LDSC 1.0.1, ρ-HESS 0.5.4, LCV, mtCOJO 1.9.3.2 beta, SMR 1.03, PLINK 1.9, ShinyGo 0.76 and Ricopili 1118b. All methodological details can be found in the STAR Methods, and all statistical tests are named as they are used. All statistical tests are two-sided, with the exception of the heterogeneity test among instrumental SNPs in the MR sensitivity analyses, which were one-sided.
Acknowledgements
We acknowledge funding support from the Australian National Health and Medical Research Council (GNT1127440 and GNT2013281 to J.G.; GNT20098389 to B.V.T.; GNT1173155 to Y.Z.), Mater Foundation (Y.Y., C.X.Y., R.K.T., J.G.), MS Australia (B.V.T.), and the Macquarie Foundation paired senior research fellowship (B.V.T.). We also thank the International Multiple Sclerosis Genetics Consortium (IMSGC) and the IHGC for providing access to GWAS summary data. This research was enabled using the UK Biobank Resource under application 12505. We are grateful to Professor Naomi Wray for insightful comments and discussion.
Author contributions
Y.Y. and J.G. designed the study and wrote the manuscript. Y.Y. performed the primary analyses, with assistance from Z.Z. (MR analyses) and Y. Wu (SMR analyses). J.G. supervised the study. Y.Y., Y.Z., D.R.N., C.X.Y., R.K.T., Y. Wu, Y. Wang, Z.Z., B.V.T., and J.G. contributed to the interpretation of results and the critical revision of the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a gender minority in their field of research. We support inclusive, diverse, and equitable conduct of research.
Published: January 25, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2022.100249.
Contributor Information
Yuanhao Yang, Email: yuanhao.yang@mater.uq.edu.au.
Jacob Gratten, Email: jacob.gratten@mater.uq.edu.au.
Supplemental information
Data and code availability
-
•
GWAS summary statistics for MS are available by application from https://imsgc.net/?page_id=31. GWAS summary statistics for migraine are available from the International Headache Genetics Consortium (IHGC, http://www.headachegenetics.org/content/datasets-and-cohorts) by request to Professor Dale Nyholt (d.nyholt@qut.edu.au). All other data are publicly available and listed in the key resources table.
-
•
This study did not generate any unique datasets or code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Sealock J.M., Lee Y.H., Moscati A., Venkatesh S., Voloudakis G., Straub P., Singh K., Feng Y.C.A., Ge T., Roussos P., et al. Use of the PsycheMERGE network to investigate the association between depression polygenic scores and white blood cell count. JAMA Psychiatr. 2021;78:1365–1374. doi: 10.1001/jamapsychiatry.2021.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medema S., Mocking R.J.T., Koeter M.W.J., Vaz F.M., Meijer C., de Haan L., van Beveren N.J.M., GROUP;Genetic Risk and Outcome of Psychosis investigators. Kahn R., de Haan L., et al. Levels of red blood cell fatty acids in patients with psychosis, their unaffected siblings, and healthy controls. Schizophr. Bull. 2016;42:358–368. doi: 10.1093/schbul/sbv133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheremata W.A., Jy W., Horstman L.L., Ahn Y.S., Alexander J.S., Minagar A. Evidence of platelet activation in multiple sclerosis. J. Neuroinflammation. 2008;5:27. doi: 10.1186/1742-2094-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furlan J.C., Vergouwen M.D.I., Fang J., Silver F.L. White blood cell count is an independent predictor of outcomes after acute ischaemic stroke. Eur. J. Neurol. 2014;21:215–222. doi: 10.1111/ene.12233. [DOI] [PubMed] [Google Scholar]
- 5.Abbott R.D., Ross G.W., Tanner C.M., Andersen J.K., Masaki K.H., Rodriguez B.L., White L.R., Petrovitch H. Late-life hemoglobin and the incidence of Parkinson's disease. Neurobiol. Aging. 2012;33:914–920. doi: 10.1016/j.neurobiolaging.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reay W.R., Kiltschewskij D.J., Geaghan M.P., Atkins J.R., Carr V.J., Green M.J., Cairns M.J. Genetic estimates of correlation and causality between blood-based biomarkers and psychiatric disorders. Sci. Adv. 2022;8:eabj8969. doi: 10.1126/sciadv.abj8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Astle W.J., Elding H., Jiang T., Allen D., Ruklisa D., Mann A.L., Mead D., Bouman H., Riveros-Mckay F., Kostadima M.A., et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harshfield E.L., Sims M.C., Traylor M., Ouwehand W.H., Markus H.S. The role of haematological traits in risk of ischaemic stroke and its subtypes. Brain. 2020;143:210–221. doi: 10.1093/brain/awz362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couturier N., Bucciarelli F., Nurtdinov R.N., Debouverie M., Lebrun-Frenay C., Defer G., Moreau T., Confavreux C., Vukusic S., Cournu-Rebeix I., et al. Tyrosine kinase 2 variant influences T lymphocyte polarization and multiple sclerosis susceptibility. Brain. 2011;134:693–703. doi: 10.1093/brain/awr010. [DOI] [PubMed] [Google Scholar]
