In eukaryotic cells, proteins are transported between membrane-bound organelles by vesicles (1). To deliver cargo, a vesicle must merge with its target organelle in a membrane fusion process catalyzed by soluble N-ethylmaleimide–sensitive factor attachment protein receptors (SNAREs) (2, 3). When a vesicle finds its target membrane, vesicle-anchored v-SNAREs pair with target membrane–associated t-SNAREs to form trans-SNARE complexes that zipper progressively toward the membranes (1). Energy derived from SNARE complex assembly is used to overcome the energy barrier of membrane merging (4). SNAREs, however, do not work alone and must be assisted by Sec1/Munc18 (SM) proteins, soluble molecules of 60 to 70 kDa (1). Despite intense studies, SM proteins stubbornly resist fully revealing their molecular secrets, baffling and fascinating researchers for decades. The prototypical isoform used in most SM protein studies is Munc18-1, which mediates synaptic vesicle fusion in mammalian neurons (5, 6). A molecular function of Munc18-1 was first discovered using reconstituted fusion assays, in which Munc18-1 grabs both v- and t-SNAREs and promotes their assembly into fusion-competent trans-SNARE complexes (7). This chaperone function of Munc18-1 was genetically confirmed in the cell (8–10) and later found to be conserved in other SM proteins (11, 12). In PNAS, Yang et al. present novel insights into how SM proteins chaperone SNARE complex assembly using high-resolution optical tweezers (13).
To drive membrane fusion, the SNARE motifs of cognate v- and t-SNAREs must align precisely in a parallel manner and assemble into a four-helix bundle (3, 14). However, SNAREs alone tend to misassemble into kinetically trapped dead-end structures unable to drive membrane fusion (14, 15). In retrospect, requirement of chaperones for SNARE assembly is hardly surprising as protein complex assembly is a highly challenging process, especially in the crowded cellular environment. How do SM proteins chaperone SNARE assembly? In 2015, two crystal structures solved by Baker et al. gave the first glimpse of a SM protein engaged in an initial stage of SNARE assembly (12). The yeast SM protein Vps33 was observed interacting with either Vam3 (Qa subunit of t-SNAREs) or Nyv1 (v-SNARE, also known as R-SNARE). Superposition of the two binary structures suggests a ternary complex, in which the N-terminal regions (membrane distal) of SNARE motifs align in parallel when bound to Vps33, while the C-terminal regions are kept separate (12). Based on these observations, it was proposed that a SM protein simultaneously binds to Qa- and v-SNAREs to form a template complex, which subsequently recruits Qbc subunits of t-SNAREs to complete SNARE complex assembly (Fig. 1) (12). Recent cryoelectron microscopic (cryo-EM) structures of the synaptic template complex strongly support the template model and revealed new binding modes of Munc18-1, syntaxin-1 (synaptic Qa-SNARE), and VAMP2 (synaptic v-SNARE) (16).
Fig. 1.
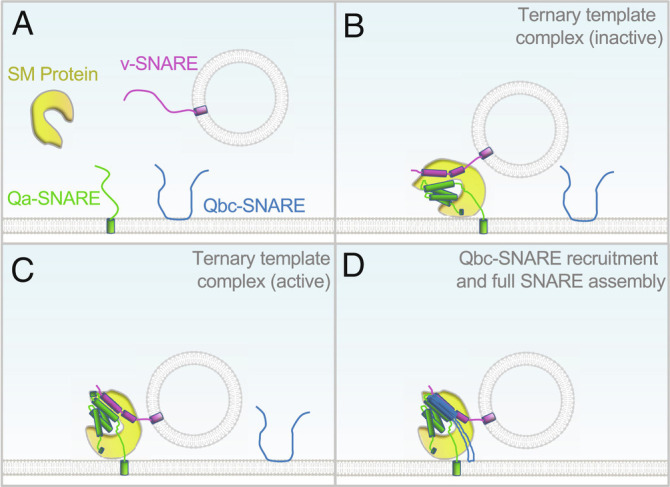
SM protein–chaperoned SNARE assembly. According to the template model (4, 12, 13, 16, 17), a SM protein first interacts with Qa- and R-SNAREs to form a ternary template complex, which switches between inactive and active states (A–C). The active template complex rapidly recruits a specific Qbc-SNARE to complete SNARE complex assembly (D). For clarity, the diagram depicts exocytic vesicle fusion, in which a Qbc-SNARE contains two SNARE motifs connected by a linker. In other vesicle fusion pathways, Qb- and Qc-SNAREs are usually two separate proteins (14).
