Abstract
Objective
A previous study, PheMAP, combined independent, online resources to enable high-throughput phenotyping (HTP) using electronic health records (EHRs). However, online resources offer distinct quality descriptions of diseases which may affect phenotyping performance. We aimed to evaluate the phenotyping performance of single resource-based PheMAPs and investigate an optimized strategy for HTP.
Materials and Methods
We compared how each resource produced top-ranked concept unique identifiers (CUIs) by term frequency—inverse document frequency with Jaccard matrices comparing single resources and the original PheMAP. We correlated top-ranked concepts from each resource to features used in established Phenotype KnowledgeBase (PheKB) algorithms for hypothyroidism, type II diabetes mellitus (T2DM), and dementias. Using resources separately, we calculated multiple phenotype risk scores for individuals from Vanderbilt University Medical Center’s BioVU DNA Biobank and compared phenotyping performance against rule-based eMERGE algorithms. Lastly, we implemented an ensemble strategy which classified patient case/control status based upon PheMAP resource agreement.
Results
Jaccard similarity matrices indicate that the similarity of CUIs comprising single resource-based PheMAPs varies. Single resource-based PheMAPs generated from MedlinePlus and MedicineNet outperformed others but only encompass 81.6% of overall disease phenotypes. We propose the PheMAP-Ensemble which provides higher average accuracy and precision than the combined average accuracy and precision of single resource-based PheMAPs. While offering complete phenotype coverage, PheMAP-Ensemble significantly increases phenotyping recall compared to the original iteration.
Conclusions
Resources comprising the PheMAP produce different phenotyping performance when implemented individually. The ensemble method significantly improves the quality of PheMAP by fully utilizing dissimilar resources to capture accurate phenotyping data from EHRs.
Keywords: electronic health records, high-throughput phenotyping, natural language processing, online health information
INTRODUCTION
Electronic health records (EHRs) hold an abundance of real-world clinical data; however, their use in medical research has proven to be a challenge as EHRs are primarily designed for clinical care rather than research.1,2 A major barrier to the successful use of EHR data for biomedical research is quality phenotyping—that is, to efficiently and accurately identify phenotypic information from large, fragmented datasets.3,4 Current phenotyping algorithms often require clinical informaticians and domain experts to create and take considerable resources to develop,5 which is time consuming. With the rapid accumulation of clinical and omics data in biobanks, high-throughput phenotyping (HTP) approaches are becoming increasingly necessary to conduct large-scale analysis efficiently and effectively.6 Researchers have proposed using phecodes, a grouped relevant International Classification of Diseases (ICD) codes to represent clinically meaningful phenotypes for identifying phenotypes.7 Phecodes have been widely used in phenome-wide association studies (PheWAS) using EHRs.8 PheCAP (common semi-supervised approach) is another tool that utilizes machine learning to model phenotyping representations from EHR data and calculate a patient's probability of exhibiting certain phenotypes.6
Unlike the diagnosis-code-based and EHR-derived approaches, free online content, which includes descriptions of phenotypes or human diseases curated from trusted and reliable sources, provides valuable phenotypic information to enable HTP.9,10 Recently, we developed PheMAP—a HTP tool that learned the representation of a phenotype through six independent online resources (ie, Mayo Clinic, MedlinePlus, MedicineNet, Medscape, Wikipedia, and WikiDoc).11 Utilizing a VUMC natural language processing (NLP) pipeline, PheMAP parsed phenotype descriptions from extracted documents and quantified the relationships between phenotypes and relevant clinical concepts represented by standard medical terminology. PheMap is composed of medical concepts with quantified relationships to 841 disease phenotypes. It enables an automatic search of EHRs for each phenotype's quantified concepts and uses them to calculate an individual's probability of having this phenotype. Then, Gaussian mixture models were fitted to phenotype scores, and a posterior probability was calculated to determine if each individual was a case or control. PheMAP demonstrated comparable or better performance in identifying phenotypes compared to multiple established algorithms created by domain experts.12–14
The original PheMAP combines documents from the six online resources as a single corpus. Given that each resource has distinctive styles of descriptions, for example, MedlinePlus documents are typically concise while Wikipedia often contains longer documentation and more sections such as history of disease, the following questions arise: (1) Whether PheMAP constructed using each resource yields different phenotyping results. (2) What are the best ways to combine documents from diverse sources precisely? In this study, we quantified the phenotype-concept relationship by each resource and applied these relationships to EHRs to calculate the phenotype risk scores (PheRS). We evaluated the difference of PheMAPs derived from each single resource intrinsically and extrinsically. We compared the similarity of top-ranked concepts for certain phenotypes and compared it with conventional Phenotype KnowledgeBase (PheKB) algorithms.15 We also compared the phenotyping accuracy by applying our algorithms to EHR data.16 In addition, we developed an ensemble method that leverages the six corpora and compared it with our original implementation of PheMAP as well as PheMAPs learned from each individual resource.