- 10.Scherzer C.R., Grass J.A., Liao Z., Pepivani I., Zheng B., Eklund A.C., Ney P.A., Ng J., McGoldrick M., Mollenhauer B., et al. GATA transcription factors directly regulate the Parkinson's disease-linked gene alpha-synuclein. Proc. Natl. Acad. Sci. USA. 2008;105:10907–10912. doi: 10.1073/pnas.0802437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies N.M., Holmes M.V., Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chahine L.M., Stern M.B., Chen-Plotkin A. Blood-based biomarkers for Parkinson's disease. Park. Relat. Disord. 2014;20(Suppl 1):S99–S103. doi: 10.1016/S1353-8020(13)70025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hampel H., O'Bryant S.E., Molinuevo J.L., Zetterberg H., Masters C.L., Lista S., Kiddle S.J., Batrla R., Blennow K. Blood-based biomarkers for Alzheimer disease: mapping the road to the clinic. Nat. Rev. Neurol. 2018;14:639–652. doi: 10.1038/s41582-018-0079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henriksen K., O'Bryant S.E., Hampel H., Trojanowski J.Q., Montine T.J., Jeromin A., Blennow K., Lönneborg A., Wyss-Coray T., Soares H., et al. The future of blood-based biomarkers for Alzheimer's disease. Alzheimers Dement. 2014;10:115–131. doi: 10.1016/j.jalz.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai C.Y., Scarr E., Udawela M., Everall I., Chen W.J., Dean B. Biomarkers in schizophrenia: a focus on blood based diagnostics and theranostics. World J. Psychiatr. 2016;6:102–117. doi: 10.5498/wjp.v6.i1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ning Z., Pawitan Y., Shen X. High-definition likelihood inference of genetic correlations across human complex traits. Nat. Genet. 2020;52:859–864. doi: 10.1038/s41588-020-0653-y. [DOI] [PubMed] [Google Scholar]
- 17.Shi H., Mancuso N., Spendlove S., Pasaniuc B. Local genetic correlation gives insights into the shared genetic architecture of complex traits. Am. J. Hum. Genet. 2017;101:737–751. doi: 10.1016/j.ajhg.2017.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrison J., Knoblauch N., Marcus J.H., Stephens M., He X. Mendelian randomization accounting for correlated and uncorrelated pleiotropic effects using genome-wide summary statistics. Nat. Genet. 2020;52:740–747. doi: 10.1038/s41588-020-0631-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgess S., Butterworth A., Thompson S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013;37:658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgess S., Thompson S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017;32:377–389. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartwig F.P., Davey Smith G., Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 2017;46:1985–1998. doi: 10.1093/ije/dyx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowden J., Davey Smith G., Haycock P.C., Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Z., Zheng Z., Zhang F., Wu Y., Trzaskowski M., Maier R., Robinson M.R., McGrath J.J., Visscher P.M., Wray N.R., Yang J. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 2018;9:224. doi: 10.1038/s41467-017-02317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Connor L.J., Price A.L. Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat. Genet. 2018;50:1728–1734. doi: 10.1038/s41588-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R., Powell J.E., Montgomery G.W., Goddard M.E., Wray N.R., Visscher P.M., Yang J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 26.Vuckovic D., Bao E.L., Akbari P., Lareau C.A., Mousas A., Jiang T., Chen M.H., Raffield L.M., Tardaguila M., Huffman J.E., et al. The polygenic and monogenic basis of blood traits and diseases. Cell. 2020;182:1214–1231.e11. doi: 10.1016/j.cell.