Abstract
BACKGROUND:
We sought to characterize methylation changes in brain and blood associated with major depressive disorder (MDD). As analyses of bulk tissue may obscure association signals and hamper the biological interpretation of findings, these changes were studied on a cell type–specific level.
METHODS:
In 3 collections of human postmortem brain (n = 206) and 1 collection of blood samples (N = 1132) of MDD cases and controls, we used epigenomic deconvolution to perform cell type–specific methylome-wide association studies within subpopulations of neurons/glia for the brain data and granulocytes/T cells/B cells/monocytes for the blood data. Sorted neurons/glia from a fourth postmortem brain collection (n = 58) were used for validation purposes.
RESULTS:
Cell type–specific methylome-wide association studies identified multiple findings in neurons/glia that were detected across brain collections and were reproducible in physically sorted nuclei. Cell type–specific analyses in blood samples identified methylome-wide significant associations in T cells, monocytes, and whole blood that replicated findings from a past methylation study of MDD. Pathway analyses implicated p75 neurotrophin receptor/nerve growth factor signaling and innate immune toll-like receptor signaling in MDD. Top results in neurons, glia, bulk brain, T cells, monocytes, and whole blood were enriched for genes supported by genome-wide association studies for MDD and other psychiatric disorders.
CONCLUSIONS:
We both replicated and identified novel MDD–methylation associations in human brain and blood samples at a cell type–specific level. Our results provide mechanistic insights into how the immune system may interact with the brain to affect MDD susceptibility. Importantly, our findings involved associations with MDD in human samples that implicated many closely related biological pathways. These disease-linked sites and pathways represent promising new therapeutic targets for MDD.
Keywords: Depression, Epigenetics, Immune deconvolution, Methylation, Nerve growth factor
Major depressive disorder (MDD) has high prevalence (1), can start early in life, and is often chronic, making it the leading cause of disability worldwide (2). DNA methylation studies offer unique opportunities to improve the understanding and treatment of MDD by identifying molecular signatures of disease features or the traces of environmental insults that influence susceptibility (3–5). These studies also have profound translational potential, as methylation is modifiable and may potentially serve as a diagnostic biomarker.
There exists good evidence that MDD has a systemic component that involves both the brain and peripheral immune system (6,7). Therefore, we sought to characterize MDD-linked methylation changes in both brain and blood. Methylation studies are typically performed in bulk tissue that contains multiple cell types. Failure to account for multiple cell types has several drawbacks (8). First, false-positives may occur when cell type abundances vary between cases and controls (9,10). Second, associations may not be detectable in bulk when case-control differences are in opposite directions between cell types or when the most common cell types obscure signals in cells of low abundance. Third, knowing which cell type harbors an association is key for the biological interpretation of results and the design of follow-up experiments.
We examined methylation differences between MDD cases and controls in 3 collections of brain samples totaling 264 individuals, as well as in 1132 independent blood samples. To perform cell type–specific methylome-wide association studies (MWASs) for neurons/glia and granulocytes/T cells/B cells/monocytes, we applied epigenomic deconvolution (11,12) and studied physically sorted neuron/glia nuclei from case and control brains. Methylation findings that were detected across the brain collections were identified by meta-analysis, and findings in blood were compared with those of previous methylation study of MDD. Finally, we tested for over-representation of top MWAS findings among top results from genome-wide association studies (GWASs) for MDD and related disorders.
METHODS AND MATERIALS
Samples
We used methylation enrichment-based sequencing data from 206 postmortem brain samples from 3 collections, as previously described (13). The sample collections were predominantly from Australia (AUS) (30 MDD, 31 control; Brodmann area [BA] 25), United States of America (USA) (44 MDD, 37 control; BA 10) and Canada (CAN) (39 MDD, 25 control; BA 10). Additionally, we used array-based methylation data from physically sorted neurons (28 MDD, 29 control) and glia (29 MDD, 29 control) from an independent sample collection (14). Methylation enrichment-based sequencing data from whole blood of 812 cases and 320 controls was from the Netherlands Study of Depression and Anxiety (NESDA) (13,15). Descriptions of study participants are presented in Supplement 1.
Cell Type–Specific MWASs
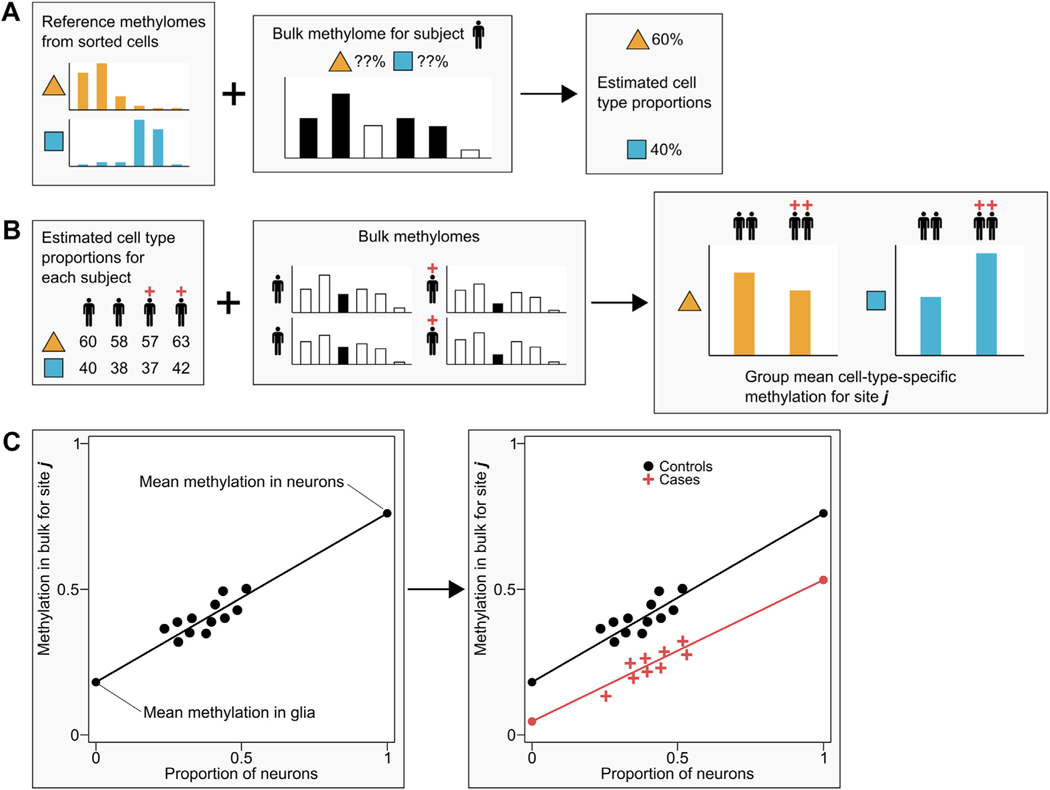
It is impractical to sort and assay methylation in each cell type for every sample in large-scale studies. Therefore, we used epigenomic deconvolution (Figure 1) to perform cell type–specific MWASs using the bulk brain and whole blood data. Deconvolution is commonly used in gene expression studies (12,16) and may readily be applied to methylation data (11,17,18). The approach has been validated by showing that it can detect known associations in artificial data as well as associations observed with empirical data from purified cells (12,17). We further showed through simulations (Code S1 in Supplement 3) 1) that if there are no effects, the model properly controls the type I error, or 2) that if the cell type proportion estimates have errors, the model is fairly robust. We also performed cell type–specific MWASs after permuting case-control labels. The resulting quantile-quantile plots for brain (Figure S3 in Supplement 1) and blood (Figure S8 in Supplement 1) showed that the p values were close to the main diagonal with average lambdas that were not significantly different from 1. Cell type–specific MWASs are performed by first estimating cell type proportions in all study samples with bulk data using a reference panel (9,19). These reference panels comprise methylation profiles of physically sorted subtypes of cells generated from a small subset of subjects. We sorted nuclei/cells from 5 frozen postmortem brain and 6 fresh whole blood samples. Simulations (Figure S1 in Supplement 1) showed that whereas the total number of sites used to estimate cell type proportions is critical, the number of subjects per panel has little impact on the precision of the estimates. Thus, the simulations show that our panel was sufficiently large (see Supplement 1). These predicted cell type proportions are then used to test case-control differences in methylation on a cell type–specific level using all study samples with available bulk data. To declare methylome-wide significance we applied an appropriate (20,21) false discovery rate (FDR) threshold of 0.1.
Figure 1.
