Abstract
Deleterious mutations decrease reproductive fitness and are ubiquitous in genomes. Given that many organisms face ongoing threats of extinction, there is interest in elucidating the impact of deleterious variation on extinction risk and optimizing management strategies accounting for such mutations. Quantifying deleterious variation and understanding the effects of population history on deleterious variation are complex endeavors because we do not know the strength of selection acting on each mutation. Further, the effect of demographic history on deleterious mutations depends on the strength of selection against the mutation and the degree of dominance. Here we clarify how deleterious variation can be quantified and studied in natural populations. We then discuss how different demographic factors, such as small population size, nonequilibrium population size changes, inbreeding, and gene flow, affect deleterious variation. Lastly, we provide guidance on studying deleterious variation in nonmodel populations of conservation concern.
Keywords: deleterious mutations, conservation biology, genetic load, inbreeding depression, simulations
INTRODUCTION
As a result of errors in DNA replication or environmental DNA damage, genomes accumulate mutations. Some mutations may have no effect on reproductive fitness and be evolutionarily neutral. Other mutations may be beneficial, conferring some fitness advantage to those that carry them. Lastly, some mutations may be deleterious, resulting in lower fitness of individuals that carry them. Mutation accumulation studies, conceptual models of evolution, and molecular population genetic studies all suggest that deleterious mutations are abundant in genomes (1–4).
Deleterious variation is relevant for conservation biology, which is the focus of this review. Specifically, theoretical models as well as empirical case studies have shown that an accumulation of deleterious mutations can reduce the health of individuals within a species, potentially leading to extinction (5–7). Researchers are now studying deleterious variation to inform management of small and endangered populations and making predictions about the future health of the population. For example, one management strategy often employed to mitigate the risks of a small population size is to translocate individuals from another population into the small population (8). This strategy is called genetic rescue. Knowledge of the role of deleterious variation in extinction and the dynamics of evolutionary processes on deleterious variation will enable more effective genetic rescue strategies (9).
The study of deleterious variation has benefited greatly from advances in whole-genome sequencing technology. In the past decade, it has become possible to build reference genomes for nonmodel organisms and survey genome-wide patterns of variation in tens to hundreds of individuals (10). As a result, researchers have attempted to quantify the amounts of deleterious variation in different populations with differing demographic histories and life history traits. Some general trends are emerging, which we describe below. However, along with the general trends, new challenges with respect to interpretation of deleterious variation are also arising.
In this review, we discuss some essential facets of deleterious variation in natural populations. We begin by introducing the concept of genetic load and the theory behind it. We then discuss empirical strategies aimed at quantifying load in populations. We next turn to discussing how different evolutionary forces, such as small population size and inbreeding, can affect deleterious variation. We close by discussing these new challenges of interpretation in studies of deleterious variation.
PRINCIPLES OF DELETERIOUS VARIATION
Quantifying Fitness Effects of Mutations: The Distribution of Fitness Effects
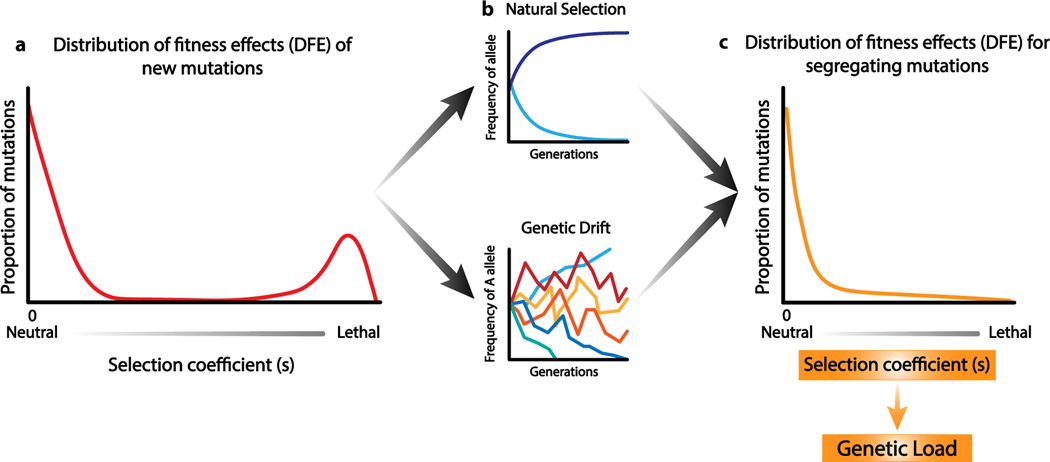
Model-based inferences of natural selection using molecular population genetic data have provided estimates of fitness effects of mutations. Specifically, if s denotes the fitness effect of a mutation from A to a, then the three genotypes AA, Aa, and aa have fitnesses of 1, 1 + hs, and 1 + s, respectively. Here h is the dominance coefficient, reflecting the fitness of the heterozygous genotype relative to that of the two homozygous genotypes. The distribution of fitness effects (DFE) refers to the distribution of s for new mutations in a particular component of the genome (Figure 1). Population genetic inferences from many organisms consistently find that new nonsynonymous mutations have fitness effects ranging from neutral to strongly deleterious (2, 11–16). Very few new mutations are beneficial. In humans, approximately 20% of amino acid– changing mutations are nearly neutral with s > −0.001. Approximately 17% have s < −0.01. The remainder have fitness effects that are weakly deleterious (11).
Figure 1.
The distribution of fitness effects and the fate of mutations in populations. (a) Each new mutation that occurs in an individual has a selection coefficient (s) describing its effect on fitness. The distribution of values of s is called the distribution of fitness effects (DFE) and has been estimated for nonsynonymous mutations in several species. (b) Once a mutation enters the population, its fate is affected by natural selection and genetic drift. Natural selection will push beneficial alleles to higher frequency and deleterious alleles to lower frequency, whereas drift could allow for random fluctuations in allele frequency. (c) The distribution of selection coefficients for variants segregating in the population differs from the DFE of new mutations. The DFE for segregating variants (c) has been affected by selection and drift. Typically, the distribution of selection coefficients for segregating mutations will be shifted toward more neutral variants because the most deleterious mutations will have been eliminated by natural selection. The selection coefficients for the segregating variants are most directly relevant for assessing the genetic load of a population.
The Fate of Deleterious Mutations in Populations: Selection and Drift
The DFE described above refers to new mutations entering the population. These mutations comprise the raw material on which evolutionary forces can act. The dynamics of mutations in populations are shaped by a combination of deterministic and nondeterministic evolutionary forces (Figure 1b). Selection is a deterministic force, driving beneficial alleles to increase in frequency and deleterious alleles to decrease in frequency and having no impact on the frequency of neutral alleles in the absence of linkage. In contrast, genetic drift is a stochastic force, leading to fluctuations in allele frequencies by chance due to the random sampling of alleles each generation. The magnitude of drift each generation depends on the population size, having a larger effect on allele frequencies in smaller populations.
We can think about the interplay of these evolutionary forces by considering the life cycle of a deleterious mutation. When the mutation first enters the population, it has a frequency of 1/2N, where N is the number of diploid individuals in the population. The mutation will also have a selective (s) and dominance (h) effect. These two quantities are properties of each mutation and are what DFE-inference methods estimate (Figure 1a). Then, selection and drift will affect what happens to the new deleterious mutation over time (Figure 1b). The deleterious mutation may be removed from the population due to selection, or it may persist due to the effects of drift.
Surveys of genetic variation detect those mutations that are segregating in the population at that particular point in time in the individuals that were sampled (Figure 1c). This set of mutations is a nonrandom subset of the mutations that initially entered the population, biased toward those that are less deleterious and have escaped removal by selection. Nevertheless, as we discuss below, there are conditions under which deleterious mutations may be segregating in populations and have biological consequences.
Genetic Load
In the 1950s, Mueller (17) emphasized the idea that mutations could lead to a decrease in fitness when he coined the term genetic load, building on Haldane’s (18) earlier mutation-selection balance theory. Crow (19) later defined load more precisely as the mean fitness of the population relative to a population free of deleterious mutations. The load (L) can be specified as
| 1. |
where is the mean fitness of the population and wmax is the fitness of a population free of deleterious mutations. The mean fitness, , is a function of the number of deleterious mutations in the population, their allele frequencies, and their selection and dominance effects. Specifically, at locus i, the mean fitness can be written as
| 2. |
where qi is the frequency of the deleterious allele, si is the selection coefficient, and hi is the dominance coefficient. When considering multiple loci, the mean fitness is the product of the fitnesses across all loci.
In the simplest models of viability selection, L, or the genetic load, refers to the reduction in mean fitness of the population compared to the optimal genotype. For example, L = 0.4 implies that the average individual has a fitness that is 40% of that of an individual without any deleterious mutations. Under such a scenario, the individual would survive with a probability of 60%. Further, 40% of individuals leave no descendants. This model makes several assumptions, and its biological applicability is questionable because it postulates a huge burden of genetic deaths (7, 20). Indeed, load has been interpreted in other models as involving differences in relative fitnesses between individuals. More work remains to be done in understanding the ecological implications of genetic load.
Regardless of the precise biological interpretation of genetic load, it still may be relevant to natural populations and conservation biology. If demographic processes (discussed below) lead to an accumulation of deleterious variation, this could increase genetic load, increasing the probability of extinction of the population. Because conservation biology focuses on preserving small populations, determining the extent to which small population size may lead to higher genetic load is an important area of research that we discuss below. Further, management strategies of small populations may focus on trying to minimize or decrease genetic load (6).
