Abstract
Background.
Blockchain distributed ledger technology is just starting to be adopted in genomics and healthcare applications. Despite its increased prevalence in biomedical research applications, skepticism regarding the practicality of blockchain technology for real-world problems is still strong and there are few implementations beyond proof-of-concept. We focus on benchmarking blockchain strategies applied to distributed methods for sharing records of gene-drug interactions. We expect this type of sharing will expedite personalized medicine.
Basic Procedures.
We generated gene-drug interaction test datasets using the Clinical Pharmacogenetics Implementation Consortium (CPIC) resource. We developed three blockchain-based methods to share patient records on gene-drug interactions: Query Index, Index Everything, and Dual-Scenario Indexing.
Main Findings.
We achieved a runtime of about 60 seconds for importing 4,000 gene-drug interaction records from four sites, and about 0.5 seconds for a data retrieval query. Our results demonstrated that it is feasible to leverage blockchain as a new platform to share data among institutions.
Principal Conclusions.
We show the benchmarking results of novel blockchain-based methods for institutions to share patient outcomes related to gene-drug interactions. Our findings support blockchain utilization in healthcare, genomic and biomedical applications. The source code is publicly available at https://github.com/tsungtingkuo/genedrug.
Keywords: Blockchain Distributed Ledger Technology, Pharmacogenetics, Gene-Drug Interaction, Data Sharing, Smart Contract
1. INTRODUCTION
1.1. Gene-Drug Interaction
Genetic variation is known to affect drug response. Presence of specific genetic variants can result in variability of drug efficacy and adverse drug reactions (ADR) through alternate pharmacokinetic (PK) and pharmacodynamic (PD) pathways. One such example is warfarin, an anticoagulant commonly used to prevent or treat blood clots. It is notoriously challenging to correctly adjust warfarin doses due to inter-patient variability resulting from both clinical data (e.g., age, sex, race, body mass index, conditions, and other medications) and genetics (e.g., variants in VKORC1, CYP2C9, and CYP4F2 genes) [1]. While patients with AA genotypes in SNP rs9923231 of the VKORC1 gene are sensitive to warfarin and require lower doses, those with AG or GG genotypes are less sensitive. Complications arising from inadequate warfarin dosing constitute some of the most common ADRs reported to the Food and Drug Administration (FDA) [2]. For this reason, warfarin has been added to the FDA list of drugs with pharmacogenomics labeling; the recent list has 304 unique drugs [3].
Gene-drug relationship data are very important for clinicians and researchers. There are several publicly available gene-drug interaction datasets, such as the one produced by the Clinical Pharmacogenetics Implementation Consortium (CPIC) [4]. Based on these datasets, researchers may evaluate and investigate interactions for associations with specific patient outcomes (e.g., improved, unchanged, or deteriorated), suspected gene-outcome-relations (e.g., yes, or no), and serious side-effects (e.g., yes, or no). However, these evaluation results may be siloed within an institution. A mechanism for institutions to share the evaluation results of the gene-drug interactions they obtained locally could help speed up research.
With the advance of sequencing technology, genetic testing is becoming more available, making pharmacogenetic-based drug dosing more viable in clinical practice. CPIC is one such effort to provide peer-reviewed, updated, and evidence-based guidelines for gene-drug pairs. However, a level 1 quality guideline in CPIC requires consistent evidence, with large sample sizes in well-designed and well-conducted studies. Gathering sufficient and high-quality evidence of gene-drug outcomes is still a daunting task due to technical, economic, administrative, and ethical reasons.
1.2. Traditional Methods and Threat Models
Intuitively, we can adopt a centralized method that uses a central server and collect the evaluation results (Figure 1A) via a traditional local software program performing logging/querying operations (Figure 2A). However, this setting could introduce multiple threats. As shown in previous studies [5 6], a central server and traditional program can present the barriers/challenges listed below:
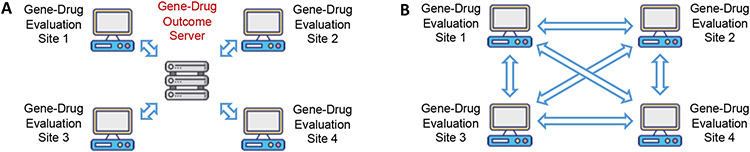
Figure 1.
Architecture of storing the patient outcome of gene-drug pairs. A. Centralized architecture (central server) where the centralized gene-drug outcome server can lead to a single-point-of-failure. The central server can change the records from other sites and can even modify the source of evaluation results. B. Decentralized architecture (blockchain) without a central server that can eliminate the possibility of a single point-of-failure. By adopting blockchain technology, the data are immutable and source-verifiable.
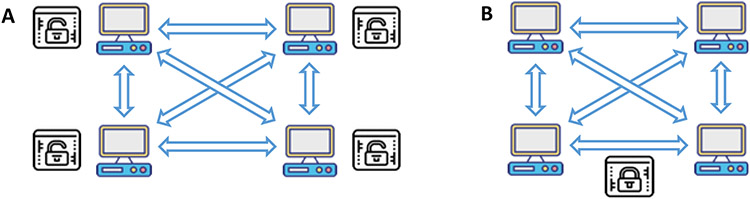
Figure 2.
Programs used to store and query patient outcomes for gene-drug pairs. A. Traditional off-chain program that is non-transparent and mutable. B. On-chain smart contracts that are transparent and immutable among the sites.
