Abstract
Objective
Many genetic variants are classified, but many more are variants of uncertain significance (VUS). Clinical observations of patients and their families may provide sufficient evidence to classify VUS. Understanding how long it takes to accumulate sufficient patient data to classify VUS can inform decisions in data sharing, disease management, and functional assay development.
Materials and Methods
Our software models the accumulation of clinical evidence (and excludes all other types of evidence) to measure their unique impact on variant interpretation. We illustrate the time and probability for VUS classification when laboratories share evidence, when they silo evidence, and when they share only variant interpretations.
Results
Using conservative assumptions for frequencies of observed clinical evidence, our models show the probability of classifying rare pathogenic variants with an allele frequency of 1/100 000 increases from less than 25% with no data sharing to nearly 80% after one year when labs share data, with nearly 100% classification after 5 years. Conversely, our models found that extremely rare (1/1 000 000) variants have a low probability of classification using only clinical data.
Discussion
These results quantify the utility of data sharing and demonstrate the importance of alternative lines of evidence for interpreting rare variants. Understanding variant classification circumstances and timelines provides valuable insight for data owners, patients, and service providers. While our modeling parameters are based on our own assumptions of the rate of accumulation of clinical observations, users may download the software and run simulations with updated parameters.
Conclusions
The modeling software is available at https://github.com/BRCAChallenge/classification-timelines.
Keywords: genetic variation, benign, pathogenic, classification, modeling
OBJECTIVE
Genomic testing is now widely used for patients to determine if their genetics put them at increased risk of heritable disorders and to enable them to manage this risk clinically. For example, a patient with a known pathogenic variant in BRCA1 or BRCA2 should be screened more often for breast, ovarian, and pancreatic cancer.1 Similarly, asymptomatic patients with familial cardiomyopathy might consider certain lifestyle changes such as losing weight, reducing stress, and sleeping well.2
The American College of Medical Genetics (ACMG) and the Association for Molecular Pathology (AMP) define qualitative, evidence-based guidelines for classifying genetic variants. Evidence for variant classification can come from many sources including clinical data, functional assays, and in silico predictors. Clinical data, typically derived through genetic testing reports, includes family history, cosegregation, cooccurrence, and de novo status. When sufficient evidence is present, a variant curation expert panel (VCEP) may classify the variant as Likely Benign (LB), Benign (B), Likely Pathogenic (LP), or Pathogenic (P) using the ACMG/AMP rules for combining evidence. Variants with little or no evidence to support classification, called variants of uncertain significance (VUS), create stress for patients and may lead to improper care. Because VUS do not yield medically actionable information, patients with VUS do not benefit from clinical management of their heritable disease risk. Ultimately, the significance of a variant remains uncertain until there is sufficient evidence to classify it. Although computational and functional predictions are helpful, some clinical data linking genotype and phenotype are usually needed to classify most variants.3 However, there is no centrally available repository of clinical data that can be used for variant classification. Molecular testing laboratories and sequencing centers are the largest sources of variant data. Many, but not all, clinical laboratories and sequencing centers actively share variant interpretations through ClinVar; however, they hold most of the clinical data they collect privately, due in large part to patient privacy and regulatory concerns. The shared interpretations for many genetic variants vary or even conflict between laboratories depending on the amount and nature of the evidence provided.4
One solution to these problems is for clinical laboratories to develop approaches to centrally share their clinical data associated with specific variants. Widespread sharing of variant pathogenicity evidence would lead to more rapid variant interpretation, greater scientific reproducibility, and novel discoveries.4,5 Indeed, the National Institutes of Health recently mandated the sharing of all data for the research which it funds.6
While there is no question that data sharing would lead to expedited variant interpretation, and better patient outcomes by extension, under what circumstances is data sharing the most impactful? We have addressed this question by developing open-source software to model the probability of variant classification over time under various forms of data sharing.
Our model not only quantifies the value of sharing clinical patient data, the understanding of likely timelines and mechanisms of classification that this modeling illustrates could guide genetics organizations in prioritizing their efforts, inform strategies for functional assay development, improve variant classification guidelines, and enable healthcare providers to develop better strategies for managing specific patients with VUS. Furthermore, the model serves as a platform for testing hypotheses on factors including the rates of gathering clinical evidence on the variant interpretation timeline. While we have informed the model with data according to the scientific literature and our own clinical experience, these factors are modeling parameters that can be modified easily as new evidence emerges, or to test the impact of clinical assumptions on the variant classification rate.
MATERIALS AND METHODS
This section outlines a statistical model that combines clinical information from multiple sequencing centers to create an aggregate, pooled center so that VUS may be classified faster.
