Significance
Cleavage of the flavivirus premembrane (prM) structural protein is an essential, yet inefficient, process during virion maturation. The contribution of partially mature flavivirus virions that retain uncleaved prM to pathogenesis during primary infection is unknown. Newly-generated dengue virus prM-reactive antibodies were characterized using both functional and structural methods. In vivo administration of prM-reactive antibodies modified to be incapable of promoting antibody-dependent enhancement resulted in protection from dengue virus (DENV) infection. Our results establish that in addition to mature forms of DENV, partially mature infectious prM+ virions generated in vivo can also contribute to pathogenesis.
Keywords: dengue virus, virion maturation, antibody-dependent enhancement, prM antibody
Abstract
Cleavage of the flavivirus premembrane (prM) structural protein during maturation can be inefficient. The contribution of partially mature flavivirus virions that retain uncleaved prM to pathogenesis during primary infection is unknown. To investigate this question, we characterized the functional properties of newly-generated dengue virus (DENV) prM-reactive monoclonal antibodies (mAbs) in vitro and using a mouse model of DENV disease. Anti-prM mAbs neutralized DENV infection in a virion maturation state–dependent manner. Alanine scanning mutagenesis and cryoelectron microscopy of anti-prM mAbs in complex with immature DENV defined two modes of attachment to a single antigenic site. In vivo, passive transfer of intact anti-prM mAbs resulted in an antibody-dependent enhancement of disease. However, protection against DENV-induced lethality was observed when the transferred mAbs were genetically modified to inhibit their ability to interact with Fcγ receptors. These data establish that in addition to mature forms of the virus, partially mature infectious prM+ virions can also contribute to pathogenesis during primary DENV infections.
Flaviviruses are positive-sense RNA viruses that cause significant disease worldwide (1, 2). Among this group, dengue viruses (DENV) are the greatest threat to public health, as reflected by an estimated 100 million human infections each year (3). Four DENV serotypes circulate in tropical and subtropical climates in transmission cycles involving humans and mosquitoes. The outcome of DENV infection may be clinically inapparent or result in syndromes ranging from severe febrile illness to potentially fatal disease manifestations that meet the WHO classification for severe dengue (4). In children and adults, the principal risk factor for severe dengue is a secondary infection by a heterologous DENV serotype (5). Antibodies with limited neutralizing activity are hypothesized to contribute to severe disease via a phenomenon called antibody-dependent enhancement (ADE) of infection (6, 7). ADE describes an increase in the efficiency of infection of cells expressing Fcγ receptors by viruses decorated with antibodies at a stoichiometry insufficient for virus neutralization (8, 9).
Flaviviruses are composed of three structural proteins, a host-derived lipid envelope, and a genomic RNA molecule. The envelope (E) protein is an elongated three-domain structural protein that has critical roles in virion assembly and entry (10, 11) and is the major target of neutralizing antibodies (12). Flaviviruses are assembled as noninfectious immature virions that display on their surface spikes composed of three heterodimers of the E protein and a second structural protein, premembrane (prM) (11). The association of prM and E proteins prevents pH-dependent conformational changes associated with membrane fusion (13, 14). In contrast, cryoelectron microscopy (cryo-EM) studies of mature flaviviruses reveal a relatively smooth virion with E proteins arranged as 90 antiparallel dimers lying flat against the viral membrane (15, 16). The transition from an immature to mature virion structure is enabled by the pH-dependent changes in E protein arrangement and cleavage of prM by host furin-like serine proteases that occurs during viral egress (17). Cleavage of prM by furin results in the formation of a 75–amino acid M peptide anchored in the viral membrane and a 93–amino acid, 14-kDa soluble “pr” fragment that remains associated with the mature virus particle until the release of the virion into the neutral pH environment of the extracellular space (13).
Although virion maturation is an essential step in the flavivirus life cycle, cleavage of prM may be inefficient, particularly for DENV (18–20). Flavivirus-infected cells can release a heterogeneous mixture of virus particles that retain uncleaved prM with an unknown stoichiometry. Partially mature viruses are defined as having biochemical features of both mature and immature viruses. The structure(s) of this heterogeneous class of virions are not fully understood. Cryoelectron tomography reconstructions of partially mature DENV indicate that the E proteins on prM+ virions are partitioned into regions with mature- and immature-like organization in varying proportions (21, 22). Several lines of evidence suggest that partially mature virions can be infectious (20, 23). The virion maturation state also has been shown to impact the accessibility of epitopes recognized by many E protein–specific antibodies (24, 25). Furthermore, the specific infectivity of prM+ virions on cells expressing Fcγ receptors may be enhanced in the presence of anti-prM antibody (6, 26). Little is known regarding how uncleaved prM is distributed among infectious and noninfectious virions in vitro or in vivo. A recent study reported that a DENV serotype 1 (DENV1) strain isolated from human plasma was more structurally mature than the same virus strain passaged in cell culture (27). The maturation state of virions found in other anatomical compartments and the contribution of partially mature virions to flavivirus disease remain unknown.
Here, we demonstrate that prM-reactive antibodies can efficiently neutralize infection of DENV virions that retain prM. The proportion of neutralization-sensitive virions correlates inversely with the cleavage efficiency of prM in virus preparations and thus provides insight into the size of the population of infectious prM+ virions. Cryo-EM studies of prM monoclonal antibodies (mAbs) bound to immature DENV virions identified two modes of attachment to a single antigenic site. While intact prM-reactive mAbs were capable of enhancing disease in a murine model of DENV infection, we demonstrate that the passive transfer of aglycosyl forms of prM mAbs that are incapable of interacting with Fcγ receptors conferred protection against primary DENV infection and disease. These data provide direct evidence that partially mature prM+ virions can also contribute to DENV pathogenesis during primary infection independently of their potential to contribute to ADE.
Results
Generation and Characterization of prM-Reactive mAbs.
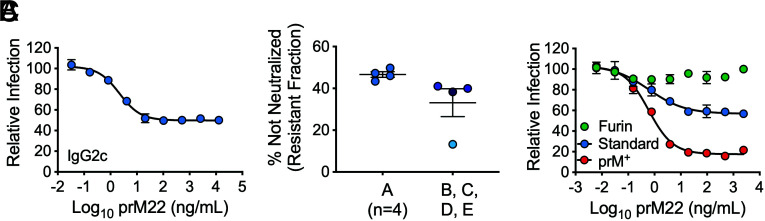
DENV prM-reactive antibodies were produced in mice following infection with DENV2 strain New Guinea C and boosting with recombinant pr protein (residues 1 to 86 of prM) derived from the same virus strain. Antibodies from three hybridomas, designated prM12, prM13, and prM22, were selected for further study (SI Appendix, Table S1). Each cell line produced antibody of the IgG2c subclass. The neutralizing activity of these mAbs was evaluated using DENV reporter virus particles (RVPs) (28, 29). DENV2 strain 16681 RVPs were incubated with serial dilutions of anti-prM mAbs and used to infect Raji cells expressing the C-type lectin DC-SIGNR (Raji-DCSIGNR), a flavivirus attachment factor (30, 31). Antibody dose–response studies revealed sigmoidal-shaped neutralization curves characterized by a significant nonneutralized plateau at the highest concentrations of antibody tested (Fig. 1A and SI Appendix, Fig. S1 A and D). This flat portion of the curve identifies virions in each genetically homogeneous preparation of RVPs that is insensitive to neutralization. Similar patterns of neutralization by some E protein–specific mAbs have been observed when the number of accessible epitopes on individual virions does not exceed the stoichiometric requirements of neutralization (25, 32). Some variation in the resistant fraction was observed when four additional independent RVP stocks were compared (Fig. 1B and SI Appendix, Fig. S1 B and E).
Fig. 1.
Neutralization of DENV2 by a prM-specific mAb. GFP-expressing DENV2 RVPs were incubated with serial dilutions of prM22 for 1 h at 37 °C, followed by infection of Raji-DCSIGNR cells. Infectivity was assessed 48 h later by flow cytometry. (A) Representative dose–response curve of standard DENV2 RVPs (stock A) neutralized by mAb prM22. Error bars indicate the range of duplicate infections. A resistant fraction of nonneutralized virions is apparent at saturating concentrations of antibody. (B) The resistant fraction (calculated as the percentage of virions not neutralized at saturation) is consistent when four independent prM22 neutralization assays are performed with aliquots from the same RVP preparation (stock A) but varies when the assay is repeated with four distinct preparations of RVPs collected from independent transfections (stocks B, C, D, and E). Horizontal line and error bars indicate the mean and SEM, respectively. (C) Representative prM22 neutralization curves of DENV2 RVPs with varying levels of uncleaved prM (furin < standard < prM+). Error bars indicate the range of duplicate infections. Data were confirmed in two additional experiments performed with DENV2 furin and prM+ RVPs only.
Because virion maturation reduces the number of prM proteins on individual virions directly, and thus the number of epitopes displayed for antibody recognition, we investigated whether changes in the prM content of virions account for differences in the size of the resistant fraction among independent stocks of viruses. DENV2 16681 RVPs that vary with respect to the efficiency of prM cleavage were produced (33). RVPs produced in cells that overexpress the human furin protease (DENV furin) were less sensitive to neutralization by prM mAbs than RVPs produced using standard conditions (DENV standard) or in the presence of an inhibitor of human furin (DENV prM+) (Fig. 1C and SI Appendix, Fig. S1 C and F). These results demonstrate that the neutralization activity of prM-reactive antibodies, as measured by the size of the resistant fraction, is modulated by the extent of virion maturation.
DENV prM-reactive mAbs inhibit virus infection primarily by blocking attachment to target cells (SI Appendix, Fig. S2) and do not cross-react with the related flavivirus ZIKV (SI Appendix, Fig. S3). Neutralization studies revealed an inability of all three prM antibodies to neutralize DENV2 RVPs when assayed on Vero cells as compared with Raji-DCSIGNR cells (SI Appendix, Fig. S4), which may explain the poor performance of prM antibodies in conventional neutralization tests on Vero cells (6, 34). In agreement with this interpretation, we previously showed that cell-type–dependent patterns of flavivirus neutralization are influenced by effects of virion-associated prM on virus attachment and epitope accessibility (33). Finally, the addition of the complement component C1q to assays performed with DENV2 RVPs and prM-reactive mAbs resulted in increased neutralization and decreased ADE (SI Appendix, Fig. S5), a phenomenon demonstrated previously with anti-E antibodies (35).
Epitope Mapping of prM-Reactive Antibodies.