Single-point-of-failure (e.g., the whole system stops working when the server stops due to a routine maintenance or a malicious attack).
Mutable data (e.g., the information on the server may be altered by the “root” user).
Unverifiable data source (e.g., the sources of the evaluation results may also be changed on the central server).
Non-transparent software (e.g., unspecified changes and thus inconsistent code).
Alterable programs (e.g., the deployed program can still be altered locally).
1.3. Blockchain Smart Contracts
To overcome these issues, we consider a decentralized architecture to solve the above-mentioned risks brought by a central server and traditional program. This architecture enables consistent and large-scale evidence gathering from multiple participating hospitals and individuals. Among the decentralized data storage methods, blockchain [7-10] is one of the more promising candidates (Figure 1B). The latest blockchain platforms, such as Ethereum [9], Hyperledger Fabric [12], or R3 Corda [13], support smart contracts, (Figure 2B) which are computer programs running on blockchain [14]. The desired technical properties of blockchain with smart contracts [14-16] include:
No single-point-of-failure (i.e., it is peer-to-peer).
Immutable data (i.e., it is very difficult to change the data on the chain).
Data provenance (i.e., the source of data is confirmed and therefore cannot be falsified).
Transparent software (e.g., each software change can be verified and confirmed).
Unchangeable program code (e.g., the deployed program is not alterable, and new versions of the program are recorded and visible to all nodes) [17].
Therefore, using smart contracts on blockchain to store and query patient outcomes related to gene-drug data pairs could further improve the transparency and immutability of the software among the participating institutions.
Blockchain has been proposed in various healthcare, genomic and biomedical applications [5 6 16-19], such as medical record management [16 20 21], dynamic consent in biobanking [21], genomic data access logging [18 23], pharmaceutical supply chain [24-26], and privacy-preserving predictive modeling [27-29]. Meanwhile, applications in pharmacogenetics are still limited [19]. While blockchain has been the underlying infrastructure for crypto-currencies such as Bitcoin [8] for more than a decade, the design and usability of blockchain have yet to be well-understood in health sciences as they currently are in the world of finance.
Although the idea of adopting blockchain and smart contracts for sharing gene-drug evaluation results may conceptually be feasible, practical issues in implementing such a system have yet to be investigated. Many blockchain-based solutions are still in early stages [23 34] and the resources to support blockchain and smart contract developers are also scarce [35 36]. Therefore, we aim at benchmarking the potential of a decentralized gene-drug system on blockchain, with smart contracts.
2. MATERIAL AND METHODS
2.1. Competition
University of California San Diego (UCSD) adopted a community-based approach to benchmarking, and organized Track 1 of the iDASH Secure Genome Analysis Competition in 2019 [17]. There were 30 teams from 11 countries, including China, Germany, India, Japan, Luxembourg, Netherlands, Singapore, Switzerland, Turkey, United Kingdom, and the USA. The development phase lasted three months, after which five teams submitted solutions. We requested that each solution be able to store all patient outcomes for gene-drug pair records on-chain (i.e., no off-chain local storage of data was allowed). For querying the records, the solution was required to support searching records by any combination of gene name, variant number, and drug name. Results had to contain counts and percentages of outcomes, suspected-gene-outcome-relations, and serious-side-effects. The solution was also required to make the records searchable from any site (e.g., Institution 1 should be able to search any record from Institution 2 and so on).
Existing blockchain and smart contract studies have demonstrated their features and advantages, such as immutability/robustness [8 37 38], either by mathematical proof or empirical analyses, along with thorough comparisons with centralized or redundant solutions [17 35 36]. In this competition, we aimed to demonstrate the feasibility of adopting blockchain and smart contracts to share patient outcomes related to gene-drug interactions among institutions. Of the five submitted solutions, one was unable to complete within the competition timeline and another published their results separately [41]. Therefore, in this study, we focus on the benchmarking and comparison of three solutions.
The blockchain platform we selected based on prior review [15] was Ethereum [9], which is an open source platform that supports smart contracts and that is maintained by the community. We configured the Ethereum blockchain network as a permissioned network, so that the evaluations could be executed independently of the public blockchain, and the testing environment would not be tied to the concept of crypto-currency. We adopted the Proof-of-Authority (PoA) consensus protocol using the Clique algorithm [42], which is suitable for permissioned networks that do not need intensive computation like the one needed for the Proof-of-Work (PoW) Ethash algorithm [37] to secure the network. Compared to other platforms (e.g., Hyperledger Fabric [12] or R3 Corda [13]) that also support smart contracts, Ethereum does not require additional ordering or notary services, thus it is appropriate for our purpose. We adopted Solidity [43], one of the most popular smart contract languages running on Ethereum, to implement the solutions.
2.2. Data
The dataset for benchmarking was generated using the gene-drug relationship data from CPIC [4]. Each record contained the following six fields (Table 1): gene name, variant number, drug name, outcome, suspected gene outcome relation, and serious side effect. First, we obtained 127 unique gene names and 226 unique drug names from CPIC and randomly chose one gene name and one drug name as a pair to generate a record. Next, for each record, we selected a variant number [1 – 99], an outcome status [Improved, Unchanged, Deteriorated], a suspected gene outcome relation [Yes, No], and a serious side effect [Yes, No], all randomly. For the development process the teams were provided with four patient outcomes of gene-drug pair files, each of which with 10,000 records representing the observed patient outcome for a gene-drug pair from four institutions. During the evaluation process we utilized 200 and 1,000 records from each of the four sites.