Epigenomic deconvolution for testing case-control differences in subpopulations of cells. (A) In step 1 of the deconvolution method, reference methylomes from purified samples of sorted cells are used to estimate cell type proportions. For one subject at a time, bulk methylation data are regressed on the most informative sites in the reference methylomes to obtain predicted proportions of each cell type. (B) After cell type proportions have been predicted for each subject in the study, step 2 uses these proportions to estimate case-control differences at each CpG site. (C) To illustrate how cell type–specific differences are predicted, we present a simple example. Since bulk methylation and proportions of neurons/glia will differ between subjects, we can regress bulk methylation levels (y-axis) on the proportion of neuronal cells (x-axis). Thus, extrapolating the regression line to the point where the proportion of neurons is 0 (i.e., there are only glia cells) estimates the group mean methylation in glia, and extrapolation to the point where the proportion of neurons is 1 estimates the group mean methylation in neurons. By allowing the regression lines to differ between controls (black dots) and cases (red crosses), we obtain different predicted cell type–specific group means that can be tested for significance using standard statistical tests. See Methods and Materials and Supplement 1 for discussion of the statistical models.
Reference Panels for Brain and Blood Cell Types.
For methylation enrichment-based sequencing references for neurons (NeuN+) and glia (NeuN−), we sorted nuclei (22) from 5 control samples from brain banks in AUS, CAN, The Netherlands, and USA (sources of MWAS samples). To generate methylation enrichment-based sequencing references for blood cell types, we obtained fresh whole blood samples from 6 USA subjects (23). Granulocytes (CD15+), T cells (CD3+), B cells (CD19+), and monocytes (CD14+) were isolated from whole blood using EasySep magnetic separation (Stemcell Technologies, Cambridge, MA). DNA extracted from sorted nuclei/cells was then assayed using methyl-CG binding domain sequencing (24–26). Details are discussed in Supplement 1.
Model.
While Figure 1 and its legend explain the basic principles underlying the deconvolution method, here we present the statistical model [also see equation 2 in the Methods section of Zheng et al. (17)]:
Thus, methylation measurements in bulk tissue Ybulk are regressed on c = 1 to nc, cell type proportions Pc, and the product of disease status for MDD coded as 0 or 1 by cell type proportions (MDD × Pc). The model allows for covariates (not shown) and residual effects E. Coefficient mc is the effect of cell type c. MDD is coded 0 or 1, is the case-control difference for cell type c that is used to test the null hypothesis that cell type methylation means are equal for cases and controls. Note that the model has no constant, because . Expanded discussion and robustness analyses are presented in Supplement 1.
Covariates in Methylome-wide Association Testing
To test each CpG site for association with MDD, we performed multiple regression analyses that included several classes of covariates. We included measured technical variables including the quantity of methylation-enriched DNA captured, batch, and peak location (26), as well as demographic variables for age and sex. For NESDA blood samples, we also included smoking status, alcohol use, body mass index, and 3 principal components (PCs) from a GWAS to capture ancestral differences. For brain, we included postmortem interval. PC analysis on the methylation data was performed on the covariate-adjusted data to capture any remaining unmeasured sources of variation. We used a scree test to select the methylation PCs to include in the final MWAS (1 PC for AUS, CAN, USA, and NESDA). In the bulk brain and whole blood MWASs, predicted cell type proportions were also included as covariates to control for cell type heterogeneity (23). Raw array-based data were processed following Lehne et al. (27), and demographics, technical variables, and PCs were included as covariates. For all analyses, covariates and PCs were included in the final MWASs.
Meta-analyses
To identify top bulk brain and cell type–specific MWAS findings that replicated among the 3 sets of brain collections (AUS, CAN, USA), we performed a meta-analysis using Stouffer’s weighted z score method (28) after transforming t statistics from each MWAS into z statistics.
Pathway, Colocalization, and Gene Overlap Testing
Pathway, colocalization, and gene overlap testing were performed using circular permutations (29) that generate the empirical test-statistic distribution under the null hypothesis while preserving the correlational structure of the data (see Supplement 1). Pathway analyses were performed with the Reactome (30) database using 10,000 circular permutations. Pathway tests were restricted to only the top 1000 findings to prevent oversaturation. We also tested whether our MWAS findings colocalized with UCSC Genome Browser genomic feature tracks, Roadmap Epigenomics Project chromHMM 15-state chromatin model tracks (31), Genotype-Tissue Expression (GTEx) expression quantitative trait loci (eQTLs), known expression quantitative trait methylation (eQTMs) (32,33), or colocalized and/or shared top genes from previous MDD MWASs and GWASs using 100,000 circular permutations. In general, we tested the top 0.5%, 0.1%, or 0.05% of sites from our MWASs for enrichment testing across asymmetrical datasets (i.e., methylation enrichment-based sequencing vs. methylation arrays) and corrected for testing multiple thresholds. Expanded details can be found in Supplement 1.
RESULTS
Cell Type–Specific MWASs in Brain
Predicted proportions of neurons to glia (approximately 1:3) matched expectations for cortex (34) and showed no significant case-control differences (Table S1 in Supplement 1). Quantile-quantile plots for the cell type–specific MWASs (Figure S3 in Supplement 1) suggested that multiple CpGs had discernible effects within individual cell types. MWAS of permuted case-control status for each analysis yielded average lambdas of approximately 1 (Figure S4 in Supplement 1), which indicated no evidence of test-statistic inflation under the empirical null. To identify top findings in neurons, glia, and bulk brain, we performed meta-analyses across the AUS, CAN, and USA brain sample collections for 17,321,920 CpGs.
Neurons and Glia.
No CpGs reached methylome-wide significance (FDR 0.1) among the meta-analyses for the MWASs for neurons or glia (Tables S2 and S3 in Supplement 2). The top site in neurons (p value = 8.52 × 10−8) was found in an intergenic region situated between the serotonin receptor gene HTR4 and the beta-adrenergic receptor gene ADRB2. In glia, the top site (p value = 5.18 × 10−8) was also intergenic in a region that was most near TPRA1. Many top results were shared between neurons and glia (i.e., 419 of the top 1000 from both) and were found at genes such as RBFOX2, HERC2, TRIM3, and RNF111. Encoding splicing regulators that are important for neuronal development (35), the RBFOX regulatory network has been implicated in previous genetic (36) and methylation (37) studies of depression. HERC2, TRIM3, and RNF111 all encode ubiquitin ligases that influence neurodevelopment and synaptic plasticity (38–43).
The top findings that were unique to neurons included those at TRAPPC9 (p value = 1.04 × 10−6) and HRH4 (p value = 1.77 × 10−6). Highly expressed in neurons and involved in nuclear factor-κB and nerve growth factor (NGF) signaling (44), rare variants in TRAPPC9 have been associated with intellectual disability (45,46). Encoding a histamine receptor, HRH4 is located in a region implicated in a pharmacogenomic study of citalopram response (47).
Finally, among top unique glia findings were sites (top p value = 4.43 × 10−7) located in an intergenic region nearest to ZC3H12A, which is involved in innate immune regulation (48), and GRIK3, which is involved in glutamate signaling. Notably, GRIK3 has been linked to recurrent MDD (49).
The top findings for neurons were enriched at DNase I hypersensitivity sites, and both neuron and glia results were enriched at introns and transcription factor binding sites (Tables S4 and S5 in Supplement 2). Results for neurons and glia were not overrepresented at chromHMM chromatin states (31) nor eQTLs for brain (Tables S4 and S5 in Supplement 2). However, as chromatin states and eQTLs for brain were generated from bulk tissue, they may not accurately capture cell type–specific states. For neurons, 81 top CpGs mapped to known eQTMs (Table S6 in Supplement 2), and for glia, 71 sites mapped to eQTMs (Table S7 in Supplement 2).
Complementary Analyses in Sorted Neurons and Glia.
To check the results obtained via epigenomic deconvolution, we performed MWASs on array-based methylation data from sorted neuronal (28 MDD, 29 control) and glial (29 MDD, 29 control) nuclei of independent samples (14) (Tables S8 and S9 in Supplement 2; Figures S6 and S7 in Supplement 1). We tested for CpG colocalization between the top sites from deconvoluted neurons/glia and nominally significant MWAS findings (p value <.05) from the sorted nuclei. The top 0.05% of results for deconvoluted neurons were significantly enriched among nominally significant results for sorted neurons (70 CpGs, p value = 4.20 × 10−3). Results for deconvoluted glia colocalized with 270 CpGs among nominally significant results for sorted glia but were not enriched (p value = .48).
Bulk Brain.
Bulk brain analysis may have better power for sites that are affected in similar fashion across multiple cell types. Therefore, we also performed meta-analyses among the MWASs of bulk brain methylation data. However, no CpG reached methylome-wide significance (FDR 0.1) in the meta-analysis for bulk brain (Table S10 in Supplement 2), where the top site (p value = 1.03 × 10−7) was situated in an intergenic region between the glucose transporter SLC5A4 and human telomerase reverse transcriptase regulator RFPL3. This finding is notable given the abnormal glucose tolerance, decreased telomere length, and increased epigenetic aging that is observed in patients with MDD (50–53).