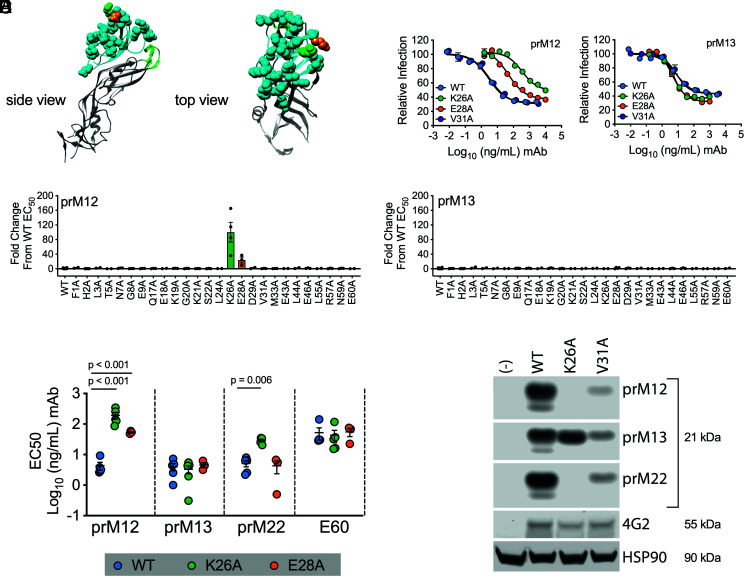
To identify the binding sites for the prM antibodies, we generated a panel of RVPs incorporating alanine substitutions at residues predicted to be solvent accessible on the surface of prM. Twenty-six RVP variants capable of infecting Raji-DCSIGNR cells were assayed for neutralization by prM mAbs (Fig. 2A). These experiments revealed contributions of residues K26 and E28 to inhibition by prM mAbs to varying extents (Fig. 2 B–D and SI Appendix, Fig. S6). Mutation of K26 reduced the neutralization activity of prM12 and prM22 but did not impact sensitivity to prM13. These findings agree with a prior study that identified K26 as an important residue contacted by human-derived DENV prM-reactive antibodies (36). Mutation of position E28 had a modest impact on neutralization sensitivity to prM12 but not prM13 or prM22 (Fig. 2E). To demonstrate that the overall antigenicity and prM content of the variants remained similar to wild-type (WT), the pan-flavivirus E domain II (DII) maturation state–sensitive mAb E60 was used as a control (Fig. 2E and SI Appendix, Fig. S6) (33). We further confirmed the role of K26A in binding by western blotting of cell lysates (Fig. 2F). Alanine substitution at position K26 abrogated binding to prM12 and prM22 but not to prM13. Binding of all three prM mAbs to cells expressing WT or V31A prM, which did not impact neutralization sensitivity, remained intact. Altogether, these experiments indicate that residue K26, and to a lesser extent E28, contributes to prM antibody recognition.
Fig. 2.
DENV prM alanine scanning to identify amino acids contacted by prM mAbs. (A) The structure of the DENV2 prM protein (cyan) in association with domain II (DII) of the E protein (gray) is shown in a ribbon form from the top and side views relative to the arrangement on the immature virion (adapted from PDB: 3C6E). The conserved hydrophobic fusion loop within E DII is highlighted in green. prM amino acids predicted to be surface exposed are designated in space-filling representation, and residues K26 and E28 are highlighted in green and orange, respectively. (B–E) A panel of variant RVPs individually incorporating an alanine at each of the 26 surface-exposed residues was generated. Wild-type (WT) and variant RVPs were incubated with serial dilutions of the indicated mAbs for 1 h at 37 °C, followed by infection of Raji-DCSIGNR cells. Infectivity was assessed 48 h later by flow cytometry. (B) Representative dose–response curves of WT and selected prM variant RVPs (K26A, E28A, and V31A) neutralized by prM12 and prM13 mAbs. Error bars indicate the range of duplicate infections. (C and D) EC50 values for mAb prM12 (C) and mAb prM13 (D) against the panel of 26 variant RVPs incorporating an alanine at the indicated prM residue. Data are expressed as the fold change from the average WT EC50 value (2 to 4 independent experiments per mutant; WT EC50 value calculated from 20 independent experiments for each mAb). The horizontal line and error bars indicate the mean and standard error, respectively. (E) Confirmatory neutralization assays were performed with the indicated mAbs against WT and prM K26A and E28A variant RVPs. EC50 values from 3 to 5 independent experiments are shown. E60 is a maturation state–sensitive E DII-specific antibody used here as a control to demonstrate that the overall antigenicity and prM content of the variants were similar to WT. The horizontal line and error bars represent the mean and SEM, respectively. Statistical differences in log10-transformed mean EC50 values were determined for each mAb using ANOVA with multiple comparison correction; p values are displayed when significant. (F) Lysates from 293T cells transfected to produce WT, prM K26A, or V31A variant DENV2 RVPs were subjected to SDS–PAGE and western blotting analysis with the indicated prM mAbs. HSP90 bands indicate loading of equivalent amounts of cell lysate. 4G2 is a pan-flavivirus–specific control mAb. Data are representative of n = 2 independent experiments for each mAb.
Cryo-EM Structures of Immature DENV2 Complexed with prM12 and prM13 Fab Fragments.
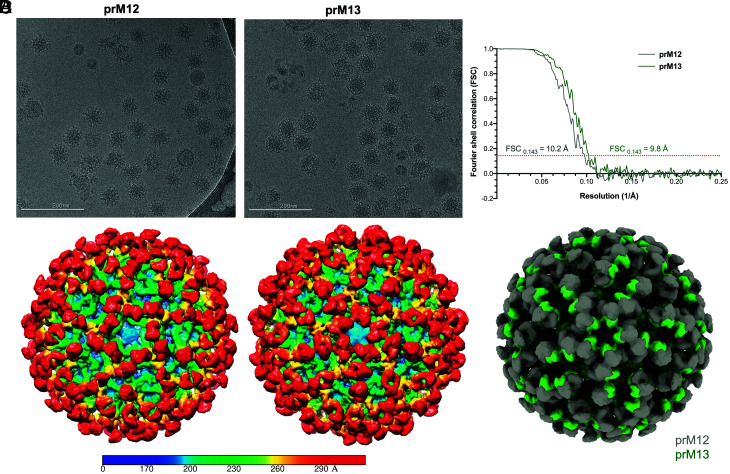
To understand how prM12 and prM13 antibodies engage and neutralize the virion, the binding of their respective Fab fragments to DENV2 strain 16681 was studied by cryo-EM. These two antibodies were selected due to their differential sensitivity to substitution of prM residue K26 (Fig. 2). Structural analysis by single-particle reconstruction requires a homogeneous population of prM-bearing virus. The prM+ immature virus was produced in C6/36 Aedes albopictus cells using NH4Cl, subsequently purified, and cryo-frozen on grids with a >1.5-fold excess of prM12 or prM13 Fab fragments. Cryo-EM micrographs for immature virus bound to prM12 compared with prM13 Fab showed morphologically distinct virus–antibody complexes, suggesting that these antibodies differ in their binding mode (Fig. 3A). Single-particle reconstructions yielded models with an average resolution of 10.2 and 9.8 Å for prM12- and prM13-complexed immature viruses, respectively (Fig. 3B and SI Appendix, Table S2). The Fab density was seen clearly in both the models surrounding the icosahedral immature virus shell, atop all three independent positions (P1, P2, and P3) of the asymmetric trimeric prM–E spike. The Fab density, however, was not superimposable between the two models, confirming that these antibodies interact with the virus in distinct ways (Fig. 3 C and D).
Fig. 3.
Cryo-EM single-particle reconstruction of immature DENV2 virions bound to prM12 or prM13 Fabs. (A) Representative cryo-EM micrographs for immature DENV2 virions complexed with saturating concentrations of prM12 (Left) or prM13 (Right) Fabs. (B) The resolution for the three-dimensional (3D) cryo-EM maps was determined using the Fourier shell correlation (FSC) vs. resolution plot. FSC measures the cross-correlation between two 3D maps independently generated from half datasets (gold standard method). Resolution of the maps, assigned when the FSC crosses the 0.143 threshold, was estimated to be 10.2Å and 9.8Å, respectively, for prM12- and prM13-complexed viruses. (C) Surface-shaded radial density maps for immature virion–Fab complexes (prM12 complex, Left, and prM13 complex, Right) viewed down the icosahedral fivefold axis. Low-pass filter was applied to prM12 (10.2 Å) and prM13 (9.8 Å) maps to reduce noise. The color key depicts variation of color as a function of radial distance from the particle center: blue, up to 170Å; cyan, up to 200 Å; green, up to 230 Å; yellow, up to 260 Å; and red, 290 Å and above. The Fab density (red color) is visible on all the three prM molecules constituting an immature asymmetric trimeric spike in both prM12 and prM13 maps. (D) Superimposed density maps of prM12 (gray) and prM13 (green) complexes viewed down the icosahedral fivefold axis reveal differences in the mode of binding of the two Fabs.
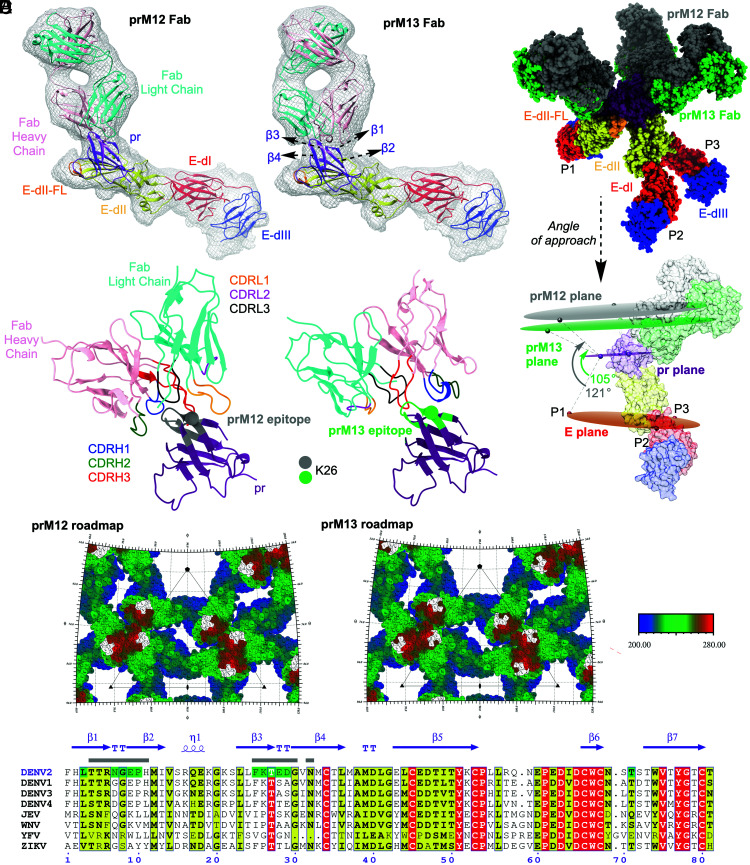
Pseudoatomic models for immature DENV bound to prM12 or prM13 Fabs were created by fitting the crystal structure of soluble prM–E (PDB ID: 3C6E) and homology models of Fab (SI Appendix, Fig. S7) into the cryo-EM density maps (Fig. 4A). The occupancy of Fab per virion was estimated by using the “sumf” function in the program EMfit, which measures the average density around the fitted atoms. The sumf values were similar at the P1, P2, and P3 positions of the trimeric spike for both prM12 and prM13 Fabs (SI Appendix, Tables S3 and S4). This indicates that all the available epitopes can be engaged in the presence of saturating concentrations of these Fabs, resulting in 100% occupancy of the 180 antigenic sites per immature virus (3 antigenic sites on each trimeric spike × 60 trimeric spikes per virion). The prM12 Fabs approach the virion at a steeper angle compared with the relatively flat angle of engagement by prM13 Fabs. The contrast in their mode of binding is readily apparent when the virus is centered at the 5-fold axis (Fig. 3 C and D). The angular difference in approach for these Fabs was estimated to be around 16° (Fig. 4B).