Table 1.
Description of a record in our dataset. The dataset is available in [17].
2.3. Methods Overview
We developed three methods to solve the distributed data sharing problem: Query Index (hashing-based mapping), Index Everything (comprehensive mapping), and Dual-Scenario Indexing (complete/wildcard mapping). The three solutions were developed by the following three teams, respectively: Emory Team, formed by members from Emory University and Kyoto University (1st place winner of the competition), Team Genigma from Sandia National Laboratories (2nd place), and Omics for all from BGI-Shenzhen (Honorable Mention). The details of these promising solutions are introduced in the following subsections.
2.4. Query Index
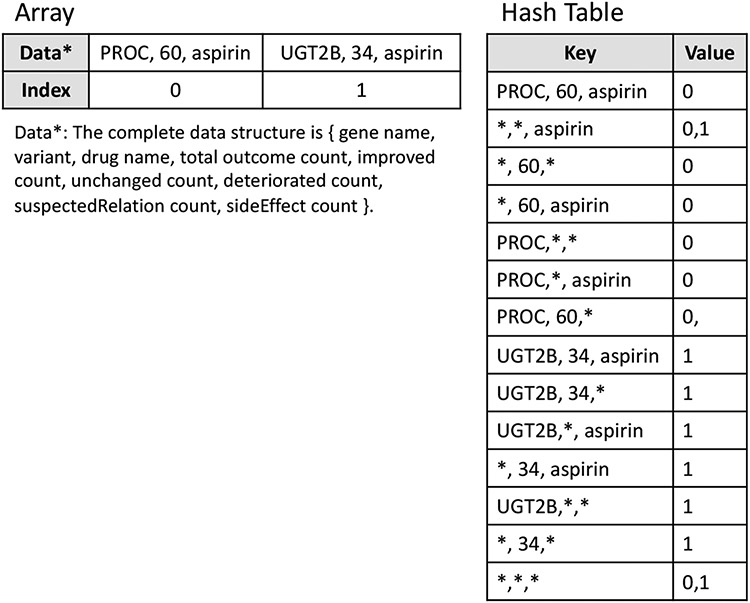
The first method, Query Index, was a domain knowledge-based approach to implement a storage and query efficient solution. The following two kinds of domain knowledge in the gene-drug interaction data sharing were utilized in the design of an efficient solution: (1) the query output is the accumulated statistics of the gene-drug interaction data, and (2) the amount of unique gene-drug relations (i.e., approximately 106 in CPIC specification) is much smaller than the amount of raw gene-drug interaction records. This implementation utilized the above two facts, stored the statistical information of all unique gene-drug relations (i.e., gene-variant-drug triples) in an upper-bounded size array and cached all indices in a hash table for fast insertion and query. Figure 3 illustrates an example of the array and hash table data structure of Query Index. Every gene-variant-drug triple could be invoked in 8 different types of queries (i.e., a query specifying gene name, drug name, and variant number and 7 queries with wildcard characters in different fields). For example, the result of GBA-nicotine-74 will be returned in query (GBA, nicotine, 74), query (GBA, *, *), query (*, *, *), and so on. Based on this small number of query fields, a key-value hash table was built to support all possible queries. In the hash table, the keys were gene-variant-drug tuples and their wildcard alternatives, and the values were the indices of the actual information in the array. Upon receiving a query request, the Query Index method first found the matching index list in the hash table if the record existed, then traversed the indices to retrieve the actual information from the array. For the insertion, with the help of the hash table, the method could locate the index of the gene-variant-drug tuple in the array in O(1) time and update the counts. If the record did not exist, the method would append the record at the end of the array and insert corresponding entries in the hash table.
Figure 3.
Example of two records for the Query Index method.
2.5. Index Everything
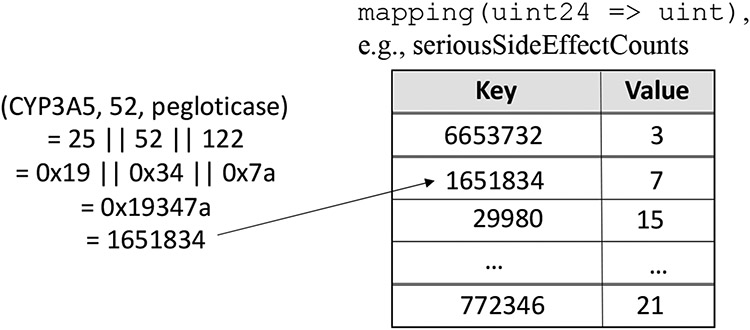
The second method, Index Everything, was a straightforward implementation approach. Since there were only a few hundred distinct genes and drugs, a unique 8-bit unsigned integer (uint8) value was assigned to each distinct gene (respectively, drug) value. These values were assigned lazily, i.e., the next available ascending value was assigned upon the first insert containing that gene or drug. As such, a unique 24-bit unsigned integer (uint24) could be trivially derived for each gene-variant-drug triple, specifically by concatenating the corresponding three uint8s. Thus, for any observation, this uint24 derived by concatenation was used as an index into various outcome counts stored in the Solidity mapping structures. This indexing/storage scheme is illustrated in Figure 4. The two query modalities (entryExists and query) implementations were similarly straightforward. Specifically, given the wildcard value (‘*’) in any position, all possible values were searched for that position, expressed as a triple for which any non-wildcard search value collapsed the specific dimension.