Combining multiple forms of variant classification evidence
The evidence that the ACMG/AMP uses to classify variants encompasses several sources of data, including the type of variant (eg, nonsense or frameshift), in vitro functional studies, in trans cooccurrence with a pathogenic variant, cosegregation in family members, allele frequency, and in silico predictions. They are divided into four levels of strength: “Supporting” (or “Predictive”), “Moderate,” “Strong,” and “Very Strong.” For example, PP1, which represents cosegregation of the disease with multiple family members, is considered “Supporting” evidence for a Pathogenic interpretation. Another form of evidence called BS4 represents the lack of segregation of the disease with the variant in affected family members. The BS4 evidence is considered “Strong” evidence for a Benign interpretation.7 Tavtigian et al8 showed that the rule-based ACMG/AMP guidelines can be modeled as a quantitative Bayesian classification framework. Specifically, the ACMG/AMP classification criteria were translated into a naive Bayes classifier, assuming the four levels of evidence and exponentially scaled odds of pathogenicity. While the ACMG/AMP guidelines define rules for the combinations of evidence which lead to variant classifications, the Bayesian framework assigns points to each form of evidence. These points are summed and compared to thresholds to determine the variant’s pathogenicity. We leverage this Bayesian framework to model calculating odds of pathogenicity conditioned on the presence of one or more pieces of evidence for a given variant. For more detail regarding the combination of evidence, see Supplementary Equations S1–S3.
To model the impact of clinical data sharing on variant classification, we exclude all other forms of evidence besides clinical evidence, as described in the following sections. For each variant, our model calculates two odds of pathogenicity: the odds of a VUS being benign and the odds of a VUS being pathogenic, both of which are conditioned on statistically sampled evidence.
Selecting categories of variant evidence for model
Some sources of variant classification evidence are not impacted by data sharing, such as in silico prediction scores and functional assay scores. We will not use those categories of evidence in our model so we can specifically quantify the unique contribution of cumulative clinical data to variant interpretation.
Several sources of clinical case and family information will contribute to variant classification over time. As clinical databases grow and data are shared more effectively across institutions, more variants will be classified. Increased clinical information is the major source for variant reclassification as well.9 We selected the following categories of clinical pathogenic evidence for our model:
de novo variants without paternity and maternity confirmation (PM6);
cosegregation in family members affected with the disease (PP1); and
de novo variants with both paternity and maternity confirmed (PS2).
Similarly, we selected the following categories of benign evidence criteria that relate to clinical information:
in trans cooccurrence with a known pathogenic variant (BP2);
disease with an alternate molecular basis (BP5); and
lack of segregation in affected family members (BS4).s
The more evidence that is gathered over time, the sooner and more likely a VUS will be classified. However, not all the evidence that is gathered over this time will be concordant.10 Patients who have a pathogenic variant may occasionally present evidence from one or more benign categories, for example, lack of segregation in affected family members due to disease heterogeneity. This presentation of conflicting evidence for a given variant occurs at a low, nonzero frequency. Therefore, we use a combination of pathogenic and benign evidence in the classification of every VUS.
Parameters affecting clinical observations
To model the accumulation of clinical evidence, we defined certain modeling parameters according to the literature and to our own clinical experience. While the values that we have assigned to these parameters constitute well-informed assumptions, these values can be modified to test hypotheses, or as new knowledge emerges over time. In our software, these parameters are encapsulated in a single JSON file, so rerunning the model with revised parameter values requires modifying only one file.
Frequency distribution for evidence
Tavtigian et al calculated the corresponding odds of pathogenicity for each category of evidence and showed that the numerical-based odds are consistent with the rule-based ACMG/AMP guidelines for combining evidence. Those odds are shown in the “Pathogenicity odds of evidence” column of Table 1. Specifically, they determined that, for pathogenic evidence, the odds for “Strong” evidence is 18.7, for “Moderate” is 4.3, and for “Supporting” is 2.08. For benign evidence, the odds for “Strong” evidence is 1/18.7, for “Moderate” it is 1/4.3, and for “Supporting” it is 1/18.7. We derived the estimates for the evidence frequencies from the literature.11–14
Table 1.