Fig. 4.
Angle of approach and predicted footprint of prM12 and prM13 Fabs on immature DENV2. (A) Atomic structure of DENV2 prM–E ectodomain (PDB: 3C6E) and modeled structure of prM12 or prM13 Fab were fitted into the cryo-EM density (prM12 complex, Left, and prM13 complex, Right). Only one molecule each of prM–E ectodomain and Fab, corresponding to the P2 position of the asymmetric trimer, is shown. The coloring scheme of E protein domains is per convention (domain 1 [E-dI], red; domain 2 [E-dII], yellow with fusion loop (FL) depicted in orange; and domain 3 [E-dIII], blue). pr is shown in purple, and the heavy and light chains of Fabs are shown in pink and cyan, respectively. Cryo-EM density, within 5 Å radius of the fitted coordinates, is represented as a gray mesh. (B) The different modes of engagement by prM12 and prM13 Fabs are apparent in the superimposed and molecular surface–rendered view of the fitted Fab-bound trimeric spike (Top) and angle of approach (Bottom). prM12 and prM13 Fabs are shown in gray and green, respectively. The pr protein and E protein domains are colored as in A. For clarity, the molecular surfaces of P1 and P3 are omitted in the Bottom panel. Planes drawn using the center of mass of the three E, pr, and Fabs of the trimeric spike are shown as ovals and colored orange, magenta, and gray (prM12)/green (prM13), respectively. (C) The Fab–pr interaction interface for prM12 (Left) and prM13 (Right) is displayed in a ribbon representation. The epitope residues (within ~5 Å of Fabs) for prM12 and prM13 are displayed in gray and green, respectively. Cα of the residue K26 is displayed as a sphere. pr and Fab are colored as in A. CDRH1, CDRH2, CDRH3, CDRL1, CDRL2, and CDRL3 loops are shown in blue, green, red, orange, magenta, and black, respectively. (D) The epitopes of prM12 (Left) and prM13 (Right) are displayed on a stereographic road map of the immature DENV2, which is generated by projecting the prM–E surface residues onto a plane using the program RIVEM. To enable depth perception, the prM–E residues are radially colored relative to distance from the center as shown in the color key: proximal (200 Å), blue, and distal (280 Å), red. The residues that form the epitope are colored in white. The road map is zoomed on the asymmetric unit of the virus (represented as a triangle). The 5-, 3-, and 2-fold axes of symmetry are denoted by pentagon, triangle, and oval shapes, respectively. (E) Sequence alignment of DENV2 strain 16681 prM (zoomed on the prM12 and prM13 epitopes) with other mosquito-transmitted flaviviruses. Residues shaded in red and yellow reflect conserved amino acids: identical (red) and similar (yellow). Amino acid residues predicted to comprise the prM12 epitope are indicated by a gray rectangle above them, while those predicted to form the prM13 epitope are highlighted in green. Secondary structure elements of DENV2 prM (PDB:3C6E) are shown in purple with β, TT, and η referring to β-strand, β-turn, and 310-helix, respectively. The figure was generated using ESPript 3.0.
The interactions of Fab complementarity-determining region (CDR) loops with the prM protein could only be predicted at a coarse level, given the inherent limitations associated with a ~10 Å resolution map and computational modeling. As such, emphasis was placed on identifying the antigen–CDR interaction interface rather than fine-mapping of amino acids within the epitope. The CDR loops of both the heavy and light chains of prM12 Fab appeared to interact with the pr antigen, whereas the interaction seemed to be guided primarily by the CDRs of the heavy chain in the case of prM13. Both prM12 and prM13 Fabs appeared to bind a similar region on prM, i.e., β1–4-strands and associated turns. To predict amino acids that may be involved in epitope–paratope interactions, energy-based minimization and modeling of the CDR loops were performed. The epitope shared by both Fabs was predicted to include amino acids 7 to 10 and 25 to 29, including K26. Residues 4 to 6, 11, 30, and 32 may be unique for prM12, whereas residues 3 and 71 appeared to be distinctive for prM13 (Fig. 4 A and C–E). The amino acids within the epitope share high similarity among the different DENV serotypes but not across the flavivirus genus (Fig. 4E). This supports the observation that these antibodies are DENV specific but do not cross-react with other mosquito-borne flaviviruses such as ZIKV (SI Appendix, Table S1 and Fig. S3).
Intact prM Antibodies Promote ADE and Do Not Protect against DENV Infection in Mice.
Administration of prM-reactive antibodies can exacerbate disease in immunocompromised mouse models of DENV infection via ADE (37, 38). The ability of our newly generated prM antibodies to protect against or enhance DENV infection was assessed in susceptible mice lacking both type I and II interferon (IFN) signaling responses using the well-characterized mouse-adapted DENV2 strain D2S20 (39). D2S20 differs from the DENV2 16681 strain at pr residues 28 and 83 (SI Appendix, Fig. S8). Residue 28 was identified as a minor contact for mAb prM12 but not for prM13 or prM22 (Fig. 2C). ADE assays performed in vitro demonstrated that prM12 was modestly less efficient at enhancing D2S20 infection of Fcγ-receptor–expressing K562 cells as compared with prM13 and prM22 (SI Appendix, Fig. S8), a difference not observed in studies with DENV2 16681 (SI Appendix, Figs. S5 and S9). These studies confirm that prM cleavage of D2S20 can be inefficient when the virus is propagated in vitro.
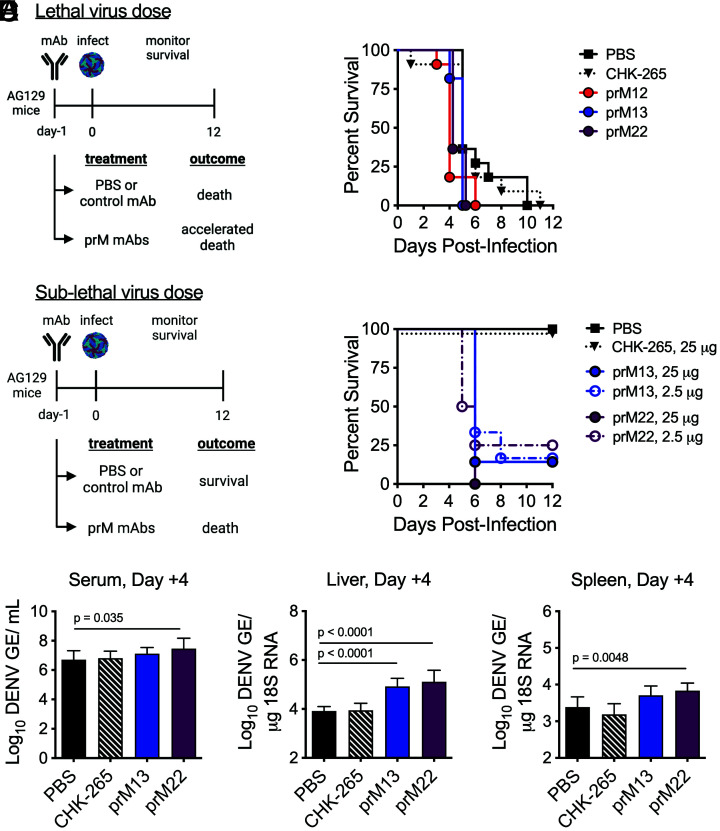
AG129 (Ifnar1−/− Ifngr−/−) mice were administered 250 μg (~10 mg/kg) of murine prM12, prM13, or prM22 1 d prior to infection with a lethal dose of D2S20 (Fig. 5A). Compared with control animals administered phosphate-buffered saline (PBS) or an equivalent dose of an irrelevant chikungunya virus (CHIKV)–reactive isotype-matched control mAb, protective effects of prM mAbs were not observed. Instead, mice that received prM mAbs succumbed to infection more rapidly (P < 0.05 for prM12, prM13, or prM22 vs. PBS control). Enhanced disease was also observed in a sublethal mouse model of infection. In these studies, AG129 mice were administered 25 or 2.5 μg of prM13 or prM22 1 d prior to a lower dose of D2S20 virus (Fig. 5B). Whereas control animals administered PBS or 25 μg isotype control mAb showed 100% survival after DENV infection, the majority of mice receiving prM mAb succumbed to infection by day 6. To evaluate effects on viral burden caused by prM mAb–induced ADE, additional groups of AG129 mice were administered 25 μg of prM13, prM22, or irrelevant CHIKV-reactive mAb 1 d prior to sublethal DENV infection, followed by serum and organ harvest at day 4 after infection (Fig. 5 C–E). Virological analysis was performed by qRT-PCR. Compared with control mice that received PBS, increased viral burden was observed in the serum, liver, and spleen from mice administered prM22 and in the liver from mice administered prM13. Together, and as expected, these studies demonstrate that intact prM-reactive antibodies capable of interacting with Fcγ–receptors exacerbate disease in vivo.
Fig. 5.