Figure 4.
Visual depiction of the scheme of the Index Everything method (∣∣ denotes integer concatenation) on the left, and an example mapping data structure counting side effects for each unique gene/variant/drug triple on the right. Structures like the one on the right exist for all observation categories: improved, unchanged, deteriorated, suspected relation, and side effect.
2.6. Dual-Scenario Indexing
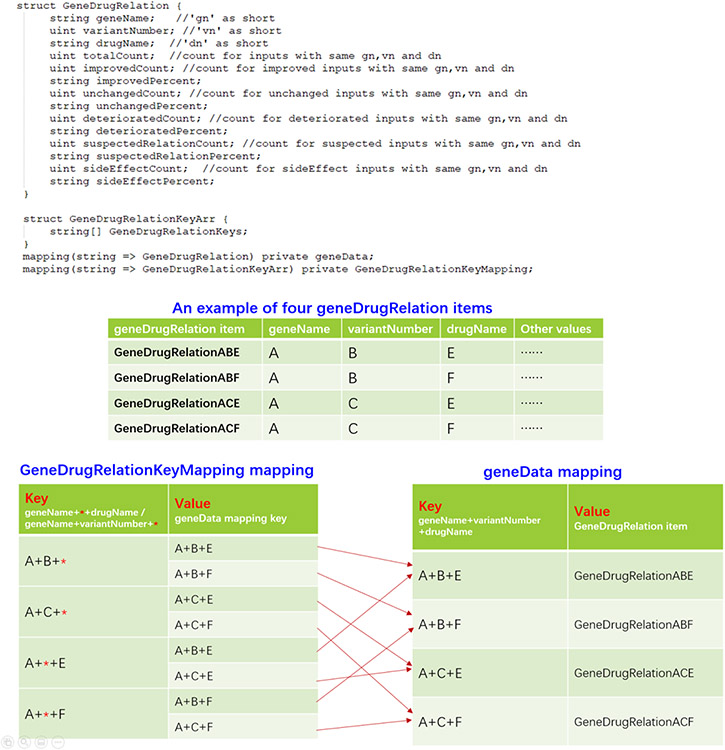
The third method, Dual-Scenario Indexing, adopted a special data structure to store gene-drug relationship data. It was also assumed here that query operations (such as query and entryExists) were more frequently invoked than insert operations, thus the team focused on query performance optimizations. Two different data structures were used to support the precise search with all three given inputs (gene name, variant number and drug name) and the search with wildcard inputs under two scenarios: complete (i.e., gene-variant-drug ) and wildcard searches. For the complete search scenario, a mapping structure named geneData mapping was used to store all GeneDrugRelation items with a key that was the concatenation of gene name A, variant number B and drug name E. Therefore, the geneData map could easily support all queries with “ABE” inputs. For the wildcard search scenario, the team built a special mapping structure GeneDrugRelationKeyMapping with keys of wildcard search strings (e.g., “AB*”) and values of the complete search strings (e.g., “ABE”, the keys of the geneData data structure). The algorithm then pre-generated all possible combinations of geneData mapping keys for each wildcard input, and stored these combinations into the GeneDrugRelationKeyMapping data structure . For querying, the algorithm first searched GeneDrugRelationKeyMapping by “AB*” to get all geneData keys (e.g., “ABE” and others) that correspond to GeneDrugRelation items with A and B. Then, it searched geneData mapping to get the detailed GeneDrugRelation items. An example explaining how GeneDrugRelationKeyMapping supports wildcard query operations is shown in Figure 5.
Figure 5.
Key data store structure of the Dual-Scenario Indexing method.
3. RESULTS
3.1. Evaluation
To evaluate the solutions, we inserted the two datasets (i.e., 200 and 1,000) to the blockchain either 1 or 200 records at a time to simulate different insertion speeds and generated 60 queries to compute the query time required by each solution. Our evaluation criteria specified that: (a) a solution must complete the insertion of all records, (b) a solution must provide 100% correct query results, and (c) the speed of insertion and query is the most important feature, followed by storage and memory cost, and then scalability. Therefore, after checking the completeness and correctness of the solutions, we measured the insertion times, query times, disk storage, and memory usage, and then normalized these measurements to raw scores from 0 to 100. The raw scores were then weighted-summed to a subtotal score (insertion time = 35%, query time = 35%, disk usage = 15%, and memory usage = 15%). Next, the subtotal scores were weighted-summed to an overall score, with the weights corresponding to the number of test records (i.e., 200 and 1,000) to account for scalability. Finally, the overall scores for inserting 1 and 200 records at a time were averaged to generate the final scores.
The compute environment for evaluation was iDASH 2.0 [44], a Health Insurance Portability and Accountability Act (HIPAA) compliant platform based on Amazon Web Services (AWS) and supported by the UCSD Health Information Services and Department of Biomedical Informatics. We set up 24 Virtual Machines (VMs) to evaluate the solutions. Each VM had 2 CPU cores, 8 GB of RAM and 100 GB of storage; Ubuntu was the operating system.