MATERIALS AND METHODS
Retrieving phenotype information from publicly available resources
We gathered document describing diagnoses, symptoms, and treatments relating to diseases of interests (phenotypes) from six publicly available resources that offer consumer health information, including Mayo Clinic Patient Care and Health Information website, MedlinePlus, MedicineNet, Medscape, WikiDoc, and Wikipedia.11 We parsed the articles and mapped them to phenotypes by matching article titles to concept unique identifiers (CUIs) in the Unified Medical Language System (UMLS). We utilized an NLP pipeline—the KnowledgeMap Concept Indexer to identify CUIs found in each document.17 We then linked CUIs to ICD codes, Current Procedural Terminology (CPT) codes, Logical Observation Identifiers, Names, and Codes (LOINC), Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT), and RxNorm Normalized Names and Codes (RxNORM).8,18–20 We utilized bar graphs and Venn diagrams to visualize the phenotype and CUI coverage among the six online resources.21
Constructing single resource-based PheMAP
In the previous PheMAP article, we concatenated all articles from different resources describing one phenotype. We then calculated term frequency—inverse document frequency (TF-IDF) to quantify the importance of the relationship between a concept and a phenotype.11 In the original version of PheMAP, we limited the implementation to the top 100 CUIs. In this study, we calculated the TF-IDF separately by each resource, and thus we constructed six different PheMAPs (ie, PheMAP-Mayo, PheMAP-MedlinePlus, PheMAP-Mednet, PheMAP-Medscape, PheMAP-WikiDoc, and PheMAP-Wikipedia) that were learned from each resource.
Source of data
This study used the data from VUMC Synthetic Derivative, which contains over 2.2 million unique individuals’ rich and dense EHR data.22
Evaluation tasks and metrics
We evaluated different PheMAP implementations by measuring similarity among resource CUIs, comparing to established phenotype algorithms (eg, PheKB), and calculating phenotyping prediction agreement within EHR data. In this study, we selected three algorithms from PheKB (ie, T2DM, hypothyroidism, and dementia) to evaluate PheMAP implementations. We chose these three algorithms because they (1) are disease phenotypes, (2) are rule-based, (3) have been validated across institutes, (4) have been demonstrated with highly consistent overall performance, and (5) have been recently updated. The eMERGE case–control definition standards for type II diabetes mellitus required information on diagnoses, lab results, medication orders, and physician encounter dates; ICD-9, LOINC, and RxNorm codes were utilized for implementation. For hypothyroidism, the eMERGE algorithm utilized ICD-9 codes, lab results, medications, and CPT codes to identify patients with hypothyroidism without a secondary cause of surgical removal or radiological ablation; the search was designed to remove subclinical hypothyroidism as part of the case definition. Lastly, the dementias eMERGE algorithm utilized ICD-9 codes, medications, and visit history to classify cases and controls.
Similarity measurement
The similarity between each PheMAP is measured by calculating Jaccard similarity coefficients between their learned top N-ranked CUIs (N = 10, 30, 50, 70, 90) according to the TF-IDF score for each phenotype. The Jaccard similarity coefficient is a statistic metric used for gauging the similarity or diversity between sample sets. The metric is defined as the size of the intersection of two sets divided by the size of the union of two sets:
| (1) |
where A and B are two sets. The value of the index ranges between 0 and 1. A Jaccard index of 1 indicates complete equivalence while an index of 0 indicates complete diversity. The Jaccard similarity coefficients between individual resources and/or the original implementation of the PheMAP were calculated for all phenotypes via phecodes by utilizing the top N CUIs ranked by TF-IDF (N = 10, 30, 50, 70, 90). The mean Jaccard similarity coefficient between two given resources was calculated by averaging Jaccard coefficients determined from the phenotypes covered by both resources. For any given resource comparison, if one or both resources lacked a given phenotype to be incorporated into the mean Jaccard score, then the given resource comparison was ignored as one or both sets of CUIs to be compared do not exist.
Comparison to established phenotyping algorithms
For each resource, the number of top features found in both well-known phenotyping algorithms and the top CUIs ranked by TF-IDF (N = 50) was identified for selected phenotypes which include hypothyroidism, type II diabetes, and dementia.13,23,24 We did not compare overlap at different thresholds of N CUIs as little to no overlap occurred at lower values of N. The number of matches on the concept level was recorded for each resource, and the amount of overlap among resources was visualized using UpSet plots.25
Phenotyping prediction
We applied each PheMAP to EHR data to compare the phenotyping predictions of each single resource and the PheMAP-Original. We utilized the kappa statistic—a statistical measure of inter-rater reliability for categorical variables—to quantify the relationships between each resource and the original PheMAP implementation.26 The kappa statistic can be measured with the following equation:
| (2) |
where po is the relative observed agreement among raters and pe is the hypothetical probability of chance agreement. The number of CUIs (N = 20, 50, 100) utilized from each resource when creating predictions was varied to determine how phenotyping performance changed and compared with other single resource implementations.
Ensemble approach to PheMAP implementation
We utilized an ensemble approach which leveraged the outcomes of each individual resource PheMAP to determine if patients were a case or control for a selected phenotypes—hypothyroidism, type II diabetes, and dementias. If the majority of resources, that is, more than half the resources, identified a patient as a case, the patient was labeled as a case; otherwise, the patient was labeled as a control. If a phenotype was not covered by a given resource, the resource would not contribute to the ensemble approach. Both the PheMAP-Ensemble and the PheMAP-Original utilize all resources available, that is, if a phenotype was not covered by a given resource, the resource would not contribute. The PheMAP-Original calculates phe-scores for patients by combining phenotype documents from all available resources and then calculating TF-IDF and ranking CUIs from one corpus. In contrast, the PheMAP-Ensemble calculates phe-scores for patients by calculating TF-IDF and ranking CUIs from each individual resource’s corresponding phenotype documents. For example, if five of the six resources cover a phenotype; the PheMAP-Ensemble should utilize five separate phe-scores from the five corresponding resources to diagnose patients based on majority agreement.