The top findings in bulk brain also included sites located in ASTN2 (p value = 1.29 × 10−6) and RBFOX1 (p value = 2.09 × 10−6). Variants in both the neuronal–glial adhesion molecule ASTN2 and the RNA-slicing regulator RBFOX1 obtained genome-wide significance in a recent meta-analysis of large MDD GWASs (36). Bulk brain results were not enriched for any particular genomic feature consensus track or eQTLs but were significantly enriched at predicted chromatin states for “Bivalent-Poised TSS” and “Flanking Bivalent TSS-Enh” (Table S11 in Supplement 2) with 86 CpGs mapping to known eQTMs (Table S12 in Supplement 2).
Cell Type–Specific MWASs in Blood
Mean predicted cell type proportions were 55.8%, 31.3%, 9.3%, and 3.6% for granulocytes (CD15), T cells (CD3), B cells (CD19), and monocytes (CD14), respectively. With the exception of monocytes, the predicted cell type proportions differed significantly between cases and controls (Table S13 in Supplement 1). Cases tended to have increased myeloid cell (granulocytes, monocytes) and decreased lymphocyte (T cells/B cells) levels, as generally expected (54,55).
After quality control, 21,869,561 CpGs were available for testing. The quantile-quantile plots (Figure S8 in Supplement 1) suggested that associations mainly involved monocytes and T cells. No methylome-wide significant findings (FDR 0.1) were observed for granulocytes or B cells. Permutations of case-control status for each MWAS (Figure S9 in Supplement 1) again suggested that the observed effects were not inflation artifacts.
Monocytes.
The MWAS for monocytes yielded 904 methylome-wide significant CpGs (Table S14 in Supplement 2). Top genic findings for monocytes included ITPR2, SHANK2, KATNAL2, and GRIA1. Also present among methylome-wide significant results for monocytes were sites within CDC42BPB that was implicated by a previous methylation meta-analysis of depressive symptoms (56). Findings for monocytes were significantly enriched at 5′- and 3′-UTRs, CpG islands and shores, brain-specific DNase I hypersensitivity sites, genes, transcription factor binding sites, and promoters (Table S15 in Supplement 2). Overlap with chromatin state tracks showed enrichment at “Active TSS,” “Strong Transcription,” “Weak Transcription,” and “ZNF Genes and Repeats,” along with significant enrichment at blood eQTLs (Table S15 in Supplement 2) and mapping of 137 top results to known eQTMs (Table S16 in Supplement 2).
T Cells.
In T cells, 9 CpGs passed methylome-wide significance (Table S17 in Supplement 2) with the top site (p value = 3.57 × 10−9) found in an intergenic region located most near GRIA2 and FAM198B. Significant genic findings for T cells involved MTA3, DAB2IP, and STRADB. Top results for T cells were enriched for a number of genomic features including 5′-UTRs, DNase I hypersensitivity sites, and introns, as well at chromatin states for “Flanking Bivalent TSS-Enhancer” and “Bivalent Enhancer” (Table S18 in Supplement 2) with 142 top sites mapping to eQTMs (Table S19 in Supplement 2).
Whole Blood.
Only one intergenic site in whole blood passed the FDR threshold of 0.1 employed in the current analysis (top site p = 2.54 × 10−9) that was most proximal to CYP2J2 and HOOK1 (Table S20 in Supplement 2). As the cell types showing the largest signals in our MWASs are of relatively low abundance (T cells and monocytes), statistical power may be lacking to detect many of these differences in whole blood, as they represent a minority of cells. Nonetheless, top results from whole blood were significantly enriched at features like CpG islands, exons, splice sites, promoters, and virtually all regulatory chromatin states (Table S21 in Supplement 2), with 145 top sites mapping to known eQTMs (Table S22 in Supplement 2).
Top Findings Are Replicated at Loci Implicated in Past Methylation Studies of Depression
Story Jovanova et al. (56) reported significant associations at CDC42BPB and ARHGEF3 from a large multicohort MWAS meta-analysis for depressive symptoms in whole blood. We tested for enrichment between our top results versus the top 41 loci from the meta-analysis of Story Jovanova et al. The most significant enrichment was observed for the top 0.05% of our whole blood results (6 CpGs, p value = 2.50 × 10−3), which was driven by the CpG at CDC42BPB. In contrast, our results for T cells/monocytes and neurons/glia/bulk brain were not enriched for CpGs implicated by Story Jovanova et al. However, their study was performed in whole blood rather than on a cell type–specific level, included some cohort data that was not corrected for cell type heterogeneity, and again relied on sparse methylation array data.
Thus, we additionally tested whether genes implicated by our top results were significantly overrepresented for genes at the top 41 loci reported by Story Jovanova et al. Results showed that genes implicated by the top 0.5% sites of our MWASs for whole blood (43 genes, p value = 3.00 × 10−5), T cells (37 genes, p value = 2.03 × 10−2), monocytes (40 genes, p value = 2.10 × 10−4), neurons (37 genes, p value = 1.25 × 10−2), glia (35 genes, p value = 2.56 × 10−2), and bulk brain (37 genes, p value = 5.04 × 10−3) were indeed over-represented for those that had been previously reported to harbor methylation associations (including CDC42BPB and ARHGEF3) with depressive symptoms. Thus, different methylation sites may influence the same disease-related genes in multiple tissues and cell types.
Top Findings Are Overrepresented at Genes From GWAS of MDD and Other Neuropsychiatric Disorders
We tested for overlap of genes between the top findings of our cell type–specific MWASs and the top 10,000 variants from GWASs for attention-deficit/hyperactivity disorder (57), anxiety disorders (58), autism spectrum disorder (59), bipolar disorder (60), MDD (36), and schizophrenia (60). We also tested against 869 variants for neurodegenerative disease collected from GWAS Catalog data (61). To check for specificity, we also tested our findings versus the top 10,000 variants for breast cancer (62).
Given the large differences in power and strength of association signals between our different methylation studies, we conservatively selected only the top 1000 findings from each MWAS to test for gene overlap against GWAS. As expected, none of our MWAS results were enriched at genes associated with breast cancer.
In contrast, our results (Table 1) showed very robust and highly significant overrepresentation of genes implicated by MDD GWASs and the top 1000 sites from MWASs of neurons, glia, bulk brain, T cells, monocytes, and whole blood. Results for neuron and bulk brain MWASs were further over-represented for genes associated with bipolar disorder. Genes associated with anxiety disorders were overrepresented among top MWAS results from monocytes and whole blood. Finally, neuron and glia results significantly overlapped genes associated with neurodegenerative disease.
Table 1.
Enrichment of Top MWAS Findings Versus GWAS Findings of Neuropsychiatric Disorders
| MWAS Neurons | MWAS Glia | MWAS Bulk Brain | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| GWAS | Gene Overlap | Odds Ratio | p Value | Gene Overlap | Odds Ratio | p Value | Gene Overlap | Odds Ratio | p Value |
| Attention-Deficit/Hyperactivity Disorder | 18 | 1.06 | .43 | 17 | 1.02 | .49 | 24 | 1.42 | .06 |
|
| |||||||||
| Anxiety Disorders | 43 | 0.93 | .70 | 37 | 0.82 | .92 | 55 | 1.19 | .10 |
|
| |||||||||
| Autism Spectrum Disorder | 23 | 1.32 | .11 | 17 | 0.99 | .54 | 24 | 1.38 | .08 |
|
| |||||||||
| Bipolar Disorder | 29 | 1.45 | .03 | 24 | 1.22 | .19 | 31 | 1.56 | .01 |
|
| |||||||||
| Major Depressive Disorder | 166 | 2.71 | <.00001 | 157 | 2.61 | <.00001 | 156 | 2.56 | <.00001 |
|
| |||||||||
| Schizophrenia | 10 | 1.57 | .16 | 6 | 0.96 | .54 | 1.89 | 12 | .06 |
|
| |||||||||
| Neurodegenerative Disease | 33 | 1.48 | .02 | 33 | 1.50 | .01 | 27 | 1.21 | .17 |
|
| |||||||||
| Breast Cancer | 9 | 1.72 | .09 | 9 | 1.76 | .09 | 9 | 1.73 | .09 |
|
| |||||||||
| MWAS CD3 T cells | MWAS CD14 Monocytes | MWAS Whole Blood | |||||||
|
|
|
|
|||||||
| GWAS | Gene Overlap | Odds Ratio | p Value | Gene Overlap | Odds Ratio | p Value | Gene Overlap | Odds Ratio | p Value |
|
| |||||||||
| Attention-Deficit/Hyperactivity Disorder | 18 | 1.09 | .38 | 12 | 1.33 | .20 | 18 | 1.22 | .24 |
|
| |||||||||
| Anxiety Disorders | 52 | 1.17 | .14 | 34 | 1.40 | .03 | 65 | 1.63 | .0001 |
|
| |||||||||
| Autism Spectrum Disorder | 21 | 1.25 | .18 | 7 | 0.76 | .80 | 16 | 1.06 | .44 |
|
| |||||||||
| Bipolar Disorder | 23 | 1.19 | .22 | 14 | 1.33 | .18 | 24 | 1.39 | .08 |
|
| |||||||||
| Major Depressive Disorder | 181 | 3.03 | <.00001 | 77 | 2.40 | <.00001 | 160 | 3.01 | <.00001 |
|
| |||||||||
| Schizophrenia | 6 | 0.97 | .53 | 4 | 1.18 | .41 | 6 | 1.08 | .45 |
|
| |||||||||
| Neurodegenerative Disease | 18 | 0.83 | .82 | 16 | 1.34 | .12 | 23 | 1.19 | .22 |
|
| |||||||||
| Breast Cancer | 4 | 0.79 | .72 | 1 | 0.36 | .92 | 7 | 1.54 | .18 |
GWAS, genome-wide association study; MWAS, methylome-wide association study.