The structures described above represent snapshots of the template complex, but its dynamics and functional roles in SNARE assembly were still unclear. Single-molecule optical tweezers have proven to be very powerful in studying the dynamics and energetics of SNARE assembly (4). In a typical optical tweezers experiment, Qa- and v-SNAREs are cross-linked at the N termini of their SNARE motifs through a disulfide bond, whereas their C-terminal ends are conjugated to two micron-sized polystyrene beads held in two optical traps (17, 18). This setup enables researchers to exert force on single SNARE complexes and measure their conformational changes at submillisecond and subnanometer resolution (17, 18). When Munc18-1 was added to syntaxin-1 and VAMP2 held at an appropriate force range, a ternary template complex was observed (17, 18). The template complex subsequently recruits SNAP-25, a synaptic Qbc-SNARE, to complete SNARE complex assembly (17, 18). Another two SM proteins exhibited similar behaviors (17), suggesting that the template complex represents a universal intermediate in SM protein–chaperoned SNARE assembly. These single-molecule studies also identified additional elements required for the formation of the template complex including the N-terminal regulatory domain of the Qa-SNARE.
“In PNAS, Yang et al. present novel insights into how SM proteins chaperone SNARE complex assembly using high-resolution optical tweezers.”
In PNAS, Yang et al. further investigated the mechanism by which the template complex recruits the Qbc-SNARE using optical tweezers (13). They measured the kinetics of SNAP-25 binding to the synaptic template complex over a range of SNAP-25 concentrations. From these measurements, they inferred that the synaptic template complex switches between two configurations: one is compatible with SNAP-25 binding (active state), whereas the other is not (inactive state) (Fig. 1). SNAP-25 binds rapidly to the active template complex, likely due to its long-range electrostatic interactions with Qa- and v-SNAREs in the template complex. Mutations of a conserved hairpin region in domain 3a of Munc18-1 abolish both template complex formation and SNAP-25 recruitment. Since the hairpin region is critical for SM protein function in the cell (19, 20), these mutational studies established a physiological connection to the optical tweezer findings. SNAP-25 recruitment requires a poorly characterized linker region located between the N-terminal regulatory domain and the SNARE motif of syntaxin-1 (13). In cryo-EM structures of the synaptic template complex, the linker region of syntaxin-1 forms a helical bundle with the SNARE motifs of syntaxin-1 and VAMP2 (16). Mutations of two conserved residues in the linker region, M183 and D184, abolish reconstituted liposome fusion (16). Surprisingly, truncation of the linker region of syntaxin-1 does not significantly influence the stability of the template complex in optical tweezer measurements, and M183 and D184 mutations have little effect on template complex formation or SNAP-25 recruitment (13). The reason for these discrepancies is unclear.