As can be seen in Equation 2, any process that would lead to more deleterious alleles segregating in the population (increase in the number of loci potentially experiencing deleterious mutations, increase in q, increase in s) would lead to an increase in genetic load. As we discuss below, genetic drift can lead to an increase in the frequency of weakly deleterious alleles in the population (21–23). Thus, the term drift load was coined to refer to the increase in load in small populations. Typically, drift load is driven by the increase in frequency of weakly deleterious alleles that are codominant or have additive effects (i.e., h = 0.5). Drift load also includes those deleterious alleles that reach fixation (i.e., frequency of 100% in the population). Drift load is the type of load that underlies the “mutational meltdown” scenario (23, 24; see below).
Another type of genetic load that is relevant for populations of conservation concern is the inbreeding load (B). The inbreeding load refers to the decrease in fitness that would result from inbreeding occurring in the population (25–27). The inbreeding load for locus i can be written as
| 3. |
To gain some intuition, would be the load if all mutations were homozygous in the population. The second two terms, , represent the actual expressed load in a randomly mating population. Thus, the inbreeding load is the total possible load minus the currently expressed load. Note, the inbreeding load is masked if there is no inbreeding in the population. As such, in the absence of inbreeding, the inbreeding load has no impact on fitness. However, if inbreeding were to occur, this load would be unmasked and would decrease fitness. As we discuss below, certain demographic processes can affect drift load and inbreeding load in different ways.
The applicability of genetic load theory to actual natural populations is not straightforward. First, the hypothetical mutation-free genotype does not actually exist in a real population. All individuals carry some number of deleterious mutations. Thus, selection may operate on a relative scale, not an absolute one, where individuals with a higher load reproduce less than those with a lower load (soft selection; 7, 28). Under this scenario, L may not have the same meaning as Crow proposed originally. Typically, in conservation biology, load can be compared between several closely related populations to measure whether the one of conservation concern has a higher load (see below). A second challenge is that calculations of load focus only on deleterious mutations, without regard for ecological processes occurring in real populations. For example, if density-dependent processes are limiting population size, then load may not matter for the dynamics of the population because many individuals would likely die anyway. Finally, calculations of load make several simplifying assumptions, such as multiplicative effects across loci. Multiplicative effects may not hold if there is epistasis. For example, after a certain threshold, a continued increase in deleterious mutations may no longer continue to decrease fitness.
Perhaps the biggest challenge in applying genetic load theory to natural populations is that we do not know the selection coefficients and dominance coefficients for individual variants segregating in genomes. Thus, it is hard to directly estimate load for particular individuals or populations. Below, we describe some existing approaches to estimating load in natural populations, highlighting the strengths and limitations of each.
QUANTIFYING GENETIC LOAD IN NATURAL POPULATIONS
Method 1: Empirical Characterization of Deleterious Variation
Genome sequencing provides a way to directly quantify variation across the genome and is increasingly applied to populations of conservation concern. If one could distinguish harmful, neutral, and beneficial variants in genomes, one could simply tabulate the number of harmful variants per genome to estimate genetic load. However, in practice, differentiating between harmful, neutral, and beneficial variants is challenging. Nonetheless, several computational approaches attempt to predict which variants are likely to be deleterious.
One straightforward approach is to identify mutations that are predicted to alter gene structure or function, such as those that encode premature stop codons, induce frameshifts, disrupt sites involved in messenger RNA transcript splicing, or modify encoded amino acid(s). SnpEff (29) and ANNOVAR (30) are two of the most commonly used computational tools for identifying such mutations. The underlying assumption of this approach is that amino acid–changing (nonsynonymous) and other mutations predicted to impact gene structure or function tend to be deleterious, whereas silent (synonymous) mutations are effectively neutral. These may be reasonable assumptions on average, but there are undoubtedly many cases where they are violated. For instance, there is accumulating evidence that selection acts on synonymous variants, particularly in functionally important and/or highly expressed genes (e.g., 31–34). Conversely, some mutations that strongly impact gene structure or function may ultimately have neutral or even adaptive effects. Mutations encoding premature stop codons that lead to truncated gene transcripts, so-called loss-of-function (LOF) or protein-truncating variants, are generally assumed to be strongly deleterious. However, such mutations can occasionally be adaptive (35) and are segregating at appreciable frequency in the population without negative phenotypic consequences (36). Thus, although mutations impacting gene structure or function may have deleterious effects on average, the effect of a particular variant remains uncertain.
A second approach to identify putatively deleterious variants in genomes incorporates the physicochemical properties of amino acid substitutions to infer deleteriousness. As amino acids vary in size/weight and polarity, mutations that introduce substitutions between highly dissimilar amino acids are more likely to impact protein structure/function than those that substitute an amino acid with similar properties. Grantham (37) and Miyata et al. (38), among others, developed scoring matrices to reflect the degree of dissimilarity between all possible amino acid pairs, and one application of these scores has been to assign probable deleteriousness to nonsynonymous mutations. Again, this approach assumes that greater disruptions to protein structure/function are more likely to be deleterious, which may not always be true; therefore, the caveats noted above also apply here.
A third approach uses sequence conservation across species to distinguish putatively neutral from putatively deleterious variants. This approach assumes that mutations in sites under strong evolutionary constraint are likely to be deleterious, as evolutionary constraint may reflect strong negative selection. Here, evolutionary constraint is defined by a lack of diversity in alignments of homologous nucleotide or protein sequences from many species. Methods that use this framework to infer the deleteriousness of mutations in studies of nonhuman species include the GERP (Genomic Evolutionary Rate Profiling; 38, 40) and SIFT (Sorting Intolerant from Tolerant; 41, 42) algorithms. Other methods, such as PolyPhen-2 (43), PROVEAN (44), and CADD (45), are designed for use in studies of humans and a small number of model organisms but occasionally are adapted for use in a wider range of species.
All these approaches have notable limitations in addition to those already mentioned. Most importantly, none of these methods can reliably discern strongly from weakly deleterious mutations (46–48) or reveal whether mutations are additive or recessive. Also, in many cases, these methods can be applied only to simple variants (single-nucleotide variants and small indels) within protein-coding sequences. Evaluating more complex variation or variation outside protein-coding genes is typically not feasible for nonmodel species. Third, counting the number of deleterious alleles in a genome requires knowing which allele is deleterious at every variant site. Typically, we would assume that the ancestral allele (i.e., the type before the mutation) is not likely to be deleterious, whereas a derived (i.e., the mutant) allele might be. However, most variant annotation tools use variant call format files as input and apply classifications only to “nonreference” alleles (i.e., those that differ from the allele in the underlying reference genome), effectively confounding the reference with the ancestral allele and making it difficult to evaluate fixed mutations. To overcome this problem, one potential strategy is to use a reference genome from a closely related outgroup species to assign the ancestral and derived states. However, this can introduce biases when different populations have differing levels of divergence from the reference, as variant predictors are less likely to predict that the reference allele is deleterious.
Moreover, once putatively deleterious variants have been identified, it is not obvious how to estimate genetic load. Load is typically defined as the difference between the mean fitness and a theoretical optimal fitness. Immediately, two fundamental questions arise: First, what is the maximal or theoretically optimal fitness, and second, how do quantities of putatively deleterious mutations translate to fitness? A common method for circumventing the first problem is to simply compare the number of deleterious alleles between populations to determine which has higher or lower load, producing a relative rather than an absolute measure. If deleterious mutations are additive and have identical selection coefficients, then the number of deleterious alleles per genome will be proportional to the load (49). Addressing the second problem is far more challenging because fitness and genetic load cannot be inferred directly from levels of putatively deleterious alleles in the absence of information about s, h, epistasis, and the effects of complex or noncoding variation.
Consequently, various metrics are often used as proxies for load, such as the number, frequency, or proportion of deleterious alleles and/or genotypes. Further distinctions can be incorporated to count deleterious alleles found in only one population and not another [e.g., RXY (50)], or to account for segregating versus fixed mutations, or to normalize quantities with respect to the amount of putatively neutral variation (e.g., the ratio of nonsynonymous to synonymous variation). There is currently no agreed-upon standard for quantifying deleterious variation to approximate load or fitness. Calculating and interpreting many of these metrics is complicated by technical issues, as noted above, as well as varying sensitivity to demographic factors, such as fluctuating population size or inbreeding (51, 52). Lastly, the methods described above are proxies for realized genetic load and do not incorporate the burden of recessive deleterious heterozygotes. As shown in Equation 3, such mutations have no impact on fitness until they become homozygous through inbreeding (6). In summary, no single metric encapsulates the fitness of an individual or population from sequence data alone, and there are many pitfalls and challenges in estimating suitable proxies.
Method 2: Model-Based Studies of Deleterious Variation
Population genetic models implemented through computer simulations provide a powerful complementary approach to empirical studies of genetic load. Simulations are computer programs designed to emulate simplified real-world processes in silico. The goal of genetic simulations is to produce pseudogenetic data sets under a model of the evolutionary process. The model specifies key variables (parameters) such as mutation rate, recombination rate, number of loci or genome size, and numbers of individuals in the population at various times. Many evolutionary processes are inherently random (e.g., Mendelian inheritance, drift, mutation); therefore, simulation outcomes can vary from one iteration to the next. To quantify this stochasticity, simulations should be run repeatedly under a given set of parameters to bracket the range of potential outcomes. Though some evolutionary dynamics can be studied analytically (i.e., using mathematical equations), evolutionary models can be infinitely complex, and analytical derivations often become intractable beyond the simplest cases. Simulations, however, can be arbitrarily complex and are limited only by their feasibility in terms of computational resources and runtime, and by our ability to design models that accurately capture the key features of the evolutionary process.