Phenotyping performance
For each phenotype, we applied the PheMAP to calculate the PheRS using EHR data. Gaussian mixture models were fitted to the phenotype score under the assumption that the phenotype scores follow a roughly bimodal distribution for cases and controls; the Gaussian mixture models allowed us to ascertain the posterior probability that a patient is a case or control for a phenotype of interest.11 We used clinician-validated Electronic Health Records and Genomics (eMERGE) algorithms as a reference standard for cases and controls. The eMERGE algorithms were designed for a high positive predictive value and leave many patients unclassified while PheMap assigns a continuous score to all patients. Each PheMAP implementation (single resource and ensemble methods) were compared to each other using eMERGE case–control definitions as reference standards. The following metrics were recorded: accuracy—the number of correctly predicted data points out of all data points; precision—total number of true positives divided by number of positive predictions; recall—the percentage of a given class that is correctly identified; F1—a measure of binary classification accuracy; AUROC—area under receiver operating characteristics curve. One-tailed t tests were performed between the PheMAP-Original and all other implementations to determine if any of the implementation types improved metrics by a statistically significant margin.
RESULTS
Characteristics of resources comprising the PheMAP
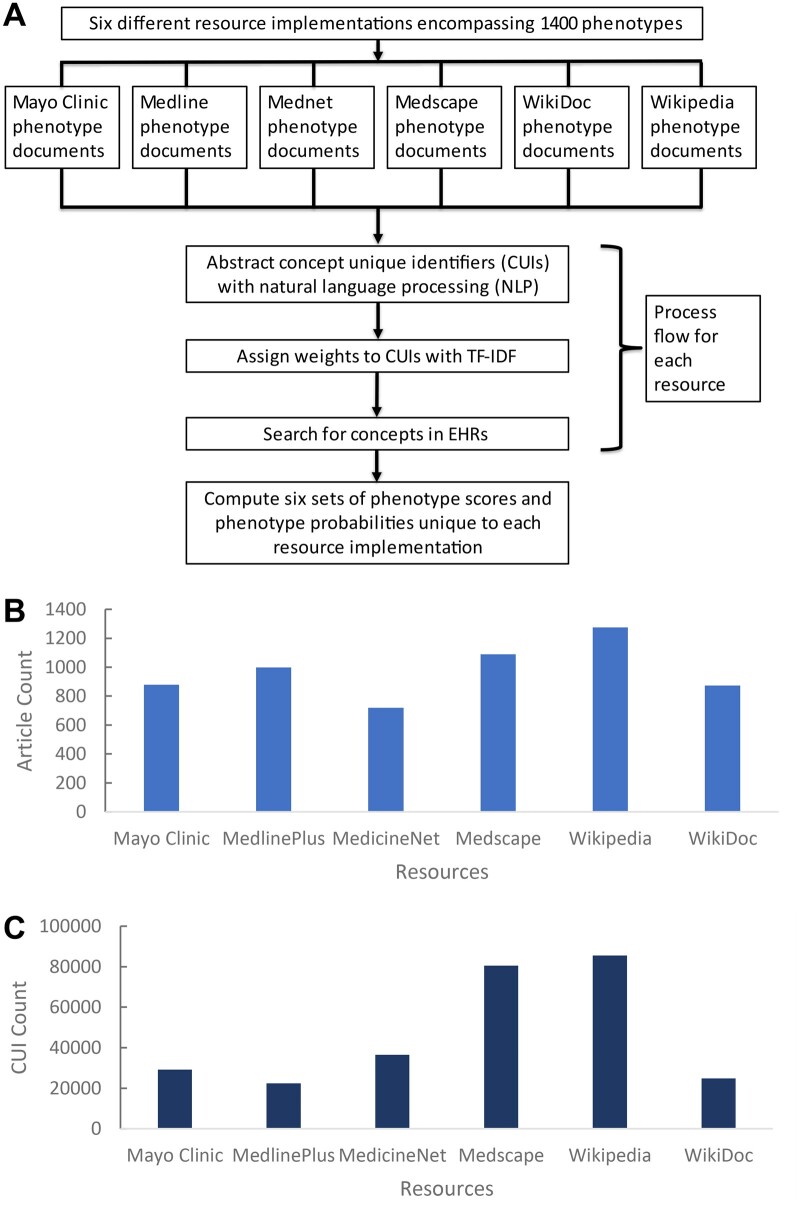
The six online resources covered 1400 phenotypes in total. The number of phenotypes covered by each resource can be visualized with a bar graph (Figure 1). Overlap visualized with Venn diagrams (Supplementary Material S1) indicated 384 phenotypes of 1400 total phenotypes were covered by each of the resources. A total of 1303 phenotypes were covered by two or more resources. All phenotypes found in WikiDoc are also found in at least one other resource. In contrast, Wikipedia contains 33 phenotypes that were not found in any other resource. In a similar manner, the number of CUIs identified in phenotype documents from different resources was visualized in Figure 1. A total of 9936 of 127 271 total CUIs can be found in each resource. Wikipedia had the greatest number of CUIs that are unique to its own set (26 377 CUIs) while Medline only had 1299 unique CUIs.