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jansen I.E., Savage J.E., Watanabe K., Bryois J., Williams D.M., Steinberg S., Sealock J., Karlsson I.K., Hägg S., Athanasiu L., et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer's disease risk. Nat. Genet. 2019;51:404–413. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicolas A., Kenna K.P., Renton A.E., Ticozzi N., Faghri F., Chia R., Dominov J.A., Kenna B.J., Nalls M.A., Keagle P., et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron. 2018;97:1268–1283.e6. doi: 10.1016/j.neuron.2018.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gormley P., Anttila V., Winsvold B.S., Palta P., Esko T., Pers T.H., Farh K.H., Cuenca-Leon E., Muona M., Furlotte N.A., et al. Meta-analysis of 375, 000 individuals identifies 38 susceptibility loci for migraine. Nat. Genet. 2016;48:856–866. doi: 10.1038/ng.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.International Multiple Sclerosis Genetics Consortium Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science. 2019;365:eaav7188. doi: 10.1126/science.aav7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nalls M.A., Blauwendraat C., Vallerga C.L., Heilbron K., Bandres-Ciga S., Chang D., Tan M., Kia D.A., Noyce A.J., Xue A., et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malik R., Chauhan G., Traylor M., Sargurupremraj M., Okada Y., Mishra A., Rutten-Jacobs L., Giese A.K., van der Laan S.W., Gretarsdottir S., et al. Multiancestry genome-wide association study of 520, 000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018;50:524–537. doi: 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demontis D., Walters R.K., Martin J., Mattheisen M., Als T.D., Agerbo E., Baldursson G., Belliveau R., Bybjerg-Grauholm J., Bækvad-Hansen M., et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grove J., Ripke S., Als T.D., Mattheisen M., Walters R.K., Won H., Pallesen J., Agerbo E., Andreassen O.A., Anney R., et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019;51:431–444. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stahl E.A., Breen G., Forstner A.J., McQuillin A., Ripke S., Trubetskoy V., Mattheisen M., Wang Y., Coleman J.R.I., Gaspar H.A., et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 2019;51:793–803. doi: 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wray N.R., Ripke S., Mattheisen M., Trzaskowski M., Byrne E.M., Abdellaoui A., Adams M.J., Agerbo E., Air T.M., Andlauer T.M.F., et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trubetskoy V., Pardiñas A.F., Qi T., Panagiotaropoulou G., Awasthi S., Bigdeli T.B., Bryois J., Chen C.Y., Dennison C.A., Hall L.S., et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–508. doi: 10.1038/s41586-022-04434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam M., Awasthi S., Watson H.J., Goldstein J., Panagiotaropoulou G., Trubetskoy V., Karlsson R., Frei O., Fan C.C., De Witte W., et al. RICOPILI: rapid imputation for COnsortias PIpeLIne. Bioinformatics. 2020;36:930–933. doi: 10.1093/bioinformatics/btz633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeller J.A., Frahm K., Baron R., Stingele R., Deuschl G. Platelet-leukocyte interaction and platelet activation in migraine: a link to ischemic stroke? J. Neurol. Neurosurg. Psychiatry. 2004;75:984–987. doi: 10.1136/jnnp.2003.019638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prat A., Biernacki K., Lavoie J.F., Poirier J., Duquette P., Antel J.P. Migration of multiple sclerosis lymphocytes through brain endothelium. Arch. Neurol. 2002;59:391–397. doi: 10.1001/archneur.59.3.391. [DOI] [PubMed] [Google Scholar]
- 41.Lim Z.W., Elwood E., Naveed H., Galea I. Lymphopenia in treatment-naive relapsing multiple sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 2016;3:e275. doi: 10.1212/NXI.0000000000000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drexhage R.C., Hoogenboezem T.A., Cohen D., Versnel M.A., Nolen W.A., van Beveren N.J.M., Drexhage H.A. An activated set point of T-cell and monocyte inflammatory networks in recent-onset schizophrenia patients involves both pro- and anti-inflammatory forces. Int. J. Neuropsychopharmacol. 2011;14:746–755. doi: 10.1017/S1461145710001653. [DOI] [PubMed] [Google Scholar]