Odds and frequency estimate confidence intervals per ACMG/AMP clinical data evidence category
| Evidence category | Estimated benign evidence frequency (low, medium, high) | Estimated pathogenic evidence frequency (low, medium, high) | Pathogenicity odds of evidence |
|---|---|---|---|
| PS2 | (0.0001, 0.0015, 0.005) | (0.0006, 0.003, 0.02) | 18.7 |
| PM6 | (0.0007, 0.0035, 0.01) | (0.0014, 0.007, 0.025) | 4.3 |
| PP1 | (0.005, 0.01, 0.0625) | (0.05, 0.23, 0.67) | 2.08 |
| BS4 | (0.025, 0.1, 0.4863) | (0.0001, 0.001, 0.17) | 1/18.7 |
| BP5 | (0.038, 0.099, 0.36) | (0.00002, 0.0001, 0.00215) | 1/2.08 |
| BP2 | (1.0 * f, 1.0 * f, 1.0 * f) | (0.001 * f, 0.005 * f, 0.02 * f) | 1/2.08 |
Note: The variable f represents the frequency of the variant itself.
Abbreviations: ACMG: American College of Medical Genetics; AMP: Association for Molecular Pathology.
Table 1 depicts the odds and estimated frequency confidence intervals for the ACMG/AMP evidence categories that correspond only to clinical evidence. There may be pathogenic evidence observed for benign variants and benign evidence observed for pathogenic variants, though such observations generally occur at a low rate. For example, the frequency of BP2 for pathogenic variants is very unusual, except in tumors or in the case of rare diseases such as Fanconi anemia. Conversely, we assume that the frequency of BP2 evidence for benign variants is quite common and so occurs at the same rate (f) as the variant itself.
Thresholds for odds of pathogenicity
Tavtigian et al defined four threshold ranges for the odds of pathogenicity for each of the four ACMG/AMP variant classifications (B, LB, LP, and P), as shown in Table 2.
Table 2.
Odds of pathogenicity per ACMG/AMP classification
| Classification | Threshold for odds of pathogenicity |
|---|---|
| Benign | (−∞, 0.001) |
| Likely Benign | [0.001, 1/18.07) |
| Uncertain significance | [1/18.07, 18.07] |
| Likely Pathogenic | (18.07, 100] |
| Pathogenic | (100, +∞) |
Abbreviations: ACMG: American College of Medical Genetics; AMP: Association for Molecular Pathology.
These thresholds correspond to the values from Table 1 and are consistent with the ACMP/AMP rules for combining evidence. For example, having one piece of strong evidence (eg, BS4) and one piece of supporting evidence (eg, BP2) is sufficient to classify a variant as “LB.”
Data from participating sequencing centers
For generating simulated clinical data, we define three categories of sequencing centers: small, medium, and large as shown in Table 3.
Table 3.
Current number of tests in database and testing rate per center type
| Center type | Current number of patients tested | Tests per year |
|---|---|---|
| Small | 15 000 | 3000 |
| Medium | 150 000 | 30 000 |
| Large | 1 000 000 | 450 000 |
We estimated the large database size and testing rate from the online publications of relatively large sequencing labs,15,16 and we estimated the small database size and testing rate based on our own experience at the University of Washington Department of Laboratory Medicine (a relatively small laboratory). We estimated the medium database size and testing rate by interpolating between the large and small database values. Our model permits that these estimated sizes be replaced with other hypothetical or real sizes to predict outcomes under different scenarios. In relatively rare circumstances, the same patient may be tested at multiple facilities. In our model, we are assuming that each center has entirely distinct patient populations.
Ascertainment bias
Healthy people from healthy families are underrepresented in many forms of genetic testing.17 Accordingly, patients with pathogenic variants are observed (or ascertained) more often than those with benign variants, and the forms of evidence that support a pathogenic interpretation accumulate more quickly. How much more likely a person is to present pathogenic evidence than benign evidence is captured in our model as a configurable real-valued constant. We conservatively estimated this term to be 2 based on our experience at the University of Washington Department of Laboratory Medicine.
Prior odds of pathogenicity
The Bayesian prior odds of a variant’s pathogenicity represents all other criteria that are not clinical and do not change much, if at all, over time. For this implementation, we sampled a random value from a uniform distribution between 1/18.07 and 18.07 which is the lower and upper bound of the odds of pathogenicity for VUS.
Paradigms for sharing
There are three sharing paradigms, which we use in our model: sharing nothing, sharing classifications, and sharing evidence. Of the three, we anticipate that sharing nothing will make variant classification most protracted and least probable. The paradigm of sharing classifications is what ClinVar currently enables, and we anticipate that sharing classifications will lead to shorter timelines with higher probabilities of variant classification than sharing nothing. Last, the essence of this research is to model the impact of sharing clinical evidence on the timeline of variant interpretation. We anticipate that sharing clinical data will lead to the shortest timelines with the highest probabilities of classification.
Implementing the simulation
Our statistical model contains one variable: the allele frequency of the VUS of interest. Parameters of the simulation software include the number and types of each of the participating sequencing centers and the number of years for which to run the simulation. Because the variant is of uncertain significance, we gather evidence for both benign and pathogenic classifications simultaneously.