Pathway and Cross-Tissue Analyses
To investigate the possible biological mechanisms and processes underlying our findings, we performed pathway analyses for the top 1000 sites from our MWASs.
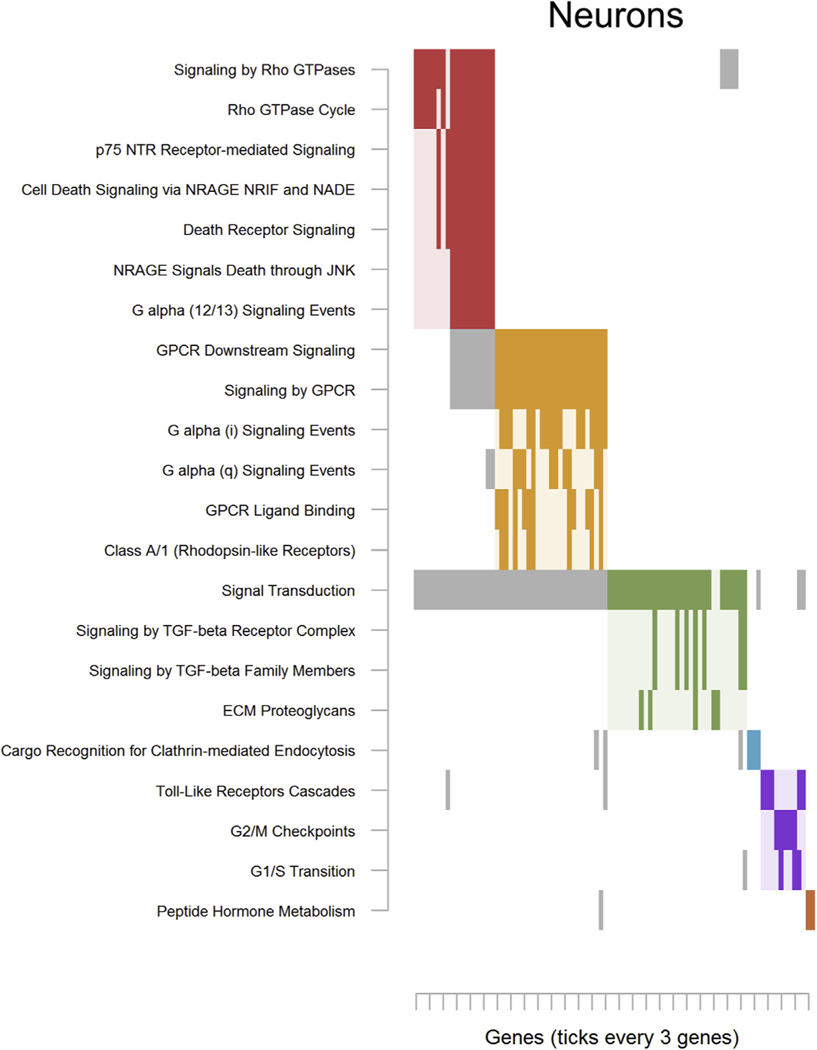
For neurons, top findings were enriched for 22 pathways (Table S23 in Supplement 2) that clustered into 6 groups. The first major cluster (Figure 2; red) was populated for terms related to “Signaling by Rho GTPases” and neurotrophin-mediated cell death like “p75 NTR Receptor-mediated Signaling” and “Death Receptor Signaling.” Other clusters centered on G protein–coupled receptor signaling (Figure 2; orange) and transforming growth factor β signaling (Figure 2, green).
Figure 2.
Significantly enriched pathways for neurons. As pathways often share genes, the raster plot visualizes the clustering of pathways (y-axis) determined on the basis of their overlapping genes (x-axis). The solid rectangles indicate genes that both were among the top methylome-wide association study results and were members of the listed pathway. Note that only genes that were among the top methylome-wide association study results, rather than all possible pathway members, are plotted. Only pathways containing a minimum of 3 overlapping genes and those passing nominal significance (α <.05) were retained. Complete pathway names, gene names, odds ratios, and p values are presented in Table S5 in Supplement 2 for deconvoluted neurons and Table S9 in Supplement 2 for sorted neurons. ECM, extracellular matrix protein; G1/S, G1 phase/S phase; G2/M, G2 phase/mitosis; GPCR, G protein–coupled receptor; GTPase, guanosine triphosphate hydrolase enzyme; JNK, c-Jun N-terminal kinase; NADE, p75NTR-associated cell death executor; NRAGE, neurotrophin receptor–interacting MAGE homolog; NRIF, neurotrophin receptor interacting factor; p75 NTR, neurotrophin receptor p75; TGF, transforming growth factor.
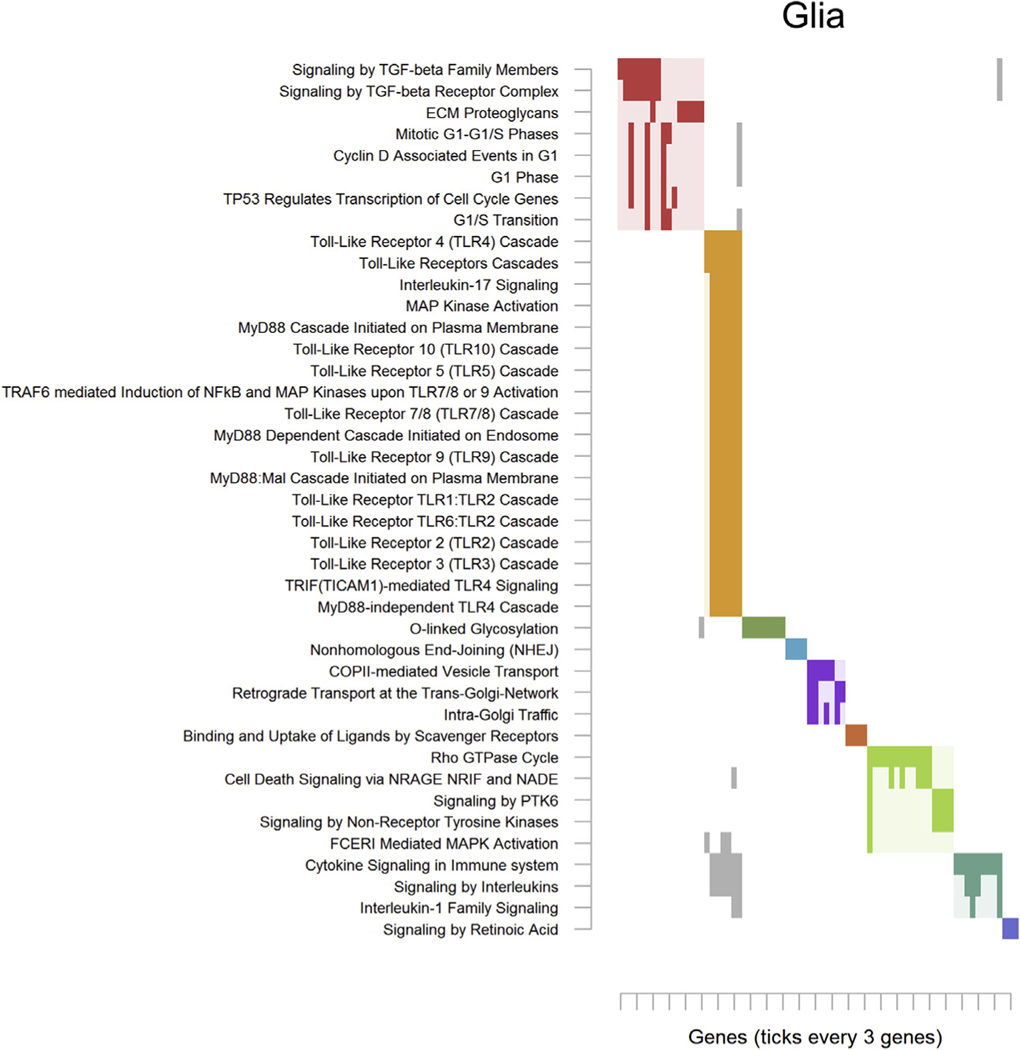
Results in glia drove enrichment of 41 pathways (Table S24 in Supplement 2) that segregated into 9 clusters. The first cluster (Figure 3; red) involved transforming growth factor β signaling and cell cycle processes. The second and predominant cluster (Figure 3; yellow) contained many overlapping pathways related to innate immune response via toll-like receptors (TLRs) and inflammatory cytokine signaling.