Figure 1.
Process of single resource PheMAP constructions and phenotype/CUI coverage by resource. (A) Flow diagram of single resource PheMAP implementations; six unique sets of phenotype probabilities for relevant EHR patients are calculated. (B) Phenotype coverage by online resource. Overall, 1400 phenotypes are encompassed. (C) CUI coverage by online resource. Medscape and Wikipedia exhibit the greatest number of CUIs while MedlinePlus and WikiDoc exhibit the lowest number. CUIs: concept unique identifiers; EHR: electronic health record.
Wikipedia captured the most phenotypes with 1275 total. In contrast, Mednet covered only 719 phenotypes—the lowest among any resource (Table 1). MedlinePlus had the lowest average number of CUIs per phenotype document (ie, shortest articles averagely) while Wikipedia contains the largest average number of CUIs per document (ie, longest articles averagely). The large standard deviations associated with each CUI average indicate that the distribution of the number of CUIs per document was skewed. Each document from a given resource significantly varied in CUI count, for example, documents from the Medscape resource range in a CUI count from 59 CUIs to 21 707 CUIs.
Table 1.
Description of resource composition based on disease phenotypes and CUI counts in phenotype documents
| Number of phenotypes | Mean number of CUIs per document (minimum CUI count–maximum CUI count) | |
|---|---|---|
| Mayo Clinic | 879 | 991.6 ± 908.4 (79–11 115) |
| MedlinePlus | 998 | 374.4 ± 374.6 (14–5512) |
| Mednet | 719 | 1269.6 ± 1152.6 (108–13 441) |
| Medscape | 1090 | 1810.9 ± 1893.4 (59–21 707) |
| Wikipedia | 1275 | 1881.0 ± 1986.5 (13–21 072) |
| WikiDoc | 873 | 479.6 ± 501.4 (50–6452) |
CUIs: concept unique identifiers.
Jaccard similarity coefficient
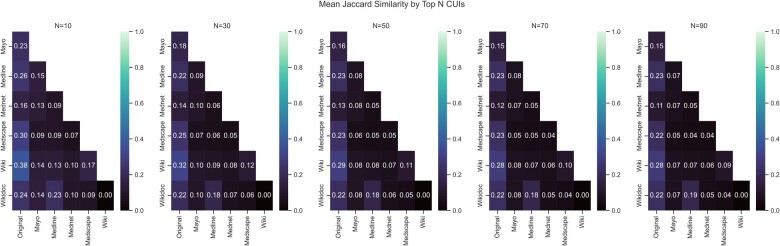
Based on TF-IDF ranked CUIs, each resource is most like the original PheMAP implementation which leveraged and combined every resource, that is, each resource shares a higher Jaccard similarity coefficient based upon CUIs with the PheMAP-Original than with any other resource regardless of the number of CUIs being compared. With more CUIs being compared, the Jaccard similarity coefficient decreased, so the resources became more dissimilar (Figure 2).
Figure 2.
Jaccard similarity matrices. Jaccard similarity coefficients or scores (indicated in matrix boxes) are shown as they change as the top N CUIs from each resource and the original PheMAP relating to a given phenotype are compared. For each resource comparison, the Jaccard similarity score was calculated by averaging the Jaccard similarity scores obtained from resources sharing the same phenotype articles and phecodes, that is, if two resources did not share a phecode, then the corresponding phenotype was ignored when calculating the average Jaccard score for the two resources. CUIs: concept unique identifiers.
PheKB algorithm comparison
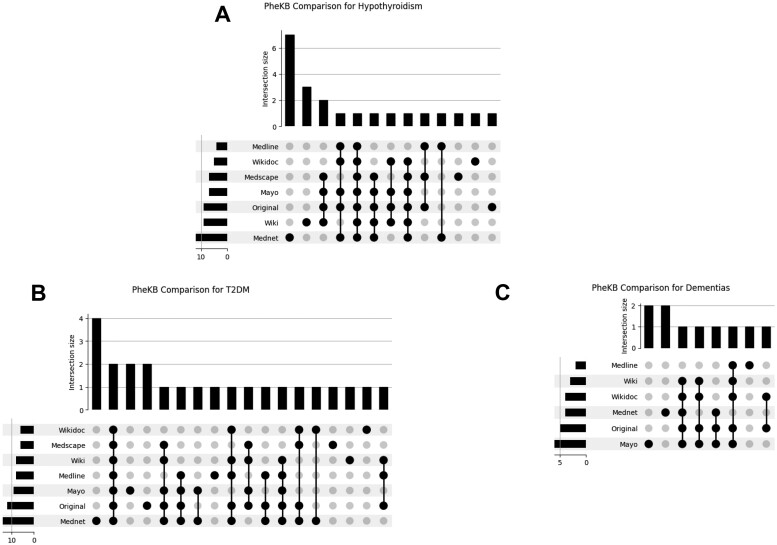
We compared the top 50 CUIs ranked by TF-IDF from each resource to features from established phenotyping algorithms.13,23,24 Although the features or variables of interest in each unique PheKB algorithm differs, among the selected phenotypes—hypothyroidism, T2DM, and dementias—MedicineNet CUIs exhibited the greatest overlap with PheKB algorithms. For example, 12 of the top 50 CUIs ranked by TF-IDF in the MedicineNet resource were also features or variables of interest in the PheKB algorithm for hypothyroidism (Figure 3).