For the first year of our simulation, all the evidence that is assumed to be currently present at each of the individual testing centers is initialized and aggregated. We use the Poisson distribution sampling method when determining how many times the variant is observed, given the VUS frequency. For each year in the simulation, we generate new observations for variants assumed to be benign and assumed to be pathogenic at each sequencing center. We aggregate those observations across participating centers into a single collection to simulate the sharing of data.
We ran simulations as described above 1000 times to simultaneously generate data points for VUS which occur at the rate of one in every 100 000 people (1e−05), combining data from 10 small centers, 7 medium centers, and 3 large centers generated over 5 years. We then ran simulations for a VUS of frequency 1e−06 (one in every 1 000 000 people) in the same grouping of centers.
We created histograms and scatter plots that show the distribution and progression of the evidence over time. For each year, we plot the probability that each center classifies the variants individually using siloed data or if they collectively pool their data. We calculated the probability of a variant being classified at any sequencing center using the inclusion-exclusion principle in probability18 assuming all centers would share all variant interpretations. This is a conservative estimate: not all sequencing centers share all their variant interpretations. We performed a sensitivity analysis to show the impact that each of the evidence types has on the probability of being either benign or pathogenic.
RESULTS
In this section, we discuss the results of our simulation with variants over the course of 5 years at 20 participating sequencing centers. We first examine the histograms of the evidence for pathogenic and benign variants after 5 years of observations. Second, we examine the trajectory of evidence over the course of 5 years in scatter plots. Third, we examine the probability scatter plots over the course of 5 years. Fourth, we analyze the sensitivity of our results with respect to each type of evidence. These four sets of results were generated using a variant of 1e−05 frequency. Last, we examine the probability scatter plots over the course of 5 years for a 1e−06 (one-in-a-million) variant.
Histogram plots of variants occurring at 1e−05 frequency
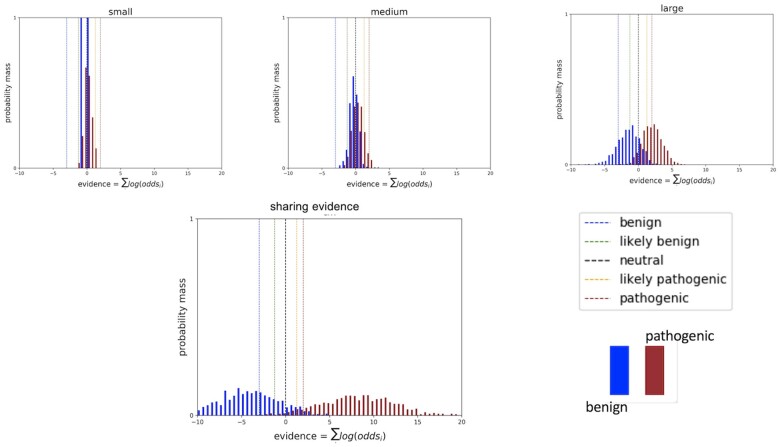
The distribution of evidence gathered individually and combined across all sequencing centers is plotted in Figure 1. As expected, increasing the number of classification data points for the many different variants results in wider Gaussian distributions that increasingly separate from the null assumption of no clinical evidence. More evidence provides more certainty in classifications as evidence exceeds the classification thresholds for an increasing number of variants.
Figure 1.
Histograms of cumulative log odds for classifying each of 1000 simulated variants present at a 1e−05 frequency in the population. Classification thresholds are demarcated as vertical hash lines.
Trajectories of evidence for variants at 1e−05 frequency
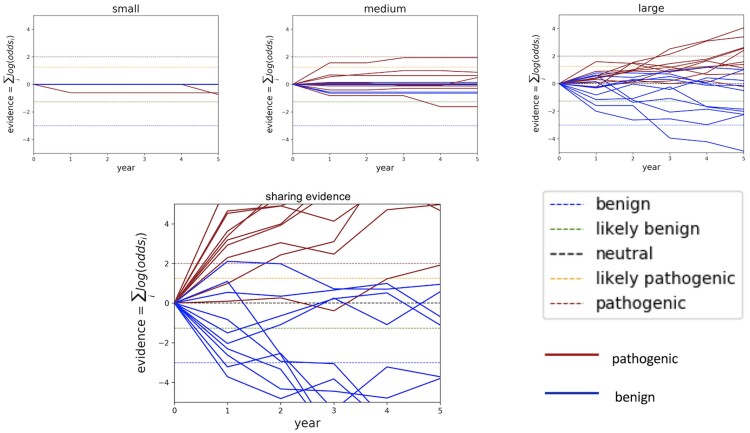
The classification trajectory for individual variants can vary depending on which observations are made and when those are made. Although data accumulation increases the likelihood of classification and the likelihood of correct classification for variants as a group, evidence for individual variants may rise and fall. Figure 2 plots a subset of 20 classification trajectories (10 benign and 10 pathogenic) at a small, medium, and large sequencing center as compared to the combined data across all sequencing centers assumed to be sharing evidence. Trajectories in these scatter plots mimic real-world phenomena: variants may accumulate contradictory evidence; and long time periods may pass with insufficient evidence.