Effect of prM-reactive mAbs on DENV2 infection in AG129 mice. (A) Groups of AG129 mice (n = 11, combined from three experiments) were administered 250 µg of the indicated prM mAb or CHIKV-specific isotype-matched control mAb (CHK-265) intraperitoneally 1 d prior to retro-orbital infection with a lethal dose (5 × 105 FFU) of DENV2 D2S20. Animals were monitored for survival. Treatment with prM mAbs, but not CHK-265, resulted in more rapid death compared with PBS control (P < 0.05 for prM12, prM13, or prM22 vs. PBS control, log-rank test). (B) Groups of AG129 mice (n = 6 to 8, combined from two experiments) were administered 25 or 2.5 µg of the indicated mAb intraperitoneally 1 d prior to retro-orbital infection with a sublethal dose (5 × 103 FFU) of DENV2 D2S20. Animals were monitored for survival. Mice receiving prM mAbs, but not CHK-265, displayed evidence of enhanced disease compared with PBS control (P < 0.005 for prM13 or prM22 vs. PBS control, log-rank test). (C–E) Groups of AG129 mice (n = 8) were administered 25 µg prM13, prM22, or CHK-265 mAb intraperitoneally 1 d prior to retro-orbital infection with a sublethal dose (5 × 103 FFU) of DENV2 D2S20. Virologic analysis was performed on the serum, spleen, and liver harvested 4 d postinfection by qRT-PCR. The resulting data were analyzed by one-way ANOVA; multiplicity-adjusted P values are denoted when significant (P < 0.05) for comparisons against PBS control animals. GE = genome equivalents. Images in panels A and B were created with BioRender.com.
N297Q-Modified prM Antibodies Confer Protection against DENV Infection in Mice.
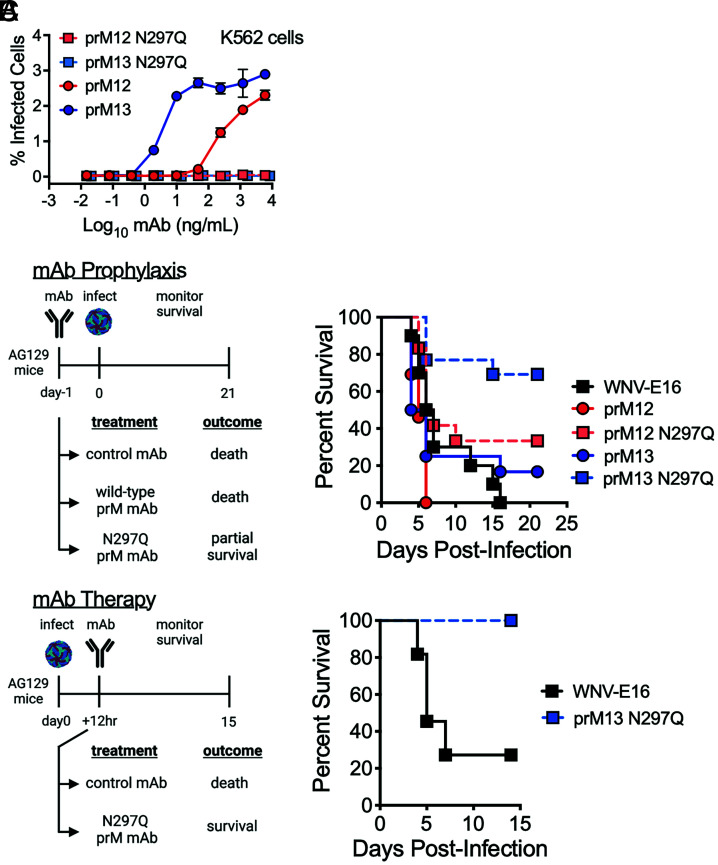
prM-reactive antibodies neutralize infection of partially mature virions that retain prM with a stoichiometry that allows binding by a neutralizing quantity of antibody. Virions decorated by a nonneutralizing quantity of antibody support ADE in vitro (SI Appendix, Figs. S3, S5, S8, and S9) and in vivo (Fig. 5). To uncouple the ability of prM antibodies to both neutralize and enhance DENV infection, we generated WT and aglycosyl (N297Q) Fc-modified chimeric (human Fc) mAb variants of prM12 and prM13 to use in mouse studies (Fig. 6). This mutation abrogates Fcγ receptor binding and inhibits the ability of an antibody to promote ADE (40, 41), as demonstrated in vitro with D2S20 virus (Fig. 6A) and 16681 RVPs (SI Appendix, Fig. S9). Additional experiments with 16681 RVPs confirmed that aglycosyl prM12 and prM13 mAbs maintained the ability to neutralize virus infection, although slightly less efficiently than the WT counterparts (SI Appendix, Fig. S9). To investigate whether prM antibodies can protect animals when ADE is not possible, AG129 mice were administered 250 μg of WT or N297Q prM chimeric mAbs 1 d prior to infection with a lethal dose of DENV2 D2S20 (Fig. 6B). As seen with the parental murine mAbs (Fig. 5A), WT prM12 and prM13 were not protective and did not prolong survival. In contrast, both N297Q-modified prM mAbs resulted in partial protection from DENV infection. That prM12 provides less protection than prM13 may be explained by the decreased reactivity to D2S20 due to variation at prM residue 28 (Fig. 6A and SI Appendix, Fig. S8). To rule out the possibility that direct neutralization of the prM+ challenge virus stock contributed to protection in this model, N297Q-modified prM13 was administered 12 h after challenge to allow virus infection and replication in the host (Fig. 6C). Therapeutic prM mAb administration was similarly protective, demonstrating that prM+ partially mature virions produced in vivo can contribute to the pathogenesis of primary DENV infection.
Fig. 6.
Effect of aglycosyl prM-reactive mAbs on DENV2 infection in AG129 mice. (A) WT or aglycosyl (N297Q) chimeric human Fc prM12 and prM13 mAbs were used in an in vitro ADE assay with the DENV2 D2S20 virus stock used to infect mice. After 24 h, infected cells were detected by intracellular staining with an E-specific antibody conjugated to Alexa Fluor 488. Error bars indicate the range of duplicate infections. (B) Groups of AG129 mice (n = 6 to 13, combined from two experiments) were administered 250 µg of the indicated WT or aglycosyl (N297Q) chimeric mAb intraperitoneally 1 d prior to retro-orbital inoculation with a lethal dose (1 × 104 FFU) of DENV2 D2S20. Survival was monitored. WNV-E16 is an isotype-matched control chimeric mAb. N297Q-modified mAbs demonstrated partial protection in comparison with their unmodified counterparts (P < 0.005 for prM12 vs. prM12 N297Q, and prM13 vs. prM13 N297Q comparisons, log-rank test). (C) Groups of AG129 mice (n = 11, combined from two experiments) were infected with a lethal dose (2 × 105 FFU) of DENV2 D2S20 via retro-orbital infection and 12 h later were administered 500 µg of aglycosyl human chimeric prM13 or an isotype control mAb. Survival was monitored. Therapeutic administration of prM13 N297Q was protective (P = 0.0005 for prM13 N297Q vs. isotype control, log-rank test). Images in panels B and C were created with BioRender.com.
Discussion
In this study, we isolated murine DENV prM-reactive mAbs to investigate the role of infectious prM+ partially mature virions in a mouse model of DENV pathogenesis. These prM mAbs exhibit considerable neutralizing activity that is governed by the efficiency of prM cleavage on infectious virions. Cryo-EM structures solved for two of the prM mAbs in complex with immature DENVs indicated distinct modes of engagement of an epitope on the prM molecule. Finally, AG129 mouse studies performed with N297Q prM mAb variants engineered to inhibit Fc–Fcγ receptor interactions resulted in protection from lethal DENV infection, demonstrating in vivo the ability of partially mature virions to contribute to pathogenesis independent of a role in ADE. The inability of aglycosyl antibodies to coordinate effector functions suggests that the protective effects of prM antibodies are due to their capacity to directly neutralize partially mature virus infection in vivo.
While structural studies demonstrate the presence of uncleaved prM on flavivirus virions (21, 22), these images cannot distinguish between infectious and noninfectious forms of the virus. In vitro manipulation of the extent of virus maturation impacts virus–antibody interactions and neutralization sensitivity (25, 33, 42), as well as cell entry and fusion (30, 43), providing considerable functional evidence that prM+ virions can be infectious. Prior studies of maturation state–sensitive E protein–reactive antibodies provided an indirect measure of the prM content of infectious virions as many epitopes on the E protein are poorly/nonaccessible on structurally mature virions. Similarly, neutralization by prM-reactive mAbs correlates inversely with the degree of prM cleavage on infectious virions. The nonneutralizable fraction of virions observed in our studies corresponds to the fraction of virions with little to no uncleaved prM, in accordance with a multiple-hit threshold model of neutralization (9). prM mAbs were surprisingly potent when only the sensitive fraction of virions was considered, with EC50 values of ~5 ng/mL for prM12, prM13, and prM22. These values are comparable with previously described potently neutralizing flavivirus E protein–specific mAbs (24, 41, 44). Prior reports that prM mAbs have limited neutralization activity may be explained by the maturation state of virus stocks used in experiments and cell-type–dependent patterns of neutralization arising from a reduced capacity of prM+ virions to bind some cell types when compared with structurally mature virions (6, 36, 45).
The binding of two of the three murine prM-reactive mAbs studied (prM12 and prM22) was sensitive to alanine substitution at prM position K26. Consistent with these results, a prior study of human prM-reactive mAbs isolated from DENV-infected or vaccinated subjects identified a single immunodominant antigenic site for antibody binding that often included residue K26 (36). In a separate study, BALB/c mice immunized with linear peptides spanning the prM protein elicited an antibody response only to a peptide spanning amino acids 19 to 34 (46). Thus, a similar epitope on prM may be targeted by both humans and mice, of which residue K26 contributes to the energetics of antibody binding. In contrast, the repertoire of E protein–reactive antibodies differs among mice and humans (47). The relatively small size of prM and the accessible display of this region in the context of prM–E heterodimers may contribute to the similar targeting of human and murine prM mAbs.
Neutralization assays performed with our DENV2 prM alanine variants also identified a minor role of prM residue E28 in the binding of mAb prM12. Unlike K26, which is highly conserved among all DENV serotypes, the amino acid incorporated at position 28 is variable among DENV strains. Because prM12 was reactive against serotypes other than DENV2 in binding studies, K26 likely represents the critical contact residue for this mAb. Neutralization studies performed with mAb prM13 were not impacted by any of the DENV2 RVP alanine mutants in our panel, including residue L3, which was previously identified as a common secondary target of human prM-reactive mAbs (36). Additional studies will be required to determine the critical contact residue(s) for this antibody.