Figure 3.
UpSet plots visualizing PheKB feature comparisons. The plots quantify the amount of overlap between the top 50 ranked CUIs from each PheMAP resource and the features used in PheKB algorithms. The leftmost bars indicate the number of PheKB features found in the corresponding resource set of CUIs. The bottom right plot of dots and lines indicate what set or sets of resources are being compared. The size of the intersection indicates the number of concepts belonging to a particular intersection. Intersections can relate multiple resources or only one resource as denoted by the black dots below each intersection size bar, for example, the first intersection size of 4 only corresponds to Mednet—this means that of the entire Mednet categorical values, 4 categorical values are unique to Mednet. The lines connecting dots in the lower right-hand portion of the graph have no significance. CUIs: concept unique identifiers; PheKB: Phenotype KnowledgeBase.
Comparing the agreement of phenotype prediction
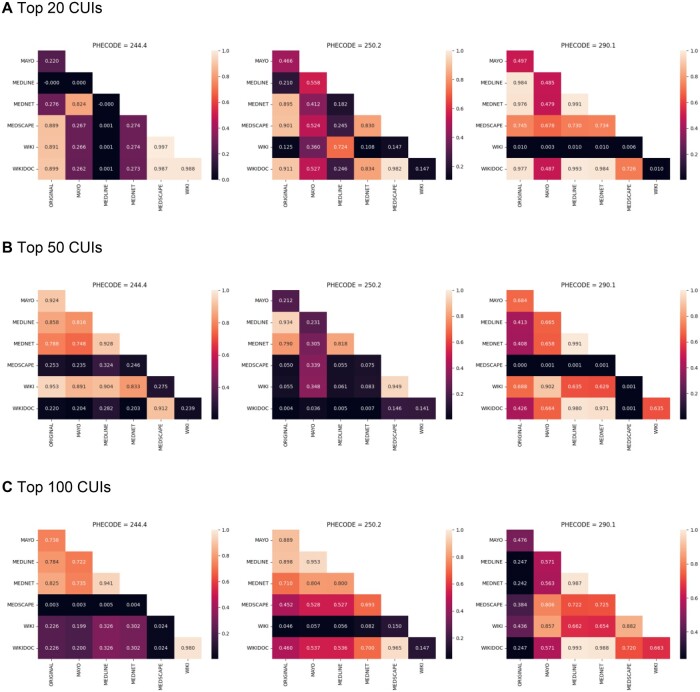
The kappa heatmaps indicate how frequently the resources agree on a specific outcome for a patient. A total of 129 016 patients were utilized for evaluation; about 13 000 individuals appeared to be cases or controls across all three disease types. As the threshold (N = 20, 50, 100) of CUIs used in predicting a patient as a case or control increases, inter-rater agreement increases between some resources but remains low for others (Figure 4); for example, as the threshold increases for type II diabetes mellitus (phecode = 250.2) from N = 20 to N = 100, the kappa statistic increases between several resources and a maximum kappa statistic of 0.97 is observed between the PheMAP-WikiDoc and PheMAP-Medscape at N = 100. In contrast, inter-rater agreement remains relatively low between PheMAP-Medscape and other resources for hypothyroidism (phecode = 244.4) as the threshold increases to N = 100; at N = 100, the highest kappa value relating to Medscape is 0.024 and is shared between PheMAP-Medscape, PheMAP-WikiDoc, and PheMAP-Wikipedia while the lowest kappa value relating to Medscape of 0.0027 is shared between PheMAP-Medscape and PheMAP-Mayo.
Figure 4.
Cohen’s kappa coefficient heatmaps. Phecodes 244.4, 250.2, and 290.1 represent hypothyroidism, type II diabetes mellitus, and dementias, respectively. The heatmaps visualize how frequently the resources agree on a specific outcome for a patient. The heatmaps illustrate how inter-rater agreement can change as the number (N = 20, 50, 100) of CUIs (ranked by TF-IDF) used in assessing phenotype risk increases. CUIs: concept unique identifiers; TF-IDF: term frequency—inverse document frequency.