Figure 2.
Classification trajectories for 20 randomly selected variants at 1e−05 frequency in the population. Classification thresholds are demarcated as horizontal hash lines in the timeline plots.
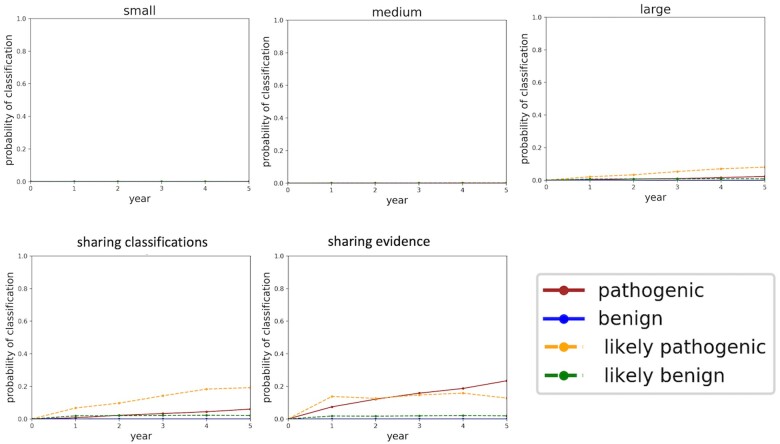
Probabilities of classifying variants at 1e−05 frequency
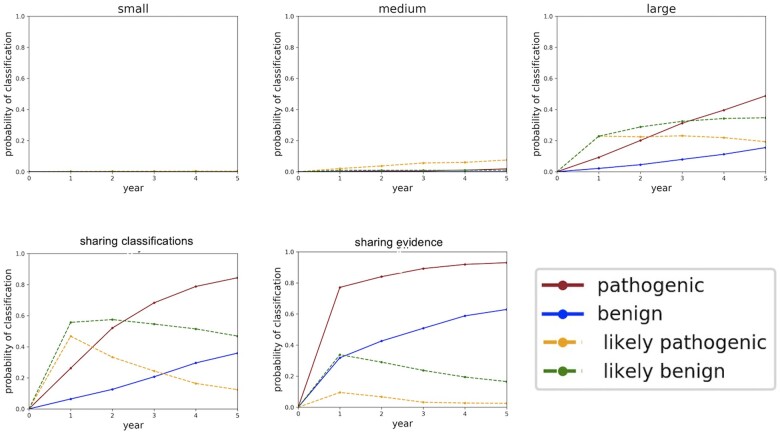
Figure 3 shows the probability of classifying a variant which occurs at 1e−05 frequency in the population over the course of 5 years under different sharing paradigms. We show a small, medium, and large sequencing center not sharing anything as compared to two forms of sharing: centers sharing their all their variant interpretations but none of their clinical data (labeled “sharing classifications”); and centers sharing all their clinical data (labeled “sharing evidence”). From these graphs, we see that any data sharing increases the likelihood of variant classification. We also see that sharing evidence rather than sharing classifications makes variant interpretation more certain by moving “LB” variants into the “B” classification and similarly moving “LP” variants into the “P” classification. Moreover, sharing evidence rather than sharing classifications reduces the amount of time required to classify variants.
Figure 3.
Classification probabilities over the course of 5 years. The y-axis of these plots is the probability of classifying the variant, converted from the aggregated likelihoods of pathogenicity generated in the simulations. Year 0 constitutes the time just before the sequencing centers share their data and all the variants are unclassified. Year 1 constitutes the moment just after the sequencing centers share their data. As time progresses and more evidence becomes available, some of the variants which were LB get “promoted” to B, and similarly some of the variants which were LP get “promoted” to P. B: Benign; LB: Likely Benign; LP: Likely Pathogenic; P: Pathogenic.
In the Supplementary Material, we explore changing the distribution of sequencing centers and the number of years sharing data. In Supplementary Figure S1, we see that after 20 years of data sharing, almost all benign and pathogenic variants are classified using clinical data alone. In Supplementary Figure S7, we see that reducing the number of participating sequencing centers from 10, 7, and 3 (small, medium, and large) to 5, 3, and 1 significantly reduces the probability of classifying variants using clinical data alone.