3.2. Measurement Results and Final Scores
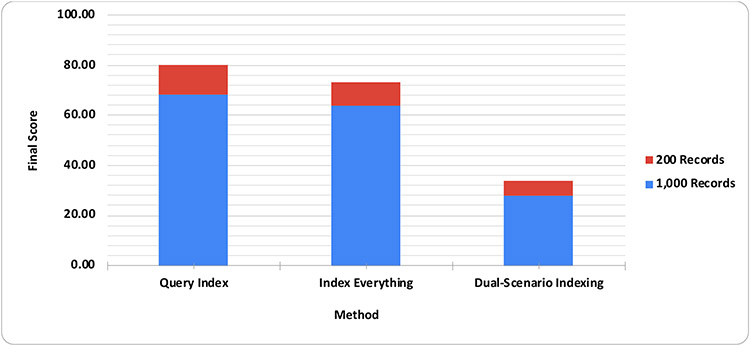
Results and the scores are summarized in Table 2 and Figure 6, respectively. As shown in the tables, inserting 200 records at a time reduced insertion time per record significantly. Also, while the insertion time increased linearly with the number of records in the test data, query times were more consistent, which could reflect the blockchain characteristic that writing is relatively slow (because it requires consensus block creation), while reading is fast (only local blocks are searched). The required disk space (< 40 MB) and memory (< 300 MB) were relatively small. In terms of final scores, the Query Index method performed the best, followed by the Index Everything method. The Dual-Scenario Indexing method used more memory, and its insertion/query time and disk usage were comparable with those of other solutions.
Table 2.
Results of each solution with different combination scenarios of records in test data (i.e., 200 versus 1,000) and number of records inserted at a time (i.e., 1 versus 200). The Runtime of Query is the time to execute 60 different queries. Note: A software update of the Dual-Scenario Indexing (marked with “*”) was applied after the competition deadline to produce correct results with performance no worse than that of the original submission on one record per insert, and a negligible increase in insertion speed on 200 records per insert; measured query speed increased in all cases since the correct results had smaller size.
| Number of Total Records per Site |
Number of Records per Insert |
Solution | Complete | Correct | Runtime of Insertion (s) |
Runtime of Query (s) |
Storage Usage (MB) |
Memory Usage (MB) |
|---|---|---|---|---|---|---|---|---|
| 200 | 1 | Query Index | Yes | Yes | 212.75 | 23.00 | 21.18 | 56.54 |
| Index Everything | Yes | Yes | 226.75 | 28.00 | 21.23 | 56.44 | ||
| Dual-Scenario Indexing | Yes | Yes* | 203.00 | 30.00 | 21.19 | 106.11 | ||
| 200 | Query Index | Yes | Yes | 13.25 | 23.00 | 19.49 | 90.32 | |
| Index Everything | Yes | Yes | 4.75 | 28.00 | 19.46 | 73.26 | ||
| Dual-Scenario Indexing | Yes | Yes* | 11.25 | 29.50 | 19.50 | 106.06 | ||
| 1,000 | 1 | Query Index | Yes | Yes | 1006.75 | 24.00 | 31.44 | 59.41 |
| Index Everything | Yes | Yes | 1157.25 | 29.00 | 32.98 | 58.87 | ||
| Dual-Scenario Indexing | Yes | Yes* | 1003.50 | 53.00 | 31.36 | 226.14 | ||
| 200 | Query Index | Yes | Yes | 51.50 | 24.00 | 24.99 | 110.10 | |
| Index Everything | Yes | Yes | 12.25 | 29.00 | 24.09 | 103.72 | ||
| Dual-Scenario Indexing | Yes | Yes* | 49.75 | 53.00 | 25.11 | 225.73 |
Figure 6.
Final scores for each solution. The results were weighted based on the number of records in the test data (i.e., 200 records in red and 1,000 records in blue) and were averaged from the results of inserting 1 or 200 records at a time.
3.3. Comparison of the Three Proposed Methods
To further understand the differences between our three proposed methods, we analyzed the results in Table 2 for each of our proposed methods as follows. The storage usage for all solutions is similar (approximately 20 – 35 MB) and negligible when considering modern storage devices (e.g., 100 GB in our experiments). Therefore, our analysis focused on the other three measurements (i.e., runtime of insertion, runtime of query, and memory usage).
Query Index. This method constructed a hash table for the queries and provided superior runtimes of query (23 – 24 seconds for 60 queries, or about 0.5 seconds per query, the fastest in all different scenarios regardless of the number of records per insertion). It also had relatively small memory usage (like the best solution, Index Everything, in all scenarios). For the runtime of insertion, it performed better when one record at a time was inserted, while it was comparatively slower when multiple records were inserted at a time.
Index Everything. This approach indexed all possible queries ahead in a mapping table and performed extremely well when multiple records at a time were inserted (only 24% - 42% of the time used by the other two methods). It also used the least memory in all combination scenarios. However, this method required more insertion time when one record at a time was inserted. Also, the query time was slightly slower than that for the Query Index method.
Dual-Scenario Indexing. This solution created two mapping structures to store the complete and wildcard queries and provided the shortest insertion time when one record at a time was inserted. The runtimes of insertion for multiple records at a time were comparable to those for the Query Index method. It required more time to query and more memory usage when compared to the other two methods.
To summarize, different methods can be more suitable for different applications and scenarios. To reach a fast insertion time, Index Everything (inserting multiple records at a time) and the Dual-Scenario Indexing (inserting one record at a time) would be more appropriate. To optimize query time, Query Index would be the best method. To preserve memory usage, both Index Everything and Query Index approaches could be considered.