Cryo-EM structures solved for Fab fragments of prM12 and prM13 bound to immature DENV2 resolved epitopes including both shared and distinct amino acid residues. The limited resolution of these reconstructions prevented the identification of prM residues that contribute to the energetics of antibody binding with confidence. The epitope for prM12 was predicted to include residues 4 to 11, 25 to 29, and 32, which contains the K26 and E28 residues identified by alanine scanning mutagenesis. The epitope for prM13 was predicted by cryo-EM to include residues 3, 7 to 10, 25 to 30, and 71, yet neither K26 nor L3 was identified as a critical contact residue for this mAb in biochemical or neutralization assays. The cryo-EM structure of Fab fragments of a human-derived non-K26 prM-reactive mAb, 1H10, has been solved in complex with immature DENV3 virions to 12 Å (48). Binding studies indicated that this particular mAb was sensitive to mutation at prM residues L3 and S5, but not K26, despite the inclusion of all three of these residues in the 1H10 structure-predicted epitope (36, 48). Thus, identifying contact residues critical for prM antibody binding may not be predicted by these prM–Fab structures alone.
DENV-infected humans elicit a significant anti-prM antibody response ranging from ~35 to 60% of the total virion-reactive antibody response (6, 49). Because DENV prM-reactive antibodies are generally cross-reactive against multiple serotypes and are reported to have limited capacity to neutralize virus, they have been hypothesized to contribute little to protection and instead promote DENV pathogenesis via ADE. In immunocompromised mouse models of DENV infection, the passive transfer of prM-reactive antibodies results in enhanced disease (37, 38). prM-reactive antibodies may enhance infection of otherwise poorly infectious, immature populations of virions in vitro and in vivo (50, 51). Our in vitro neutralization studies demonstrate that prM antibodies are potently neutralizing only against partially mature virions that retain high enough levels of uncleaved prM. We and others have shown this to be true for E protein–specific mAbs that bind conformationally restricted cryptic epitopes as well (20, 22, 25). Thus, the prM content of individual virions tunes the proportion of virions that is directly sensitive to neutralization by prM-reactive antibodies and those with the potential to contribute to severe disease outcomes by infecting Fcγ–receptor–expressing cells via ADE (20). Relatively efficient cleavage of prM on virions in vivo may limit the contribution of intact prM antibodies to protection; virions that retain prM at a level below the stoichiometric requirements for neutralization will be enhanced even at very high concentrations of an otherwise neutralizing prM antibody.
Flavivirus-infected cells in vitro release infectious virions that retain uncleaved prM in unknown proportions (18–20). Little is known regarding the prM content of virus produced in vivo. The one existing study of DENV1 isolated directly from human plasma from six subjects revealed structurally mature forms of the virus predominate (27). Whether the prM content of virus isolated from plasma is representative of virus found in tissues is not known, nor the degree to which virion maturation efficiency varies among primary DENV strains. Here, we demonstrate that the passive transfer of aglycosyl variants of prM-specific mAbs conferred significant protection in a lethal mouse model of DENV infection, even when provided therapeutically. Because these antibodies cannot bind Fcγ–receptors or C1q, their antiviral activity likely is restricted to mechanisms that involve direct neutralization of virions. While additional studies will be required to characterize the structural heterogeneity of DENV in vivo, our data provide compelling evidence that partially mature virions are produced in vivo and can also contribute to pathogenesis independent of their potential to contribute to ADE. Because the prM content of virus particles impacts the biology, antigenicity, and immunogenicity of flaviviruses, methods to probe the efficiency of virion maturation of infectious or attenuated virus will have significant utility in future studies.
Materials and Methods
Study Approval.
All mouse study protocols were approved and performed according to the guidelines of the Washington University School of Medicine Animal Safety Committee (assurance number A3381-01). Ifnar1−/− mice were originally obtained from J. Sprent (The Scripps Research Institute), backcrossed ten generations on a C57BL/6 background, and used for mAb generation. Ifnar1−/− Ifngr−/− AG129 mice (Marshall BioResources) were used in challenge studies.
Cell Lines.
Vero and HEK-293T/17 cells were cultured in a high-glucose formulation of Dulbecco’s modified Eagle medium (DMEM) containing GlutaMax and HEPES (Thermo Fisher Scientific) that was supplemented with 7% fetal bovine serum (FBS) (Thermo Fisher Scientific) and 100 U/mL penicillin–streptomycin (Thermo Fisher Scientific). HEK-293T-FIRB cells (33) that stably express the human furin protease were maintained in complete DMEM described above with the addition of 5 to 10 μg/mL Blasticidin S HCl for continued selection (Thermo Fisher Scientific). Raji B lymphoblast cells engineered to stably express the flavivirus attachment factor DC-SIGNR (Raji-DCSIGNR) (30) were cultured in RPMI 1640 medium containing GlutaMax (Thermo Fisher Scientific) and supplemented with 7% FBS and 100 U/mL penicillin-streptomycin. The A. albopictus mosquito cell line C6/36 (ATCC) was cultured in minimum essential medium (MEM) containing glutamine and supplemented with 10% FBS, 25 mM HEPES, and 1X nonessential amino acids. All cell lines were maintained in the presence of 5 to 7% CO2, mammalian cells at 37 °C, and insect cells at 30 °C.
Viruses.
For hybridoma screening and binding studies, Vero cells were infected with DENV1 strain 16007, DENV2 strain New Guinea C, DENV3 strain 16652, and DENV4 strain 1036. For in vivo studies, the mouse-adapted strain DENV2 D2S20 was used (39) (a gift of S. Shresta, La Jolla, CA). Viruses were propagated in C6/36 cells or Vero cells after inoculating at a multiplicity of infection (MOI) of 0.01 and incubating at 37 °C for 96 h. DENV stocks were titrated by a focus-forming assay by infecting Vero cell monolayers with virus for 1 h at 37 °C followed by overlaying with 1% (w/v) methylcellulose in MEM. After 40 h, cells were fixed with 1% paraformaldehyde (PFA), followed by sequential incubations with the flavivirus cross-reactive mouse mAb E60 (500 ng/mL) (41) and horseradish peroxidase–conjugated goat anti-mouse IgG (1:5,000) (Sigma) diluted in PBS supplemented with 0.1% (w/v) saponin and 0.1% bovine serum albumin. Foci were detected by addition of TrueBlue peroxidase substrate (SeraCare) and counted with a CTL Immunospot instrument (Cellular Technology Limited). DENV2 strain 16681 used for cryo-EM imaging of virus–Fab complexes was produced in C6/36 insect cells; the virus purification process is described in detail below.
Recombinant pr Production and Purification.
A recombinant form of pr, encoding residues 1 to 86 of the prM protein from DENV2 strain New Guinea C, was produced as previously described (52). Briefly, a plasmid encoding the pr sequence with a C-terminal tandem Strep tag was transfected into 293T cells. Culture medium was collected after 48 and 72 h, and the pr protein was purified by affinity chromatography on a 1-mL Strep-Tactin Sepharose column (IBA BioTAGnology) and by gel filtration using a Sephadex G-75 column (GE Healthcare Life Sciences).
Monoclonal Antibody Production.
To generate anti-pr mAbs, Ifnar1−/− C57BL/6 mice were inoculated with 105 PFU of DENV2 strain New Guinea C via an intraperitoneal route and challenged a second time 3 wk later with the same strain and dose. Several weeks later, mice were boosted with 25 μg of recombinant pr protein complexed with Freund’s incomplete adjuvant using pr that was expressed in 293T cells and purified as described above. Three weeks later, mice were immunized again with 25 μg of purified pr in PBS as a final intravenous boost. Three days later, splenocytes were fused to P3X63Ag8.6.5.3 myeloma cells using polyethylene glycol 1500. Hybridomas producing anti-pr mAbs were subcloned by limiting dilution, isotyped using an enzyme-linked immunosorbent assay kit (Southern Biotech), and purified by protein A or protein G immunoaffinity chromatography. For screening experiments, Vero cells were infected with DENV1, DENV2, DENV3, or DENV4, fixed with 4% PFA, permeabilized with 0.1% (w/v) of saponin, and stained with anti-prM mAbs or isotype controls and processed by flow cytometry (SI Appendix, Table S1).
Chimerization of Anti-prM mAbs.
Heavy- and light-chain RNA was isolated from hybridoma cells after guanidinium thiocyanate and phenol–chloroform extraction and converted to cDNA by reverse transcription. The VH and VL segments were amplified by PCR using the 5′ rapid amplification of cDNA ends (RACE) system (Invitrogen) as described previously (41) and inserted into the plasmid pCR2.1-TOPO using the TopoTA kit (Invitrogen). The mouse VH was joined to a human Cγ1 constant region and an Ig leader sequence and inserted into pCI-neo for mammalian expression. The mouse VL was joined to a human Cκ segment and an Ig leader sequence and also cloned into pCI-neo for mammalian expression of chimeric human Fc mAbs. Site-directed mutagenesis was performed to change heavy-chain residue 297 from asparagine to glutamine (N297Q) to eliminate the single glycosylation site on the γ1 Fc.
Reporter Virus Particle Production.
DENV2 and ZIKV RVPs were produced as described previously (28, 29, 53). Briefly, HEK-293T cells were transfected with two plasmids, one encoding a subgenomic West Nile virus (WNV) lineage II replicon that expresses GFP, and the other that encodes the C–prM–E structural gene cassette from DENV2 strain 16681 or ZIKV strain H/PF/2013 at a ratio of 1:3 by mass using Lipofectamine 3000 reagent according to the manufacturer’s instructions (Thermo Fisher Scientific). Transfected cells were incubated at 30 °C in a low-glucose formulation of DMEM containing GlutaMax and 25 mM HEPES (Thermo Fisher Scientific) supplemented with 7% FBS and 100 U/mL PS. Standard RVP preparations were harvested on days 3 to 6 posttransfection, clarified through a 0.22-μM Millex-GS Syringe Filter (Millipore), and frozen at −80 °C until further use. To generate a panel of DENV prM alanine variants, the DENV2 16681 C–prM–E plasmid was used as a template for site-directed mutagenesis using the Quikchange II Site-Directed Mutagenesis Kit (Agilent Technologies). Primers were designed using the Agilent QuikChange Primer Design tool (agilent.com/store/primerDesignProgram.jsp). Mutated plasmids were fully sequenced prior to use in transfection studies. All plasmids were propagated at 30 °C in MAX Efficiency Stbl2 competent bacteria (Thermo Fisher Scientific). To determine the infectious titer of DENV2 and ZIKV RVP preparations, two-fold serial dilutions of virus stocks were used to infect Raji-DCSIGNR cells in duplicate. After 48 h of incubation at 37 °C, cells were fixed with 2% PFA (Electron Microscopy Sciences) and analyzed on a BD Celesta flow cytometer with a High Throughput Sampler (HTS) attachment. The percentage of GFP-positive infected cells was determined using FACSDiva software version 8.0.1. In subsequent neutralization and ADE experiments, virus was sufficiently diluted to ensure antibody excess at informative points of the dose–response curve.