Sensitivity analysis for variants at 1e−05 frequency
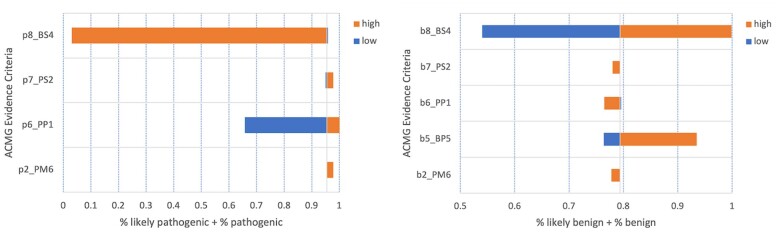
We estimated conservative confidence intervals around the evidence observation frequencies defined in our model to determine how sensitive the probabilities of classification were to each type of ACMG/AMP evidence. We held all other parameters constant (equal to their expected values) while changing one frequency at a time to the low and high value in their respective interval to determine how sensitive the model is to changes in the frequencies observing different types of clinical data. Based on the assumptions of our experiments, classification of pathogenic variants is most sensitive to BS4 and PP1 evidence criteria (Figure 4A). Classification of benign variants is most sensitive to BS4 and BP5 evidence criteria (Figure 4B). Classifications were not affected by the change in BP2 evidence frequencies for either B or P variants. P variant classification was not affected by changes in BP5 evidence frequency and was therefore dropped from Figure 4A.
Figure 4.
Sensitivity of variant classification to the frequency of observing ACMG/AMP evidence criteria. These “high” and “low” values are taken from the confidence intervals in Table 1. (A) Tornado plot for the sensitivity of Pathogenic and Likely Pathogenic variants. (B) Tornado plot for the sensitivity of Benign and Likely Benign variants. ACMG: American College of Medical Genetics; AMP: Association for Molecular Pathology.
Probabilities of classifying variants at 1e−06 frequency
For comparison, we evaluated the probability of gathering data for a one-in-a-million variant through data sharing. Figure 5 shows the probability of classifying a 1e−06 variant over the course of 5 years.
Figure 5.
Probabilities of classifying variants at 1e−06 frequency plotted over the course of 5 years. The y-axis of these plots is the probability of classifying the variant, converted from the aggregated likelihoods of pathogenicity generated in the simulations. Year 0 constitutes the time just before the sequencing centers share their data and all the variants are unclassified. Year 1 constitutes the moment just after the sequencing centers share their data.
In addition to these probability plots, we also performed analysis of 1e−06 variants to generate cumulative odds histograms (Supplementary Figure S2) and classification trajectories (Supplementary Figure S3). These illustrate similar results. To further explore variant classification timelines for 1e−06 variants, we evaluated classification over 20 years of data sharing (Supplementary Figures S4–S6). Sharing evidence is predicted to help classify a minority of 1e−06 variants even after 20 years of data sharing.
DISCUSSION
These simulations illustrate that clinical data sharing reduces the time and increases the certainty in classifying VUS. Sharing only variant interpretations rather than clinical data, however, results in longer timelines and lower certainty. For example, the same variant could be interpreted as LP at one laboratory and as a VUS at a different laboratory based on evidence seen at the two respective laboratories. Similarly, the simulations show that evidence for a given variant can, at times, be contradictory. As defined in the ACMG/AMP classification standards, evidence of pathogenicity may be presented for benign variants (and vice versa), though less frequently than for pathogenic variants. Importantly, our simulations demonstrate that discordant evidence resolves more quickly and with higher certainty when centers share their clinical data rather than only sharing their variant interpretations. These are critical results: mis-classified variants mis-inform healthcare providers and may lead to disastrous patient outcomes.19 Variants originally classified as LP or LB more readily become classified as P and B, respectively, when data are shared.
Our simulations show that, using clinical evidence alone, classifying pathogenic variants has a higher probability and quicker timeline than for classifying benign. Those ACMG/AMP evidence criteria and classification guidelines that rely on patient clinical data, which we have modeled, require more evidence for benign classification7 which results in longer timelines. Models indicate that improved guidelines could balance pathogenic or benign evidence categories or alternatively create a new “lack of pathogenic evidence despite sufficient observations” category of benign evidence.
In addition, our model can guide functional assay developers as to which variants they should include in their panels. Very rare variants for which we expect insufficient clinical data under any sharing model will need a functional assay to classify it. Functional assays are expensive and require expert interpretation, and this information can maximize the impact of those efforts by identifying variant frequencies and sharing scenarios in which data sharing by itself is insufficient for classification.