Figure 3.
Significantly enriched pathways for glia. See Figure 2 for an explanation of the plot. Complete pathway names, gene names, odds ratios, and p values are presented in Table S6 in Supplement 2 for deconvoluted glia and Table S10 in Supplement 2 for sorted glia. COPII, coat protein complex II; ECM, extracellular matrix protein; FCERI, Fc epsilon receptor; G1/S, G1 phase/S phase; MAPK, mitogen-activated protein kinase; NFκB, nuclear factor-κB; PTK6, protein-tyrosine kinase 1; TGF, transforming growth factor; TRAFC, tumor necrosis factor receptor-associated factor family protein.
Bulk brain findings showed enrichment of only 6 unrelated pathways that involved “Reproduction,” cell cycle processes, posttranslational protein modification, ribosomal RNA expression, and hormone metabolism (Table S25 in Supplement 2).
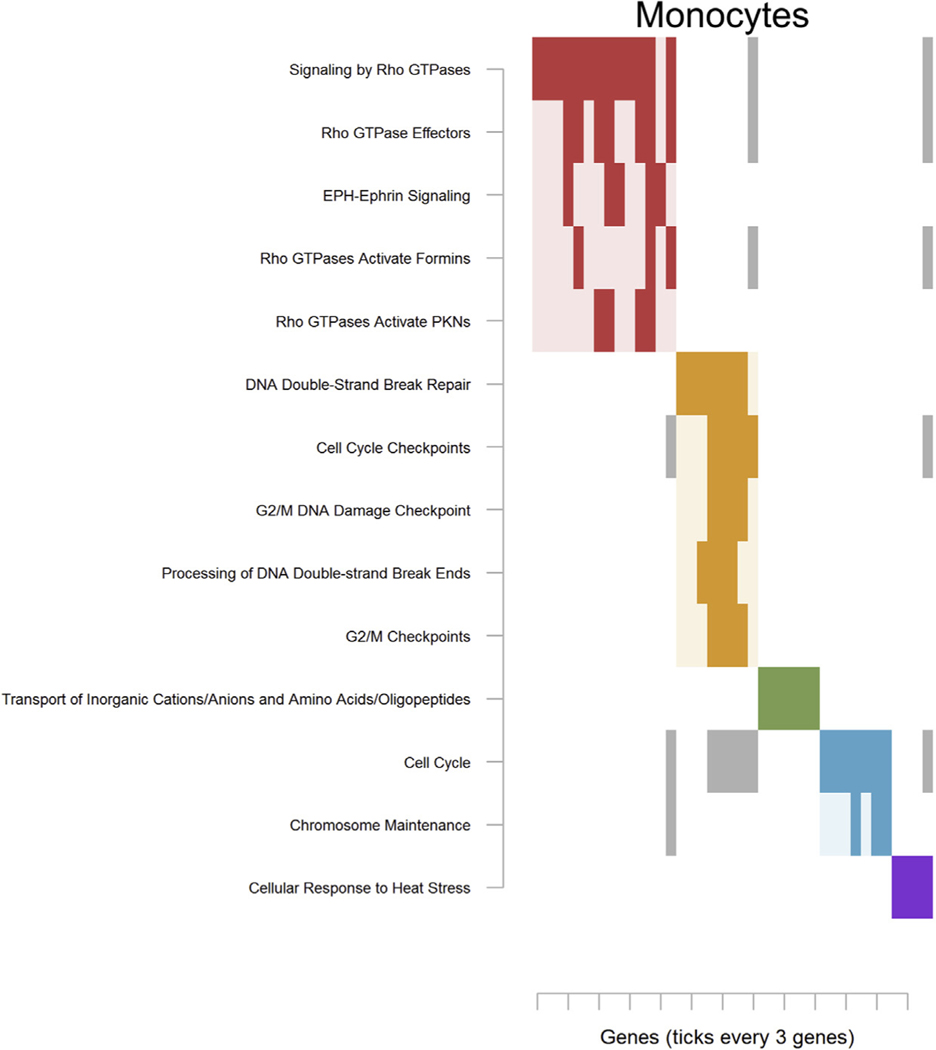
Findings for monocytes drove enrichment of 14 pathways that grouped into 5 clusters (Table S26 in Supplement 2). The first major cluster (Figure 4; red) involved “Signaling by Rho GTPases,” and a second cluster (Figure 4; orange) centered around cell cycle processes.
Figure 4.
Significantly enriched pathways for monocytes (CD14). See Figure 2 for an explanation of the plot. Complete pathway names, gene names, odds ratios, and p values are presented in Table S14 in Supplement 2 for monocytes. EPH, ephrin receptor; G2/M, G2 phase/mitosis; PKN, protein kinase N.
T cell results contributed to the enrichment of only 4 pathways for “Keratinization” (driven by blood-expressed keratins KRT72, KRT73, KRT86), cellular respiration, and integrin signaling (Table S27 in Supplement 2).
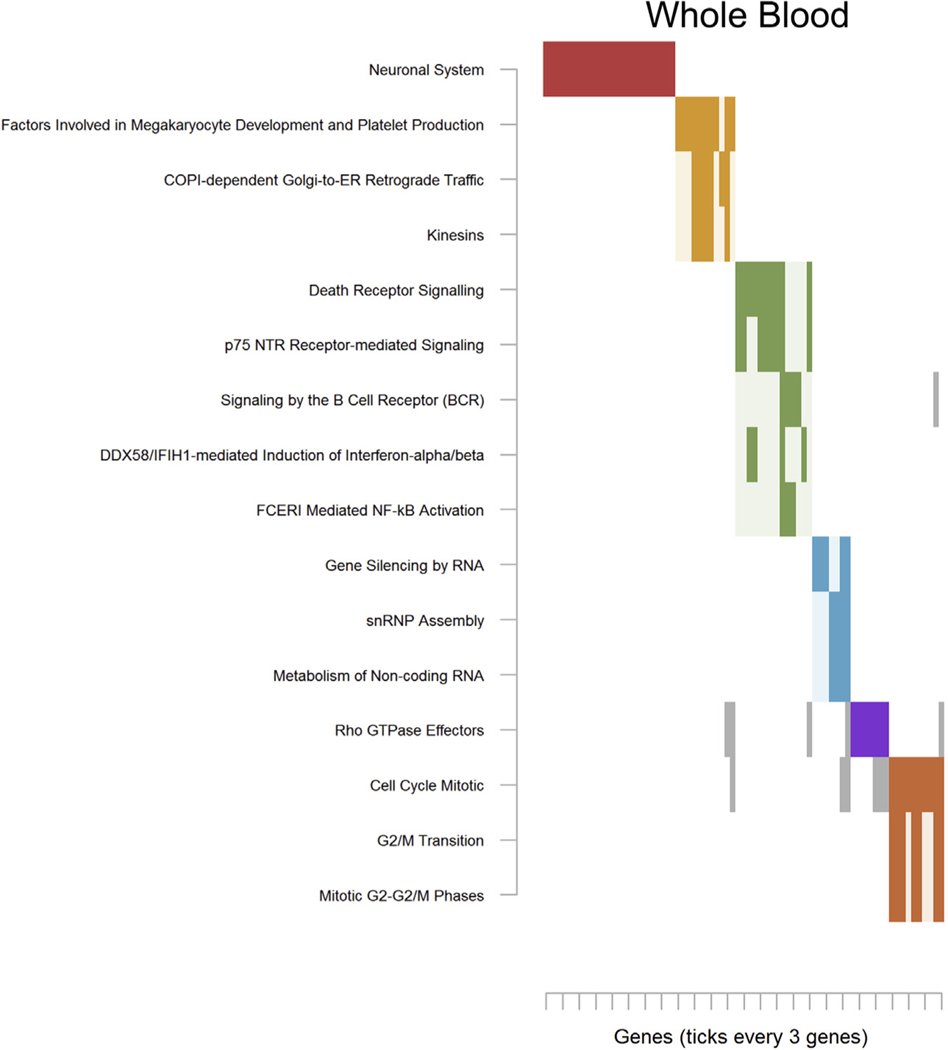
Lastly, top results from whole blood led to enrichment of 16 pathways that gathered into 6 smaller clusters (Table S28 in Supplement 2). The third cluster for pathways whole blood (Figure 5; green) implicated “p75 NTR Receptor-mediated Signaling” and “Death Receptor Signaling,” which were also key pathways in neurons. Other notable pathways included “Neuronal System,” and a sixth cluster (Figure 5; orange) again involving cell cycle processes.