4. DISCUSSION
To benchmark and understand the potential of the decentralized gene-drug relationship sharing system on blockchain with smart contracts, we developed three methods: Query Index, Index Everything, and Dual-Scenario Indexing. These methods applied different techniques (hash, comprehensive, and complete/wildcard mapping) to index the queries. The concepts of the proposed methods were straightforward, and we demonstrated their feasibility. Our results can serve as the basis for future researchers to improve their blockchain-based solutions in different applications (e.g., requiring faster insertion time, needing shorter query time, or preferring smaller memory usage).
Although the speed of logging and querying gene-drug outcome records on blockchain via smart contract is not comparable with that of a traditional database and may limit the real-world applications, we believe the benefits of our proposed solution (i.e., no single-point-of-failure, immutable data, guaranteed data provenance, transparent software, and an unchangeable program) are important to the sharing of the gene-drug evaluation results. Our work also provides a contribution to the broader perspective of benchmarking blockchain platforms for non-healthcare applications and implementations [45 46].
During the development and evaluation of solutions, we identified that the rapidly evolving blockchain and smart contract platform could create challenges. Looking at the example of Ethereum, the platform is implemented in using the GO programming language and has had more than 150 releases since its first release in 2014 (i.e., about 2 weeks per release on average) [47]. Therefore, the performance of our methods may be improved when the underlying blockchain platform becomes more mature.
Our observations are limited to the results based on Ethereum smart contract implementation using PoA consensus protocol. Although the general concept of the simulated evaluation for the pharmacogenetics gene-drug sharing application can be adopted by using other blockchain platforms such as Hyperledger Fabric and R3 Corda, more experiments need to be conducted to compare the speed and scalability of different blockchain platform options. Also, evaluations on a larger dataset and more blockchain nodes can further reveal the scalability performance of this application.
Moving forward, this benchmark study only simulated multiple-site record sharing, and real deployments of the suggested solutions can be the next step. For example, the implementations can be packaged into Docker [48] image files to simplify the process of adopting our proposed approaches. Additionally, our benchmarking is limited to evaluating the performance of our methods on pharmacogenetics data; investigating other aspects of blockchain (e.g., governance, adjudication, and permission controls) could also extend this study.
5. CONCLUSION
We demonstrated that sharing gene-drug interaction data using smart contracts on blockchain technology is feasible. Specifically, we can store 4,000 gene-drug evaluation results from 4 sites within 1 minute and query all these pairs within 0.5 seconds. We believe these results can serve as benchmarks for future blockchain-based healthcare, genomic and biomedical applications.
HIGHLIGHTS.
We developed blockchain-based methods to share gene-drug interactions.
We showed the feasibility to using blockchain to share data among institutions.
Our results suggested that blockchain can enhance the process of drug development.
SUMMARY TABLE.
| What was already known on the topic |
|
| What this study added to our knowledge |
|
ACKNOWLEDGEMENTS.
The authors would like to acknowledge the support from PlatON International Limited. We would also like to acknowledge Dr. Heidi J. Sofia, Dr. Haixu Tang, Dr. XiaoFeng Wang, Dr. Xiaoqian Jiang, Dr. Arif Harmanci, and Dr. Miran Kim for their support in co-organizing the competition and the workshop. The use of the integrating Data for Analysis, Anonymization, and SHaring (iDASH) 2.0 Amazon Web Services (AWS) cloud network is supported by Dr. Michael Hogarth, Andrew Greaves and Jit Bhattacharya.
FUNDING.
The competition is funded by the U.S. National Institutes of Health (NIH) (R13HG009072). T.-T. Kuo is partly funded by the National Human Genome Research Institute (NHGRI) of the U.S. NIH under Award Number R00HG009680, the U.S. NIH (R01HL136835 and R01GM118609), and UCSD Academic Senate Research Grant RG084150. L. Ohno-Machado is funded by the U.S. NIH (R01GM118609, R01HL136835, R01HG011066). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. C. M. Hudson and N. Pattengale are partly supported by the Laboratory Directed Research and Development program at Sandia National Laboratories, a multi-mission laboratory managed and operated by National Technology and Engineering Solutions of Sandia, LLC, a wholly owned subsidiary of Honeywell International, Inc., for the U.S. Department of Energy's National Nuclear Security Administration under contract DE-NA0003525. This paper describes objective technical results and analysis. Any subjective views or opinions that might be expressed in the paper do not necessarily represent the views of the U.S. Department of Energy or the United States Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The author M. Yang is affiliated with BGI-Shenzhen. All the other authors declare no competing interests.