We see that highly rare variants (one-in-a-million or less) may be unlikely to be classified by aggregating clinical information alone. Because most variants are highly rare,20 it is essential that we invest in strategies for the interpretation of highly rare VUS. One strategy is additional investment in cascade testing for highly rare variants in high-penetrance genes. This is an effective strategy because the variant may be rare in the general population but can still be enriched in the family.9 Moreover, cascade testing is part of the PP1 and BS4 classification categories, and Table 4 indicates that both benign and variant classifications are quite sensitive to those categories. Another effective strategy is investment in large-scale functional assays, such as Multiplexed Assays of Variant Effect, which can assay thousands of variants at once.21
Most importantly, variant classification timelines will guide prevention, diagnosis, and treatment decisions for patients and their healthcare teams. For example, a patient with a known pathogenic variant in BRCA1 or BRCA2 may elect to have a prophylactic mastectomy which, according to the National Cancer Institute, reduces the risk of breast cancer in women who carry a pathogenic BRCA1 variant by 95%.22 A patient with a BRCA1 VUS, on the other hand, may choose to wait if their variant is likely to be classified in the near-term (eg, within 2 years) but seek alternative options, such as family cosegregation analysis, if that variant will not likely get classified for another 10 years or more. More than half of the variants in the BRCA1 and BRCA2 genes are VUS, even though these are two of the most widely studied genes in the human genome. Other Mendelian diseases with highly penetrant alleles have a significantly larger proportion of VUS, so understanding timelines and probabilities of variant classification will have an even higher impact on those genes.
With sufficient clinical data from cooperating sequencing laboratories, these estimates enumerate tangible outcomes that may result from data sharing. It is clear that more variants will be classified and patients will benefit from robust data sharing. This is particularly important over longer time horizons (see the Supplementary Material for 20-year modeling). There are several mature privacy mechanisms that may be leveraged to share data responsibly; differential privacy, secure multi-party computation, homomorphic encryption, blockchain, and federated computing are approaches that have matured and are available today to protect the privacy of those individuals who have shared their data as well as protect the business interests of the institutions which own the data.23–26
CONCLUSION
It is assumed that sharing clinical patient data should improve variant interpretation using the ACMG/AMP variant classification guidelines. Our research provides a framework to explicitly quantify how much and under what circumstances it improves. We have built and made available a model that simulates the generation and sharing of clinical evidence over time. The software provides graphical results to compare sharing clinical data with sharing only interpretations and sharing nothing. Our experiments were based on data estimates from the literature and from our own experience, but readers can define their own values for the frequencies of observations of various ACMG/AMP evidence criteria and experiment with different combinations of centers, different sizes and testing rates, and with different allele frequencies.
Supplementary Material
ACKNOWLEDGMENTS
The authors are very grateful to David Goldgar and Sean Tavtigian for providing their expert insight in the approximation of ACMG/AMP evidence criteria observation frequencies.
CONFLICT OF INTEREST STATEMENT
None declared.
Contributor Information
James Casaletto, Genomics Institute, University of California, Santa Cruz, Santa Cruz, California, USA.
Melissa Cline, Genomics Institute, University of California, Santa Cruz, Santa Cruz, California, USA.
Brian Shirts, Department of Laboratory Medicine and Pathology, University of Washington, Seattle, Washington, USA.
FUNDING
JC is supported by NHGRI grant U54HG007990 and NHLBI grant U01HL137183. MC is supported by NCI grant U01CA242954 and BioData Catalyst fellowship OT3 HL147154 from the NHLBI through UNC-CH 5118777. BS is supported by a grant from the Brotman Baty Institute for Precision Medicine and by the NIH 1OT2OD002748.
AUTHOR CONTRIBUTIONS
BS conceived the initial statistical model and wrote a draft in R. JC developed the model software in Python. MC conceived the “sharing classifications” sharing paradigm. JC wrote the manuscript. JC, MC, and BS reviewed, revised, and approved the final version of the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
DATA AVAILABILITY
There are no external data associated with this manuscript. All the data are generated synthetically in the modeling software, which is available at https://github.com/BRCAChallenge/classification-timelines.