Simulations can be used to explore how deleterious mutations are impacted by evolutionary forces and to quantify genetic load. Specifically, if one has inferred a model of population history and has a rough approximation of mutation rates, DFE, and dominance coefficients for new mutations, then the evolutionary history of the population can be simulated and evolutionary forces, such as a bottleneck, can be observed directly with regard to their impact on deleterious variation. In a simulation, the values of s and h for each mutation are known; therefore, load can be computed from Equations 1 and 2 directly. This approach to measuring load obviates the need to know s and h for individual mutations in empirical sequence data and may be more accurate than empirical measures for detecting subtle differences in load (53).
Computer simulations are also not limited to understanding past evolutionary processes; they can be used to explore hypothetical scenarios or make predictions, which may be especially important in applied contexts such as conservation. Examples of questions that could be answered with simulations include (a) how will population fitness (or some other trait) evolve under a given management scenario; (b) what is the probability of extinction/recovery/persistence as a function of population size or inbreeding; and (c) what is the expected level of deleterious variation in a hypothetical historical or future scenario?
Computing capabilities have increased dramatically over time, and population genetic simulators have also become increasingly sophisticated. In particular, the program SLiM combines high flexibility with recent innovations to enhance the computational tractability of complex models (54), making it an especially powerful tool for genetic simulations under more realistic ecological models (55).
Despite considerable advances in the sophistication of genetic simulation software, several challenges remain. Simulations involving large amounts of sequence, long time spans, and/or large population sizes may not be feasible with finite computational resources and time. Modifications such as rescaling parameter values or narrowing the scope of simulations can help reduce computational overhead, though the implications of these changes are not fully known for models of genetic load (56). Designing models and choosing appropriate parameter values can also be difficult, especially when the “true” values are unknown. No model can ever be entirely realistic, and all models involve simplifications and assumptions. Sensitivity analysis, which involves perturbing parameters and rerunning simulations to evaluate outcomes, should be done whenever possible to assess the robustness of results. Thus, although simulations have their limitations, a suite of powerful tools is available to supplement and enhance empirical analyses.
HOW DO DEMOGRAPHIC FORCES IMPACT LOAD?
Next, we discuss how different demographic forces affect patterns of deleterious variation, summarizing some trends in Table 1. However, as we discuss below, the specific patterns of how demography impacts deleterious variation may depend on the particular parameter values. Thus, the predictions in Table 1 are meant to provide some intuition but not serve as absolute rules that will apply in all circumstances. We recommend that simulations be used to test these predictions for specific empirical applications.
Table 1.
Summary of how demographic forces affect different types of genetic variation
| Demographic scenario | Predictions for neutral heterozygosity | Predictions for weakly deleterious additive mutations | Predictions for recessive strongly deleterious mutations |
|---|---|---|---|
| Long-term small population | Low across the genome | May accumulate due to drift | Few segregating due to increased drift and purifying selection |
| Long-term large population | High across the genome | Efficiently removed by selection | Many segregating due to being masked as heterozygotes |
| Recent population contraction | Slight decrease across the genome | Slight increase due to drift | Increased homozygosity; decreased total number of alleles after selection |
| Recent population expansion/growth | Little effect | Depends on parameters; likely little effect | Possible increases on timescale of hundreds of generations |
| Recent inbreeding in small population | Little effect | Little effect | Little effect because few are segregating |
| Recent inbreeding in large population | Regions of low heterozygosity mixed with regions of high heterozygosity | Little effect | Can become exposed due to inbreeding and decrease fitness |
| Gene flow from very heterozygous population | Increase | Increase in population size should decrease effects of drift, preventing fixation | Can help mask existing recessive deleterious mutations but also introduce new recessive deleterious mutations |
| Gene flow from moderately heterozygous population | Slight increase | Increase in population size should decrease effects of drift, preventing fixation | Can help mask existing recessive deleterious mutations without introducing many new recessive deleterious mutations |
Small Population Size and Drift
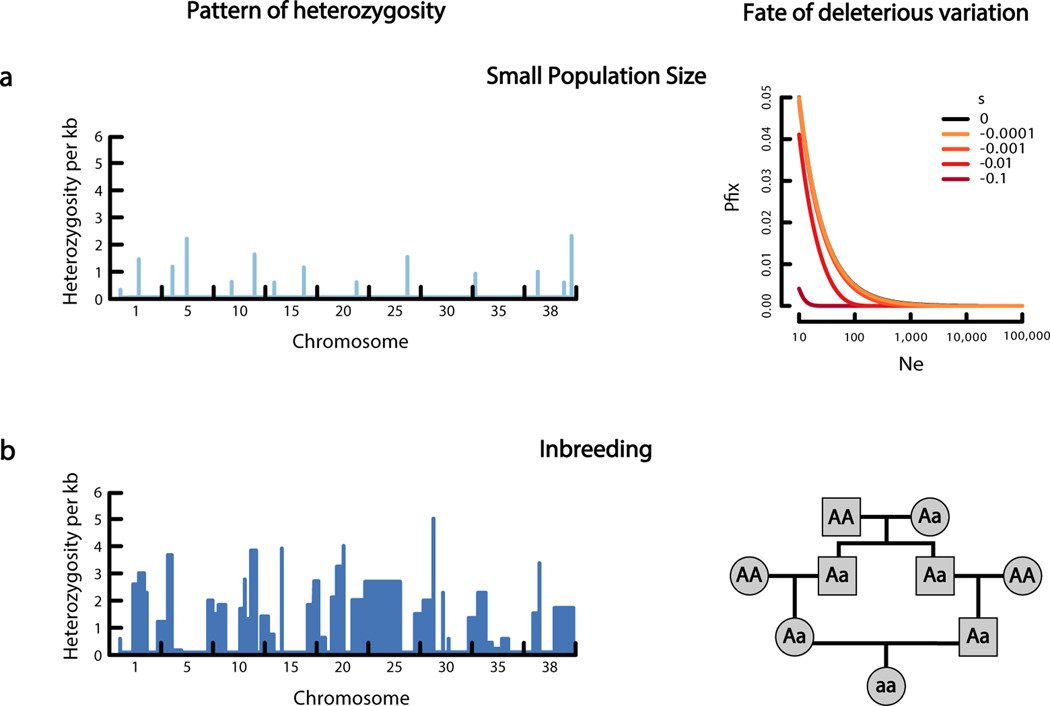
Small populations will experience an increased amount of genetic drift. One effect of increased genetic drift is decreased heterozygosity across the genome (Figure 2a, left). Additionally, genetic drift in small populations may cause deleterious alleles to increase in frequency or even become fixed (i.e., reach a frequency of 100% in the population) unless selection is sufficiently strong (21, 57–59) (Figure 2a, right). For selection to outweigh the effects of drift and prevent the fixation of deleterious alleles, the |s| against a deleterious allele must be >>1/(2Ne), where Ne is the effective population size. In large populations (i.e., Ne > 1,000), the value of 1/(2Ne) is small, so selection will ultimately remove even weakly deleterious alleles (s>−5×10−4). In small populations (i.e., Ne < = 1,000), selection against such weakly deleterious alleles is ineffectual, and therefore these alleles can persist, reach fixation, and accumulate over time. Note that selection will prevent the fixation of strongly deleterious mutations (s < = −0.01) so long as Ne >> 50 (Figure 2a, right).
Figure 2.
Genetic drift and inbreeding have different effects on the genome and on deleterious variation. (a, left) Genetic drift associated with a small population size leads to low heterozygosity across the genome. (right) Genetic drift also allows weakly deleterious mutations to become fixed in the population, contributing to drift load. The right-hand panel shows the probability of fixation for a new mutation with s specified by the different lines in a population of the size denoted on the x-axis. Probabilities were calculated using equation 10 in Reference 128. (b, left) Inbreeding can lead to a sawtooth pattern of heterozygosity across the genome. Regions of low heterozygosity correspond to the regions of the genome that likely are copies of the same ancestral chromosome inherited from both parents. (right) Inbreeding results in an increase in the probability of an ancestral chromosome (denoted by a) being inherited from both the maternal and paternal lineage, increasing homozygosity. Thus, recessive deleterious mutations have a higher probability of becoming homozygous and thereby affecting fitness.
Increased drift in populations with small effective sizes is predicted to result in higher frequencies, higher fixation rates, higher total numbers, and higher homozygosity of deleterious alleles. The increased frequency and accumulation of weakly deleterious mutations due to drift (called drift load) is predicted to reduce fitness in small populations. Studies have explored the impacts of drift on genetic load by investigating deleterious variation in populations with varying historical population sizes.
Many studies of deleterious variation have focused on domesticated species, which are hypothesized to have elevated load due to the effects of reduced population size and increased drift (60). Consistent with the predicted effects of drift, many domesticated species harbor lower genetic diversity than their wild counterparts (60, 61). Further, studies of domesticated plants [e.g., rice (62–64), barley (65), tomatoes (66), and sunflowers (67)] and animals [e.g., dogs (68), horses (69), chickens (70), and rabbits (71)] have found elevated rates of putatively deleterious variation in domesticated relative to wild species. One notable exception is European pigs, which do not have less diversity or increased levels of deleterious variation relative to wild European boars (72), suggesting that the effects of domestication on load may be more complex in some cases (60, 61; see the section titled Nonequilibrium Population Size: Bottlenecks and Range Expansions). Overall, many studies show that domestication is associated with increased numbers and/or proportions of putatively deleterious alleles, consistent with the predicted effects of drift and the “cost of domestication” (62).