REFERENCES
- 1.Dean L. Warfarin therapy and VKORC1 and CYP genotype. Medical Genetics Summaries [Internet]: National Center for Biotechnology Information (US), 2018. [PubMed] [Google Scholar]
- 2.WarfarinDosing.org. http://warfarindosing.org/Source/Home.aspx Accessed March 26, 2020
- 3.Table of Pharmacogenomic Biomarkers in Drug Labeling. https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling Accessed April 20, 2021
- 4.Clinical Pharmacogenetics Implementation Consortium (CPIC) Genes-Drugs. https://cpicpgx.org/genes-drugs/ Accessed November 12, 2019
- 5.Mackey TK, Kuo T-T, Gummadi B, et al. ‘Fit-for-purpose?’ – challenges and opportunities for applications of blockchain technology in the future of healthcare. BMC Medicine 2019; 17 (1): 68 doi: 10.1186/s12916-019-1296-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo T-T, Kim H-E, Ohno-Machado L. Blockchain distributed ledger technologies for biomedical and health care applications. Edited by Ohno-Machado Lucila. Published by Oxford University Press, Kettering, Northants, UK. Journal of the American Medical Informatics Association (JAMIA) 2017; 24 (6): 1211–20 doi: 10.1093/jamia/ocx068. September 8, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yue X, Wang H, Jin D, Li M, Jiang W. Healthcare Data Gateways: Found Healthcare Intelligence on Blockchain with Novel Privacy Risk Control. Journal of medical systems 2016; 40 (10): 218. [DOI] [PubMed] [Google Scholar]
- 8.Nakamoto S. Bitcoin: A peer-to-peer electronic cash system. 2008. https://bitcoin.org/bitcoin.pdf Accessed January 4, 2021 [Google Scholar]
- 9.Buterin V. A next-generation smart contract and decentralized application platform. 2014. https://ethereum.org/en/whitepaper/ Accessed January 4, 2021 [Google Scholar]
- 10.TheLinuxFoundation. Hyperledger Architecture, Volume I: Introduction to Hyperledger Business Blockchain Design Philosophy and Consensus. 2017. https://www.hyperledger.org/wp-content/uploads/2017/08/HyperLedger_Arch_WG_Paper_1_Consensus.pdf Accessed July 5, 2019
- 11.Swan M. Blockchain: Blueprint for a new economy. Sebastopol, CA, United States: O'Reilly Media, Inc., 2015. [Google Scholar]
- 12.Androulaki E, Barger A, Bortnikov V, et al. Hyperledger fabric: a distributed operating system for permissioned blockchains. Proceedings of the thirteenth EuroSys conference; 2018. [Google Scholar]
- 13.Brown RG. The Corda Platform: An Introduction. Retrieved 2018; 27: 2018. [Google Scholar]
- 14.Yu H, Sun H, Wu D, Kuo T-T. Comparison of Smart Contract Blockchains for Healthcare Applications. AMIA Annual Symposium. : American Medical Informatics Association, Bethesda, MD, 2019. [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo T-T, Zavaleta Rojas H, Ohno-Machado L. Comparison of blockchain platforms: a systematic review and healthcare examples. Edited by Bakken Suzanne. Published by Oxford University Press, Kettering, Northants, UK. Journal of the American Medical Informatics Association (JAMIA) 2019; 26 (5): 462–78 doi: 10.1093/jamia/ocy185. March 25, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenspan G. MultiChain Private Blockchain - White Paper. 2015. http://www.multichain.com/download/MultiChain-White-Paper.pdf Accessed January 4, 2021 [Google Scholar]
- 17.iDASH Privacy & Security Workshop - Secure Genome Analysis Competition 2019. 2019. http://www.humangenomeprivacy.org/2019/ Accessed May 29, 2020
- 18.Azaria A, Ekblaw A, Vieira T, Lippman A. MedRec: Using Blockchain for Medical Data Access and Permission Management. International Conference on Open and Big Data (OBD); 2016; Vienna, Austria. IEEE. [Google Scholar]
- 19.Roehrs A, da Costa CA, da Rosa Righi R, da Silva VF, Goldim JR, Schmidt DC. Analyzing the performance of a blockchain-based personal health record implementation. Journal of biomedical informatics 2019; 92: 103140. [DOI] [PubMed] [Google Scholar]
- 20.Vazirani AA, O’Donoghue O, Brindley D, Meinert E. Blockchain vehicles for efficient Medical Record management. npj Digital Medicine 2020; 3 (1): 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mamo N, Martin GM, Desira M, Ellul B, Ebejer J-P. Dwarna: a blockchain solution for dynamic consent in biobanking. European Journal of Human Genetics 2020; 28 (5): 609–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuo T-T, Jiang X, Tang H, et al. iDASH Secure Genome Analysis Competition 2018: Blockchain Genomic Data Access Logging, Homomorphic Encryption on GWAS, and DNA Segment Searching. BMC Medical Genomics 2020; 13 (7): 98 doi: 10.1186/s12920-020-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.TeamO2 DCPPC. Towards a Sustainable Commons: The Role of Blockchain Technology. 2018. https://public.nihdatacommons.us/Blockchain/ Accessed May 14, 2020
- 24.Sylim P, Liu F, Marcelo A, Fontelo P. Blockchain technology for detecting falsified and substandard drugs in distribution: pharmaceutical supply chain intervention. JMIR research protocols 2018; 7 (9): e10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackey TK, Nayyar G. A review of existing and emerging digital technologies to combat the global trade in fake medicines. Expert opinion on drug safety 2017; 16 (5): 587–602. [DOI] [PubMed] [Google Scholar]
- 26.Clauson KA, Breeden EA, Davidson C, Mackey TK. Leveraging Blockchain Technology to Enhance Supply Chain Management in Healthcare. Blockchain in Healthcare Today 2018; 1: doi: 10.30953/bhty.v1.20· [DOI] [Google Scholar]