- 43.Liu M., Jiang Y., Wedow R., Li Y., Brazel D.M., Chen F., Datta G., Davila-Velderrain J., McGuire D., Tian C., et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 2019;51:237–244. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okbay A., Wu Y., Wang N., Jayashankar H., Bennett M., Nehzati S.M., Sidorenko J., Kweon H., Goldman G., Gjorgjieva T., et al. Polygenic prediction of educational attainment within and between families from genome-wide association analyses in 3 million individuals. Nat. Genet. 2022;54:437–449. doi: 10.1038/s41588-022-01016-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hill W.D., Davies N.M., Ritchie S.J., Skene N.G., Bryois J., Bell S., Di Angelantonio E., Roberts D.J., Xueyi S., Davies G., et al. Genome-wide analysis identifies molecular systems and 149 genetic loci associated with income. Nat. Commun. 2019;10:5741. doi: 10.1038/s41467-019-13585-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen M., Kachadoorian M., Quicksall Z., Zou F., Chai H.S., Younkin C., Crook J.E., Pankratz V.S., Carrasquillo M.M., Krishnan S., et al. Association of MAPT haplotypes with Alzheimer's disease risk and MAPT brain gene expression levels. Alzheimer's Res. Ther. 2014;6:39. doi: 10.1186/alzrt268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghetti B., Oblak A.L., Boeve B.F., Johnson K.A., Dickerson B.C., Goedert M. Invited review: frontotemporal dementia caused by microtubule-associated protein tau gene (MAPT) mutations: a chameleon for neuropathology and neuroimaging. Neuropathol. Appl. Neurobiol. 2015;41:24–46. doi: 10.1111/nan.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lloyd-Jones L.R., Zeng J., Sidorenko J., Yengo L., Moser G., Kemper K.E., Wang H., Zheng Z., Magi R., Esko T., et al. Improved polygenic prediction by Bayesian multiple regression on summary statistics. Nat. Commun. 2019;10:5086. doi: 10.1038/s41467-019-12653-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burgess S., Thompson S.G. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am. J. Epidemiol. 2015;181:251–260. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Võsa U., Claringbould A., Westra H.-J., Bonder M.J., Deelen P., Zeng B., Kirsten H., Saha A., Kreuzhuber R., Kasela S., et al. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. bioRxiv. 2018 doi: 10.1101/447367. Preprint at. [DOI] [Google Scholar]
- 51.Kammers K., Taub M.A., Rodriguez B., Yanek L.R., Ruczinski I., Martin J., Kanchan K., Battle A., Cheng L., Wang Z.Z., et al. Transcriptional profile of platelets and iPSC-derived megakaryocytes from whole-genome and RNA sequencing. Blood. 2021;137:959–968. doi: 10.1182/blood.2020006115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McRae A.F., Marioni R.E., Shah S., Yang J., Powell J.E., Harris S.E., Gibson J., Henders A.K., Bowdler L., Painter J.N., et al. Identification of 55, 000 replicated DNA methylation QTL. Sci. Rep. 2018;8:17605. doi: 10.1038/s41598-018-35871-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qi T., Wu Y., Zeng J., Zhang F., Xue A., Jiang L., Zhu Z., Kemper K., Yengo L., Zheng Z., et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat. Commun. 2018;9:2282. doi: 10.1038/s41467-018-04558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ge S.X., Jung D., Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kerr D., Krishnan C., Pucak M.L., Carmen J. The immune system and neuropsychiatric diseases. Int. Rev. Psychiatry. 2005;17:443–449. doi: 10.1080/0264830500381435. [DOI] [PubMed] [Google Scholar]
- 56.Khan S.Z., Dosluoglu H.H., Pourafkari L., Rivero M., Nader N.D. High plateletcrit is associated with early loss of patency after open and endovascular interventions for chronic limb ischemia. J. Vasc. Surg. 2020;71:2089–2097. doi: 10.1016/j.jvs.2019.08.258. [DOI] [PubMed] [Google Scholar]
- 57.Marto J.P., Lambrou D., Eskandari A., Nannoni S., Strambo D., Saliou G., Maeder P., Sirimarco G., Michel P. Associated factors and long-term prognosis of 24-hour worsening of arterial patency after ischemic stroke. Stroke. 2019;50:2752–2760. doi: 10.1161/STROKEAHA.119.025787. [DOI] [PubMed] [Google Scholar]