Manipulating the Maturation State of DENV RVPs.
To produce a more homogeneous population of RVPs with reduced uncleaved prM (referred to as DENV furin), RVPs were created by transfection of a HEK-293T cell line that expresses very high levels of human furin (HEK-293T-FIRB) (33). Transfected HEK-293T-FIRB cells were incubated at 30 °C and DENV furin RVPs harvested on days 3 to 6 posttransfection, clarified through a 0.22-μM filter, and frozen at −80 °C until further use. RVPs that retain significant uncleaved prM (DENV prM+) were produced including an inhibitor of furin-like protease activity during RVP production in WT HEK-293T cells. Briefly, transfection of replicon and structural gene plasmids was performed as described above. At 4 h after transfection, the media of transfected cells were exchanged with media containing 25 μM furin inhibitor Dec-RVKR-CMK (Enzo Life Sciences). Transfected cells were incubated at 37 °C and DENV prM+ RVPs harvested on days 1 to 2 posttransfection, clarified through a 0.22-μM filter, and frozen at −80 °C until further use. The efficiency of virion maturation was characterized functionally via neutralization studies with the maturation state–sensitive E DII fusion loop (FL)–reactive mAb E60 (25).
DENV Structural Protein Immunoblotting.
DENV2 prM was detected in transfected cell lysates by western blotting. 293T cells transfected to produce DENV2 16681 WT, prM K26A or prM V31A variant RVPs or WNVII replicon-only negative control, were pelleted and resuspended in RIPA cell lysis buffer (Thermo Fisher) supplemented with protease inhibitors (Roche) and 15 units/mL recombinant DNase I (Millipore Sigma) and incubated on ice for 30 min. Lysed cells were clarified at 10,000 × g for 15 min at 4 °C, and the supernatant was transferred to a new tube. Lysates were run on four identical NuPAGE 4 to 12% Bis-Tris gels (Thermo Fisher Scientific) under nonreducing conditions, transferred to nitrocellulose membranes, and blocked with Odyssey blocking buffer in PBS (Li-Cor). Following a wash with PBS + 0.1% Tween-20 (PBS-T), individual membranes were incubated with either 3 μg/mL mouse mAb prM12, prM13, or prM22, or 6 μg/mL of the pan-flavivirus E protein–reactive mouse mAb 4G2 for 1 h. All membranes were simultaneously incubated with 1:500 rabbit polyclonal anti-HSP 90α/β IgG (Santa Cruz Biotechnology). After primary antibody incubation, membranes were washed three times with PBS-T. All four membranes were then incubated with two secondary antibodies for 1 h: 1) goat monoclonal anti-mouse IgG IRDye 800CW (Li-Cor) to detect the prM or E-specific mouse mAbs and 2) goat monoclonal anti-rabbit IgG IRDye 680RD (Li-Cor) to detect the rabbit anti-HSP 90 control mAb. Following five additional PBS-T washes, images were read using an Odyssey CLx infrared imaging system (Li-Cor), with detection of DENV prM and E signals in the 800-nm channel and cellular HSP 90 housekeeping gene signal in the 700-nm channel.
Mechanism of Neutralization RVP Binding Assay.
Standard DENV2 16681 RVPs were diluted and incubated in the presence or absence of 30 μg/mL prM12, prM13, prM22, or DV2-77 mAb for 1 h at 37 °C to allow binding to reach equilibrium. Half of the resulting immune complexes was used to infect quadruplicate wells of Raji-DCSIGNR cells, incubated at 37 °C for 48 h, and analyzed by flow cytometry to determine the percentage of GFP-positive infected cells. The other half was added to quadruplicate wells of Raji-DCSIGNR cells that had been pretreated for 30 min at 37 °C with 30 mM NH4Cl to allow cell entry but inhibit low pH–mediated fusion and subsequent RNA replication. NH4Cl was supplemented to maintain a concentration of 30 mM. After a 3-h incubation at 37 °C, cells were washed twice with PBS to remove unbound virus, after which cell-associated viral RNA was extracted using the Qiagen RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions, including the QIAshredder (Qiagen) for cell disruption and the optional on-column DNase treatment (Qiagen). Viral RNA remaining after incubation in the presence or absence of mAb was quantified by qRT-PCR in triplicate per sample. Briefly, WNVII replicon RNA packaged into DENV RVPs was quantified using the SuperScript III Platinum One-Step qRT-PCR Kit (Thermo Fisher Scientific), with primers and probe specific for the WNVII 3′ UTR as previously described (54). Each sample was run with and without reverse transcriptase to rule out contaminating plasmid DNA from the 293T transfection, and the results were normalized to a standard curve generated using the WNVII replicon plasmid. Cycling was performed on an Applied Biosystems QuantStudio 3 Real-Time PCR System (QuantStudio Design and Analysis Software v1.5.1) with the following conditions: reverse transcription, 25 °C for 10 min, 48 °C for 30 min, and 95 °C for 5 min, and PCR, 40× cycles of 95 °C for 15 s and 60 °C for 1 min. Using values obtained in the absence of mAb as a reference, the percentage remaining RNA and percentage remaining infectivity were calculated after incubation with each mAb. Equal values for infectivity and RNA indicate a mechanism of neutralization primarily dependent on blocking attachment. DV2-77 is an E protein–reactive mAb previously shown to neutralize virus at a postattachment step (viral RNA is detected despite near-complete neutralization) used here as a control (55).
Neutralization and ADE Assays.
RVP neutralization and ADE experiments were performed as previously described (25, 33, 56). Briefly, serial dilutions of purified mAbs or hybridoma supernatants were incubated with DENV or ZIKV RVPs for 1 h at 37 °C to allow for steady-state binding. For experiments with C1q (Complement Technologies), 50 µg/mL was included during the 1-h incubation. Thereafter, immune complexes were used to infect target cells in duplicate technical replicates and incubated for 48 h at 37 °C. Neutralization studies were performed using Raji-DCSIGNR or Vero cells, whereas ADE experiments were performed with Fcγ receptor–expressing K562 cells. Infection of K562 cells occurs through Fcγ receptor–mediated uptake of antibody–virus immune complexes; in the absence of antibody, these cells exhibit minimal infection due to an inability to efficiently bind virus directly. Following infection with RVP–antibody immune complexes for 48 h at 37 °C, Raji-DCSIGNR, K562, or trypsinized Vero cells were fixed with 2% PFA (Electron Microscopy Sciences) and analyzed on a BD Celesta cytometer with a HTS attachment. The percentage of GFP-positive infected cells was determined using FACSDiva software version 8.0.1. For neutralization assays, dose–response neutralization curves were analyzed by nonlinear regression analysis, with a variable slope, to estimate the concentration of antibody that results in half-maximal infectivity (EC50). To account for the presence of nonneutralizable fractions of virus observed with prM-reactive mAbs, curve fitting was not constrained to 0. For representative dose–response curves depicted in figures, data were normalized to RVP infectivity observed in the absence of antibody.
ADE assays with infectious D2S20 virus were performed similar to RVP assays, with some modifications (53, 57). At 24 h after infection, cells were pelleted, washed in PBS, and incubated for 20 min in 100 μL Fix/Perm Solution (BD Biosciences). After the fixation step, cells were pelleted, washed twice in Perm/Wash Solution (BD Biosciences), and incubated for 30 min at 4 °C with 50 μL of the flavivirus cross-reactive anti-E mAb E60 conjugated to Alexa Fluor 488. Cells were pelleted again, washed twice with Perm/Wash solution, and resuspended in 150 μL PBS + 2% PFA. Infection was monitored by flow cytometry.
Production and Purification of Immature DENV2 Strain 16681 for Cryo-EM.
C6/36 A. albopictus cells were used for the propagation of virus. Cells were grown in 2- and 10-chambered cell stacks (Corning) with cell growth areas of 1,272 and 6,360 cm2, respectively. The cells were infected at a high multiplicity of infection (MOI = 10) when they achieved ~90% confluence. At 48 h after infection (t = 0), the media were replaced with media supplemented with 30 mM NH4Cl. NH4Cl inhibits the structural transformation of the DENV virion in the trans-Golgi network and furin cleavage of prM, leading to accumulation of unprocessed immature virus in the culture media (58). At t = 12 and t = 36 h, the culture supernatant was again replaced with fresh media containing NH4Cl. The culture supernatant (volume ~ 1,500 mL), harvested at t = 60 h, was used for the purification of immature virus as published before (59) and described below. The supernatant was clarified first by low-speed centrifugation (6,371 g for 15 min at 4 °C) to eliminate cell debris. This was followed by sterile filtration through a 0.22-µM polyethersulfone membrane–equipped Steritop bottle top filter assembly (Millipore Sigma). The filtered supernatant was precipitated by 8% polyethylene glycol 8000 overnight at 4 °C with gentle stirring. The precipitated virus was pelleted by centrifugation (15,971 g for 50 min at 4 °C). The pellet was resuspended in NT buffer containing 20 mM Tris and 120 mM NaCl, pH 8.0, and subjected to sucrose cushion (24%) centrifugation (175,587 × g for 2 h at 4 °C). The pellet was again resuspended in NT buffer and loaded on the top of a discontinuous potassium tartrate (35 to 10%)–glycerol (26.3 to 7.5%) gradient with 5% potassium tartrate–3.8% glycerol density interval between the six layers. After centrifugation at 175,587 × g for 2 h at 4 °C, the virus band located at 20% potassium tartrate–15% glycerol was extracted by puncturing the tube with a 22-gauge needle (BD Biosciences). The virus was concentrated in NT buffer using Amicon Ultra-4 Centrifugal Filter Unit with 100-kDa cutoff regenerated cellulose membrane (Millipore Sigma). The quality and the concentration of the virus sample were analyzed by SDS–PAGE. The amount of E glycoprotein was predicted using a standard curve generated with BSA. The concentration of virus was estimated based on the amount of E glycoprotein (~1 µg E per 6 × 1010 virions).
Generation and Cryo-Freezing of Virus–Fab Complexes.