Evaluation of phenotyping performance within EHR
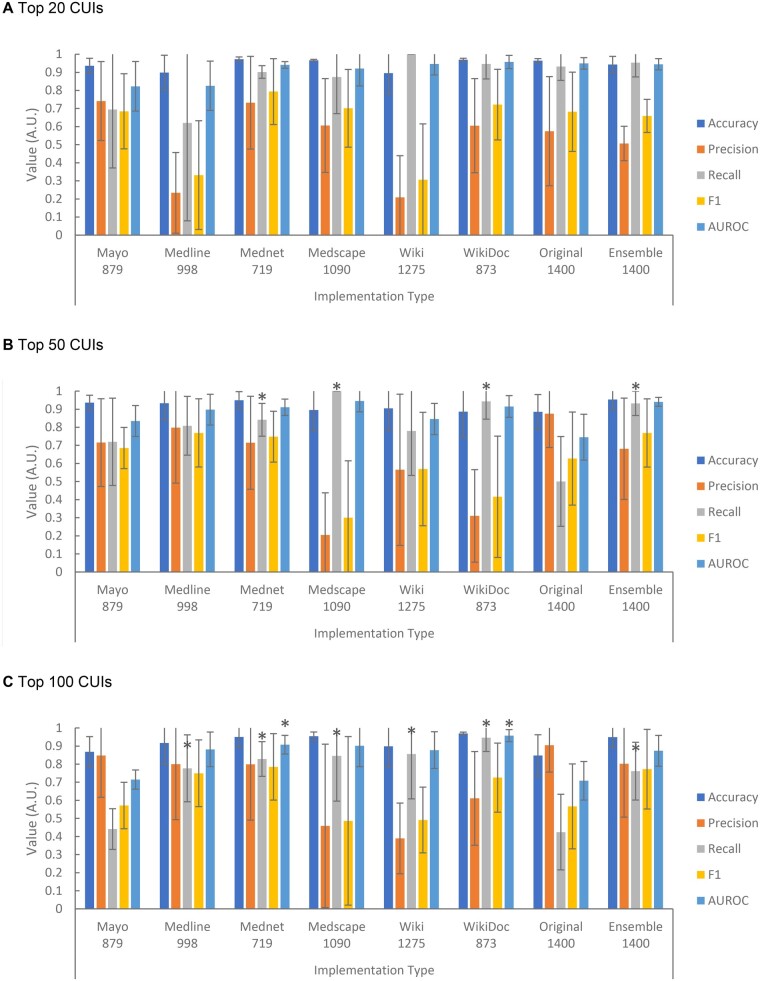
We compared the PheMAPs implemented by using individual resources and using the ensemble approach (PheMAP-Ensemble) to the original implementation (PheMAP-Original) (Figure 5). Overall, 33 106, 27 830, and 80 677 patients linked to eMERGE data were utilized for each implementation evaluation for type II diabetes mellitus, hypothyroidism, and dementias, respectively. A single patient could act as a control for all three phenotypes, but inclusion in each evaluation subset was not guaranteed. The individual resource PheMAPs, PheMAP-Ensemble, and PheMAP-Original showed high levels of accuracy in diagnosing a patient. The PheMAP-Ensemble, however, achieved significantly higher recall as well as accuracy, F1, and AUROC compared to the PheMAP-Original (Figure 5). While not statistically significant in all cases, the PheMAP-Original offered better mean precision in diagnosing the three selected phenotypes—hypothyroidism, type II diabetes mellitus, and dementias compared to all other implementation types.
Figure 5.
Individual resource PheMAPs and PheMAP-Ensemble accuracy, precision, recall, F1, and area under ROC curve for selected phenotypes when utilizing top N (N = 20, 50, 100) CUIs for implementation (A, B, C). The ensemble approach exhibits higher values than the original implementation for every metric except for precision at N = 50 and N = 100. The PheMAP-MedlinePlus and PheMAP-Mednet have high metric values similar in magnitude with the ensemble approach across all metrics at N = 50 and N = 100. At N = 20, PheMAP-Ensemble performance is comparable to PheMAP-Original performance. The number under each implementation type represents the number of phenotypes covered by the corresponding implementation resource or method. One tailed t tests were performed with each implementation’s metric means and the PheMAP-Original metric means; each star or asterisk associated with a given metric bar indicates that the metric statistically outperformed the PheMAP-Original. Note that at N = 20, no metrics were significantly different from the PheMAP-Original. CUIs: concept unique identifiers.
As the number of CUIs utilized for PheMAP implementation (top N CUIs) increases, PheMAP-Ensemble accuracy remains relatively high, precision increases, recall decreases, F1 nominally increases, and AUROC remains relatively high. For the same increase in top N CUIs, PheMAP-Original accuracy remains relatively high, precision increases, recall decreases, F1 remains relatively constant, and AUROC decreases. Overall, the PheMAP-Ensemble nominally outperforms the original implementation’s accuracy, recall, F1, and AUROC.
DISCUSSION
In this study, we calculated the weights of CUIs for phenotypes from each resource, respectively. We compared the weighted CUIs corresponding to all single-resource phenotype documents and measured the similarity among each resource to visualize how each single-resource phenotype document’s weighted CUI composition varies and contributes to the PheMAP-Original. The analysis showed that Wikipedia, Mayo Clinic, and WikiDoc appear to contribute the most to the PheMAP-Original CUI composition. We also found that similarity among resources and the PheMAP-Original at the concept level decreased as the N threshold (number of top-ranked CUIs being compared) increased. This finding was expected as these corpora are extracted from public, independent online resources with unique formats and differing target audiences. For example, the six independent resources may differ in emphasis on risk factors, symptoms, and detection—topics that are often accurately covered on online health websites. Coverage of prognosis may also differ—a topic that most websites do not cover.27
In addition, we compared the top-ranked CUIs from each resource with features of well-known PheKB algorithms for T2DM, hypothyroidism, and dementias. While the features of these algorithms differed based on phenotype, each algorithm utilized a combination of medications, laboratory tests and procedures, or diagnosis codes. At least one factor of overlap was found between the top-ranked CUIs corresponding to each resource for each phenotype and the features of the PheKB algorithms except in one instance: no overlap was found between the top-ranked CUIs corresponding to Medscape for Dementias and the features of the Dementias PheKB algorithm. While not statistically significant, MedicineNet CUIs appeared to show the greatest amount of overlap with PheKB algorithm features; the MedicineNet resource may have a high degree of overlap with PheKB features because MedicineNet captures and displays a high number of medications—a common phenotyping algorithm variable of interest that can increase phenotyping performance.10,28
Through kappa statistical analysis, we visualized resource performance in calculating a PheRS. Kappa heatmaps can be used to indicate the ideal number of CUIs necessary to maintain accurate performance and inter-rater agreement.26 In this study, we found that inter-rater agreement among resources varied by resource comparison, phenotype, and the level of CUI filtering, that is, each resource offers differing phenotyping predictions depending on the phenotype identified. In addition, inter-rater agreement for one specific resource and other resources may be close to zero for all comparisons. This low kappa score, which results in a dark line pattern within a heatmap as seen in Figure 4, is caused by data that may not follow a roughly bimodal distribution as assumed so the applied Gaussian mixture model for predicting cases and controls cannot efficiently diagnose patients; this means that some resources, when applied to certain diseases, cannot be used to accurately predict cases and controls.