- 58.Tirozzi A., Izzi B., Noro F., Marotta A., Gianfagna F., Hoylaerts M.F., Cerletti C., Donati M.B., de Gaetano G., Iacoviello L., Gialluisi A. Assessing genetic overlap between platelet parameters and neurodegenerative disorders. Front. Immunol. 2020;11:02127. doi: 10.3389/fimmu.2020.02127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koupenova M., Clancy L., Corkrey H.A., Freedman J.E. Circulating platelets as mediators of immunity, inflammation, and thrombosis. Circ. Res. 2018;122:337–351. doi: 10.1161/CIRCRESAHA.117.310795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morrell C.N., Aggrey A.A., Chapman L.M., Modjeski K.L. Emerging roles for platelets as immune and inflammatory cells. Blood. 2014;123:2759–2767. doi: 10.1182/blood-2013-11-462432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leiter O., Walker T.L. Platelets in neurodegenerative conditions-friend or foe? Front. Immunol. 2020;11:747. doi: 10.3389/fimmu.2020.00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rawish E., Nording H., Münte T., Langer H.F. Platelets as mediators of neuroinflammation and thrombosis. Front. Immunol. 2020;11:548631. doi: 10.3389/fimmu.2020.548631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hirsch E.C., Hunot S. Neuroinflammation in Parkinson's disease: a target for neuroprotection? Lancet Neurol. 2009;8:382–397. doi: 10.1016/S1474-4422(09)70062-6. [DOI] [PubMed] [Google Scholar]
- 64.Wang Q., Liu Y., Zhou J. Neuroinflammation in Parkinson's disease and its potential as therapeutic target. Transl. Neurodegener. 2015;4:19. doi: 10.1186/s40035-015-0042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.San Luciano M., Tanner C.M., Meng C., Marras C., Goldman S.M., Lang A.E., Tolosa E., Schüle B., Langston J.W., Brice A., et al. Nonsteroidal anti-inflammatory use and LRRK2 Parkinson's disease penetrance. Mov. Disord. 2020;35:1755–1764. doi: 10.1002/mds.28189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ligthart S., Vaez A., Võsa U., Stathopoulou M.G., de Vries P.S., Prins B.P., Van der Most P.J., Tanaka T., Naderi E., Rose L.M., et al. Genome analyses of >200, 000 individuals identify 58 loci for chronic inflammation and highlight pathways that link inflammation and complex disorders. Am. J. Hum. Genet. 2018;103:691–706. doi: 10.1016/j.ajhg.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park S.M., Jung H.Y., Kim H.O., Rhim H., Paik S.R., Chung K.C., Park J.H., Kim J. Evidence that alpha-synuclein functions as a negative regulator of Ca(++)-dependent alpha-granule release from human platelets. Blood. 2002;100:2506–2514. doi: 10.1182/blood.V100.7.2506. [DOI] [PubMed] [Google Scholar]
- 68.Siewert K.M., Klarin D., Damrauer S.M., Chang K.M., Tsao P.S., Assimes T.L., Davey Smith G., Voight B.F., The International Headache Genetics Consortium Cross-trait analyses with migraine reveal widespread pleiotropy and suggest a vascular component to migraine headache. Int. J. Epidemiol. 2020;49:1022–1031. doi: 10.1093/ije/dyaa050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jooss N.J., De Simone I., Provenzale I., Fernández D.I., Brouns S.L.N., Farndale R.W., Henskens Y.M.C., Kuijpers M.J.E., Ten Cate H., van der Meijden P.E.J., et al. Role of platelet glycoprotein VI and tyrosine kinase syk in thrombus formation on collagen-like surfaces. Int. J. Mol. Sci. 2019;20:2788. doi: 10.3390/ijms20112788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumari S., Dash D. Regulation of beta-catenin stabilization in human platelets. Biochimie. 2013;95:1252–1257. doi: 10.1016/j.biochi.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 71.Huigsloot M., Nijtmans L.G., Szklarczyk R., Baars M.J.H., van den Brand M.A.M., Hendriksfranssen M.G.M., van den Heuvel L.P., Smeitink J.A.M., Huynen M.A., Rodenburg R.J.T. A mutation in C2orf64 causes impaired cytochrome c oxidase assembly and mitochondrial cardiomyopathy. Am. J. Hum. Genet. 2011;88:488–493. doi: 10.1016/j.ajhg.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schapira A.H., Cooper J.M., Dexter D., Jenner P., Clark J.B., Marsden C.D. Mitochondrial complex I deficiency in Parkinson's disease. Lancet. 1989;1:1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- 73.Boudreau L.H., Duchez A.C., Cloutier N., Soulet D., Martin N., Bollinger J., Paré A., Rousseau M., Naika G.S., Lévesque T., et al. Platelets release mitochondria serving as substrate for bactericidal group IIA-secreted phospholipase A2 to promote inflammation. Blood. 