The single-molecule data of Yang et al. also shed light on the role of SM proteins in determining the specificity of SNARE assembly (13). Intracellular vesicle fusion requires specific SNARE association, but SNAREs themselves pair promiscuously (7). Specificity is achieved when a SM protein recognizes and chaperones its cognate v- and t-SNAREs (7, 11). Formation of the template complex determines the initial pairing specificity of Qa- and v-SNAREs, but it was unclear whether the template complex selects specific Qbc-SNAREs. Yang et al. observed that the Munc18c:syntaxin-4:VAMP2 template complex, which is involved in GLUT4 exocytosis, recruits the Qbc-SNARE SNAP-23 but not SNAP-25 (13). By contrast, the synaptic template complex recruits both SNAP-25 and SNAP-23, consistent with the observation that SNAP-23 supports significant levels of synaptic vesicle fusion when SNAP-25 is absent (21). The Munc18c data clearly demonstrate that a template complex can determine the specificity of Qbc-SNARE binding.
Yang et al. (13), along with earlier single-molecule measurements (17, 18), yielded rich information on SM protein–chaperoned SNARE assembly with at least seven kinetic stages observed. These findings filled a major gap in our knowledge of SNARE assembly. It should be noted that the challenges faced by SNARE assembly in the cell are shared by all protein complexes in membrane trafficking. Thus, other trafficking complexes are likely also dependent on dedicated chaperones to guide their assembly. The principles established by Yang et al. and other SM protein studies offer a blueprint for dissecting other assembly processes.
Looking forward, key questions remain to be answered. First, what is the conformation of the template complex without SNARE cross-linking? Due to their limited stability, template complexes were prepared and characterized with artificial cross-linking between Qa- and v-SNAREs (13, 17, 18). Yang et al. demonstrated that the choice of cross-linking sites influences the behaviors of the template complex (13). Thus, new methods are needed to prepare template complexes without SNARE cross-linking. Second, what are the alternative routes in SM protein–chaperoned SNARE assembly? The template model described by Yang et al. correlates well with reconstituted fusion assays in which Qa- and Qbc-SNAREs were added separately (11, 12, 22). However, when t-SNAREs were preassembled, both Munc18-1 and Munc18c potently stimulated SNARE assembly and fusion kinetics with stringent specificity (7, 11, 23), suggesting that t-SNARE formation can precede v-SNARE binding in certain vesicle fusion events. Moreover, in yeast vacuole fusion, a template complex was formed by a SM protein, Qb-, Qc-, and R-SNAREs (24). Further studies of these alternative SNARE assembly routes will expand the scope of the template model. Finally, how do SM proteins act in concert with other regulators of SNARE assembly such as Munc13? Besides its role in upstream vesicle tethering, Munc13 promotes SNARE assembly by recruiting SNARE subunits and regulating Qa-SNARE conformation (18, 22, 25, 26). Interestingly, Munc13 is not expressed in adipocytes (27), raising the question of whether Munc18c regulates GLUT4 exocytic SNAREs by itself. However, it is more likely that another tethering factor (e.g., exocyst) assists the assembly of GLUT4 exocytic SNAREs analogous to the role of Munc13 in synaptic SNARE assembly. The single-molecule optical tweezers methods described by Yang et al. are well positioned to address these questions when complemented with reconstituted fusion assays and genetic analyses.
Acknowledgments
This work was supported by the National Natural Science Foundation of China grants 31871425 and 32270735 (H.Y.) and the NIH grants DK124431 and GM126960 (J.S.).
Author contributions
H.Y. and J.S. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
See companion article, “A dynamic template complex mediates Munc18-chaperoned SNARE assembly,” 10.1073/pnas.2215124119.
Contributor Information
Haijia Yu, Email: yuhaijia@njnu.edu.cn.
Jingshi Shen, Email: jingshi.shen@colorado.edu.