The impacts of drift on genetic load have also been studied in island populations and other populations with historically low Ne. These studies typically include one or more outgroups with historically larger Ne to provide relative rather than absolute measures of load. Many of these studies have shown that long-term small population size is associated with reduced overall diversity, as well as increased homozygosity and other measures of deleterious alleles, consistent with the predicted effects of drift. Examples of species and populations with historically low Ne and genomic evidence of elevated load include gorillas from East Africa (72, 73), Channel Island foxes (74, 75), Indian lions (76), Sumatran rhinoceroses (77), pygmy hogs (78), kākāpō (79), and vaquita porpoises (80). In a study comparing 14 island and 11 continental songbird species, Leroy et al. (81) found a clear relationship between population size and the ratio of nonsynonymous to synonymous diversity, which is a statistic commonly used as a proxy measure of selection. Importantly, increased deleterious variation due to drift tends to be associated with long-term small population size on the coalescent or evolutionary timescale (i.e., on the order of thousands of generations or more) and might not be detected in populations or species that have experienced very recent declines, or that have more complex demographic histories. Nonetheless, there is now abundant genomic evidence that long-term small Ne is associated with elevated measures of genetic load, consistent with the increased frequency and accumulation of weakly deleterious alleles due to genetic drift.
Nonequilibrium Population Size: Bottlenecks and Range Expansions
Changes in population size cause allele frequencies to shift toward new equilibrium values. Such shifts proceed at different rates according to population size, mutation rate, and selection strength. Some statistics used to measure deleterious variation, such as ratios between deleterious and neutral variation, can be affected by the unequal rates at which deleterious and neutral variants equilibrate following population size changes (50, 52, 82). Understanding the impacts of nonequilibrium demography on deleterious variation is therefore essential for measuring load in real-world populations that are likely not at equilibrium. Below, we discuss load in the context of two nonequilibrium scenarios: population bottlenecks and range expansions.
A population bottleneck is a reduction in population size, potentially followed by growth. Population decline is predicted to reduce diversity through the loss of rare alleles, increased homozygosity, and higher fixation rates induced by drift (83). Population decline can particularly impact recessive deleterious mutations, which will initially increase in homozygosity and contribute to higher load before being removed from the population by negative selection (84, 85). If the population remains small, drift can drive the accumulation of weakly deleterious mutations while overall diversity declines to a new equilibrium. These effects caused by population contraction differ from the predicted effects of growth. Growth increases the number of deleterious mutations, but effective selection at higher Ne maintains deleterious alleles at low frequencies, primarily as heterozygotes. So, if the population contracts and then later expands, the patterns of deleterious variation will be a function of both the contraction and the growth and are therefore hard to predict without modeling the specific demographic scenario (49, 51, 86, 87).
The duration of bottlenecks also determines their impact on genetic diversity (52). Bottlenecks of short duration (on the coalescent timescale) are predicted to minimally impact diversity, even if they involve extreme changes in population size, because the population size changes too rapidly for allele frequencies to re-equilibrate. However, low-frequency deleterious alleles may be lost in such scenarios. Conversely, low diversity may persist following long-lasting bottlenecks because the replenishment of diversity through the introduction of new mutations can lag behind population growth. In such long bottlenecks, weakly deleterious variation may start to approximate the equilibrium values established by the small population during the bottleneck, potentially leading to an increase in drift load. Thus, different stages of the bottleneck have contrasting and potentially persistent effects depending on the parameters of selection, dominance, and demography (84).
Empirical studies investigating the impacts of population bottlenecks on deleterious variation have yielded complex results. Reduced neutral diversity is found in virtually all cases, but impacts on patterns of deleterious variation vary across studies. Evidence for an accumulation of load due to drift and/or inbreeding has been observed in the genomes of Apennine brown bears (88), sea otters (89), crested ibises (90), Alpine ibexes (91), and Himalayan red pandas (92). In contrast, some studies have found reduced levels of deleterious variation in bottlenecked populations, suggesting enhanced purging of (recessive) deleterious alleles. Evidence of purging has been found in gorillas from East Africa (72), Alpine ibexes (91), Massasauga rattlesnakes (93), Iberian lynxes (94), and Chinese crocodile lizards (95). In some cases, evidence of purging has been found among presumably recessive, strongly deleterious variants (e.g., LOF mutations), along with an accumulation in categories of more weakly deleterious, and presumably less recessive, variants (e.g., nonsynonymous mutations; 72, 91, 93). As exemplified in some studies (e.g., 91, 94–96), simulations can be used to examine the dynamics of deleterious variation in complex demographic models. In summary, population bottlenecks can impact genetic load in many, sometimes contrasting, ways.
A second nonequilibrium scenario predicted to impact the load of deleterious variation is range expansion. In contrast to a simple growth model, the theoretical range expansion model is spatially explicit, such that local population densities are lower at the expansion front relative to the population core, leading to reduced Ne at range margins. Reduced Ne induces stronger genetic drift, leading to an accumulation of deleterious alleles (known as expansion load) and reduced fitness among individuals at the expansion front (97, 98). Empirical support for this model has been found in humans (87, 99), Arabidopsis lyrata (100, 101), snowshoe hares (102), and Pacific salmon (103). Thus, even in a scenario of population growth, the dynamics of load may be impacted locally by drift due to population structure.
Inbreeding and Purging
Reproduction between closely related individuals is known as inbreeding. In the strict sense, inbreeding refers to reproduction between related individuals beyond what would be expected under random mating. In a broader sense, inbreeding can refer to any reproduction between individuals that share a recent common ancestor, even if individuals are mating randomly, as might occur in very small populations where effectively all individuals may be closely related to each other. Inbreeding leads to deviations in genotype frequencies from those expected under random mating by causing an increase in homozygosity and a corresponding decrease in heterozygosity. Inbreeding can be detected in genomes as long stretches of the genome devoid of heterozygosity, reflecting the recent common ancestry of the two chromosomes carried by the individual, interspersed with regions of high heterozygosity, reflecting the regions of the genome that do not share a recent common ancestor (Figure 2b, left). An example of how inbreeding increases homozygosity is shown by the pedigree in Figure 2b (right). Here, we see how a single allele can be passed down from a single ancestor to multiple descendants and ultimately become homozygous in the offspring of those descendants. In this way, a deleterious mutation can manifest in the inbred offspring of unaffected parents that each carried one copy of a recessive allele inherited from a shared ancestor.
The main consequence of inbreeding on deleterious variation is that it will lead to homozygosity of deleterious mutations. Recessive deleterious mutations will then become expressed, leading to a reduction in reproductive fitness, known as inbreeding depression. Inbreeding leads to inbreeding depression via two potential mechanisms (104, 105). The first is that inbreeding depression results from the increased homozygosity of recessive deleterious alleles. The second is that inbreeding depression results from the loss of heterozygosity at loci where the heterozygote is the fittest genotype. Most theoretical and empirical evidence suggests the former mechanism is the primary driver of inbreeding depression. The severity of inbreeding depression is determined by the load of recessive deleterious alleles hidden in heterozygous genotypes that would cause fitness decline under inbreeding (inbreeding load). Populations or individuals may possess a high inbreeding load and still have high fitness, because recessive deleterious mutations will exist primarily as heterozygotes in a randomly mating population. The reduction in fitness occurs only through the increased probability of becoming homozygous when inbred.
Inbreeding can drive substantial fitness declines in just a few generations by rapidly shifting the zygosity of recessive deleterious mutations. However, increased homozygosity of recessive deleterious alleles induced by inbreeding also exposes these alleles to negative selection and may ultimately reduce their frequency below that in the ancestral population. Enhanced selection against recessive deleterious mutations due to excess homozygosity induced by inbreeding (or drift) is known as purging. Thus, in the long run, inbreeding can reduce the load of recessive deleterious alleles through purging, but rapid fitness declines are predicted in the short term. Empirical and theoretical experiments have suggested that purging may be effective against strongly deleterious mutations but is unlikely to be effective against weakly deleterious mutations (75, 85, 106, 107). Consequently, some part of the inbreeding load may be purged, but overall fitness may still be compromised by excess homozygosity of weakly deleterious recessive alleles that are not purged effectively.
Genomic studies have shown how inbreeding leads to large runs of homozygosity and elevated homozygosity of deleterious alleles. An extreme example is that of the Isle Royale gray wolf population, which became highly inbred following several generations of isolation at very small population size (<50 individuals) and suffered from severe inbreeding depression (108, 109). A severe founding bottleneck and subsequent inbreeding led to long runs of homozygosity across large fractions of Isle Royale wolf genomes but an equal number of deleterious alleles per genome relative to the nearby mainland wolf population (110). Thus, severe inbreeding depression in Isle Royale wolves was driven by increased homozygosity of deleterious alleles, rather than by a change in the number of deleterious alleles per genome. These results conform to the predicted effects of very recent inbreeding, in which heterozygous recessive deleterious alleles present among standing variation can be converted rapidly to homozygotes through inbreeding, leading to inbreeding depression.
Genomic studies have also found evidence for purging occurring as a consequence of inbreeding. For example, a study of Indian tigers, which exist in patchy habitats of varying sizes and connectivity, found that LOF mutations were depleted in individuals from isolated and highly inbred populations relative to larger and less inbred populations (111). Other studies of bottlenecked populations have also found evidence of purging along with increased homozygosity caused by small population size or inbreeding, as discussed previously (72, 91, 93–95, 112).