Fab fragments of prM12 and prM13 were generated with Pierce Fab Preparation Kit (Thermo Scientific) using 500 µg of the respective antibody. The Fab fragments were concentrated in NT buffer using Amicon Ultra-4 Centrifugal Filter Unit with 10-kDa cutoff membrane (Millipore Sigma). Complexes for cryo-EM were prepared by incubating immature DENV with saturating concentrations (~1.5- to 2-fold excess) of prM12 or prM13 Fabs in NT buffer (pH 8.0) overnight at 4 °C. Samples were plunge-frozen in liquid ethane using a Cryoplunge 3 System (Gatan). Briefly, liquid nitrogen was used to liquefy ethane at −190 °C. A 2.5 µL volume of sample was spotted on lacey carbon grids (Ultrathin Carbon Film on Lacey Carbon Support film, 400 mesh, Copper; Ted Pella, Inc.) and blotted with Whatman grade 1, 200-mm circle filter paper (GE Healthcare Life Sciences) for 2.5 s. Blotting air pressure setting used was 100 psi.
Cryo-EM Single-Particle Reconstruction.
Micrographs were acquired on a Titan Krios electron microscope (Thermo Fisher Scientific) equipped with an energy filter and a Gatan K2 Summit direct electron detector at a nominal magnification of 81,000×. Movies were collected in superresolution mode, resulting in an effective pixel size of 0.865Å. A total of 927 and 991 micrographs were collected for prM12 and prM13 complexes, respectively, using Leginon software package (60) and processed using the Appion pipeline (61). Each micrograph was corrected for beam-induced sample motion using MotionCor2 (62), and CTF parameters for each were estimated using CTFFIND4 (63). Particles were picked in an automated manner using a template-based approach employed by FindEM (64). Particles were then subjected to nonreference, 2D classification using RELION (65). Classes showing virus bound to Fab were manually selected and combined resulting in 5,881 and 5,458 particles for prM12 and prM13 complexes, respectively. Single-particle reconstruction was performed assuming icosahedral symmetry and following the “gold standard” method. The generation of the initial model and the iterative refinement of orientation and centers until convergence were done using jspr (66, 67). Upon correcting for astigmatism, defocus and elliptical distortion and soft masking of the disordered core, the final cryo-EM models for prM12 and prM13 virus complexes had an average resolution of 10.2 Å and 9.8 Å, respectively, using the 0.143 Fourier shell correlation criterion (68). These parameters and statistics are summarized in SI Appendix, Table S2.
Fitting Atomic or Homology Models into Cryo-EM Maps.
The pseudoatomic model was created by fitting the structures of DENV glycoproteins (prM–E), as well as the variable and constant domains of prM12 or prM13 Fab into the density corresponding to the three independent positions (termed P1, P2, and P3) of the Fab-bound trimeric spike of the immature DENV2. The crystal structure of the soluble DENV2 prM–E heterodimers (PDB ID: 3C6E) was used for model building (13). The densities for the transmembrane helices of prM–E were not well resolved in the cryo-EM maps, so this region was not included in the atomic model. As the atomic structure of prM12 or prM13 Fab was unavailable, the model used for fitting was created as follows: The homology models of variable light (VL) and variable heavy (VH) domains of Fab were generated based on their amino acid sequence using I-TASSER (69, 70) (SI Appendix, Fig. S7). The homology model of the variable domain was then pieced together with the crystal structure of the constant domain, derived from ZV-67, which belongs to murine IgG2c isotype similar to prM12 and prM13 (PDB ID: 5KVG) (71). The structures of soluble prM–E heterodimers and Fab were fitted sequentially into each of the three positions in the trimeric spike using the “Fit in Map” function in UCSF Chimera (72). The program phenix.map_box was used to extract density within 5 Å of the prM–E–Fab fitted into the P1, P2, and P3 positions of the trimeric spike to create a portion of the map for further model refinement. The fit was optimized by one at a time, single structure fitting using colors (73) followed by simultaneous, multibody refinement using the collage tool (74) in situs 3.0 package (75).
Map Interpretation: Epitope Prediction, Occupancy Determination, and Angle of Approach Measurements.
At the map resolution of ~10 Å, the density for prM and CDR loops of the antibodies was not clearly discernable. Furthermore, as the side chain interactions cannot be mapped, it was not possible to pinpoint the interacting residues (epitope) with certainty. The interacting interface was predicted by visual inspection and by identifying the residues of prM protein located within 5 Å of prM12/13 Fab using PYMOL. The program Radial Interpretation of Viral Electron density Maps (RIVEM) was used to display the “predicted” antibody epitope on the icosahedral DENV surface (76). The conservation of the predicted epitopes across different mosquito-transmitted flaviviruses was evaluated using ESPript3.0 (77). The prM protein sequences used for analysis are as follows: DENV2 (strain 16681, GenBank accession: NC_001474), DENV1 (strain SG/07K3640DK1/2008, GenBank accession: GQ398255), DENV3 (strain D3/SG/05K863DK1/2005, GenBank accession: EU081190), DENV4 (strain SG/06K2270DK1/2005, GenBank accession: GQ398256), Japanese Encephalitis virus (JEV) (strain P3, GenBank accession: U47032), WNV (strain NY99, GenBank accession: DQ211652), yellow fever virus (YFV) (strain Asibi, GenBank accession: AY640589), and ZIKV (strain H/PF/2013, GenBank accession: KJ776791). The program EMfit was used to determine the occupancy of Fab on the three independent positions in the trimer as described earlier (78, 79). The quality of fit factor sumf was calculated for Fab and prM–E (SI Appendix, Tables S3 and S4). The sumf represents the average density around all fitted atoms normalized to the maximum density in the map that is assigned a value of 100. Comparable sumf values for Fab at the three independent positions (P1, P2, and P3) indicate equivalent occupancies. (As expected, the sumf for prM–E was similar for all three positions.) The angle of approach for the Fabs was calculated by determining the center of mass of E ectodomain (X), prM ectodomain (Y), and Fab (Z). The angle of approach is defined as angle XYZ. The figures were prepared using the programs UCSF Chimera (72) and UCSF ChimeraX (80, 81).
Mouse Challenge Experiments.
Groups of AG129 (Ifnar1−/− Ifngr−/−) mice were administered prM-specific mAb, isotype control mAb, or PBS intraperitoneally 1 d prior to or, where stated, 12 h after retro-orbital infection with a lethal or sublethal dose of the mouse-adapted DENV2 strain D2S20. Antibody amount and virus dose are detailed in the text and figure legends for individual experiments. Animals were either monitored for survival or in some experiments killed at day 4 for serum and organ harvest. Virological analysis was performed by qRT-PCR. RNA was extracted from serum and tissues (spleen and liver) using the Qiagen RNeasy Mini Kit (Qiagen). Viral RNA levels were measured using the QuantaBio qScript one-step qRT-PCR kit on a CFX96 Touch real-time PCR detection system (Bio-Rad CFX Manager 3.1) and normalized to volume for serum and to 18S RNA levels for spleens and livers. Primers and probe have been previously described (82). A standard curve was generated using serial dilutions of an infectious stock of DENV2 D2S20.
Statistical Analysis.
Data were analyzed using GraphPad Prism, version 8.1.2 (GraphPad Software). Statistical tests are indicated in each of the figure legends.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This research was funded by the NIH grant R01AI073755 to R.J.K. and M.S.D, NIH contracts HHSN272201400018C to M.S.D., R01 AI011219 to R.J.K., and R01AI075647 to M.K., and the NIH Division of Intramural Research to T.C.P. We thank Sujan Shresta for her gift of the D2S20 virus.
Author contributions
K.A.D., D.S., A.Z., M.K., R.J.K., M.S.D., and T.C.P. designed research; K.A.D., D.S., S.D.S., L.A.V., R.E.C., S.M., B.M.W., J.G., M.A., B.L., S.S.-P., M.S., A.S.M., T.K., A.Z., and M.K. performed research; A.Z., S.K., and M.K. contributed new reagents/analytic tools; K.A.D., D.S., S.D.S., L.A.V., R.E.C., S.M., B.M.W., J.G., M.A., B.L., S.S.-P., M.S., A.S.M., T.K., A.Z., M.K., R.J.K., M.S.D., and T.C.P. analyzed data; and K.A.D., D.S., R.J.K., M.S.D., and T.C.P. wrote the paper.
Competing interest statement
The authors have organizational affiliations to disclose, M.S.D. is a consultant for Inbios, Vir Biotechnology, Moderna, Novavax, Senda Biosciences and Immunome. Yes, the authors have research support to disclose. The Diamond laboratory has received unrelated funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Richard J. Kuhn, Email: kuhnr@purdue.edu.
Michael S. Diamond, Email: diamond@borcim.wustl.edu.
Theodore C. Pierson, Email: piersontc@mail.nih.gov.
Data, Materials, and Software Availability
All data supporting the findings of this study are available within the paper. The cryo-EM density maps of immature DENV in complex with prM12 and prM13 Fabs are available at the Electron Microscopy Data Bank (EMDB) with the accession codes EMD-29020 and EMD-29021, respectively. The fitted pseudoatomic models are available at the Protein Data Bank (PDB) with the accession codes PDB 8FE3 and PDB 8FE4 for prM12 and prM13 complexes, respectively.