Figure 5.
Significantly enriched pathways for whole blood. See Figure 2 for an explanation of the plot. Complete pathway names, gene names, odds ratios, and p values are presented in Table S14 in Supplement 2 for monocytes. COPI, tethering coat protein complex I; DDX58/IFIH1, DExD/H-box helicase 58/interferon induced with helicase C domain 1; FCERI, Fc epsilon receptor; G2/M, G2 phase/mitosis; p75 NTR, neurotrophin receptor p75; RNP, ribonucleoprotein particle.
Based on the apparent overlap of implicated pathways between our results in brain and blood, we investigated whether top results were enriched across tissues and cell types. We did not observe evidence of overlap between the top 1000 findings across tissues on a CpG-to-CpG level. However, when we examined results on a gene level, we observed significant overlap between genes associated with the top 1000 sites from T cells and bulk brain MWASs (33 genes, p value = 2.99 × 10−2). Notably, the overlap included both ASTN2 and RBFOX1. While these results do not suggest that top cell type–specific methylation signals are typically shared across tissues, they do imply that methylation may impact disease-related processes in multiple tissues and cell types via a related networks of genes.
DISCUSSION
In one of the most comprehensive methylation studies of MDD to date, we characterized methylome-wide associations with MDD in large collections of brain and blood samples at a bulk tissue and a cell type–specific level. Using epigenomic deconvolution, we identified novel findings in neurons and glia in a meta-analysis of 3 brain collections and replicated our findings in a fourth sample of sorted nuclei. A cell type–specific MWAS in blood also uncovered associations in CD14 monocytes and CD3 T cells that were not detected in whole blood. Whereas we did not observe much cross-tissue overlap in our findings on a CpG-to-CpG level, we did find evidence that similar genes from our own results and those from Story Jovanova et al. (56) were consistently implicated in multiple tissues/cell types. Strong overlap with GWASs of MDD and some related disorders further supported the robustness of our findings and highlighted the shared liabilities among neuropsychiatric disorders.
Our results implicated NGF receptor (p75NTR) signaling in MDD that aligned with previous methylation investigations of depressive symptoms (56) and chronic MDD (63). A fine balance of signaling through the generally prosurvival Trk receptors and apoptotic p75NTR is needed for normal neuro-development and neuron survival (64). Notably, our results suggested differences in both neurons and whole blood of MDD cases and controls, suggesting that systemic perturbations of NGF or p75NTR signaling are present in MDD. This seems plausible considering that psychosocial stress, a major risk factor for MDD, alters both circulating and brain concentrations of NGF (65–69).
Our glial MWAS yielded results that also strongly implicated TLR activation in MDD. TLRs are key components of the innate immune system (70), which itself has long been suspected to play a role in the pathogenesis of MDD (71). Importantly, both acute and chronic stress affect the innate immune system via TLRs in both blood and brain (6,72–74). Therefore, our findings also suggest that methylation changes in glia could be indicative of the effects of stress in the brains of patients with MDD, especially when we consider that both astrocytes and microglia mediate the innate immune system in the brain through TLRs (75). Further, innate immune activation and neuroinflammation can induce microglia to increase production of NGF (76,77) and can interact with transforming growth factor β signaling (78), which was also implicated by our neural and glial MWASs. Finally, we did not find substantial evidence of human leukocyte antigen involvement in any of our analyses. Altogether, our findings suggest that MDD is characterized by stress-linked activation of the innate immune system leading to systemic inflammatory responses that affect cells in both brain and blood.
As antidepressant treatment, smoking status, alcohol use, and body mass index information was available for the NESDA blood samples, we checked for or controlled for the influences of these confounders in our case-control analyses in blood. This information was not available for the 3 postmortem brain sample collections. However, top MDD associations for neurons, glia, and bulk brain contained none of the sites that were associated with antidepressant use (Supplement 1), smoking status, alcohol use, or body mass index detected in the NESDA sample, nor methylation sites in the AHRR gene previously associated with smoking (79). Therefore, these variables were unlikely to be major confounders in our analyses.
Patients with MDD are at increased risk for suicide. Although suicide has been associated with changes in methylation in the brain (80,81), it is unclear whether suicide was a confounder in our analyses, as these changes may have preexisted death. The nonexperimental nature of research with human subjects makes disentangling these effects difficult. However, observing overlap with GWASs of MDD improves causal inferences since genotype antecedes disease, as does observing some overlap with results from blood of living patients. Ultimately, follow-up experiments using in vitro epigenetic editing (82,83) in appropriate cellular or animal models to study causality and translational potential (84) are a necessary future goal.
In conclusion, our cell type–specific MWASs revealed associations otherwise obscured in bulk brain and whole blood, and they provided mechanistic insights into underlying disease processes. Collectively, results pointed toward stress-related neuroinflammation, potentially via p75NTR/NGF and innate immune TLR signaling, as key components of MDD. Critically, these results appear to corroborate and provide links between the neurotrophic (85) and neuroimmune hypotheses of depression (86). Whereas peripheral-neuroimmune interactions are known to influence behavior in animal models, our findings involved actual associations in human patients that implicated a synergistic set of biological pathways plausibly linked to MDD pathology. As both these methylation sites and the biological processes they affect are modifiable, our findings represent promising novel targets for improving MDD treatment.
Supplementary Material
ACKNOWLEDGMENTS AND DISCLOSURES
This project was supported by the National Institute of Mental Health (Grant No. R01MH099110 [to EJCGvdO]). Postmortem brain tissues were received from the Victorian Brain Bank, which is supported by the Florey Institute of Neuroscience and Mental Health and the Alfred and Victorian Forensic Institute of Medicine and funded by Australia’s National Health and Medical Research Council and Parkinson’s Victoria; the Stanley Medical Research Institute; the Netherlands Brain Bank, Netherlands Institute of Neuroscience, Amsterdam; the Harvard Brain Tissue Resource Center; and the Douglas-Bell Canada Brain Bank, Douglas Institute Research Center, Canada. The infrastructure for the NESDA study is funded through the Geestkracht program of the Netherlands Organisation for Health Research and Development (ZonMw) (Grant No. 10-000-1002) and via financial contributions from participating universities and mental health care organizations (VU University Medical Center, GGZ inGeest, Leiden University Medical Center, Leiden University, GGZ Rivierduinen, University Medical Center Groningen, University of Groningen, Lentis, GGZ Friesland, GGZ Drenthe, and Rob Giel Onderzoekcentrum).
RFC, KAA, and EJCGvdO conceived the concept of the study and established the design. KAA and EJCGvdO executed supervision of the study. RFC, JG, MZ, LYX, and ZAK generated the methylation data. RFC, AAS, JG, ZAK, KAA, and EJCGvdO analyzed the data. GvG, GT, BD, and BWJHP provided expertise on biomaterial and phenotype information. RFC and EJCGvdO wrote the manuscript. All authors contributed important intellectual content to and critically reviewed the manuscript.
This article was published as a preprint on bioRxiv: doi: https://doi.org/10.1101/432088.
BP has received research funding (nonrelated) from Jansen Research and Boehringer Ingelheim. All other authors report no biomedical financial interests or potential conflicts of interest.
Complete test statistics for each MWAS are available for download at http://www.people.vcu.edu/~ejvandenoord/. Raw data are available on request to ejvandenoord@vcu.edu.
Footnotes
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.biopsych.2019.10.014.