Gene Flow
Gene flow or migration refers to the movement of individuals from one population to another. Gene flow generally decreases genetic load. However, the extent to which this decrease occurs depends on several factors. For example, the effect of gene flow decreasing genetic load can be very pronounced in small populations. This is due to small populations accumulating a substantial load of deleterious mutations in the form of both drift load (fixed or high-frequency weakly deleterious mutations) and inbreeding depression (recessive deleterious mutations exposed by inbreeding), as described above. Gene flow results in the introduction of novel genetic variants that can reduce the drift load by counteracting the fixation of weakly deleterious variants and reduce inbreeding depression by masking recessive deleterious mutations (22).
The effect of masking recessive deleterious variants, known as dominance heterosis, is often strong in wild and domesticated species (105, 113). For example, substantial heterosis is commonly observed as an outcome of crosses between different crop strains, and most evidence suggests that this is due to masking of recessive deleterious variants (105). Moreover, simulation studies have shown that the effects of dominance heterosis can be substantial enough to mimic the effects of adaptive introgression (114) and swamp native ancestry (115). In addition, dominance heterosis probably explains much of the beneficial effects of genetic rescue (9, 116). As noted above, genetic rescue is a conservation management approach whereby novel genetic variation introduced by gene flow may increase population growth rates and/or reduce extinction probabilities in small and inbred populations (8, 117). Simulation studies have shown that large genetic rescue effects are observed even in the absence of adaptive genetic variation, suggesting that dominance heterosis may largely explain the positive effects of genetic rescue (9).
A key aspect of dominance heterosis, however, is that its effects are typically short-lived, implying that the effects of genetic rescue may also be short-lived (116, 117). Specifically, the masking effects of dominance heterosis are maximized in the F1 generation but are expected to diminish quickly in subsequent generations after inbreeding again exposes recessive deleterious alleles and genetic drift enables them to reach high frequency or fixation (116, 117). Although some evidence suggests that the beneficial effects of genetic rescue may persist to the F3 generation (118), empirical or experimental studies that track the long-term outcomes of genetic rescue remain rare. A dramatic example of the ephemeral effects of genetic rescue came from the Isle Royale wolf population. This population experienced a genetic rescue after a single male migrant arrived in the late 1990s, though it subsequently declined nearly to extinction within 2.5 generations of the initial genetic rescue (109, 119). Although this example is extreme, similar short-lived effects of genetic rescue have also been documented in long-term studies of Arctic foxes (120). Theoretical simulations have also supported the view that the beneficial effects of gene flow in decreasing genetic load are likely to be short-lived in many cases (9).
In What Scenarios Will Genetic Load Lead to Extinction?
The ultimate goal for studies of genetic load in species of conservation concern is to avert population declines and extinction. However, the specific ways in which genetic load is most likely to cause extinction remain uncertain. The classic view of extinction due to genetic load focuses on the role of weakly deleterious mutations accumulating in populations with long-term small size, a process often termed “mutational meltdown” (23, 24, 121). These models emphasize the role of weakly deleterious mutations gradually drifting to fixation due to increased genetic drift in small populations (Ne < 100), a process that may eventually lead to extinction over the long term (hundreds or thousands of generations).
Although studies on mutational meltdown have been highly influential in shaping thinking about genetic load and extinction, examples of a population going extinct due to mutational meltdown remain scarce (though see 122). There are several reasons for why this may be. First, extinction events due to mutational meltdown may be inherently difficult to observe, given that they are expected to occur gradually over long timescales and interact with other demographic and environmental factors. Moreover, over such long timescales, compensatory adaptive mutations may become relevant as a means to rescue populations with a high drift load from extinction (123), perhaps lessening the threat of extinction due to mutational meltdown. Another key reason why mutational meltdown might not often drive extinction in populations of conservation concern is that the demographic conditions required for mutational meltdown to occur (Ne < 100 for hundreds or thousands of generations) may be rare for such populations. Instead, many endangered or threatened populations have only recently declined to small size due to habitat destruction, fragmentation, and overharvesting that occurred largely over the past 100 years (124, 125), far less than 100 generations for long-lived species. Furthermore, many populations with the greatest conservation concern have dwindled to very small size (Ne < 25), at which point close inbreeding becomes inevitable.
In populations that have recently declined to very small size (Ne < 25), the threat of inbreeding depression due to exposure of recessive strongly deleterious mutations is likely to be far more impactful than mutational meltdown. Striking empirical examples of the importance of recessive strongly deleterious mutations as drivers of extinction come from the Florida panther and Isle Royale wolf populations, two of the more iconic examples of near-extinction due to genetic factors. In both cases, these populations were driven to the brink of extinction by severe inbreeding depression in the form of widespread congenital deformities, consistent with the impact of strongly deleterious recessive mutations (110, 126). This importance of strongly deleterious recessive variation is further supported by simulation studies that find the threat of extinction due to weakly deleterious variation is negligible relative to recessive strongly deleterious variation (9, 80). These studies confirm that, in scenarios where populations have experienced recent and severe declines, the exposure of recessive strongly deleterious alleles via inbreeding can quickly drive extinction, with little or no impact from weakly deleterious variation. These studies also demonstrate that historical population size greatly influences the levels of recessive strongly deleterious variation segregating in a population and resulting risk of extinction due to inbreeding depression. These considerations suggest that populations that have recently declined from a very large historical population size (Ne > 10,000) to a very small current size (Ne < 25) are likely at the greatest risk of extinction due to deleterious genetic variation.
OUTSTANDING PROBLEMS AND FUTURE DIRECTIONS
Although tremendous progress has been made over the previous two decades in understanding deleterious variation, challenges remain for using this knowledge to improve conservation of small populations. One major remaining challenge is inferring the fitness effects of individual variants. As this currently is a central question in human genetics, progress is being made using new computational approaches that integrate different types of genomic and functional data. Efforts should be made to transfer and apply these new techniques for nonhuman species. Further, from the population genetics side, more work is needed to quantify fundamental evolutionary parameters, such as mutation rates, DFE, and dominance coefficients for many nonmodel taxa. Such parameter estimates will enable more realistic simulations and models of evolution, which will be essential for conservation purposes.
Beyond these technical challenges, conceptual challenges of integrating deleterious variation in conservation practice remain. First, the practical significance of elevated genetic load in a population is not always clear. For example, the California Channel Island foxes show an elevated burden of derived deleterious nonsynonymous alleles compared to the mainland gray foxes yet show no phenotypic signs of inbreeding depression (74, 75). Similarly, not detecting an elevated genetic load does not mean that a population is impervious to risk from deleterious mutations. For example, the iconic Isle Royale wolf population that shows extensive phenotypic evidence of inbreeding depression and has gone extinct does not show an elevated count of derived deleterious nonsynonymous alleles (110). Instead, strongly deleterious recessive alleles, which are not easily counted in genome-wide surveys of variation, are likely responsible for this species decline. More generally, conservation genetics would benefit from additional empirical and theoretical studies quantifying the relative importance of drift load compared to inbreeding load at affecting extinction risk at different timescales and under different conditions. Additional field-based studies of fitness in natural populations would support such studies.
CONSIDERATIONS FOR CONSERVATION PRACTICE
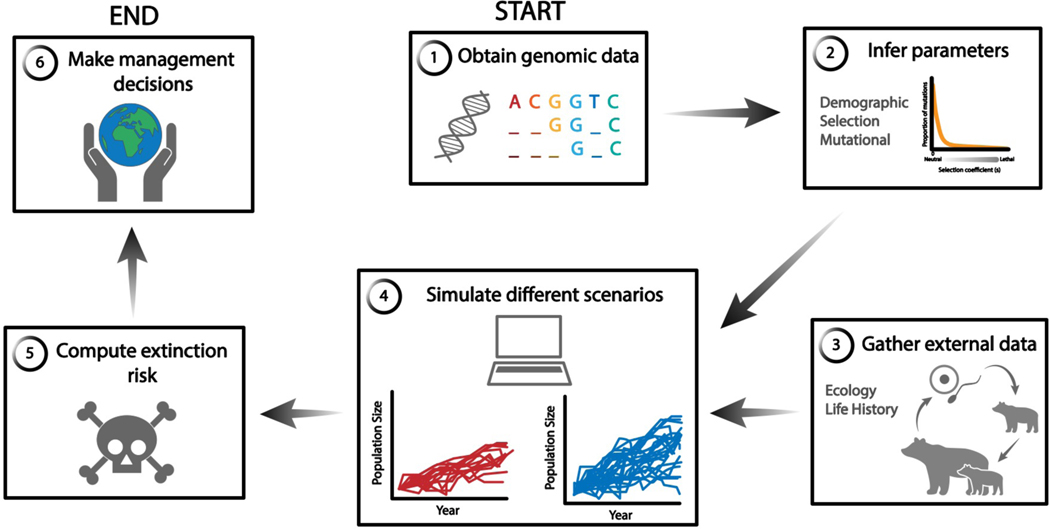
Despite these challenges, deleterious variation can be integrated into conservation genetics. Given the many nuances of how demography can impact deleterious variation, we recommend researchers employ a simulation-based strategy to examine the effects of deleterious variation (Figure 3). Demographic, selection, and mutational parameters can be estimated from genome-wide variation data, and then these parameter estimates can be used to inform simulations. Advances in ecologically realistic simulation models can both incorporate additional biological realism and model the process that is often important in conservation biology: extinction probability (55, 127). In these models, extinction probabilities are an emergent property of fundamental genetic and demographic processes, thus providing researchers with valuable information for management strategies. Researchers and practitioners should simulate under different parameter values to assess the sensitivity of their conclusions to various parameter values. Such genomic forecasting has been used to show that the vaquita porpoise, with a current population of 10 individuals, is not doomed to extinction from inbreeding depression, and recovery is possible if death from gillnet fishing ceases (80). This genomic forecasting framework provides a powerful means to integrate genomic study of deleterious variation into management of small and endangered populations.