- 27.Kuo T-T, Gabriel RA, Cidambi KR, Ohno-Machado L. Expectation Propagation LOgistic REgRession on permissioned blockCHAIN (ExplorerChain): decentralized online healthcare/genomics predictive model learning. Edited by Bakken Suzanne. Published by Oxford University Press, Kettering, Northants, UK. Journal of the American Medical Informatics Association (JAMIA) 2020; 27 (5): 747–56 doi: 10.1093/jamia/ocaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X, Ji J, Luo C, Liao W, Li P. When Machine Learning Meets Blockchain: A Decentralized, Privacy-preserving and Secure Design. 2018 IEEE International Conference on Big Data (Big Data); 2018; December 10, 2018 - December 13, 2018. Seattle, WA, United States. IEEE. [Google Scholar]
- 29.Kuo T-T. The Anatomy of a Distributed Predictive Modeling Framework: Online Learning, Blockchain Network, and Consensus Algorithm. Journal of the American Medical Informatics Association Open (JAMIA Open). 2020; 3 (2): 201–08 doi: 10.1093/jamiaopen/ooaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo T-T, Kim J, Gabriel RA. Privacy-Preserving Model Learning on Blockchain Network-of-networks. Edited by Bakken Suzanne. Published by Oxford University Press, Kettering, Northants, UK. Journal of the American Medical Informatics Association (JAMIA) 2020; 27 (3): 343–54 doi: 10.1093/jamia/ocz214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Liu J, Hao J, Wang H, Xian M. CrowdSFL: A Secure Crowd Computing Framework Based on Blockchain and Federated Learning. Electronics 2020; 9 (5): 773. [Google Scholar]
- 32.Kuo T-T, Gabriel RA, Ohno-Machado L. Fair compute loads enabled by blockchain: sharing models by alternating client and server roles. Edited by Bakken Suzanne. Published by Oxford University Press, Kettering, Northants, UK. Journal of the American Medical Informatics Association (JAMIA) 2019; 26 (5): 392–403 doi: 10.1093/jamia/ocy180. March 20, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo T-T, Hsu C-N, Ohno-Machado L ModelChain: Decentralized Privacy-Preserving Healthcare Predictive Modeling Framework on Private Blockchain Networks. ONC/NIST Use of Blockchain for Healthcare and Research Workshop. September 26, 2016 - September 27, 2016. Gaithersburg, Maryland, United States, 2016. [Google Scholar]
- 34.Johnson M, Jones M, Shervey M, Dudley JT, Zimmerman N. Building a Secure Biomedical Data Sharing Decentralized App (DApp): Tutorial. Journal of medical Internet research 2019; 21 (10): e13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griggs KN, Ossipova O, Kohlios CP, Baccarini AN, Howson EA, Hayajneh T. Healthcare Blockchain System Using Smart Contracts for Secure Automated Remote Patient Monitoring. Journal of medical systems 2018; 42 (7): 130 doi: 10.1007/s10916-018-0982-x. [DOI] [PubMed] [Google Scholar]
- 36.Singhal B, Dhameja G, Panda PS. Beginning Blockchain: A Beginner's Guide to Building Blockchain Solutions: Springer, 2018. [Google Scholar]
- 37.Wood G. Ethereum: A secure decentralised generalised transaction ledger (Petersburg version). 2019. https://ethereum.github.io/yellowpaper/paper.pdf Accessed September 30, 2020 [Google Scholar]
- 38.BigchainDBGmbH. BigchainDB 2.0: The Blockchain Database. 2018. https://www.bigchaindb.com/whitepaper/bigchaindb-whitepaper.pdf Accessed September 30, 2020
- 39.Chowdhury MJM, Colman A, Kabir MA, Han J, Sarda P. Blockchain versus database: a critical analysis. 2018 17th IEEE International Conference On Trust, Security And Privacy In Computing And Communications/12th IEEE International Conference On Big Data Science And Engineering (TrustCom/BigDataSE): IEEE, 2018: 1348–53. [Google Scholar]
- 40.Chen S, Zhang J, Shi R, Yan J, Ke Q. A comparative testing on performance of blockchain and relational database: Foundation for applying smart technology into current business systems. International Conference on Distributed, Ambient, and Pervasive Interactions: Springer, 2018: 21–34. [Google Scholar]
- 41.Gursoy G, Bjornson R, Green ME, Gerstein M. Using blockchain to log genome dataset access: Efficient storage and query. Accepted by BMC Medical Genomics 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szilágyi P. Ethereum Improvement Proposals (EIP) 225: Clique proof-of-authority consensus protocol. 2017. http://eips.ethereum.org/EIPS/eip-225 Accessed August 9, 2019 [Google Scholar]
- 43.TheEthereumCommunity. The Solidity Contract-Oriented Programming Language. https://github.com/ethereum/solidity Accessed January 4, 2021
- 44.Ohno-Machado L, Bafna V, Boxwala Aa, et al. iDASH. Integrating data for analysis, anonymization, and sharing. Edited by Ohno-Machado Lucila. Published by Oxford University Press, Kettering, Northants, UK. Journal of the American Medical Informatics Association 2012; 19: 196–201 doi: 10.1136/amiajnl-2011-000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rouhani S, Deters R. Performance analysis of ethereum transactions in private blockchain. 2017 8th IEEE International Conference on Software Engineering and Service Science (ICSESS); 2017. IEEE. [Google Scholar]
- 46.Nasir Q, Qasse IA, Abu Talib M, Nassif AB. Performance Analysis of Hyperledger Fabric Platforms. Security and Communication Networks 2018; 2018: 3976093 doi: 10.1155/2018/3976093. [DOI] [Google Scholar]
- 47.Go-Ethereum Releases. https://github.com/ethereum/go-ethereum/releases Accessed November 25, 2019
- 48.Merkel D. Docker: lightweight linux containers for consistent development and deployment. Linux Journal 2014; 2014 (239): 2. [Google Scholar]