To leverage each dissimilar resource and cover as many phenotypes as possible, we propose the PheMAP-Ensemble. The PheMAP-Ensemble method proved to provide better or comparable accuracy, recall, F1, and AUROC compared to the PheMAP-Original at all levels of filtering. While PheMAP-Mednet and PheMAP-MedlinePlus implementations resulted in similar phenotyping efficacy as the PheMAP-Ensemble across most levels of CUI filtering, these single-resource PheMAP implementations cover a limited number of disease phenotypes, that is, Mednet covers 719 phenotypes, MedlinePlus covers 998 phenotypes, and both resources combined only cover 1142 phenotypes of 1400 current phenotypes. The proposed PheMAP-Ensemble offers better coverage than the Mednet and MedlinePlus implementations by utilizing all six disparate resources. In the end, the PheMAP-Ensemble increases the already limited power of the PheMAP to capture rare diseases compared to single-resource implementations.11
There are several limitations to this analysis of PheMAP composition and individual resource accuracy. The PheKB algorithm comparison looks at only three phenotypes and the features from each algorithm are not standardized across each phenotype, that is, the dementias PheKB algorithm may focus solely on medications while the hypothyroidism PheKB algorithm may emphasize both medications and laboratory tests.13,23,24 Although the current PheKB hosts 80 distinct phenotyping algorithms, many algorithms are not diseases, for example, MACE on statin, WBC, and BMI. Some PheKB algorithms are machine learning or NLP based, which require extra training data and servers to implement. In addition, their performances are less consistent across institutes than rule-based ones. Furthermore, several algorithms have not been updated to current ICD-10-CM codes and medication/procedure lists since they were published. Therefore, we used the three most updated and well-defined algorithms. Nevertheless, the conclusion that the original implementation underperformed compared to the ensemble approach regarding accuracy metrics, held across each phenotype. A future study would aim to include additional phenotype algorithms for assessment. In addition, the kappa analysis is also limited in that it does not compare inter-rater agreement among the resources for less well-defined phenotypes or diseases; when less-well-defined phenotypes are compared, it is likely that fewer resources will cover these phenotypes so fewer comparisons of inter-rater agreement would be made. Lastly, the ensemble method does not adjust for rarer phenotypes that have fewer resources overviewing such phenotypes; in future iterations of the ensemble method, the case/control selection will need to account for decreasing resource count for rarer or less well-known phenotypes.
In the future, phe-scores from individual implementations could be summed into a cumulative score which could then be applied to a Gaussian mixture model to determine if patients fall in case or control distributions. Moreover, as more phenotypes are encompassed by the PheMAP knowledge base, comparison of implementation methods will offer more direction in increasing phenotyping accuracy; the ensemble approach may be improved by automatically removing resources that are not following a bimodal distribution. The algorithms may be optimized by removing resources that have an equivalent inter-rater agreement to other resources. If resources strongly agree, then only one of the given resources is required for classification. This reduction in the number of resources utilized for the PheMAP would decrease algorithm runtime while maintaining equivalent accuracy and precision. In addition, additional CUIs may be leveraged when calculating phenotype probabilities (N = 150, 200, etc., for the number of top-ranked CUIs) to evaluate the effects on accuracy, precision, recall, and AUROC. Lastly, the method of leveraging individual resource implementations may be optimized to produce case and control populations even when some resource implementations may lack given phenotype coverage.
CONCLUSIONS
Each resource utilized by the PheMAP is unique and distinct from one another. Through the proposed ensemble approach, we may leverage each resource to outperform the original implementation of the PheMAP in diagnosing a patient as a case or control.
Supplementary Material
Contributor Information
Nicholas C Wan, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee, USA.
Ali A Yaqoob, Department of Biomedical Informatics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Henry H Ong, Department of Biomedical Informatics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Juan Zhao, Department of Biomedical Informatics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Wei-Qi Wei, Department of Biomedical Informatics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
FUNDING
This study was supported by National Institutes of Health grant numbers R01 HL133786, R01 GM139891, R01AG069900, U01 HG011181, P50HD106446, and R01 LM012806. The data set used for the analyses described was obtained from Vanderbilt University Medical Center's resources, the Synthetic Derivative, which are supported by institutional funding and by the National Center for Advancing Translational Science grant number UL1 TR002243. The funders had no role in the study design, data collection, analysis, decision to publish, or preparation of the article.