Figure 3.
Simulation workflow for genomic forecasting including genomic data and deleterious mutations. This framework was used to show that the vaquita is not doomed to extinction from inbreeding depression (83).
Recent simulation and empirical studies of deleterious variation may inform conservation practice. First, not all species who have small populations today are at equal risk of extinction due to genetic factors. Populations which became small recently and quickly, especially from large ancestral populations, may be at greatest risk of extinction due to inbreeding depression (e.g. the Isle Royale wolf; 110). Populations that have been small for a long time (e.g. the Channel island fox, vaquita, and sea otter; 74, 75, 80, 89, 96) may carry fewer strongly deleterious recessive mutations and be better able to withstand current reductions in population size. For such populations, conservation efforts should especially focus on reducing the risk of extinction due to non-genetic factors (like predation, hunting, habitat destruction).
Additionally, simulations offer potential solutions to enhance the long-term efficacy of gene flow at reducing genetic load in small populations. First and foremost is ensuring that the small population can grow following the initiation of gene flow, such that the impacts of future inbreeding and genetic drift are lessened. However, the unfortunate reality is that many small populations are small because much of their original habitat has been destroyed, which may leave little room for a population to grow. In such cases where populations are destined to remain small, another option for enhancing the long-term effects of gene flow is to use source populations or individuals that are relatively purged of recessive deleterious variation (9). This approach can help alleviate the threat of inbreeding depression in the small population after inbreeding resumes. However, the extent to which purged source populations or purged individuals exist for species of conservation concern is uncertain in many cases or can be readily identified remains an open question.
FUTURE ISSUES
Can genomic predictions of the fitness effects of mutations be improved?
How can we validate methods that predict the fitness effects of individual mutations?
What are the selection and dominance coefficients for deleterious mutations in different species?
To what extent do weakly deleterious additive mutations matter for extinction risk of small populations?
To what extent do mutations in noncoding regions of the genome or structural variants matter for inbreeding depression, genetic load, and extinction risk?
How can we best use genome sequence data for conservation purposes?
ACKNOWLEDGMENTS
We thank the Lohmueller and Wayne labs as well as our collaborators for helpful discussions on these topics over the previous years. This work was supported by National Institutes of Health grant R35GM119856 to K.E.L.
GLOSSARY
- Ancestral/derived allele
the ancestral allele is the original state that existed before mutation, whereas the derived allele is the new mutation
- Bottleneck
a reduction in population size followed by an increase in population size
- Distribution of fitness effects
a probability distribution describing the proportions of mutations that are strongly, moderately, or weakly deleterious; neutral; or adaptive
- Dominance coefficient
the effect of the heterozygous genotype relative to that of the two homozygous genotypes
- Dominance heterosis
an increase in fitness after hybridization if different populations have distinct sets of recessive deleterious mutations
- Drift load
the genetic load coming from weakly deleterious mutations that have increased in frequency or become fixed in small populations
- Effective population size
a population size that allows approximation of the amount of genetic diversity in a real population by the Wright-Fisher model of genetic drift
- Fitness
the ability of an individual in a population with a given genotype to reproduce
- Gene flow
the movement of genetic material from one population into another as a result of mating across populations
- Genetic drift
random changes in allele frequency as a consequence of finite population size
- Genetic load
reduction in reproductive fitness due to deleterious mutations
- Genetic rescue
movement of individuals from a population into a smaller population to increase the population size and decrease extinction risk
- Inbreeding
mating among close relatives that increases homozygosity compared to that expected under random mating
- Inbreeding load
genetic load carried as heterozygotes that becomes expressed only when inbreeding increases homozygosity
- Mutational meltdown
a process in which weakly deleterious mutations become fixed in small populations, resulting in fitness reductions, further decreases in population size, further fixations of deleterious mutations, and ultimately extinction
- Purging
process by which deleterious recessive mutations are eliminated from the population by negative selection after becoming homozygous due to inbreeding
- Reference genome
a genome sequence that is well-assembled and annotated to which sequencing reads from other individuals can be mapped
- Runs of homozygosity
long stretches of homozygosity across the genome caused by the two chromosomes sharing a recent common ancestor
- Selection coefficient
the change in reproductive fitness associated with a particular mutation, averaged over environments and genetic backgrounds
- Simulation
generation of outcomes of a random process from a probabilistic model
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Kimura M. 1977. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature 267(5608):275–76 [DOI] [PubMed] [Google Scholar]
- 2.Boyko AR, Williamson SH, Indap AR, Degenhardt JD, Hernandez RD, et al. 2008. Assessing the evolutionary impact of amino acid mutations in the human genome. PLOS Genet 4(5):e1000083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eyre-Walker A, Keightley PD. 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet 8(8):610–18 [DOI] [PubMed] [Google Scholar]
- 4.Bataillon T, Bailey SF. 2014. Effects of new mutations on fitness: insights from models and data. Ann. N.Y. Acad. Sci 1320(1):76–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hedrick PW, Kalinowski ST. 2000. Inbreeding depression in conservation biology. Annu. Rev. Ecol. Evol. Syst 31:139–62 [Google Scholar]
- 6.Bertorelle G, Raffini F, Bosse M, Bortoluzzi C, Iannucci A, et al. 2022. Genetic load: genomic estimates and applications in non-model animals. Nat. Rev. Genet 23(8):492–503 [DOI] [PubMed] [Google Scholar]
- 7.Agrawal AF, Whitlock MC. 2012. Mutation load: the fitness of individuals in populations where deleterious alleles are abundant. Annu. Rev. Ecol. Evol. Syst 43:115–35 [Google Scholar]
- 8.Whiteley AR, Fitzpatrick SW, Funk WC, Tallmon DA. 2015. Genetic rescue to the rescue. Trends Ecol. Evol 30(1):42–49 [DOI] [PubMed] [Google Scholar]
- 9.Kyriazis CC, Wayne RK, Lohmueller KE. 2021. Strongly deleterious mutations are a primary determinant of extinction risk due to inbreeding depression. Evol. Lett 5(1):33–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paez S, Kraus RHS, Shapiro B, Gilbert MTP, Jarvis ED, Vertebr. Genomes Proj. Conserv. Group. 2022. Reference genomes for conservation. Science 377(6604):364–66 [DOI] [PubMed] [Google Scholar]
- 11.Kim BY, Huber CD, Lohmueller KE. 2017. Inference of the distribution of selection coefficients for new nonsynonymous mutations using large samples. Genetics 206(1):345–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eyre-Walker A, Keightley PD. 2009. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol. Biol. Evol 26(9):2097–108 [DOI] [PubMed] [Google Scholar]
- 13.Huber CD, Kim BY, Marsden CD, Lohmueller KE. 2017. Determining the factors driving selective effects of new nonsynonymous mutations. PNAS 114(17):4465–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Glémin S, Lascoux M. 2017. Genetic diversity and the efficacy of purifying selection across plant and animal species. Mol. Biol. Evol 34(6):1417–28 [DOI] [PubMed] [Google Scholar]
- 15.Tataru P, Mollion M, Glémin S, Bataillon T. 2017. Inference of distribution of fitness effects and proportion of adaptive substitutions from polymorphism data. Genetics 207(3):1103–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castellano D, Macià MC, Tataru P, Bataillon T, Munch K. 2019. Comparison of the full distribution of fitness effects of new amino acid mutations across great apes. Genetics 213(3):953–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller HJ. 1950. Our load of mutations. Am. J. Hum. Genet 2:111–76 [PMC free article] [PubMed] [Google Scholar]
- 18.Haldane JBS. 1937. The effect of variation on fitness. Am. Nat 71:337–49 [Google Scholar]
- 19.Crow JF. 1970. Genetic loads and the cost of natural selection. In Mathematical Topics in Population Genetics, ed. Kojima K, pp. 128–77. Berlin, Heidelberg: Springer [Google Scholar]
- 20.Lesecque Y, Keightley PD, Eyre-Walker A. 2012. A resolution of the mutation load paradox in humans. Genetics 191(4):1321–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura M, Maruyama T, Crow JF. 1963. The mutation load in small populations. Genetics 48:1303–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitlock MC, Ingvarsson PK, Hatfield T. 2000. Local drift load and the heterosis of interconnected populations. Heredity 84(4):452–57 [DOI] [PubMed] [Google Scholar]
- 23.Lynch M, Gabriel W. 1990. Mutation load and the survival of small populations. Evolution 44(7):1725–37 [DOI] [PubMed] [Google Scholar]
- 24.Lynch M, Conery J, Burger R. 1995. Mutation accumulation and the extinction of small populations. Am. Nat 146(4):489–518 [Google Scholar]