REFERENCES
- 1. Couch FJ, Nathanson KL, Offit K.. Two decades after BRCA: setting paradigms in personalized cancer care and prevention. Science 2014; 343 (6178): 1466–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wexler RK, Elton T, Pleister A, Feldman D.. Cardiomyopathy: an overview. Am Fam Physician 2009; 79 (9): 778–84. [PMC free article] [PubMed] [Google Scholar]
- 3. Berg JS. Exploring the importance of case-level clinical information for variant interpretation. Genet Med 2017; 19 (1): 3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harrison SM, Dolinsky JS, Knight Johnson AE, et al. Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genet Med 2017; 19 (10): 1096–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cline MS, Liao RG, Parsons MT. et al. ; BRCA Challenge Authors. BRCA Challenge: BRCA Exchange as a global resource for variants in BRCA1 and BRCA2. PLoS Genet 2018; 14 (12): e1007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kozlov M. NIH issues a seismic mandate: share data publicly. Nature 2022; 602 (7898): 558–9. [DOI] [PubMed] [Google Scholar]
- 7. Richards S, Aziz N, Bale S. et al. ; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17 (5): 405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tavtigian SV, Greenblatt MS, Harrison SM. et al. ; ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI). Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet Med 2018; 20 (9): 1054–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsai GJ, Rañola JMO, Smith C, et al. Outcomes of 92 patient-driven family studies for reclassification of variants of uncertain significance. Genet Med 2019; 21 (6): 1435–42. [DOI] [PubMed] [Google Scholar]
- 10. Balmaña J, Digiovanni L, Gaddam P, et al. Conflicting interpretation of genetic variants and cancer risk by commercial laboratories as assessed by the prospective registry of multiplex testing. J Clin Oncol 2016; 34 (34): 4071–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hampel H, Pearlman R, Beightol M. et al. ; Ohio Colorectal Cancer Prevention Initiative Study Group. Assessment of tumor sequencing as a replacement for lynch syndrome screening and current molecular tests for patients with colorectal cancer. JAMA Oncol 2018; 4 (6): 806–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Susswein LR, Marshall ML, Nusbaum R, et al. Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genet Med 2016; 18 (8): 823–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mannan AU, Singh J, Lakshmikeshava R, et al. Detection of high frequency of mutations in a breast and/or ovarian cancer cohort: implications of embracing a multi-gene panel in molecular diagnosis in India. J Hum Genet 2016; 61 (6): 515–22. [DOI] [PubMed] [Google Scholar]
- 14. Santos C, Peixoto A, Rocha P, et al. Pathogenicity evaluation of BRCA1 and BRCA2 unclassified variants identified in portuguese breast/ovarian cancer families. J Mol Diagn 2014; 16 (3): 324–34. [DOI] [PubMed] [Google Scholar]
- 15. Ambry Genetics. Genetic testing for clinicians. https://www.ambrygen.com/providers. Accessed November 23, 2022.
- 16. Invitae. Invitae Real World Data. https://www.invitae.com/en/partners/real-world-data. Accessed November 23, 2022.
- 17. Ranola JMO, Tsai GJ, Shirts BH.. Exploring the effect of ascertainment bias on genetic studies that use clinical pedigrees. Eur J Hum Genet 2019; 27 (12): 1800–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kahn J, Linial N, Samorodnitsky A.. Inclusion-exclusion: exact and approximate. Combinatorica 1996; 16 (4): 465–77. [Google Scholar]
- 19. Manrai AK, Funke BH, Rehm HL, et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med 2016; 375 (7): 655–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shirts BH, Jacobson A, Jarvik GP, Browning BL.. Large numbers of individuals are required to classify and define risk for rare variants in known cancer risk genes. Genet Med 2014; 16 (7): 529–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Starita LM, Ahituv N, Dunham MJ, et al. Variant interpretation: functional assays to the rescue. Am J Hum Genet 2017; 101 (3): 315–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rebbeck TR, Friebel T, Lynch HT, et al. Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers: the PROSE study group. J Clin Oncol 2004; 22 (6): 1055–62. [DOI] [PubMed] [Google Scholar]
- 23. Dwork C. Differential privacy: a survey of results. In: Agrawal M, Du D, Duan Z, Li A, eds. Theory and Applications of Models of Computation. Vol. 4978. Berlin/Heidelberg: Springer; 2008: 1–19. [Google Scholar]
- 24. Acar A, Aksu H, Uluagac A, S, Conti M.. A survey on homomorphic encryption schemes: theory and implementation. ACM Comput Surv 2019; 51 (4): 1–35. [Google Scholar]
- 25. Ozercan HI, Ileri AM, Ayday E, et al. Realizing the potential of blockchain technologies in genomics. Genome Res 2018; 28 (9): 1255–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sheller MJ, Edwards B, Reina GA, et al. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci Rep 2020; 10 (1): 12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
There are no external data associated with this manuscript. All the data are generated synthetically in the modeling software, which is available at https://github.com/BRCAChallenge/classification-timelines.