REFERENCES
- 1.Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR, et al. (2003): The epidemiology of major depressive disorder: Results from the National Comorbidity Survey Replication (NCS-R). JAMA 289:3095–3105. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization (2017): Depression and Other Common Mental Disorders: Global Health Estimates. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 3.Kaffman A, Meaney MJ (2007): Neurodevelopmental sequelae of postnatal maternal care in rodents: Clinical and research implications of molecular insights. J Child Psychol Psychiatry 48:224–244. [DOI] [PubMed] [Google Scholar]
- 4.Szyf M, Weaver IC, Champagne FA, Diorio J, Meaney MJ (2005): Maternal programming of steroid receptor expression and phenotype through DNA methylation in the rat. Front Neuroendocrinol 26:139–162. [DOI] [PubMed] [Google Scholar]
- 5.Abdolmaleky HM, Smith CL, Faraone SV, Shafa R, Stone W, Glatt SJ, et al. (2004): Methylomics in psychiatry: Modulation of gene-environment interactions may be through DNA methylation. Am J Med Genet B Neuropsychiatr Genet 127B:51–59. [DOI] [PubMed] [Google Scholar]
- 6.Iwata M, Ota KT, Duman RS (2013): The inflammasome: Pathways linking psychological stress, depression, and systemic illnesses. Brain Behav Immun 31:105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sotelo JL, Nemeroff CB (2017): Depression as a systemic disease. Pers Med Psychiatry 1–2:11–25. [Google Scholar]
- 8.Shen-Orr SS, Gaujoux R (2013): Computational deconvolution: Extracting cell type-specific information from heterogeneous samples. Curr Opin Immunol 25:571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. (2012): DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahmani E, Zaitlen N, Baran Y, Eng C, Hu D, Galanter J, et al. (2016): Sparse PCA corrects for cell type heterogeneity in epigenome-wide association studies. Nat Methods 13:443–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onuchic V, Hartmaier RJ, Boone DN, Samuels ML, Patel RY, White WM, et al. (2016): Epigenomic deconvolution of breast tumors reveals metabolic coupling between constituent cell types. Cell Rep 17:2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM, et al. (2010): Cell type-specific gene expression differences in complex tissues. Nat Methods 7:287–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aberg KA, Dean B, Shabalin AA, Chan RF, Han LKM, Zhao M, et al. (2018): Methylome-wide association findings for major depressive disorder overlap in blood and brain and replicate in independent brain samples [published online ahead of print Sep 21]. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guintivano J, Aryee MJ, Kaminsky ZA (2013): A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics 8:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penninx B, Beekman A, Smit J (2008): The Netherlands Study of Depression and Anxiety (NESDA): Rationales, Objectives and Methods. Int J Methods Psychiatr Res 17:121–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venet D, Pecasse F, Maenhaut C, Bersini H (2001): Separation of samples into their constituents using gene expression data. Bioinformatics 17(suppl 1):S279–S287. [DOI] [PubMed] [Google Scholar]
- 17.Zheng SC, Breeze CE, Beck S, Teschendorff AE (2018): Identification of differentially methylated cell types in epigenome-wide association studies. Nat Methods 15:1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montano CM, Irizarry RA, Kaufmann WE, Talbot K, Gur RE, Feinberg AP, et al. (2013): Measuring cell-type specific differential methylation in human brain tissue. Genome Biol 14:R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koestler DC, Christensen B, Karagas MR, Marsit CJ, Langevin SM, Kelsey KT, et al. (2013): Blood-based profiles of DNA methylation predict the underlying distribution of cell types: A validation analysis. Epigenetics 8:816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Oord EJCG (2008): Controlling false discoveries in genetic studies. Am J Med Genet B Neuropsychiatr Genet 147B:637–644. [DOI] [PubMed] [Google Scholar]
- 21.van den Oord EJ, Sullivan PF (2003): False discoveries and models for gene discovery. Trends Genet 19:537–542. [DOI] [PubMed] [Google Scholar]
- 22.Spalding KL, Bergmann O, Alkass K, Bernard S, Salehpour M, Huttner HB, et al. (2013): Dynamics of hippocampal neurogenesis in adult humans. Cell 153:1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hattab MW, Shabalin AA, Clark SL, Zhao M, Kumar G, Chan RF, et al.(2017): Correcting for cell-type effects in DNA methylation studies: Reference-based method outperforms latent variable approaches in empirical studies. Genome Biol 18:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan RF, Shabalin AA, Xie LY, Adkins DE, Zhao M, Turecki G, et al. (2017): Enrichment methods provide a feasible approach to comprehensive and adequately powered investigations of the brain methylome. Nucleic Acids Res 45:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aberg KA, Xie L, Chan RF, Zhao M, Pandey AK, Kumar G, et al. (2015): Evaluation of methyl-binding domain based enrichment approaches revisited. PLoS One 10:e0132205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aberg KA, Chan RF, Shabalin AA, Zhao M, Turecki G, Staunstrup NH, et al. (2017): A MBD-seq protocol for large-scale methylome-wide studies with (very) low amounts of DNA. Epigenetics 12:743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehne B, Drong AW, Loh M, Zhang W, Scott WR, Tan S-T, et al. (2015): A coherent approach for analysis of the Illumina Human-Methylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol 16:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stouffer S, DeVinney L, Suchmen E (1949): The American soldier: Adjustment during army life. Princeton, New Jersey: Princeton University Press. [Google Scholar]
- 29.Cabrera CP, Navarro P, Huffman JE, Wright AF, Hayward C, Campbell H, et al. (2012): Uncovering networks from genome-wide association studies via circular genomic permutation. G3 (Bethesda) 2:1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, et al. (2016): The Reactome Pathway Knowledgebase. Nucleic Acids Res 44:D481–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. (2015): Integrative analysis of 111 reference human epigenomes. Nature 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hannon E, Spiers H, Viana J, Pidsley R, Burrage J, Murphy TM, et al. (2015): Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat Neurosci 19:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hannon E, Gorrie-Stone TJ, Smart MC, Burrage J, Hughes A, Bao Y, et al. (2018): Leveraging DNA-methylation quantitative-trait loci to characterize the relationship between methylomic variation, gene expression, and complex traits. Am J Hum Genet 103:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Bartheld CS, Bahney J, Herculano-Houzel S (2016): The search for true numbers of neurons and glial cells in the human brain: A review of 150 years of cell counting. J Comp Neurol 524:3865–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacko M, Weyn-Vanhentenryck SM, Smerdon JW, Yan R, Feng H, Williams DJ, et al. (2018): Rbfox splicing factors promote neuronal maturation and axon initial segment assembly. Neuron 97:853–868.e856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. (2018): Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 50:668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crawford B, Craig Z, Mansell G, White I, Smith A, Spaull S, et al. (2018): DNA methylation and inflammation marker profiles associated with a history of depression. Hum Mol Genet 27:2840–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morice-Picard F, Benard G, Rezvani HR, Lasseaux E, Simon D, Moutton S, et al. (2016): Complete loss of function of the ubiquitin ligase HERC2 causes a severe neurodevelopmental phenotype. Eur J Hum Genet 25:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bekker-Jensen S, Danielsen JR, Fugger K, Gromova I, Nerstedt A, Lukas C, et al. (2009): HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol 12:80–86. [DOI] [PubMed] [Google Scholar]
- 40.Poulsen SL, Hansen RK, Wagner SA, van Cuijk L, van Belle GJ, Streicher W, et al. (2013): RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J Cell Biol 201:797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagano Y, Mavrakis KJ, Lee KL, Fujii T, Koinuma D, Sase H, et al. (2007): Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-β signaling. J Biol Chem 282:20492– 20501. [DOI] [PubMed] [Google Scholar]