2014;124:2173–2183. doi: 10.1182/blood-2014-05-573543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Torchy M.P., Hamiche A., Klaholz B.P. Structure and function insights into the NuRD chromatin remodeling complex. Cell. Mol. Life Sci. 2015;72:2491–2507. doi: 10.1007/s00018-015-1880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Torretta S., Rampino A., Basso M., Pergola G., Di Carlo P., Shin J.H., Kleinman J.E., Hyde T.M., Weinberger D.R., Masellis R., et al. NURR1 and ERR1 modulate the expression of genes of a DRD2 coexpression network enriched for schizophrenia risk. J. Neurosci. 2020;40:932–941. doi: 10.1523/JNEUROSCI.0786-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu H., Sun Y., Zhang X., Li S., Hu D., Xiao L., Chen Y., He L., Wang D.W. Integrated analysis of summary statistics to identify pleiotropic genes and pathways for the comorbidity of schizophrenia and cardiometabolic disease. Front. Psychiatry. 2020;11:256. doi: 10.3389/fpsyt.2020.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ma C., Gu C., Huo Y., Li X., Luo X.J. The integrated landscape of causal genes and pathways in schizophrenia. Transl. Psychiatry. 2018;8:67. doi: 10.1038/s41398-018-0114-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gregory G.D., Miccio A., Bersenev A., Wang Y., Hong W., Zhang Z., Poncz M., Tong W., Blobel G.A. FOG1 requires NuRD to promote hematopoiesis and maintain lineage fidelity within the megakaryocytic-erythroid compartment. Blood. 2010;115:2156–2166. doi: 10.1182/blood-2009-10-251280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hauberg M.E., Zhang W., Giambartolomei C., Franzén O., Morris D.L., Vyse T.J., Ruusalepp A., CommonMind Consortium. Sklar P., Schadt E.E., et al. Large-scale identification of common trait and disease variants affecting gene expression. Am. J. Hum. Genet. 2017;101:157. doi: 10.1016/j.ajhg.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Siskind D., McCartney L., Goldschlager R., Kisely S. Clozapine v. first- and second-generation antipsychotics in treatment-refractory schizophrenia: systematic review and meta-analysis. Br. J. Psychiatry. 2016;209:385–392. doi: 10.1192/bjp.bp.115.177261. [DOI] [PubMed] [Google Scholar]
- 81.Myles N., Myles H., Xia S., Large M., Kisely S., Galletly C., Bird R., Siskind D. Meta-analysis examining the epidemiology of clozapine-associated neutropenia. Acta Psychiatr. Scand. 2018;138:101–109. doi: 10.1111/acps.12898. [DOI] [PubMed] [Google Scholar]
- 82.Yu H., Cheng W., Zhang X., Wang X., Yue W. Integration analysis of methylation quantitative trait loci and GWAS identify three schizophrenia risk variants. Neuropsychopharmacology. 2020;45:1179–1187. doi: 10.1038/s41386-020-0605-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pouget J.G., Schizophrenia Working Group of the Psychiatric Genomics Consortium. Han B., Wu Y., Mignot E., Ollila H.M., Barker J., Spain S., Dand N., Trembath R., et al. Cross-disorder analysis of schizophrenia and 19 immune-mediated diseases identifies shared genetic risk. Hum. Mol. Genet. 2019;28:3498–3513. doi: 10.1093/hmg/ddz145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Deutsch S.I., Rosse R.B., Lakshman R.M. Dysregulation of tau phosphorylation is a hypothesized point of convergence in the pathogenesis of Alzheimer's disease, frontotemporal dementia and schizophrenia with therapeutic implications. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2006;30:1369–1380. doi: 10.1016/j.pnpbp.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 85.Majerova P., Zilkova M., Kazmerova Z., Kovac A., Paholikova K., Kovacech B., Zilka N., Novak M. Microglia display modest phagocytic capacity for extracellular tau oligomers. J. Neuroinflammation. 2014;11:161. doi: 10.1186/s12974-014-0161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burgess S., Davies N.M., Thompson S.G. Bias due to participant overlap in two-sample Mendelian randomization. Genet. Epidemiol. 2016;40:597–608. doi: 10.1002/gepi.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Schizophrenia Working Group of the Psychiatric Genomics. C., Patterson N., Daly M.J., Price A.L., Neale B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., ReproGen Consortium. Psychiatric Genomics Consortium. Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3. Duncan L., et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.GTEx Consortium The genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fromer M., Roussos P., Sieberts S.K., Johnson J.S., Kavanagh D.H., Perumal T.M., Ruderfer D.M., Oh E.C., Topol A., Shah H.R., et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ng B., White C.C., Klein H.U., Sieberts S.K., McCabe C., Patrick E., Xu J., Yu L., Gaiteri C., Bennett D.A., et al. An xQTL map integrates the genetic architecture of the human brain's transcriptome and epigenome. Nat. Neurosci. 2017;20:1418–1426. doi: 10.1038/nn.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hannon E., Spiers H., Viana J., Pidsley R., Burrage J., Murphy T.M., Troakes C., Turecki G., O'Donovan M.C., Schalkwyk L.C., et al. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat. Neurosci. 2016;19:48–54. doi: 10.1038/nn.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jaffe A.E., Gao Y., Deep-Soboslay A., Tao R., Hyde T.M., Weinberger D.R., Kleinman J.E. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat. Neurosci. 2016;19:40–47. doi: 10.1038/nn.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.1000 Genomes Project Consortium An integrated map of genetic variation from 1, 092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carter A.R., Harrison S., Gill D., Davey Smith G., Taylor A.E., Howe L.D., Davies N.M. Educational attainment as a modifier for the effect of polygenic scores for cardiovascular risk factors: cross-sectional and prospective analysis of UK Biobank. Int. J. Epidemiol. 2022;51:885–897. doi: 10.1093/ije/dyac002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pollitt R.A., Kaufman J.S., Rose K.M., Diez-Roux A.V., Zeng D., Heiss G. Early-life and adult socioeconomic status and inflammatory risk markers in adulthood. Eur. J. Epidemiol. 2007;22:55–66. doi: 10.1007/s10654-006-9082-1. [DOI] [PubMed] [Google Scholar]
- 97.Hahad O., Daiber A., Michal M., Kuntic M., Lieb K., Beutel M., Münzel T. Smoking and neuropsychiatric disease-associations and underlying mechanisms. Int. J. Mol. Sci. 2021;22:7272. doi: 10.3390/ijms22147272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bates M.E., Barry D., Labouvie E.W., Fals-Stewart W., Voelbel G., Buckman J.F. Risk factors and neuropsychological recovery in clients with alcohol use disorders who were exposed to different treatments. J. Consult. Clin. Psychol. 2004;72:1073–1080. doi: 10.1037/0022-006X.72.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Comes A.L., Senner F., Budde M., Adorjan K., Anderson-Schmidt H., Andlauer T.F.M., Gade K., Hake M., Heilbronner U., Kalman J.L., et al. The genetic relationship between educational attainment and cognitive performance in major psychiatric disorders. Transl. Psychiatry. 2019;9:210. doi: 10.1038/s41398-019-0547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dohrenwend B.P., Levav I., Shrout P.E., Schwartz S., Naveh G., Link B.G., Skodol A.E., Stueve A. Socioeconomic status and psychiatric disorders: the causation-selection issue. Science. 1992;255:946–952. doi: 10.1126/science.1546291. [DOI] [PubMed] [Google Scholar]
- 101.Berisa T., Pickrell J.K. Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics. 2016;32:283–285. doi: 10.1093/bioinformatics/btv546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Byrne E.M., Zhu Z., Qi T., Skene N.G., Bryois J., Pardinas A.F., Stahl E., Smoller J.W., Rietschel M., et al. Bipolar Working Group of the Psychiatric Genomics Consortium. Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium Conditional GWAS analysis to identify disorder-specific SNPs for psychiatric disorders. Mol. Psychiatr. 2021;26:2070–2081. doi: 10.1038/s41380-020-0705-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Burgess S., Bowden J., Fall T., Ingelsson E., Thompson S.G. Sensitivity analyses for robust causal inference from mendelian randomization analyses with multiple genetic variants. Epidemiology. 2017;28:30–42. doi: 10.1097/EDE.0000000000000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
GWAS summary statistics for MS are available by application from https://imsgc.net/?page_id=31. GWAS summary statistics for migraine are available from the International Headache Genetics Consortium (IHGC, http://www.headachegenetics.org/content/datasets-and-cohorts) by request to Professor Dale Nyholt (d.nyholt@qut.edu.au). All other data are publicly available and listed in the key resources table.
-
•
This study did not generate any unique datasets or code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.