AUTHOR CONTRIBUTIONS
W-QW, JZ, AAY, and NCW conceived of and designed the study. AAY and JZ constructed the PheMap implementations and executed the phenotyping with W-QW and HHO’s assistance and guidance. NCW and AAY contributed to the statistical analyses. W-QW, JZ, and HHO assisted with the interpretation of results. W-QW acquired funding for the study. NCW wrote the article with W-QW and the participation of all authors.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The PheMap knowledge base of quantified concepts is made freely available for download at https://www.vumc.org/cpm/phemap. Example scripts that calculate PheMap phenotype scores and phenotype probabilities from EHRs that are structured following the OMOP Common Data Model are also provided.29 These scripts do not calculate individual resource implementations—these implementations require code adjustment.
REFERENCES
- 1. Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet 2011; 12 (6): 417–28. [DOI] [PubMed] [Google Scholar]
- 2. Bowton E, Field JR, Wang S, et al. Biobanks and electronic medical records: enabling cost-effective research. Sci Transl Med 2014; 6 (234): 234cm3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Delude CM. Deep phenotyping: the details of disease. Nature 2015; 527 (7576): S14–5. doi: 10.1038/527S14a [DOI] [PubMed] [Google Scholar]
- 4. Wei W-Q, Denny JC.. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med 2015; 7 (1): 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc 2013; 20 (e1): e147–e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang Y, Cai T, Yu S, et al. High-throughput phenotyping with electronic medical record data using a common semi-supervised approach (PheCAP). Nat Protoc 2019; 14 (12): 3426–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu P, Gifford A, Meng X, et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: workflow development and initial evaluation. JMIR Med Inform 2019; 7 (4): e14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wei W-Q, Bastarache LA, Carroll RJ, et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS One 2017; 12 (7): e0175508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zheng NS, Kerchberger VE, Borza VA, et al. An updated, computable MEDication-Indication resource for biomedical research. Sci Rep 2021; 11 (1): 18953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wei W-Q, Cronin RM, Xu H, et al. Development and evaluation of an ensemble resource linking medications to their indications. J Am Med Inform Assoc 2013; 20 (5): 954–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng NS, Feng Q, Kerchberger VE, et al. PheMap: a multi-resource knowledge base for high-throughput phenotyping within electronic health records. J Am Med Inform Assoc 2020; 27 (11): 1675–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc 2012; 19 (2): 212–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. PheKB. Dementia. https://phekb.org/phenotype/dementia. Accessed January 18, 2022.
- 14. Denny JC, Crawford DC, Ritchie MD, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet 2011; 89 (4): 529–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O’Callaghan D, Greene D, Carthy J, et al. An analysis of the coherence of descriptors in topic modeling. Expert Syst Appl 2015; 42 (13): 5645–57. [Google Scholar]
- 16. Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc 2016; 23 (6): 1046–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Denny JC, Smithers JD, Miller RA, et al. “Understanding” medical school curriculum content using KnowledgeMap. J Am Med Inform Assoc 2003; 10 (4): 351–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics 2010; 26 (9): 1205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol 2013; 31 (12): 1102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ritchie MD, Denny JC, Zuvich RL. et al. ; Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) QRS Group. Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation 2013; 127 (13): 1377–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pérez-Silva JG, Araujo-Voces M, Quesada V.. nVenn: generalized, quasi-proportional Venn and Euler diagrams. Bioinformatics 2018; 34 (13): 2322–4. [DOI] [PubMed] [Google Scholar]
- 22. Roden D, Pulley J, Basford M, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008; 84 (3): 362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. PheKB. Hypothyroidism. https://phekb.org/phenotype/hypothyroidism. Accessed December 21, 2021.
- 24. PheKB. Type 2 diabetes mellitus. https://phekb.org/phenotype/type-2-diabetes-mellitus. Accessed January 18, 2022.
- 25. Lex A, Gehlenborg N, Strobelt H, et al. UpSet: visualization of intersecting sets. IEEE Trans Vis Comput Graph 2014; 20 (12): 1983–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McHugh ML. Interrater reliability: the kappa statistic. Biochem Med 2012; 22: 276–82. [PMC free article] [PubMed] [Google Scholar]
- 27. Li JZH, Kong T, Killow V, et al. Quality assessment of online resources for the most common cancers. J Cancer Educ 2021; doi: 10.1007/s13187-021-02075-2. [DOI] [PubMed] [Google Scholar]
- 28. Wei W-Q, Teixeira PL, Mo H, et al. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J Am Med Inform Assoc 2016; 23 (e1): e20–e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Voss EA, Makadia R, Matcho A, et al. Feasibility and utility of applications of the common data model to multiple, disparate observational health databases. J Am Med Inform Assoc 2015; 22 (3): 553–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PheMap knowledge base of quantified concepts is made freely available for download at https://www.vumc.org/cpm/phemap. Example scripts that calculate PheMap phenotype scores and phenotype probabilities from EHRs that are structured following the OMOP Common Data Model are also provided.29 These scripts do not calculate individual resource implementations—these implementations require code adjustment.