References
- 1.Sudhof T. C., Rothman J. E., Membrane fusion: Grappling with SNARE and SM proteins. Science 323, 474–477 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sollner T., et al. , SNAP receptors implicated in vesicle targeting and fusion. Nature 362, 318–324 (1993). [DOI] [PubMed] [Google Scholar]
- 3.Sutton R. B., Fasshauer D., Jahn R., Brunger A. T., Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y., Hughson F. M., Chaperoning SNARE folding and assembly. Annu. Rev. Biochem. 90, 581–603 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hata Y., Slaughter C. A., Sudhof T. C., Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature 366, 347–351 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Verhage M., et al. , Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science 287, 864–869 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Shen J., Tareste D. C., Paumet F., Rothman J. E., Melia T. J., Selective activation of cognate SNAREpins by Sec1/Munc18 Proteins. Cell 128, 183–195 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Shen C., et al. , The trans-SNARE-regulating function of Munc18-1 is essential to synaptic exocytosis. Nat. Commun. 6, 8852 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou P., et al. , Syntaxin-1 N-peptide and H(abc)-domain perform distinct essential functions in synaptic vesicle fusion. EMBO J. 32, 159–71 (2012), 10.1038/emboj.2012.307 emboj2012307 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khvotchev M., et al. , Dual modes of Munc18-1/SNARE interactions are coupled by functionally critical binding to syntaxin-1 N terminus. J. Neurosci. 27, 12147–12155 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu H., et al. , Comparative studies of Munc18c and Munc18-1 reveal conserved and divergent mechanisms of Sec1/Munc18 proteins. Proc. Natl. Acad. Sci. U.S.A. 110, E3271–E3280 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker R. W., et al. , A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science 349, 1111–1114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J., Jin H., Liu Y., Guo Y., Zhang Y., A dynamic template complex mediates Munc18-chaperoned SNARE assembly. Proc. Natl. Acad. Sci. U.S.A. 119, e2215124119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunger A. T., Structure and function of SNARE and SNARE-interacting proteins. Q. Rev. Biophys. 38, 1–47 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Yu H., et al. , SNARE zippering requires activation by SNARE-like peptides in Sec1/Munc18 proteins. Proc. Natl. Acad. Sci. U.S.A. 115, E8421–E8429 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stepien K. P., Xu J., Zhang X., Bai X. C., Rizo J., SNARE assembly enlightened by cryo-EM structures of a synaptobrevin-Munc18-1-syntaxin-1 complex. Sci. Adv. 8, eabo5272 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiao J., et al. , Munc18-1 catalyzes neuronal SNARE assembly by templating SNARE association. Elife 7, e41771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shu T., Jin H., Rothman J. E., Zhang Y., Munc13-1 MUN domain and Munc18-1 cooperatively chaperone SNARE assembly through a tetrameric complex. Proc. Natl. Acad. Sci. U.S.A. 117, 1036–1041 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin S., et al. , The Munc18-1 domain 3a loop is essential for neuroexocytosis but not for syntaxin-1A transport to the plasma membrane. J. Cell Sci. 126, 2353–2360 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Munch A. S., et al. , Extension of Helix 12 in Munc18-1 induces vesicle priming. J. Neurosci. Off. J. Soc. Neurosci. 36, 6881–6891 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delgado-Martinez I., Nehring R. B., Sorensen J. B., Differential abilities of SNAP-25 homologs to support neuronal function. J. Neurosci. 27, 9380–9391 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma C., Su L., Seven A. B., Xu Y., Rizo J., Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science 339, 421–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma L., et al. , Munc18-1-regulated stage-wise SNARE assembly underlying synaptic exocytosis. eLife 4, e09580 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song H., Orr A. S., Lee M., Harner M. E., Wickner W. T., HOPS recognizes each SNARE, assembling ternary trans-complexes for rapid fusion upon engagement with the 4th SNARE. Elife 9, e53559 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai Y., et al. , Molecular mechanisms of synaptic vesicle priming by Munc13 and Munc18. Neuron 95, 591–607.e510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X., et al. , Munc13 activates the Munc18-1/syntaxin-1 complex and enables Munc18-1 to prime SNARE assembly. EMBO J. 39, e103631 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Humphrey S. J., et al. , Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 17, 1009–1020 (2013), 10.1016/j.cmet.2013.04.010 S1550-4131(13)00152-6 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]