- 42.Schreiber J, Végh MJ, Dawitz J, Kroon T, Loos M, Labonté D, et al. (2015): Ubiquitin ligase TRIM3 controls hippocampal plasticity and learning by regulating synaptic γ-actin levels. J Cell Biol 211:569–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hung AY, Sung CC, Brito IL, Sheng M (2010): Degradation of post-synaptic scaffold GKAP and regulation of dendritic spine morphology by the TRIM3 ubiquitin ligase in rat hippocampal neurons. PLoS One 5: e9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu W-H, Pendergast JS, Mo X-M, Brambilla R, Bracchi-Ricard V, Li F, et al. (2005): NIBP, a novel NIK and IKKβ-binding protein that enhances NF-κB activation. J Biol Chem 280:29233–29241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mochida GH, Mahajnah M, Hill AD, Basel-Vanagaite L, Gleason D, Hill RS, et al. (2009): A truncating mutation of trappc9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am J Hum Genet 85:897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marangi G, Leuzzi V, Manti F, Lattante S, Orteschi D, Pecile V, et al.(2013): TRAPPC9-related autosomal recessive intellectual disability: Report of a new mutation and clinical phenotype. Eur J Hum Genet 21:229–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adkins DE, Åberg K, McClay JL, Hettema JM, Kornstein SG, Bukszár J, et al. (2010): A genomewide association study of citalopram response in major depressive disorder—A psychometric approach. Biol Psychiatry 68:e25–e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miao R, Huang S, Zhou Z, Quinn T, Van Treeck B, Nayyar T, et al.(2013): Targeted disruption of MCPIP1/Zc3h12a results in fatal inflammatory disease. Immunol Cell Biol 91:368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schiffer HH, Heinemann SF (2007): Association of the human kainite receptor GluR7 gene (GRIK3) with recurrent major depressive disorder. Am J Med Genet B Neuropsychiatr Genet 144B:20–26. [DOI] [PubMed] [Google Scholar]
- 50.Garcia-Rizo C, Fernandez-Egea E, Miller BJ, Oliveira C, Justicia A, Griffith JK, et al. (2013): Abnormal glucose tolerance, white blood cell count, and telomere length in newly diagnosed, antidepressant-naïve patients with depression. Brain Behav Immun 28:49–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhoeven JE, Révész D, Epel ES, Lin J, Wolkowitz OM, Penninx BWJH (2014): Major depressive disorder and accelerated cellular aging: Results from a large psychiatric cohort study. Mol Psychiatry 19:895–901. [DOI] [PubMed] [Google Scholar]
- 52.Mamdani F, Rollins B, Morgan L, Myers RM, Barchas JD, Schatzberg AF, et al. (2015): Variable telomere length across postmortem human brain regions and specific reduction in the hippocampus of major depressive disorder. Transl Psychiatry 5:e636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han LKM, Aghajani M, Clark SL, Chan RF, Hattab MW, Shabalin AA, et al. (2018): Epigenetic aging in major depressive disorder. Am J Psychiatry 175:774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Demir S, Atli A, Bulut M, İbiloğlu AO, Güneş M, Kaya MC, et al. (2015): Neutrophil–lymphocyte ratio in patients with major depressive disorder undergoing no pharmacological therapy. Neuropsychiatr Dis Treat 11:2253–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weber MD, Godbout JP, Sheridan JF (2017): Repeated social defeat, neuroinflammation, and behavior: Monocytes carry the signal. Neuropsychopharmacology 42:46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Story Jovanova O, Nedeljkovic I, Spieler D, Walker RM, Liu C, Luciano M, et al. (2018): DNA methylation signatures of depressive symptoms in middle-aged and elderly persons: Meta-analysis of multiethnic epigenome-wide studies. JAMA Psychiatry 75:949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. (2019): Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet 51:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Otowa T, Hek K, Lee M, Byrne EM, Mirza SS, Nivard MG, et al. (2016): Meta-analysis of genome-wide association studies of anxiety disorders. Mol Psychiatry 21:1391–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grove J, Ripke S, Als TD, Mattheisen M, Walters R, Won H, et al. (2019): Identification of common genetic risk variants in autism spectrum disorder. Nat Genet 51:431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruderfer DM, Ripke S, McQuillin A, Boocock J, Stahl EA, Pavlides JMW, et al. (2018): Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell 173:1705–1715, e1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, et al. (2017): The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Michailidou K, Lindström S, Dennis J, Beesley J, Hui S, Kar S, et al. (2017): Association analysis identifies 65 new breast cancer risk loci. Nature 551:92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clark SL, Hattab MW, Chan RF, Shabalin AA, Han LKM, Zhao M, et al. (2019): A methylation study of long-term depression risk [published online ahead of print Sep 9]. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hempstead BL (2002): The many faces of p75NTR. Curr Opin Neurobiol 12:260–267. [DOI] [PubMed] [Google Scholar]
- 65.Aloe L, Bracci-Laudiero L, Alleva E, Lambiase A, Micera A, Tirassa P (1994): Emotional stress induced by parachute jumping enhances blood nerve growth factor levels and the distribution of nerve growth factor receptors in lymphocytes. Proc Natl Acad Sci U S A 91:10440–10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kamezaki Y, Katsuura S, Kuwano Y, Tanahashi T, Rokutan K (2012): Circulating cytokine signatures in healthy medical students exposed to academic examination stress. Psychophysiology 49:991–997. [DOI] [PubMed] [Google Scholar]
- 67.Hadjiconstantinou M, McGuire L, Duchemin A-M, Laskowski B, Kiecolt-Glaser J, Glaser R (2001): Changes in plasma nerve growth factor levels in older adults associated with chronic stress. J Neuroimmunol 116:102–106. [DOI] [PubMed] [Google Scholar]
- 68.Filho CB, Jesse CR, Donato F, Giacomeli R, Del Fabbro L, da Silva Antunes M, et al. (2015): Chronic unpredictable mild stress decreases BDNF and NGF levels and Na+,K+-ATPase activity in the hippocampus and prefrontal cortex of mice: Antidepressant effect of chrysin. Neuroscience 289:367–380. [DOI] [PubMed] [Google Scholar]
- 69.Kucharczyk M, Kurek A, Detka J, Slusarczyk J, Papp M, Tota K, et al. (2016): Chronic mild stress influences nerve growth factor through a matrix metalloproteinase-dependent mechanism. Psychoneuroendocrinology 66:11–21. [DOI] [PubMed] [Google Scholar]
- 70.Medzhitov R (2001): Toll-like receptors and innate immunity. Nat Rev Immunol 1:135–145. [DOI] [PubMed] [Google Scholar]
- 71.Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW (2008): From inflammation to sickness and depression: When the immune system subjugates the brain. Nat Rev Neurosci 9:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y, Woodruff M, Zhang Y, Miao J, Hanley G, Stuart C, et al. (2008): Toll-like receptor 4 mediates chronic restraint stress-induced immune suppression. J Neuroimmunol 194:115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gárate I, Garcia-Bueno B, Madrigal JLM, Caso JR, Alou L, Gomez-Lus ML, et al. (2013): Stress-induced neuroinflammation: Role of the toll-like receptor 4 pathway. Biol Psychiatry 73:32–43. [DOI] [PubMed] [Google Scholar]
- 74.Breen MS, Beliakova-Bethell N, Mujica-Parodi LR, Carlson JM, Ensign WY, Woelk CH, et al. (2016): Acute psychological stress induces short-term variable immune response. Brain Behav Immun 53:172–182. [DOI] [PubMed] [Google Scholar]
- 75.Jack CS, Arbour N, Manusow J, Montgrain V, Blain M, McCrea E, et al. (2005): TLR signaling tailors innate immune responses in human microglia and astrocytes. J Immunol 175:4320–4330. [DOI] [PubMed] [Google Scholar]
- 76.Kannan Y, Bienenstock J, Ohta M, Stanisz AM, Stead RH (1996): Nerve growth factor and cytokines mediate lymphoid tissue-induced neurite outgrowth from mouse superior cervical ganglia in vitro. J Immunol 157:313–320. [PubMed] [Google Scholar]
- 77.Heese K, Hock C, Otten U (1998): Inflammatory signals induce neuro-trophin expression in human microglial cells. J Neurochem 70:699–707. [DOI] [PubMed] [Google Scholar]
- 78.De Simone R, Ambrosini E, Carnevale D, Ajmone-Cat MA, Minghetti L (2007): NGF promotes microglial migration through the activation of its high affinity receptor: Modulation by TGF-β. J Neuroimmunol 190:53–60. [DOI] [PubMed] [Google Scholar]
- 79.Gao X, Jia M, Zhang Y, Breitling LP, Brenner H (2015): DNA methylation changes of whole blood cells in response to active smoking exposure in adults: A systematic review of DNA methylation studies. Clin Epigenetics 7:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Murphy TM, Crawford B, Dempster EL, Hannon E, Burrage J, Turecki G, et al. (2017): Methylomic profiling of cortex samples from completed suicide cases implicates a role for PSORS1C3 in major depression and suicide. Transl Psychiatry 7:e989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nagy C, Suderman M, Yang J, Szyf M, Mechawar N, Ernst C, et al. (2015): Astrocytic abnormalities and global DNA methylation patterns in depression and suicide. Mol Psychiatry 20:320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vojta A, Dobrinić P, Tadić V, Bočkor L, Korać P, Julg B, et al. (2016): Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res 44:5615–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morita S, Noguchi H, Horii T, Nakabayashi K, Kimura M, Okamura K, et al. (2016): Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat Biotechnol 34:1060–1065. [DOI] [PubMed] [Google Scholar]
- 84.Jeffries MA (2018): Epigenetic editing: How cutting-edge targeted epigenetic modification might provide novel avenues for autoimmune disease therapy. Clin Immunol 196:49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Duman RS, Li N (2012): A neurotrophic hypothesis of depression: Role of synaptogenesis in the actions of NMDA receptor antagonists. Philos Trans R Soc Lond B Biol Sci 367:2475–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hodes GE, Kana V, Menard C, Merad M, Russo SJ (2015): Neuroimmune mechanisms of depression. Nat Neurosci 18:1386–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.