Supporting Information
References
- 1.Gould E. A., Solomon T., Pathogenic flaviviruses. Lancet 371, 500–509 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Pierson T. C., Diamond M. S., The continued threat of emerging flaviviruses. Nat. Microbiol. 5, 796–812 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng Z., Zhan J., Chen L., Chen H., Cheng S., Global, regional, and national dengue burden from 1990 to 2017: A systematic analysis based on the global burden of disease study 2017. EClin. Med. 32, 100712 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization, Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control: New Edition, World Health Organization, Ed., (World Health Organization, Geneva, 2009). [PubMed] [Google Scholar]
- 5.Guzman M. G., Alvarez M., Halstead S. B., Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: An historical perspective and role of antibody-dependent enhancement of infection. Arch. Virol. 158, 1445–1459 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Dejnirattisai W., et al. , Cross-reacting antibodies enhance dengue virus infection in humans. Science 328, 745–748 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katzelnick L. C., et al. , Antibody-dependent enhancement of severe dengue disease in humans. Science 358, 929–932 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halstead S. B., Dengue antibody-dependent enhancement: Knowns and unknowns. Microbiol. Spectr. 2 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Pierson T. C., et al. , The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host. Microbe. 1, 135–145 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rey F. A., Heinz F. X., Mandl C., Kunz C., Harrison S. C., The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375, 291–298 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Hasan S. S., Sevvana M., Kuhn R. J., Rossmann M. G., Structural biology of Zika virus and other flaviviruses. Nat. Struct. Mol. Biol. 25, 13–20 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Rey F. A., Stiasny K., Vaney M. C., Dellarole M., Heinz F. X., The bright and the dark side of human antibody responses to flaviviruses: Lessons for vaccine design. EMBO Rep. 19, 206–224 (2017), 10.15252/embr.201745302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L., et al. , The flavivirus precursor membrane-envelope protein complex: Structure and maturation. Science 319, 1830–1834 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Yu I. M., et al. , Association of the pr peptides with dengue virus at acidic pH blocks membrane fusion. J. Virol. 83, 12101–12107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn R. J., et al. , Structure of dengue virus: Implications for flavivirus organization, maturation, and fusion. Cell 108, 717–725 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X., et al. , Cryo-EM structure of the mature dengue virus at 3.5-A resolution. Nat. Struct. Mol. Biol. 20, 105–110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elshuber S., Allison S. L., Heinz F. X., Mandl C. W., Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J. General Virol. 84, 183–191 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Junjhon J., et al. , Influence of pr-M cleavage on the heterogeneity of extracellular dengue virus particles. J. Virol. 84, 8353–8358 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keelapang P., et al. , Alterations of pr-M cleavage and virus export in pr-M junction chimeric dengue viruses. J. Virol. 78, 2367–2381 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierson T. C., Diamond M. S., Degrees of maturity: The complex structure and biology of flaviviruses. Curr. Opin. Virol. 2, 168–175 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plevka P., et al. , Maturation of flaviviruses starts from one or more icosahedrally independent nucleation centres. EMBO Rep. 12, 602–606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cherrier M. V., et al. , Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody. EMBO J. 28, 3269–3276 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodenhuis-Zybert I. A., Wilschut J., Smit J. M., Partial maturation: An immune-evasion strategy of dengue virus? Trends Microbiol. 19, 248–254 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Dejnirattisai W., et al. , A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 16, 170–177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson S., et al. , Maturation of West Nile virus modulates sensitivity to antibody-mediated neutralization. PLoS Pathog. 4, e1000060 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukherjee S., Lin T. Y., Dowd K. A., Manhart C. J., Pierson T. C., The infectivity of prM-containing partially mature West Nile virus does not require the activity of cellular furin-like proteases. J. Virol. 85, 12067–12072 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raut R., et al. , Dengue type 1 viruses circulating in humans are highly infectious and poorly neutralized by human antibodies. Proc. Natl. Acad. Sci. U.S.A. 116, 227–232 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dowd K. A., DeMaso C. R., Pierson T. C., Genotypic differences in Dengue Virus neutralization are explained by a single amino acid mutation that modulates virus breathing. MBio 6, e01559-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ansarah-Sobrinho C., Nelson S., Jost C. A., Whitehead S. S., Pierson T. C., Temperature-dependent production of pseudoinfectious dengue reporter virus particles by complementation. Virology 381, 67–74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis C. W., et al. , West Nile virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection. J. Virol. 80, 1290–1301 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierson T. C., et al. , A rapid and quantitative assay for measuring antibody-mediated neutralization of West Nile virus infection. Virology 346, 53–65 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Mehlhop E., et al. , Complement protein C1q reduces the stoichiometric threshold for antibody-mediated neutralization of West Nile virus. Cell Host. Microbe. 6, 381–391 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukherjee S., et al. , Mechanism and significance of cell type-dependent neutralization of flaviviruses. J. Virol. 88, 7210–7220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaufman B. M., et al. , Monoclonal antibodies for dengue virus prM glycoprotein protect mice against lethal dengue infection. Am. J. Trop. Med. Hyg. 41, 576–580 (1989). [DOI] [PubMed] [Google Scholar]
- 35.Mehlhop E., et al. , Complement protein C1q inhibits antibody-dependent enhancement of flavivirus infection in an IgG subclass-specific manner. Cell Host. Microbe. 2, 417–426 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith S. A., et al. , Dengue Virus prM-specific human monoclonal antibodies with virus replication-enhancing properties recognize a single immunodominant antigenic site. J. Virol. 90, 780–789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M., et al. , Anti-idiotypic antibodies specific to prm monoantibody prevent antibody dependent enhancement of dengue virus infection. Front. Cell Infect. Microbiol. 7, 157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zellweger R. M., Prestwood T. R., Shresta S., Enhanced infection of liver sinusoidal endothelial cells in a mouse model of antibody-induced severe dengue disease. Cell Host. Microbe. 7, 128–139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makhluf H., et al. , Tracking the evolution of dengue virus strains D2S10 and D2S20 by 454 pyrosequencing. PloS one 8, e54220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tao M. H., Morrison S. L., Studies of aglycosylated chimeric mouse-human IgG. Role of carbohydrate in the structure and effector functions mediated by the human IgG constant region. J. Immunol. 143, 2595–2601 (1989). [PubMed] [Google Scholar]
- 41.Oliphant T., et al. , Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 11, 522–530 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dowd K. A., Mukherjee S., Kuhn R. J., Pierson T. C., Combined effects of the structural heterogeneity and dynamics of flaviviruses on antibody recognition. J. Virol. 88, 11726–11737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guirakhoo F., Bolin R. A., Roehrig J. T., The Murray Valley encephalitis virus prM protein confers acid resistance to virus particles and alters the expression of epitopes within the R2 domain of E glycoprotein. Virology 191, 921–931 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edeling M. A., et al. , Potent dengue virus neutralization by a therapeutic antibody with low monovalent affinity requires bivalent engagement. PLoS Pathog. 10, e1004072 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mukherjee S., et al. , Mechanism and significance of cell type-dependent neutralization of flaviviruses. J. Virol. 88, 7210–7220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo Y., et al. , Comprehensive mapping infection-enhancing epitopes of dengue pr protein using polyclonal antibody against prM. Appl. Microbiol. Biotechnol. 99, 5917–5927 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wahala W. M., Kraus A. A., Haymore L. B., Accavitti-Loper M. A., de Silva A. M., Dengue virus neutralization by human immune sera: role of envelope protein domain III-reactive antibody. Virology 392, 103–113 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wirawan M., et al. , Mechanism of enhanced immature Dengue Virus attachment to endosomal membrane induced by prM antibody. Structure 27, 253–267.e258 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith S. A., et al. , Human monoclonal antibodies derived from memory B cells following live attenuated dengue virus vaccination or natural infection exhibit similar characteristics. J. Infect. Dis. 207, 1898–1908 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colpitts T. M., et al. , prM-antibody renders immature West Nile virus infectious in vivo. J. General Virol. 92, 2281–2285 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodenhuis-Zybert I. A., et al. , Immature dengue virus: A veiled pathogen? PLoS Pathog. 6, e1000718 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng A., Umashankar M., Kielian M., In vitro and in vivo studies identify important features of dengue virus pr-E protein interactions. PLoS Pathog. 6, e1001157 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dowd K. A., et al. , Broadly neutralizing activity of Zika virus-Immune sera identifies a single viral serotype. Cell Rep. 16, 1485–1491 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geiss B. J., Pierson T. C., Diamond M. S., Actively replicating West Nile virus is resistant to cytoplasmic delivery of siRNA. Virol. J. 2, 53 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sukupolvi-Petty S., et al. , Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J. Virol. 84, 9227–9239 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dowd K. A., Jost C. A., Durbin A. P., Whitehead S. S., Pierson T. C., A dynamic landscape for antibody binding modulates antibody-mediated neutralization of West Nile virus. PLoS Pathog. 7, e1002111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Alwis R., de Silva A. M., Measuring antibody neutralization of dengue virus (DENV) using a flow cytometry-based technique. Methods Mol. Biol. 1138, 27–39 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Randolph V. B., Winkler G., Stollar V., Acidotropic amines inhibit proteolytic processing of flavivirus prM protein. Virology 174, 450–458 (1990). [DOI] [PubMed] [Google Scholar]
- 59.Prasad V. M., et al. , Structure of the immature Zika virus at 9 A resolution. Nat. Struct. Mol. Biol. 24, 184–186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suloway C., et al. , Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Lander G. C., et al. , Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roseman A. M., FindEM–A fast, efficient program for automatic selection of particles from electron micrographs. J. Struct. Biol. 145, 91–99 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo F., Jiang W., Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol. Biol. 1117, 401–443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu G., et al. , An algorithm for estimation and correction of anisotropic magnification distortion of cryo-EM images without need of pre-calibration. J. Struct. Biol. 195, 207–215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rosenthal P. B., Henderson R., Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Roy A., Kucukural A., Zhang Y., I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y., I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao H., et al. , Structural basis of Zika Virus-specific antibody protection. Cell 166, 1016–1027 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Chacon P., Wriggers W., Multi-resolution contour-based fitting of macromolecular structures. J. Mol. Biol. 317, 375–384 (2002). [DOI] [PubMed] [Google Scholar]
- 74.Birmanns S., Rusu M., Wriggers W., Using Sculptor and Situs for simultaneous assembly of atomic components into low-resolution shapes. J. Struct. Biol. 173, 428–435 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wriggers W., Conventions and workflows for using Situs. Acta Crystallogr. D. Biol. Crystallogr. 68, 344–351 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiao C., Rossmann M. G., Interpretation of electron density with stereographic roadmap projections. J. Struct. Biol. 158, 182–187 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rossmann M. G., Bernal R., Pletnev S. V., Combining electron microscopic with x-ray crystallographic structures. J. Struct. Biol. 136, 190–200 (2001). [DOI] [PubMed] [Google Scholar]
- 79.Wang Z., et al. , Obstruction of dengue virus maturation by Fab fragments of the 2H2 antibody. J. Virol 87, 8909–8915 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pettersen E. F., et al. , UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein. Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein. Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Prestwood T. R., Prigozhin D. M., Sharar K. L., Zellweger R. M., Shresta S., A mouse-passaged dengue virus strain with reduced affinity for heparan sulfate causes severe disease in mice by establishing increased systemic viral loads. J. Virol. 82, 8411–8421 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All data supporting the findings of this study are available within the paper. The cryo-EM density maps of immature DENV in complex with prM12 and prM13 Fabs are available at the Electron Microscopy Data Bank (EMDB) with the accession codes EMD-29020 and EMD-29021, respectively. The fitted pseudoatomic models are available at the Protein Data Bank (PDB) with the accession codes PDB 8FE3 and PDB 8FE4 for prM12 and prM13 complexes, respectively.