- 25.Morton NE, Crow JF, Muller HJ. 1956. An estimate of the mutational damage in man from data on consanguineous marriages. PNAS 42(11):855–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedrick PW, Garcia-Dorado A. 2016. Understanding inbreeding depression, purging, and genetic rescue. Trends Ecol. Evol 31(12):940–52 [DOI] [PubMed] [Google Scholar]
- 27.Keller LF, Waller DM. 2002. Inbreeding effects in wild populations. Trends Ecol. Evol 17(5):230–41 [Google Scholar]
- 28.Wallace B. 1975. Hard and soft selection revisited. Evolution 29(3):465–73 [DOI] [PubMed] [Google Scholar]
- 29.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, et al. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 6(2):80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Resch AM, Carmel L, Mariño-Ramírez L, Ogurtsov AY, Shabalina SA, et al. 2007. Widespread positive selection in synonymous sites of mammalian genes. Mol. Biol. Evol 24(8):1821–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrie DS, Messer PW, Hershberg R, Petrov DA. 2013. Strong purifying selection at synonymous sites in D. melanogaster. PLOS Genet. 9(5):e1003527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machado HE, Lawrie DS, Petrov DA. 2020. Pervasive strong selection at the level of codon usage bias in Drosophila melanogaster. Genetics 214(2):511–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou T, Gu W, Wilke CO. 2010. Detecting positive and purifying selection at synonymous sites in yeast and worm. Mol. Biol. Evol 27(8):1912–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monroe JG, McKay JK, Weigel D, Flood PJ. 2021. The population genomics of adaptive loss of function. Heredity 126(3):383–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, et al. 2012. A systematic survey of loss-of-function variants in human protein-coding genes. Science 335(6070):823–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grantham R. 1974. Amino acid difference formula to help explain protein evolution. Science 185(4154):862–64 [DOI] [PubMed] [Google Scholar]
- 38.Miyata T, Miyazawa S, Yasunaga T. 1979. Two types of amino acid substitutions in protein evolution. J. Mol. Evol 12(3):219–36 [DOI] [PubMed] [Google Scholar]
- 39.Cooper GM, Stone EA, Asimenos G, NISC Comp. Seq. Prog., Green ED, et al. 2005. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15(7):901–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. 2010. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLOS Comput. Biol 6(12):e1001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar P, Henikoff S, Ng PC. 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc 4(7):1073–81 [DOI] [PubMed] [Google Scholar]
- 42.Ng PC, Henikoff S. 2001. Predicting deleterious amino acid substitutions. Genome Res. 11(5):863–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. 2010. A method and server for predicting damaging missense mutations. Nat. Methods 7(4):248–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. 2012. Predicting the functional effect of amino acid substitutions and indels. PLOS ONE 7(10):e46688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet 46(3):310–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huber CD, Kim BY, Lohmueller KE. 2020. Population genetic models of GERP scores suggest pervasive turnover of constrained sites across mammalian evolution. PLOS Genet. 16(5):e1008827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandell L, Sharp NP. 2022. Fitness effects of mutations: an assessment of PROVEAN predictions using mutation accumulation data. Genome Biol. Evol 14(1):evac004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawrie DS, Petrov DA. 2014. Comparative population genomics: power and principles for the inference of functionality. Trends Genet. 30(4):133–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simons YB, Turchin MC, Pritchard JK, Sella G. 2014. The deleterious mutation load is insensitive to recent population history. Nat. Genet 46(3):220–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Do R, Balick D, Li H, Adzhubei I, Sunyaev S, Reich D. 2015. No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nat. Genet 47(2):126–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henn BM, Botigué LR, Bustamante CD, Clark AG, Gravel S. 2015. Estimating the mutation load in human genomes. Nat. Rev. Genet 16(6):333–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brandvain Y, Wright SI. 2016. The limits of natural selection in a nonequilibrium world. Trends Genet. 32(4):201–10 [DOI] [PubMed] [Google Scholar]
- 53.Pedersen C-ET, Lohmueller KE, Grarup N, Bjerregaard P, Hansen T, et al. 2017. The effect of an extreme and prolonged population bottleneck on patterns of deleterious variation: insights from the Greenlandic Inuit. Genetics 205(2):787–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Messer PW. 2013. SLiM: simulating evolution with selection and linkage. Genetics 194(4):1037–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haller BC, Messer PW. 2019. SLiM 3: forward genetic simulations beyond the Wright-Fisher model. Mol. Biol. Evol 36(3):632–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adrion JR, Cole CB, Dukler N, Galloway JG, Gladstein AL, et al. 2020. A community-maintained standard library of population genetic models. eLife 9:e54967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohta T. 1976. Role of very slightly deleterious mutations in molecular evolution and polymorphism. Theor. Popul. Biol 10(3):254–75 [DOI] [PubMed] [Google Scholar]
- 58.Ohta T. 1973. Slightly deleterious mutant substitutions in evolution. Nature 246(5428):96–98 [DOI] [PubMed] [Google Scholar]
- 59.Ohta T. 1992. The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Evol. Syst 23:263–86 [Google Scholar]
- 60.Moyers BT, Morrell PL, McKay JK. 2018. Genetic costs of domestication and improvement. J. Hered 109(2):103–16 [DOI] [PubMed] [Google Scholar]
- 61.Makino T, Rubin C-J, Carneiro M, Axelsson E, Andersson L, Webster MT. 2018. Elevated proportions of deleterious genetic variation in domestic animals and plants. Genome Biol. Evol 10(1):276–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu J, Tang T, Tang H, Huang J, Shi S, Wu C-I. 2006. The accumulation of deleterious mutations in rice genomes: a hypothesis on the cost of domestication. Trends Genet. 22(3):126–31 [DOI] [PubMed] [Google Scholar]
- 63.Nabholz B, Sarah G, Sabot F, Ruiz M, Adam H, et al. 2014. Transcriptome population genomics reveals severe bottleneck and domestication cost in the African rice (Oryza glaberrima). Mol. Ecol 23(9):2210–27 [DOI] [PubMed] [Google Scholar]
- 64.Liu Q, Zhou Y, Morrell PL, Gaut BS. 2017. Deleterious variants in Asian rice and the potential cost of domestication. Mol. Biol. Evol 34(4):908–24 [DOI] [PubMed] [Google Scholar]
- 65.Kono TJY, Fu F, Mohammadi M, Hoffman PJ, Liu C, et al. 2016. The role of deleterious substitutions in crop genomes. Mol. Biol. Evol 33(9):2307–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koenig D, Jiménez-Gómez JM, Kimura S, Fulop D, Chitwood DH, et al. 2013. Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. PNAS 110(28):E2655–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Renaut S, Rieseberg LH. 2015. The accumulation of deleterious mutations as a consequence of domestication and improvement in sunflowers and other Compositae crops. Mol. Biol. Evol 32(9):2273–83 [DOI] [PubMed] [Google Scholar]
- 68.Marsden CD, Ortega-Del Vecchyo D, O’Brien DP, Taylor JF, Ramirez O, et al. 2016. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. PNAS 113(1):152–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schubert M, Jónsson H, Chang D, Sarkissian CD, Ermini L, et al. 2014. Prehistoric genomes reveal the genetic foundation and cost of horse domestication. PNAS 111(52):E5661–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang M-S, Zhang J-J, Guo X, Li M, Meyer R, et al. 2021. Large-scale genomic analysis reveals the genetic cost of chicken domestication. BMC Biol. 19(1):118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frantz LAF, Schraiber JG, Madsen O, Megens H-J, Cagan A, et al. 2015. Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat. Genet 47(10):1141–48 [DOI] [PubMed] [Google Scholar]
- 72.Xue Y, Prado-Martinez J, Sudmant PH, Narasimhan V, Ayub Q, et al. 2015. Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science 348(6231):242–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van der Valk T, Díez-del-Molino D, Marques-Bonet T, Guschanski K, Dalén L. 2019. Historical genomes reveal the genomic consequences of recent population decline in eastern gorillas. Curr. Biol 29(1):165–70.e6 [DOI] [PubMed] [Google Scholar]
- 74.Robinson JA, Ortega-Del Vecchyo D, Fan Z, Kim BY, vonHoldt BM, et al. 2016. Genomic flatlining in the endangered island fox. Curr. Biol 26(9):1183–89 [DOI] [PubMed] [Google Scholar]
- 75.Robinson JA, Brown C, Kim BY, Lohmueller KE, Wayne RK. 2018. Purging of strongly deleterious mutations explains long-term persistence and absence of inbreeding depression in island foxes. Curr. Biol 28(21):3487–94.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Manuel M, Barnett R, Sandoval-Velasco M, Yamaguchi N, Garrett Vieira F, et al. 2020. The evolutionary history of extinct and living lions. PNAS 117(20):10927–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.von Seth J, Dussex N, Díez-del-Molino D, van der Valk T, Kutschera VE, et al. 2021. Genomic insights into the conservation status of the world’s last remaining Sumatran rhinoceros populations. Nat. Commun 12(1):2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu L, Bosse M, Megens H-J, de Visser M, Groenen MAM, Madsen O. 2021. Genetic consequences of long-term small effective population size in the critically endangered pygmy hog. Evol. Appl 14(3):710–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dussex N, van der Valk T, Morales HE, Wheat CW, Díez-del-Molino D, et al. 2021. Population genomics of the critically endangered kākāpō. Cell Genom. 1(1):100002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robinson JA, Kyriazis CC, Nigenda-Morales SF, Beichman AC, Rojas-Bracho L, et al. 2022. The critically endangered vaquita is not doomed to extinction by inbreeding depression. Science 376(6593):635–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leroy T, Rousselle M, Tilak M-K, Caizergues AE, Scornavacca C, et al. 2021. Island songbirds as windows into evolution in small populations. Curr. Biol 31(6):1303–10.e4 [DOI] [PubMed] [Google Scholar]
- 82.Gravel S. 2016. When is selection effective? Genetics 203(1):451–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nei M, Maruyama T, Chakraborty R. 1975. The bottleneck effect and genetic variability in populations. Evolution 29(1):1–10 [DOI] [PubMed] [Google Scholar]
- 84.Balick DJ, Do R, Cassa CA, Reich D, Sunyaev SR. 2015. Dominance of deleterious alleles controls the response to a population bottleneck. PLOS Genet. 11(8):e1005436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kirkpatrick M, Jarne P. 2000. The effects of a bottleneck on inbreeding depression and the genetic load. Am. Nat 155(2):154–67 [DOI] [PubMed] [Google Scholar]
- 86.Lohmueller KE. 2014. The impact of population demography and selection on the genetic architecture of complex traits. PLOS Genet. 10(5):e1004379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Henn BM, Botigué LR, Peischl S, Dupanloup I, Lipatov M, et al. 2016. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. PNAS 113(4):E440–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Benazzo A, Trucchi E, Cahill JA, Maisano Delser P, Mona S, et al. 2017. Survival and divergence in a small group: the extraordinary genomic history of the endangered Apennine brown bear stragglers. PNAS 114(45):E9589–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beichman AC, Koepfli K-P, Li G, Murphy W, Dobrynin P, et al. 2019. Aquatic adaptation and depleted diversity: a deep dive into the genomes of the sea otter and giant otter. Mol. Biol. Evol 36(12):2631–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feng S, Fang Q, Barnett R, Li C, Han S, et al. 2019. The genomic footprints of the fall and recovery of the crested ibis. Curr. Biol 29(2):340–49.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grossen C, Guillaume F, Keller LF, Croll D. 2020. Purging of highly deleterious mutations through severe bottlenecks in Alpine ibex. Nat. Commun 11(1):1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu Y, Thapa A, Fan H, Ma T, Wu Q, et al. 2020. Genomic evidence for two phylogenetic species and long-term population bottlenecks in red pandas. Sci. Adv 6(9):eaax5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ochoa A, Gibbs HL. 2021. Genomic signatures of inbreeding and mutation load in a threatened rattlesnake. Mol. Ecol 30(21):5454–69 [DOI] [PubMed] [Google Scholar]
- 94.Kleinman-Ruiz D, Lucena-Perez M, Villanueva B, Fernández J, Saveljev AP, et al. 2022. Purging of deleterious burden in the endangered Iberian lynx. PNAS 119(11):e2110614119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xie H-X, Liang X-X, Chen Z-Q, Li W-M, Mi C-R, et al. 2022. Ancient demographics determine the effectiveness of genetic purging in endangered lizards. Mol. Biol. Evol 39(1):msab359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Beichman AC, Kalhori P, Kyriazis CC, DeVries AA, Nigenda-Morales S, et al. 2021. Genomic analyses reveal range-wide devastation of sea otter populations. Mol. Ecol In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Peischl S, Dupanloup I, Kirkpatrick M, Excoffier L. 2013. On the accumulation of deleterious mutations during range expansions. Mol. Ecol 22(24):5972–82 [DOI] [PubMed] [Google Scholar]
- 98.Peischl S, Kirkpatrick M, Excoffier L. 2015. Expansion load and the evolutionary dynamics of a species range. Am. Nat 185(4):E81–93 [DOI] [PubMed] [Google Scholar]
- 99.Peischl S, Dupanloup I, Foucal A, Jomphe M, Bruat V, et al. 2018. Relaxed selection during a recent human expansion. Genetics 208(2):763–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Willi Y, Fracassetti M, Zoller S, Van Buskirk J. 2018. Accumulation of mutational load at the edges of a species range. Mol. Biol. Evol 35(4):781–91 [DOI] [PubMed] [Google Scholar]
- 101.Perrier A, Sánchez-Castro D, Willi Y. 2020. Expressed mutational load increases toward the edge of a species’ geographic range. Evolution 74(8):1711–23 [DOI] [PubMed] [Google Scholar]
- 102.Jones MR, Mills LS, Jensen JD, Good JM. 2020. The origin and spread of locally adaptive seasonal camouflage in snowshoe hares. Am. Nat 196(3):316–32 [DOI] [PubMed] [Google Scholar]
- 103.Rougemont Q, Moore J-S, Leroy T, Normandeau E, Rondeau EB, et al. 2020. Demographic history shaped geographical patterns of deleterious mutation load in a broadly distributed Pacific salmon. PLOS Genet. 16(8):e1008348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Charlesworth D, Charlesworth B. 1987. Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Evol. Syst 18:237–68 [Google Scholar]
- 105.Charlesworth D, Willis JH. 2009. The genetics of inbreeding depression. Nat. Rev. Genet 10(11):783–96 [DOI] [PubMed] [Google Scholar]
- 106.Wang J, Hill WG, Charlesworth D, Charlesworth B. 1999. Dynamics of inbreeding depression due to deleterious mutations in small populations: mutation parameters and inbreeding rate. Genet. Res 74(02):165–78 [DOI] [PubMed] [Google Scholar]
- 107.Hedrick PW. 1994. Purging inbreeding depression and the probability of extinction: full-sib mating. Heredity 73(4):363–72 [DOI] [PubMed] [Google Scholar]
- 108.Räikkönen J, Vucetich JA, Peterson RO, Nelson MP. 2009. Congenital bone deformities and the inbred wolves (Canis lupus) of Isle Royale. Biol. Conserv 142(5):1025–31 [Google Scholar]
- 109.Hedrick PW, Peterson RO, Vucetich LM, Adams JR, Vucetich JA. 2014. Genetic rescue in Isle Royale wolves: genetic analysis and the collapse of the population. Conserv. Genet 15(5):1111–21 [Google Scholar]
- 110.Robinson JA, Räikkönen J, Vucetich LM, Vucetich JA, Peterson RO, et al. 2019. Genomic signatures of extensive inbreeding in Isle Royale wolves, a population on the threshold of extinction. Sci. Adv 5(5):eaau0757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Khan A, Patel K, Shukla H, Viswanathan A, van der Valk T, et al. 2021. Genomic evidence for inbreeding depression and purging of deleterious genetic variation in Indian tigers. PNAS 118(49):e2023018118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kyriazis CC, Beichman AC, Brzeski KE, Hoy SR, Peterson RO, et al. 2022. Genomic underpinnings of population persistence in Isle Royale moose. bioRxiv. 10.1101/2022.04.15.488504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Crow JF. 1948. Alternative hypotheses of hybrid vigor. Genetics 33(5):477–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim BY, Huber CD, Lohmueller KE. 2018. Deleterious variation shapes the genomic landscape of introgression. PLOS Genet. 14(10):e1007741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Harris K, Zhang Y, Nielsen R. 2019. Genetic rescue and the maintenance of native ancestry. Conserv. Genet 20(1):59–64 [Google Scholar]
- 116.Tallmon DA, Luikart G, Waples RS. 2004. The alluring simplicity and complex reality of genetic rescue. Trends Ecol. Evol 19(9):489–96 [DOI] [PubMed] [Google Scholar]
- 117.Bell DA, Robinson ZL, Funk WC, Fitzpatrick SW, Allendorf FW, et al. 2019. The exciting potential and remaining uncertainties of genetic rescue. Trends Ecol. Evol 34(12):1070–79 [DOI] [PubMed] [Google Scholar]
- 118.Frankham R. 2016. Genetic rescue benefits persist to at least the F3 generation, based on a meta-analysis. Biol. Conserv 195:33–36 [Google Scholar]
- 119.Hedrick PW, Robinson JA, Peterson RO, Vucetich JA. 2019. Genetics and extinction and the example of Isle Royale wolves. Anim. Conserv 22(3):302–9 [Google Scholar]
- 120.Lotsander A, Hasselgren M, Larm M, Wallén J, Angerbjörn A, Norén K. 2021. Low persistence of genetic rescue across generations in the Arctic fox (Vulpes lagopus). J. Hered 112(3):276–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lande R. 1994. Risk of population extinction from fixation of new deleterious mutations. Evolution 48(5):1460–69 [DOI] [PubMed] [Google Scholar]
- 122.Rogers RL, Slatkin M. 2017. Excess of genomic defects in a woolly mammoth on Wrangel Island. PLOS Genet. 13(3):e1006601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schultz ST, Lynch M. 1997. Mutation and extinction: the role of variable mutational effects, synergistic epistasis, beneficial mutations, and degree of outcrossing. Evolution 51(5):1363–71 [DOI] [PubMed] [Google Scholar]
- 124.Beyer RM, Manica A. 2020. Historical and projected future range sizes of the world’s mammals, birds, and amphibians. Nat. Commun 11(1):5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ceballos G, Ehrlich PR, Raven PH. 2020. Vertebrates on the brink as indicators of biological annihilation and the sixth mass extinction. PNAS 117(24):13596–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Johnson WE, Onorato DP, Roelke ME, Land ED, Cunningham M, et al. 2010. Genetic restoration of the Florida panther. Science 329(5999):1641–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Haller BC, Messer PW. 2019. Evolutionary modeling in SLiM 3 for beginners. Mol. Biol. Evol 36(5):1101–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kimura M. 1962. On the probability of fixation of mutant genes in a population. Genetics 47:713–19 [DOI] [PMC free article] [PubMed] [Google Scholar]